Nonsense mutations account for ~11.3% of reported hemophilia A (HA) mutations, based on the Center for Disease Control Hemophilia A Mutation Project (CHAMP) database. Rescue of nonsense mutations by drug-induced ribosomal readthrough over premature termination codons (PTC) is a potentially attractive therapy that has increasingly gained attention in recent years.1 Aminoglycosides were among the first compounds to show readthrough effects. A new compound, ataluren (PTC124),2 was granted conditional approval in the European Union for the treatment of the nonsense mutation Duchenne muscular dystrophy and has also shown early promise in treating other PTC-mediated genetic disorders. However, recent clinical studies revealed a wide variability in outcomes.3,4
In theory, hemophilias are ideally suited for drug-induced readthrough therapy, because even a tiny increase in coagulation factor levels could significantly improve bleeding symptoms. A small-molecule drug also has strong advantage over the costly intravenous infusion of recombinant proteins. Gentamicin generally showed poor responses in several hemophilia patients.5 Recent studies found significant G418-induced readthrough for nonsense hemophilia B (HB) mutations,6,7 as well as a therapeutic response to G418 in a mouse model.8 A Phase 2a trial (NCT00947193) was initiated to evaluate the safety and efficacy of ataluren in treating hemophilia A (HA) and HB with nonsense mutations in 2009, but was suspended in 2011. Over 330 nonsense mutations are reported in HA databases (CHAMP and the factor VIII (FVIII) variant database), making it a challenging task to identify candidate mutations amenable to readthrough therapy. In this study, we developed a cell-based system to evaluate candidate drugs for nonsense rescue and identified candidate mutations for readthrough therapy.
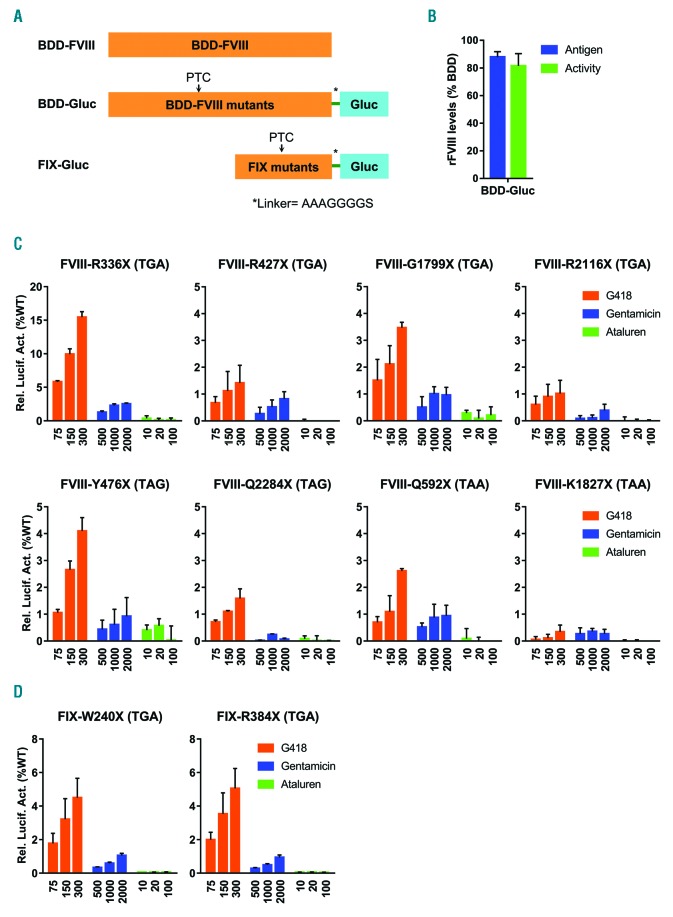
B domain-deleted-FVIII (BDD) with nonsense mutations was fused at its C-terminus with a humanized Gaussia luciferase (Gluc)9 and stably expressed from a retrovirus construct in HEK293 cells (Figure 1A). Gluc fusion provides a tag for detecting the full readthrough product secreted from cells and a rapid assay for drug and mutation screening. FVIII cofactor activity and fusion protein levels in conditioned media were comparable to HEK293 cells stably expressing BDD-FVIII (Figure 1B), indicating that Gluc fusion did not significantly increase or impair the activity or secretion of BDD-FVIII. No significant decrease in cell proliferation or increase in wild- type (WT) BDD-Gluc secretion was observed 48 hr after treatment of various concentrations of G418, gentamicin and ataluren (Online Supplementary Figure S1). Therefore, the BDD-Gluc expression system is suitable for monitoring the secretion of FVIII in response to treatment of these drugs. Luciferase fusion assays have previously been used to study readthrough of PTC of other genes, and observed readthrough was not a result of the stabilization of the luciferase reporter,10 which is consistent with our results.
Figure 1.
Responses of FVIII and FIX nonsense mutations to readthrough drugs. (A) Schematic representation of the BDD-FVIII, BDD-Gluc and FIX-Gluc fusion protein constructs. The mature Gaussia luciferase was joined with BDD-FVIII or FIX at the C terminus, separated by a glycine-rich linker. PTC were introduced into BDD-FVIII or FIX by site-directed mutagenesis. (B) Gluc fusion did not significantly affect the secretion and cofactor activity of BDD-FVIII. Recombinant FVIII (rFVIII) antigen and activity levels in the cultured media of HEK293 cells stably expressing the BDD-Gluc fusion were compared with HEK293 cells stably expressing the BDD-FVIII. Results are reported as mean ± standard deviation and are representative of at least three independent experiments. (C) (D) HEK293 cells stably expressing FVIII BDD-Gluc fusions (C) or FIX-Gluc fusions (D) of the indicated mutations were treated with candidate readthrough drugs at the indicated concentrations (μM). Secreted luciferase activities of drug-treated mutant fusion proteins were compared with activities of the wild-type (WT) fusion protein (relative luciferase activities: 12,008 ± 1,052 for FVIII BDD-Gluc and 626,610 ± 70,829 for FIX-Gluc) and plotted as percentages of WT levels. Background luciferase activity values of DMSO-treated cells (Online Supplementary Figure S2) were subtracted. Results are reported as mean ± standard deviation from three independent experiments.
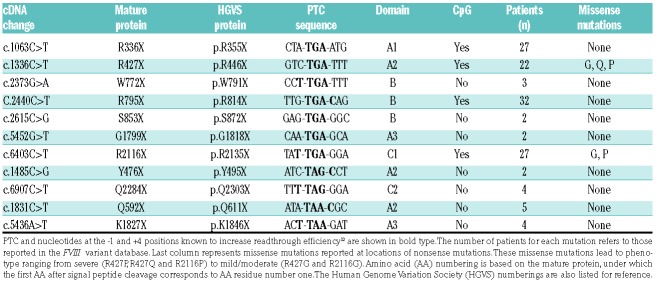
We introduced eight FVIII nonsense mutations (Table 1) into the BDD-Gluc fusion construct. These mutations include two (R427X and R2116X) that were previously tested in a small clinical trial.5 The other six nonsense mutations include two of each type of PTC selected from the heavy chain and the light chain, respectively. Each mutation has been substantiated by at least two independent reports and no missense mutations were reported at the same positions in FVIII variant databases. Cells were treated with ascending concentrations of G418, gentamicin and ataluren. Among these drugs, G418 treatment led to variable dose-dependent increases in secreted luciferase activities in seven of the eight nonsense mutant BDD-Gluc fusions tested (Figure 1C). Best response occurred with R336X, reaching 10-15% of luciferase activity of WT BDD-Gluc fusion with 150 and 300 μM G418 treatments. K1827X was the only mutant that did not respond to G418 treatment. In contrast, ataluren failed to significantly increase secreted luciferase activities in conditioned media of any mutants at all tested doses. Although shown to induce readthrough in other diseases, gentamicin led to only slight increases in secreted luciferase activities of several mutants at high doses. Poor responses of R427X and R2116X mutants to gentamicin treatment may explain their negative clinical trial results.5
Table 1.
Features of FVIII nonsense mutations.
Similarly, we used our system to test two nonsense mutants (p.W240X and p.R384X) factor IX (FIX) that were recently shown to be rescued by G418 treatment.6 We confirmed that G418 elicited a rescue of both mutants (Figure 1D). Again, ataluren and gentamicin failed to significantly increase secretion of these mutants. A recent report also showed response of PTC in the leptin receptor gene to G418, but not to ataluren.11 Readthrough mechanisms of aminoglycosides and ataluren are different,1 potentially contributing to discrepancies in effectiveness between different genes.
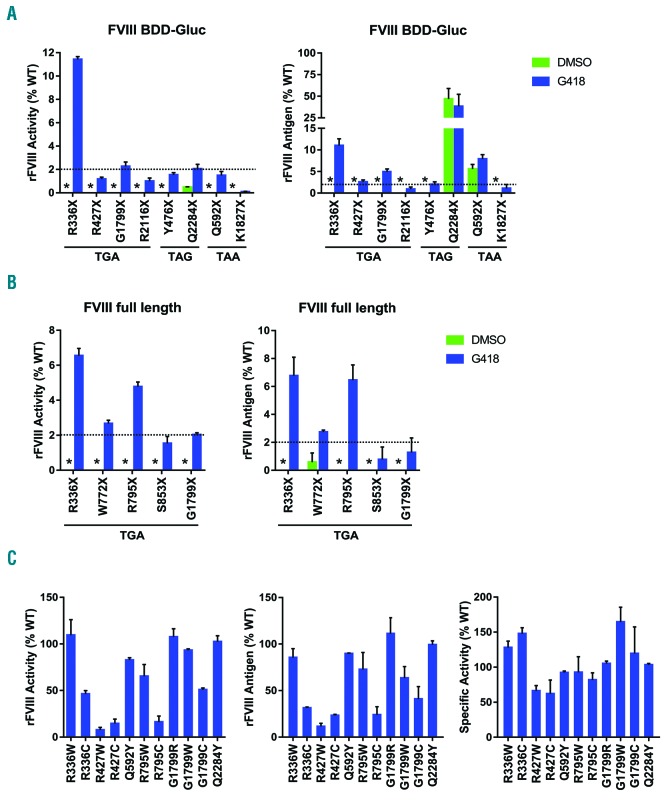
Next we measured FVIII cofactor activity and antigen levels of the nonsense BDD-Gluc mutants in conditioned media after G418 treatment. The R336X mutant displayed a strong rescue after G418 treatment, reaching 11-12% of the WT activity and antigen levels (Figure 2A). All other mutants except K1827X had a measurable increase in activities after G418 treatment, with the G1799X mutant (2.3%) also exceeding the 2% therapeutic threshold. We also detected variant levels of FVIII antigen after G418 treatment, which generally agreed with their activity levels with the exception of the Q592X and Q2284X mutants. Despite the lack of FVIII activity (Figure 2A) and luciferase activity (Online Supplementary Figure S2), substantial antigen levels were detected in conditioned media of the Q592X (5.6%) and Q2284X (46.8%) mutants without G418 treatment. These mutants are located near the C-termini of the A2 and C2 domains and may produce truncated inactive proteins that are partially secreted.
Figure 2.
FVIII cofactor activities and antigen levels in BDD-Gluc and FL-FVIII after G418-induced readthrough. (A) (B) FVIII cofactor activities and antigen levels of mutant BDD-Gluc fusion proteins from stably expressed HEK293 cell lines (A) or mutant full-lenght FVIII (FL-FVIII) from transiently transfected HEK293 cells (B) secreted into conditioned media. Cells were challenged with 300 μM G418 or DMSO for 48 hr. Recombinant FVIII (rFVII) cofactor activity and antigen levels were measured and plotted as percentages relative to the WT activity level. WT BDD-Gluc and FL-FVIII antigen levels in conditioned media were determined to be 45.9 ± 7.3 ng/mL and 13.2 ± 4.7 ng/mL, respectively, on the assumption of a concentration of 150 ng/ml in FVIII standard (1 IU/mL). Asterisks indicate activities and antigens undetectable from DMSO-treated cells. Dashed lines represent the 2% therapeutic threshold. (C) FL-FVIII cofactor activities, antigen levels and specific activities of potential nonnative readthrough products of indicated nonsense mutants secreted into conditioned media from transiently transfected HEK293 cells. rFVIII cofactor activities and antigens were plotted as percentages relative to the WT levels. Specific activity for a given mutant was calculated as the ratio of the activity level and the antigen level in conditioned media. Relative activities are shown as percentages of the WT FVIII relative activity. Results in all three panels are reported as mean ± standard deviation from three independent experiments.
To further validate nonsense rescue results, we introduced the R336X and G1799X mutations that responded to G418 treatment into a full-length FVIII (FL-FVIII) construct. In addition, we also introduced three TGA mutants (Table 1) in the B domain into this construct. FL-FVIII mutants were transiently expressed in HEK293 cells and treated with G418. Robust rescue was again observed for the R336X mutant, reaching 6.6% of the WT activity level (Figure 2B). The G1799X mutant and the two B domain mutants also displayed significant rescue (2.0% for G1799X, 2.7% for W772X and 4.8% for R795X). FVIII antigen levels were similar to activity levels after G418 treatment for all tested mutants (Figure 2B).
Cytosine at the +4 position and to a lesser extent a thyrosine at the −1 position were found to associate with a higher readthrough efficiency.12 B domain mutations W772X and R795X that responded well to G418 have one of these favorable sequence contexts (Table 1). However, the R336X and G1799X mutations are suboptimal PTC for readthrough, suggesting that the sequence context alone is not sufficient to predict the nonsense rescue efficiency. Different replacement amino acids (AA) inserted at PTC may affect FVIII secretion and cofactor activity. For the TGA codon, various proportions of arginine, cysteine and tryptophan are the most commonly found replacement AA inserted by near-cognate tRNAs in HEK293 cells after G418-induced readthrough.10
Underlying AA of nonsense mutations range from highly to poorly conserved residues (Figure S3). We expressed the most likely non-native readthrough products of the R336X, R427X, Q592X, R795X, G1799X, and Q2284X mutants as FL-FVIII in HEK293 cells (Figure 2C). All replacement AA resulted in >45% cofactor activity for R336X, Q592X, G1799X and Q2284X mutations, suggesting that nonsense rescue levels of these mutants are primarily determined by readthrough efficiencies. Both replacement AA for R427X decreased FVIII cofactor activity to <15%, consistent with reports of multiple missense mutations at this location (Table 1). Poor rescue of the R427X may be because readthrough is inefficient and/or replacement AA other than the native arginine are associated with poor secretion and function of FVIII. For the B domain mutation R795X, one of the nonnative replacement (R795C) decreased FVIII cofactor activity and antigen levels to <25%, while the other one (R795W) moderately decreased the activity. Decreased secretion and activity of the R795C mutant is surprising, given the lack of pathological missense mutations in the B domain. Perhaps the addition of a free cysteine interferes with normal disulfide bond formation. Readthrough with the combination of replacement AA is apparently sufficient to allow significant rescue of the R795X mutant. Tolerance of replacement AA was also associated with higher rescue of FIX nonsense mutations.6,7 Location-dependent variations in efficacy may at least partially explain the variations in reported clinical outcomes by ataluren.3,4 Prescreening of patients for susceptible mutations should maximize the effectiveness of readthrough therapies.
This is the first report to comprehensively evaluate drugs and mutations for HA nonsense readthrough therapy. The Gluc fusion allows convenient screening of readthrough compounds and identification of nonsense mutations of secreted proteins that are susceptible to readthrough therapy. Consistent with FIX,6,7 our results suggest that only a small number of FVIII nonsense mutations may be amenable to readthrough. Most responsive PTC are likely TGA mutations that, 1) with favorable sequence contexts for efficient readthrough; 2) a substantial replacement of the native residue is likely, or at locations where replacement AA are compatible with secretion and function. Despite being the first drug approved for readthrough therapy, our results suggest that ataluren may not be effective in treating hemophilias. Aminoglycoside derivatives with higher readthrough potency and less toxicity profiles under development13,14 or targeted delivery into endothelial cells15 may hold promise in treating patients carrying certain nonsense HA mutations in the future.
Acknowledgments
The authors thank Dr. Bakhos Tannous (Massachusetts General Hospital) for providing the humanized Gaussia luciferase cDNA.
Footnotes
Funding: this study was supported by grants from the National Institutes of Health (R01HL094505 and R03CA202131) to BZ and National Natural Science Foundation of China (81860430) to MZ.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Keeling KM, Xue X, Gunn G, Bedwell DM. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet. 2014;15:371–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welch EM, Barton ER, Zhuo J, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447(7140):87–91. [DOI] [PubMed] [Google Scholar]
- 3.Bushby K, Finkel R, Wong B, et al. Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle Nerve. 2014; 50(4):477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonald CM, Campbell C, Torricelli RE, et al. Ataluren in patients with nonsense mutation Duchenne muscular dystrophy (ACT DMD): a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390(10101):1489–1498. [DOI] [PubMed] [Google Scholar]
- 5.James PD, Raut S, Rivard GE, et al. Aminoglycoside suppression of nonsense mutations in severe hemophilia. Blood. 2005;106(9):3043–3048. [DOI] [PubMed] [Google Scholar]
- 6.Branchini A, Ferrarese M, Campioni M, et al. Specific factor IX mRNA and protein features favor drug-induced readthrough over recurrent nonsense mutations. Blood. 2017;129(16):2303–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrarese M, Testa MF, Balestra D, Bernardi F, Pinotti M, Branchini A. Secretion of wild-type factor IX upon readthrough over F9 pre-peptide nonsense mutations causing hemophilia B. Hum Mutat. 2018;39(5):702–708. [DOI] [PubMed] [Google Scholar]
- 8.Yang C, Feng J, Song W, et al. A mouse model for nonsense mutation bypass therapy shows a dramatic multiday response to geneticin. Proc Natl Acad Sci U S A. 2007;104(39):15394–15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11(3):435–443. [DOI] [PubMed] [Google Scholar]
- 10.Roy B, Friesen WJ, Tomizawa Y, et al. Ataluren stimulates ribosomal selection of near-cognate tRNAs to promote nonsense suppression. Proc Natl Acad Sci U S A. 2016;113(44):12508–12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolze F, Mocek S, Zimmermann A, Klingenspor M. Aminoglycosides, but not PTC124 (Ataluren), rescue nonsense mutations in the leptin receptor and in luciferase reporter genes. Sci Rep. 2017;7(1):1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manuvakhova M, Keeling K, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6(7):1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bidou L, Bugaud O, Belakhov V, Baasov T, Namy O. Characterization of new-generation aminoglycoside promoting premature termination codon readthrough in cancer cells. RNA Biol. 2017;14(3):378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moosajee M, Tracey-White D, Smart M, et al. Functional rescue of REP1 following treatment with PTC124 and novel derivative PTC-414 in human choroideremia fibroblasts and the nonsense-mediated zebrafish model. Hum Mol Genet. 2016;25(16):3416–3431. [DOI] [PubMed] [Google Scholar]
- 15.Howard MD, Hood ED, Zern B, Shuvaev VV, Grosser T, Muzykantov VR. Nanocarriers for vascular delivery of anti-inflammatory agents. Annu Rev Pharmacol Toxicol. 2014;54:205–226. [DOI] [PMC free article] [PubMed] [Google Scholar]