Abstract
We report relative bioactivities of extracts prepared from a large collection of plants from three national parks in Panama. Over 181 plants were collected, taxonomically identified and their detannified dichloromethane (DCM)-methanolic extracts were used for profiling selected bioactivities. Assays were performed to evaluate the antioxidant activity of the extracts for Antioxidant Response Element (ARE) induction, total non-enzymatic antioxidant potential, anti-inflammatory and anticancer properties. The high throughput analysis of 280 extracts resulted in identification of 57.5% of the extracts that could induce ARE at one or more concentrations tested, 93.5% that harbored total antioxidant capacity, and 2.1% of the extracts that showed lung cancer cell line-specific cytotoxicity. Data from our profiling experiments indicate that a large number of extracts could be a source for further isolation and chemical identification of compounds that could serve as leads for discovery of antioxidant, anticancer and anti-inflammatory agents to prevent or treat complex diseases like cancer and neurodegenerative disorders.
Keywords: Bioprospection, Plants, Panama, Biomedicine, Screening, Cancer, Alzheimer, Anti-inflammation, Total Antioxidant Capacity, Antioxidant Response Element, Cytotoxicity, IL-2
Panama, located on the isthmus linking Central and South America, ranks fourth in plant biodiversity in North and Central America, and is considered a biodiversity “hot spot”. Many of its plants are the basis for ethnomedicine, especially by the Amerindians of Panama. In spite of this, a systematic bioprospection of Panamanian flora in funneling therapeutic leads to accelerate mainstream drug discovery efforts has been rather limited. Likewise, modern drug discovery is full of promise, yet leaves much to be desired. While the discovery of new therapeutic drug targets in the post-humangenome-sequencing era is at an all-time high, the introduction of new molecular entities (NMEs) against therapeutic targets is at an all-time low. Industry-style probe discovery has now gained unparalleled momentum in academia with ready access to institutional high-throughput screening (HTS) laboratories. Both academia and pharma are engaged in screening large compound libraries, corporate and suppliers’ databases, and virtual compound collections to identify lead drug candidates. However, the wealth of chemical diversity that has evolved with biological diversity is underrepresented in the commercial chemical library offerings, but needs to be expanded to strategically cover available chemical space and include drug-like compounds with improved pharmacologic, pharmacodynamic, and pharmacokinetic properties as compared to their current nitrogen-rich counterparts. Although natural plant extracts are a treasure trove of novel chemical matter with significant therapeutic potential, application of HTS for identification of biologically active natural products has also not sufficiently progressed due to inherent difficulties associated in profiling biological samples. The University of Kansas has heavily been involved in bridging this gap, and has collaborated extensively with Professor Mahabir Gupta, a pioneering figure in the field of Panamanian ethnomedicine and the commemorative honoree of this issue.
Since native plants have been used extensively across cultures for their medicinal properties and are a rich source for novel chemical scaffold identification and as chemical probes, we profiled 280 Panamanian plant extracts for their antioxidant, anti-inflammatory and anticancer properties. Three parcels of 0.1 ha in three national parks: ParqueNacional Chagres (PNC), ParqueNacional General de Division Omar Torrijos Herrera (PNGDOTH), and ParqueNacionalSarigua (PNS) were established. Two thousand five hundred thirty-seven registers of plants distributed in 1536 (PNC), 708 (PNGDOTH) and 293 (PNS) were made. Of all the individual specimens collected in the three national parks, a total of 176 species, 230 genera, and 95 families were identified. Some plants could only be identified upto family, or genus, or species.
State of the art high throughput technologies were utilized for assaying bioactivities of all extracts. The extracts were transferred acoustically, with high precision, using Echo 550 (Labcyte Inc.) at the required concentrations directly into the assay plates. The assays were adapted for screening in miniaturized 384 well microplate formats. HTS automation technologies enabled precise and robust handling of the 280 extracts under identical conditions and very small volumes of extracts were used for all bioassays. All extracts were handled and assayed at the same time under identical experimental conditions. In comparison to manual sample processing, the high throughput analysis facilitated faster processing and identification of extract activities when dealing with a large number of samples in this bioprospecting campaign.
The de-tannified plant extracts were evaluated for their ability to induce the binding of Nrf2 transcription factor (NF-E2-related factor)-ARE signaling pathway, that regulates expression of genes encoding over 250 antioxidant and detoxification proteins, thereby protecting tissues via enhanced antioxidant capacity [1]. The extracts were added to AREc32 cell line that stably expresses luciferase under the control of regulatory cis-acting enhancer, the Antioxidant Response Element (ARE) [2]. Out of 280 extracts, 161 extracts (57.5%) induced ARE significantly to greater than 10-fold at one or more concentrations tested. Figure 1 shows the scatterplot of fold-ARE induction at 25 µgs/mL of the extracts from various plant sources. A large number of extracts showed potent 30–70 fold ARE induction activity at high extract concentrations: 38 extracts induced the AREs at 50 µgs/mL, 6 extracts at 25 µgs/mL and one at 12.5 µgs/mL. Some of the extracts that induced the ARE to greater than 50 fold at 25µgs/mL are: leaves of Aegiphila anomala Pittierand Sinclairia polyantha (Klatt) Rydb, stems of Witheringia solanacea L.Hér.and Erythroxylumsp. The extracts that induced the ARE to >50 fold at 50 µgs/mL include fruits of Cordia bicolor A.DC., stems of Erythroxylum sp., leaves of Tetrochidium sp., stems of Elaegianitidifolia Dwyer, pads of Opuntia, leaves and stems of Licaria sp., stems of Piper sancti-felicis, and leaves of Ovedaeaver besinoidesDC. There were at least two extracts that showed 26-fold ARE-induction at low concentrations of 2.5 µgs/mL: leaves of Casearia corymbosa(Kunth)and above ground biomass of Brickellia diffusa(Vahl) A. Gray. Table 1 lists fold ARE induction of top active extracts. Further fractionation of plant extracts, followed by isolation of purified active components will be critical in elucidating the mechanism of action for activation of Nrf2-ARE transcriptional pathway which plays an important role in the detoxication and elimination of reactive oxygen species and electrophiles.
Figure 1.
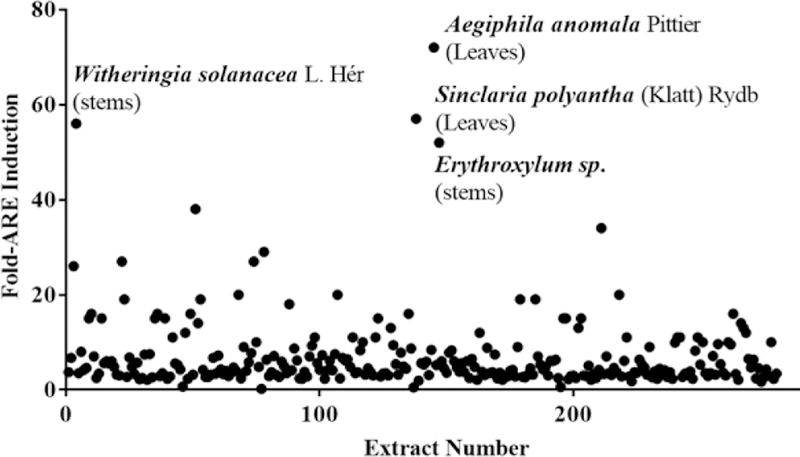
Scattergram of fold-increase in ARE activity shown by Panamanian plant extracts.
Table 1:
Fold-ARE Induction of top active plant extracts.
| Plant Species | 50 µg/mL | 25 µg/mL | 12.5 µg/mL | 6 µg/mL |
|---|---|---|---|---|
| Sinclaria polyantha (Klatt) Rydb (leaves) | 15 | 57 | 10 | 3 |
| Witheringia solanacea L.Hér (stems) | 1 | 56 | 71 | 23 |
| Aegiphila anomala Pittier (leaves) | 1 | 72 | 22 | 8 |
| Cordia bicolor A. DC. (fruits) | 77 | 15 | 3 | 2 |
| Erythroxylum sp. (stems) | 64 | 52 | 18 | 3 |
| Tetrochidium sp. (leaves) | 58 | 15 | 5 | 2 |
| Elaegia nitidifolia Dwyer (stems) | 56 | 13 | 5 | 3 |
| Opuntia sp.(pads) | 55 | 11 | 4 | 3 |
| Licaria sp. (leaves) | 54 | 15 | 4 | 2 |
| Piper sancti-felicis Trel.(stems) | 54 | 15 | 7 | 3 |
| Licaria sp. (stems) | 52 | 15 | 4 | 3 |
| Ocetea sp. (leaves) | 47 | 38 | 6 | 2 |
| Ilex sp. (stems) | 46 | 27 | 8 | 3 |
| Machaerium biovulatum Micheli (stems) | 12 | 34 | 15 | 6 |
In addition to the ARE induction activity, we also tested the extracts for total antioxidant capacity [3], which is a measure of overall extract capability to counteract reactive oxygen species, and capability to prevent oxidative damage in oxidative stress-related diseases. Table 2 lists extracts that exhibited high total antioxidant capacity. Of the 280 samples, 262 or 93.5% of the extracts showed significant total antioxidant capacity ranging from 10 to 200 fold at one or more concentrations tested. As shown in Figure 2, the following five extracts showed >30 TAC units at 12.5 µgs/mL: leaves and stems of Caesalpinia coriaria (Jacq.) Wild, leaves of Theobroma bicolor Bonpl., leaves and stems of Mimosa pigraL., stems and leaves of Ilex sp., leaves and stems of Hymenaea coubari L., stems of Licaria sp. and Andira inermis (W.Wright) Kunth. At 100 µgs/mL, very high trolox equivalent units were obtained for leaves of Ocotea sp.and stems of Licaria sp.
Table 2:
Plant extracts exhibiting high total antioxidant capacity.
| Plant Species | 100 µg/ mL | 50 µg/ mL | 25 µg/ mL | 12.5 µg/ mL |
|---|---|---|---|---|
| Ocotea sp. (leaves) | 215 | 27 | 12 | 3 |
| Malpighia sp. (stems) | 9 | 35 | 24 | 23 |
| Coupia sp. (leaves) | 21 | 67 | 21 | 8 |
| Maclura tinctoria (L.) D. Don ex Steud (leaves) | 6 | 37 | 18 | 2 |
| Myrcia tomentosa (Aubl.)Amshoff (leaves) | 36 | 29 | 18 | 3 |
| Caesalpinia coriaria (Jacq.) Wild (leaves) | 12 | 11 | 11 | 2 |
| Caesalpinia coriaria (Jacq.) Wild (stems) | 6 | 12 | 10 | 3 |
| Theobroma bicolor Bonpl. (leaves) | 7 | 11 | 10 | 3 |
| Mimosa pigra L. (leaves) | 8 | 10 | 12 | 2 |
| Ilex sp. (stems) | 10 | 10 | 11 | 3 |
| Hymenaea courbaril L. (stems) | 2 | 6 | 9 | 3 |
| Hymenaea courbaril L. (stems) | 0 | 8 | 8 | 2 |
| Mimosa pigra L. (leaves) | 8 | 10 | 10 | 3 |
| Licaria sp. (stems) | 86 | 13 | 14 | 6 |
| Miconia argentea (Sw.) DC. (leaves) | 69 | 9 | 12 | 13 |
| Licaria sp. (stems) | 4 | 5 | 4 | 42 |
| Arrabiddaea patellifera (Schltdl.) Sandwith (leaves) | 60 | 5 | 12 | 9 |
| Aegiphyla anomala Pittier (leaves) | 50 | 4 | 4 | 9 |
| Calophyllum longifolium Wild (leaves) | 45 | 12 | 11 | 9 |
| Stemmadenia pubescens Benth (leaves) | 32 | 1 | 3 | 9 |
Figure 2.
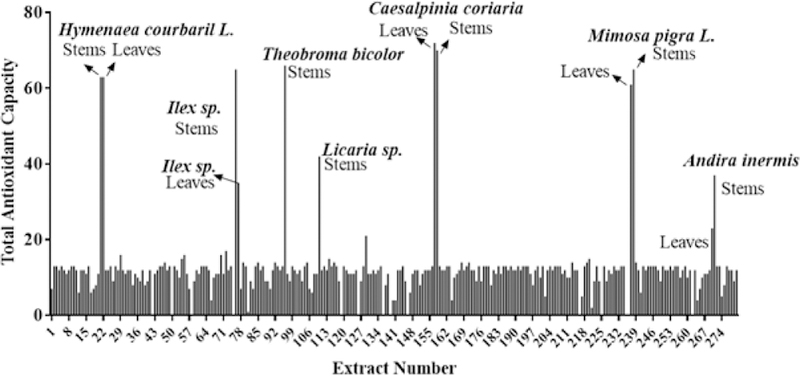
Distribution of Total antioxidant capacity of plant extracts.
In generalized immune response to foreign antigens, the proinflammatory cytokine, IL-2 mediates T-cell response to many biological stimuli (e.g. bacterial infection) and activates T-cells to proliferate and differentiate. Suppression of IL-2 activity has implications in immune suppression and preventing graft rejections. The ability of the plant extracts to inhibit secretion of IL-2 was also evaluated [4] using the Jurkat cells, an immortalized human T-cell line. Of the 280 extracts, only 8 extracts or 2.8% of the total plant extracts inhibited IL2 secreted response of phytohaemagglutinin (PHA)/phorbolmyristate acetate (PMA) stimulated Jurkat cells significantly. The eight extracts that inhibited IL2 secretion to greater than 50% at either of the two concentrations tested (Table 3) are the stems of Stemmadenia pubescens Benth, Caesalpinia bonduc(L.) Roxb.,Marcgravia nervosa Triana & Planch, Otoba novogranatense (Moldenke), leaves of Parkia nitida Miq., fruits ofCavaponiaracemosa(Mill.)Cogn., leaves of Garcinia madruno(Kunth) Hamme and stems of Pithecellobium dulce(Roxb.)Benth. The IL-2 inhibitory activity of the extracts was not associated with direct cytotoxicity of the extracts to Jurkat cells.
Table 3:
Panamanian plant extracts inhibiting IL2 production in Jurkat cells activated with PMA/PHA.
| Plant Species | % Inhibition 25 µgs/mL | % Inhibition 12.5 µgs/mL |
|---|---|---|
| Stemmadenia pubescens Benth (stems) | 86 | 81 |
| Caesalpinia bonduc (L.) Roxb.(stems) | 50 | 67 |
| Marcgravia nervosa Triana (stems) | 25 | 56 |
| Otoba novogranatense (stems) | 31 | 55 |
| Garcinia madruno (Kunth) Hammel (leaves) | 53 | 46 |
| Pithecellobium dulce (Roxb.)Benth.(stems) | 53 | 43 |
| Cavaponia racemosa (Mill.)Cogn.(fruits) | 56 | 38 |
| Parkia nitida Miq.(leaves) | 59 | 28 |
The human lung fibroblast MRC-5 and the human A549 alveolar epithelial cell lines were used to represent normal lung and lung cancers respectively. To identify the extracts that showed cytotoxicity against A549 cell line and not the normal MRC-5 lung fibroblasts, all extracts were added to the cells in an eight-concentration dose-response assay for 72h [5].Six extracts that were found to be efficacious in preferentially inhibiting the growth of lung cancer cell line (EC50< 10 µM) and not of the normal lung fibroblasts are: leaves of Aegiphila anomalaPittier and stems of Guarea rhopalocarpa Radlk., stems of Hortia colombiana Gleason , leaves and stems of Matisia cf. Exalata W.S. Alverson, and roots of Erythrina fuscaLour. The cytotoxicity profiles of the top six extracts against the lung cancer and control cell line models i s shown in Figure 3. The EC50 values for the cytotoxicity of the extracts is shown in Table 4. Data is not shown for another 106 extracts that were relatively less potent (EC50 ~12–75 µM) but still showed selective killing of the lung cancer cells and did not s how any activity against the MRC-5 cell line. More than 156 extract s were equally cytotoxic against both cell lines over 72h of exposure.
Figure 3.
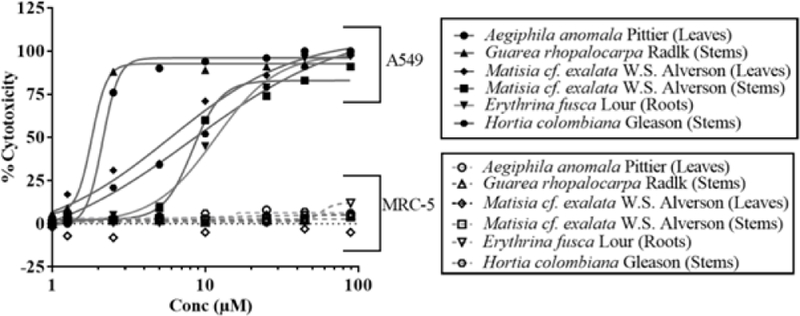
Non-linear regression analysis of dose-responsive cytotoxicity of most active extracts against lung cancer cell line A549. The representative extracts did not show any activity against the control MRC-5 cell line.
Table 4:
Cytotoxicity profile of Panamanian plant extracts preferentially to A549 cell line.
| Plant Species | A549 EC50 (µM) | MRC5 EC50 (µM) |
|---|---|---|
| Aegiphila anomala Pittier (leaves) | 1.9 | No Inhibition |
| Guarea rhopalocarpa Radlk. (stems) | 1.7 | No Inhibition |
| Matisia cf. exalata W.S. Alverson (leaves) | 5.2 | No Inhibition |
| Matisia cf. exalata W.S. Alverson (stems) | 8.2 | No Inhibition |
| Erythrina fusca Lour (roots) | 8 | No Inhibition |
| Hortia colombiana Gleason (stems) | 8.4 | No Inhibition |
| Erythrina fusca Lour (roots) | 10 | No Inhibition |
| Castilla elastica Sesse ex Cerv. (stems) | 0.9 | 45 |
| Mosquitoxylum jamaicense Krug & Urb (stems) | 2.9 | 32 |
In summary, the high throughput analysis of 280 extracts from selected Panamanian plants resulted in the identification of 161 extracts (57.5%) that significantly induced ARE at 1 or more concentrations tested. A majority of the extracts (93.5%) harbored total antioxidant capacity, and a small percentage of the extracts (2.1%) exhibited lung cancer cell line-specific cytotoxicity. These results indicate that a large number of extracts could be a source for further isolation and chemical identification of compounds with significant therapeutic potential, especially as leads for the discovery of antioxidant, anticancer and anti-inflammatory properties.
Experimental
Sample Preparation and Plant Material:
All Panamanian plant materials were collected and authenticated by Alex Espimosa and Carlos Guerra and identified by Prof Mireya Correa, Emerirus Professor of Botany at the University of Panama. The voucher specimens are deposited at the Herbarium of the University of Panama (PMA). Plant materials (whole plants and plant organs e.g fruits, stems, leaves, and roots) were air dried under shade and pulverized in a Wiley Mill to obtain a powder of 40 mesh. The pulverized powders were extracted for 72 h at room temperature in a 1:1 dichloromethane (DCM)-methanol solvent (Fisher Scientific Co., NJ). Since tannins are known to interefere with the several enzyme activities, we did not test the un-detannified extracts in the biochemical assays. All extracts were detannified according to the method of Wall et al. (6). The 280 extracts derived from 181 plants were solubilized in 100% DMSO and stored at −800C at 10 and 20 mg/mL concentrations and utilized for bioassays. DMSO vehicle was used as a control in all the assays.
Cell Growth and Maintenance:
The cell lines used for all cell- based assays were grown at 37°C in the presence of 5% CO2. The cell lines and the media used for growth and maintenance is as follows: (1) AREc32 (CRX Biosciences, Scotland, UK) in Dulbecco’s Modified Eagle’s medium (DMEM), GlutaMAX, 10% FBS and G418 (ThermoFisher Scientific, Waltham, MA, USA). (2) Jurkat cells (ATCC # TIB-152 lymphoblast Jurkat Clone E6–1) in RPMI-1640 Medium (ATCC, Manassas, VA) and 10% FBS. (3) A549 (ATCC # CCL-185 lung carcinoma) in F-12K Medium and 10% FBS. (4) MRC-5 (ATCC # CCL-171 lung fibroblast) in Eagle’s Minimum Essential Medium and 10% FBS. For assays, the cells were seeded into 384-well plates using a Wellmate bulk dispenser (Thermo Fisher Scientific, Waltham, MA) in relevant complete media.
Antioxidant Response Element (ARE) Activation:
AREc32 cells were exposed to library compounds for 24 hrs, followed by the addition of Steady-Glo luciferase assay reagent (Promega, Madison, WI). The luminescence was read 30 min later on a Tecan Safire2 microplate reader (Mannedorf, Switzerland). The two known ARE activators tBHQ and CDDO-Im were used as positive controls and the cells in media containing 0.35% DMSO served as uninduced ARE control. Extracts that exhibited ARE fold induction to 10-fold above DMSO vehicle controls in one or more concentrations were considered to have significant ARE induction activity.
Total Antioxidant Capacity:
Plant extracts were assayed for combined nonenzymatic antioxidant capacity using the Total Antioxidant Capacity assay (K274–100, BioVision, CA) following the manufacturer’s instructions. In brief, an equal volume (12.5 µl) of Cu2+ ion probe working solution was added to the protein extracts in 384 well plates and incubated at room temperature for 1.5h before reading the absorbance at 570nm in a microplate reader (Tecan Safire2). Absorbance values were used to obtain the total antioxidant capacity relative to Trolox control. Extracts that exhibited TAC values of greater than 5 trolox units at any one concentration tested was considered significant. The data was normalized to DMSO vehicle controls in the assay.
Cytotoxicity Assay:
A549 and MRC-5 cells were exposed to the plant extracts for 72 hours at 37°C, 5% CO2 in a 95% humidified incubator followed by the addition of the CellTiter-Glo Luminescent assay reagent (Promega, Madison, WI). The plates were shaken for 2 minutes at 1000 rpm and luminescence was read 20 min later on a Tecan Safire2 microplate reader (Mannedorf, Switzerland). Percent cytotoxicity was normalized to DMSO and no cell controls. Non-linear regression analysis of the dose-response data was performed using GraphPad Prizm and EC50 or concentrations for 50% cytotoxicity was reported for each extract. The extracts that exhibited EC50 values of less than 10 uM in A549 cell line and also were not cytotoxic (no cytoxicity or >100 fold cytotoxicity window) to the control cell line, MRC-5 were considered significant.
IL-2 Cytokine Inhibition:
The pro-inflammatory cytokine IL-2 was evaluated in Jurkat cells (ATCC TIB-152 Jurkat, Clone E6–1), an immortalized T lymphocyte cell line. The cells were stimulated with PHA (20 ngs/mL) a plant lectin and mitogen, and PMA (2.5µgs/mL), a tumor promoter from the croton plant. The 280 plant extracts were added to the stimulated cells (60,000 cells/well) for 24 hours, after which IL-2 production (pgs/mL) was measured using the using the DuoSet ELISA (R&D Systems) kits. The cells left untreated with PMA or PHA served as a negative control. The percent inhibition data was normalized to PMA/PHA treated DMSO vehicle controls and the untreated negative controls in the assay. The extracts that inhibited IL-2 production to >50% at at least one concentration tested was considered significant.
Acknowledgments
MPG acknowledges National Secretariat of Science, Technologogy, and Innovation of Panama for the grant No. COL-06–24 and National System of Investigators (SNI). The KUHTS laboratory is supported in part by NIGMS Chemical Biology of Infectious Disease grant [P20GM113117 03] and the NCI Cancer Center Support grant P30 CA16852407. BT acknowledges the partial support by grants from NIH Center for Cancer Experimental Therapeutics [P30GM103495] as well as IND 0061464 from the Kansas Bioscience Authority and KU 2506014–910/099 from the University of Kansas. The authors wish to acknowledge the contribution of Byron Taylor, Ashleigh Price, Juan Jose Araya, Gemma O’Donnell, Robert Gallagher and Huaping Zhang.
References
- [1].Surh YJ, Kundu JK, Na HK. (2008) Nrf2 as a master redox switch in turning on the cellular signaling involved in the induction of cytoprotective genes by some chemopreventive phytochemicals. PlantaMedica 74, 1526–1539. [DOI] [PubMed] [Google Scholar]
- [2].Wang XJ, Hayes JD, Wolf CR (2006) Generation of a stable antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of nrf2 by cancer chemotherapeutic agents. Cancer Research, 66, 10983–10994. [DOI] [PubMed] [Google Scholar]
- [3].Bartosz G (2003) Total antioxidant capacity. Advances in Clinical Chemistry, 37, 219–292. Review. [DOI] [PubMed] [Google Scholar]
- [4].Pawelec G, Borowitz A, Krammer PH, Wernet P. (1982) Constitutive interleukin 2 production by the JURKAT human leukemic T cell line. EuropeanJournal of Immunology, 12, 387–392. [DOI] [PubMed] [Google Scholar]
- [5].Farah IO, Lewis VL, Ayensu WK, Cameron JA. (2013) Assessing the survival of MRC5 and a549 cell lines upon exposure to pyruvic Acid, sodium citrate and sodium bicarbonate. Biomedical Sciences Instrumentation, 49, 109–116. [PubMed] [Google Scholar]
- [6].Wall ME, Wani MC, Brown DM, Fullas F, Olwald JB, Josephson FF, Thornton NM, Pezzuto JM, Beecher CW, Farnsworth NR, Cordell GA, Kinghorn AD. (1996) Effect of tannins on screening of plant extracts for enzyme inhibitory activity and techniques for their removal. Phytomedicine, 3, 281–285. [DOI] [PubMed] [Google Scholar]


