Abstract
There are no validated molecular biomarkers to identify newly-diagnosed individuals with chronic-phase chronic myeloid leukemia likely to respond poorly to imatinib and who might benefit from first-line treatment with a more potent second-generation tyrosine kinase inhibitor. Our inability to predict these ‘high-risk’ individuals reflects the poorly understood heterogeneity of the disease. To investigate the potential of genetic variants in epigenetic modifiers as biomarkers at diagnosis, we used Ion Torrent next-generation sequencing of 71 candidate genes for predicting response to tyrosine kinase inhibitors and probability of disease progression. A total of 124 subjects with newly-diagnosed chronic-phase chronic myeloid leukemia began with imatinib (n=62) or second-generation tyrosine kinase inhibitors (n=62) and were classified as responders or non-responders based on the BCRABL1 transcript levels within the first year and the European LeukemiaNet criteria for failure. Somatic variants affecting 21 genes (e.g. ASXL1, IKZF1, DNMT3A, CREBBP) were detected in 30% of subjects, most of whom were non-responders (41% non-responders, 18% responders to imatinib, 38% non-responders, 25% responders to second-generation tyrosine kinase inhibitors). The presence of variants predicted the rate of achieving a major molecular response, event-free survival, progression-free survival and chronic myeloid leukemia-related survival in the imatinib but not the second-generation tyrosine kinase inhibitors cohort. Rare germline variants had no prognostic significance irrespective of treatment while some pre-leukemia variants suggest a multi-step development of chronic myeloid leukemia. Our data suggest that identification of somatic variants at diagnosis facilitates stratification into imatinib responders/non-responders, thereby allowing earlier use of second-generation tyrosine kinase inhibitors, which, in turn, may overcome the negative impact of such variants on disease progression.
Introduction
Although tyrosine kinase inhibitors (TKI) have profoundly changed the prognosis of chronic-phase chronic myeloid leukemia (CML-CP), some 10-15% of affected individuals do not respond and need other therapies.1 Four TKI are approved for use in newly-diagnosed CML, including imatinib and the second-generation TKI (2G-TKI) dasatinib, nilotinib and bosutinib. In randomized studies, 2G-TKI induce faster, deeper molecular responses than imatinib with a lower risk of progression to blast phase but no convincing evidence of better survival.2–4 Consequently, there is controversy as to which TKI to use as initial therapy, although imatinib remains the first choice for many people because of the low incidence of serious life-threatening side-effects and recent availability of less expensive generic formulations.5,6
Clinical risk scores can be used to direct initial therapy but are often inaccurate at the subject level.7–10 Early molecular responses analyzed by reverse transcription-quantitative PCR (RT-qPCR) at 3, 6 and 12 months are widely used to direct therapy.11–13 Individuals failing to achieve these landmarks can be switched to a different TKI but there are no convincing data that the change of therapy changes their outcome. Because most progressions occur within two years of starting TKI-therapy14 early identification of those at high risk of progression would facilitate more rapid decision-making regarding more aggressive therapy.
In other hematologic neoplasms, the presence of somatically mutated genes involving signaling, RNA splicing, transcriptional control, DNA damage response and epigenetic regulation is correlated with survival and sometimes drives treatment.15,16 However, no ‘mutator’ phenotype associated with clinical outcome has been described in CML, particularly in chronic phase, as earlier studies focused on blastic phase.17–20 Four recent whole-exome sequencing (WES) and/or whole-transcriptome (RNA-Seq) studies identified somatic variants in 24, 19, 13 and 65 exomes (or transcriptomes in one study) from newly-diagnosed CML-CP21,22 and in chronic phase and blast transformation.23,24 Studies using deeper, targeted sequencing described genetic variants in CML-CP,25–28 especially in genes associated with epigenetic regulation. These variants were also present in Philadelphia (Ph)-chromosome-negative cells, suggesting that they antedated the BCRABL1 translocation event as an early step in developing CML.29 Finally, variants in hematologically normal elderly individuals are described, although the risk of conversion from age-related clonal hematopoiesis (ARCH) to leukemia was modest.30,31
We recently identified differences in genome-wide DNA methylation patterns in CD34+ cells of CML-CP compared with normal subjects. These differences were not observed at the time of complete cytogenetic remission,32 suggesting a role for epigenetic regulation in CML-CP. In this study, we interrogated genetic variants in pre-therapy CML-CP using a targeted panel of genes enriched in epigenetic modifiers and Ion Torrent Personal-Genome-Machine (PGM) next-generation sequencing (NGS). We then assessed the predictive value of these variants for diverse therapy outcomes in the context of different TKI therapies.
Methods
Study participants
We studied 124 untreated subjects with CML-CP and 14 normal individuals as negative controls, selecting CD34+ cells. Subjects were non-consecutive and selected for optimal response (n=69) or non-response (n=55) to TKI-therapy.11 Evolution of the somatic variants after treatment, was investigated using CD34+ (n=11) and whole-blood cells (n=4) from subjects in major molecular remission (MR3; 3-log reduction in BCRABL1-transcripts from baseline) and after progression to blast phase, respectively. We also used CD34+ cells from three subjects with somatic variants at diagnosis, to establish liquid cultures with in vitro TKI-treatment, as a biological validation of our findings (Online Supplementary Methods and Results, and Online Supplementary Figure S1). All subjects gave written informed consent and the local research Ethics Committee approved the study.
Definitions
Response was defined as BCRABL1/ABL1 transcript levels according to the International Scale (IS) ≤10%, ≤1% and ≤0.1% at 3, 6 and 12 months after initiating TKI-therapy, respectively.11 Because some subjects did not have real-time qualitative polymerase chain reaction (RT-qPCR) sampling at pre-specified times during the first year, responders were required to have at least ≥2 of these results available. Non-responders satisfied the European LeukemiaNet criteria for failure.11
DNA preparation
In 103 subjects, paired leukemia/control DNA was analyzed; in 44 and 59 subjects control DNA was obtained from diagnostic T cells expanded in vitro and from samples in MR4-molecular remission (4-log reduction from baseline),33 respectively. We measured BCRABL1/ABL1 in 13 T-cell samples and confirmed very low expression. In seven subjects with somatic variants at diagnosis we compared detection of somatic variants in whole-blood and CD34+ cell populations (Online Supplementary Methods and Results).
Targeted gene panel design
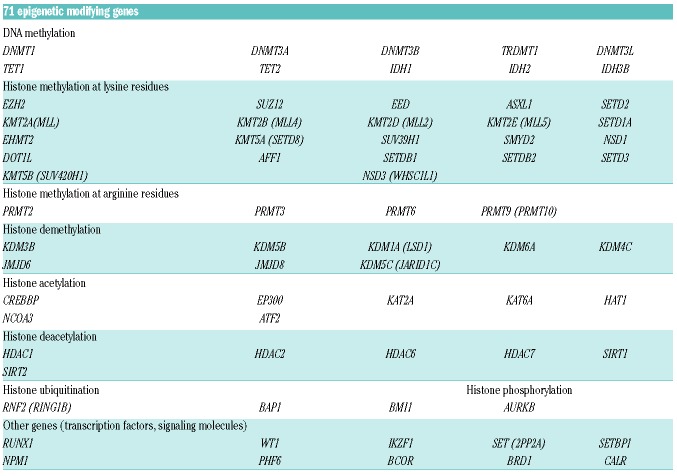
Based on preliminary analyses investigating gene expression for epigenetic modifiers in CML-CP (Online Supplementary Methods and Results, and Online Supplementary Figure S2), and a literature review for frequently mutated genes in leukemia, we generated a custom panel of 71 genes enriched for modifiers of DNA methylation and histone methylation/acetylation (2002 amplicons) (Table 1).
Table 1.
Seventy-one epigenetic modifiers grouped according to gene function.
Semi-conductor-based targeted sequencing
Amplicon library preparation, templating and sequencing using the Ion Torrent PGM (Thermo Fisher Scientific, Waltham, MA, USA) were performed in line with the manufacturer’s instructions (Online Supplementary Methods). To validate somatic variants, we re-ran newly-prepared libraries with validation rate of 70% (49 of 70).
Ion PGM sequencing informatics
Base-calling, mapping, alignment and further quality filtering were performed using Torrent-Suite_v4.0.2 and the Cloud-based Ion-Reporter_v5-software (Thermo Fisher Scientific; analysis pipeline available in Online Supplementary Methods and Online Supplementary Figure S3). The data have been deposited at the European Variation Archive under accession n. PRJEB32264.
Statistical analysis
Results were analyzed in R_v3.2.2. Event-free-survival (EFS),34 progression-free-survival (PFS) and CML-related-survival probabilities were estimated by the Kaplan-Meier method and compared by the log-rank test at six or eight years (2G-TKI and imatinib, respectively) from starting therapy. A Cox proportional hazard regression model was used to estimate hazard ratios (HR) and 95% confidence intervals (95%CI) in univariate/multivariate analyses. The rate of MR3 at five years was estimated by the cumulative incidence function with groups compared by the Gray test (univariate), and the Fine-Gray model (multivariate analysis; see Online Supplementary Methods). Logistic regression and Fisher’s exact test were used to calculate associations of variables and probability of greater-than-random overlaps, respectively. P<0.05 were considered significant.
Results
Subjects
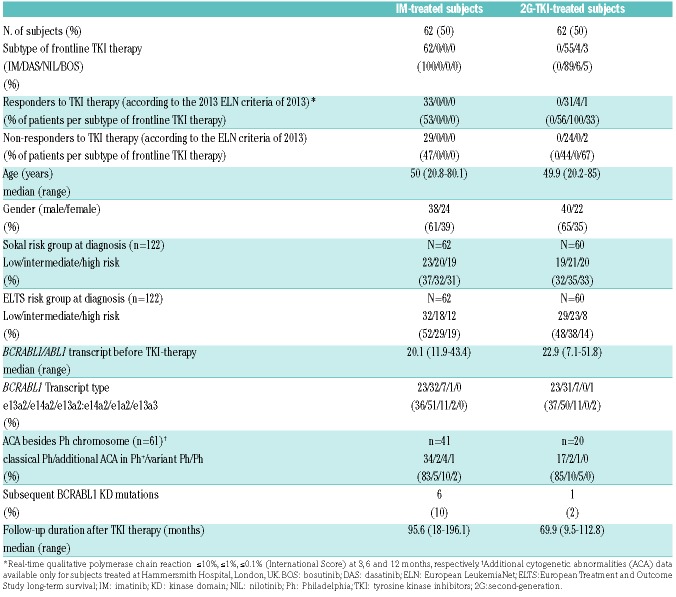
Demographic, clinical and molecular data of the subjects are shown in Table 2. Sixty-two, 55, 4 and 3 subjects started treatment with imatinib, dasatinib, nilotinib and bosutinib. Subjects treated initially with 2G-TKI did so because they were enrolled in clinical trials. Among subjects treated with initial imatinib, 33 were responders (R) and 29 non-responders (NR). Of those who received 2G-TKI, 36 and 26 were classified as responders and non-responders, respectively.
Table 2.
Demographics and clinical/molecular characteristics of subjects with chronic phase-chronic myeloid leukemia at diagnosis.
Sequencing data
A mean depth of coverage of 302× (range: 85×-1088×) was achieved yielding a limit of detection of 4% variant allele frequency (VAF). After filtering (Online Supplementary Figure S3), 142 non-synonymous variants remained (in 51 of 71 genes), of which 43 were somatically acquired variants. Of these, 40 were classed as somatic if they were present only in leukemia DNA, and three as pre-leukemia (before BCRABL1) if VAF in leukemia DNA was >20% greater than VAF in control DNA. The remaining 99 were present at similar VAF (about 50%) in leukemia and control/matched DNA and were most likely germline variants.
Incidence of somatic variants in chronic-phase chronic myeloid leukemia
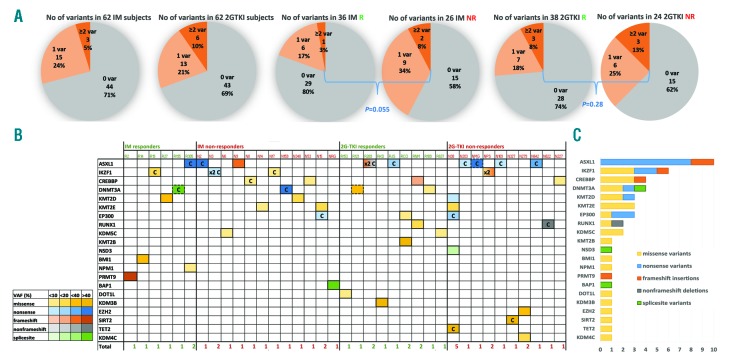
Forty-three somatic variants were observed 49 times (5 variants >1) in 37 of 124 subjects [30% (95%CI: 23, 39%)], including 18 of 62 subjects [29%, (95%CI:20, 43%)] in the imatinib cohort and 19 of 62 subjects [31% (95%CI:21, 45%)] in the 2G-TKI cohort. The incidence of subjects with at least one somatic variant (1 or ≥2 grouped together) was higher in non-responders [22 of 55; 40% (95%CI:28, 53%)] compared with responders from both imatinib- and 2G-TKI-treated cohorts [15 of 69; 22% (95%CI: 14, 33%); P=0.031) (Figure 1A). More than one somatic variant in the same or different genes was seen in three subjects in the imatinib cohort and in six of the 2G-TKI cohort and occurred more often in non-responders (Figure 1A).
Figure 1.
Landscape of somatic variants in individuals with chronic-phase chronic myeloid leukemia (CML-CP) at diagnosis. (A) Pie charts show the percentage of somatic variants in imatinib (IM)- and second-generation tyrosine kinase inhibitor (2G-TKI)-treated subjects and per responders (R) + non-responders (NR) group. Gray: no variants; light orange: one variant; dark orange: ≥2 variants. P-value from Fisher’s exact test comparing the incidence of subjects with variants (1 or ≥2 grouped together), compared with no variants, in R versus NR from the IM and 2G-TKI groups. (B) Somatic variants number and type in each patient (n=37) sorted in IM-R, IM-NR, 2G-TKI-R and 2G-TKI-NR. Number of variants/subject are reported at the bottom of each column. Yellow: missense; blue: nonsense; orange: frameshift insertions; gray: non-frameshift deletions; green: splice-site variants. Intensities of each color cell indicate the variant allele frequency (VAF) of each somatic variant with darker colors associated with higher VAF. Pre-leukemia variants are depicted in boxes in dashed lines, COSMIC with “C” and 2 variants affecting the same gene with “2x”. (C) Bar plots indicate the number of variants affecting each gene. Genes (rows) ordered by prevalence of variants/gene in CML-CP.
Most of the 49 variants (26 missense, 14 nonsense, 3 splice-site, 5 frameshift insertions and 1 non-frameshift deletion) identified in 21 of 71 genes were in non-responders (Figure 1B and Online Supplementary Table S1). The most frequently altered genes were ASXL1 (n=10 in 9 subjects), IKZF1 (n=6 in 4 subjects), DNMT3A and CREBBP (n=4), KMT2D (MLL2), KMT2E (MLL5), and EP300 (n=3) (Figure 1C). VAF were 4.6-64% and for 28 of 49 variants were <20% (Figure 1B). In three subjects, two variants occurred in the same gene. Fifteen of 43 variants (35%) are currently listed in the COSMIC v86 database (Figure 1B).
None of these variants were found in the paired-control DNA, apart from three variants, classed as pre-leukemia and detected in the diagnostic CD34+ cells at a markedly higher VAF than in control DNA. Two variants occurred in DNMT3A: a missense (p.Arg899Gly) and a splice-site (c.1123-2A>G) variant with VAF of 50% and 48%, respectively, at diagnosis, but reduced to 22% and 6% in paired remission samples collected at 55 and 47 months from starting therapy, respectively (with BCRABL1/ABL1 of 0.0012% and 0.0009%, respectively). An ASXL1 nonsense variant (p.Tyr591*) occurred at 52% and 16% in leukemia and paired T cells, respectively.
Evolution of somatic variants after imatinib treatment
We next examined somatic variants in follow-up samples from the imatinib-treated subjects. In four subjects with somatic variants detected in CD34+ cells at diagnosis who progressed to blast phase (BP) we compared paired samples from whole-blood cells (as opposed to CD34+ cells) at diagnosis and in BP (median follow up 25 months). The somatic variants identified in diagnostic CD34+ cells (ASXL1 p.Gln780*, ASXL1 p.Gln594fs) at high level (VAF 51% and 40%, respectively) were also found in diagnostic whole-blood cells at similar VAF. On the contrary, those identified in diagnostic CD34+ cells (IKZF1 p.Arg184Trp and IKZF1 p.Arg213*/IKZF1 p.Tyr348*) at low levels (VAF 5.9%, and 6.9%/4.9%) were undetectable in diagnostic whole-blood cells. In one case of low-level variants (IKZF1 p.Arg213*/IKZF1 p.Tyr348*) identified in diagnostic CD34+ cells, these were undetectable in whole-blood samples from CP and BP; however, the clone with the low-level variant IKZF1 p.Arg184Trp, expanded during progression (from undetectable to 17% VAF). As for the high-level variants, in one case the variant ASXL1 p.Gln594fs remained at similar levels (from 40% to 43%) in both CP and BP, whereas in the second case, the variant ASXL1 p.Gln780* dropped to lower, but still high, levels in BP (from 45% to 27%) (Online Supplementary Table S2).
Of the 11 patients who achieved MR3 and in whom we had paired samples, only three had somatic variants at diagnosis (KMT2D p.Gln3946Leu, PRMT9 p.Phe591fs, IKZF1 p.Arg184Trp) with VAF of 54%, 42% and 16%, respectively. These variants were undetectable in the follow-up samples collected at a median of 20 months from starting therapy. In two subjects achieving MR3 we identified variants (DNMT3A p.Cys497Tyr and EHMT2 p.Pro196Gln) in follow-up samples in MR3 with median follow-up 18 months with VAF of 5% and 10%. These variants were undetectable at diagnosis, so the possibility arises that they are due to clonal evolution in Philadelphia negative clone (Online Supplementary Table S2). The same DNMT3A variant was identified at 141 months from diagnosis, when the patient was in durable MR4 and the sample was collected as a control. The presence of eight missense variants identified only in the paired deep remission samples (with median follow up 72 months) was associated with increased age (P=0.029) (Online Supplementary Results and Online Supplementary Table S3).
Somatic variants and outcomes in subjects treated with imatinib
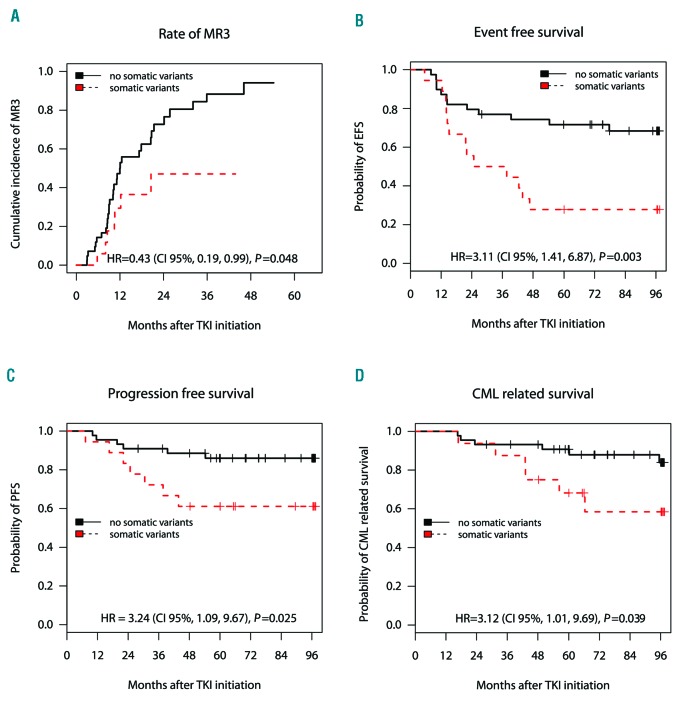
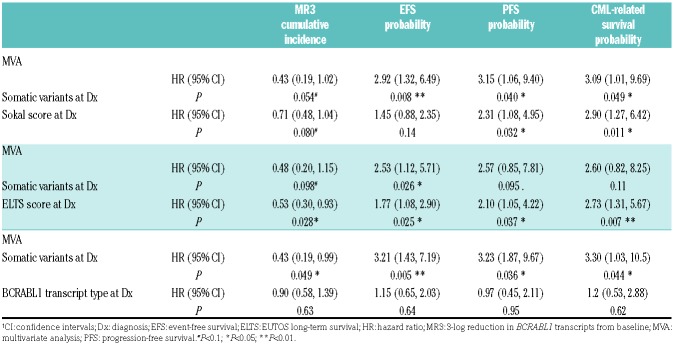
Imatinib-treated subjects with somatic variants had lower rates of 5-year MR3 compared to subjects without variants at diagnosis [47% (95%CI: 11, 68%) vs. 94% (67, 99%); P=0.048) (Figure 2A). Similarly they had lower rates of 8-year EFS, PFS and CML-related survival compared with the non-variant subjects [28% (13, 59%) vs. 68% (55, 85%); P=0.003 for EFS; 61% (42, 88%) vs. 85% (74, 97%); P=0.025 for PFS; 58% (37, 92%) vs. 84% (73, 97%); P=0.039 for CML-related survival] (Figure 2 B-D). Somatic variants and Sokal score were independently predictive for PFS and CML-related survival whereas only somatic variants predicted EFS (Table 3). When compared with European Treatment and Outcome Study (EUTOS) long-term survival (ELTS) score, only ELTS score predicted MR3, PFS and CML-related survival while both ELTS and somatic variants predicted EFS (Table 3). Subjects with a low Sokal score or a low ELTS score and somatic variants had worse EFS (P=0.015 for Sokal; P=0.021 for ELTS) and PFS (P=0.040 for Sokal; P=0.031 for ELTS) than those without somatic variants (Online Supplementary Figure S4A and B). The trends towards poorer outcomes in subjects with somatic variants was also evident for intermediate-and high-Sokal/ELTS subjects. Somatic variants were a more accurate predictor of outcomes compared with BCRABL1 transcript type (Table 3) and similarly compared with BCRABL1/ABL1 transcript levels before TKI-therapy, age and gender (data not shown).
Figure 2.
Association of occurrence of somatic variants with clinical outcome of chronic-phase chronic myeloid leukemia (CML-CP) patients starting on imatinib (IM) treatment. Kaplan-Meier survival analyses in IM-treated subjects with somatic variants (red dashed line) versus non-variant (black solid line). The end points used were cumulative incidence of major molecular response (3-log reduction in BCRABL1 transcripts from baseline; MR3) at five years (A) and probabilities of event-free survival (EFS) (B), progression-free survival (PFS) (C) and CML-related survival at eight years after start of therapy (D). Hazard R (95%CI) derived from Cox proportional hazard regression models and the P-value calculated by the Log Rank test also shown. Number of subjects (N) per group is also shown. Notably, two subjects have been excluded from the survival analysis due to non-CML-related deaths, whereas five subjects have been excluded from the EFS and two from the major molecular response (MR3) analyses, because of IM failure due to intolerance.
Table 3.
Multivariate analysis of somatic variants with Sokal score, European Treatment and Outcome Study long-term survival (ELTS) score and BCRABL1 transcript type in the imatinib cohort for cumulative incidence of 3-log reduction in BCRABL1 transcripts from baseline (MR3) (by the Fine-Gray model) and probabilities of event-free survival (EFS), progression-free survival (PFS) and chronic myeloid leukemia (CML)-related survival (by the Cox proportional hazard regression model).
Molecular response within the 1st year (calculated by BCRABL1/ABL1 transcripts at 3, 6, 12 months) (P<0.001) and somatic variants (P=0.044) were independently predictive for EFS. Non-responders without somatic variants had a better EFS [23% (9, 59%)] compared to non-responders with somatic variants at 0% (Online Supplementary Figure S5A). No association was detected between somatic variants in epigenetic modifiers at diagnosis and subsequent BCRABL1 kinase domain (KD) mutations (P=0.81).
In all the above analyses, we included the subject with pre-leukemia variant. Excluding this subject did not alter our conclusions (Online Supplementary Table S4A).
Somatic variants and outcomes in the second-generation tyrosine kinase inhibitor cohort
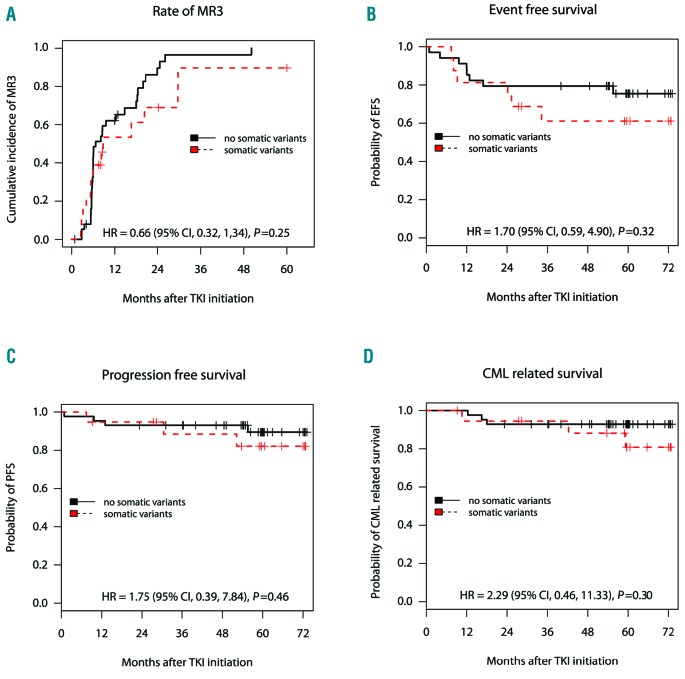
There was no association between the presence of somatic variants and the 5-year rates of MR3 and 6-year rates of EFS, PFS and CML-related survival in 2G-TKI-treated subjects: 90% (39, 98%) vs. 100%, P=0.25 for MR3; 61% (41, 91%) vs. 75% (62, 92%), P=0.32 for EFS; 82% (66, 99%) vs. 89% (80, 99%), P=0.46 for PFS; 81% (63, 99%) vs. 93% (85, 99%), P=0.29 for CML-related survival in variant versus non-variant subjects (Figure 3). We found no association of the somatic variants with outcomes in multivariate analysis with Sokal score, ELTS score or type of BCRABL1 transcript (Online Supplementary Table S5A). When combined with molecular response within the 1st year, only BCRABL1/ABL1 transcripts within the 1st year but not somatic variants were predictive for MR3 and EFS. Non-responders with and without variants had no significant difference in rates of EFS: 38% (16, 87%) vs. 48% (27, 85%), respectively; P=0.69 (Online Supplementary Figure S5B).
Figure 3.
Association of occurrence of somatic variants with clinical outcome of individuals starting on second-generation tyrosine kinase inhibitor (2G-TKI) treatment. Kaplan-Meier survival analyses in 2G-TKI-treated subjects with somatic variants (red-dashed line) versus non-variant (black-solid line). The end points used were cumulative incidence of major molecular response MR3 at five years (A) and probabilities of event-free survival (EFS) (B), progression-free survival (PFS) (C) and chronic myeloid leukemia (CML)-related survival at six years after start of therapy (D). HR (95% CI) derived from Cox proportional hazard regression models and the P-value calculated by the Log Rank test also shown. Number of subjects (N) per group is also shown. Notably, one subject has been excluded from the survival analysis due to non-CML-related death, whereas 12 subjects have been excluded from the EFS and five from the major molecular response (MR3) analyses because of 2G-TKI failure due to intolerance.
Excluding the two subjects with somatic pre-leukemia variants did not alter our conclusions (Online Supplementary Table S4B).
To reduce potential heterogeneity conferred by different 2G-TKI, we next investigated the association of somatic variants with outcomes in the dasatinib-treated cohort only (n=55). No association with outcomes was detected (Online Supplementary Table S5B).
Germline variants in chronic-phase chronic myeloid leukemia subjects
We identified 99 missense variants classified as rare germline (with frequency <1% in the general population) (Online Supplementary Results, Online Supplementary Table S6 and Online Supplementary Figure S6), which did not affect clinical outcome in either cohort (data not shown).
Functional associations of variants with chronic myeloid leukemia
To further investigate any association of the altered genes with CML a protein-protein-interaction (PPI) network of P210BCRABL1 with the 21 coded proteins affected by the somatic variants was constructed. Twenty-one of 23 proteins were parts of the network with six proteins including those encoded by ASXL1, IKZF1, EP300 and RUNX1 interacting directly with P210BCRABL1 suggesting a functional association (Online Supplementary Table S7 and Online Supplementary Figure S7).
Finally, we studied whether the presence of a somatic variant in epigenetic modifiers influenced the DNA methylation signature in the imatinib cohort. Hierarchical clustering based on 1,028 differentially methylated positions (DMP) (see criteria in the Online Supplementary Results) clearly separated the 12 variant and 30 non-variant CMP-CP subjects (Online Supplementary Figure S8). Functional annotation of DMP showed the imatinib pharmacokinetics/pharmacodynamics pathway being among the top ten hits of over-represented pathways (P=0.0016) (Online Supplementary Table S8).
Discussion
The successful introduction of TKI in CML therapy has resulted in an excellent outcome for approximately 90% of individuals, who have a life expectancy approaching that of unaffected individuals.35 However, the remaining 10% should ideally be identified at diagnosis and offered more potent TKI immediately or early allogeneic-stem cell transplantation if they demonstrate TKI resistance. The most widely used biomarker for outcomes in CML-CP, namely BCRABL1 transcript levels after three months on TKI (BCRABL1 ≤10%), identifies a cohort with an excellent prognosis. However, patients with BCRABL1 >10% at three months may or may not respond to 2G-TKI. Our aim was to investigate associations between somatic variants in epigenetic modifiers and response to imatinib and 2G-TKI given from diagnosis of CML-CP.
Others have explored the predictive value of a number of different biomarkers at the time of diagnosis including gene expression,36–38 protein expression,39 DNA methylation,40 miRNA expression,41 and SNP analysis,42–44 but none has proved sufficiently accurate and precise for clinical decision-making. We previously identified different genome-wide DNA methylation and gene expression patterns between CML-CP and normal individuals,32 but this did not correlate clearly with TKI response. However, this prompted us to investigate the role of genetic variants in epigenetic modifiers in greater detail.
We used targeted amplicon sequencing to detect genetic variants that might correlate with TKI response. To optimize the opportunity to assess differences in genetic variants between responders and non-responders to TKI-therapy, we selected a cohort enriched for non-responders (approx. 45% in each of the imatinib and 2G-TKI cohorts), which explains why the outcomes of patients in this study are inferior to those seen in unselected subjects.3,4,14 We believed our strategy of deliberately choosing equal proportions of responders and non-responders would maximize our chances of detecting a biomarker, if such existed. Moreover, the use of CD34+ progenitor cells from patients at diagnosis, the population in which the clone capable of progression resides, would maximize our chance of yielding meaningful results and reduce heterogeneity. We are aware that, for clinical utilization, our data should be validated in whole-blood samples and in a larger cohort of newly-diagnosed individuals with CML.
Progression to blast crisis in CML is often attributed to underlying ‘genetic instability’, in part because this is increased in stem/progenitor cells from individuals with CML (especially in blast phase) compared to normal individuals. BCRABL1-induced genomic aberrations and/or BCRABL1-independent pre-existing genetic lesions may then function as “amplifiers” of a genetically unstable phenotype and thereby predispose to blastic transformation.45 However, our results, and those of others, suggest that the ‘mutator’ phenotype of CML is moderate compared with other cancers, particularly in chronic phase. We included genes in which somatic variants have been identified by WES21–24 including ASXL1, RUNX1, IKZF1, KDM2D, BCOR, IDH1/2, PHF6, TET2, KDM1A, KAT6A, SETBP1, SETD2 and found somatic variants in approximately 30% of newly-diagnosed individuals with CML-CP, similar to previous reports.25,27,28 We identified overlap with other studies of 14-30% but feel this can be explained, at least in part, by our focus on epigenetic regulators that resulted in the omission of some genes that have frequently been found mutated in CML, such as TP53, and also by the readily available technology of targeted NGS at the start of this project.18,27,28 Concordance with other studies regarding specific variants was also limited, while 15 somatic variants in our study were COSMIC, mostly identified in other hematologic neoplasms. Because most of the variants we identified affect epigenetic modifiers and genome-wide DNA methylation changes are reported in CML,32,46 a better understanding of the role of such epigenetic alteration should be complemented by genome-wide landscape of histone marks.
In this study, we report for the first time a correlation between somatic variants and survival of individuals with CML-CP. Others have reported the presence of somatic variants but have so far been unable to directly associate these with clinical outcome21–24,28 or limited the assessment to achievement of major molecular remission.25 One study found that a subset of variants (16 of 73) affecting epigenetic modifiers had an adverse impact on cytogenetic/molecular responses.27 Because of our use of extreme responders, and because of the availability of prolonged follow up, our cohort contains patients who had experienced disease progression, with 20 patients developing blast crisis over the period of observation. Eleven of these (8 of 13 and 3 of 7 on imatinib and 2G-TKI, respectively) had somatic variants. Absence of variants in the remaining subjects does not exclude their presence in genes absent from our panel, who could have structural variants/copy number variations such as IKZF1 deletions that would be identified by WES/whole-genome sequencing whole-genome sequencing (WGS).
Although the two patient cohorts (treated with imatinib or 2G-TKI) were similar in their clinical characteristics at diagnosis and in the proportion of responders and non-responders, somatic variants impacted clinical outcome only in those treated with imatinib. This mirrors clinical experience in which the 2G-TKI can result in deep and durable responses in patients who were resistant to imatinib, and induce these responses more rapidly and in a larger proportion of patients when used as first-line therapy. One possible explanation is that the increased potency of the 2G-TKI results in the rapid eradication of the mutated clones and thus overcome the adverse prognostic impact. This hypothesis is supported by the fact that 90% of 2G-TKI-treated subjects with somatic variants achieved MR3 compared with <50% of subjects treated with imatinib. Data from in vitro liquid cultures corroborated our original findings, since at least cells containing some variants were eradicated on treatment with dasatinib but persisted or were eradicated more slowly on treatment with imatinib. We also have additional indirect evidence that our findings may predict response to imatinib. First, we were able to show distinct methylation patterns between imatinib-treated subjects with and without variants, and second, the PPI network indicates close interactions between p210BCRABL1 and proteins affected by somatic variation.
Somatic variants had better predictive power for outcomes than other widely-used predictive variables such as the Sokal score4,14,47 and BCRABL1 transcript type48,49 but less compared with the newly defined ELTS score.10 Previous clinical risk scores identified individuals at high risk of early progression but were less successful in predicting poor-risk subjects in the low/intermediate cohorts. Combining a clinical risk score with somatic variants is a potentially promising approach, and may be particularly valuable in those with low Sokal/ELTS scores, who are heavily influenced by age, such that a young patient with inherently poor prognosis may be inappropriately classified as non-high risk.
By using paired leukemia and control DNA, we found most somatic variants were part of a Ph+ clone. Therapy with imatinib eradicated the Ph+ clones with somatic variants in responders achieving MR3 but not in non-responders who progressed. These persistent somatic variants may be implicated in disease evolution or may be passenger mutations and require confirmation in larger cohorts.
The presence of pre-leukemia variants was implicated in three subjects. DNMT3A, ASXL1 variants were found in Ph+ and, albeit at lower levels, in Ph− cells. This suggests that these variants preceded the acquisition of BCRABL1. Variants in DNMT3A, ASXL1 and TET2 are thought to be latent initiating mutations31 and are described in CML.27,28 However, ASXL1 mutations have also been found in children and young adults with CML.26 Rare germline variants had no impact on clinical outcomes in either of the imatinib or 2G-TKI cohorts, contrary to other reports.43 Variants detected only in Ph− cells from subjects aged >60 years in molecular remission may have developed during therapy, or have been present at diagnosis and unmasked in remission.
In summary, we showed potentially pathogenic somatic variants of epigenetic modifiers are common in CML-CP at diagnosis, and when combined with other risk factors may be promising predictive biomarkers determining which is the best TKI for each individual.
Our study has some limitations, the most important of which are small sample size (although it is the largest study to date assessing the effect of genetic variants on survival), the potential exaggeration of the effect size due to the selection of the extreme responders / non-responders, the limited number of target genes, and the retrospective nature of our observations. Furthermore, we were unable to assess the impact of additional chromosomal abnormalities (ACA)50 due to the absence of cytogenetic data at diagnosis. The intriguing question remains as to whether any of the variants identified has more or less impact on prognosis, but because of the small numbers of each variant we were unable to explore this in more detail. We now wish to see our panel enriched by the addition of genes found to be altered in CML-CP by targeted, exome or WGS, and validated in larger, unselected, consecutive cohorts of individuals with CML. Our findings, if confirmed in a prospective study, could assist in distinguishing individuals who would benefit starting therapy with a more potent 2G-TKI rather than imatinib.
Acknowledgments
We would also like to thank the Molecular Pathology Laboratory and the John Goldman Centre for Cellular Therapy for providing us with guanidinium thiocyanate (GTC) lysates and cells from CML and normal subjects’ samples. We are grateful to individuals with CML for participating in this study.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/12/2400
Funding
JFA is a NIHR Senior Investigator and we are grateful for the support from the Imperial College NIHR Biomedical Research Centre.
References
- 1.Innes AJ, Milojkovic D, Apperley JF. Allogeneic transplantation for CML in the TKI era: striking the right balance. Nat Rev Clin Oncol. 2016;13(2):79–91. [DOI] [PubMed] [Google Scholar]
- 2.Brummendorf TH, Cortes JE, de Souza CA, et al. Bosutinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukaemia: results from the 24-month follow-up of the BELA trial. Br J Haematol. 2015;168(1):69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortes JE, Saglio G, Kantarjian HM, et al. Final 5-Year Study Results of DASISION: The Dasatinib Versus Imatinib Study in Treatment-Naive Chronic Myeloid Leukemia Patients Trial. J Clin Oncol. 2016; 34(20):2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hochhaus A, Saglio G, Hughes TP, et al. Long-term benefits and risks of frontline nilotinib vs imatinib for chronic myeloid leukemia in chronic phase: 5-year update of the randomized ENESTnd trial. Leukemia. 2016;30(5):1044–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aichberger KJ, Herndlhofer S, Schernthaner GH, et al. Progressive peripheral arterial occlusive disease and other vascular events during nilotinib therapy in CML. Am J Hematol. 2011;86(7):533–539. [DOI] [PubMed] [Google Scholar]
- 6.Montani D, Bergot E, Gunther S, et al. Pulmonary arterial hypertension in patients treated by dasatinib. Circulation. 2012;125 (17):2128–2137. [DOI] [PubMed] [Google Scholar]
- 7.Hasford J, Baccarani M, Hoffmann V, et al. Predicting complete cytogenetic response and subsequent progression-free survival in 2060 patients with CML on imatinib treatment: the EUTOS score. Blood. 2011; 118(3):686–692. [DOI] [PubMed] [Google Scholar]
- 8.Hasford J, Pfirrmann M, Hehlmann R, et al. A new prognostic score for survival of patients with chronic myeloid leukemia treated with interferon alfa. Writing Committee for the Collaborative CML Prognostic Factors Project Group. J Natl Cancer Inst. 1998;90(11):850–858. [DOI] [PubMed] [Google Scholar]
- 9.Sokal JE, Cox EB, Baccarani M, et al. Prognostic discrimination in “good-risk” chronic granulocytic leukemia. Blood. 1984;63(4):789–799. [PubMed] [Google Scholar]
- 10.Pfirrmann M, Baccarani M, Saussele S, et al. Prognosis of long-term survival considering disease-specific death in patients with chronic myeloid leukemia. Leukemia. 2016;30(1):48–56. [DOI] [PubMed] [Google Scholar]
- 11.Baccarani M, Deininger MW, Rosti G, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122(6):872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanfstein B, Muller MC, Hehlmann R, et al. Early molecular and cytogenetic response is predictive for long-term progression-free and overall survival in chronic myeloid leukemia (CML). Leukemia. 2012;26(9): 2096–2102. [DOI] [PubMed] [Google Scholar]
- 13.Marin D, Ibrahim AR, Lucas C, et al. Assessment of BCR-ABL1 transcript levels at 3 months is the only requirement for predicting outcome for patients with chronic myeloid leukemia treated with tyrosine kinase inhibitors. J Clin Oncol. 2012;30(3): 232–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochhaus A, Larson RA, Guilhot F, et al. Long-Term Outcomes of Imatinib Treatment for Chronic Myeloid Leukemia. N Engl J Med. 2017;376(10):917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoi K, Cross NC. Molecular pathogenesis of atypical CML, CMML and MDS/MPN-unclassifiable. Int J Hematol. 2015; 101(3):229–242. [DOI] [PubMed] [Google Scholar]
- 16.Zent CS, Burack WR. Mutations in chronic lymphocytic leukemia and how they affect therapy choice: focus on NOTCH1, SF3B1, and TP53. Hematology Am Soc Hematol Educ Program. 2014;2014(1):119–124. [DOI] [PubMed] [Google Scholar]
- 17.Boultwood J, Perry J, Pellagatti A, et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 2010;24(5):1062–1065. [DOI] [PubMed] [Google Scholar]
- 18.Grossmann V, Kohlmann A, Zenger M, et al. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia. 2011;25(3):557–560. [DOI] [PubMed] [Google Scholar]
- 19.Makishima H, Jankowska AM, McDevitt MA, et al. CBL, CBLB, TET2, ASXL1, and IDH1/2 mutations and additional chromosomal aberrations constitute molecular events in chronic myelogenous leukemia. Blood. 2011;117(21):e198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soverini S, de Benedittis C, Mancini M, Martinelli G. Mutations in the BCR-ABL1 Kinase Domain and Elsewhere in Chronic Myeloid Leukemia. Clin Lymphoma Myeloma Leuk. 2015;15 Suppl:S120–128. [DOI] [PubMed] [Google Scholar]
- 21.Togasaki E, Takeda J, Yoshida K, et al. Frequent somatic mutations in epigenetic regulators in newly diagnosed chronic myeloid leukemia. Blood Cancer J. 2017;7(4):e559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mologni L, Piazza R, Khandelwal P, Pirola A, Gambacorti-Passerini C. Somatic mutations identified at diagnosis by exome sequencing can predict response to imatinib in chronic phase chronic myeloid leukemia (CML) patients. Am J Hematol. 2017;92(10):E623–E625. [DOI] [PubMed] [Google Scholar]
- 23.Branford S, Wang P, Yeung DT, et al. Integrative genomic analysis reveals cancer-associated mutations at diagnosis of CML in patients with high-risk disease. Blood. 2018;132(9):948–961. [DOI] [PubMed] [Google Scholar]
- 24.Kim T, Tyndel MS, Zhang Z, et al. Exome sequencing reveals DNMT3A and ASXL1 variants associate with progression of chronic myeloid leukemia after tyrosine kinase inhibitor therapy. Leuk Res. 2017;59: 142–148. [DOI] [PubMed] [Google Scholar]
- 25.Elena C, Gallì A, Bianchessi A, et al. Somatic Mutations Are Frequently Detected in Chronic Myeloid Leukemia in Chronic Phase and Do Not Affect Response to Tyrosine-Kinase Inhibitors. Blood. 2016;128(22):1117. [Google Scholar]
- 26.Ernst T, Busch M, Rinke J, et al. Frequent ASXL1 mutations in children and young adults with chronic myeloid leukemia. Leukemia. 2018;32(9):2046–2049. [DOI] [PubMed] [Google Scholar]
- 27.Kim T, Tyndel MS, Kim HJ, et al. Spectrum of somatic mutation dynamics in chronic myeloid leukemia following tyrosine kinase inhibitor therapy. Blood. 2017; 129(1):38–47. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt M, Rinke J, Schafer V, et al. Molecular-defined clonal evolution in patients with chronic myeloid leukemia independent of the BCR-ABL status. Leukemia. 2014;28(12):2292–2299. [DOI] [PubMed] [Google Scholar]
- 29.Fialkow PJ, Martin PJ, Najfeld V, Penfold GK, Jacobson RJ, Hansen JA. Evidence for a multistep pathogenesis of chronic myelogenous leukemia. Blood. 1981;58(1):158–163. [PubMed] [Google Scholar]
- 30.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bazeos A, Lowe R, Nteliopoulos G, et al. The CML epigenome shows dynamic changes in the CD34+ compartment in patients who achieve complete cytogenetic response on tyrosine kinase inhibitors [abstract]. Haematologica. 2013;98(suppl 1):255.22929980 [Google Scholar]
- 33.Cross NC, White HE, Colomer D, et al. Laboratory recommendations for scoring deep molecular responses following treatment for chronic myeloid leukemia. Leukemia. 2015;29(5):999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Brien SG, Guilhot F, Larson RA, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348(11):994–1004. [DOI] [PubMed] [Google Scholar]
- 35.Bower H, Bjorkholm M, Dickman PW, Hoglund M, Lambert PC, Andersson TM. Life Expectancy of Patients With Chronic Myeloid Leukemia Approaches the Life Expectancy of the General Population. J Clin Oncol. 2016;34(24):2851–2857. [DOI] [PubMed] [Google Scholar]
- 36.Alonso-Dominguez JM, Grinfeld J, Alikian M, et al. PTCH1 expression at diagnosis predicts imatinib failure in chronic myeloid leukaemia patients in chronic phase. Am J Hematol. 2015;90(1):20–26. [DOI] [PubMed] [Google Scholar]
- 37.Kok CH, Leclercq T, Watkins DB, et al. Elevated PTPN2 expression is associated with inferior molecular response in de-novo chronic myeloid leukaemia patients. Leukemia. 2014;28(3):702–705. [DOI] [PubMed] [Google Scholar]
- 38.McWeeney SK, Pemberton LC, Loriaux MM, et al. A gene expression signature of CD34+ cells to predict major cytogenetic response in chronic-phase chronic myeloid leukemia patients treated with imatinib. Blood. 2010;115(2):315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucas CM, Harris RJ, Holcroft AK, et al. Second generation tyrosine kinase inhibitors prevent disease progression in high-risk (high CIP2A) chronic myeloid leukaemia patients. Leukemia. 2015; 29(7):1514–1523. [DOI] [PubMed] [Google Scholar]
- 40.Jelinek J, Gharibyan V, Estecio MR, et al. Aberrant DNA methylation is associated with disease progression, resistance to imatinib and shortened survival in chronic myelogenous leukemia. PLoS One. 2011;6(7):e22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Machova Polakova K, Lopotova T, Klamova H, et al. Expression patterns of microRNAs associated with CML phases and their disease related targets. Mol Cancer. 2011;10:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grinfeld J, Gerrard G, Alikian M, et al. A common novel splice variant of SLC22A1 (OCT1) is associated with impaired responses to imatinib in patients with chronic myeloid leukaemia. Br J Haematol. 2013;163(5):631–639. [DOI] [PubMed] [Google Scholar]
- 43.Marum JE, Yeung DT, Purins L, et al. ASXL1 and BIM germ line variants predict response and identify CML patients with the greatest risk of imatinib failure. Blood Adv. 2017;1(18):1369–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng KP, Hillmer AM, Chuah CT, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18(4):521–528. [DOI] [PubMed] [Google Scholar]
- 45.Perrotti D, Jamieson C, Goldman J, Skorski T. Chronic myeloid leukemia: mechanisms of blastic transformation. J Clin Invest. 2010;120(7):2254–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heller G, Topakian T, Altenberger C, et al. Next-generation sequencing identifies major DNA methylation changes during progression of Ph+ chronic myeloid leukemia. Leukemia. 2016;30(9):1861–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castagnetti F, Gugliotta G, Breccia M, et al. Long-term outcome of chronic myeloid leukemia patients treated frontline with imatinib. Leukemia. 2015;29(9):1823–1831. [DOI] [PubMed] [Google Scholar]
- 48.Pfirrmann M, Evtimova D, Saussele S, et al. No influence of BCR-ABL1 transcript types e13a2 and e14a2 on long-term survival: results in 1494 patients with chronic myeloid leukemia treated with imatinib. J Cancer Res Clin Oncol. 2017;143(5):843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain P, Kantarjian H, Patel KP, et al. Impact of BCR-ABL transcript type on outcome in patients with chronic-phase CML treated with tyrosine kinase inhibitors. Blood. 2016;127(10):1269–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fabarius A, Kalmanti L, Dietz CT, et al. Impact of unbalanced minor route versus major route karyotypes at diagnosis on prognosis of CML. Ann Hematol. 2015; 94(12):2015–2024. [DOI] [PubMed] [Google Scholar]