Abstract
Sleep is a behavior that exists broadly across animal phyla, from flies to humans, and is necessary for normal brain function. Recent studies in both vertebrates and invertebrates have suggested a role for glial cells in sleep regulatory processes. Changes in neural-glial interactions have been shown to be critical for synaptic plasticity and circuit function. Here, we wanted to test the hypothesis that changes in sleep pressure alters neural-glial interactions. In the fruit fly, Drosophila melanogaster, sleep is known to be regulated by mushroom body (MB) circuits. We used the technique GFP Reconstitution Across Synaptic Partners (GRASP) to test whether changes in sleep pressure affect neural-glial interactions between MB neurons and astrocytes, a specialized glial cell type known to regulate sleep in flies and mammals. The MB-astrocyte GRASP signal was reduced after 24 h of sleep deprivation, whereas the signal returned to baseline levels following 72 h of recovery. Social enrichment, which increases sleep drive, similarly reduced the MB-astrocyte GRASP signal. We did not observe any changes in the MB-astrocyte GRASP signal over time-of-day, or following paraquat exposure or starvation. These data suggest that changes in sleep pressure are linked to dynamic changes in neural-glial interactions between astrocytes and neuronal sleep circuits, which are not caused by normal rest-activity cycles or stressors.
Keywords: Glia, Astrocyte, Sleep Homeostasis, Sleep Deprivation, Fruit Fly
Introduction
Sleep is a ubiquitous behavior exhibited broadly throughout animal phyla, yet its functions remain enigmatic. Fundamental properties of sleep function may be revealed by determining phylogenetically conserved mechanisms associated with sleep behavior across evolutionarily distant species. Sleep is affected both by the amount of time spent awake, as well as experience-dependent changes in synaptic plasticity. Sleep regulatory processes have largely focused on the role of neurons; however, glial cells also play an important role in sleep, across invertebrates and vertebrates alike, including humans [1] [2] [3] [4] [5] [6] [7].
The fruit fly Drosophila melanogaster is a powerful model for exploring neural and glial processes that regulate sleep. For example, sleep in Drosophila has been shown to be regulated independently by mushroom body (MB) neural circuits [8] [9] as well as glial cells [1] [4] [6] [10] [11]. However, it is currently unclear how MB neurons and glial cells physically interact as a result of changes in sleep pressure. The GFP Reconstitution Across Synaptic Partners (GRASP) technique is a method that labels the membrane contact of two cell types by complementary fragments of the green fluorescent protein (GFP) molecule [12]. Here we use the GRASP technique to label interactions between MB neurons and astrocytes, a specific glial cell type known to regulate sleep in flies and mammals [1] [2] [5]. Determining that changes in neural-glial interactions are associated with sleep pressure would provide a new model system to test functional aspects of sleep-regulatory molecular events that may be conserved across species.
Objective
To determine whether changes in sleep pressure regulate mushroom body neural-glial interactions in flies.
Results & Discussion
Our GRASP studies utilized flies harboring mb247-splitGFP11 (specific to MB neurons) [13] [14] and the UAS-splitGFP1–10 expressed in glia, using the astrocyte-directed Alrm- GAL4 driver [15]. MB neural-glial interactions are quantified by the presence of a GFP fluorescent signal, which is triggered upon contact of the individual GRASP components [12]. MB-astrocyte GRASP flies also express Ds-Red under control of the mb247 MB neuronal promoter, which labels the whole MB region of the fly brain. MB-astrocyte GRASP flies were subjected to 24 h of mechanical sleep deprivation [16] and brains were harvested and examined for changes in GFP signal compared to control (undisturbed) flies using confocal microscopy (Fig. 1A-C). All flies were examined at the same time-of-day (ZTo, lights-on). MB-astrocyte GRASP flies showed a significant reduction in GFP signal (Corrected Total Cell Fluorescence, see methods) following sleep deprivation compared to control flies (Fig. 1D). Following 72 h of recovery after 24 h of sleep deprivation, we observed that the GFP signal returned to control levels (Fig. 1D). This MB-astrocyte GRASP response to sleep deprivation and recovery closely resembled the amount of sleep observed under these conditions (Fig. 1E).
Figure 1. Increased sleep pressure decreases neural-glial interactions between mushroom body neurons and astrocytes in Drosophila melanogaster.
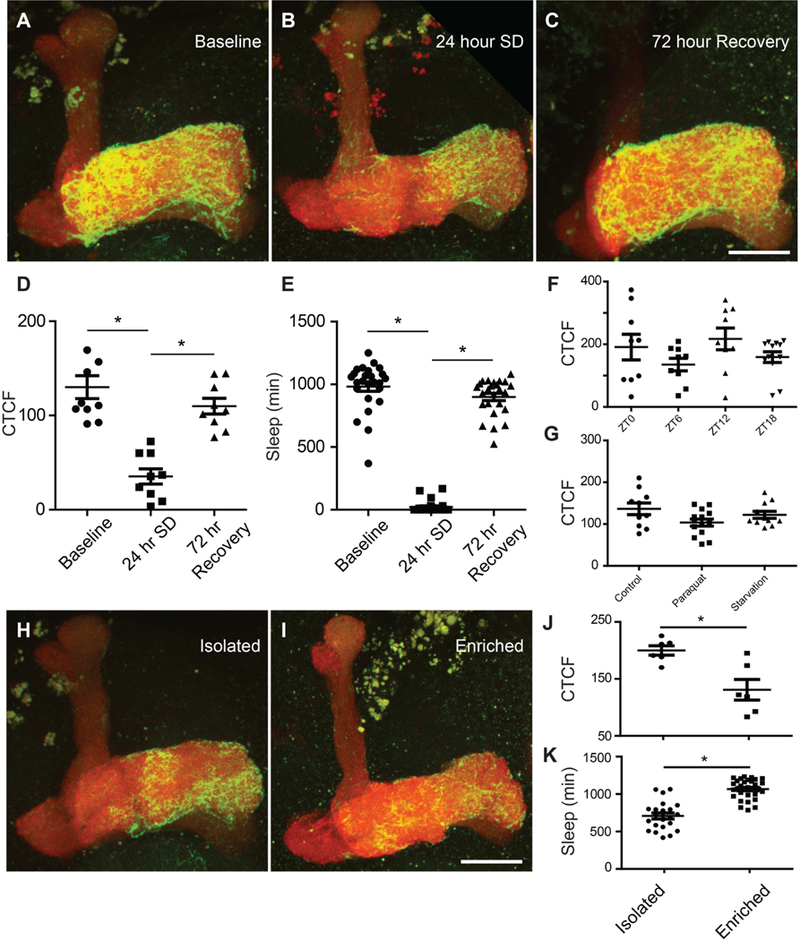
(A) Representative image of MB-astrocyte GRASP signal in baseline (non-sleep deprived) condition.
(B) Representative image of MB-astrocyte GRASP signal after 24 h of sleep deprivation (SD).
(C) Representative image of MB-astrocyte GRASP signal after 72 h of recovery following 24 h of SD. Scale bar 25 μm.
(D) Average CTCF signal in groups from the 24 h of baseline (non-sleep deprived) condition, after 24 h of sleep deprivation (SD), and after 72 h of recovery following 24 h of SD. One-way ANOVA revealed a significant effect of sleep loss on GRASP signal (P<0.0001). Tukey multiple comparisons test showed a significant reduction in GRASP signal after 24 h of SD when compared to both baseline (*P<0.001) and after 72 h of recovery following 24 h of SD (*P<0.001).
(E) Average sleep amount (min) per 24 h day in groups from the 24 h of baseline (non-sleep deprived) condition, during 24 h of sleep deprivation (SD), and for the final 24 h period after 72 h of recovery following 24 h of SD. One-way ANOVA confirmed a significant effect of sleep loss (P<0.0001). Tukey multiple comparisons test showed a significant reduction in sleep during SD when compared to baseline (*P<0.001) and during 72 h of recovery following 24 h of SD (*P<o.oo1).
(F) Diurnal measures of MB-astrocyte GRASP signal in flies on a 12:12 light-dark cycle. One-way ANOVA indicated no significant changes across time-of-day (P=o.22). ZTo (zeitgeber time 0) = lights on, ZT12 = lights off.
(G) Stress induced by 24 h of paraquat treatment (0.1 mM) or by 24 h of starvation did not affect MB-astrocyte GRASP signal compared to control flies. One-way ANOVA (P=0.08); Tukey multiple comparisons tests among conditions (n.s.).
(H) Representative image of MB-astrocyte GRASP signal in control (isolated flies).
(I) Representative image of MB-astrocyte GRASP signal in flies following social enrichment (enriched flies). Scale bar 25 μm.
(J) Effect of social enrichment (enriched flies) on MB-astrocyte GRASP signal compared to control (isolated flies). T-test (*P<0.01).
(K) Total sleep amount (min) in the 24 h day from MB-astrocyte GRASP flies following social enrichment (enriched flies) compared to control (isolated flies). T-test (*P<0.0001).
To determine whether the changes in MB-astrocyte GRASP signal may have been influenced by the normal sleep-wake cycle, we performed brain dissections on MB-astrocyte GRASP flies at multiple times-of-day. We did not find any differences in the diurnal profile of GFP signal (Fig. 1F), indicating that the observed differences in MB-astrocyte GRASP signal were due to changes in sleep pressure, and not normal sleep-wake cycles. To rule out the possibility that the differences in GFP signal were caused by stress effects, we examined MB-astrocyte GRASP flies for changes in GFP signal following paraquat treatment and starvation stressors. We did not observe any differences in GFP signal in stressor-treated MB-astrocyte GRASP flies compared to control flies (Fig. 1G). To determine whether the decreases in MB-astrocyte GRASP signal were due to increases in sleep pressure, and not a nonspecific artifact of mechanical sleep deprivation, we tested flies following a social enrichment paradigm, which is known to increase sleep drive in flies [17]. Compared to control (isolated flies), MB-astrocyte GRASP flies subjected to social enrichment (enriched flies) showed a significant reduction in GFP signal (Fig. 1H-J) in tandem with a significant increase in sleep (Fig. 1K), pointing to sleep pressure as the specific driver of the changes in MB-astrocyte GRASP signal.
Sleep deprivation is detrimental to cognitive functioning [18], including learning and memory [19], as well as overall health [20]. Yet, we lack a fundamental understanding of sleep function [21]. The fly model is a powerful tool to understand sleep regulatory processes and functions [22]. Here, we used the GRASP technique to show that increases in sleep pressure decrease neural-glial interactions between MB neurons and astrocytes. Dynamic changes in neural-glial interactions may influence sleep homeostasis through glial uptake of neurotransmitters, efficiency of gliotransmission, and glial processes underlying metabolic support of neurons. Future studies examining phylogenetically conserved molecular pathways in regulating the dynamic relationship of sleep pressure with sleep deprivation- and experience-dependent changes in neural-glial interactions will help to better understand the functional roles for sleep behavior.
Conclusions
This study shows that changes in sleep pressure regulate MB neural-glial interactions in flies.
Limitations
We used Drosophila melanogaster to test whether changes in sleep pressure regulate changes in neural-glial interactions. The current studies are limited to MB neurons and astrocytes, and we have not determined whether other sleep circuits or other glial cells are responsive to changes in sleep pressure. Our studies are also limited to the GRASP technique, and alternative strategies, such as immunohistochemistry against neural-glial extra-cellular matrix proteins or cell adhesion molecules [23] or electron microscopy-based morphological studies [24] may provide alternative means to examine sleep-dependent changes in neural-glial interactions. More research is needed to fully understand the time constants of the changes at the neural-glial interface. These changes may take place at time scales ranging from milliseconds to hours, and perhaps even days. Our study was limited to assessing the effects of acute (24 h) sleep deprivation. Our studies are also limited to the fly model, and testing vertebrate models is desirable in order to determine whether the observed changes in neural-glial interactions are phylogenetically conserved. Recent studies demonstrated sleep-dependent changes in the astrocyte glutamate transporter, GLT1, apposition to hypothalamic neurons in mice [25]. However, these studies did not conclude whether the changes were due to a redistribution of GLT1 protein in astrocytes, or to structural changes of astrocyte processes (e.g., extension/retraction) onto these neurons.
Alternative Explanations
Conjectures
Future studies using, e.g., RNAi-based strategies to knock down cell type-specific target proteins involved in cell growth or morphology or the maintenance of the extracellular matrix in our model will be needed to determine mechanisms underlying sleep- dependent changes in neural-glial interactions. In addition, since the MB is an important brain region for learning and memory processes in flies [26], testing whether changes in neural-glial interactions are important for cognitive function would be a worthwhile direction for future studies.
Additional Information
Methods
Fly stocks
Alrm-Gal4 stocks were obtained from the Bloomington Drosophila Stock Center (Indiana University). The mb247-DsRed; mb247-splitGFP11, UAS-splitGFP1–10/TM3sb flies were obtained from T. Riemensperger and A. Fiala (University of Göttingen).
Fly husbandry
Flies were cultured at 25°C, 60% humidity, maintained on a 12:12 Light:Dark cycle, on Nutri-fly Bloomington Formulation fly food (Genesee Scientific, San Diego, CA). Newly eclosed virgin female flies were collected from culture vials daily under CO2 anesthesia and housed in groups of approximately 30 prior to experimentation.
Sleep analysis
Female flies 4–7 days after eclosion were used for all sleep studies. Flies were mouth aspirated into 5 mm × 65 mm (outside diameter × length) polycarbonate recording tubes (Trikinetics, Waltham, MA) with food (Bloomington Nutri-fly formula) on one end and yarn plugs on the other. Sleep parameters were continuously evaluated using the Trikinetics Drosophila activity monitoring system (DAMS; Trikinetics, Waltham, MA) as described previously [27]. One acclimatization day was followed by 2 days of baseline sleep recording, one 24 h period of mechanical sleep deprivation, and 72 h of recovery sleep. Sleep deprivation was performed using a Sleep Nullifying Apparatus (SNAP), which produces waking without nonspecifically activating stress responses [16], as described previously [27].
Imaging
Drosophila brains were dissected in phosphate-buffered saline (0.9% NaCl, 10 mm NaH2PO4, pH 7.2) containing 0.3% Triton X-100 (PBS-T) and fixed in 4% paraformaldehyde, washed, and mounted on cover slips. Optical sections were collected with a Leica DMi8 laser scanning confocal microscope. For each experiment, calibration on the microscope was held constant by establishing a signal threshold value for the control group. GRASP intensity levels were measured using Corrected Total Cell Fluorescence (CTCF). The corrected total fluorescence = Integrated Fluorescence density – (Area of ROI multiplied by Mean Fluorescence of background) and was calculated in max projected image stacks with the region of interest (ROI) around the mushroom bodies.
Stress and starvation
Virgin female flies were collected as described above. 4–7 days after eclosion, animals were mouth aspirated into 5 mm × 65 mm (outside diameter × length) polycarbonate recording tubes (Trikinetics, Waltham, MA) containing 0.1 mM Paraquat in minimal media (2% agar, 5% sucrose in ddH2O) or starvation food (2% agar in ddH2O). Animals were housed for 24 h on these media and afterwards were rapidly dissected at ZT0 for imaging of GRASP signal.
Social enrichment
To standardize the environmental conditions during critical periods of brain development, virgin female flies were collected upon eclosion and maintained in same-sex vials containing approximately 30 flies for 2 days. This protocol kept environmental conditions constant between subsequently isolated and enriched flies for the first 2 days of adult life. 3 day old flies were then divided into a socially isolated group, in which flies were individually housed in 5 mm × 65 mm plastic tubes, and a socially enriched group, consisting of 50 female flies housed in a single vial. After 5 days of social enrichment/isolation, flies were placed into clean 5 mm × 65 mm plastic tubes and sleep was recorded for 3 days using the Trikinetics DAMS.
Data analysis
Statistics were calculated using Graphpad Prism software. Student’s t-test, one-way ANOVA, two-way ANOVA, and Tukey post-hoc analysis were used for analyses. Sleep data were analyzed by averaging across multiple experiments. Flies that did not survive the entire experimental paradigm were removed from data analysis.
Triple Blind Peer Review
The handling editor, the reviewers,and the authors are all blinded during the review process.
Creative Commons 4.0
This observation is distributed under the terms of the Creative Commons Attribution 4.0 International License
Acknowledgements
We would like to thank the Washington State University Microscopy Core Facility for use of core facility microscopes and T. Riemensperger and A. Fiala for providing the mb247-DsRed; mb247-splitGFP11, UAS-splitGFP1-10/TM3sb flies. We would also like to thank R. Taylor for technical assistance.
Funding Statement
HVD and MGF were supported by CDMRP grant W81XWH-18-1-0100. This work was also supported by NIH grant MH099544 (MGF).
Footnotes
Ethics Statement
Not Applicable.
Citations
- [1].Gerstner Jason R. et al. “Normal sleep requires the astrocyte brain-type fatty acid binding protein FABP7”. In: Science Advances 3.4 (2017), ei602663. Doi: 10.1126/sciadv.1602663 URL: 10.1126/sciadv.1602663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Halassa Michael M. et al. “Astrocytic Modulation of Sleep Homeostasis and Cognitive Consequences of Sleep Loss”. In: Neuron 61.2 (2009), pp. 213–219. doi: 10.1016/j.neuron.2008.11.024 URL: 10.1016/j.neuron.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Katz Menachem et al. “Glia Modulate a Neuronal Circuit for Locomotion Suppression during Sleep in C. elegans”. In: Cell Reports 22.10 (2018), pp. 2575–2583. doi: 10.1016/j.celrep.2018.02.036 URL: 10.1016/j.celrep.2018.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fropf Robin, Zhou Hong, and Yin Jerry C.P. “The clock gene period differentially regulates sleep and memory in Drosophila”. In: Neurobiology of Learning and Memory 153 (2018), pp. 2–12. doi: 10.1016/j.nlm.2018.02.016 URL: https://doi.org/10.1016Zj.nlm.2018.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Clasadonte Jerome et al. “Connexin 43-Mediated Astroglial Metabolic Networks Contribute to the Regulation of the Sleep-Wake Cycle”. In: Neuron 95.6 (2017), 1365–1380.E5. doi: 10.1016/j.neuron.2017.08.022 URL: 10.1016/j.neuron.2017.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen W-F et al. “A neuron-glia interaction involving GABA transaminase contributes to sleep loss in sleepless mutants”. In: Molecular Psychiatry 20.2 (2014), pp. 240–251. doi: 10.1038/mp.2014.11 URL: 10.1038/mp.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Vanderheyden William M. et al. “Astrocyte expression of the Drosophila TNF-alpha homologue, Eiger, regulates sleep in flies”. In: PLOS Genetics 14.10 (2018), e1007724. doi: 10.1371/journal.pgen.1007724 URL: 10.1371/journal.pgen.1007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pitman Jena L. et al. “A dynamic role for the mushroom bodies in promoting sleep in Drosophila”. In: Nature 441.7094 (2006), pp. 753–756. doi: 10.1038/nature04739 URL: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- [9].Joiner William J. et al. “Sleep in Drosophila is regulated by adult mushroom bodies”. In: Nature 441.7094 (2006), pp. 757–760. Doi: 10.1038/nature04811 URL: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- [10].Artiushin Gregory et al. “Endocytosis at the Drosophila blood-brain barrier as a function for sleep”. In: eLife 7 (2018), e43326. doi: 10.7554/elife.43326 URL: 10.7554/elife.43326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stahl Bethany A. et al. “The Taurine Transporter Eaat2 Functions in Ensheathing Glia to Modulate Sleep and Metabolic Rate”. In: Current Biology 28.22 (2018), 3700–3708.e4. doi: 10.1016/j.cub.2018.10.039 URL: 10.1016/j.cub.2018.10.039. [DOI] [PubMed] [Google Scholar]
- [12].Feinberg Evan H. et al. “GFP Reconstitution Across Synaptic Partners (GRASP) Defines Cell Contacts and Synapses in Living Nervous Systems”. In: Neuron 57.3 (2008), pp. 353–363. Doi: 10.1016/j.neuron.2007.11.030 URL: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- [13].Pech Ulrike et al. “Localization of the contacts between Kenyon cells and aminergic neurons in theDrosophila melanogasterbrain using splitGFP reconstitution”. In: Journal of Comparative Neurology 521 (2013), pp. 3992–4026. doi: 10.1002/cne.23388 URL: 10.1002/cne.23388. [DOI] [PubMed] [Google Scholar]
- [14].Pech Ulrike et al. “Mushroom body miscellanea: transgenic Drosophila strains expressing anatomical and physiological sensor proteins in Kenyon cells”. In: Frontiers in Neural Circuits 7 (2013), p. 147. doi: 10.3389/fncir.2013.00147 URL: 10.3389/fncir.2013.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Doherty Johnna et al. “Ensheathing Glia Function as Phagocytes in the Adult Drosophila Brain”. In: Journal ofNeuroscience 29.15 (2009), pp. 4768–4781. Doi: 10.1523/jneurosci.5951-08.2009 URL: 10.1523/jneurosci.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shaw Paul J. et al. “Stress response genes protect against lethal effects of sleep deprivation in Drosophila”. In: Nature 417.6886 (2002), pp. 287–291. doi: 10.1038/417287a URL: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- [17].Ganguly-Fitzgerald Indrani, Donlea Jeff, and Shaw Paul J. “Waking Experience Affects Sleep Need in Drosophila”. In: Science 313.5794 (2006), pp. 1775–1781. Doi: 10.1126/science.1130408 URL: 10.1126/science.1130408. [DOI] [PubMed] [Google Scholar]
- [18].Jackson Melinda L. et al. “Deconstructing and reconstructing cognitive performance in sleep deprivation”. In: Sleep Medicine Reviews 17.3 (2013), pp. 215–225. doi: 10.1016/j.smrv.2012.06.007 URL: 10.1016/j.smrv.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Diekelmann Susanne and Born Jan. “The memory function of sleep”. In: Nature Reviews Neuroscience 11.2 (2010), pp. 114–126. doi: 10.1038/nrn2762 URL: 10.1038/nrn2762. [DOI] [PubMed] [Google Scholar]
- [20].Medic Goran, Wille Micheline, and Hemels Michiel E. H. “Short- and long-term health consequences of sleep disruption”. In: Nature and Science of Sleep 9 (2017), pp. 151–161. doi: 10.2147/nss.s134864 URL: 10.2147/nss.s134864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Krueger James M. et al. “Sleep function: Toward elucidating an enigma”. In: Sleep Medicine Reviews 28 (2016), pp. 46–54. Doi: 10.1016/j.smrv.2015.08.005 URL: 10.1016/j.smrv.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Donlea Jeffrey M “Roles for sleep in memory: insights from the fly”. In: Current Opinion in Neurobiology 54 (2019), pp. 120–126. doi: 10.1016/j.conb.2018.10.006 URL: 10.1016/j.conb.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Matsuno Motomi et al. “Long-Term Memory Formation in Drosophila Requires Training-Dependent Glial Transcription”. In: Journal ofNeuroscience 35.14 (2015), pp. 5557–5565. Doi: 10.1523/jneurosci.3865-14.2015 URL: 10.1523/jneurosci.3865-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bellesi Michele et al. “Effects of sleep and wake on astrocytes: clues from molecular and ultrastructural studies”. In: BMC Biology 13.1 (2015), p. 66 Doi: 10.1186/s12915-015-0176-7 URL: 10.1186/s12915-015-0176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Briggs Chantalle, Hirasawa Michiru, and Semba Kazue. “Sleep Deprivation Distinctly Alters Glutamate Transporter 1 Apposition and Excitatory Transmission to Orexin and MCH Neurons”. In: Journal of Neuroscience 38.10 (2018), pp. 2505–2518. doi: 10.1523/jneurosci.2179-17.2018 URL: 10.1523/jneurosci.2179-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cognigni Paola, Felsenberg Johannes, and Waddell Scott. “Do the right thing: neural network mechanisms of memory formation, expression and update in Drosophila”. In: Current Opinion in Neurobiology 49 (2018), pp. 51–58. Doi: 10.1016/j.conb.2017.12.002 URL: 10.1016/j.conb.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shaw Paul J. et al. “Correlates of Sleep and Waking in Drosophila melanogaster”. In: Science 287.5459 (2000), pp. 1834–1837. Doi: 10.1126/science.287.5459.1834 URL: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]


