ABSTRACT
The specific and rapid formation of protein complexes, involving IQGAP family proteins, is essential for diverse cellular processes, such as adhesion, polarization, and directional migration. Although CDC42 and RAC1, prominent members of the RHO GTPase family, have been implicated in binding to and activating IQGAP1, the exact nature of this protein-protein recognition process has remained obscure. Here, we propose a mechanistic framework model that is based on a multiple-step binding process, which is a prerequisite for the dynamic functions of IQGAP1 as a scaffolding protein and a critical mechanism in temporal regulation and integration of cellular pathways.
KEYWORDS: CDC42, IQGAP1, RAC1, RHO effectors, RHO GTPases
The RHO family GTPases, most prominently CDC42, RAC1, and RHOA, are known to play an important role in diverse cellular processes and progression of different diseases, such as cardiovascular diseases, developmental and neurological disorders, as well as in tumor invasion and metastasis.1 RHO GTPases share two common functional characteristics, membrane anchorage and an on/off switch cycle.2 Thus, membrane-associated RHO GTPases act, with some exceptions,3 as molecular switches by cycling between an inactive GDP-bound state and an active GTP-bound state. This cycle underlies 2 critical intrinsic functions, GDP-GTP exchange and GTP hydrolysis3 and is controlled by at least three classes of regulatory proteins, including guanine nucleotide dissociation inhibitors (GDIs), guanine nucleotide exchange factors (GEFs), and GTPase activating proteins (GAPs)4. The formation of the active GTP-bound state of RHO GTPases is accompanied by a conformational change in two regions, known as switch I and II (encompassing amino acids or aa 29–42 and 62–68, respectively)4, which provide a platform for the selective interaction with structurally and functionally diverse effectors, e.g., PAK1, WASP, and IQGAP1. This class of proteins activates a wide variety of downstream signaling cascades5-8 thereby regulating many important physiological and pathophysiological processes in eukaryotic cells.9,10
IQGAP1 is a ubiquitously expressed scaffold protein. It has been assigned to multiple subcellular sites and implicated in multiple functions by most probably safeguarding the strength, efficiency, and specificity of signal transduction (reviewed in refs. 7, 8, 10-13). Notably, IQGAP1 has been implicated as a drug target due to its vital regulatory roles in cancer development14-17 although the molecular mechanism of its functions is unclear. IQGAP1 possesses distinct protein interaction domains and motifs to achieve its scaffolding functions. A prerequisite to understand its cellular properties is the dissection of its distinct domains and the analysis of their interactions with desired protein partners. It contains an N-terminal calponin homology domain (CHD), a coiled-coil repeat region (CC), a tryptophan-containing proline-rich motif-binding region (WW), 4 isoleucine/glutamine-containing motifs (IQ), a RASGAP-related domain (GRD), RASGAP C-terminal domain (RGCT) and very C-terminal domain (CT)6 as illustrated in Fig. 1.
Figure 1.
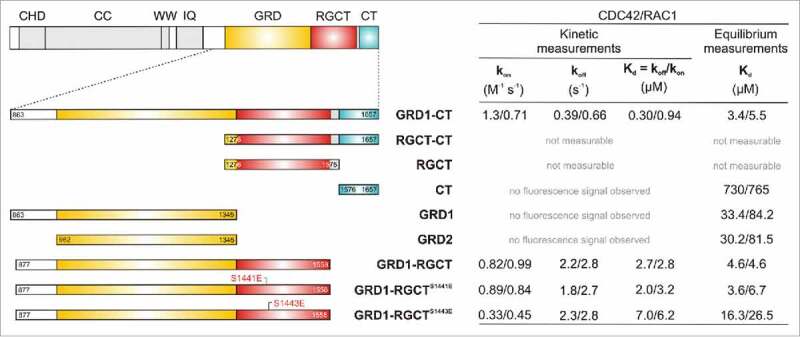
Domain organization of IQGAP1 and different constructs of C-terminal half (left panel) along with individual rate constants of interaction of corresponding proteins with active RAC1 and CDC42 (right panel). In the left panel IQGAP1 domain organization and White et al.6 and different constructs used in our previous study (ref. 42) is shown. Right panel shows the binding affinity of IQGAP1 proteins for RAC1/CDC42 proteins analyzed by stopped-flow fluorometry (kinetic measurements) and fluorescence polarization (equilibrium measurements) (adapted from Nouri et al.42). Kinetic conditions provide individual association and dissociation rate constants (kon and koff) and determine the dissociation constants (Kd) which is obtained from the ratio koff/kon. Equilibrium conditions determine the equilibrium dissociation constants (Kd) directly.
A multitude of IQGAP1 interacting partners have been reported,6,18 among them RAC1 and CDC42,7,19,20 which are crucial for the controlling of IQGAP's activities. For instance, it is well accepted that RAC1 and CDC42 facilitate the function of IQGAP1 in intercellular adhesion sites in epithelial cells, in actin-crosslinking, and determining its subcellular localization.7,21 IQGAP1 is localized in the sites of cell-cell contact22 and its overexpression decreases E-cadherin mediated cell-cell adhesion by interacting with β-catenin and causing the dissociation of α-catenin from the cadherin-catenin complex in epithelial cells.22 Active forms of RAC1 and CDC42 positively regulate E-cadherin-mediated cell-cell adhesion by inhibiting the interaction of IQGAP1 with β-catenin.23 When the amount of active RAC1 and CDC42 increase, they interact with IQGAP1, thus crosslinking actin filaments. Under these conditions, IQGAP1 is not able to bind β-catenin and thus cannot dissociate α-catenin from the cadherin-catenin complex, leading to strong adhesion. On the other hand, when the amount of GDP-bound inactive RAC1/CDC42 increases, IQGAP1 is released from RAC1/CDC42 and then interacts with β-catenin to dissociate α-catenin from the cadherin-catenin complex and this result in weak adhesion.
Another example is in the case of IQGAP1 binding to actin filaments and crosslinking actin filaments.24 This ability to crosslink depends on IQGAP1 dimerization and/or oligomerization.25 In addition, it has been shown that binding of active forms of CDC42 or RAC1 enhance IQGAP1 dimerization or oligomerization.25,26 IQGAP1 directly binds to actin filaments through its N-terminal CHD. The IQGAP1 dimer or CDC42/RAC1-mediated IQGAP1 oligomer utilizes multiple CHDs to crosslink and bundle actin filaments.
While modulation of the cytoskeletal architecture was initially thought to be the primary function of the interaction of IQGAP1 with RHO proteins, it is now clear that they have critical physiological roles beyond the cytoskeleton.10 One example is its neuronal function. IQGAP1 is observed throughout neuronal cells, along neuritis and the developing axon, and also at the growth cone.27 It has been shown that an interaction between IQGAP1 and protein-tyrosine phosphatase PTPµ is necessary for neurite outgrowth in ganglion cells.28 PTPµ forms a complex with IQGAP1, N-cadherin, E-cadherin, and β-catenin.28 Active GTP-bound CDC42 by interacting IQGAP1, promotes the interaction of PTPμ with IQGAP1 to stimulate actin remodeling and, eventually, neurite outgrowth.10,27
In spite of knowing the importance of these interactions, the molecular mechanisms are still obscure and remains to be clearly investigated in vitro. Physical interaction of IQGAP1 with these members of RHO family under cell-free conditions was first reported by Hart et al..29 They showed that the C-terminal half of IQGAP1, encompassing aa 915–1657 and containing the GRD, RGCT, and CT bound to CDC42 and RAC1 in a GTP-dependent fashion, but other deletion fragments did not, most probably due to their instability. Joyal et al. have shown a disruption of the interaction between CDC42 and full-length IQGAP1 in the presence of calmodulin (CaM) and calcium ions.30 It has been suggested that CaM-bound IQGAP1 adopts a different conformation that is less accessible to CDC42 (see also ref. 31). Two interesting observations were made by Swart-Mataraza et al.: On the one hand, CDC42 binding was also shown with IQGAP1-ΔGRD (lacking aa 1122–1324) and on the other hand, IQGAP1 was still able to bind GDP-bound CDC42 although with a lower affinity.32 This suggested that additional regions of IQGAP1, other than GRD, are responsible for CDC42 binding and that IQGAP1 is able to bind CDC42 outside the switch regions in a nucleotide-independent manner. Ho et al. have reported that increasing the calcium concentration enhanced the interaction between calmodulin and IQGAP1, with a concomitant decrease in the association of CDC42 with IQGAP1.33 Zhang et al. have determined an inhibitory constants of 0.4 and 2.1 µM for the inhibition of GTP hydrolysis of CDC42 and RAC1, respectively, in the presence of increasing concentrations of the C-terminal half of IQGAP1 (GRD1-CT, aa 863-1657, Fig. 1).34 Li et al. have mapped the IQGAP1-binding regions on CDC42.35 Switch I and its surrounding regions (aa 29–55) as well as the insert region (aa 122–133) of CDC42 have been suggested as essential determinants for the IQGAP1 binding. Owen et al. have studied the interaction of GRD1-CT with a large panel of CDC42 and RAC1 variants with point mutations within and around switch I/II, α helices 1, 3 and 5, β stands 1, 2 and 3, and the insert region.36 These analyses were conducted in the background of a constitutively active variant of both CDC42 and RAC1 (Gln-61 to leucine or Q61L). This comprehensive study has provided various suggestions for the IQGAP1 interaction with CDC42 vs. RAC1. Accordingly, IQGAP1 reveals only partially overlapping contact sites for CDC42 and RAC1. Despite a sequence identity of more than 70%, these GTPases apparently use different regions to achieve high-affinity interactions with GRD1-CT. Tyr-32 and Val-36 of switch I are critical for both CDC42 and RAC1, whereas Asp-63 and Arg-68 of switch II are critical for only RAC1 and optionally Asn-132 of the insert region only for CDC42. The impact of the latter may be insignificant since CDC42 lacking the insert region still efficiently binds GRD1-CT. However, Owen et al. have determined equilibrium dissociation constants (Kd) of 0.024 and 0.018 µM for the binding of GRD1-CT to CDC42 and RAC1, respectively.36 Moreover, a shorter, mainly GRD-containing fragment, encompassing aa 950-1407, has shown a different binding behavior to the analyzed RHO GTPases. It binds CDC42 with significantly lower affinity (0.14 µM) as compared with GRD1-CT, but does not bind RAC1.36
The crystal structures of a GRD fragment (aa 962–1345; Fig. 1) alone and in complex with the constitutively active variant of CDC42 have been determined.37 Kurella et al. have shown that the GRD adopts a RASGAP-like structure with a conserved central domain (GAPc) that is coupled to the variable flanking regions forming an extra domain (GAPex).37-39 IQGAP1 GRD, however, is functionally an inactive RASGAP due to the lack of critical catalytic and structural fingerprints.37,40,41 Very recently, LeCour et al. have reported that IQGAP1 GRD dimerizes upon binding to CDC42 but not to RAC1.26 Accordingly, four CDC42 molecules interact differently with GAPc and GAPex domains of the GRD dimer by using in both cases various regions, including switch I, II, α3, and the insert region.26 The binding constants obtained for the CDC42 interaction with the GRD were 1.3 and 0.1 µM in the two studies by Kurella et al. and LeCour et al., respectively.26 The CDC42-GAPc interaction, resembling the RAS-RASGAP binding mode,38 has previously been proposed by Owen et al. and Kurella et al..36,37 Such a role of the GRD in associating with CDC42 is astonishing considering aforementioned studies on both GRD1-CT that binds CDC42 with a higher affinity as compared with GRD and an IQGAP1 variant, lacking the GRD, which equally interacts with CDC42 as compared with IQGAP1 wild type. Consistently, Nouri et al. did not observe any fluorescence signal upon mixing CDC42 with the GRD1 or GRD2 which strongly suggest a binding of GRD adjacent to or outside the switch regions of CDC42.42
The studies by Elliot et al. and Nouri et al. have clearly disclosed the critical role of RGCT for the RHO GTPase interactions.42,43 They have shown that an IQGAP1 fragment, containing the GRD and RGCT (aa 877-1558, GRD1-RGCT), is significantly compromised in its ability to tightly bind CDC42 if using phosphomimetic mutations at Ser-1441 and Ser-1443,42,43 originally identified by Li et al..27 Nouri et al. have obtained a very low binding affinity for the interaction between the GRD and CDC42 (Fig. 1) that is 20- to 200-fold lower than that determined by Owen et al., Kurella et al. and LeCour et al. 26,36,37 A principal explanation for this discrepancy is the use of the constitutive active CDC42Q61L by the latter groups that strongly increase the binding affinity for effector proteins, such as IQGAP1.42 Nouri et al. have shown, in addition, that GRD1-RGCT, lacking the CT, dissociates significantly faster from CDC42 as compared with GRD1-CT. This strongly indicates that CT itself may physically contact CDC42 as determined by equilibrium measurements (Fig. 1). Very similar data have been obtained by Nouri et al. for RAC1 interaction with GRD1-CT, GRD1-RGCT and CT (Fig. 1). However, RAC1 interaction with the GRD is different as compared with that of CDC42. All binding constants for the interaction between various IQGAP1 fragments and RHO GTPases along with the techniques applied are tabularly summarized in Nouri et al.42
The switch regions of the RHO family proteins have been previously proposed as the first binding site for the downstream effectors and if this first contact is achieved then additional contacts outside the switch regions are required to fulfill effector activation.4 It is still unclear how RHOGTPases, such as CDC42, activate IQGAP1. However, in vitro studies reported above clearly point to the importance of the C-terminal half of IQGAP1 to achieve the interaction with RAC1- and CDC42-like proteins. It utilizes at least three functionally distinct units, including GRD, RGCT, and CT.42 GRD undergoes a low-affinity, GDP-/GTP-independent complex with RAC1 and CDC42 proteins outside their switch regions. RGCT only binds to the RAC1 and CDC42 proteins if they are active and exist in the GTP-bound forms, and the C-terminal region of IQGAP1 may potentiate the IQGAP1 interaction with RAC1 and CDC42 proteins probably by extending the resident time of the respective proteins complexes. The rationales underlying a multiple-step binding mechanism for the IQGAP1 interaction with CDC42 are summarized in Fig. 2A. First, the RGCT recognize CDC42-GTP and associates with its switch regions with high affinity. Second, the GRD binds CDC42 with a very low affinity adjacent to and/or outside the switch regions and induces an active conformation of full-length IQGAP1 that is now accessible for additional downstream interactions. Third, the CT contacts, although with extremely low affinity, CDC42 outside the switch regions of the whole evolutionary process, which stabilizes this bimolecular interaction and prolongs residence time of IQGAP1 on CDC42. This scenario may be a prerequisite for the dynamic functions of IQGAP1 as a scaffolding protein and a critical mechanism in temporal regulation and integration of IQGAP1-mediated cellular responses.42 In a different scenario, IQGAP1 acts as a scaffold protein upstream of the CDC42-WASP Pathway. The formation of such a ternary complex is likely initiated by phosphorylation of IQGAP1 at Ser-1443 leading to the release of its autoinhibited state that allows, as previously proposed,44,45 the exposed C-terminal half to activate N-WASP in a CDC42-dependent manner and consequently leading to actin polymerization at the leading edge of the cells. The ability of GRD in binding outside the switch regions of CDC42 may facilitate the scaffolding function of IQGAP1 in synergistically localizing CDC42 and WASP at specific sites of the cell.
Figure 2.
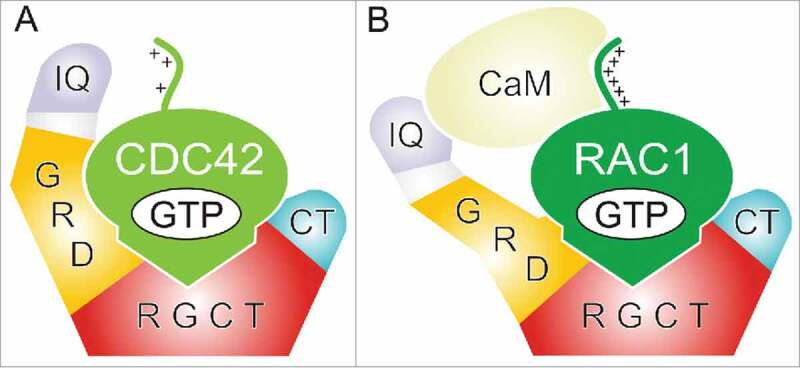
A proposed multiple-step mechanistic model of IQGAP1 interaction with CDC42 and RAC1. IQGAP1 harbors at least two distinct binding domains. The molecular nature of IQGAP1 interaction with CDC42 partially differs from that with RAC1 particularly with regard to the role of GRD. RGCT contributes with a high affinity binding to the switch regions of the GTP-bound, active (A) CDC42 and (B) RAC1. GRD more selectively recognizes active forms of CDC42 and RAC1 but also binds to other regions adjacent to the switch regions obviously in a nucleotide-independent manner. Equilibrium measurements using fluorescence polarization by Nouri et al.42 demonstrated that GRD undergoes a low-affinity interaction with CDC42 but its binding in contrast to RGCT is partially nucleotide dependent. The very C-terminal domain (CT) of IQGAP1 may potentiate the IQGAP1 interaction with RAC1 and CDC42 proteins by probably extending the resident time of the respective proteins complexes. Lack of a tight GRD interaction with RAC1 (B) may be compensated by calmodulin as an accessory protein,46,47 which has been reported to bind the polybasic region of RAC1 and IQ motifs of IQGAP1.
Despite a high sequence identity between RAC1 and CDC42, they obviously differ in regard to their IQGAP1 binding. RAC1 binds to GRD1-RGCT and CT as tight as CDC42 but exhibits different affinities for the GRD and GRD1-CT (Fig. 1). Unlike CDC42, an extremely low affinity was determined for the RAC1-GRD interaction.42 This strongly suggests that the molecular nature of IQGAP1 interaction with CDC42 partially differs from that of RAC1. Given the hypothesis that the GRD does not play a prime role in the recognition of CDC42 and its binding outside the switch regions induces a conformational change and activation of IQGAP1, the functional impact of the interaction of RAC1 remains rather ambiguous. If a direct impact of RAC1 on IQGAP1 activity does not follow the same trend as in the case of CDC42 then we need to think about an alternative regulatory mechanism. One hypothetical scenario is that CaM potentiates RAC1-mediated activation of IQGAP1 by acting as a scaffold to assemble these accessory proteins (Fig. 2B).47 The dual roles of IQGAP1 as an effector downstream and a scaffold protein upstream of CDC42 and alternatively RAC1 remains to be experimentally explored.
Abbreviations
- CaM
calmodulin
- CC
coiled-coil repeat region
- CHD
calponin homology domain
- CT
C-terminal domain
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- GDI
guanine nucleotide dissociation inhibitor
- GRD
GAP-related domain
- GTPase
guanosintriphosphatase
- IQ
4 isoleucine/glutamine-containing motifs
- IQGAP1
IQ-domain GTPase-activating protein 1
- PAK1
p21- activated kinase 1
- RGCT
RASGAP C-terminus
- WASP
Wiskott-Aldrich syndrome protein
- WW
tryptophan-containing proline-rich motif-binding region
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We appreciate the comments and discussions provided by our group members Ehsan Amin, Eyad K. Fansa, Radovan Dvorsky, and Saeideh Nakhaei-Rad. We apologize for not being able to cite the work of many colleagues owing to space constraints.
Funding
This work was supported by the Research committee of the Medical Faculty of the Heinrich-Heine University Düsseldorf “Illuminating the activation mechanisms of IQGAP1, a multifaceted scaffold protein underlying tumorigenesis” (grant Number 9772617), the ERA-Net E-Rare program NSEuroNet “European network on Noonan syndrome and related disorders” (grant 01GM1602B), the Biotechnology and Biological Sciences Research Council (BBSRC, UK) “Biochemical analysis of human IQGAP proteins” (BB/D000394/1) and Action Cancer (Northern Ireland) “Functional analysis of cancer-causing mutations in human IQGAP1” (PG2 2005).
References
- [1].Hall A. Rho family GTPases. Biochem Soc Trans 2012; 40:1378-82; PMID:23176484; https://doi.org/ 10.1042/BST20120103 [DOI] [PubMed] [Google Scholar]
- [2].Wennerberg K, Der CJ. Rho-family GTPases: It's not only Rac and Rho (and I like it). J Cell Sci 2004; 117:1301-12; PMID:15020670; https://doi.org/ 10.1242/jcs.01118 [DOI] [PubMed] [Google Scholar]
- [3].Jaiswal M, Fansa EK, Dvorsky R, Ahmadian MR. New insight into the molecular switch mechanism of human Rho family proteins: Shifting a paradigm. Biol Chem 2013; 394:89-95; PMID:23096567; https://doi.org/ 10.1515/hsz-2012-0207 [DOI] [PubMed] [Google Scholar]
- [4].Dvorsky R, Ahmadian MR. Always look on the bright site of Rho: Structural implications for a conserved intermolecular interface. EMBO Rep 2004; 5:1130-6; PMID:15577926; https://doi.org/ 10.1038/sj.embor.7400293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem J 2000; 2:241-55; https://doi.org/ 10.1042/bj3480241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].White CD, Erdemir HH, Sacks DB. IQGAP1 and its binding proteins control diverse biological functions. Cell Signal 2012; 24:826-34; PMID:22182509; https://doi.org/ 10.1016/j.cellsig.2011.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Watanabe T, Wang S, Kaibuchi K. IQGAPs as key regulators of actin-cytoskeleton dynamics. Cell Struct Funct 2015; 40:69-77; PMID:26051604; https://doi.org/ 10.1247/csf.15003 [DOI] [PubMed] [Google Scholar]
- [8].Abel AM, Schuldt KM, Rajasekaran K, Hwang D, Riese MJ, Rao S, Thakar MS, Malarkannan S. IQGAP1: Insights into the function of a molecular puppeteer. Mol Immunol 2015; 65:336-49; PMID:25733387; https://doi.org/ 10.1016/j.molimm.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Heasman SJ, Ridley AJ. Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat Rev Mol Cell Biol 2008; 9:690-701; PMID:18719708; https://doi.org/ 10.1038/nrm2476 [DOI] [PubMed] [Google Scholar]
- [10].Hedman AC, Smith JM, Sacks DB. The biology of IQGAP proteins: Beyond the cytoskeleton. EMBO Reports 2015; 16:427-46; PMID:25722290; https://doi.org/ 10.15252/embr.201439834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Smith JM, Hedman AC, Sacks DB. IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol 2015; 25:171-84; PMID:25618329; https://doi.org/ 10.1016/j.tcb.2014.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Choi S, Anderson RA. IQGAP1 is a phosphoinositide effector and kinase scaffold. Adv Biol Regul 2016; 60:29-35; PMID:26554303; https://doi.org/ 10.1016/j.jbior.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tanos BE, Yeaman C, Rodriguez-Boulan E. An emerging role for IQGAP1 in tight junction control. Small GTPases 2016; 23:1-9; PMID:27880081; https://doi.org/ 10.1080/21541248.2016.1244440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Choi S, Hedman AC, Sayedyahossein S, Thapa N, Sacks DB, Anderson RA. Agonist-stimulated phosphatidylinositol-3,4,5-trisphosphate generation by scaffolded phosphoinositide kinases. Nat Cell Biol 2016; 18:1324-35; PMID:27870828; https://doi.org/ 10.1038/ncb3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jameson KL, Mazur PK, Zehnder AM, Zhang J, Zarnegar B, Sage J, Khavari PA. IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nat Med 2013; 19:626-30; PMID:23603816; https://doi.org/ 10.1038/nm.3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu C, Billadeau DD, Abdelhakim H, Leof E, Kaibuchi K, Bernabeu C, Bloom GS, Yang L, Boardman L, Shah VH, et al.. IQGAP1 suppresses TbetaRII-mediated myofibroblastic activation and metastatic growth in liver. J Clin Invest 2013; 123:1138-56; PMID:23454766; https://doi.org/ 10.1172/JCI63836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].White CD, Li Z, Dillon DA, Sacks DB. IQGAP1 protein binds human epidermal growth factor receptor 2 (HER2) and modulates trastuzumab resistance. J Biol Chem 2011; 286:29734-47; PMID:21724847; https://doi.org/ 10.1074/jbc.M111.220939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Malarkannan S, Awasthi A, Rajasekaran K, Kumar P, Schuldt KM, Bartoszek A, Manoharan N, Goldner NK, Umhoefer CM, Thakar MS. IQGAP1: A regulator of intracellular spacetime relativity. J Immunol 2012; 188:2057-63; PMID:22345702; https://doi.org/ 10.4049/jimmunol.1102439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brown MD, Sacks DB. IQGAP1 in cellular signaling: Bridging the GAP. Trends Cell Biol 2006; 16:242-9; PMID:16595175; https://doi.org/ 10.1016/j.tcb.2006.03.002 [DOI] [PubMed] [Google Scholar]
- [20].Briggs MW, Sacks DB. IQGAP1 as signal integrator: Ca2+, calmodulin, Cdc42 and the cytoskeleton. FEBS Lett 2003; 542:7-11; PMID:12729888; https://doi.org/ 10.1016/S0014-5793(03)00333-8 [DOI] [PubMed] [Google Scholar]
- [21].Kuroda S, Fukata M, Kobayashi K, Nakafuku M, Nomura N, Iwamatsu A, Kaibuchi K. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J Biol Chem 1996; 271:23363-7; PMID:8798539; https://doi.org/ 10.1074/jbc.271.49.31029 [DOI] [PubMed] [Google Scholar]
- [22].Kuroda S, Fukata M, Nakagawa M, Fujii K, Nakamura T, Ookubo T, Izawa I, Nagase T, Nomura N, Tani H, et al.. Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin- mediated cell-cell adhesion. Science 1998; 281:832-5; PMID:9694656; https://doi.org/ 10.1126/science.281.5378.832 [DOI] [PubMed] [Google Scholar]
- [23].Noritake J, Watanabe T, Sato K, Wang S, Kaibuchi K. IQGAP1: A key regulator of adhesion and migration. J Cell Sci 2005; 118:2085-92; PMID:15890984; https://doi.org/ 10.1242/jcs.02379 [DOI] [PubMed] [Google Scholar]
- [24].Bashour AM, Fullerton AT, Hart MJ, Bloom GS. IQGAP1, a Rac- and Cdc42-binding protein, directly binds and cross-links microfilaments. J Cell Biol 1997; 137:1555-66; PMID:9199170; https://doi.org/ 10.1083/jcb.137.7.1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fukata M, Kuroda S, Fujii K, Nakamura T, Shoji I, Matsuura Y, Okawa K, Iwamatsu A, Kikuchi A, Kaibuchi K. Regulation of cross-linking of actin filament by IQGAP1, a target for Cdc42. J Biol Chem 1997; 272:29579-83; PMID:9368021; https://doi.org/ 10.1074/jbc.272.47.29579 [DOI] [PubMed] [Google Scholar]
- [26].LeCour L, Jr, Boyapati VK, Liu J, Li Z, Sacks DB, Worthylake DK. The structural basis for Cdc42-induced dimerization of IQGAPs. Structure (London, England: 1993) 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li Z, McNulty DE, Marler KJ, Lim L, Hall C, Annan RS, Sacks DB. IQGAP1 promotes neurite outgrowth in a phosphorylation-dependent manner. J Biol Chem 2005; 280:13871-8; PMID:15695813; https://doi.org/ 10.1074/jbc.M413482200 [DOI] [PubMed] [Google Scholar]
- [28].Phillips-Mason PJ, Gates TJ, Major DL, Sacks DB, Brady-Kalnay SM. The receptor protein-tyrosine phosphatase PTPmu interacts with IQGAP1. J Biol Chem 2006; 281:4903-10; PMID:16380380; https://doi.org/ 10.1074/jbc.M506414200 [DOI] [PubMed] [Google Scholar]
- [29].Hart MJ, Callow MG, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. Embo J 1996; 15:2997-3005; PMID:8670801 [PMC free article] [PubMed] [Google Scholar]
- [30].Joyal JL, Annan RS, Ho YD, Huddleston ME, Carr SA, Hart MJ, Sacks DB. Calmodulin modulates the interaction between IQGAP1 and Cdc42. Identification of IQGAP1 by nanoelectrospray tandem mass spectrometry. J Biol Chem 1997; 272:15419-25; PMID:9182573; https://doi.org/ 10.1074/jbc.272.24.15419 [DOI] [PubMed] [Google Scholar]
- [31].Li Z, Sacks DB. Elucidation of the interaction of calmodulin with the IQ motifs of IQGAP1. J Biol Chem 2003; 278:4347-52; PMID:12446675; https://doi.org/ 10.1074/jbc.M208579200 [DOI] [PubMed] [Google Scholar]
- [32].Swart-Mataraza JM, Li Z, Sacks DB. IQGAP1 is a component of Cdc42 signaling to the cytoskeleton. J Biol Chem 2002; 277:24753-63; PMID:11948177; https://doi.org/ 10.1074/jbc.M111165200 [DOI] [PubMed] [Google Scholar]
- [33].Ho YD, Joyal JL, Li Z, Sacks DB. IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J Biol Chem 1999; 274:464-70; PMID:9867866; https://doi.org/ 10.1074/jbc.274.1.464 [DOI] [PubMed] [Google Scholar]
- [34].Zhang B, Chernoff J, Zheng Y. Interaction of Rac1 with GTPase-activating proteins and putative effectors. A comparison with Cdc42 and RhoA. J Biol Chem 1998; 273:8776-82; PMID:9535855; https://doi.org/ 10.1074/jbc.273.15.8776 [DOI] [PubMed] [Google Scholar]
- [35].Li R, Debreceni B, Jia B, Gao Y, Tigyi G, Zheng Y. Localization of the PAK1-, WASP-, and IQGAP1-specifying regions of Cdc42. J Biol Chem 1999; 274:29648-54; PMID:10514434; https://doi.org/ 10.1074/jbc.274.42.29648 [DOI] [PubMed] [Google Scholar]
- [36].Owen D, Campbell LJ, Littlefield K, Evetts KA, Li Z, Sacks DB, Lowe PN, Mott HR. The IQGAP1-Rac1 and IQGAP1-Cdc42 interactions: Interfaces differ between the complexes. J Biol Chem 2008; 283:1692-704; PMID:17984089; https://doi.org/ 10.1074/jbc.M707257200 [DOI] [PubMed] [Google Scholar]
- [37].Kurella VB, Richard JM, Parke CL, Lecour LF, Jr, Bellamy HD, Worthylake DK. Crystal structure of the GTPase-activating protein-related domain from IQGAP1. J Biol Chem 2009; 284:14857-65; PMID:19321438; https://doi.org/ 10.1074/jbc.M808974200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 1997; 277:333-8; PMID:9219684; https://doi.org/ 10.1126/science.277.5324.333 [DOI] [PubMed] [Google Scholar]
- [39].Scheffzek K, Ahmadian MR, Wiesmuller L, Kabsch W, Stege P, Schmitz F, Wittinghofer A. Structural analysis of the GAP-related domain from neurofibromin and its implications. Embo J 1998; 17:4313-27; PMID:9687500; https://doi.org/ 10.1093/emboj/17.15.4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol 1997; 4:686-9; PMID:9302992; https://doi.org/ 10.1038/nsb0997-686 [DOI] [PubMed] [Google Scholar]
- [41].Ahmadian MR, Kiel C, Stege P, Scheffzek K. Structural fingerprints of the Ras-GTPase activating proteins neurofibromin and p120GAP. J Mol Biol 2003; 329:699-710; PMID:12787671; https://doi.org/ 10.1016/S0022-2836(03)00514-X [DOI] [PubMed] [Google Scholar]
- [42].Nouri K, Fansa EK, Amin E, Dvorsky R, Gremer L, Willbold D, Schmitt L, Timson DJ, Ahmadian MR. IQGAP1 interaction with RHO family proteins revisited: Kinetic and equilibrium evidence for multiple distinct binding sites. J Biol Chem 2016; 291:26364-76; PMID:27815503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Elliott SF, Allen G, Timson DJ. Biochemical analysis of the interactions of IQGAP1 C-terminal domain with CDC42. World J Biol Chem 2012; 3:53-60; PMID:22451851; https://doi.org/ 10.4331/wjbc.v3.i3.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Grohmanova K, Schlaepfer D, Hess D, Gutierrez P, Beck M, Kroschewski R. Phosphorylation of IQGAP1 modulates its binding to Cdc42, revealing a new type of rho-GTPase regulator. J Biol Chem 2004; 279:48495-504; PMID:15355962; https://doi.org/ 10.1074/jbc.M408113200 [DOI] [PubMed] [Google Scholar]
- [45].Le Clainche C, Schlaepfer D, Ferrari A, Klingauf M, Grohmanova K, Veligodskiy A, Didry D, Le D, Egile C, Carlier MF, et al.. IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J Biol Chem 2007; 282:426-35; PMID:17085436; https://doi.org/ 10.1074/jbc.M607711200 [DOI] [PubMed] [Google Scholar]
- [46].Elsaraj SM, Bhullar RP. Regulation of platelet Rac1 and Cdc42 activation through interaction with calmodulin. Biochim Biophys Acta 2008; 1783:770-8; PMID:18328269; https://doi.org/ 10.1016/j.bbamcr.2008.01.022 [DOI] [PubMed] [Google Scholar]
- [47].Xu B, Chelikani P, Bhullar RP. Characterization and functional analysis of the calmodulin-binding domain of Rac1 GTPase. PLoS One 2012; 7:e42975; PMID:22905193; https://doi.org/ 10.1371/journal.pone.0042975 [DOI] [PMC free article] [PubMed] [Google Scholar]


