Abstract
During storage in the silk gland, the N-terminal domain (NT) of spider silk proteins (spidroins) keeps the aggregation-prone repetitive region in solution at extreme concentrations. We observe that NTs from different spidroins have co-evolved with their respective repeat region, and now use an NT that is distantly related to previously used NTs, for efficient recombinant production of the amyloid-β peptide (Aβ) implicated in Alzheimer’s disease. A designed variant of NT from Nephila clavipes flagelliform spidroin, which in nature allows production and storage of β-hairpin repeat segments, gives exceptionally high yields of different human Aβ variants as a solubility tag. This tool enables efficient production of target peptides also in minimal medium and gives up to 10 times more isotope-labeled monomeric Aβ peptides per liter bacterial culture than previously reported.
Subject terms: Biochemistry, Biophysics, Biosynthesis, Peptides, Proteins, Neurodegeneration, Neurodegenerative diseases
Introduction
Orb-weaving spiders manufacture up to seven different silks, e.g. dragline silk derived from major ampullate silk proteins (spidroins, MaSp) and flagelliform silk derived from flagelliform spidroins (FlSp). The various spidroins share a common architecture - a large core repetitive region capped by globular N- and C-terminal domains (NT and CT)1. The divergent and large aggregation-prone repetitive regions of the spidroins determine the mechanical properties of the respective spider silks, while the terminal domains regulate silk fiber formation2,3. Despite their high aggregation propensity the spidroins can be stored at extremely high concentrations (30–50% w/v) in the spider silk gland, solubilized by the NT domain1,4.
The NT dimerizes upon a drop in pH, which is crucial for silk fiber formation1,5. To ensure solubility also at low pH and widen the applicability of NT as a solubility enhancing fusion partner, a charged-reversed mutant has been designed (referred to as NT*MaSp)6. The previously reported NT*MaSp tag is derived from the NT domain of Euprosthenops australis MaSp1 and folds as a five-helix bundle6,7. NT*MaSp is a pH insensitive constitutive monomer, highly stable and extremely soluble, and has been successfully applied for efficient production and purification of, among others, lung surfactant protein analogs, cholecystokinin-58, human antimicrobial cathelicidin and a designed β-sheet protein6,8.
Aggregation-prone proteins and peptides are associated with several neurodegenerative disorders, e.g. Alzheimer’s disease (AD), the most prevalent form of dementia9,10. These proteins/peptides often exhibit high β-sheet propensity, which make them prone to aggregate and form insoluble amyloid fibrils11. These intrinsic properties of amyloid-forming proteins make high-yield biochemical production challenging, yet the availability of pure protein samples is crucial for studying protein self-assembly and its associated neurotoxicity in vitro and in vivo. This is probably one important reason behind the fact that, despite immense efforts, the exact mechanisms of Aβ self-assembly are still unknown9–11. Recent advances have however revealed new insights into the nucleation mechanism of Aβ in vitro12,13, structural details of the fibril morphology14 and biological mechanisms implicated in the AD etiopathology15,16. These experiments typically require access to very pure and homogeneous Aβ peptides as small impurities or preformed seeds have a great impact on the aggregation behavior17. In particular for structural studies of amyloid fibrils, but also for certain in vivo experiments, the availability of large quantities of isotope-labeled Aβ is essential.
Studies of Aβ aggregation in vitro have often been conducted with synthetically produced peptides18,19. Synthetic preparations have several drawbacks including batch-to-batch variations, intrinsic impurities and relatively high cost, especially for isotope labeling. As a consequence, several recombinant expression systems have been established. These production protocols either result in peptides with an initiating non-native methionine residue20,21 or are based on solubility tags that require proteolytic cleavage to obtain the native human Aβ sequence22–24. The main disadvantage of having methionine as the first residue is that it might affect processes such as posttranslational modifications, e.g. pyroGlu formation25–27, and metal ion binding since the metal ion-binding site is located in the N-terminus28–30. Here we describe a useful solubility tag for production of aggregation-prone proteins and peptides, and demonstrate that this tool enables very efficient production of native and isotope-labeled Aβ peptides.
Results and Discussion
Evolutionary relationships of NT and repetitive regions of different spidroins
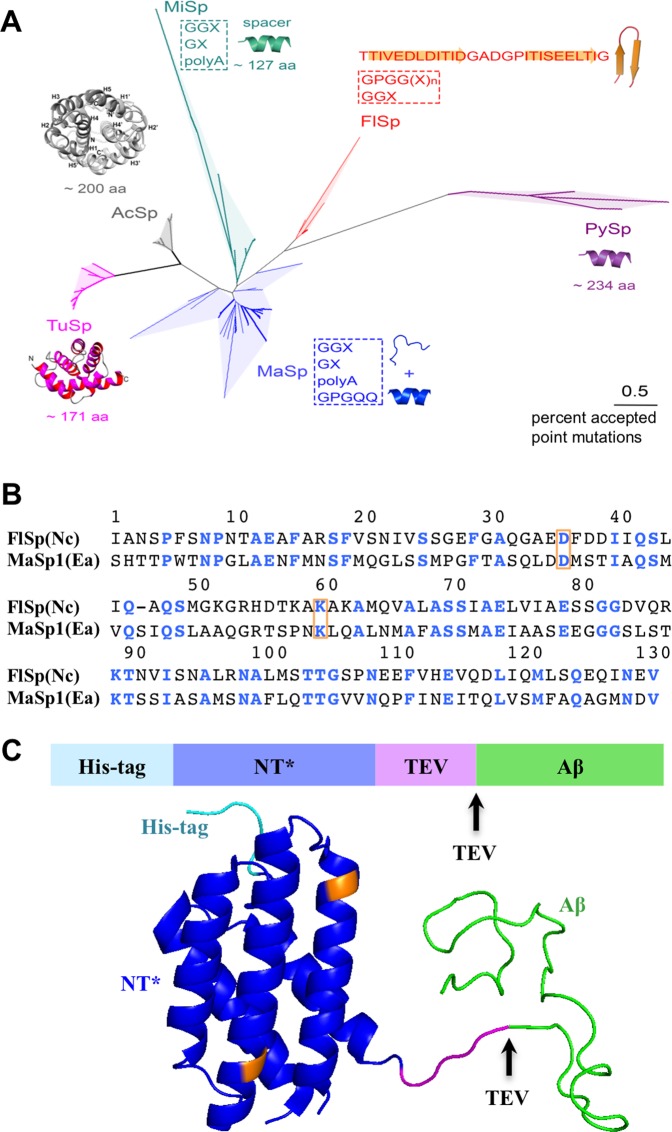
A phylogenetic tree based on sequence alignment of 67 NTs found in GenBank (Supplementary Fig. S1) reveal evolutionary relationships between NT and their respective repetitive regions (Fig. 1A). The NT domains cluster according to silk type, as previously reported7. Hence, the NTs of different spidroin types, which are defined by the nature of their respective repetitive regions, have been conserved through evolution of different spider species. Structural characteristics of the repetitive regions appear to co-vary with the evolution of NTs, e.g. for the tubuliform (TuSp) and aciniform (AcSp) NTs, which are evolutionarily close, the corresponding repetitive regions stand out by forming globular folded domains31,32 (Fig. 1A). NT from FlSp is linked to a unique repetitive region that contains several embedded spacers (each 27 residues), which are predicted to form β-hairpins33 (Fig. 1A). NTFlSp exhibits distant evolutionary relationship (<35% sequence identity) to the previously reported NTMaSp6 (Fig. 1B) and MaSps contain repetitive regions with predicted α-helical and random coil structures34–38. We speculate that different NTs may have evolved to facilitate optimal solubility of their respective repeat region in the silk gland during storage conditions, where pH is neutral and NT monomeric4,5. Irrespective of any potential evolutionary co-variation between NTs and the repetitive regions, we aimed to explore whether NTFlSp could work in protein expression in an equivalent way to the previously investigated and distantly related NTMaSp6.
Figure 1.
(A) Evolutionary relationships of the NTs of different spidroins. The analysis involved 67 NT amino acid sequences (Supplementary Fig. 1), revealing that spidroins from different spider species cluster according to the silk type in the phylogenetic tree. The typical repetitive regions of the respective spidroins and their known structures or main secondary structure propensities are displayed. (B) Sequence alignment of NTFlSp and NTMaSp where strictly conserved residues are colored in blue. The residues marked in orange display the mutated sites in NT*. (C) Schematic representation and structure of the NT*-Aβ fusion protein where the arrows indicate the TEV protease cleavage site. The mutated D and K residues are marked by yellow colour in the NT structure (pdb 4FBS).
Design of the novel solubility tag NT*FlSp
To prevent dimerization of NTFlSp at low pH we introduced the D40K and K65D mutations6 in NTFlSp from Nephila clavipes (Nc) (Fig. 1B) (numbering as described previously6, wherefore the mutations correspond to positions 36 and 60 in Fig. 1B). NT*FlSp has a larger number of charged residues (25 vs. 11) and stronger net charge (−7 vs. −5) compared to NT*MaSp, which potentially enhance its solubility properties. In contrast to NT*MaSp, NT*FlSp has no tryptophan, whose absorbance at 280 nm would cover the intrinsic low absorbance at 280 nm of the target peptide Aβ. Thus, for size exclusion chromatography (SEC), where detection relies on the protein absorbance at 280 nm, NT*FlSp enables clearly separated intensity peaks.
Efficient expression and purification of Aβ monomers using NT*FlSp
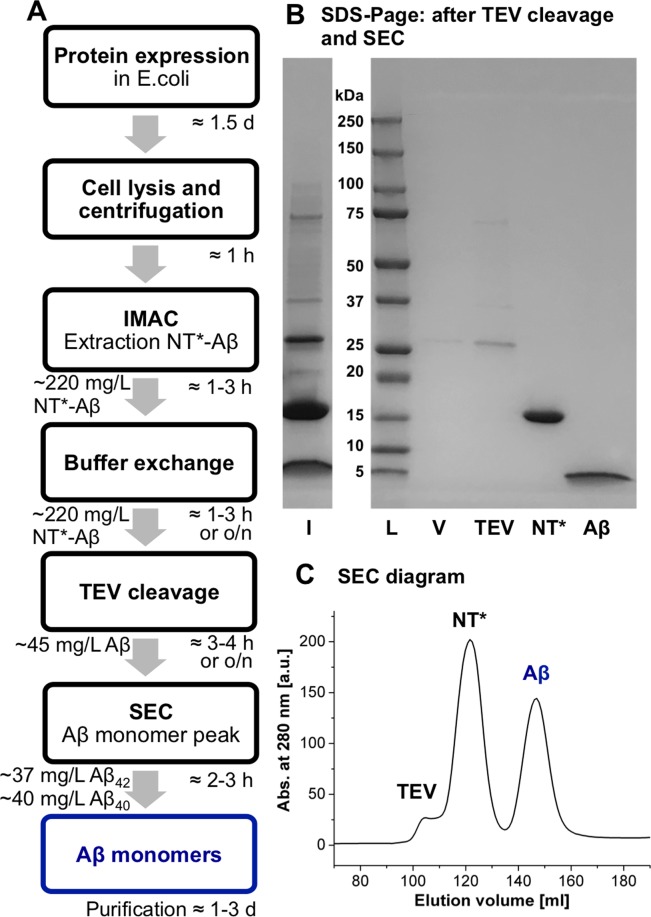
We designed the fusion protein NT*FlSp-Aβ by fusing the genetic codes of the solubility tag NT*FlSp and Aβ with a TEV recognition site in-between (Fig. 1C). An overview of the expression and purification scheme is given in Fig. 2. The fusion protein was expressed in BL21(DE3) E.coli cells grown in rich or minimal medium, dissolved in 8 M urea after cell lysis and purified using immobilized metal ion affinity chromatography (IMAC). Urea was added as denaturant to increase binding to IMAC column. For optimal cleavage of the fusion protein by TEV, a buffer exchange was conducted, either by overnight dialysis or by column chromatography. TEV cleavage can alternatively be conducted during buffer dialysis to speed-up the purification, yet a short dialysis step to decrease the urea concentration below 2 M is recommended before the addition of TEV protease. Finally, the solution was applied to SEC with a Superdex 30 column, whereby monomeric Aβ monomers were isolated.
Figure 2.
(A) Schematic expression and purification protocol, including typical times for performance. Yields of NT*-Aβ42/40 are derived from 1 L expression cultures and extrapolated from purification from 100 and 500 mL, resulting in very similar values. (B) SDS-PAGE gel, with protein ladder (L), void (V), before (I) and after SEC yielding pure Aβ. An uncropped full-length gel is presented in Supplementary Fig. S2. (C) SEC diagram showing separation of TEV, NT* and monomeric Aβ.
The expression and purification protocol presented here results in highly pure Aβ40 and Aβ42 monomers within 2.5–4.5 days, depending on buffer exchange and cleavage method. The yields in rich and minimal medium are listed in Table 1. The example shown in Fig. 2 represents purifications from 100 and 500 mL culture medium, yielding very similar amounts of 37 ± 7 mg of pure Aβ42 monomers if extrapolated to one-liter culture. Notably, the present scheme gives by far the highest yields, both in rich and minimal medium, compared to other reported protocols (Table 2).
Table 1.
Average yields of fusion proteins and monomeric Aβ peptides in rich (LB) and minimal (M9) medium in mg per liter culture.
| Protein/peptide | Rich medium [mg/L] | M9 medium [mg/L] |
|---|---|---|
| NT*FlSp-Aβ40 | 216 ± 29 | 74 ± 22 |
| NT*FlSp-Aβ42 | 223 ± 42 | 88 ± 10 |
| Monomeric Aβ40 | 40 ± 5 | 13 ± 4 |
| Monomeric Aβ42 | 37 ± 7 | 14 ± 2 |
Errors were estimated as standard deviations from 5 replicates by western blot analysis (see Methods).
Table 2.
Yields of Aβ40 and Aβ42 variants reported in literature and herein.
| Aβ variant | Fusion partner/expression method | Purified Aβ peptide yield in mg/L in rich medium | Purified Aβ peptide yield in mg/L in minimal medium | Reference |
|---|---|---|---|---|
| Aβ(1–40) | NT*FlSp | 40 ± 5 | 13 ± 4 | here |
| (NANP)19 | 22 | — | 22 | |
| IFABP | 4 | — | 24 | |
| GST | 7 | 1.5 | 23 | |
| Aβ(M1–40) | Directly from inclusion bodies | 10–20 | — | 21 |
| Directly from inclusion bodies | — | 10–15 | 51 | |
| Co-expression with ZAβ3 | — | 4 | 20 | |
| Aβ(1–42) | NT*FlSp | 37 ± 7 | 14 ± 2 | here |
| (NANP)19 | 19 | — | 22 | |
| IFABP | 3 | — | 24 | |
| IFABP | — | 6 | 52 | |
| GST | 15 | — | 53 | |
| Ub | 4 | — | 54 | |
| Aβ(M1–42) | Directly from inclusion bodies | 8 | — | 21 |
| Co-expression with ZAβ3 | — | 3 | 20 |
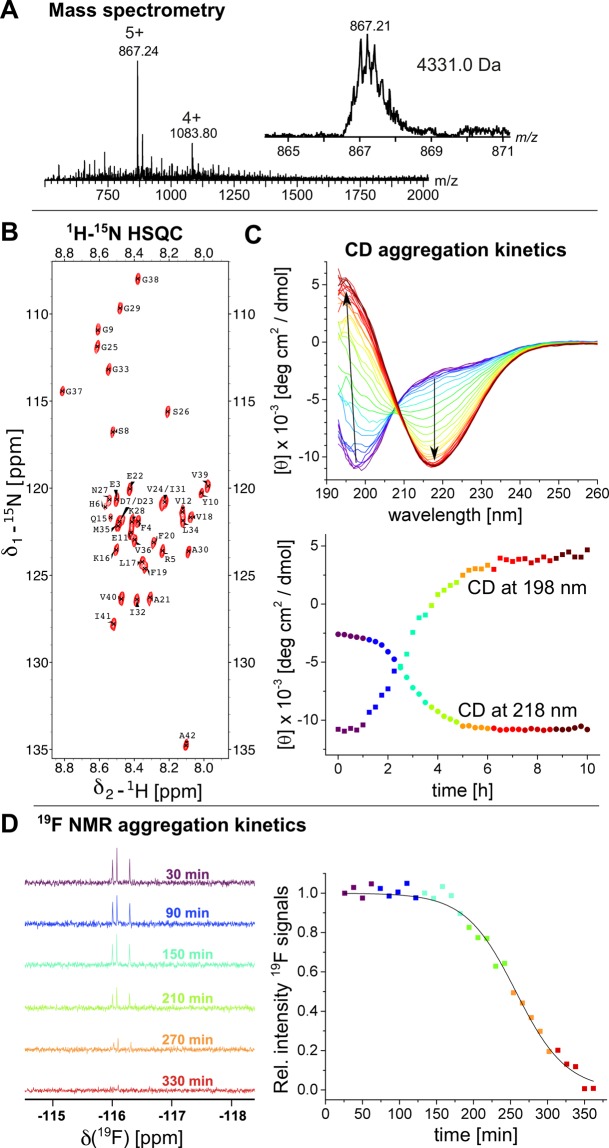
The purified peptides were investigated using mass spectrometry, confirming the expected masses for Aβ, here shown for Aβ40 (Fig. 3A). Using 13C-15N-double-labeled Aβ40 and Aβ42 we performed nuclear magnetic resonance (NMR) experiments to confirm the purity and structural state of the purified peptides. We recorded 1H-15N-HSQC experiments (Fig. 3B and Supplementary Fig. S3) where the chemical shifts of the cross-peaks coincide with previous assignments reported in the literature, revealing a monomeric, predominantly unstructured conformation of the purified Aβ peptides39,40. To analyze the secondary structure of monomeric Aβ42 we applied circular dichroism (CD) spectroscopy. The initial CD spectra (Fig. 3C) indicated a predominantly unstructured conformation as previously reported20,21,41,42. Taken together, these experiments confirm that our method results in monomeric Aβ40 and Aβ42 peptides.
Figure 3.
(A) Mass spectrum of Aβ40 showing a pure peptide with an average mass of 4331 Da. The inset shows a zoom of the 5 + charged ion. (B) 1H-15N-HSQC spectrum of 15 μM Aβ42 recorded at 5 °C, revealing monomeric peptide. (C) Aggregation kinetics of 10 μM Aβ42 at 37 °C under continuous stirring recorded by CD spectroscopy. The spectra exhibit a structural transition from a predominantly unstructured state to a β-structure. The lower panel shows the time dependence of the CD extremes at 198 nm (squares) and 218 nm (circles), with the same color code as used for the CD spectra. (D) Aggregation kinetics of 50 μM 4FF-Aβ42 at 25 °C monitored by 19F-NMR spectra of the signals around −116 ppm, exhibiting attenuation of 4FF-signals. The color code represents the same time points in both panels.
Production of 4-fluoro-Phe-labeled Aβ peptides
The present approach also opens new opportunities for NMR studies that require more complex isotope labeling approaches associated with reduced protein yields. For example, we have used the NT*FlSp tag to produce monomeric Aβ42 incorporating 4-fluoro-Phe (4FF-Aβ42) in milligram yields. The expression was performed similarly as described above, but glyphosate and DL-tyrosine was supplemented to bacterial cultures at an OD600nm value of 0.6. Further, DL-4-fluorophenylalanine was added when the OD600nm value reached 0.8 and cell expression was induced. The 1H-15N-HSQC spectrum of 15N-labeled 4FF-Aβ42 revealed again a monomeric peptide (Supplementary Fig. S3).
Aggregation kinetics of native and isotope-labeled Aβ42
To ensure that the isolated peptides behave as expected, we investigated the aggregation kinetics starting from monomeric Aβ peptides. Recording CD signals under continuous stirring at 37 °C of 10 μM Aβ42, a structural conversion from an unstructured to a β-structured conformation was observed (Fig. 3C), where the isodichroic point at 208 nm indicates a two-state transition. Furthermore, we used 50 μM 4FF-Aβ42 for real-time aggregation 19F-NMR studies at 25 °C, revealing a decrease of 4FF-signals over time (Fig. 3D). The signal loss can be fitted to a sigmoidal decline, with an aggregation half time of 258 ± 5 min under the conditions used.
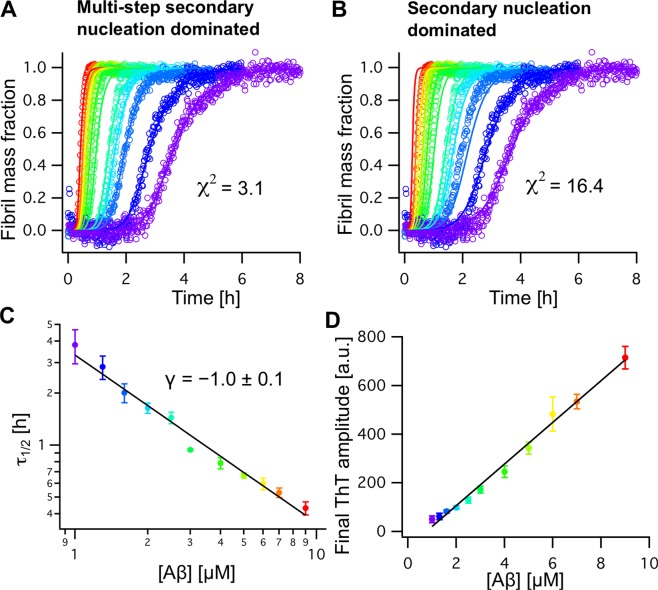
Alternatively, Aβ aggregation kinetics can be monitored using the fluorescence dye thioflavin T (ThT), for a detailed elucidation of the nucleation mechanism. Here, we conducted ThT experiments on Aβ42 in 20 mM sodium phosphate buffer, pH 8.0, at 37 °C under quiescent conditions at different initial Aβ42 monomer concentrations, [Aβ] (Fig. 4). The final fluorescence intensity exhibits a linear dependence on the initial monomer concentration (Fig. 4D), suggesting that the total amount of initially monomeric peptides forms ThT-active fibril material, as previously shown for Aβ12,13,42. The aggregation half times of Aβ42 used here exhibit a simple relation τ1/2 ∝ [Aβ]γ, with γ = −1.0 ± 0.1, corresponding to the slope in a double logarithmic plot (Fig. 4C). This value is in the same range as found for AβM42 with an initial methionine, where γ = −1.3 was reported12. For γ = −1.0 a multi-step secondary nucleation model describes better the observed aggregation traces compared to a single-step secondary nucleation model (Fig. 4A,B). The multi-step model additionally includes saturation of secondary nucleation and was previously shown to be applicable for the shorter Aβ40 and AβM40 variants13,29,42 and for AβM42 at pH 7.443, which all exhibit higher γ-values, but also describes well the kinetics of AβM42 at pH 8.044. Hence, these results confirm that the native and isotope-labeled peptides obtained herein are highly pure and in a monomeric state, which is essential for accurate and reproducible aggregation kinetics experiments.
Figure 4.
(A,B) Aggregation kinetics of Aβ42 at different concentrations from 1.0 (violet) to 9.0 μM (red) recorded by ThT fluorescence experiments fitted with a multi-step (A) and simple secondary nucleation model (B). The kinetic traces fit best to the multi-step secondary nucleation model, reflected by a lower χ2 value. (C) Aggregation half times, 𝜏1/2, plotted against the initial peptide concentration, [Aβ], exhibit a γ coefficient of γ = −1.0 ± 0.1. (D) The final ThT intensity of the normalized aggregation traces in (A) exhibits a linear relation to [Aβ].
Conclusions
Taken together, we have developed a biomimetic tool that provides facile, fast and inexpensive production of pure and monomeric Aβ40 and Aβ42 peptides. The high yield obtained also in minimal medium enables efficient generation of isotope-labeled Aβ peptides. Peptides produced by our protocol recapitulate the behavior of Aβ peptides obtained by other means, which indicate the applicability of using NT*FlSp for generating functional Aβ peptides. The NT*FlSp-tag holds great potential, also when compared to NT*MaSp6, for efficient production of medically relevant aggregation-prone peptides and proteins. This is important since the majority of new pharmaceuticals are biologics and facile protocols for efficient production of proteins that are difficult to produce are needed.
Methods
Expression and purification protocol
The synthetic gene coding for NT FlSp from Nephila clavipes with the D40K and K65D mutations (NT*FlSp) was ordered from GenScript (GenScript Biotech, Netherlands). The NT*FlSp gene was ligated into pT7 plasmid containing TEV recognintion site (TRS)-Aβ40/Aβ42 as described previously6. The plasmids were transformed into chemically competent E. coli BL21 (DE3) cells and expressed as described previously45. In short, 1 mL overnight culture was inoculated to 100 mL LB medium (1/100) or 100 mL M9 overnight culture was inoculated to 1 L M9 minimal medium (10/100) with 70 mg/l kanamycin. Cells were grown at 30 °C at 120 RPM to OD600nm around 0.8–0.9, where the temperature was lowered to 20 °C, and 0.1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added and the cells were incubated overnight. To isolate the cells from media, the bacterial culture was centrifuged at 5,000 × g for 20 minutes at 4 °C and the cell pellets resuspended in 40 mL 20 mM Tris-HCl pH 8.0, split in to two 50 mL falcon tubes and stored at −20 °C. The frozen cells were thawed and urea was added to a concentration of 8 M. The cells were sonicated to obtain a clear solution. The lysate was then loaded on 2 × 5 mL Ni-NTA column (GE Healthcare). Unbound proteins were washed away with 15 mM Imidazole in 20 mM Tris-HCl, pH 8 and 8 M urea. The fusion protein was eluted with 300 mM imidazole in 20 mM Tris-HCl, pH 8 and 8 M urea. The fractions containing the fusion protein were pooled and then dialyzed overnight against 20 mM Tris-HCl pH 8.0 at 4 °C. To remove the His6-NT*FlSp part, the fusion protein was cleaved with TEV protease (1:20–1:30, enzyme to substrate, w/w) at 4 °C overnight in 20 mM Tris-HCl pH 8, 0.5 mM EDTA and 1 mM DTT. After TEV cleavage, the sample was dissolved in 15 mL 7 M guanidine-HCl and separated on a Superdex 30 26/600PG size exclusion column (Fig. 2C). The correct size of Aβ, NT*FlSp and TEV was confirmed by SDS/PAGE in a 4–20% polyacrylamide gel, stained with Coomassie brilliant blue dye (Fig. 2B). For expression of 15N- and 13C- labelled NT* FlSp -Aβ, the same procedure was used except that M9 minimal medium containing 15NH4Cl (1 g/L M9) and 13C-glucose (4 g/L M9) was used. The plasmid pRK793 for TEV expression was obtained from addgene (addgene.org, deposited by David S. Waugh) and was expressed as described above and purified as described previously46.
Determination of yields
Both NT*FlSp-Aβ40 and NT*FlSp-Aβ42 was transformed into BL21 (DE3) E. coli cells and spread onto an agar plate with kanamycin. 5 starting cultures of LB and M9 were inoculated with individual colonies and incubated at 31 °C overnight. The expression was performed as described above in 100 mL LB and M9 media. 100 μL of each culture was taken before and after induction, lyophilized, dissolved in SDS loading buffer and boiled for 10 minutes at 96 °C. 1 μL of each induced sample and 1 μL uninduced sample from each condition was loaded on a 4–20% mini protean TGX gel (BioRad) and blotted on a PVDF membrane (GE healthcare). 5% w/v non-fat dry milk/PBS was used to block the membrane after blotting for 1 h, followed by incubation with 6E10 primary antibody in 5% w/v non-fat dry milk, 0.1% Tween/PBS overnight at 4 °C. The membranes were washed three times with 0.1% Tween/PBS, and ECL anti-mouse secondary antibodies in 5% w/v non-fat dry milk and 0.1% Tween/PBS were added for 1 h at room temperature. Enhanced chemiluminescence detection reagent (GE Healthcare) was added and images were acquired using an AI600 imaging system (GE healthcare). The concentration of each sample was calculated by integration of the peaks from IMAC (fusion protein) and SEC (monomeric Aβ) with an extinction coefficient ε280 = 2,980 M−1cm−1 for the fusion protein and 1,424 M−1 cm−1 for Aβ. Western blot intensities were analyzed by ImageJ software47 and average and standard deviation from 5 replicates was calculated using yields from one full purification of each condition. Values are listed in Table 1.
Expression protocol of 4FF-Aβ42
The plasmid pT7His6NT*FlSp-TEV recognition site -Aβ42 was transformed into chemically competent E. coli BL21(DE3) cells. Colonies were inoculated to 10 mL LB medium with 70 mg/L kanamycin and grown at 30 °C and 200 r.p.m. to OD600nm > 1.0. 0.5 mL day culture was inoculated to 25 mL M9 medium with 70 mg/l kanamycin and grown at 30 °C and 200 r.p.m. overnight. 10 mL overnight culture was inoculated to 1 L M9 medium and cells were further grown at 30 °C. Uniform labeling with 4-fluorophenylalanine was achieved by the introduction of 1 g/L glyphosate and 75 mg/L DL-tyrosine to shaking bacterial cultures at 30 °C which had reached an OD600nm of 0.6. Once cells achieve an OD600nm of 0.8, 30 mg/L DL-4-fluorophenylalanine was added. The incubation temperature was lowered to 20 °C and expression was induced with the addition of IPTG to 0.1 mM, the cells were incubated overnight and were harvested by 7,000 × g centrifugation at 4 °C.
Evolutionary relationships of the NT domains of different spidroins
The evolutionary history of the NT domains from different spidroins was inferred by the Neighbor-Joining method with the Poisson correction. Evolutionary analyses were conducted in MEGA748. The analysis involved 67 amino acid sequences. In the spider silk gland (liquid protein), the repetitive region of MaSp, consisting of GGX, polyA, GX and GPGQQ, is disordered and partially helical34–38, and MiSp and FlSp share identical motifs33. However, there are ∼127-aa spacer in MiSp, which adopt α-helical conformation, whereas the 27-aa spacer in FlSp is predicted to fold to β-hairpin33. The large repetitive domains of AcSp and TuSp adopt α-helical conformation31,32, and the repetitive domains of PySp is also predicted to adopt α-helical conformation by PSIPRED v3.3 (http://bioinf.cs.ucl.ac.uk/psipred/).
Mass spectrometry
Purified Aβ40 was diluted 1:10 in H2O/acetonitrile/formic acid (70:30:0.2) and directly infused into a Waters LCT Time of flight mass spectrometer (MS Vision, NL) equipped with an offline nanospray source using borosilicate capillaries (Thermo Scientific). The capillary voltage was 1.5 kV and the cone voltage was 200 V. Spectra were acquired between m/z 500 and 4000 and the mass scale was calibrated with Cesium Iodide. Data were analyzed using MassLynx 4.1 (Waters).
Nuclear magnetic resonance (NMR)
1H-15N HSQC spectra were recorded on a 500 MHz or 700 MHz Bruker Avance spectrometer equipped with cryogenic probes. The HSQC spectrum of Aβ40 was recorded at 500 MHz at 8 °C using 75 μM peptide concentration in 16 mM sodium phosphate buffer, pH 7.4, with 0.02% NaN3 and 0.2 mM EDTA. For Aβ42 the peptide concentration was 15 μM in 20 mM sodium phosphate buffer, pH 6.8, recorded at 5 °C and 700 MHz. The spectra were recorded using 2048 × 128 complex points and 32 scans per transient. For Aβ42 we recorded the HSQC at 15 μM directly after the SEC purification, ensuring the monomeric state of the peptide.
19F-NMR experiments were recorded using 50 μM 4FF-Aβ42 in 20 mM sodium phosphate buffer, pH 7.4 with 0.03% NaN3 and 1 mM EDTA at 25 °C and 565 MHz. 19F spectrum was acquired with 512 transients and 1.0 s pulse delay between each transient. Line broadening of 1.0 Hz was used to process the final spectrum. The 1H-15N HSQC spectrum of 15 μM 4FF-Aβ42 in 20 mM sodium phosphate buffer, pH 7.4, with 0.02% NaN3 and 0.2 mM EDTA, was recorded at 4 °C on a 600 MHz Bruker Avance Neo spectrometer equipped with a cryogenic probe.
Circular dichroism (CD)
CD measurements of 10 μM Aβ42 in 20 mM sodium phosphate buffer, pH 8.0, at 37 °C were performed in a quartzglass Suprasil 10 × 4 mm CD cuvette (Hellma Analytics) where the optical path length was 4 mm, using a Chirascan CD spectrometer (Applied Photophysics). A resolution of 1.0 nm and a bandwidth of 1 nm were chosen for the aggregation kinetics experiments42. During the enire measurement the sample was continuously stirred at around 1200 r.p.m and each 3 min a new CD spectrum was recorded to follow the aggregation kinetics.
Thioflavin T (ThT) fluorescence kinetics experiments
For ThT aggregation kinetics experiments 1 to 9 μM monomeric Aβ42 was used, which was obtained after SEC purification45. ThT fluorescence was measured as described previously45 using 96-well microplates, where each well contained 80 μl sample solution with 10 μM ThT.
Analysis ThT aggregation kinetics
Aggregation traces were first analyzed using a fit to a sigmoidal function, revealing the aggregation half time, τ1/229,42,45. Subsequently, the aggregation traces were normalized and averaged over six replicates. The averaged aggregation half times are related to the initial monomer concentration, [Aβ], by τ1/2 ∝ [Aβ]γ where γ reflects the slope in a double-logarithmic plot (Fig. 4C). Further, we applied a nucleation model including primary and secondary nucleation in addition to fibril-end elongation12,49,50. In order to account for saturation of secondary nucleation an equilibrium constant (Michaelis constant) KM can be introduced, referring to a multi-step secondary nucleation model13. The kinetic equations for the time dependence of the fibril mass fractions for the two models can be found in refs. 12,13,29,45. The models were applied to describe the kinetic traces using a global fit analysis12,13. The kinetic fitting parameter are listed in Supplementary Table S1.
Supplementary information
Acknowledgements
This study was supported by the Magnus Bergvall foundation (A.A., H.B.), FLPP/Latvia lzp-2018/1-0275 project support (H.B., J.P.), Swedish Research Council (A.R., J.J.), Center for Innovative Medicine (CIMED) (A.R. J.J.), FORMAS (A.R.), Vinnova (J.J.), Olle Engkvists Foundation (G.C.), Swedish Alzheimer foundation (G.C.), Åhlen Foundation (A.A., G.C., H.B.), Stiftelsen för Gamla Tjänarinnor (A.A., H.B., G.C.), P. & A. Hedlunds Stiftelse (G.C.), Instruct R&D pilot project grant APPID 272 (A.A.), Loo and Hans Osterman Foundation (A.A., G.C.), Geriatric Diseases Foundation at Karolinska Institutet (A.A., G.C.), Norsk forskningsrådet 251290, 260786 (J.P.), JPco-fuND/EU PROP-AD 643417/Horizon 2020/EC 643417 (H.B., J.P.).
Author contributions
A.A., J.J. and H.B. conceived and designed the project. A.A., G.C. and H.B. developed the experimental protocol for Aβ production and performed and analyzed the CD, 15N-NMR and ThT kinetics experiments. G.C. performed the evolutionary relationship analysis. K.K., R.A., F.O. and K.J. performed and analyzed the 19F-labelling protocol and 19F-NMR experiments. M.L. performed and analyzed the mass spectrometry experiments. M.S., J.P., K.N., N.K., A.R. contributed additional data and analysis tools. H.B. supervised the project. A.A., A.R., J.J. and H.B. wrote the first draft of the manuscript. All authors provided comments on the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-57143-x.
References
- 1.Rising A, Johansson J. Toward spinning artificial spider silk. Nat. Chem. Biol. 2015;11:309–315. doi: 10.1038/nchembio.1789. [DOI] [PubMed] [Google Scholar]
- 2.Babb PL, et al. The Nephila clavipes genome highlights the diversity of spider silk genes and their complex expression. Nat. Genet. 2017;49:895–903. doi: 10.1038/ng.3852. [DOI] [PubMed] [Google Scholar]
- 3.Eisoldt L, Thamm C, Scheibel T. Review the role of terminal domains during storage and assembly of spider silk proteins. Biopolymers. 2012;97:355–361. doi: 10.1002/bip.22006. [DOI] [PubMed] [Google Scholar]
- 4.Andersson M, et al. Carbonic anhydrase generates CO2 and H+ that drive spider silk formation via opposite effects on the terminal domains. PLoS Biol. 2014;12:e1001921. doi: 10.1371/journal.pbio.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kronqvist N, et al. Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation. Nat. Commun. 2014;5:3254. doi: 10.1038/ncomms4254. [DOI] [PubMed] [Google Scholar]
- 6.Kronqvist N, et al. Efficient protein production inspired by how spiders make silk. Nat. Commun. 2017;8:15504. doi: 10.1038/ncomms15504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rising A, Hjalm G, Engstrom W, Johansson J. N-terminal nonrepetitive domain common to dragline, flagelliform, and cylindriform spider silk proteins. Biomacromolecules. 2006;7:3120–3124. doi: 10.1021/bm060693x. [DOI] [PubMed] [Google Scholar]
- 8.Sarr M, et al. A spidroin-derived solubility tag enables controlled aggregation of a designed amyloid protein. FEBS J. 2018;285:1873–1885. doi: 10.1111/febs.14451. [DOI] [PubMed] [Google Scholar]
- 9.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 10.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 11.Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 12.Cohen SIA, et al. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl Acad. Sci. USA. 2013;110:9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meisl G, et al. Differences in nucleation behavior underlie the contrasting aggregation kinetics of the Aβ40 and Aβ42 peptides. Proc. Natl Acad. Sci. USA. 2014;111:9384–9389. doi: 10.1073/pnas.1401564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fändrich M, et al. Amyloid fibril polymorphism: a challenge for molecular imaging and therapy. J. Intern. Med. 2018;283:218–237. doi: 10.1111/joim.12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masters CL, et al. Alzheimer’s disease. Nat. Rev. Dis. Prim. 2015;1:15056. doi: 10.1038/nrdp.2015.56. [DOI] [PubMed] [Google Scholar]
- 16.Aleksis R, Oleskovs F, Jaudzems K, Pahnke J, Biverstal H. Structural studies of amyloid-beta peptides: Unlocking the mechanism of aggregation and the associated toxicity. Biochimie. 2017;140:176–192. doi: 10.1016/j.biochi.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Arosio P, Cukalevski R, Frohm B, Knowles TP, Linse S. Quantification of the concentration of Abeta42 propagons during the lag phase by an amyloid chain reaction assay. J. Am. Chem. Soc. 2014;136:219–225. doi: 10.1021/ja408765u. [DOI] [PubMed] [Google Scholar]
- 18.Tickler AK, Barrow CJ, Wade JD. Improved preparation of amyloid-beta peptides using DBU as Nalpha-Fmoc deprotection reagent. J. Pept. Sci. 2001;7:488–494. doi: 10.1002/psc.342. [DOI] [PubMed] [Google Scholar]
- 19.Zarandi M, et al. Synthesis of Abeta[1-42] and its derivatives with improved efficiency. J. Pept. Sci. 2007;13:94–99. doi: 10.1002/psc.801. [DOI] [PubMed] [Google Scholar]
- 20.Macao B, et al. Recombinant amyloid beta-peptide production by coexpression with an affibody ligand. BMC Biotechnol. 2008;8:82. doi: 10.1186/1472-6750-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh DM, et al. A facile method for expression and purification of the Alzheimer’s disease-associated amyloid beta-peptide. FEBS J. 2009;276:1266–1281. doi: 10.1111/j.1742-4658.2008.06862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finder VH, Vodopivec I, Nitsch RM, Glockshuber R. The recombinant amyloid-beta peptide Abeta1-42 aggregates faster and is more neurotoxic than synthetic Abeta1-42. J. Mol. Biol. 2010;396:9–18. doi: 10.1016/j.jmb.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 23.Long F, Cho W, Ishii Y. Expression and purification of 15N- and 13C-isotope labeled 40-residue human Alzheimer’s beta-amyloid peptide for NMR-based structural analysis. Protein Expr. Purif. 2011;79:16–24. doi: 10.1016/j.pep.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garai K, Crick SL, Mustafi SM, Frieden C. Expression and purification of amyloid-beta peptides from Escherichia coli. Protein Expr. Purif. 2009;66:107–112. doi: 10.1016/j.pep.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dammers C, et al. Purification and Characterization of Recombinant N-Terminally Pyroglutamate-Modified Amyloid-beta Variants and Structural Analysis by Solution NMR Spectroscopy. PLoS One. 2015;10:e0139710. doi: 10.1371/journal.pone.0139710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunn AP, Masters CL, Cherny RA. Pyroglutamate-Abeta: role in the natural history of Alzheimer’s disease. Int. J. Biochem. Cell Biol. 2010;42:1915–1918. doi: 10.1016/j.biocel.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Jawhar S, Wirths O, Bayer TA. Pyroglutamate amyloid-beta (Abeta): a hatchet man in Alzheimer disease. J. Biol. Chem. 2011;286:38825–38832. doi: 10.1074/jbc.R111.288308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wärmländer S, et al. Biophysical studies of the amyloid beta-peptide: interactions with metal ions and small molecules. ChemBioChem. 2013;14:1692–1704. doi: 10.1002/cbic.201300262. [DOI] [PubMed] [Google Scholar]
- 29.Abelein A, Gräslund A, Danielsson J. Zinc as chaperone-mimicking agent for retardation of amyloid beta peptide fibril formation. Proc. Natl Acad. Sci. USA. 2015;112:5407–5412. doi: 10.1073/pnas.1421961112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danielsson J, Pierattelli R, Banci L, Graslund A. High-resolution NMR studies of the zinc-binding site of the Alzheimer’s amyloid beta-peptide. FEBS J. 2007;274:46–59. doi: 10.1111/j.1742-4658.2006.05563.x. [DOI] [PubMed] [Google Scholar]
- 31.Wang S, Huang W, Yang D. Structure and function of C-terminal domain of aciniform spidroin. Biomacromolecules. 2014;15:468–477. doi: 10.1021/bm401709v. [DOI] [PubMed] [Google Scholar]
- 32.Lin Z, Huang W, Zhang J, Fan JS, Yang D. Solution structure of eggcase silk protein and its implications for silk fiber formation. Proc. Natl Acad. Sci. USA. 2009;106:8906–8911. doi: 10.1073/pnas.0813255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen G, et al. Full-length minor ampullate spidroin gene sequence. PLoS One. 2012;7:e52293. doi: 10.1371/journal.pone.0052293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hijirida DH, et al. 13C NMR of Nephila clavipes major ampullate silk gland. Biophys. J. 1996;71:3442–3447. doi: 10.1016/S0006-3495(96)79539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hronska M, van Beek JD, Williamson PT, Vollrath F, Meier BH. NMR characterization of native liquid spider dragline silk from Nephila edulis. Biomacromolecules. 2004;5:834–839. doi: 10.1021/bm0343904. [DOI] [PubMed] [Google Scholar]
- 36.Jenkins JE, Holland GP, Yarger JL. High resolution magic angle spinning NMR investigation of silk protein structure within major ampullate glands of orb weaving spiders. Soft Matter. 2012;8:1947–1954. doi: 10.1039/C2SM06462F. [DOI] [Google Scholar]
- 37.Xu D, Yarger JL, Holland GP. Exploring the backbone dynamics of native spider silk proteins in Black Widow silk glands with solution-state NMR spectroscopy. Polymer. 2014;55:3879–3885. doi: 10.1016/j.polymer.2014.06.018. [DOI] [Google Scholar]
- 38.Oktaviani NA, et al. Conformation and dynamics of soluble repetitive domain elucidates the initial beta-sheet formation of spider silk. Nat. Commun. 2018;9:2121. doi: 10.1038/s41467-018-04570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roche J, Shen Y, Lee JH, Ying J, Bax A. Monomeric Abeta(1-40) and Abeta(1-42) Peptides in Solution Adopt Very Similar Ramachandran Map Distributions That Closely Resemble Random Coil. Biochemistry. 2016;55:762–775. doi: 10.1021/acs.biochem.5b01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danielsson J, Andersson A, Jarvet J, Gräslund A. 15N relaxation study of the amyloid beta-peptide: structural propensities and persistence length. Magn. Reson. Chem. 2006;44:S114–121. doi: 10.1002/mrc.1814. [DOI] [PubMed] [Google Scholar]
- 41.Danielsson J, Jarvet J, Damberg P, Gräslund A. The Alzheimer beta-peptide shows temperature-dependent transitions between left-handed 3-helix, beta-strand and random coil secondary structures. FEBS J. 2005;272:3938–3949. doi: 10.1111/j.1742-4658.2005.04812.x. [DOI] [PubMed] [Google Scholar]
- 42.Abelein A, Jarvet J, Barth A, Gräslund A, Danielsson J. Ionic Strength Modulation of the Free Energy Landscape of Abeta40 Peptide Fibril Formation. J. Am. Chem. Soc. 2016;138:6893–6902. doi: 10.1021/jacs.6b04511. [DOI] [PubMed] [Google Scholar]
- 43.Meisl G, Yang X, Frohm B, Knowles TP, Linse S. Quantitative analysis of intrinsic and extrinsic factors in the aggregation mechanism of Alzheimer-associated Abeta-peptide. Sci. Rep. 2016;6:18728. doi: 10.1038/srep18728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X, et al. On the role of sidechain size and charge in the aggregation of Abeta42 with familial mutations. Proc. Natl Acad. Sci. USA. 2018;115:E5849–E5858. doi: 10.1073/pnas.1803539115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen G, et al. Bri2 BRICHOS client specificity and chaperone activity are governed by assembly state. Nat. Commun. 2017;8:2081. doi: 10.1038/s41467-017-02056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kapust RB, et al. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 47.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knowles TPJ, et al. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326:1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 50.Cohen SIA, Vendruscolo M, Dobson CM, Knowles TPJ. From macroscopic measurements to microscopic mechanisms of protein aggregation. J. Mol. Biol. 2012;421:160–171. doi: 10.1016/j.jmb.2012.02.031. [DOI] [PubMed] [Google Scholar]
- 51.Bertini I, Gonnelli L, Luchinat C, Mao J, Nesi A. A new structural model of Abeta40 fibrils. J. Am. Chem. Soc. 2011;133:16013–16022. doi: 10.1021/ja2035859. [DOI] [PubMed] [Google Scholar]
- 52.Sharma SC, et al. A facile method for expression and purification of (15)N isotope-labeled human Alzheimer’s beta-amyloid peptides from E. coli for NMR-based structural analysis. Protein Expr. Purif. 2015;116:82–89. doi: 10.1016/j.pep.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chhetri G, Pandey T, Chinta R, Kumar A, Tripathi T. An improved method for high-level soluble expression and purification of recombinant amyloid-beta peptide for in vitro studies. Protein Expr. Purif. 2015;114:71–76. doi: 10.1016/j.pep.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 54.Lee EK, Hwang JH, Shin DY, Kim DI, Yoo YJ. Production of recombinant amyloid-beta peptide 42 as an ubiquitin extension. Protein Expr. Purif. 2005;40:183–189. doi: 10.1016/j.pep.2004.12.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.