Abstract
Recent evidence indicates that the long non-coding RNA (lncRNA) cancer susceptibility candidate 2 (CASC2) is involved in tumorigenesis of several types of cancer through targeting microRNAs (miRs); however, the molecular mechanism of CASC2 in pancreatic cancer remains elusive. In the present study, the expression levels of CASC2, miR-24 and mucin 6 (MUC6) were measured in pancreatic cancer specimens and cell lines by reverse transcription-quantitative PCR. Western blotting was used to determine the protein expression levels of MUC6, Integrin β4 (ITGB4), phosphorylated (p)-focal adhesion kinase (FAK) and several epithelial-to-mesenchymal transition markers in pancreatic cancer cells. MTT, colony formation, wound healing, Transwell and flow cytometry assays were performed to detect cell proliferation, colony formation, migration, invasion and apoptosis, respectively,in vitro. Morphological changes of pancreatic cancer cells were assessed by light microscopy. The interactions between CASC2, miR-24 and MUC6 were assessed by the dual-luciferase reporter assay. A tumor xenograft model was generated to investigate tumor growth in vivo. CASC2 and MUC6 were downregulated, and miR-24 was upregulated in pancreatic cancer specimens and cell lines. Functionally, CASC2 overexpression or miR-24 knockdown suppressed pancreatic cancer cell proliferation, colony formation, migration and invasion, and promoted apoptosis. Additionally, they altered cell-cell adhesion as demonstrated by the attenuated ITGB4, p-FAK and N-cadherin protein levels, as well as morphological changes. Mechanistically, CASC2 sponged miR-24 and activated its downstream target MUC6 to suppress pancreatic cancer growth and progression. CASC2 exerted tumor-suppressive functions in pancreatic cancer through the miR-24/MUC6 axis, which may be a promising target for pancreatic cancer therapy.
Keywords: pancreatic cancer, metastasis, cancer susceptibility candidate 2, microRNA-24, mucin 6, Integrin β4/focal adhesion kinase pathway
Introduction
Pancreatic cancer was the seventh leading cause of cancer-related death worldwide in 2012 (1), with a 5-year overall survival rate of <8% (2). Disease progression and metastasis are major barriers in the survival of patients with pancreatic cancer (3). Despite the improvement in diagnosis and treatment, surgical resection is the only effective therapy for pancreatic cancer. However, because of local invasion and/or metastasis, only 15-20% of patients with pancreatic cancer qualify for surgical intervention (4). Understanding the potential molecular mechanisms contributing to metastasis could therefore be of great value for effective pancreatic cancer therapy.
Long non-coding RNAs (lncRNAs) have been implicated in the regulation of disease progression in numerous types of cancer by functioning as tumor suppressors or promoters (5,6). Increasing evidence indicates that lncRNA cancer susceptibility candidate 2 (CASC2) exhibits tumor-suppressive functions by acting as a competing endogenous RNA (ceRNA) for microRNAs (miRNAs/miRs) (7-10). CASC2 was shown to be downregulated and suppress tumor progression in pancreatic cancer (11). miR-24 has been reported to be upregulated in pancreatic cancer cells (12,13). While CASC2 was demonstrated to act as a sponge for miR-24 and regulated tumorigenesis of hepatocellular carcinoma (8,14), little is known about the interaction between CASC2 and miR-24 in pancreatic cancer.
Mucins (MUCs) are a group of heavily glycosylated proteins, which are involved in the regulation of proliferation, migration and invasion of pancreatic cancer (15,16). MUC6, a high molecular weight secretory polymeric MUC, has been shown to be downregulated in pancreatic cancer (17,18), and exerts tumor-suppressive effects by interfering with cell-matrix adhesion in the local tumor microenvironment (19). Bioinformatics analysis using Targetscan 7.2 predicted that MUC6 is a direct target of miR-24. However, to the best of our knowledge, no studies have been conducted that focus on the association between MUC6 and miR-24 in pancreatic cancer. Integrin β4 (ITGB4), one of the main mediators of cell-matrix adhesion of epithelial cells, is actively involved in regulating progression of pancreatic cancer through the focal adhesion kinase (FAK) signaling pathway (20). Moreover, MUC5AC, a MUC subtype, regulates cell migration through the ITGB4/FAK signaling pathway in lung cancer (21). It remains unknown if a similar crosstalk exists between MUC6 and the ITGB4/FAK pathway in pancreatic cancer.
The present study demonstrated that CASC2 was downregulated in pancreatic cancer tissues and cell lines, and served crucial roles in pancreatic cancer growth and progression. Conversely, miR-24 was upregulated and had a pro-metastatic role in pancreatic cancer. Furthermore, to the best of our knowledge, this study is the first to demonstrate that CASC2 may function as a ceRNA for miR-24 and may exert tumor-suppressive effects in part through the MUC6/ITGB4/FAK signaling pathway. These findings may provide promising therapeutic targets for pancreatic cancer.
Materials and methods
Clinical samples
A total of 20 pancreatic cancer tissues and adjacent normal tissues were collected from patients (age, 45-70 years; 12 female and 8 male patients) that underwent surgical resection between June 2016 and June 2018 at the Zhongda Hospital Affiliated with Southeast University (Nanjing, China). Histologic diagnosis was confirmed for each specimen. This study was approved by the Ethical Committee of Zhongda Hospital Affiliated with Southeast University and written informed consent was obtained from all patients. Samples were frozen immediately after being surgically resected and were stored in liquid nitrogen.
Cell culture
The human pancreatic normal epithelial cell line (hTERT-HPNE; cat. no. ATCC® CRL-4023™), which is comprised of intermediary cells formed during acinar-to-ductal metaplasia, and the human pancreatic cancer cell lines AsPC-1 (cat. no. ATCC® CRL-1682™) and PANC-1 (cat. no. ATCC® CRL-1469™) were purchased from the American Type Culture Collection. The human pancreatic cancer cell line PATU8988T was provided by Cell Biology Research Center of Central South University. Cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS; both Gibco; Thermo Fisher Scientific, Inc.) in the presence of 100 U/ml penicillin and streptomycin (Beyotime Institute of Biotechnology) at 37°C with a humidified atmosphere (5% CO2).
Cell transfection and transduction
For in vitro assays, the human CASC2 sequence was cloned into the pIRES2-EGFP vector (cat. no. GV146; Shanghai GeneChem Co., Ltd.) to create the CASC2 overexpression vector. The empty pIRES2-EGFP vector served as a negative control (NC). miR-24 mimics, miR-24 inhibitor and their negative controls (NCs) were purchased from Shanghai GenePharma Co., Ltd. AsPC-1 or PANC-1 cells (5×105/well) were cultured in 6-well plates for 24 h and were then transfected with 5 µg/ml pIRES2-EGFP-CASC2 overexpression vector, pIRES2-EGFP empty vector, 10 nM miR-24 mimics, miR-24 inhibitor or their NCs at 40-60% confluence using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The transfected cells were harvested after 48 h at 37°C. Morphological changes of pancreatic cancer cells were assessed by light microscopy. The sequences were as follows: miR-24 mimics, sense 5′-UGG CUC AGU UCA GCA GGA ACA G-3′, antisense 5′-GUU CCU GCU GAA CUG AGC CAU U-3′; mimics NC, sense 5′-UUC UCC GAA CGU GUC ACG UTT-3′, antisense 5′-ACG UGA CAC GUU CGG AGA ATT-3′; miR-24 inhibitor, 5′-CUG UUC CUG CUG AAC UGA GCC A-3′; inhibitor NC, 5′-CAG UAC UUU UGU GUA GUA CAA-3′.
For in vivo studies, AsPC-1 cells were transduced with lentivirus (LV)-CASC2 (LV5-EF1a-GFP/Puro vector; Shanghai GenePharma Co., Ltd.) and LV-miR-24 (LV3-pGLV-h1-GFP-puro vector; Shanghai GenePharma Co., Ltd.), or LV-NC vectors (LV-CASC2-NC and LV-miR-24 NC; Shanghai GenePharma Co., Ltd.) as previously described (22). Briefly, AsPC-1 cells (5×105 per well) were plated in 6-well plates for 24 h; the medium was then replaced with fresh medium containing 8 µg/ml polybrene. The lentiviral vectors were transduced into AsPC-1 cells for 24 h at 37°C at a multiplicity of infection of 50, followed by puromycin (5 µg/ml) antibiotic selection for 3 days.
Immunohistochemistry
Tumor tissues were fixed with 3.7% paraformaldehyde at room temperature for 12 h, and were then embedded in paraffin, cut into 5-µm sections and washed with PBS. After blocking with 5% FBS and 0.3% Triton X-100 in PBS for 1 h at room temperature, MUC6 antibody (cat. no. sc-33668; Santa Cruz Biotechnology, Inc.) was applied at a dilution of 1:200 and the sections were incubated at 4°C overnight. After incubation with the secondary antibody (anti-mouse biotinylated; 1:1,000; cat. no. 14709; Cell Signaling Technology, Inc.) for 1 h at room temperature, the avidinbiotin peroxidase method (avidin peroxidase conjugate, 1:2,000, cat. no. ab59653; Abcam) was adopted for 1 h at room temperature to determine the location and relative expression level of the target protein. Finally, the visualization signal was developed with 3,3′-diaminobenzidine tetrahydrochloride, and the slides were counterstained in hematoxylin diluted at 1:5 in H2O for 1 min at room temperature and observed by light microscopy at ×100 magnification.
Dual-luciferase reporter assay
TargetScan 7.2 software (http://www.targetscan.org/vert_72/) was applied to predict the miR-24 binding sites on MUC6. RNA Interactome Database (http://www.rna-society.org/raid/home.html) was applied to predict the miR-24 binding sites on CASC2. The wild type and mutant human MUC6 3′-untranslated regions (UTRs) and the CASC2 sequence containing putative binding sites for miR-24 were designed and synthesized by Shanghai GeneChem Co., Ltd. The wild type and mutant constructs were cloned into the pGL3-promoter vector (Promega Corporation). For the dual-luciferase reporter assay, AsPC-1 and PANC-1 cells (2×105/well) in 24-well plates were harvested at 24 h and transfected with 5 µg/ml pGL3-promoter vectors and 10 nM miR-24 mimics or miR-24 inhibitor, or their NCs using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, cells were harvested and lysed, and luciferase activity was detected using the Dual Luciferase Reporter Assay kit (Promega Corporation), according to the manufacturer’s protocol. Firefly luciferase activities were normalized to Renilla luciferase activities.
MTT assay
AsPC-1 and PANC-1 (1×104 cells/well) were seeded in 96-well plates and grown overnight. After trans-fection for 1, 2, 3 or 4 days, the medium was replaced with DMEM supplemented with 10% FBS. Subsequently, 20 µl MTT solution (5 mg/ml; Abcam) was added to each well and samples were incubated for 4 h at 37°C. DMSO (100 µl) was added and samples were agitated in the dark for 30 min at room temperature to dissolve formazan crystals. The absorbance was measured at 490 nm using a microplate reader; a 630 nm reference was subtracted.
Colony formation assay
Transfected cells were seeded in 6-well plates at 200 cells/well and cultured for 2 weeks. Colonies were washed twice with PBS, fixed with 100% methanol at 4°C for 15 min and stained with 1% crystal violet at room temparature for 60 min. Images of the number of colonies were captured with a camera. The colonies were counted by three researchers who were blinded to the experimental condition.
Apoptosis assay
Apoptotic cells were identified using the Annexin V-APC/PI Apoptosis Detection kit (Nanjing KeyGen Biotech Co., Ltd.), according to the manufacturer’s protocol. Transfected cells (2×105/well) in 24-well plates were collected, washed twice with cold PBS and resuspended in 1X binding buffer. Subsequently, cells were stained with 5 µl Annexin V-APC for 15 min and 5 µl PI for 10 min in the dark at room temperature. Cells were examined using the FACSCanto II flow cytometer (BD Biosciences). Analysis of flow cytometry data was performed using FlowJo version X.10.0.7-1 (FlowJo, LLC).
Wound healing and Transwell assays
For cell migration, transfected cells were seeded in 6-well plates (5×105/well) and cultured overnight to >95% confluence. Wounds were inflicted with the sterile 200-µl pipette tips and loose cells were washed off with culture medium. Cells were then maintained in DMEM without FBS and images were captured at 0 and 24 h using a light microscope at ×40 magnification. ImageJ version 1.52a (National Institutes of Health) was used to analyze the wound width. The migration distance (%) was determined by the following formula: (W0 h−W24 h)/W0 h ×100. W, refers to width.
For cell invasion, cells (2×104/well) were cultured in 24-well Transwell chambers with the upper chambers coated with Matrigel (BD Bioscience). Cells in the upper chamber were incubated in 200 µl DMEM containing 10% FBS; as a chemoattractant, the bottom chamber contained DMEM with 20% FBS. After 24 h at 37°C, cells remaining on the upper membrane surface were removed with a cotton swab and cells adhering to the lower membrane surface were fixed with 4% paraformaldehyde at 4°C overnight and stained with 1% crystal violet at room temparature for 60 min. The number of invaded cells in five random fields was counted under a light microscope at ×100 magnification.
Tumor xenograft using nude mice
A total of 20 male BALB/c nude (nu/nu) mice (age, 6 weeks; weight, 16-20 g) were purchased from the Model Animal Research Center of Nanjing University. Animals were acclimated under specific pathogen-free conditions. Mice were kept in cages (n=5 mice/cage), and were housed in a sterile room under a 12-h light/dark cycle at ~23°C and 50% humidity, with ad libitum access to food and water. Animals were maintained on a balanced diet for rodents and given free access to water and food. All of the animal studies were conducted in accordance with the Institutional Animal Care and Use Committee and were approved by the Medical Ethics Committee of Southeast University (Nanjing, China).
AsPC-1 cells were stably transduced with lentiviral vectors, according to the indicated groups (n=5 mice/group). Transduced AsPC-1 cells (1×106) were suspended in 100 µl PBS and subcutaneously injected into the flank of the mice under anesthesia. The xenograft tumor size was measured every 5 days with a vernier caliper, and the tumor volumes were calculated using the equation: 0.5 × length x width2. After 30 days, mice were euthanized by cervical dislocation and the tumor tissues were harvested. The tumor take rate was ~95%. Pathological and molecular expression analyses of tumor tissues were performed.
RNA extraction and reverse transcription-quantitative (RT-q) PCR
Total RNA from tumor tissues and transfected cells was extracted using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized using the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.) at 25°C for 5 min, 37°C for 30 min and 85°C for 5 sec. qPCR analysis was performed using the StepOnePlus Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with SYBR Premix EX Taq kit (Takara Biotechnology Co., Ltd.). The thermocycling conditions were as follows: Initial denaturation, 95°C for 5 sec; followed by 35 cycles of denaturation at 94°C for 15 sec, annealing at 55°C for 25 sec and extension at 70°C for 30 sec. Primers for CASC2, miR-24 and MUC6 were purchased from Sangon Biotech Co., Ltd. GAPDH and U6 were used as internal reference genes for mRNA and miRNA, respectively. Relative expression was calculated using the 2−ΔΔCq method (23). The primer sequences were as follows: CASC2, forward 5′-AGC TCA TGT GGT TGC AAG GT-3′, reverse 5′-CTG CCT GAA ACT AGG CGG AA-3′; miR-24, forward 5′-GCG TGG CTC AGT TCA GCA G-3′, reverse 5′-AGT GCA GGG TCC GAG GTA TT-3′; MUC6, forward 5′-CGT CTG TGG TGT CAA CGA CT-3′, reverse 5′-CCG GTG ACC GTG TAG TTT CT-3′; U6 (internal control for miRNA), forward 5′-CTC GCT TCG GCA GCA CA-3′, reverse 5′-AAC GCT TCA CGA ATT TGC GT-3′; and GAPDH (internal control for mRNA), forward, 5′-CCA CAG TCC ATG CCA TCA C-3′ and reverse 5′-GCT TCA CCA CCT TCT TGA TG-3′.
Western blotting
Total proteins were extracted from tissues and cultured cells by RIPA buffer (Beyotime Institute of Biotechnology) with protease inhibitor cocktail (Roche Diagnostics). Bicinchoninic acid assays were used to detect protein concentration. Equal amounts of protein (30 µg) were separated by SDS-PAGE on 10% gels and were then transferred to polyvinylidene fluoride membranes. Membranes were blocked with 5% non-fat milk for 1 h at room temperature, followed by incubation with primary antibodies at 4°C overnight. The primary antibodies used were: Anti-INTB4 (1:1,000; cat. no. 4707; Cell Signaling Technology, Inc.), anti-phosphorylated (p)-FAK (1:1,000; cat. no. 3284; Cell Signaling Technology, Inc.), anti-FAK (1:2,000; cat. no. 3285; Cell Signaling Technology, Inc.), anti-E-cadherin (1:5,000; cat. no. 3195; Cell Signaling Technology, Inc.), anti-N-cadherin (1:1,000; cat. no. 4061; Cell Signaling Technology, Inc.), anti-vimentin (1:1,000; cat. no. 5741; Cell Signaling Technology, Inc.), anti-matrix metalloproteinase (MMP)-2 (1:500; cat. no. 4022; Cell Signaling Technology, Inc.), anti-MMP-9 (1:1,000; cat. no. 3852; Cell Signaling Technology, Inc.), anti-Snail (1:2,000; cat. no. 3879; Cell Signaling Technology, Inc.), anti-fibronectin (1:1,000; cat. no. F3648; Sigma-Aldrich; Merck KGaA), anti-MUC6 (1:1,000; cat. no. ab192318; Abcam) and anti-GAPDH (1:3,000; cat. no. 60004-1-1g; ProteinTech Group, Inc.). Membranes were then incubated with goat anti-rabbit (cat. no. 7074) and a goat anti-mouse (cat. no. 7076) horseradish peroxidase-conjugated secondary antibodies (1:3,000; Cell Signaling Technology, Inc.) for 1 h at room temperature. Bands were developed using chemiluminescence substance (Thermo Fisher Scientific, Inc.). The proteins were semi-quantified using Quantity One version 4.2.1 (Bio-Rad Laboratories, Inc.).
Statistical analysis
All experiments were repeated ≥3 times and representative results are presented. Data were analyzed with Prism 6.0 (GraphPad Software, Inc.) and are expressed as the mean ± standard deviation. Comparisons between two groups were performed using Student’s t-test (both paired and unpaired tests). Comparisons among ≥3 groups were conducted using one-way ANOVA followed by Tukey’s test. P<0.05 was considered to indicate a statistically significant difference.
Results
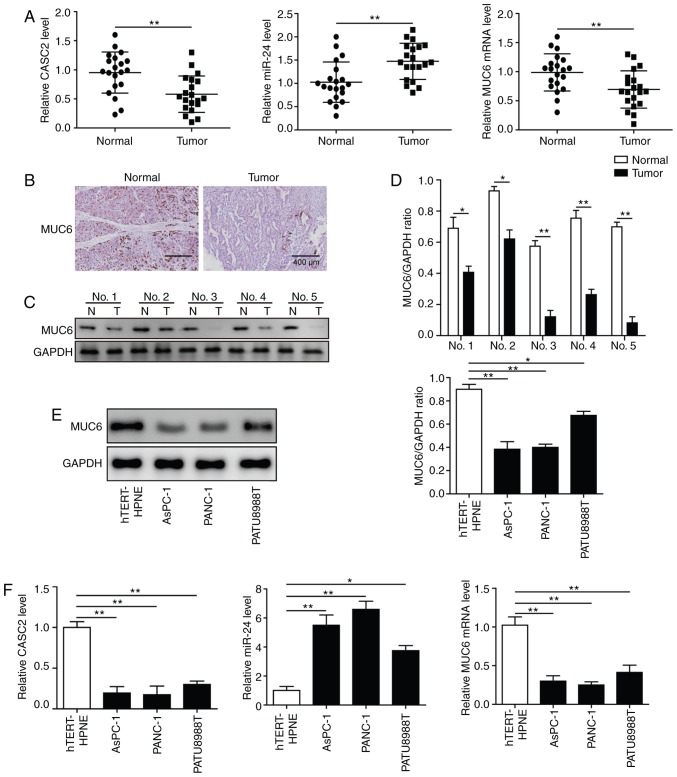
CASC2 and MUC6 are downregulated, and miR-24 is upregulated in pancreatic cancer tissues and cell lines
To investigate the clinical relevance of CASC2, miR-24 and MUC6, 20 pancreatic tumor samples and paired adjacent normal tissues were collected. RT-qPCR analysis revealed that CASC2 and MUC6 expression levels were significantly decreased, whereas miR-24 levels were significantly increased in tumor samples compared with normal tissues (Fig. 1A). Immunohistochemistry and western blot analyses demonstrated that MUC6 protein expression levels were lower in pancreatic cancer samples than in normal pancreatic tissues (Fig. 1B-D). Subsequently, the expression levels of CASC2, miR-24 and MUC6 were detected in three pancreatic cancer cell lines, namely AsPC-1, PANC-1 and PATU8988T, and in the human pancreatic normal epithelial cell line hTERT-HPNE. The results revealed that CASC2 and MUC6 expression levels were significantly lower, and miR-24 expression was significantly higher in the pancreatic cancer cell lines compared with the hTERT-HPNE cells (Fig. 1F). Western blotting further confirmed that MUC6 protein expression was significantly decreased in pancreatic cancer cell lines compared with the control cells (Fig. 1E). As AsPC-1 and PANC-1 cells had low CASC2 and MUC6 and high miR-24 expression compared with PATU8988T cells, these two cell lines were used for subsequent loss- and gain-of-function experiments. Collectively, these data suggested the involvement of CASC2, miR-24 and MUC6 in pancreatic cancer.
Figure 1.
CASC2 and MUC6 are downregulated, and miR-24 is upregulated in pancreatic cancer tissues and cell lines. (A) RT-qPCR analysis of CASC2, miR-24 and MUC6 expression in 20 paired pancreatic cancer and adjacent normal tissues. (B) Immunohistochemistry (scale bar, 400 µm) and (C and D) western blot analysis of MUC6 protein expression levels in pancreatic cancer and adjacent normal tissues. (E) Western blot analysis of MUC6 protein expression levels in AsPC-1, PANC-1 and PATU8988T pancreatic cancer cell lines, and the human pancreatic normal epithelial cell line hTERT-HPNE. (F) RT-qPCR analysis of CASC2, miR-24 and MUC6 expression levels in pancreatic cancer cell lines and hTERT-HPNE cells. *P<0.05 and **P<0.01. CASC2, cancer susceptibility candidate 2; miR, microRNA; MUC6, mucin 6; N, adjacent normal tissues; RT-qPCR, reverse transcription-quantitative PCR; T, pancreatic cancer tissues.
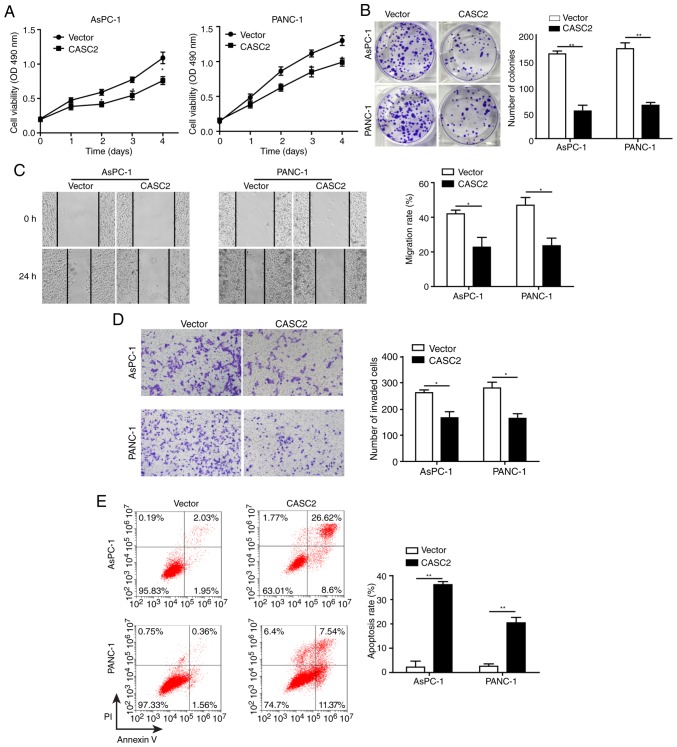
Overexpression of CASC2 inhibits pancreatic cancer cell growth and progression partially by altering cell-cell adhesion
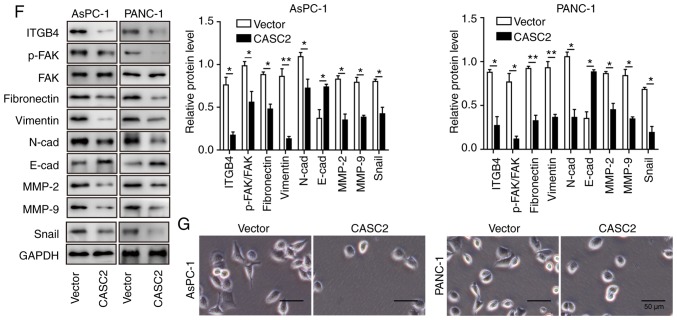
This study explored the role of CASC2 in regulating pancreatic cancer cell proliferation and colony formation. Overexpression of CASC2 significantly reduced cell proliferation and colony formation of AsPC-1 and PANC-1 cells (Fig. 2A and B). This study also assessed whether CASC2 affected other aspects of pancreatic cancer progression. To this end, the effects of CASC2 overexpression on pancreatic cancer cell migration, invasion and apoptosis were analyzed. The results revealed that overexpression of CASC2 significantly inhibited the migration and invasion of pancreatic cancer cells (Fig. 2C and D), and induced cell apoptosis (Fig. 2E). On the basis of the results that CASC2 overexpression suppressed pancreatic cell development and progression, the expression of pro- and anti-metastatic proteins was analyzed (Fig. 2F). Previous studies have reported that FAK is a key downstream signaling molecule triggered by integrin activation (24) and that FAK activity is required for the formation of cell-cell adhesion (25). Additionally, it has been shown that Snail induces the expression of MMPs that degrade the basement membrane, thereby favoring invasion (26). Consistent with these reports, upon CASC2 overexpression, the expression of ITGB4 and phosphorylation of FAK were decreased. Additionally, N-cadherin and fibronectin expression was decreased, and E-cadherin expression was increased in CASC2-overexpressing cells. CASC2 overexpression further downregulated Snail, while attenuating MMP-2 and MMP-9 expression (Fig. 2F). The effect of CASC2 on cell adhesion was further confirmed by light microscopy. AsPC-1 and PANC-1 cells appeared round and had shortened synapses following transfection with CASC2 overexpression vector compared with the spindle shape under normal condition (Fig. 2G). Also, the number of cells was decreased, which may be due to increased cell apoptosis in CASC2-overexpressing cells. These observations demonstrated that CASC2 suppressed development and progression of pancreatic cancer cells, potentially via negative regulation of the ITGB4/FAK signaling pathway.
Figure 2.
Overexpression of CASC2 inhibits pancreatic cancer cell growth and progression partially by altering cell-cell adhesion. Overexpression of CASC2 suppresses (A) proliferation of AsPC-1 and PANC-1 cells, as determined by MTT assay, (B) cell colony formation of AsPC-1 and PANC-1 cells, as determined by colony formation assay, (C) migration of AsPC-1 and PANC-1 cells, as determined by wound healing assay (×40 magnification), and (D) invasion of AsPC-1 and PANC-1 cells, as determined by Transwell assay (×100 magnification). (E) Overexpression of CASC2 promotes apoptosis of AsPC-1 and PANC-1 cells, as determined by flow cytometry. (F) Western blot analysis of indicated proteins in AsPC-1 and PANC-1 cells overexpressing CASC2. (G) Morphology of AsPC-1 and PANC-1 cells transfected with CASC2 overexpression vector; scale bar, 50 µm. *P<0.05 and **P<0.01. cad, cadherin; CASC2, cancer susceptibility candidate 2; FAK, focal adhesion kinase; ITGB4, Integrin β4; MMP, matrix metalloproteinase; NC, negative control; OD, optical density; p-, phosphorylated.
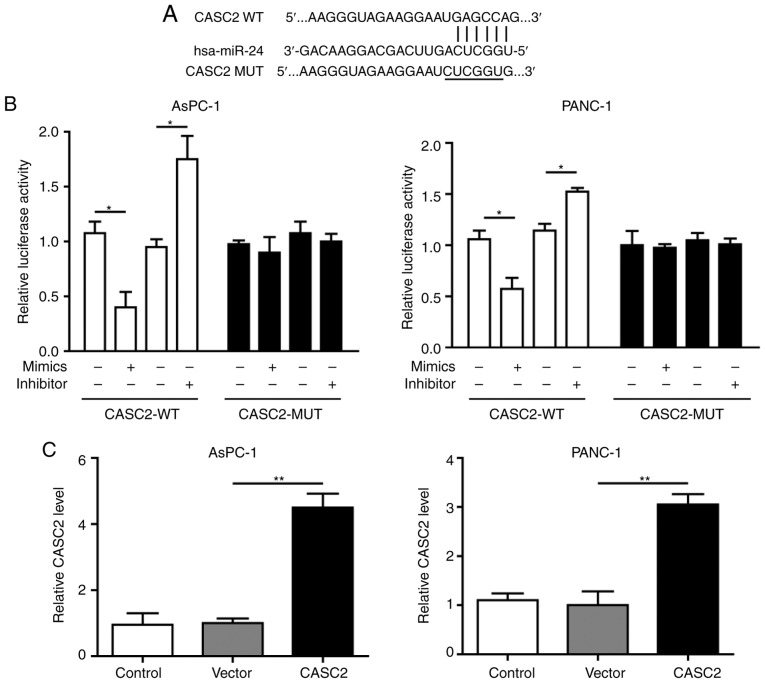
CASC2 sponges miR-24 in pancreatic cancer
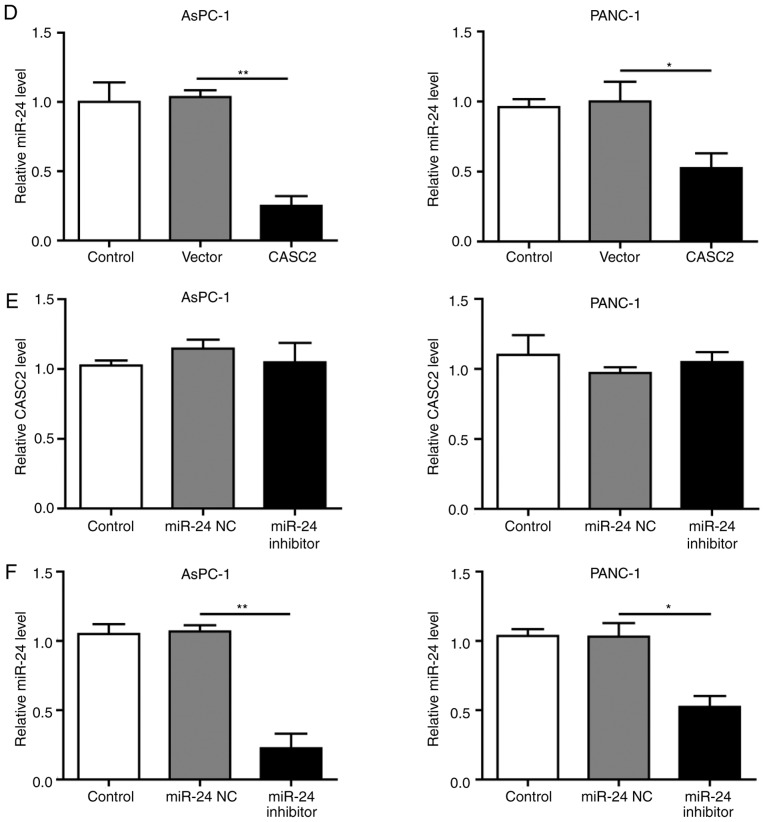
To explore the potential regulatory associations between CASC2 and miR-24 in pancreatic cancer cells, the CASC2 promoter sequence was analyzed and a binding site for miR-24 was identified (Fig. 3A). The present results revealed that miR-24 knockdown in pancreatic cancer cells promoted CASC2 promoter-driven luciferase activity, and miR-24 overexpression decreased luciferase activity; in both cases, luciferase activity was not affected when the miR-24 binding site was mutated (Fig. 3B). This interaction was further assessed by overexpression of CASC2 and inhibition of miR-24 in AsPC-1 and PANC-1 cells; the findings confirmed that upregulation of CASC2 significantly inhibited miR-24 expression (Fig. 3C and D). In addition, downregulation of miR-24 had no significant effect on CASC2 expression (Fig. 3E and F). Collectively, these results demonstrated that CASC2 sponged miR-24 in pancreatic cancer cells.
Figure 3.
miR-24 is a target of CASC2 in pancreatic cancer cells. (A) Predicted binding site of CASC2 and miR-24. (B) Relative luciferase activity of CASC2-WT and CASC2-MUT following transfection with miR-24 mimics or inhibitor. Luciferase activity was measured after 48 h and is presented as relative luciferase units. Expression levels of (C) CASC2 and (D) miR-24 in AsPC-1 and PANC-1 cells transfected with pIRES2-EGFP-CASC2 or empty vector. Expression levels of (E) CASC2 and (F) miR-24 in AsPC-1 and PNAC-1 cells transfected with miR-24 inhibitor or miR-24 negative control. *P<0.05 and **P<0.01. CASC2, cancer susceptibility candidate 2; miR, microRNA; MUT, mutant; NC, negative control; WT, wild type.
Knockdown of miR-24 inhibits pancreatic cancer cell growth and progression partially by altering cell-cell adhesion
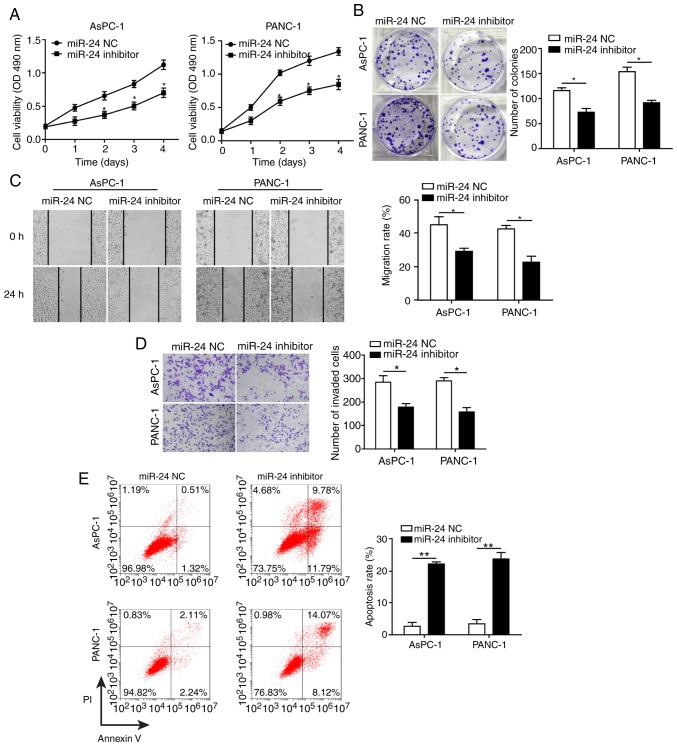
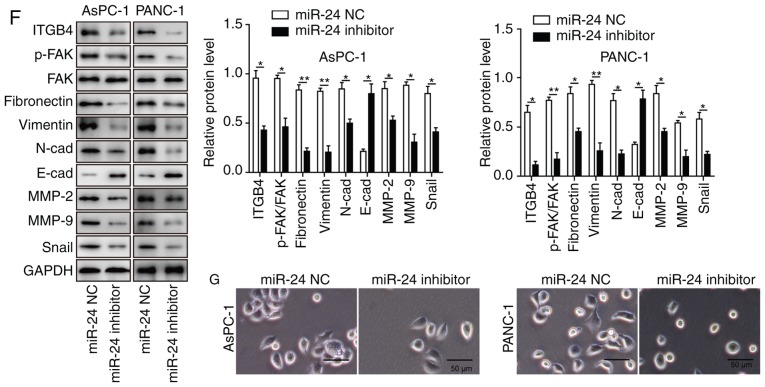
To determine the effects of miR-24 on pancreatic cancer cell development and progression, AsPC-1 and PANC-1 cells were transfected with miR-24 inhibitor and miR-24 NC. Knockdown of miR-24 (transfection success is shown in Fig. 3F) significantly reduced cell proliferation, colony formation, migration and invasion, and promoted apoptosis of AsPC-1 and PANC-1 cells (Fig. 4A-E). In agreement with the results in CASC2-overexpressing pancreatic cancer cells, decreased cell migration and invasion in the miR-24 knockdown cells were accompanied by significantly decreased levels of ITGB4, p-FAK, N-cadherin, fibronectin, Snail, MMP-2 and MMP-9, and significantly increased levels of E-cadherin (Fig. 4F). AsPC-1 and PANC-1 cells transfected with miR-24 inhibitor changed from spindle-shaped to rounded cells with shortened synapses, similar to observations in CASC2-overexpressing cells (Fig. 4G). In addition, the number of cells was decreased, potentially due to increased cell apoptosis in miR-24 inhibitor treatment group. These results demonstrated that miR-24 may act as a tumor promoter in pancreatic cancer cells, potentially via positive regulation of the ITGB4/FAK signaling pathway.
Figure 4.
Knockdown of miR-24 inhibits pancreatic cancer cell growth and progression partially by altering cell-cell adhesion. Knockdown of miR-24 suppressed (A) proliferation, as measured by MTT assay, (B) colony formation, (C) migration, as measured by wound healing assay (×40 magnification), and (D) invasion, as measured by Transwell assay (×100 magnification) in AsPC-1 and PANC-1 cells transfected with miR-24 inhibitor. (E) Knockdown of miR-24 promoted apoptosis of AsPC-1 and PANC-1 cells transfected with miR-24 inhibitor, as measured by flow cytometry. (F) Western blot analysis of indicated proteins in AsPC-1 and PANC-1 cells transfected with miR-24 inhibitor or negative control. (G) Morphology of AsPC-1 and PANC-1 cells transfected with miR-24 inhibitor; scale bar, 50 µm. *P<0.05 and **P<0.01. cad, cadherin; CASC2, cancer susceptibility candidate 2; FAK, focal adhesion kinase; ITGB4, Integrin β4; MMP, matrix metalloproteinase; NC, negative control; OD, optical density; p-, phosphorylated.
MUC6 is a target of miR-24 and is positively regulated by CASC2 in pancreatic cancer cells
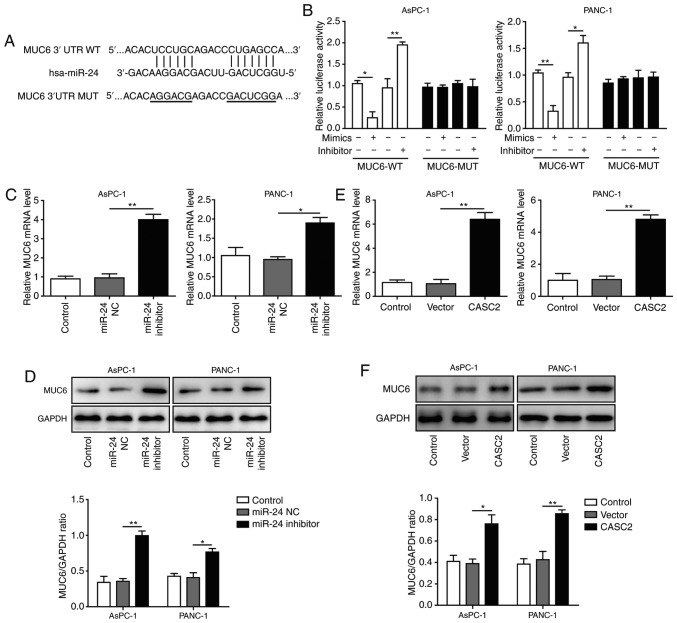
Bioinformatics analysis predicted miR-24 binding sites on the human MUC6 3’-UTR (Fig. 5A). Upon transfection, miR-24 overexpression significantly reduced the wild-type reporter activity, whereas miR-24 inhibition exhibited an opposite, promoting effect (Fig. 5B). No significant effects were observed in the mutated MUC6 group. These findings indicated that miR-24 directly targeted MUC6. To further assess the impact of miR-24 on MUC6 level, AsPC-1 and PANC-1 cells were transfected with miR-24 inhibitor. It was revealed that the mRNA and protein expression levels of MUC6 were significantly increased upon miR-24 knockdown (Fig. 5C and D), suggesting that miR-24 functioned as an antagonist to MUC6. Based on the aforementioned results that CASC2 exerted tumor-suppressive functions, the association between CASC2 and MUC6 was analyzed. To this end, MUC6 mRNA and protein expression levels in CASC2-overexpressing pancreatic cancer cells were detected. MUC6 was upregulated and accumulated in the transfected cells (Fig. 5E and F). Therefore, it was hypothesized that MUC6 was a direct target of miR-24 and was positively modulated by CASC2 in pancreatic cancer cells.
Figure 5.
MUC6 is a target of miR-24 and is positively regulated by CASC2 in pancreatic cancer cells. (A) Predicted binding site of miR-24 and MUC6. (B) Relative luciferase activity of MUC6-WT and MUC6-MUT after transfection with miR-24 mimics or inhibitor. Luciferase activity was measured after 48 h and is presented as relative luciferase units. (C) mRNA and (D) protein expression levels of MUC6 in AsPC-1 and PANC-1 cells transfected with miR-24 NC or miR-24 inhibitor. (E) mRNA and (F) protein expression levels of MUC6 in AsPC-1 and PANC-1 cells transfected with pIRES2-EGFP-CASC2 or empty vector. *P<0.05 and **P<0.01. CASC2, cancer susceptibility candidate 2; miR, microRNA; MUC6, mucin 6; NC, negative control.
miR-24 mediates suppressive effects of CASC2 in tumorigenesis of pancreatic cancer cells
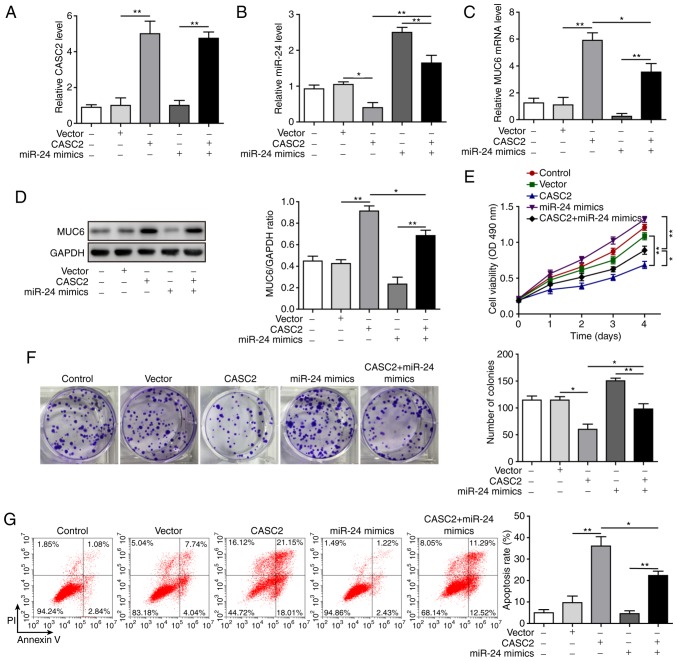
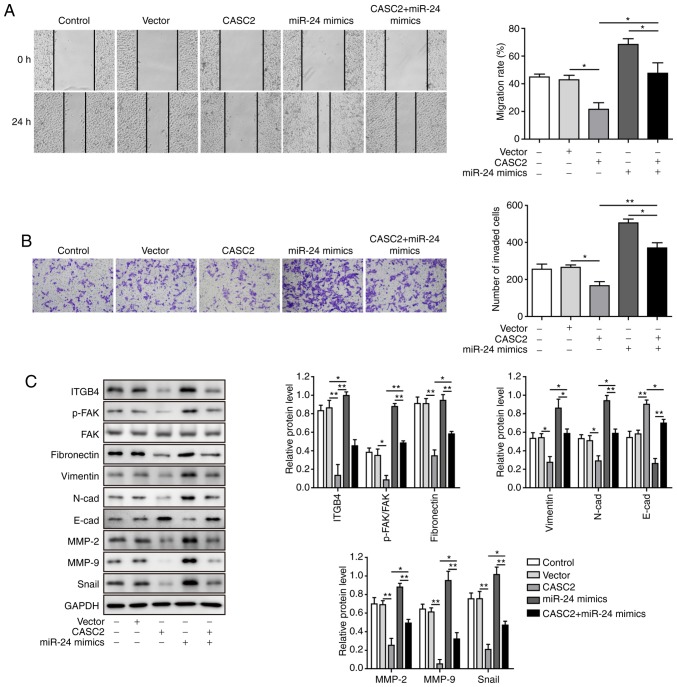
After confirming the molecular mechanism by which CASC2 functioned as a ceRNA for miR-24 and upregulated its downstream target MUC6 in tumorigenesis of pancreatic cancer cells, this study investigated the functional implication of miR-24 in CASC2 overexpression-induced tumor suppression in AsPC-1 cells. The study initially analyzed whether miR-24 mimics reversed alterations in miR-24 and MUC6 levels induced by CASC2 overexpression. The results revealed that overexpression of miR-24 significantly restored miR-24 and reduced MUC6 expression in CASC2-overexpressing cells (Fig. 6A-D). Furthermore, it was investigated as to whether overexpression of miR-24 reversed the effects of CASC2 overexpression on cell functions. The results confirmed that miR-24 overexpression promoted cell proliferation, colony formation, migration and invasion, and decreased apoptosis of AsPC-1 cells induced by CASC2 overexpression (Figs. 6E-G, 7A and B). In addition, miR-24 overexpression resulted in partial restoration of ITGB4, p-FAK, epithelial-mesenchymal transition (EMT) and cell adhesion marker protein levels (Fig. 7C). Collectively, these observations suggested that CASC2 exerted its inhibitory effects on tumorigenesis partially via the miR-24/MUC6 signaling pathway.
Figure 6.
miR-24 mediates the suppressive effects of CASC2 on proliferation and apoptosis of pancreatic cancer cells. Expression levels of (A) CASC2, (B) miR-24 and (C) MUC6, and (D) protein expression levels of MUC6 in AsPC-1 cells after indicated transfection. (E) Proliferation, as determined by MTT assay, (F) colony formation and (G) apoptosis, as measured by flow cytometry of AsPC-1 cells transfected with indicated targets. *P<0.05 and **P<0.01. CASC2, cancer susceptibility candidate 2; miR, microRNA; MUC6, mucin 6; OD, optical density.
Figure 7.
miR-24 mediates suppressive effects of CASC2 on migration and invasion of pancreatic cancer cells. (A) Migration, as determined by wound healing assay (×40 magnification), (B) invasion, as determined by Transwell assay (×100 magnification), and (C) western blot analysis of indicated proteins in AsPC-1 cells transfected with the indicated targets. *P<0.05 and **P<0.01. cad, cadherin; CASC2, cancer susceptibility candidate 2; FAK, focal adhesion kinase; ITGB4, Integrin β4; miR, microRNA; MMP, matrix metalloproteinase; p-, phosphorylated.
CASC2 inhibits and miR-24 promotes tumorigenesis of pancreatic cancer cells in vivo
To further explore whether CASC2 and miR-24 affected tumorigenesis in vivo, AsPC-1 cells were transduced with LV-CASC2, LV-miR-24 or LV-NC vectors (LV-CASC2-NC and LV-miR-24 NC), and these cells were injected subcutaneously into nude mice. Upon transduction, CASC2 expression was increased by ~5-fold and miR-24 expression by ~3.5-fold compared with the respective NC (Fig. 8A and B). Notably, the size and weight of tumors in the CASC2-overexpressing group were significantly smaller than those in the LV-NC group (Fig. 8C-E). However, miR-24 overexpression enhanced the size and weight of tumors compared with the miR-NC injected animals (Fig. 8C-E). This study further confirmed the upregulation of CASC2 and MUC6, and downregulation of miR-24, in the tumors tissues of CASC2-overexpressing animals compared with the respective control. In addition, overexpression of miR-24 suppressed mRNA and protein expression levels of MUC6 (Fig. 8F-H). These results indicated that CASC2 inhibited and miR-24 promoted tumor growth in vivo.
Figure 8.
CASC2 inhibits and miR-24 promotes tumorigenesis of pancreatic cancer in vivo. (A) Relative expression of CASC2 in AsPC-1 cells stably expressing LV-CASC2 or LV-CASC2-NC, as assessed by RT-qPCR. (B) Relative expression of miR-24 in AsPC-1 cells stably expressing LV-miR-24 or LV-miR-24-NC, as assessed by RT-qPCR. (C) Images, (D) volume and (E) weights of tumors from mice injected with LV-CASC2 and LV-miR-24 cells (n=5/group). Expression levels of (F) CASC2 and miR-24, and (G) MUC6 in tumors. (H) Protein expression levels of MUC6 in tumors. *P<0.05 and **P<0.01. CASC2, cancer susceptibility candidate 2; LV, lentivirus; miR, microRNA; MUC6, mucin 6; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR.
Discussion
This study identified CASC2 as a suppressor of cell growth and progression in pancreatic cancer. These findings resulted in the drawing of several important conclusions. Firstly, CASC2 and MUC6 were downregulated, and miR-24 was upregulated in pancreatic cancer tissues and cell lines. Secondly, CASC2 overexpression or miR-24 knockdown suppressed pancreatic cancer cell growth and progression partially by altering cell-cell adhesion. Finally, CASC2 acted as a ceRNA for miR-24 and upregulated its downstream target MUC6, in order to suppress pancreatic cancer growth and progression in vitro and in vivo. Therefore, this study suggested a novel mechanism for the progression of pancreatic cancer modulated by CASC2, and proposed the clinical implication of CASC2 as a potential biomarker or therapeutic target in pancreatic cancer.
Aggressiveness and recurrence of pancreatic cancer are closely associated with cancer cell migration and invasion (3), and increasing numbers of lncRNAs have been implicated in the regulation of these processes in pancreatic cancer (27-29). In this study, CASC2 was downregulated in pancreatic cancer tissues and cell lines, and downregulated proliferation, migration and invasion, and promoted the apoptotic abilities of pancreatic cancer cells. Furthermore, CASC2 altered cell-cell adhesion, as evidenced by the decrease in the levels of ITGB4 and p-FAK, together with attenuation of N-cadherin and MMP expression, enhancement of E-cadherin expression, and morphological alterations. These findings were consistent with previous reports in which CASC2 functioned as a tumor suppressor in numerous types of human cancer, including colorectal cancer, hepatocellular cancer, osteosarcoma and pancreatic cancer (7-11). To the best of our knowledge, this study was the first to indicate that CASC2 exerted its tumor-suppressive effects through altering cell-cell adhesion in pancreatic cancer.
lncRNAs predominantly serve the role of miRNA sponges that reduce the availability of the target miRNA, which in turn prevents miRNAs from binding and negatively regulating downstream target genes (30). Available evidence suggested that CASC2 acts as a tumor suppressor gene via interactions with several networks, including miRNAs and other elements (7-10). miR-24 has been recognized as a tumor-associated miRNA that regulates cancer-associated processes, including adhesion, migration, invasion and metastasis in colorectal, pancreatic and lung cancer (31-33). In this study, miR-24 expression levels were increased and negatively associated with CASC2 levels in pancreatic cancer tissues and cell lines. The results from loss- and gain-of-function experiments confirmed that miR-24 promoted migration and invasion, and regulated the ITGB4/FAK pathway and EMT progression of pancreatic cancer cells. Furthermore, bioinformatics analysis and luciferase reporter assay identified CASC2 sponged miR-24 in pancreatic cancer cells. A previous study reported that miR-24 functions as a tumor-promoting target that leads to increased pancreatic cancer cell migration and invasion (32). The present results demonstrated that CASC2 exerted its tumor-suppressive effects on pancreatic cancer cells via interacting with miR-24. The rescue experiments demonstrated that overexpression of miR-24 partially reversed the inhibitory effects of CASC2 on tumor cell growth and progression. Other reports have revealed that CASC2 serves as a sponge of miR-24 to suppress tumorigenesis of hepatocellular carcinoma (8,14). To the best of our knowledge, this study is the first to elaborate on the interaction between CASC2 and miR-24 in pancreatic cancer.
Bioinformatics analysis was used to identify potential downstream targets of miR-24 and identified MUC6. To the best of our knowledge, this is the first study that explored the associations between MUC6 and miR-24 in pancreatic cancer. Several other MUCs, such as MUC1 and MUC4, have been confirmed as targets of miR-200c/141 and miR-219-1 in pancreatic cancer (34,35). Previous reports also identified decreases in MUC6 expression during pancreatic cancer progression (17,36). Furthermore, a previous study suggested that MUC6 inhibits the invasion of pancreatic cancer cells by affecting basement membrane organization (19). MUC5AC, a similar secretory MUC, regulates the ITGB4/FAK signaling pathway in lung cancer (21). In line with these studies, the present findings demonstrated that miR-24 regulated the tumorigenesis of pancreatic cancer cells via targeting MUC6 and modulating the ITGB4/FAK signaling pathway. In addition, CASC2 positively regulated the expression of MUC6 via targeting miR-24 in pancreatic cancer cells, and the tumor-suppressive role of CASC2 was mediated via miR-24. Our findings demonstrated that overexpression of CASC2 inhibited miR-24, which in turn, activated MUC6 to regulate cell-cell adhesion and suppress pancreatic cancer growth and progression.
In conclusion, the present study demonstrated that CASC2 was downregulated in pancreatic cancer tissues and cell lines, and inhibited cell growth and progression. To the best of our knowledge, this study is the first to show that CASC2 suppressed progression of pancreatic cancer cells potentially via the miR-24/MUC6 axis. Given that low CASC2 expression is closely correlated with clinical progression and unfavorable prognosis in patients with pancreatic cancer (37), future studies on the diagnostic power of CASC2, including specificity and sensitivity, may facilitate investigations into CASC2 functions and its use as a potential biomarker to monitor or screen high-risk patients with pancreatic cancer. These findings further provided insight into the molecular mechanisms associated with the tumorigenesis of pancreatic cancer and supplied potential therapeutic targets for pancreatic cancer treatment.
Acknowledgments
Not applicable.
Funding
This project was supported by a grant from National Natural Science Foundation Of China (grant no. 81572408) and Natural Science Foundation of Jiangsu Province (grant no. BE2015712).
Availability of data and materials
The datasets used and/or analysed during the present study are available from the corresponding author on reasonable request.
Authors’ contributions
DFX, LSW and JHZ conceived the study. DFX and LSW collected and analyzed the data. DFX and LSW performed the experiments. DFX and JHZ provided the resources and supervised the study. DFX and LSW wrote the original draft. JHZ reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The assessment of patient-derived samples in this study was approved by the Ethical Committee of Zhongda Hospital Affiliated with Southeast University and written informed consent was obtained from all patients. All of the animal studies were conducted in accordance with the Institutional Animal Care and Use Committee and were approved by the Medical Ethics Committee of Southeast University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, Arslan AA, Bueno-de-Mesquita HB, Gross M, Helzlsouer K, Jacobs EJ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140–1151. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Huang G, Wu X, Li S, Xu X, Zhu H, Chen X. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Sci Rep. 2016;6:26524. doi: 10.1038/srep26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan JC, Zeng F, Le YG, Xin L. LncRNA CASC2 inhibited the viability and induced the apoptosis of hepatocellular carcinoma cells through regulating miR-24-3p. J Cell Biochem. 2018;119:6391–6397. doi: 10.1002/jcb.26479. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang C, Dou C, Xu M, Liu Q, Tu K. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer. 2017;16:123. doi: 10.1186/s12943-017-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ba Z, Gu L, Hao S, Wang X, Cheng Z, Nie G. Downregulation of lncRNA CASC2 facilitates osteosarcoma growth and invasion through miR-181a. Cell Prolif. 2018;51:e12409. doi: 10.1111/cpr.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu Y, Liang S, Zhou Y, Li S, Li Y, Liao W. HNF1A/CASC2 regulates pancreatic cancer cell proliferation through PTEN/Akt signaling. J Cell Biochem. 2019;120:2816–2827. doi: 10.1002/jcb.26395. [DOI] [PubMed] [Google Scholar]
- 12.Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, Hu Z, Zhuang R, Ning G, Zhang C, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58:610–618. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- 13.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin X, Cai L, Wang C, Deng X, Yi S, Lei Z, Xiao Q, Xu H, Luo H, Sun J. CASC2/miR-24/miR-221 modulates the TRAIL resistance of hepatocellular carcinoma cell through caspase-8/caspase-3. Cell Death Dis. 2018;9:318. doi: 10.1038/s41419-018-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ringel J, Löhr M. The MUC gene family: Their role in diagnosis and early detection of pancreatic cancer. Mol Cancer. 2003;2:9. doi: 10.1186/1476-4598-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suh H, Pillai K, Morris DL. Mucins in pancreatic cancer: Biological role, implications in carcinogenesis and applications in diagnosis and therapy. Am J Cancer Res. 2017;7:1372–1383. [PMC free article] [PubMed] [Google Scholar]
- 17.Sierzega M, Mlynarski D, Tomaszewska R, Kulig J. Semiquantitative immunohistochemistry for mucin (MUC1, MUC2, MUC3, MUC4, MUC5AC, and MUC6) profiling of pancreatic ductal cell adenocarcinoma improves diagnostic and prognostic performance. Histopathology. 2016;69:582–591. doi: 10.1111/his.12994. [DOI] [PubMed] [Google Scholar]
- 18.Jonckheere N, Skrypek N, Van Seuningen I. Mucins and pancreatic cancer. Cancers (Basel) 2010;2:1794–1812. doi: 10.3390/cancers2041794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leir SH, Harris A. MUC6 mucin expression inhibits tumor cell invasion. Exp Cell Res. 2011;317:2408–2419. doi: 10.1016/j.yexcr.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 20.Baril P, Gangeswaran R, Mahon PC, Caulee K, Kocher HM, Harada T, Zhu M, Kalthoff H, Crnogorac-Jurcevic T, Lemoine NR. Periostin promotes invasiveness and resistance of pancreatic cancer cells to hypoxia-induced cell death: Role of the beta4 integrin and the PI3k pathway. Oncogene. 2007;26:2082–2094. doi: 10.1038/sj.onc.1210009. [DOI] [PubMed] [Google Scholar]
- 21.Lakshmanan I, Rachagani S, Hauke R, Krishn SR, Paknikar S, Seshacharyulu P, Karmakar S, Nimmakayala RK, Kaushik G, Johansson SL, et al. MUC5AC interactions with integrin β4 enhances the migration of lung cancer cells through FAK signaling. Oncogene. 2016;35:4112–4121. doi: 10.1038/onc.2015.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou M, Huang W, Jiang W, Wu Y, Chen Q. Role of Cav-1 in HIV-1 Tat-induced dysfunction of tight junctions and Aβ-transferring proteins. Oxid Med Cell Longev. 2019;2019:3403206. doi: 10.1155/2019/3403206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Sieg DJ, Hauck CR, Ilic D, Klingbeil CK, Schaefer E, Damsky CH, Schlaepfer DD. FAK integrates growth-factor and integrin signals to promote cell migration. Nat Cell Biol. 2000;2:249–256. doi: 10.1038/35010517. [DOI] [PubMed] [Google Scholar]
- 25.Yano H, Mazaki Y, Kurokawa K, Hanks SK, Matsuda M, Sabe H. Roles played by a subset of integrin signaling molecules in cadherin-based cell-cell adhesion. J Cell Biol. 2004;166:283–295. doi: 10.1083/jcb.200312013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Liu P, Yang H, Zhang J, Peng X, Lu Z, Tong W, Chen J. The lncRNA MALAT1 acts as a competing endogenous RNA to regulate KRAS expression by sponging miR-217 in pancreatic ductal adenocarcinoma. Sci Rep. 2017;7:5186. doi: 10.1038/s41598-017-05274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng SJ, Chen HY, Ye Z, Deng SC, Zhu S, Zeng Z, He C, Liu ML, Huang K, Zhong JX, et al. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene. 2018;37:5811–5828. doi: 10.1038/s41388-018-0382-1. [DOI] [PubMed] [Google Scholar]
- 29.Giulietti M, Righetti A, Principato G, Piva F. LncRNA co-expression network analysis reveals novel biomarkers for pancreatic cancer. Carcinogenesis. 2018;39:1016–1025. doi: 10.1093/carcin/bgy069. [DOI] [PubMed] [Google Scholar]
- 30.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerimis D, Kontos CK, Christodoulou S, Papadopoulos IN, Scorilas A. Elevated expression of miR-24-3p is a potentially adverse prognostic factor in colorectal adenocarcinoma. Clin Biochem. 2017;50:285–292. doi: 10.1016/j.clinbiochem.2016.11.034. [DOI] [PubMed] [Google Scholar]
- 32.Listing H, Mardin WA, Wohlfromm S, Mees ST, Haier J. miR-23a/-24-induced gene silencing results in mesothelial cell integration of pancreatic cancer. Br J Cancer. 2015;112:131–139. doi: 10.1038/bjc.2014.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jing P, Zhao N, Xie N, Ye M, Zhang Y, Zhang Z, Li M, Lai X, Zhang J, Gu Z. miR-24-3p/FGFR3 signaling as a novel axis is involved in epithelial-mesenchymal transition and regulates lung adenocarcinoma progression. J Immunol Res. 2018;2018:2834109. doi: 10.1155/2018/2834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohr AM, Bailey JM, Lewallen ME, Liu X, Radhakrishnan P, Yu F, Tapprich W, Hollingsworth MA. MUC1 regulates expression of multiple microRNAs involved in pancreatic tumor progression, including the miR-200c/141 cluster. PLoS One. 2013;8:e73306. doi: 10.1371/journal.pone.0073306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lahdaoui F, Delpu Y, Vincent A, Renaud F, Messager M, Duchêne B, Leteurtre E, Mariette C, Torrisani J, Jonckheere N, Van Seuningen I. miR-219-1-3p is a negative regulator of the mucin MUC4 expression and is a tumor suppressor in pancreatic cancer. Oncogene. 2015;34:780–788. doi: 10.1038/onc.2014.11. [DOI] [PubMed] [Google Scholar]
- 36.Kim GE, Bae HI, Park HU, Kuan SF, Crawley SC, Ho JJ, Kim YS. Aberrant expression of MUC5AC and MUC6 gastric mucins and sialyl Tn antigen in intraepithelial neoplasms of the pancreas. Gastroenterology. 2002;123:1052–1060. doi: 10.1053/gast.2002.36018. [DOI] [PubMed] [Google Scholar]
- 37.Yu L, Chen S, Bao H, Zhang W, Liao M, Liang Q, Cheng X. The role of lncRNA CASC2 on prognosis of malignant tumors: A meta-analysis and bioinformatics. Onco Targets Ther. 2018;11:4355–4365. doi: 10.2147/OTT.S166132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the present study are available from the corresponding author on reasonable request.