Abstract
Over the years, 3D printing technologies have transformed the field of tissue engineering and regenerative medicine by providing a tool that enables unprecedented flexibility, speed, control, and precision over conventional manufacturing methods. As a result, there has been a growing body of research focused on the development of complex biomimetic tissues and organs produced via 3D printing to serve in various applications ranging from models for drug development to translational research and biological studies. With the eventual goal to produce functional tissues, an important feature in 3D printing is the ability to tune and modulate the microenvironment to better mimic in vivo conditions to improve tissue maturation and performance. This paper reviews various strategies and techniques employed in 3D printing from the perspective of achieving control over physical, chemical, and biological properties to provide a conducive microenvironment for the development of physiologically relevant tissues. We will also highlight the current limitations associated with attaining each of these properties in addition to introducing challenges that need to be addressed for advancing future 3D printing approaches.
I. INTRODUCTION
In tissue engineering, a combination of biomaterials, cells, and bioactive factors is often used to provide the necessary biophysical and biochemical cues to form a functional tissue construct. Namely, the ability to recapitulate the native cellular microenvironment is crucial as we strive to develop more physiologically relevant tissue models for the study of biological mechanisms, drug screening, and regenerative medicine. While traditional 2D planar culture systems have been commonly used, there is now growing evidence that these rigid monolayer platforms do not accurately reflect the in vivo cellular morphology, behavior, gene expression profiles, and differentiation potentials.1–3 This stems from the fact that cells naturally exist in a 3D microenvironment with contact in all directions to their surrounding matrix and neighboring cell populations. Thus, mimicking this 3D dynamic reciprocity between the cells and their local environment as well as replicating the tissue heterogeneity and complex native microarchitecture is an important strategy to more closely mimic their native function.1,4
To this end, several approaches have been developed to create 3D culture platforms following the main design principles: (1) porous microarchitecture to enable diffusion of nutrients and cellular ingrowth, (2) biodegradable biomaterials with non-cytotoxic degradation products, (3) recapitulation of the tissue-specific shape, microarchitecture, and mechanical properties, (4) biocompatible without eliciting host immune response, and (5) anatomically relevant cell distribution, density, and heterogeneity as found in vivo.5 Most current techniques employ a top down approach where cells are seeded directly onto a prefabricated scaffold and allowed to infiltrate and populate. Namely, porous 3D scaffolds of this nature have been commonly prepared using electrospinning, gas foaming, particle or porogen leaching, freeze-drying, and micro molding.6 Although promising these methods are often limited to producing simple homogeneous cell-laden scaffolds with difficult control over local cell and extracellular matrix heterogeneity as well as the formation of complex biomimetic internal microstructures such as vascular networks.
To circumvent these challenges, a bottom up approach has emerged by way of assembling tissue building blocks to form spatially organized structures. Based on these concepts, recent advances have led to the revolutionary “bioprinting” discipline in tissue engineering. By combining scaffold fabrication strategies and computer-aided technology, functional 3D tissue and organ substitutes can be produced via 3D printing. This technology first surfaced in 2000 and the general process involves translating a template created from computerized tomography (CT), magnetic resonance imaging (MRI), or computer aided design (CAD) to an automated printing apparatus to reconstruct the structure in a layer-by-layer fashion.7,8 Over the years, significant progress has been made in developing advanced printing modalities capable of coordinating the precise placement of multiple cell types, control of multimaterial properties, and assembly of intricate microarchitectures to produce a physiologically relevant tissue construct. Given the flexibility of 3D printing, it is expected that this technology will serve as an invaluable tool in many areas of application including biology, regenerative medicine, in vitro pharmaceutical drug screening, and tissue or organ-on-a-chip models for studying dynamic culture systems.9–11
Several reviews have discussed details on the different printing processes as well as biomaterial designs for printable hydrogels, in which the reader is directed to comprehensive reviews by Murphy et al., Lee et al., and Jungst et al.11–13 Here, we will focus on the strategies used by various 3D printing platforms to create biomimetic tissue constructs with an emphasis on the control of the structural and compositional parameters. More specifically, methods to alter the physical microarchitecture and mechanical properties will be discussed followed by a review of different techniques to modulate the chemical and biological properties. This paper will highlight the versatility of current 3D printing platforms in terms of their advancements and limitations in fabricating functional tissue constructs and serve as a guide for advancing future designs and approaches in 3D printing.
II. OVERVIEW OF 3D PRINTING SYSTEMS
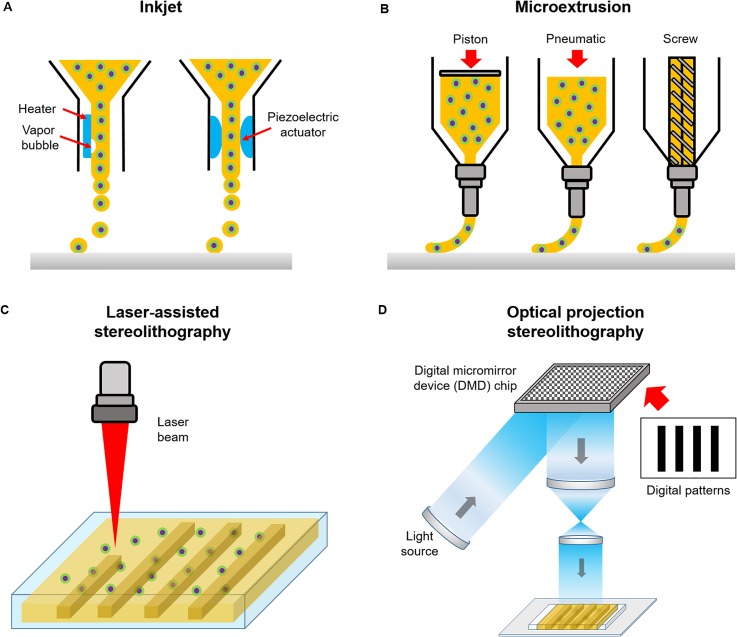
Currently, the most commonly used 3D printing system is the nozzle-based technique which works by depositing the bioink in a raster-like fashion to create a 3D structure layer-by-layer. More specifically, early inkjet printers alternatively known as drop-on-demand printers were originally built from modified commercial ink-based paper printers to dispense cells and biomaterials simultaneously [Fig. 1(a)].7 In this case, liquid droplets are ejected through a nozzle via thermal or acoustic forces onto a receiving substrate set on a movable stage.7,8 Inkjet printing is relatively low cost and its simplistic design has been utilized by many labs to fabricate 3D cellular constructs with spatial patterning control.14–16 However, drawbacks associated with this system is the poor cell viability due to mechanical or thermal stress, difficulties in creating thick constructs with physiologically relevant cell densities, droplet nonuniformity, and limited range of compatible low viscosity biomaterials to minimize clogging.11,17
FIG. 1.
Schematic of different bioprinting systems: (a) inkjet, (b) microextrusion, (c) laser-assisted stereolithography, and (d) optical projection stereolithography.
Microextrusion printing overcomes some of these issues by using mechanical or pneumatic pressure to continuously extrude liquid bioink from the nozzle tip which then solidifies immediately upon discharge for each layer [Fig. 1(b)]. Under robotic control, the nozzle can move seamlessly with submillimeter resolution, moderate speed, and quickly interchange between different materials through the use of multiple print heads. Microextrusion can accommodate a wider range of biomaterial viscosities, provide better spatial resolution, and deposit higher cell densities compared to inkjet printers.11,18 Despite these advantages, enhancing print resolution with larger nozzle gauges can lead to decreased cell viability and integrity due to higher shear stress.18
Similarly, laser-assisted stereolithography utilizes light to solidify a photopolymerizable bioink within a reservoir in an iterative contactless manner via direct laser writing [Fig. 1(c)]. With this technique, the microfabrication of intricate overhanging structures with nanoscale spatial resolutions can be achieved by two-photon polymerization using ultrafast light pulses emitted from a femtosecond laser. Due to Gaussian distribution of the laser intensity, the central region of the laser beam is highest in energy, thus a resolution below the diffraction limit can be achieved to form high quality nanostructures less than 100 nm in size.19,20
More recently, rapid optical based 3D printing platforms [Fig. 1(d)] provide substantial improvements in increasing fabrication speed as well as spatial resolution by achieving smooth features not capable with conventional nozzle-based strategies. These high-speed printers adopt a layerless approach via dynamic optical projection stereolithography (DOPsL) or continuous liquid interphase production (CLIP) based printing method. In this case, the optical projection and stage continuously move in unison as the structure is built, which in turn eliminates imperfections between adjacent layers present in iterative layer-by-layer printed structures. In DOPsL, the structure is quickly formed by illuminating multiple patterns representing slices of the object to build the final form within a photopolymerizable reservoir.21 With the use of a digital micromirror device (DMD) chip, which contains approximately one to four million micromirrors that can quickly modulate the projected light, various patterns of a single plane can be simultaneously photocrosslinked with microscale resolution.21 Similarly, the formation of structures in CLIP based printing is achieved by photopolymerizing the material at a projection window located in the bottom of a liquid prepolymer bath and drawn upwards as the construct is being solidified. A gas permeable membrane at the fabrication window ensures that the structure does not adhere to the bottom to maintain streamline construction. In this orientation, overhanging structures as well as centimeter sized constructs with resolutions of less than 100 μm can be achieved.22 Fabrication with optical 3D printers is typically performed on the order of seconds to minutes compared to hours with traditional nozzle based printers.19,21,22 Furthermore, optical based printers are now used in tissue engineering applications to encapsulate cells for the formation of intricately patterned cell-laden biomimetic tissue constructs.21,23–25
III. CONTROL OF PHYSICAL PROPERTIES
Cells maintain physical contact with their surrounding extracellular matrix (ECM) microenvironment via integrin binding mechanisms, which play a significant role in mediating cellular activities such as migration, proliferation, and differentiation.26–29 To date, several studies have demonstrated this phenomenon using rationally engineered environments possessing desired physical properties, including topographical features and mechanical stiffness, to control cell behavior as well as facilitate tissue maturation and functionality.26–37 In this section, we will review the promising applications of 3D printing technology to create biological scaffolds and biomimetic tissues with unprecedented flexibility by tuning the physical properties of the cellular microenvironment. We will highlight the current progress in bioprinting physiologically relevant microarchitectures to produce functional microphysiological systems as well as techniques used to control mechanical properties and the emergence of auxetic tissue engineering scaffolds.
A. 3D printing physiologically relevant microarchitectures
The human body contains approximately 80 organs and each organ is a complex 3D system composed of many tissues featuring intrinsically different structures to perform a specific function. For instance, the myocardium of the heart, which is the main layer providing contractile strength during systole, consists of highly aligned cardiomyocytes in parallel following the topographic cues within the microarchitecture of the heart ECM.38,39 Such intricate alignment in the cellular structure across multiple length scales, ranging from a macroscopic level to a micro/nano-scale, is the key to myocardium contraction.39,40 A growing body of studies have also shown that tissue function is closely related to their native microarchitectures and deterministically designed topographic cues have proven to regulate the structure and function of engineered tissue constructs.39–43 For example, Kim et al. engineered a myocardium model with nanoscale topographic cues mimicking in vivo ventricular organization and demonstrated that the nanoscale features of the ECM surrounding the cardiomyocytes can stipulate cell and tissue behaviors including cell adhesion, spreading, alignment, cell-cell coupling, and tissue contraction.39 In the field of neural tissue engineering, there have also been studies that showed improved neuronal behavior and axonal growth when cells were cultured on substrates possessing geometrical cues to help guide nerve development and regeneration.44–46 The traditional methods to engineer such topographic cues at the microscale or nanoscale include electron beam lithography, photolithography, molding, electrospinning, laser direct writing, and micro-stamping.39,44–49 However, with the advantage of unprecedented flexibility to achieve complex 3D designs, 3D printing technologies have recently emerged as transformational tools to create biological tissues and scaffolds with rationally designed microarchitectures and topographic cues for guiding cell behavior and tissue growth.
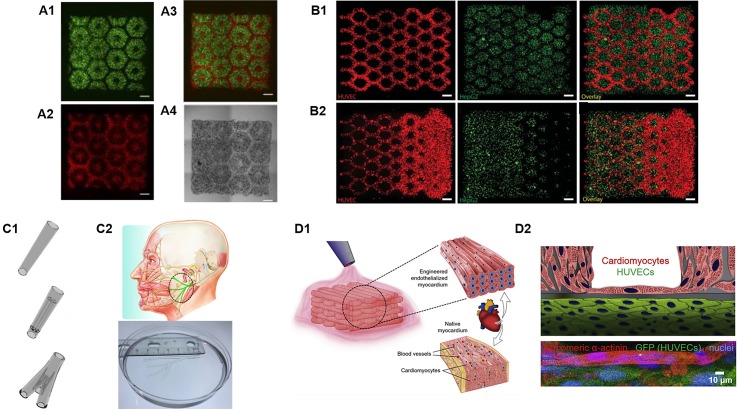
Liver plays a vital role in nearly all metabolic processes in the human body including drug metabolism and detoxification. The liver lobule features highly organized hexagonal units with closely packed plates of hepatocytes and sinusoids irradiating from the central vein. This unique structural organization provides the foundation for optimal exchange of nutrients and oxygen between the highly metabolic hepatocytes and the blood flow.23 Recently, inspired by the intricate microarchitectures of the native liver lobules, Ma et al. bioprinted a sophisticated human hepatic tissue model with liver-mimetic structures and cellular compositions [Fig. 2(A)].23 In this study, a DLP-based bioprinting platform was utilized to encapsulate iPSC-derived hepatocytes and other supportive cell types within photosensitive bioinks. The bioprinted liver tissue resembled hexagonal units of the liver lobule along with patterned supportive cells to mimic the structural organization of the sinusoids. As a result, this biomimetic liver model exhibited improved morphological organization, higher liver-specific gene expression, and increased liver metabolic functions in comparison to 2D monolayer culture of hepatocytes.23 Remarkably, this is one of the pioneering works that demonstrated the feasibility of using DLP-based 3D printers to bioprint functional tissues with multiple cell types and materials featuring complex biomimetic microarchitectures. Following this work, Zhu et al. also employed a DLP-based platform for the direct bioprinting of prevascularized tissues with gradient microvessel structures [Fig. 2(B)].24 Notably, the endothelial cells bioprinted in these tissues followed the guidance of the bioprinted microstructural features and formed functional endothelial networks that anastomosed with the host circulation after in vivo implantation.24
FIG. 2.
3D bioprinting of complex biomimetic microarchitectures. (A1) Fluorescent images of DLP-based bioprinted cellularized liver tissue constructs with human iPSC-hepatic progenitor cells (green) and (A2) supporting vascular cells (red) complementary including (A3) merged and (A4) bright field images. (Reproduced with permission from X. Ma et al., Proc. Natl. Acad. Sci. U. S. A. 113, 2206 (2016). Copyright 2016 National Academy of Sciences). (B1) DLP-based bioprinted cellularized complex prevascularized tissue constructs with uniform channel widths and (B2) gradient channel widths, scale bar = 250 μm. (Reproduced with permission from W. Zhu et al., Biomaterials 124, 106 (2017). Copyright 2017 Elsevier, Inc.). (C1) Different CAD designs and their corresponding DLP-based printed peripheral nerve conduits, scale bar = 1 mm. (C2) Life-size 3D printed facial nerve. (Reproduced with permission from W. Zhu et al., Mater. Today 21, 951–959 (2018). Copyright 2018 Elsevier, Inc.) (D1) Schematic of 3D bioprinting process to produce an endothelialized myocardium. (D2) Schematic and corresponding confocal images of the cellular organization 15 days post endothelialization of the microfibrous myocardial tissue. (Reproduced with permission from Y. S. Zhang et al., Biomaterials 110, 45 (2016). Copyright 2016 Elsevier, Inc.).
One of the noteworthy advantages of DLP-based 3D printing technology over other 3D printing modalities is its capability to print continuously with smooth topographic features as demonstrated by both the DOPsL and CLIP technologies, which could significantly improve the structural integrity of the printed scaffolds. Structural integrity is highly desired for grafts that are implanted to the lesion sites that are subject to frequent movements and impacts such as in the case for sciatic nerve repair.50,51 Most recently, a DLP-based rapid continuous 3D printing technology was utilized to print customizable peripheral nerve guidance conduits (NGCs) with a variety of designs for sciatic nerve repair [Fig. 2(C)].51 As a proof of concept, a life-size NGC was printed based on the human facial nerve anatomy within 10 min. In addition, the 3D printed NGCs featuring microchannels were grafted to repair complete sciatic nerve transection in mice. The results showed that the regenerating nerves followed the physical guidance of the 3D printed microchannels over 6 mm with promising functional recovery achieved.51
In addition to liver and nerve, the heart is another organ that features highly organized structures that have a significant role in cardiac function. In particular, the myocardium of the heart consists of highly aligned cardiomyocytes that are bundled together to enable synchronous contraction.39,52 In recent work, Zhang et al. utilized an extrusion-based 3D bioprinter to fabricate endothelialized myocardium with precisely controlled anisotropic architectures of microfibers [Fig. 2(D)].52 Specifically, a microfibrous scaffold was first printed with encapsulated endothelial cells which subsequently reorganized into the physical contours of the microfibers and formed a vascular bed.52 Cardiomyocytes were then seeded into the interstitial space of the endothelialized construct thus forming a vascularized myocardium that structurally resembles native myocardial tissue. It was further demonstrated that human iPSC derived cardiomyocytes could be printed in this tissue model to generate human heart-on-a-chip for potential drug testing and disease modeling.
B. Tuning of mechanical stiffness
In addition to the physical cues provided by topographical features, cells are also highly sensitive to the mechanical stiffness of their surrounding ECM environment, which has been demonstrated to play a crucial role in dictating cell fate.30,31,37 For instance, tissue-level matrix stiffness has been proven to significantly impact the differentiation of the mesenchymal stem cells (MSCs) and can be used to drive their commit towards a specific lineage without exogenous induction factors.31 In particular, MSCs cultured on soft matrices with mechanical stiffness similar to brain tissue were found to differentiate towards neurons, stiff matrices with mechanical stiffness close to that of muscle tissue drove differentiation toward myoblasts, and finally more rigid matrices mimicking that of bone induced lineage commitment towards osteoblasts.31 This fundamental work demonstrated the possibility to guide stem cell behavior by tuning the mechanical stiffness of scaffold substrates. Previously, other fabrication technologies such as photolithography and molding have been utilized to create biomaterial matrices with controlled mechanical stiffness to regulate cell behaviors. However, these traditional fabrication technologies involve multiple processing steps and require cleanroom facilities which can be costly, laborious, and not easily accessible for biological researchers.30,37 3D printing technologies are not only known to have the flexibility to create multi-scale arbitrary structures as introduced in the last section but are also capable of providing tissue matrices with excellent control over mechanical stiffness.
In one example, an extrusion-based 3D printer was used to fabricate anatomically accurate and mechanically heterogeneous aortic valves.53 The aortic valves featured complex 3D architecture and mechanical heterogeneity that are crucial for its dynamic physiological function. For instance, the valve leaflets are relatively soft and compliant with a modulus of ∼54 kPa, while the aortic root is significantly more rigid with a modulus of approximately 140 kPa to 180 kPa.53 To build the anatomically accurate aortic valve with biomimetic mechanical heterogeneity, a multi-nozzle 3D printer was used in combination with the photocrosslinkable hydrogel poly(ethylene glycol) diacrylate (PEGDA). By combining PEGDA with different molecular weights (i.e., 700 and 8000 kDa) at different concentrations, hydrogels with a wide range of mechanical stiffness were achieved and the resulting aortic valves retained mechanical heterogeneity post printing.53 In addition, porcine aortic valve interstitial cells were seeded on these 3D printed hydrogel aortic valves and maintained high viability over 3 weeks.53 The key instrument in this study is the multi-nozzle 3D printer that enables the simultaneous printing with multiple materials thus leading to a final 3D construct composed of heterogeneous mechanical properties. In another example, a multi-nozzle 3D printer was used to demonstrate the hybrid bioprinting of a stiffer material for structural support and another softer hydrogel material for direct cell encapsulation, which yielded tissue constructs with tailorable mechanical properties.53
DLP-based 3D printers employ photopolymerization as the principal mechanism for 3D fabrication, which in turn allows for the tuning of the mechanical stiffness by altering the prepolymer compositions and light exposure parameters. Orthogonal tuning of the mechanical stiffness and the microarchitecture has been demonstrated with a DLP-based 3D printer to decouple the effects of the mechanical stiffness and the microstructures on cancer cell migration.54 In this work, the authors compared cancer cell migration behaviors on 2D slabs and 3D log pile structures composed of polyethylene glycol (PEG) to study the influence of microstructures. Furthermore, a high-concentration of prepolymer and a low-concentration of prepolymer was used to fabricate the 2D and 3D structures to investigate the influence of stiffness.54 Interestingly, the findings indicated substantial difference in the cell response to the stiffness change in 2D versus 3D. Specifically, no significant difference in cell migration behavior including cell displacement, velocity, and path straightness parameters were observed between the 2D stiff and 2D soft substrate. However, substantial differences were found between the 3D stiff and soft structures characterized.54 These findings not only indicate that cells respond to the environmental changes differently in 2D versus 3D but also highlight the significance of using 3D printing technologies to decouple and individually study the cellular influence of geometrical cues and mechanical stiffness. Apart from changing the material concentration to control stiffness, the exposure time can also be used to control the degree of crosslinking to vary the regional stiffness in the printed constructs. This was demonstrated by Pyo et al. wherein gradient stiffness within a construct was achieved by printing green chemistry-derived isocyanate-free aliphatic polyurethane materials under different exposure times via the programmed projection of striped patterns using a continuous optical printer [Fig. 3(A)].55
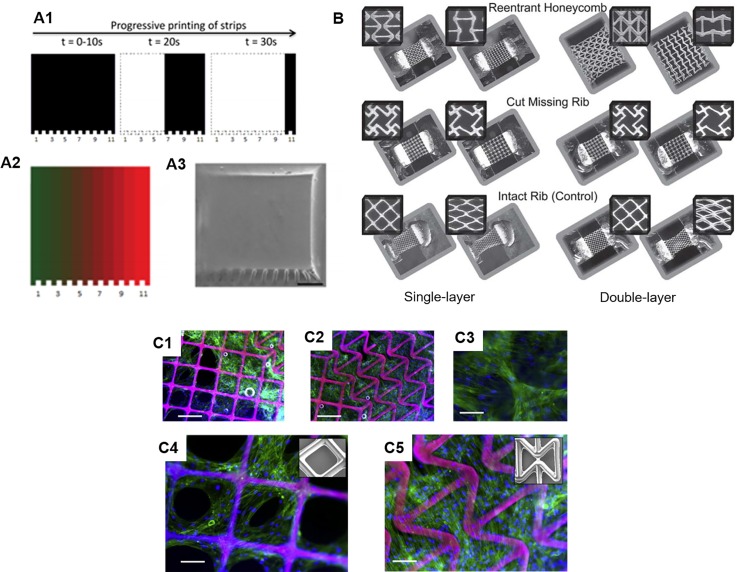
FIG. 3.
Tuning of mechanical properties via 3D printing. (A1) Schematic of programmed exposure time for production gradient stiffness. (A2) Designed gradient pattern and (A3) SEM image of the corresponding printed construct, scale bar = 500 μm. (Reproduced with permission from S. H. Pyo et al., ACS Appl. Mater. Interfaces 9, 836 (2017). Copyright 2016 American Chemical Society). 3D printed auxetic structures. (B) Optical images of various printed auxetic patterns as single-layer and double-layer PEG scaffolds showing undeformed and deformed states in response to applied axial strain. (Reproduced with permission from D. Y. Fozdar et al., Adv. Funct. Mater. 21, 2712 (2011). Copyright 2011 Wiley & Sons, Inc.). Human mesenchymal stem cells seeded on (C1 and C4) positive Poisson ratio and (C2, C3, and C5) negative Poisson ratio regions of printed PEG scaffolds. Scale bars = 250 μm (C1 and C2) and 125 μm (C3-C5). (Reproduced with permission from P. Soman et al., Acta Biomater. 8, 2587 (2012). Copyright 2012 Acta Materialia, Inc.).
C. Controlling Poisson's ratio
Poisson's ratio is another important mechanical property that has yet to be fully explored in the bioengineering field, primarily due to the limited manufacturing capabilities in controlling the Poisson's ratio of the biological constructs using traditional biofabrication techniques.56,57 In comparison to the Young's modulus, which measures a material's elastic behavior along the axial direction of the applied stress, Poisson's ratio defines the material's deformation (i.e., shrinkage or expansion) in the transverse direction that is perpendicular to the stress loading direction. In nature, a vast majority of materials possess a positive Poisson's ratio such that they experience a reduction in size in the transverse direction while being stretched in the axial direction.58 However, with the rational design of the internal pore structures, man-made scaffolds can have a negative or zero Poisson's ratio. Structures with negative Poisson's ratio, also known as “auxetic structures,” possess patterned internal structures which result in the structure becoming larger when stretched in the transverse direction while becoming thinner and condensed when compressed.58–60 As a result, the unique properties of auxetic structures can potentially offer new solutions in tissue engineering and regenerative medicine. For instance, an auxetic cardiac patch was recently introduced to treat myocardical infarction.61 The auxetic structure of the auxetic cardiac patch offers great mechanical tunability and more importantly allows it to conform better to movements during heart contraction in comparison to the control patches with positive Poisson's ratio.61 Combined with a biocompatible conductive material, this auxetic cardiac patch also allows for the electrical stimulation to interfere with the electroresponsive cardiac tissue thus offering a potential treatment for myocardical infarction.61
Tuning of the Poisson's ratio requires sophisticated alteration of the internal architecture of a scaffold. In recent years, advanced 3D printing platforms have emerged as a powerful tool for building artificial scaffolds with precise control over the Poisson's ratio for interesting biological applications.56–60,62 In one example, the fabrication of single- and multi-layer web structures with negative, zero, and positive Poisson's ratios were achieved by using DLP-based 3D printers with PEG-based biomaterials.58,59,62 Through computational simulation of the deformation mechanisms and careful design of the special geometry and arrangement of the unit pore structures, Fozdar et al. demonstrated the fabrication of auxetic web structures with tunable negative Poisson's ratio using a DLP-based 3D printer [Fig. 3(B)].58 Following this work, Soman et al. introduced the concept of hybridizing positive Poisson's ratio structure and negative Poisson's ratio structures together.59 Here, human mesenchymal stem cells (hMSCs) were able to attach and proliferate across these hybrid scaffolds which indicates the potential of using such scaffolds to build cellular patches for tissue regeneration. Interestingly, in the positive Poisson's ratio region, the hMSCs were found to gather mostly on the scaffold struts and form holes in the cellular sheet, while in the negative Poisson's ratio region, the cells were found to grow over the scaffold pores and form an intact cellular sheet [Fig. 3(C)].59 Furthermore, in a zero Poisson's ratio scaffold also fabricated by the same DLP-based 3D printing technology, hMSCs were shown to adhere and proliferate on it, which could be employed for engineering a variety of tissues that possess nearly zero Poisson's ratio such as the ligament, cartilage, and cornea.62
Although DLP-based 3D printers offer the scalable printing of structures with tunable Poisson's ratios, the unit sizes of the internal pores are usually on the order of hundreds of microns which is much larger than the individual cell size thus limiting the capability to study the impact of Poisson's ratio on individual cells.56 With resolutions at the submicron scale, laser-assisted stereolithography can provide a solution to the aforementioned challenge and provide submicroscale structures with tunable Poisson's ratios. For instance, Zhang et al. demonstrated the use of a two-photon laser direct writing system to fabricate suspended webs with positive and negative Poisson's ratios.56 The unit pore sizes of these suspending web structures were 10 μm × 10 μm which prevented single cells from passing through the pores. Interestingly, abnormal cell division behavior was observed on the web structures with a negative Poisson's ratio. In this case, some 10T1/2 cells cultured on the negative Poisson's ratio webs failed to undergo complete abscission and underwent apoptosis or became a multinucleated cell.56 These findings suggested that negative Poisson's ratio structures at the cellular level could induce unusual cell division without introducing any other biochemical manipulations to the cellular components.56 In addition to the planar web structures, 3D lattice structures with tunable Poisson's ratio having a unit size smaller than 10 μm × 10 μm × 10 μm was produced using laser direct writing, which could be potentially used to study more complex biological questions from a cell to tissue level scale.63
IV. MODULATION OF CHEMICAL COMPOSITION
A key advantage in 3D printing technology is the ability to deposit different biomaterials with high spatial control to produce complex structures with compositionally distinct regions. To date, a wide selection of 3D printable bioinks have been developed ranging from natural to synthetic to composite biomaterials which is reviewed in detail by Guvendiren et al.64 In general, the chemical properties of the selected bioinks used and their placement is critically important in dictating the functionality of the final printed construct. By engineering rationally designed chemical environments in 3D printed hydrogel structures, higher order functional properties can also be achieved such as electrical conduction, promoting desired cellular responses, and biosensing capabilities. Here, we will outline current methods used to produce heterogeneous printed constructs, incorporation of intelligent nanoparticles or bioactive molecules, and integration of chemical gradients to obtain a desired functional outcome.
A. Printing compositionally heterogeneous constructs
In the context of 3D bioprinting, the creation of biomimetic tissues resembling the heterogeneity and anatomical arrangement in vivo is of significant interest by matching the appropriate bioinks to support specific cell types to promote cellular physiological processes such as growth, proliferation, differentiation, and maturation.65 More importantly, while multimaterial 3D printing has been demonstrated by several groups most studies demonstrate at most two or three component constructs with the deposition of each material in an intermittent fashion.23,24,66,67 To be able to more fully recapitulate tissue heterogeneity, there is a now a growing need to deliver multiple components continuously at high speeds with intricate detailed patterns in order to realize the efficient fabrication of larger more complex constructs while also ensuring maximal cell viability. Currently, several advanced multimaterial fabrication techniques have been reported for extrusion-based as well as DLP-based 3D bioprinters which are highlighted in the below paragraphs.
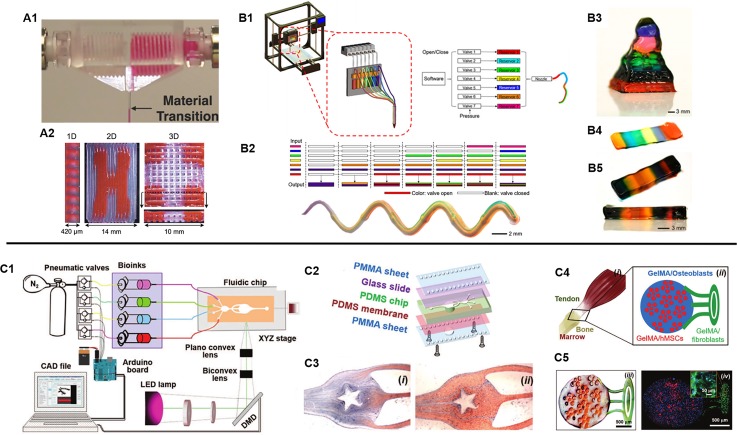
For many extrusion-based printers, multiple separate print heads with reservoirs housing individual bioinks are sequentially interchanged during the printing process to deposit different materials into predefined locations in a layer-by-layer fashion.66,67 However, due to this printing arrangement, the fabrication process is slow and limited to the deposition of a single bioink at one time, which makes it difficult to produce compositionally complex structures in a scalable and rapid manner for a larger number of materials. To circumvent these challenges, advanced approaches utilizing microfluidic-based extrusion systems to efficiently and seamlessly deposit multiple bioinks during fabrication have been reported.68–71 For instance, in a simple design, Hardin et al. demonstrated the use of a multimaterial microfluidic print head in which two opposing syringe pumps alternately control the flow of two viscoelastic PDMS-based inks through a single nozzle [Fig. 4(A)].69 As such, 3D printed structures containing sharp transitions of different materials were fabricated and this work highlights the possibility to program the assembly of various inks continuously to create sophisticated architectures with distinct local compositions. In another example, Liu et al. developed a multimaterial extrusion bioprinting platform capable of coding multiple bioinks that can smoothly transition between seven different materials to create complex tissue construct designs.71 By using digitally controlled pneumatic pressure, the release of multiple shear-thinning bioinks could be individually regulated to pass through a single nozzle-head and onto a motorized receiving stage at a speed 15 times faster than conventional extrusion-based systems.71 With this technique, the group was able to rapidly extrude one to seven bioinks at a time into a single printed microfiber with clear separation of each material [Fig. 4(B)]. Furthermore, discrete patterning of four different cell-laden GelMA-alginate composite bioinks were also bioprinted to form a vascularized multicellular tissue to demonstrate the assembly of multiple cell types with high viability maintained up to 7 days.
FIG. 4.
(A1) Microfluidic print head for seamless interchange of viscoelastic materials and (A2) printed multimaterial structures showing a 1D extruded filament single filament as well as 2D and 3D patterns. (Reproduced with permission from J. O. Hardin et al., Adv. Mater. 27, 3279 (2015). Copyright 2015 WILEY‐VCH Verlag GmbH & Co. KGaA, Weinheim). (B1) Schematic of a digitally tunable multimaterial extrusion printer with a seven-channel microfluidic print head that are pneumatically controlled. (B2) Examples of coded bioinks for continuous extrusion printing through a single microfiber and a printed serpentine multimaterial microfiber. (B3) Multimaterial pyramidal structure, and (B4) printed three and (B5) ten-layered block structures comprised of seven bioink materials. (Reproduced with permission from W. Liu et al., Adv. Mater. 29, 1604630 (2017). Copyright 2016 WILEY‐VCH Verlag GmbH & Co. KGaA, Weinheim). (C1) Schematic of a stereolithography-based microfluidics enabled multimaterial bioprinter and (C2) setup components of the fluidic chip platform. (C3) Flow streams of GelMA and food dye within the microfluidic chip to fabricate a star pattern. (C4) Schematic of a tendon-to-bone insertion model and (C5) the printed cellularized structure. (Reproduced with permission from A. K. Miri et al., Adv. Mater. 30, e1800242 (2018). Copyright 2018 WILEY‐VCH Verlag GmbH & Co. KGaA, Weinheim).
In DLP-based 3D bioprinting platforms, multimaterial printing is typically achieved by sequential UV polymerization of different bioinks followed by manual washing of the unpolymerized material in between fabrication prior to introducing the next material. This general technique has been adopted for the micron scale fabrication of bioprinted biomimetic liver and vascularized tissue structures containing two or three individual bioinks.23,24 However, while DLP-based 3D printing is rapid in nature, the scalability of this approach to produce multicomponent heterogeneous structures is highly limited by an increase in the number of materials, which will ultimately reduce the overall fabrication time. Similar to the challenges faced with extrusion-based printers, there is also a need to streamline the delivery of different bioinks during the fabrication process to enable the rapid continuous fabrication of multicomponent tissues. In a recent development, Miri et al. demonstrated the synchronization of a microfluidic device and a DLP-based 3D bioprinter to produce GelMA or PEGDA constructs with high spatial resolution and multimaterial composition [Fig. 4(C)].25 Using pneumatic valves, the injection of different bioinks into a PDMS printing chamber was controlled by flowing different materials under laminar conditions to allow for smooth transition followed by UV polymerization with the bioprinter. To generate 3D structures, a vertical movable membrane within the print chamber was used to control the height of the object during the printing process and programmed to load or unload to accommodate printing and washing regimes. This innovative platform allowed for the production of constructs consisting of two to three bioink materials in 20 s, compared to minutes in traditional manual material exchange, with resolutions as high as 25 μm in the x-y plane.25 While promising the current setup can only print under static flow conditions thus further developments to incorporate continuous dynamic printing of multiple bioinks simultaneously would represent a significant advancement in the speed as well as complexity for the future of multimaterial DLP-based 3D bioprinting technologies.
B. Incorporation of intelligent nanoparticles
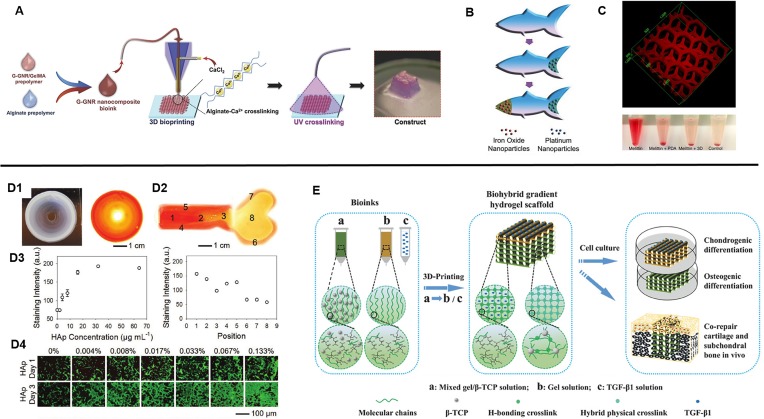
Recently, the development of new materials by way of nanoparticle-hydrogel composites has been intensely studied as an approach to impart additional functionality in existing polymer hydrogels through the improvement of stimuli responses as well as mechanical properties. There is currently a broad range of nanoparticles available which can be classified as non-metals, metals, metal oxides, and polymeric with numerous applications such as in drug delivery, environmental remediation, electronics, catalytic reactions, and biosensing.72 In consequence, the utilization of nanoparticle-hydrogel composite bioinks with 3D printing among researchers has resulted in the generation of several innovative constructs to date. For example, to enhance electrical conduction in bioprinted cardiac tissues, Zhu et al. incorporated gold nanorods into GelMA/alginate bioinks in which cardiomyocytes were encapsulated and extrusion printed into aligned fibers [Fig. 5(A)].73 They found that improved cardiomyocyte organization in addition to cell-cell coupling and synchronous beating was observed compared to GelMA/alginate bioinks absent of gold nanorods. In a similar light, Jakus et al. demonstrated the printing of graphene-polylactide-co-glycolide (PLG) composite bioinks to fabricate scaffolds with high electrical conductance while also possessing mechanical elasticity, resilience, and biocompatibility.74 Interestingly, the graphene-PLG material also provided a conducive microenvironment to promote neuronal-like phenotypic expression and morphology in seeded human mesenchymal stem cells (MSCs) in the absence of differentiation factors.74 In other applications, locomotive capabilities of printed devices can also be achieved as shown by Zhu et al. wherein printed PEGDA microfish embedded with platinum nanoparticles at the tail enabled propulsion, iron oxide nanoparticles embedded in the head enabled magnetic maneuverability, and polydiacetylene (PDA) nanoparticles embedded in the body allowed sensing and neutralization of toxins [Fig. 5(B)].75 Furthermore, a detoxification device mimicking the hexagonal liver lobule structure was fabricated by printing a PDA nanoparticle-PEGDA hydrogel ink.76 The embedded PDA nanoparticles attracted and neutralized the melittin pore-forming toxins while the printed porous structure allowed for efficient entrapment [Fig. 5(C)]. Altogether, the combined advantages of smart nanoparticle-based materials and engineered 3D printed constructs to impart higher ordered functionality is a powerful tool for advancing the development of state-of-the-art medical devices and artificial tissues in tissue engineering applications.
FIG. 5.
(A) Schematic of 3D bioprinting of GelMA-coated gold nanorod nanocomposite bioinks to form a cardiac tissue construct. (Reproduced with permission from K. Zhu et al., Adv. Funct. Mater. 27, 1605352 (2017). Copyright 2017 WILEY‐VCH Verlag GmbH & Co. KGaA, Weinheim). (B) 3D printed microfish made from PEGDA with embedded iron oxide and platinum nanoparticles to enable propulsion and magnetic control. (Reproduced with permission from W. Zhu et al., Adv. Mater. 27, 4411 (2015). Copyright 2015 WILEY‐VCH Verlag GmbH & Co. KGaA, Weinheim). (C) Bioinspired liver detoxification device comprised of PEGDA embedded with polydiacetylene (PDA) nanoparticles for toxin neutralization. (Reproduced with permission from M. Gou et al., Nat. Commun. 5, 3774 (2014). Copyright 2014 Zhu et al.). (D1) Printed concentric circle structure with gradients of hydroxyapatite nanoparticles and complementary image stained with Alizarin Red. (D2) A printed bone-like structure with varied gradient concentrations of hydroxyapatite nanoparticles stained with Alizarin Red. (D3) Corresponding quantified staining intensities of Alizarin Red in the concentric circle and bone-like structures. (D4) Seeded preosteoblasts on gradients of hydroxyapatite hydrogel slabs. (Reproduced with permission from W. Liu et al., Adv. Mater. 29, 1604630 (2017). Copyright 2016 WILEY‐VCH Verlag GmbH & Co. KGaA, Weinheim). (E) Overview of 3D printed biohybrid osteochondral gradient hydrogel scaffolds. (Reproduced with permission from F. Gao et al., Adv. Funct. Mater. 28, 1706644 (2018). Copyright 2018 WILEY‐VCH Verlag GmbH & Co. KGaA, Weinheim).
C. Integration of chemical gradients
For many tissues in the body, physical and biochemical signal gradients exist with gradual changes in mechanical and compositional properties as one tissue type transitions into another. Specifically, the study of biochemical gradients has been well-known to regulate cellular activities particularly during the stages of early development, and the presentation of these gradient signals in engineered scaffolds to control cell fate has been used for the production of novel cell-based therapies in tissue engineering and regenerative medicine.77,78 As such, leveraging 3D printing technology for the precise spatial control over the presentation of chemical gradients is an attractive approach to better mimic the dynamic microenvironment. In one example, the differentiation potential of seeded primary rat neural stem cells (NSCs) onto inkjet printed polyacrylamide-based hydrogels containing gradients of fibroblast growth factor-2 (FGF-2), ciliary neurotrophic factor (CNTF), or fetal bovine serum (FBS) was examined.79 Ilkhanizadeh et al. found that NSC differentiation into astrocytes was most efficient in CNTF with an observed trend of higher glial fibrillary acidic protein (GFAP) marker expression with increasing concentrations of CNTF, thus demonstrating a facile method to promote gradient cellular response.79 Similarly, Liu et al. studied the attachment and proliferation of MC3T3 preosteoblasts seeded onto an extrusion printed concentric ring scaffold possessing a seven level gradation of low to high concentrations of hydroxyapatite nanoparticles in the GelMA/alginate bioink from the inner to the outer rings [Fig. 5(D)].71 The results showed that increased seeding efficiency as well as proliferation correlated with regions of higher hydroxyapatite concentrations. As a proof-of-concept, this group also further demonstrated the potential to pattern localized gradients to resemble a bone shaped hydrogel to reflect the anatomical spatial composition.71 For the application of osteochondral repair, Gao et al. developed a biohybrid gradient poly(N‐acryloyl glycinamide) (PNAGA)‐based hydrogel consisting of two extrusion printed layers containing transforming growth factor beta 1 (TGF-β1) and β-tricalcium phosphate (β‐TCP) nanoparticles to promote the chondrogenic and osteogenic differentiation of seeded human bone marrow-derived stem cells (hBMSCs), respectively [Fig. 5(E)].80 This study revealed that the incorporation of these bioactive factors enhanced preferential chondrogenic and osteogenic differentiation of hBMSCs within the two regions in vitro, and upon implantation the biohybrid gradient scaffold improved the simultaneous regeneration of cartilage and subchondral bone in a rat model.80 Overall, these studies exemplify the utility of 3D printing technologies to fine tune the chemical microenvironment for modulating cell behavior and to facilitate the fabrication of interfacial tissues.
V. DIRECTING BIOLOGICAL PROPERTIES
Within the in vivo microenvironment, heterogeneous cell populations coexisting in highly organized structures work in concert to form a tissue-specific function. As such, it is well established that the interdependent relationship between cells and their surrounding extracellular matrix (ECM) as well as the crosstalk between multiple cell types occurs through a myriad of biochemical and biophysical cues.81 More specifically, a combination of direct cell-cell communication, cell-ECM interactions, paracrine signaling, cytokines, growth factors, hormones, and mechanotransduction mechanisms help maintain homeostasis and regulate function within adult tissues.81,82 As we move towards the goal of engineering complex tissue and organ systems, there is a need to be able to recapitulate these dynamic processes by guiding cellular organization within well-defined microarchitectures. As described previously, 3D printing strategies have offered several advantages by coupling multimaterial printing capabilities with the precise patterning and assembly of various cell populations. In Secs. V A–V C, we will provide an overview on the developments of different bioprinting techniques used to produce simple to complex cell-laden tissue constructs and highlight the current state of cell-based soft biorobotic devices.
A. Strategies for bioprinting cell-laden tissue constructs
The fabrication of cell-laden 3D printed tissue constructs can be divided into two general approaches: (1) post-print seeding, where the 3D scaffold is first printed followed by cell seeding, and (2) direct cell encapsulation, where cells are incorporated into the bioink and printed directly during fabrication. Generally, the post-print seeding approach has been primarily used for the production of simple homogeneous 3D printed constructs or populating hollow microchannel structures. Analogous to traditional seeding of acellular scaffolds, a porous framework is initially printed and subsequently cells are allowed to infiltrate into the construct or casted with cells suspended in a carrier biomaterial for bulk support. While this method is adequate for generating thin tissues, fabricating thick cell-laden constructs is difficult due to the limited control over even cell distribution and penetration depth into the central regions. Post-print seeding has also been used to form cell-laden interpenetrating microchannels reflective of in vivo vasculature and tubular networks. Here, a range of fugitive bioinks such as Pluronic F-127, alginate, poly(N-isopropylacrylamide), agarose, and sugar glass can be used to print the intended microchannel pattern and then subsequently removed post printing.83–86 Cells can then be perfusion seeded and cultured to generate a monolayer lining the hollow interior. This technique has been employed for creating thick vascularized perfusable tissue chips as well as perfusable convoluted renal proximal tubules of the kidney.83,84
Direct cell encapsulation has been widely adopted in nozzle and optical based printing systems as it allows greater control over spatial patterning and positioning of multiple cells types. However, maintaining high cell viability with preserved phenotype during printing is critical as several factors may have a negative impact depending on the printing procedure being used. Naturally, cells are sensitive to their mechanical environment and external forces applied have been shown to elicit changes in proliferation and differentiation potential, cytoskeletal organization, and in extreme cases excessive forces can disrupt the cellular membrane.87 For direct cell writing using nozzle-based printers, cells are inherently subjected to mechanical damage. In particular, key stress inducers include the nozzle diameter, dispensing pressure, deposition speed, and bioink viscosity, which are all important elements to consider. Furthermore, it has been demonstrated that these factors are not mutually exclusive and a careful balance between high resolution as well as cell survival and integrity must be maintained. For instance, Chang et al. observed that HepG2 cell viability and post-printing recovery increased more readily with decreasing dispensing pressure compared to increases in nozzle size.88 Blaeser et al. also found that high initial shear stress will have an immediate negative effect on cell survival in the short term but also impact long term proliferation potential during microvalve nozzle-based printing of cell-laden alginate hydrogels.18 This group also demonstrated that there is a specific shear stress level threshold cells can withstand without incurring damage during dispensing, which can be used to tune the printing process to achieve high resolution.18
Although laser writing and optical based printing systems are contactless, other factors that may contribute to decreased cell viability include the light source and photoinitiator cytotoxicity. Many of these systems utilize a UV (i.e., 315–400 nm) light source with low intensities to initiate photopolymerization, and while it is generally accepted as biocompatible under low doses prolonged exposure can lead to DNA damage through the production of free radicals.89 Similarly, the different effects of commonly used photoinitiators such as Irgacure (I2959, I819) and lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) on cell viability must also be taken into account since free radical generation is dependent on the photoinitiator concentration and light intensity.90 To circumvent these concerns, visible light photocrosslinking with cytocompatible photoinitiators have been explored in 3D printing. For instance, Wang et al. used visible light photocrosslinking with eosin Y photoinitiator to achieve high 50 μm printing resolution of composite PEGDA and gelatin methacrylate (GelMA) hydrogels with greater than 85% viability of NIH 3T3 fibroblasts for 5 days.91 Overall, the photointiating system and parameters chosen must be optimized to attain high crosslinking efficiency, structural integrity, and print quality without compromising cell viability.
B. Biomimetic cellular organization
Native tissues and organs are naturally arranged into hierarchal structures with regionally distinct organizations and compositions of cells and ECM. These features play an important role in coordinating and maintaining multicellular communication networks as well as providing the necessary mechanical performance to carry out a tissue-specific function.82,92 For instance, cardiac tissue is made up of discrete structural layers composed of the protective epicardium containing a thin layer of loose connective tissue and adipose, a thick myocardium where cardiac muscle cells as well as nerves and blood vessels reside, and a thin inner elastic endocardium comprised of endothelial cells. Concomitant to tissue architecture, the presence of supportive cell populations in in vitro studies have also shown to significantly improve the bioactivity of parenchymal cells.93 This phenomenon has been demonstrated in cardiac microtissues where cardiomyocytes exhibited a phenotype close to the native when co-cultured with cardiac fibroblasts in a 2:1 ratio (cardiomyocytes:cardiac fibroblasts) with higher expression of connexin 43 and integrin.94,95 Similarly, in a 3D printed liver tissue model, a co-culture system incorporating human iPSC-derived hepatic progenitor cells, HUVECs, and human adipose-derived stem cells (ADSCs) exhibited improved albumin and urea secretion.23 This further highlights the importance of incorporating multicellular populations and matrix heterogeneity in addition to hierarchal physiological structures as an approach to building biomimetic tissue constructs.
To date, a wide variety of tissues have been fabricated using 3D bioprinting as summarized in Table I. Control of tissue-specific microarchitecture and cell distribution can be achieved in 3D bioprinting through the precise spatial patterning of encapsulated cells. In nozzle-based printers, this is accomplished by the extrusion of various cells and biomaterials using multimaterial print heads, which can deposit the cell-laden bioinks within the desired regions of the construct. Similarly, in 3D optical printing systems, bioinks of different cellular and material compositions can be localized within a construct by assigning digital patterns to each bioink composition and photopolymerizing the defined regions. The bioink and digital pattern can then be exchanged sequentially to fabricate complementary regions of the final tissue. At the cellular level, it is also equally important to be able to guide cell orientation, since this has a direct influence on several biological processes including elongation, phenotype, differentiation, and functionality.96 More specifically, several studies have demonstrated that different cell responses can be modulated through physical cues such as pattern size, contour, and stiffness in the absence of other external stimuli.97,98 This highlights an advantage in 3D printing to utilize topographic guidance to promote cell behavior which is critical in organized tissues such as skeletal and cardiac muscle, nerve tissue, epithelium, and vascular smooth muscle.97
TABLE I.
Summary of approaches for 3D printed functional tissues.a
| Tissue | Reference | Application | 3D printer type | Bioink | Cell source | Design |
|---|---|---|---|---|---|---|
| Liver | Ma et al.23 | Heterogeneous biomimetic liver model | Optical projection printer | GelMA, GM-HA:GelMA | Human iPSC-derived hepatic progenitor cells, HUVECs, human ADSCs | Hexagonal hepatic lobule units printed with GelMA and patterned supportive cells in sinusoidal regions printed with GM-HA/GelMA |
| Faulkner-Jones et al.105 | Mini liver tissues | Valve-based printer | RGD-coupled sodium alginate | Human iPSC-derived and human ESC-derived hepatocyte-like cells | Cells encapsulated into ringed structures | |
| Lee et al.106 | Heterogeneous liver tissue | Multi-head nozzle-based printer | PCL and collagen solution | Primary rat hepatocytes, HUVECs, human lung fibroblasts | Lattice structure comprised of PCL framework with cell laden collagen in between | |
| Lee et al.107 | Liver dECM tissue | Nozzle-based printer | Pepsin solubilized liver dECM bioink, PCL | Human HepG2 | Lattice structure comprised of PCL framework with cell laden liver dECM in between | |
| Kidney | Homan et al.84 | Perfusable convoluted renal proximal tubule chip | Multimaterial nozzle-based printer | ECM solution (i.e., fibrinogen, gelatin, CaCl2, transglutaminase), Pluronic F127, silicone | Human immortalized proximal tubule epithelial cells | Silicone gasket holding printed fugitive Pluronic F127 with casted ECM solution, then perfusion seeded cells after removal of fugitive layer |
| Placenta | Kuo et al.108 | Model trophoblast migration in preeclampsia | Extrusion-based printer (3D-Bioplotter, Envision TEC) | GelMA | Human choriocarcinoma BeWo cells, human MSCs | Cylindrical hydrogel with cell-laden outer ring and chemoattractant growth factors in the center |
| Heart | Lind et al.109 | Instrumented cardiac microphysiological devices | Multimaterial nozzle-based printer | Dextran ink, TPU ink, carbon black:TPU ink, Ag:Pa ink, soft PDMS ink, rigid PDMA ink | Neonatal rat ventricular myocytes, human iPSC-CM | Cells seeded on printed device containing multilayer cantilevers with a base, embedded strain sensor, and microgrooved layer to guide cell alignment |
| Zhang et al.52 | Endothelialized-myocardium-on-a-chip | Coaxial nozzle bioprinter (Organovo, NovoGen MMX) | GelMA:alginate | Neonatal rat cardiomyocytes, human iPSC-CM | Two step crosslinking with CaCl2 followed by UV irradiation to form endothelium lattice, then cardiomyocytes were seeded onto the construct | |
| Jang et al.110 | Pre-vascularized cardiac dECM patch | Nozzle-based printer | Pepsin solubilized heart dECM bioink with/without VEGF, PCL | hCPCs, human MSCs, human endothelial cells | PCL support base with multilayered lattice structure of alternating cell-laden cardiac cells and endothelial cells in HdECM bioink | |
| Gaetani et al.111 | Cardiac patch for restoring myocardial infarction | Bioscaffolder tissue (Sys +Eng, Gladbeck, Germany) nozzle-based printer | Hyaluronic acid/gelatin | Human fetal CMPCs | Encapsulated human fetal CMPCs printed into a multilayered lattice structure | |
| Duan et al.112 | Aortic valve conduits | Nozzle-based printer | Alginate/gelatin | Porcine sinus smooth muscle cells, human aortic valve leaflet interstitial cells | Based on micro-CT scan of porcine aortic valve reconstructed heterogeneous valve root and valve leaflet regions | |
| Bone | Shim et al.113 | Osteochondral tissue regeneration | Multi-head nozzle-based printer | Mono CB[6]/DAH-hyaluronic acid, pepsin-treated collagen, PCL | hTMSCs | Printed a bottom collagenous subchondral and top superficial cartilage layer in between a PCL framework |
| Cartilage | Nguyen et al.114 | Direct 3D printed iPSCs towards chondrogenic commitment | 3D Discovery (regenHu, Switzerland) nozzle-based printer | Nanofibrillated cellulose:alginate | Human iPSCs, irradiated human chondrocytes | Multilayered lattice construct with encapsulated iPSCs co-cultured with irradiated chondrocytes |
| Vasculature | Zhu et al.24 | Prevascularized tissue construct with complex microarchitecture | Microscale continuous optical bioprinter | GelMA, GM-HA:GelMA | HUVECs, C3H/10T1/2 cells, HepG2 | Directly print endothelial cell-laden GM-HA/GelMA in hierarchal hexagonal vascular channels surrounded by printed HepG2-laden GelMA in the surrounding region |
| Kolesky et al.83 | Multicellular thick (>1 cm) vascularized perfusable tissue chips | Multi-head nozzle-based printer | Silicone, vascular ink (pluronic F127/thrombin), ECM ink (gelatin/fibrinogen/thrombin/TG) | HUVECs, hMSCs, hNDFs | Silicone supportive framework was first printed, then fugitive vascular ink is patterned in a lattice structure, cell-laden ECM ink is then cast, fugitive ink is washed, and channels perfusion seeded with endothelial cells | |
| Tseng et al.115 | Printed vascular smooth muscle rings for measuring vasoactivity in high throughput | Magnetic 3D printer | N/A | Human aortic smooth muscle cells | Incorporated magnetic particles to print cells into a ring structure, contraction and dilation monitored in response to drug treatment | |
| Mirabella et al.116 | Implantable vascular network | Nozzle-based printer | Sacrificial carbohydrate glass with fibrin gel cast | HUVECs | Patterned various microchannel geometries (straight parallel lines vs. grid) | |
| Uterus | Souza et al.117 | Printed human myometrium cell rings for measuring uterine wall contractility | Magnetic 3D printer | N/A | Human myometrial smooth muscle cells | Incorporated magnetic particles to print cells into a ring structure, contractility assessed in response to tocolytics |
| Adipose | Pati et al.118 | Soft tissue substitute with decellularized adipose tissue bioink | Nozzle-based printer | Pepsin solubilized adipose tissue dECM bioink, PCL | Human adipose-derived stem cells | Dome-shaped lattice construct consisting of a PCL framework with cell laden liver dECM in between |
| Ovary | Laronda et al.119 | Bioprosthetic ovary | EnvisionTEC 3D-Bioplotter nozzle-based printer | Gelatin | Mouse ovarian follicles | Printed lattice gelatin structures and seeded with ovarian follicles |
| Cornea | Wu et al.120 | Demonstrate printability of human corneal epithelial cells | Nozzle-based printer | Gelatin-alginate-collagen solution | Human corneal epithelial cells (HCECs) | Printed interconnected porous lattice structure with tuned degradation using sodium citrate |
| Pancreas | Farina et al.121 | Islet cell transportation | ReplicatorTM 2X (MakerBot Industries) fused deposition nozzle-based printer | PLA | Human pancreatic islet cells | Discoidal encapsulation device consisting of inner micro-reservoirs housing the cells and surrounded by square microchannels to enable vascular ingrowth |
| Sweat gland | Liu et al. | Glandular morphogenesis in printed porous constructs | Nozzle-based printer | Sodium alginate | Mouse epithelial progenitor cells and plantar dermis | Porous lattice structure printed using cell-laden sodium alginate bioink |
Abbreviations: GelMA = gelatin methacrylate; GMHA = glycidal methacrylate-hyaluronic acid; TPU = thermoplastic polyurethane; dECM = decellularized extracellular matrix; HUVECs = human umbilical vein endothelial cells; HCPCs = human c-kit+ cardiac progenitor cells; CMPCs = cardiac-derived progenitor cells; hTMSCs = human turbinate-derived mesenchymal stromal cells; CB[6] = cucurbit [6] uril; DAH = 1,6-diaminohexane; hNDFs = human neonatal dermal fibroblasts; PLA = polylactic acid.
Several researchers have applied 3D printing technology to explore directed cell organization by way of patterning cells within encapsulated hydrogels or seeding cells in printed patterned microstructures. Mozetic et al. 3D bioprinted C2C12 murine myoblast cells encapsulated in a Pluronic PF127/alginate bioink and extruded four layers of parallel-aligned fibers using a 250 μm nozzle tip.99 Cellular alignment was observed immediately post printing along the deposition direction with increased elongation and myotubular formation upon myogenic differentiation after 21 days.99 Similarly, a two-stage bioprinting technique developed by Bhuthalingam et al. was used to induce cellular alignment and cardiomyocytes lineage differentiation by precisely etching surface microgrooves onto the surface of polystyrene films followed by microextrusion of 2% gelatin containing fibroblasts or MSCs into the grooved patterns.100 Among the different shapes and channel sizes tested, they found that MSCs cultured in linear patterns with widths of 50 and 100 μm induced the best alignment and adopted an early cardiomyocyte gene expression profile compared to concentric circle or S-wave patterned controls.100 These studies highlight the importance of microgeometries as well as the need to optimize critical feature sizes to provide the necessary spatial cues to guide cellular organization.
While 3D bioprinting technology represents an important advancement towards the creation of biomimetic tissues through the artificial assembly of cells into predesigned patterns, they still remain as simplified representations of native tissues. In particular, the bottom-up approach in 3D printing makes it challenging to be able to replicate the full range and proportions of different cell types as well as their complex interactions found in vivo, which is limited by our current knowledge of organ systems. Recently, the notion of combining organoid technology and 3D bioprinting has been proposed as a potential dual strategy to impart both anatomically accurate cell populations and control over microarchitecture in engineered tissues.101 Much like in development, organoids are naturally formed by the self-organization of stem cells and through this process are able to recapitulate the cellular heterogeneity as well as interactions found in their organ of origin. However, it is often difficult to form reproducible tissue architectures in organoids and their differentiation is highly variable in vitro due to the absence of external physical, biochemical, and spatial cues normally encountered in vivo. As such, these factors limit the control over the reliable mass production of organoid cultures, maturation, and size.102 3D printing technology can potentially offer an avenue for spatial control to provide added complexity in tissue architecture when assembling organoids that is needed to create higher ordered structures at multiple length scales for preparing large intricate organoids or tissues. To date, small organoids have been incorporated within bioinks to be precisely deposited via 3D bioprinting into larger scale tissues or integrated into microfluidic chip systems. For instance, Zhang et al. demonstrated the bioprinting of human iPSC-derived myocardial organoids into vascularized cardiac tissue constructs, while Skardal et al. showed the bioprinting of heart, liver, and lung organoids into an integrated organ-on-a-chip to study the inter-tissue interaction of drug responses.52,103 Further work in this area would help towards the creation of anatomically correct and functionally relevant tissues and organs applicable to serve as more reliable drug testing platforms and modeling of biological disease mechanisms.104
C. Cell-based soft biorobotic devices
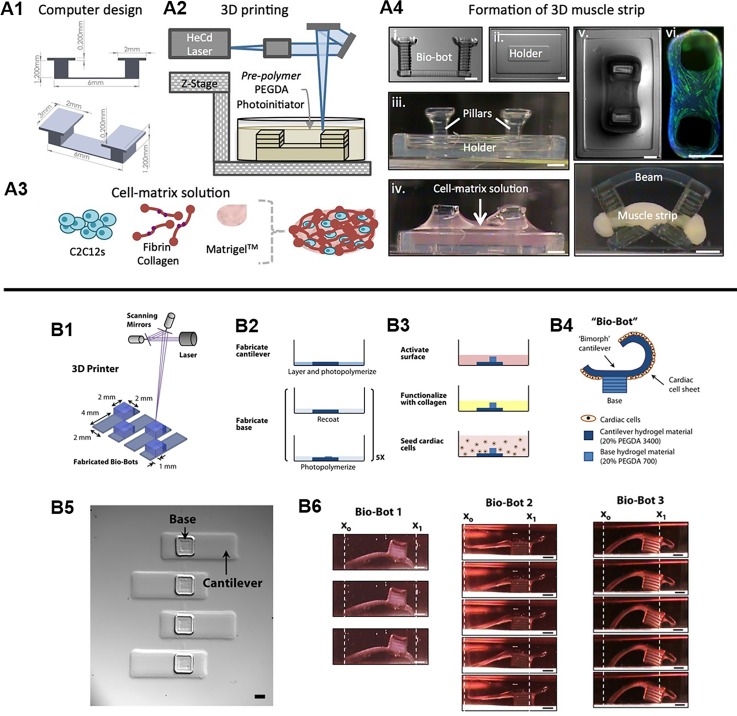
The field of soft biorobotics has recently gained growing attention as a way to integrate controlled motion complexity in 3D bioinspired devices for mimicking life-like movements that dynamically sense and respond to the environment. Unlike conventional rigid mechanical robots, soft biorobots are made up of elastic materials capable of stretching and deformation such as silicone, neoprene, and PEGDA.122–124 Although the fabrication of these soft robotic devices have been reported with molding techniques,125–127 the adoption of 3D printing technology is growing to enable faster and more reliable production of elegant biomimetic structural designs that are difficult to achieve with conventional approaches. Currently, there is a wide variety of 3D printed soft biorobotic devices in which the reader is referred to a comprehensive review by Gul et al.128 This section will focus on highlighting some current examples of cell-based soft biorobotic devices and their applications. In cell-based soft biorobotic devices, cells such as muscle cells and cardiomyocytes that naturally possess dynamic function are combined with 3D printed synthetic materials to form hybrid biological machines capable of force production and actuation. This has been demonstrated in seminal work by Cvetkovic et al. through the creation of a skeletal muscle bio-bot composed of a strip of C2C12 murine myoblasts assembled around a two PEGDA posts made using stereolithographic 3D printing [Fig. 6(A)].124 In this work, untethered symmetric and asymmetric skeletal muscle bio-bot devices were produced and upon external electrical pacing the contraction and directional locomotion was measured. Interestingly, the asymmetric designs resulted in greater net displacement and a 25-fold higher velocity relative to symmetric controls under 1 Hz electrical stimulation and points to the importance of design consideration for achieving a desired motion. A subsequent study by the same group also demonstrated the potential use of optogenetic control to drive directional locomotion of a skeletal muscle powered bioactuator by wrapping optogenetic C2C12 cell rings around a one-leg or two-leg bio-bot skeleton design.129 Key findings from this study showed that including an exercise regimen consisting of combined optogenetic and mechanical stimulation during differentiation of C2C12 cells into multinucleated myotubes improved force generation. In addition, optogenetic stimulation via illumination of a 470 nm light allowed for precision noninvasive stimulation of different muscle rings which enabled control over directional locomotion of an untethered two-leg skeletal muscle bioactuator device. In other similar works, Chan et al. used a laser stereolithographic 3D printer to fabricate an asymmetric cantilever structure composed of thin layer of flexible high molecular weight PEGDA (Mw 3400) and a solid base comprised of lower molecular weight PEGDA (Mw 700) [Fig. 6(B)].130 Cardiomyocytes were then seeded in monolayer onto the asymmetric cantilever arms and electrically stimulated to generate a net forward walking motion for three different bio-bot designs of varied cantilever thickness and curvature. The power stroke generated by the cantilever was measured and the locomotive mechanisms were evaluated to elucidate crucial design principles necessary for generating efficient actuating bio-bot devices. Overall, these soft biorobotic devices showcase the convergence of cell powered motility and rationally designed 3D printed support structures engineered to perform desired functional behaviors upon external stimuli. From the aforementioned examples, these soft biorobotic devices are on the millimeter scale and it is envisioned that 3D printing technologies will enable further miniaturization of these devices to the micron scale applicable for high throughput applications such as drug screening. 3D printing could also be used to incorporate topological patterning to provide additional guidance and organization for cells as well as the incorporation of multiple cell types to further understand complex biological interactions in biomimetic actuating tissues.
FIG. 6.
(A1) Designed pillar hydrogel structures and (A2) 3D fabrication for the generation of 3D muscle strips. (A3) Cell and matrix solution used to form the muscle strip component. (A4) The assembly and compaction of the cells and matrix around the pillars to form the skeletal muscle bio-bot device. (Reproduced with permission from C. Cvetkovic et al., Proc. Natl. Acad. Sci. U. S. A. 111, 10125 (2014). Copyright 2014 National Academy of Sciences). (B1) Stereolithographic 3D printing of cantilever structure. (B2) Photopolymerization process to form cantilever and base structures. (B3) Functionalization of cantilever surface with collagen and seeding of cardiomyocytes as a monolayer. (B4) Schematic of the actuating bio-bot. (B5) Top view images of printed cantilever bio-bot structures. (B6) Representative images of three different bio-bot designs and their power stroke. (Reproduced with permission from V. Chan et al., Sci. Rep. 2, 857 (2012). Copyright 2014 Chan et al.).
VI. CONCLUSIONS AND FUTURE PERSPECTIVES
To conclude, this review focuses on the utility of different 3D printing platforms to create biomimetic tissues by controlling the physical, chemical, and biological properties. More specifically, 3D printing has emerged as a biofabrication tool with outstanding flexibility and precision to enable the production of tissues with higher ordered functionality and structural complexity across multiple lengths scales. We have highlighted the use of 3D printing approaches to engineer topographical cues and mechanical properties to improve tissue maturation and cellular organization that recapitulate native tissue architectures. The development of smart auxetic structures via 3D printing was also introduced as a method to impart unique properties at a cell and tissue level scale to control cellular behavior and improve the mechanical performance of scaffold designs. Furthermore, next generation 3D multimaterial printers interfaced with microfluidic technology enable the generation of complex tissue patterns in a rapid manner. To provide an additional avenue for researchers to direct desired functions such as biosensing capabilities, electrical conduction, and cell differentiation, the incorporation of intelligent nanoparticles as well as chemical gradients were discussed. Finally, we reviewed strategies used to spatially pattern heterogeneous cell populations for improving phenotype, function, and maturation of 3D bioprinted biomimetic tissues as well as the generation of actuating cell-powered soft biorobotic devices.
As we aim to build functional tissues and organs, the ability to fully recapitulate the complexity of in vivo biological systems remains a challenge. This is partly attributed by the limited selection of suitable biomaterials for 3D printing since their composition and physical properties strongly dictate the cellular microenvironment such that poorly matched biomaterials may limit biological function. Furthermore, a main bottleneck is the scalability of 3D bioprinting from tissue to organ level structures. The majority of bioprinted tissues to date are miniaturized simplifications of native tissues, however, to realize the creation of full organ systems it is necessary to develop 3D bioprinters that can accommodate large tissue constructs coupled with multimaterial deposition, high resolution, tunable regional properties, and high-speed fabrication to ensure viability. It is also important to note that while there has been growing developments on prevascularized tissues to overcome diffusion limitations in large organ fabrication, the future of whole organ bioprinting will also need to integrate elements such as innervation, lymphatics, and supportive cells as a step towards building fully functioning organs. Overall, the development of 3D printing technology has led to new advances in the fields of tissue engineering and regenerative medicine and continued work to further drive innovations will come from a combined interdisciplinary effort of expertise in the fields of engineering, biology, medicine, material science, and robotics.
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health (R21HD090662, R01EB021857, and NS047101) and National Science Foundation (CMMI-1547005 and CMMI-1644967). Scholarship funding for Dr. Claire Yu was provided by the Natural Sciences and Engineering Research Council (NSERC) Postdoctoral Fellowship Scholarship of Canada.
Note: This paper is part of the Special Topic on 3D Bioprinting: Physical and Chemical Processes.
References
- 1. Edmondson R., Broglie J. J., Adcock A. F., and Yang L., Assay Drug Dev. Technol. 12, 207 (2014). 10.1089/adt.2014.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tibbitt M. W. and Anseth K. S., Biotechnol. Bioeng. 103, 655 (2009). 10.1002/bit.22361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antoni D., Burckel H., Josset E., and Noel G., Int. J. Mol. Sci. 16, 5517 (2015). 10.3390/ijms16035517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baker B. M. and Chen C. S., J. Cell Sci. 125, 3015 (2012). 10.1242/jcs.079509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O'Brien F. J., Mater. Today 14, 88 (2011). 10.1016/S1369-7021(11)70058-X [DOI] [Google Scholar]
- 6. Lu T., Li Y., and Chen T., Int. J. Nanomed. 8, 337 (2013). 10.2147/IJN.S38635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilson W. C. and Boland T., Anat. Rec. 272A, 491 (2003). 10.1002/ar.a.10057 [DOI] [PubMed] [Google Scholar]
- 8. Boland T., Xu T., Damon B., and Cui X., Biotechnol. J. 1, 910 (2006). 10.1002/biot.200600081 [DOI] [PubMed] [Google Scholar]
- 9. Ozbolat I. T., Peng W., and Ozbolat V., Drug Discov. Today 21, 1257 (2016). 10.1016/j.drudis.2016.04.006 [DOI] [PubMed] [Google Scholar]
- 10. Waheed S., Cabot J. M., Macdonald N. P., Lewis T., Guijt R. M., Paull B., Breadmore M. C., Chen Y., Adam V., Kizek R., Hanson K., Long A., Hightower B. J., Slatton G., Burnett D. C., Massey T. L., Iwai K., Lee L. P., Pisterbi K. S. J., and Lin L., Lab Chip 16, 1993 (2016). 10.1039/C6LC00284F [DOI] [PubMed] [Google Scholar]
- 11. Murphy S. V. and Atala A., Nat. Biotechnol. 32, 773 (2014). 10.1038/nbt.2958 [DOI] [PubMed] [Google Scholar]
- 12. Lee J. M. and Yeong W. Y., Adv. Healthcare Mater. 5, 2856 (2016). 10.1002/adhm.201600435 [DOI] [PubMed] [Google Scholar]
- 13. Jungst T., Smolan W., Schacht K., Scheibel T., and Groll J., Chem. Rev. 116, 1496 (2016). 10.1021/acs.chemrev.5b00303 [DOI] [PubMed] [Google Scholar]
- 14. Xu T., Zhao W., Zhu J.-M., Albanna M. Z., Yoo J. J., and Atala A., Biomaterials 34, 130 (2013). 10.1016/j.biomaterials.2012.09.035 [DOI] [PubMed] [Google Scholar]
- 15. Christensen K., Xu C., Chai W., Zhang Z., Fu J., and Huang Y., Biotechnol. Bioeng. 112, 1047 (2015). 10.1002/bit.25501 [DOI] [PubMed] [Google Scholar]
- 16. Pataky K., Braschler T., Negro A., Renaud P., Lutolf M. P., and Brugger J., Adv. Mater. 24, 391 (2012). 10.1002/adma.201102800 [DOI] [PubMed] [Google Scholar]
- 17. Gudapati H., Dey M., and Ozbolat I., Biomaterials 102, 20 (2016). 10.1016/j.biomaterials.2016.06.012 [DOI] [PubMed] [Google Scholar]
- 18. Blaeser A., Duarte Campos D. F., Puster U., Richtering W., Stevens M. M., and Fischer H., Adv. Healthcare Mater. 5, 326 (2016). 10.1002/adhm.201500677 [DOI] [PubMed] [Google Scholar]
- 19. Zhang W., Han L.-H., and Chen S., J. Manuf. Sci. Eng. 132, 030907 (2010). 10.1115/1.4001661 [DOI] [Google Scholar]
- 20. Paz V. F., Emons M., Obata K., Ovsianikov A., Peterhänsel S., Frenner K., Reinhardt C., Chichkov B., Morgner U., and Osten W., J. Laser Appl. 24, 042004 (2012). 10.2351/1.4712151 [DOI] [Google Scholar]
- 21. Zhang A. P., Qu X., Soman P., Hribar K. C., Lee J. W., Chen S., and He S., Adv. Mater. 24, 4266 (2012). 10.1002/adma.201202024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tumbleston J. R., Shirvanyants D., Ermoshkin N., Janusziewicz R., Johnson A. R., Kelly D., Chen K., Pinschmidt R., Rolland J. P., Ermoshkin A., Samulski E. T., and DeSimone J. M., Science 347, 1349 (2015). 10.1126/science.aaa2397 [DOI] [PubMed] [Google Scholar]
- 23. Ma X., Qu X., Zhu W., Li Y.-S., Yuan S., Zhang H., Liu J., Wang P., Lai C. S. E., Zanella F., Feng G.-S., Sheikh F., Chien S., and Chen S., Proc. Natl. Acad. Sci. U. S. A. 113, 2206 (2016). 10.1073/pnas.1524510113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu W., Qu X., Zhu J., Ma X., Patel S., Liu J., Wang P., Sun C., Lai E., Gou M., Xu Y., Zhang K., and Chen S., Biomaterials 124, 106 (2017). 10.1016/j.biomaterials.2017.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miri A. K., Nieto D., Iglesias L., Goodarzi Hosseinabadi H., Maharjan S., Ruiz-Esparza G. U., Khoshakhlagh P., Manbachi A., Dokmeci M. R., Chen S., Shin S. R., Zhang Y. S., and Khademhosseini A., Adv. Mater. 30, e1800242 (2018). 10.1002/adma.201800242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anselme K. and Bigerelle M., Int. Mater. Rev. 56, 243 (2011). 10.1179/1743280411Y.0000000001 [DOI] [Google Scholar]
- 27. Dalby M. J., Gadegaard N., and Oreffo R. O. C., Nat. Mater. 13, 558 (2014). 10.1038/nmat3980 [DOI] [PubMed] [Google Scholar]
- 28. Hoffman-Kim D., Mitchel J. A., and Bellamkonda R. V., Annu. Rev. Biomed. Eng. 12, 203 (2010). 10.1146/annurev-bioeng-070909-105351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Crowder S. W., Leonardo V., Whittaker T., Papathanasiou P., and Stevens M. M., Cell Stem Cell 18, 39 (2016). 10.1016/j.stem.2015.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choi Y. S., Vincent L. G., Lee A. R., Kretchmer K. C., Chirasatitsin S., Dobke M. K., and Engler A. J., Biomaterials 33, 6943 (2012). 10.1016/j.biomaterials.2012.06.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Engler A. J., Sen S., Sweeney H. L., and Discher D. E., Cell 126, 677 (2006). 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- 32. Huang T. Q., Qu X., Liu J., and Chen S., Biomed. Microdevices 16, 127 (2014). 10.1007/s10544-013-9812-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hribar K. C., Finlay D., Ma X., Qu X., Ondeck M. G., Chung P. H., Zanella F., Engler A. J., Sheikh F., Vuori K., and Chen S. C., Lab Chip 15, 2412 (2015). 10.1039/C5LC00159E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wozniak M. A. and Chen C. S., Nat. Rev. Mol. Cell Biol. 10, 34 (2009). 10.1038/nrm2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soman P., Chung P. H., Zhang a. P., and Chen S., Biotechnol. Bioeng. 110, 3038 (2013). 10.1002/bit.24957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tay C. Y., Irvine S. A., Boey F. Y. C., Tan L. P., and Venkatraman S., Small 7, 1361 (2011). 10.1002/smll.201100046 [DOI] [PubMed] [Google Scholar]
- 37. Pathak A. and Kumar S., Proc. Natl. Acad. Sci. U. S. A. 109, 10334 (2012). 10.1073/pnas.1118073109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sommer G., Schriefl A. J., Andrä M., Sacherer M., Viertler C., Wolinski H., and Holzapfel G. A., Acta Biomater. 24, 172 (2015). 10.1016/j.actbio.2015.06.031 [DOI] [PubMed] [Google Scholar]
- 39. Kim D.-H., Lipke E. A., Kim P., Cheong R., Thompson S., Delannoy M., Suh K.-Y., Tung L., and Levchenko A., Proc. Natl. Acad. Sci. U. S. A. 107, 565 (2010). 10.1073/pnas.0906504107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gopalan S. M., Flaim C., Bhatia S. N., Hoshijima M., Knoell R., Chien K. R., Omens J. H., and McCulloch A. D., Biotechnol. Bioeng. 81, 578 (2003). 10.1002/bit.10506 [DOI] [PubMed] [Google Scholar]
- 41. McDevitt T. C., Angello J. C., Whitney M. L., Reinecke H., Hauschka S. D., Murry C. E., and Stayton P. S., J. Biomed. Mater. Res. 60, 472 (2002). 10.1002/jbm.1292 [DOI] [PubMed] [Google Scholar]
- 42. Bursac N., Parker K. K., Iravanian S., and Tung L., Circ. Res. 91, e45 (2002). 10.1161/01.RES.0000047530.88338.EB [DOI] [PubMed] [Google Scholar]
- 43. Khademhosseini A., Eng G., Yeh J., Kucharczyk P. A., Langer R., Vunjak-Novakovic G., and Radisic M., Biomed. Microdevices 9, 149 (2007). 10.1007/s10544-006-9013-7 [DOI] [PubMed] [Google Scholar]
- 44. Fozdar D. Y., Lee J. Y., Schmidt C. E., and Chen S., Int. J. Nanomed. 6, 45 (2011). 10.2147/IJN.S12376 [DOI] [Google Scholar]
- 45. Fozdar D. Y., Lee J. Y., Schmidt C. E., and Chen S., Biofabrication 2, 035005 (2010). 10.1088/1758-5082/2/3/035005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Soman P., Tobe B. T. D., Lee J. W., Winquist A. M., Singec I., Vecchio K. S., Snyder E. Y., and Chen S., Biomed. Microdevices 14, 829 (2012). 10.1007/s10544-012-9662-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zong X., Bien H., Chung C.-Y., Yin L., Fang D., Hsiao B. S., Chu B., and Entcheva E., Biomaterials 26, 5330 (2005). 10.1016/j.biomaterials.2005.01.052 [DOI] [PubMed] [Google Scholar]
- 48. Alford P. W., Feinberg A. W., Sheehy S. P., and Parker K. K., Biomaterials 31, 3613 (2010). 10.1016/j.biomaterials.2010.01.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ma Z., Koo S., Finnegan M. A., Loskill P., Huebsch N., Marks N. C., Conklin B. R., Grigoropoulos C. P., and Healy K. E., Biomaterials 35, 1367 (2014). 10.1016/j.biomaterials.2013.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mobini S., Spearman B. S., Lacko C. S., and Schmidt C. E., Curr. Opin. Biomed. Eng. 4, 134 (2017). 10.1016/j.cobme.2017.10.010 [DOI] [Google Scholar]
- 51. Zhu W., Tringale K. R., Woller S. A., You S., Johnson S., Shen H., Schimelman J., Whitney M., Steinauer J., Xu W., Yaksh T. L., Nguyen Q. T., and Chen S., Mater. Today 21(9), 951–959 (2018). 10.1016/j.mattod.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y. S., Arneri A., Bersini S., Shin S.-R., Zhu K., Goli-Malekabadi Z., Aleman J., Colosi C., Busignani F., Dell'Erba V., Bishop C., Shupe T., Demarchi D., Moretti M., Rasponi M., Dokmeci M. R., Atala A., and Khademhosseini A., Biomaterials 110, 45 (2016). 10.1016/j.biomaterials.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hockaday L. A., Kang K. H., Colangelo N. W., Cheung P. Y. C., Duan B., Malone E., Wu J., Girardi L. N., Bonassar L. J., Lipson H., Chu C. C., and Butcher J. T., Biofabrication 4, 035005 (2012). 10.1088/1758-5082/4/3/035005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Soman P., Kelber J. A., Lee J. W., Wright T. N., Vecchio K. S., Klemke R. L., and Chen S., Biomaterials 33, 7064 (2012). 10.1016/j.biomaterials.2012.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pyo S. H., Wang P., Hwang H. H., Zhu W., Warner J., and Chen S., ACS Appl. Mater. Interfaces 9, 836 (2017). 10.1021/acsami.6b12500 [DOI] [PubMed] [Google Scholar]
- 56. Zhang W., Soman P., Meggs K., Qu X., and Chen S., Adv. Funct. Mater. 23, 3226 (2013). 10.1002/adfm.201202666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Warner J. J., Gillies A. R., Hwang H. H., Zhang H., Lieber R. L., and Chen S., J. Mech. Behav. Biomed. Mater. 76, 145 (2017). 10.1016/j.jmbbm.2017.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fozdar D. Y., Soman P., Lee J. W., Han L. H., and Chen S., Adv. Funct. Mater. 21, 2712 (2011). 10.1002/adfm.201002022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Soman P., Lee J. W., Phadke A., Varghese S., and Chen S., Acta Biomater. 8, 2587 (2012). 10.1016/j.actbio.2012.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee J. W., Soman P., Park J. H., Chen S., and Cho D. W., PLoS One 11, 1 (2016). 10.1371/journal.pone.0155681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kapnisi M., Mansfield C., Marijon C., Guex A. G., Perbellini F., Bardi I., Humphrey E. J., Puetzer J. L., Mawad D., Koutsogeorgis D. C., Stuckey D. J., Terracciano C. M., Harding S. E., and Stevens M. M., Adv. Funct. Mater. 28, 1800618 (2018). 10.1002/adfm.201800618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Soman P., Fozdar D. Y., Lee J. W., Phadke A., Varghese S., and Chen S., Soft Matter 8, 4946 (2012). 10.1039/c2sm07354d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bückmann T., Stenger N., Kadic M., Kaschke J., Frölich A., Kennerknecht T., Eberl C., Thiel M., and Wegener M., Adv. Mater. 24, 2710 (2012). 10.1002/adma.201200584 [DOI] [PubMed] [Google Scholar]
- 64. Guvendiren M., Molde J., Soares R. M. D., and Kohn J., ACS Biomater. Sci. Eng. 2, 1679 (2016). 10.1021/acsbiomaterials.6b00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chan B. P. and Leong K. W., Eur. Spine J. 17, 467 (2008). 10.1007/s00586-008-0745-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kolesky D. B., Truby R. L., Gladman A. S., Busbee T. A., Homan K. A., and Lewis J. A., Adv. Mater. 26, 3124 (2014). 10.1002/adma.201305506 [DOI] [PubMed] [Google Scholar]
- 67. Pati F., Jang J., Ha D.-H., Won Kim S., Rhie J.-W., Shim J.-H., Kim D.-H., and Cho D.-W., Nat. Commun. 5, 3935 (2014). 10.1038/ncomms4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Colosi C., Shin S. R., Manoharan V., Massa S., Costantini M., Barbetta A., Dokmeci M. R., Dentini M., and Khademhosseini A., Adv. Mater. 28, 677 (2016). 10.1002/adma.201503310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hardin J. O., Ober T. J., Valentine A. D., and Lewis J. A., Adv. Mater. 27, 3279 (2015). 10.1002/adma.201500222 [DOI] [PubMed] [Google Scholar]
- 70. Ober T. J., Foresti D., and Lewis J. A., Proc. Natl. Acad. Sci. U. S. A. 112, 12293 (2015). 10.1073/pnas.1509224112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu W., Zhang Y. S., Heinrich M. A., De Ferrari F., Jang H. L., Bakht S. M., Alvarez M. M., Yang J., Li Y.-C., Trujillo-de Santiago G., Miri A. K., Zhu K., Khoshakhlagh P., Prakash G., Cheng H., Guan X., Zhong Z., Ju J., Zhu G. H., Jin X., Shin S. R., Dokmeci M. R., and Khademhosseini A., Adv. Mater. 29, 1604630 (2017). 10.1002/adma.201604630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Thoniyot P., Tan M. J., Karim A. A., Young D. J., and Loh X. J., Adv. Sci. 2, 1400010 (2015). 10.1002/advs.201400010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhu K., Shin S. R., van Kempen T., Li Y.-C., Ponraj V., Nasajpour A., Mandla S., Hu N., Liu X., Leijten J., Lin Y.-D., Hussain M. A., Zhang Y. S., Tamayol A., and Khademhosseini A., Adv. Funct. Mater. 27, 1605352 (2017). 10.1002/adfm.201605352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jakus A. E., Secor E. B., Rutz A. L., Jordan S. W., Hersam M. C., and Shah R. N., ACS Nano 9, 4636 (2015). 10.1021/acsnano.5b01179 [DOI] [PubMed] [Google Scholar]
- 75. Zhu W., Li J., Leong Y. J., Rozen I., Qu X., Dong R., Wu Z., Gao W., Chung P. H., Wang J., and Chen S., Adv. Mater. 27, 4411 (2015). 10.1002/adma.201501372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gou M., Qu X., Zhu W., Xiang M., Yang J., Zhang K., Wei Y., and Chen S., Nat. Commun. 5, 3774 (2014). 10.1038/ncomms4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jeon O., Alt D. S., Linderman S. W., and Alsberg E., Adv. Mater. 25, 6366 (2013). 10.1002/adma.201302364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Singh M., Berkland C., and Detamore M. S., Tissue Eng. B 14, 341 (2008). 10.1089/ten.teb.2008.0304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ilkhanizadeh S., Teixeira A. I., and Hermanson O., Biomaterials 28, 3936 (2007). 10.1016/j.biomaterials.2007.05.018 [DOI] [PubMed] [Google Scholar]
- 80. Gao F., Xu Z., Liang Q., Liu B., Li H., Wu Y., Zhang Y., Lin Z., Wu M., Ruan C., and Liu W., Adv. Funct. Mater. 28, 1706644 (2018). 10.1002/adfm.201706644 [DOI] [Google Scholar]
- 81. Thorne J. T., Segal T. R., Chang S., Jorge S., Segars J. H., and Leppert P. C., Biol. Reprod. 92, 25 (2015). 10.1095/biolreprod.114.121368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xu R., Boudreau A., and Bissell M. J., Cancer Metastasis Rev. 28, 167 (2009). 10.1007/s10555-008-9178-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kolesky D. B., Homan K. A., Skylar-Scott M. A., and Lewis J. A., Proc. Natl. Acad. Sci. U. S. A. 113, 3179 (2016). 10.1073/pnas.1521342113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Homan K. A., Kolesky D. B., Skylar-Scott M. A., Herrmann J., Obuobi H., Moisan A., and Lewis J. A., Sci. Rep. 6, 34845 (2016). 10.1038/srep34845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lee J. B., Wang X., Faley S., Baer B., Balikov D. A., Sung H.-J., and Bellan L. M., Adv. Healthcare Mater. 5, 781 (2016). 10.1002/adhm.201500792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bégin-Drolet A., Dussault M. A., Fernandez S. A., Larose-Dutil J., Leask R. L., Hoesli C. A., and Ruel J., Addit. Manuf. 15, 29 (2017). 10.1016/j.addma.2017.03.006 [DOI] [Google Scholar]
- 87. Wells R. G., Hepatology 47, 1394 (2008). 10.1002/hep.22193 [DOI] [PubMed] [Google Scholar]
- 88. Chang R., Nam J., and Sun W., Tissue Eng. A 14, 41 (2008). 10.1089/ten.a.2007.0004z [DOI] [PubMed] [Google Scholar]
- 89. Kawabata K., Sugihara K., Sanoh S., Kitamura S., and Ohta S., J. Toxicol. Sci. 38, 215 (2013). 10.2131/jts.38.215 [DOI] [PubMed] [Google Scholar]
- 90. Mironi-Harpaz I., Wang D. Y., Venkatraman S., and Seliktar D., Acta Biomater. 8, 1838 (2012). 10.1016/j.actbio.2011.12.034 [DOI] [PubMed] [Google Scholar]
- 91. Wang Z., Abdulla R., Parker B., Samanipour R., Ghosh S., and Kim K., Biofabrication 7, 045009 (2015). 10.1088/1758-5090/7/4/045009 [DOI] [PubMed] [Google Scholar]
- 92. Xin T., Greco V., and Myung P., Cell 164, 1212 (2016). 10.1016/j.cell.2016.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Miki Y., Ono K., Hata S., Suzuki T., Kumamoto H., and Sasano H., J. Steroid Biochem. Mol. Biol. 131, 68 (2012). 10.1016/j.jsbmb.2011.12.004 [DOI] [PubMed] [Google Scholar]
- 94. Saini H., Navaei A., Van Putten A., and Nikkhah M., Adv. Healthcare Mater. 4, 1961 (2015). 10.1002/adhm.201500331 [DOI] [PubMed] [Google Scholar]
- 95. Kamkin A., Kiseleva I., Isenberg G., Wagner K. D., Günther J., Theres H., and Scholz H., Prog. Biophys. Mol. Biol. 82, 111–120 (2003). 10.1016/S0079-6107(03)00009-9 [DOI] [PubMed] [Google Scholar]
- 96. Eyckmans J., Boudou T., Yu X., and Chen C. S., Dev. Cell 21, 35 (2011). 10.1016/j.devcel.2011.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Aubin H., Nichol J. W., Hutson C. B., Bae H., Sieminski A. L., Cropek D. M., Akhyari P., and Khademhosseini A., Biomaterials 31, 6941 (2010). 10.1016/j.biomaterials.2010.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Qu X., Zhu W., Huang S., Li Y.-S., Chien S., Zhang K., and Chen S., Biomaterials 34, 9812 (2013). 10.1016/j.biomaterials.2013.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mozetic P., Giannitelli S. M., Gori M., Trombetta M., and Rainer A., J. Biomed. Mater. Res. A 105, 2582 (2017). 10.1002/jbm.a.36117 [DOI] [PubMed] [Google Scholar]
- 100. Bhuthalingam R., Lim P. Q., Irvine S., Agrawal A., Mhaisalkar P., An J., Chua C. K., and Venkatraman S., Int. J. Bioprinting 1, 57 (2015). 10.18063/IJB.2015.01.008 [DOI] [Google Scholar]
- 101. Clevers H., Lancaster M., and Takebe T., Cell Stem Cell 20, 759 (2017). 10.1016/j.stem.2017.05.014 [DOI] [Google Scholar]
- 102. Davies J. A. and Lawrence M. L., in Organs and Organoids (Elsevier, 2009), 255–259. [Google Scholar]
- 103. Skardal A., Murphy S. V., Devarasetty M., Mead I., Kang H.-W., Seol Y.-J., Shrike Zhang Y., Shin S.-R., Zhao L., Aleman J., Hall A. R., Shupe T. D., Kleensang A., Dokmeci M. R., Jin Lee S., Jackson J. D., Yoo J. J., Hartung T., Khademhosseini A., Soker S., Bishop C. E., and Atala A., Sci. Rep. 7, 8837 (2017). 10.1038/s41598-017-08879-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Weigelt B., Ghajar C. M., and Bissell M. J., Adv. Drug Deliv. Rev. 69–70, 42 (2014). 10.1016/j.addr.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Faulkner-Jones A., Fyfe C., Cornelissen D.-J., Gardner J., King J., Courtney A., and Shu W., Biofabrication 7, 044102 (2015). 10.1088/1758-5090/7/4/044102 [DOI] [PubMed] [Google Scholar]
- 106. Lee J. W., Choi Y.-J., Yong W.-J., Pati F., Shim J.-H., Kang K. S., Kang I.-H., Park J., and Cho D.-W., Biofabrication 8, 015007 (2016). 10.1088/1758-5090/8/1/015007 [DOI] [PubMed] [Google Scholar]
- 107. Lee H., Han W., Kim H., Ha D.-H., Jang J., Kim B. S., and Cho D.-W., Biomacromolecules 18, 1229 (2017). 10.1021/acs.biomac.6b01908 [DOI] [PubMed] [Google Scholar]
- 108. Kuo C.-Y., Eranki A., Placone J. K., Rhodes K. R., Aranda-Espinoza H., Fernandes R., Fisher J. P., and Kim P. C. W., ACS Biomater. Sci. Eng. 2, 1817 (2016). 10.1021/acsbiomaterials.6b00031 [DOI] [PubMed] [Google Scholar]
- 109. Lind J. U., Busbee T. A., Valentine A. D., Pasqualini F. S., Yuan H., Yadid M., Park S.-J., Kotikian A., Nesmith A. P., Campbell P. H., Vlassak J. J., Lewis J. A., and Parker K. K., Nat. Mater. 16, 303 (2017). 10.1038/nmat4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jang J., Park H.-J., Kim S.-W., Kim H., Park J. Y., Na S. J., Kim H. J., Park M. N., Choi S. H., Park S. H., Kim S. W., Kwon S.-M., Kim P.-J., and Cho D.-W., Biomaterials 112, 264 (2017). 10.1016/j.biomaterials.2016.10.026 [DOI] [PubMed] [Google Scholar]
- 111. Gaetani R., Feyen D. A. M., Verhage V., Slaats R., Messina E., Christman K. L., Giacomello A., Doevendans P. A. F. M., and Sluijter J. P. G., Biomaterials 61, 339 (2015). 10.1016/j.biomaterials.2015.05.005 [DOI] [PubMed] [Google Scholar]
- 112. Duan B., Hockaday L. A., Kang K. H., and Butcher J. T., J. Biomed. Mater. Res. A 101A, 1255 (2013). 10.1002/jbm.a.34420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Shim J.-H., Jang K.-M., Hahn S. K., Park J. Y., Jung H., Oh K., Park K. M., Yeom J., Park S. H., Kim S. W., Wang J. H., Kim K., and Cho D.-W., Biofabrication 8, 014102 (2016). 10.1088/1758-5090/8/1/014102 [DOI] [PubMed] [Google Scholar]
- 114. Nguyen D., Hägg D. A., Forsman A., Ekholm J., Nimkingratana P., Brantsing C., Kalogeropoulos T., Zaunz S., Concaro S., Brittberg M., Lindahl A., Gatenholm P., Enejder A., and Simonsson S., Sci. Rep. 7, 658 (2017). 10.1038/s41598-017-00690-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Tseng H., Gage J. A., Haisler W. L., Neeley S. K., Shen T., Hebel C., Barthlow H. G., Wagoner M., and Souza G. R., Sci. Rep. 6, 30640 (2016). 10.1038/srep30640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Mirabella T., MacArthur J. W., Cheng D., Ozaki C. K., Woo Y. J., Yang M. T., and Chen C. S., Nat. Biomed. Eng. 1, 0083 (2017). 10.1038/s41551-017-0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Souza G. R., Tseng H., Gage J. A., Mani A., Desai P., Leonard F., Liao A., Longo M., Refuerzo J. S., and Godin B., Int. J. Mol. Sci. 18, 683 (2017). 10.3390/ijms18040683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Pati F., Ha D.-H., Jang J., Han H. H., Rhie J.-W., and Cho D.-W., Biomaterials 62, 164 (2015). 10.1016/j.biomaterials.2015.05.043 [DOI] [PubMed] [Google Scholar]
- 119. Laronda M. M., Rutz A. L., Xiao S., Whelan K. A., Duncan F. E., Roth E. W., Woodruff T. K., and Shah R. N., Nat. Commun. 8, 15261 (2017). 10.1038/ncomms15261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wu Z., Su X., Xu Y., Kong B., Sun W., and Mi S., Sci. Rep. 6, 24474 (2016). 10.1038/srep24474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Farina M., Ballerini A., Fraga D. W., Nicolov E., Hogan M., Demarchi D., Scaglione F., Sabek O. M., Horner P., Thekkedath U., Gaber O. A., and Grattoni A., Biotechnol. J. 12, 1700169 (2017). 10.1002/biot.201700169 [DOI] [PubMed] [Google Scholar]
- 122. Muth J. T., Vogt D. M., Truby R. L., Mengüç Y., Kolesky D. B., Wood R. J., and Lewis J. A., Adv. Mater. 26, 6307 (2014). 10.1002/adma.201400334 [DOI] [PubMed] [Google Scholar]
- 123. McConnell A. C., Vallejo M., Moioli R. C., Brasil F. L., Secciani N., Nemitz M. P., Riquart C. P., Corne D. W., Vargas P. A., and Stokes A. A., Front. Mech. Eng. 3 (2017). 10.3389/fmech.2017.00003 [DOI] [Google Scholar]
- 124. Cvetkovic C., Raman R., Chan V., Williams B. J., Tolish M., Bajaj P., Sakar M. S., Asada H. H., Saif M. T. A., and Bashir R., Proc. Natl. Acad. Sci. U. S. A. 111, 10125 (2014). 10.1073/pnas.1401577111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Park S.-J., Gazzola M., Park K. S., Park S., Di Santo V., Blevins E. L., Lind J. U., Campbell P. H., Dauth S., Capulli A. K., Pasqualini F. S., Ahn S., Cho A., Yuan H., Maoz B. M., Vijaykumar R., Choi J.-W., Deisseroth K., Lauder G. V., Mahadevan L., and Parker K. K., Science 353, 158 (2016). 10.1126/science.aaf4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Shin S. R., Migliori B., Miccoli B., Li Y., Mostafalu P., Seo J., Mandla S., Enrico A., Antona S., Sabarish R., Zheng T., Pirrami L., Zhang K., Zhang Y. S., Wan K., Demarchi D., Dokmeci M. R., and Khademhosseini A., Adv. Mater. 30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Nawroth J. C., Lee H., Feinberg A. W., Ripplinger C. M., McCain M. L., Grosberg A., Dabiri J. O., and Parker K. K., Nat. Biotechnol. 30, 792 (2012). 10.1038/nbt.2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Gul J. Z., Sajid M., Rehman M. M., Siddiqui G. U., Shah I., Kim K.-H., Lee J.-W., and Choi K. H., Sci. Technol. Adv. Mater. 19, 243 (2018). 10.1080/14686996.2018.1431862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Raman R., Cvetkovic C., Uzel S. G. M., Platt R. J., Sengupta P., Kamm R. D., and Bashir R., Proc. Natl. Acad. Sci. U. S. A. 113, 3497 (2016). 10.1073/pnas.1516139113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Chan V., Park K., Collens M. B., Kong H., Saif T. A., and Bashir R., Sci. Rep. 2, 857 (2012). 10.1038/srep00857 [DOI] [PMC free article] [PubMed] [Google Scholar]