Abstract
Previous studies have demonstrated that STAT5 is critical for expression of granulysin and antimicrobial activity. Because the signaling pathway and the resultant microbicidal activity are defective in HIV-infected patients, the mechanism by which STAT5 leads to granulysin expression is of great interest. In the current study, IL-2–stimulated CRL-2105 CD4+ T cells expressed granulysin and killed Cryptococcus neoformans similar to primary CD4+ T cells. The enhancer activity of the upstream element of the granulysin promoter was analyzed in primary CD4+ T cells and CRL-2105 T cells with a luciferase reporter assay, and a STAT5 binding site, 18,302 to 18,177 bp upstream of the transcription start site, was identified as an enhancer. Additionally, the enhancer functioned in the context of heterologous SV40 promoter irrespective of its transcriptional orientation. Chromatin immunoprecipitation and EMSAs demonstrated that the enhancer element bound STAT5 both in vivo and in vitro, and mutation of the STAT5 binding site abrogated its enhancer activity. Furthermore, overexpression of a dominant negative STAT5a abolished the enhancer activity of the STAT5 binding site and abrogated the anticryptococcal activity of IL-2–stimulated primary CD4+ T cells. Taken together, these data provide details about the complex regulation leading to granulysin expression and anticryptococcal activity in primary CD4+ T cells.
Granulysin is a cytolytic and proinflammatory molecule expressed by activated human CTLs and NK cells (1). The human granulysin cDNA was originally isolated by subtractive hybridization in a search for T cell-specific molecules expressed “late” (3–5 d) after activation (2). Two major protein products of granulysin, 15 and 9 kDa, are detected in CTL and NK cells (3). It is apparent that the 9-kDa form is processed from the larger, precursor 15-kDa form (3), and recombinant 9-kDa granulysin disrupts artificial liposomes and cell membranes, damages mitochondria, and activates caspase 9 to induce apoptosis (4,5). The 9-kDa granulysin exhibits potent cytotoxic activity against a variety of tumors (6–8) and microbes (9, 10), including Cryptococcus neoformans (11, 12), Mycobacterium tuberculosis (13), and Plasmodium falciparum (14).
Granulysin is constitutively expressed in primary NK cells (15) and IL-2–dependent, Ag-driven T cell lines (2). By contrast, resting human peripheral blood T cells show a very low level of granulysin expression that is induced after prolonged activation (5–7 d) with mitogen, cognate Ag, CD40 ligation, or cytokine activation with IL-2 or IL-15 (2, 16–18). However, despite the importance of granulysin, the complex regulatory mechanism underlying granulysin expression in T cells or NK cells has not been well addressed. IL-2, a growth factor for Ag-stimulated T lymphocytes, is a member of the common γ chain (γc) receptor cytokine family that includes IL-4, IL-7, IL-9, IL-15, and IL-21. Within this cytokine family, IL-2, IL-15, and IL-21 promote granulysin and cytotoxic activity of CTLs (19, 20). We have demonstrated that IL-2 and IL-15 regulate the granulysin-dependent killing of the fungus C. neoformans by human peripheral blood CD4+ and CD8+ T cells (12, 18). The role of γc was extended by the observation that IL-21 and IL-15 could activate granulysin expression within CD8+ CD45RO+ T cells (21). Whereas γc cytokines share use of the MAPK, Jak/STAT, or the PI3K/AKT pathways for signal transduction, the canonical pathway involves Jak1/3 and STAT3/5 (22). We have demonstrated that granulysin production by IL-2–treated CD4+ T cells is dependent on activation of the Jak3 and STAT5 signaling pathways (23). Moreover, defects in this pathway lead to impaired anticryptococcal activity in HIV-infected patients (12). Additionally, granulysin induction occurred in CD8+ T cells via Jak/STAT-dependent signaling pathways after activation with IL-15 or IL-21 (21). Therefore, understanding the conserved and divergent pathways for regulation of granulysin in CD4+ and CD8+ T cells is critical to promoting this antimicrobial cell-mediated immune response.
It was previously demonstrated that Acholeplasma laidlawii could increase granulysin expression via the transcription factor AP-1 in a human monocytic cell line, THP-1 (24). However, the genetic regulation of granulysin expression, especially in human CTLs, is still elusive. Previous studies have demonstrated that IL-2 activated human peripheral blood CD4+ T cell signals via Jak3/STAT5, downstream of the IL-2/IL-2R signaling pathway, leading to production of granulysin (12, 23). Because the signaling pathway and the resultant microbicidal activity is defective in HIV-infected patients, the mechanism by which STAT5 leads to granulysin expression is of great interest. In this study, a functional analysis of the enhancer activity of upstream elements of the human granulysin promoter was performed, and evidence for an important role of a STAT5 binding site in increasing granulysin gene expression and microbicidal activities in primary or CRL-2105 CD4+ T cells are presented.
Materials and Methods
Cell lines and Abs
CRL-2105 cells (American Type Culture Collection, Manassas, VA), a human CD4+ T cell line, and YT cells (11), a human NK cell line, were cultured in complete medium (RPMI 1640 supplemented with 10% FCS, 100 U/ml penicillin, and 100 mg/ml streptomycin), and were stimulated with IL-2 at a concentration of 100 U/ml (R&D Systems, Minneapolis, MN) unless otherwise specified. The polyclonal anti-granulysin Ab 519/GST rabbit serum has been previously described (3), and the mouse monoclonal anti-human GAPDH Ab and an anti-STAT5 Ab were purchased from Chemicon International (Temecula, CA) and Santa Cruz Bio-technology (Santa Cruz, CA), respectively.
Primary CD4+ T cell isolation and activation
Human peripheral heparinized blood samples were collected by venipuncture from healthy young adults. PBMCs were prepared immediately after collection as previously described (12, 23). The isolation and culture of primary CD4+ T cells was also performed as previously described (12, 23). The primary CD4+ T cells were stimulated with IL-2 (100 U/ml; R&D Systems) for 7 d unless otherwise specified. All of the study subjects participated voluntarily and gave informed consent, and human research was conducted in accordance with the approval of the Board of Medical Ethics at the Wuhan Institute of Virology, in accordance with the Declaration of Helsinki.
Western blot analysis
After IL-2 stimulation of CRL-2105 T cells or primary CD4+ T cells, cold washing medium (PBS, 1% NaF, 0.5% Na3VO4, 10% sodium pyrophosphate) was added to terminate stimulation, and cells were centrifuged to generate a cell pellet that was lysed with lysis buffer (50 mM Tris [pH 6.8], 1% SDS, 0.025% bromophenol blue, 10% glycerol, 20 mM DTT) supplemented with a mixture of protease and phosphatase inhibitors (Calbiochem, Gibbstown, NJ). Western blot analysis of granulysin expression was performed as previously described (12).
Semiquantitative RT-PCR
The semiquantitative RT-PCR analysis for granulysin and GAPDH was performed as previously described (12).
The primers for granulysin (NM_012483) or GAPDH were described previously (12).
Anticryptococcal activity of CD4+ T cells
C. neoformans culture was performed as previously described (25). C. neoformans CAP67 (ATCC 52817) (2 × 103/well [200 μl]) was incubated with or without 5 × 105 CRL-2105 T cells or primary CD4+ T cells (E:T ratio of 250:1 unless otherwise specified). The number of CFU of C. neoformans per well was determined at 0, 24, or 48 h as previously described (12).
Luciferase reporter construction
The upstream element (−19,802 to −1,181 bp) of the human granulysin promoter was digested from a 186-kb human bacterial artificial chromosome clone RP11–439L14 with BglII and BstBI sites (26). The initial reporter gene construct pGL3−19,802/+62 was assembled by cloning the digested fragment into the promoterless pGL3 basic reporter vector (Promega, Madison, WI), in which the human granulysin promoter and translation initiation site (−1,181/+62) had been fused to the firefly luciferase gene (24). The progressive and internal deletion constructs (Fig. 2A) were created by corresponding restriction enzyme treatment of the fragment (−19,802 to −1,181 bp), followed by Klenow DNA polymerase (NEB, Beverly, MA) treatment, gel purification, and re-ligation. All constructs were transformed by electroporation into DH10B cells (Stratagene, La Jolla, CA). The analysis of the upstream enhancer (Fig. 2B) also involved the cloning of PCR fragments, all of which were verified by sequencing both strands of the enhancer. Mutations of the STAT5 binding site were introduced using the QuikChange site-directed mutagenesis kit (Stratagene). After verification of the sequences on both strands of the enhancers, they were recloned into a fresh vector to eliminate second-site mutations outside of the sequenced DNA. The dominant negative STAT5a expression vector was generated similarly (27).
FIGURE 2.
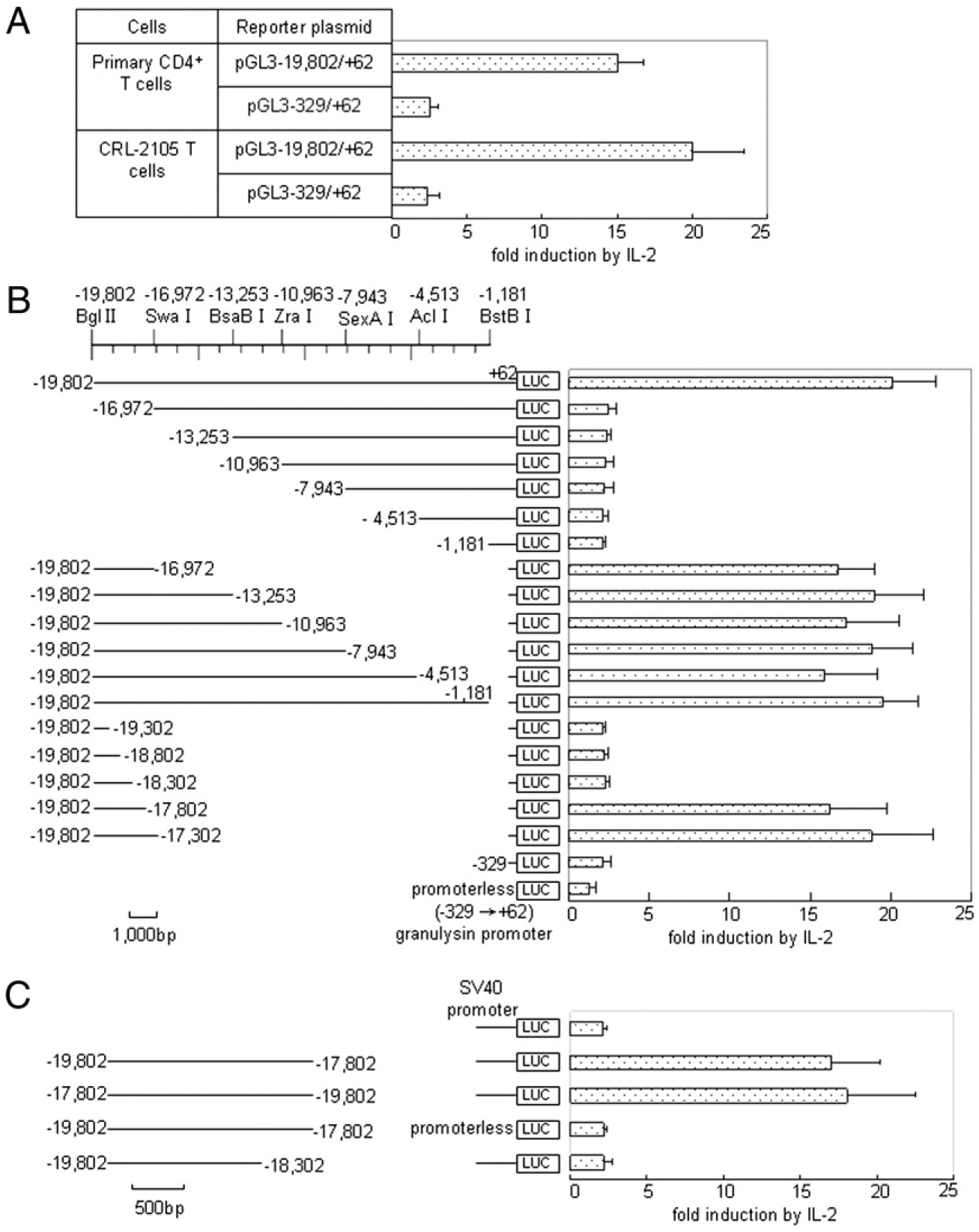
Identification and characterization of IL-2–responsive regulatory elements in the human granulysin 5′ flank. A, Transcriptional activity analysis in human primary CD4+ T cells. Transient transfection analysis of the pGL3−19,802/+62 or pGL3−329/+62 in primary CD4+ T cells was performed as described for B. B, Transient transfection analysis of granulysin 5′ (upper) and 3′ (lower) progressive flanking deletions. The genomic DNAs depicted on the left were used to control firefly luciferase expression in CRL-2105 T cells that were split 2 h after electroporation and cultured overnight for 16 h with and without IL-2. The data are given as the ratio of reporter activity of activated versus unactivated cells. DNA (2 μg) was used, comprising a 10:1 molar excess of the experimental plasmid over an internal control vector pRL–TK. The TK promoter-driven Renilla luciferase expression, which did not respond significantly to IL-2 in CRL-2105 T cells, served to correct for sample differences. In the absence of IL-2, the normalized expression of all granulysin promoter-bearing constructs was 10–40 times the levels obtained from the promoterless construct, with no reproducible correlation to any particular construct. C, Further characterization of the far-upstream enhancer of granulysin. Transient transfection analysis of the depicted DNAs in the context of the heterologous SV40 promoter was performed as described for B. Results are expressed as mean ± SEM. Data are representative of three independent experiments.
Transient transfection and luciferase assay
CRL-2105 T cells (6 ×105 cells) were transiently transfected using the Lipofectamine 2000 plus reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions, and primary CD4+ T cells were mixed with each plasmid and electroporated as previously described (12). To normalize the different transfection efficiencies of various luciferase constructs, the pRL–TK plasmid containing the Renilla luciferase gene was cotransfected into the cells in a molar ratio as indicated. After 16 h, transfected cells were stimulated with or without 100 U/ml IL-2. Luciferase activity was assayed after 24 h incubation. Cell extracts were prepared by lysing the cells with freshly diluted 1× lysis buffer (Promega) and assayed for firefly luciferase activity and Renilla luciferase activity with the Dual-Luciferase Reporter Assay System (Promega), according to the manufacturer’s manuals. Normalized reporter activity is expressed as the firefly luciferase value divided by the Renilla luciferase value. Relative fold induction is calculated as the normalized reporter activity of the test sample divided by the untreated pGL3−329/+62.
Electroporation conditions for transient transfection were also used for large m.w. DNA plasmids. After the electroporated cells were washed, they were split into two 500-μl aliquots, rested for 2 h, and then supplemented with 500 ml medium with or without 100 U/ml IL-2. After overnight incubation for 16 h, the cells were washed with PBS and lysed and extracted in 50 μl passive lysis buffer (Promega) by two rounds of freeze-thawing. Reporter gene activities were determined in three 10-μl aliquots of each extract using the Dual-Luciferase Reporter Assay System (Promega). Signals were integrated for 10 s for both luciferase activities. Their average was used to represent the analysis of each independently transfected sample.
Electrophoretic mobility shift assay
Nuclear extracts were prepared by a modified method as previously described (28). In brief, cells were washed once with ice-cold PBS and once with buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 1 mM Na3VO4, 25 mM NaF, 10 mM Na-pyrophosphate, and 25 mM p-nitrophenyl guanidinobenzoate) and lysed in buffer A containing0.05% Nonidet P-40. Lysate was placed on ice for 10 min and centrifuged at 4000 × g at 4°C for 4 min to remove cytoplasmic proteins. Nuclear proteins were extracted from the pellet in high levels of salt buffer (410 mM KCl, 25% glycerol, and 0.2 mM EDTA in buffer A). Insoluble material was removed by centrifugation at 15,000 × g for 10 min. The protein concentration was measured with a Bio-Rad protein assay (Bio-Rad, Hercules, CA), and samples were stored at −70°C until use.
Synthetic oligonucleotides were used as probe for EMSA. The following oligonucleotides containing potential STAT5 elements from the 5′-flanking region of the human granulysin gene were used: STAT5 wild-type, 5′-GCCTGGTTCCAGGAAAGGAG-3′ (−18,221 to −18,202); STAT5 mutant, 5′-GCCTGGTGCCAGCAAAGGAGAAAGC-3′ (−18,221 to −18,197); STAT5 consensus, 5′-AGATTTCTAGGAATTCAATCC-3′ (29). The double-stranded oligonucleotides were end labeled with [α−32P]dATP using the Klenow fragment of DNA polymerase I (New England Biolabs). Additionally, STAT5 wild-type oligonucleotides were unlabeled for competition assays. Labeled DNA probe was purified from a 12% polyacrylamide gel by excising the corresponding radioactive band after electrophoresis. Purified DNA probe was adjusted to 15,000 cpm/ml and stored at −20°C until use. EMSA was performed as previously described (24). For supershift analysis, nuclear extracts were incubated with 1 μl normal rabbit serum (NRS) or anti-STAT5 Ab for 2 h on ice before the addition of 32P-labeled DNA probe. Results were visualized by autoradiography after 1–4 d exposure at −70°C.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed according to the manufacturer’s recommendations (Upstate Biotechnology, Lake Placid, NY). Briefly, human primary CD4+ T cells or CRL-2105 T cells (5 × 106 cells) were stimulated with or without 100 U/ml IL-2. After overnight incubation for 16 h, the cells were fixed with 1% formaldehyde. The nuclei were isolated and sonicated 20 times on ice for 10–20 s with 90-s breaks (Sonifier 350; Branson) between each sonication interval to shear the DNA to 200–1000 bp. A small aliquot (20 μl) was saved as “input DNA” for PCR analysis by reversing histone–DNA cross-links by heating at 65°C for 4 h. Chromatin was immunoprecipitated from 200-μl aliquots at 4°C by mild agitation overnight with 5 μg Ab specific for STAT-5 (Santa Cruz Biotechnology) or with 5 μg normal rabbit IgG as negative control. Immune complexes were collected by incubation with protein A-agarose. To analyze the target region, the immunoprecipitated chromatin DNA samples were amplified by PCR with primer pairs for STAT-5 binding site containing region (231 bp) (5′-TCTCCCAAAGTGCTGGGATTAGAGGCG-3′ and 5′-TCATACCAATGTGGACTTAAAAATGGAG-3′). DNA samples or input DNA fractions were analyzed by 35 cycles of PCR (94°C for 30 s, 56°C for 30 s, and 72°C for 30 s) in 20-μl reaction mixtures. PCR products were subjected to electrophoresis by using 2% agarose gels in Tris-acetate–EDTA buffer and visualized by ethidium bromide.
Statistical analysis
Data were expressed as mean ± SEM. Statistical analysis was performed by using Student t test, and a p value <0.05 was considered significant.
Results
IL-2 promotes granulysin expression and microbicidal activity in primary CD4+ T cells and CRL-2105 T cells
To examine the regulation of granulysin, we first needed to identify a CD4+ T cell line that was activated by IL-2, expressed granulysin, and killed microbes. After screening a number of cell lines, we identified CRL-2105 T cells as such a line. CD4+ CRL-2105 T cells were stimulated with IL-2 (100 U/ml). After 24 or 48 h of incubation, total RNA was isolated, and RT-PCR was performed. Additionally, the time course of granulysin expression was examined by Western blot. There was no expression of granulysin mRNA in resting CRL-2105 T cells (Fig. 1A). Increased expression of granulysin mRNA was detected after 24 h of IL-2 stimulation and was slightly further enhanced at 48 h. By contrast, the mRNA expression of granulysin was easily detected in resting YT cells, which constitutively express granulysin (Fig. 1A) (11). Additionally, the kinetics of granulysin protein expression was similar to that of mRNA expression, and two forms of granulysin, 9 and 15 kDa, were detected (Fig. 1B) (3). Our previous study has shown that IL-2–activated primary CD4+ T cells use granulysin to kill C. neoformans (12). To determine whether the CD4+ T cell line killed C. neoformans, experiments were performed to investigate the anticryptococcal activity of CD4+ CRL-2105 T cells. When C. neoformans was cultured in medium without CRL-2105 cells, there was a 4- to 6-fold increase in the number of organisms after 48 h of incubation (Fig. 1C). There was no anticryptococcal effect when untreated CRL-2105 T cells were added to the culture (Fig. 1C). By contrast, when C. neoformans was incubated with IL-2–stimulated CRL-2105 T cells, the CFU at 24 and 48 h was similar to the starting inocula and significantly lower than in the group containing C. neoformans alone at each time (p <0.01; Fig. 1C). Additionally, primary CD4+ T cells showed similar results to CRL-2105 T cells as described previously (data not shown) (12,23). Taken together, these results demonstrate that expression of granulysin at both the level of mRNA and protein is augmented in CRL-2105 T cells similar to primary CD4+ T cells stimulated with IL-2. Additionally, the IL-2–stimulated CRL-2105 T cells acquired anticryptococcal activity similar to primary CD4+ T cells.
FIGURE 1.
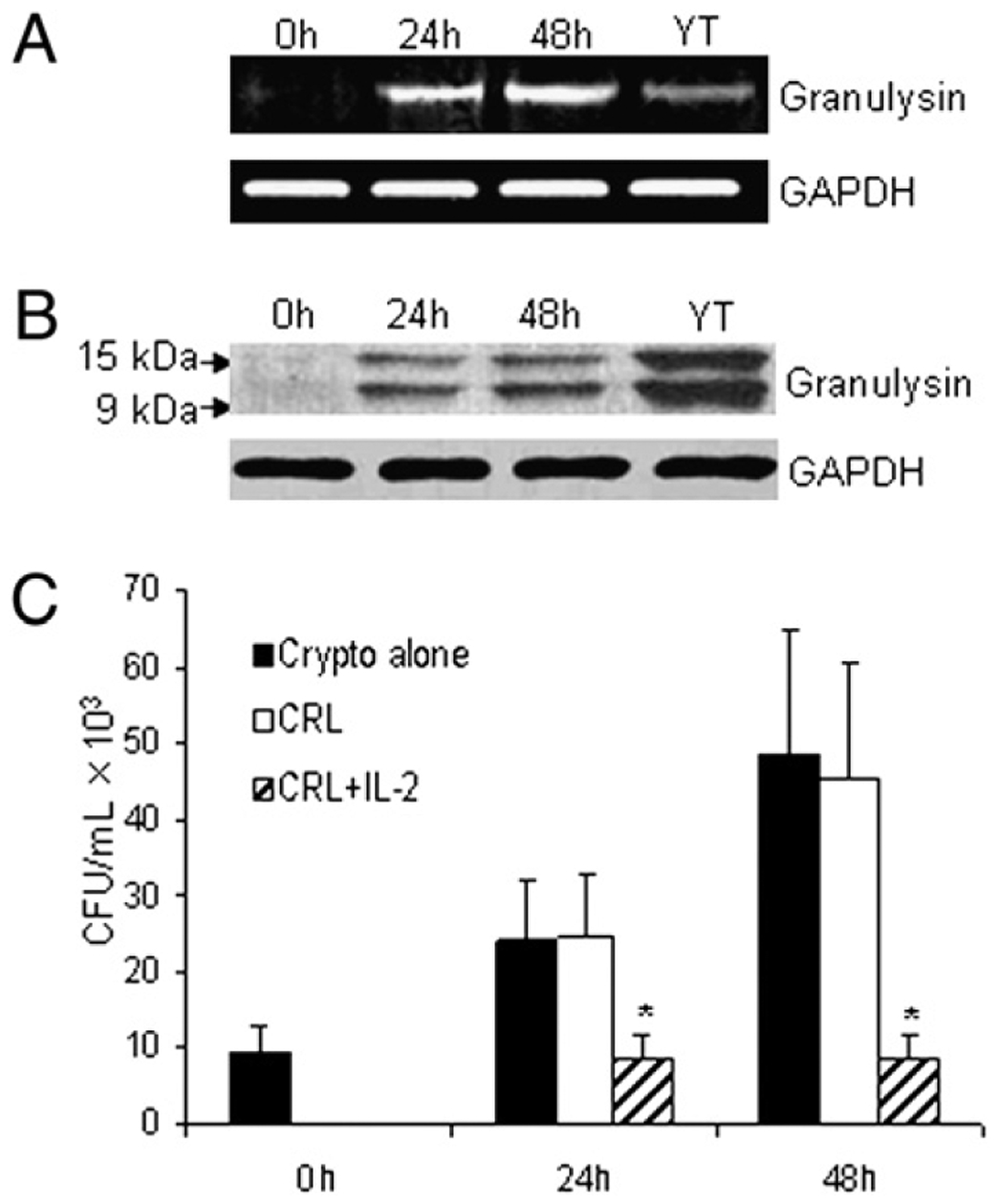
IL-2 promotes granulysin expression in CRL-2105 T cells. A, CRL-2105 T cells were stimulated for various times (0, 24, 48 h) with 100 U/ml IL-2 and compared with unstimulated YT cells. The expression of granulysin mRNA after IL-2 stimulation was assessed by RT-PCR, and GAPDH was used as an internal control. B, The time course of granulysin expression was also detected by Western blot using an Ab (519/GST) that detects both 15- and 9-kDa forms of granulysin. C, CRL-2105 T cells were treated with IL-2 and incubated with C. neoformans. The number of C. neoformans (CFU) was determined in each group as indicated. Results are expressed as mean ± SEM. Data are representative of three independent experiments. *p < 0.01 compared with the all other groups.
Identification of IL-2–responsive regulatory elements in the human granulysin 5′ flank in primary CD4+ T cells
It has been previously demonstrated that the granulysin promoter is located between −329 and +62 bp (24). To search for potential transcription factor binding sites upstream of the granulysin promoter, we initially used the TRANSFAC database (30). Based on the results searched, the −19,802 to +62 region of the granulysin promoter revealed the presence of many binding sites that could function as regulatory elements. To characterize the IL-2- responsive regulatory elements upstream of the granulysin promoter, we generated luciferase reporter constructs containing variant fragments upstream of the granulysin promoter by restriction enzyme digestion or PCR, cloned into the promoterless luciferase reporter plasmid pGL3. The size and location of the fragments used are shown in Fig. 2B. First, the transcriptional activity of pGL3−19,802/+62 was assessed in human primary CD4+ T cells (Fig. 2A). As a result, highly inducible transcriptional activities for granulysin expression were detected in both primary and CRL-2105 CD4+ T cells, whereas the transcriptional activity dropped sharply in pGL3−329/+62 (Fig. 2A). The fold induction of primary CD4+ T cells was a little lower than that of CRL-2105 T cells, which correlated with the efficiency of transfection by electroporation. Subsequent experiments were performed in CRL-2105 T cells, because the cell line has higher transfection efficiency.
Next, CRL-2105 T cells were transiently transfected with the constructs shown in Fig. 2B and were then stimulated with or without 100 U/ml IL-2. High levels of inducible enhancer activity for granulysin expression was detected in pGL3−19,802/+62 (Fig. 2B). However, the enhancer activity dropped sharply from pGL3−16,972/+62 through pGL3−1,181/+62, which were almost identical to those of pGL3−329/+62 (Fig. 2B). Additionally, the transcriptional activity of pGL3−329/+62 was ~2-fold greater than that of pGL3-basic without a promoter, suggesting the presence of basal transcriptional activity of the granulysin promoter, which is consistent with the previous report (Fig. 2B) (24). To validate these results, several reporter plasmids containing internal deletion mutants were constructed and transfected. Plasmids pGL3−19,802/−16,972 to pGL3−19,802/−1,181, which all contain the granulysin promoter, all exhibited high levels of inducible granulysin enhancer activities (Fig. 2B). Taken together, these results indicated the presence of a positive regulatory element(s) located in the granulysin 5′ flank (from −19,802 to −16,972 bp).
Characterization of an IL-2–responsive enhancer upstream of the granulysin promoter
To map and characterize further the enhancer elements upstream of the granulysin promoter, several luciferase reporter constructs derived from the region of −19,802 to −16,972 bp were generated (Fig. 2B). Levels of enhancer activity in both pGL3−19,802/−17,802 and pGL3−19,802/−17,302 (Fig. 2B) were similar to that of pGL3−19,802/−16,972 (Fig. 2B). By contrast, the plasmids from pGL3−19,802/−19,302 through pGL3−19,802/−18,302 did not show any enhancer activities compared with pGL3-basic (Fig. 2B). Therefore, the potential IL-2–responsive enhancer upstream of the granulysin promoter was demonstrated to be between −18,302 and −17,802 bp.
To investigate whether the potential enhancer (−18,302 to −17,802 bp) of granulysin could function in the context of the heterologous promoter, an SV40 promoter was introduced into reporter constructs to substitute for the granulysin promoter −329/+62 (Fig. 2C). As a result, the fragment containing the potential enhancer also functioned in the context of the heterologous SV40 promoter (Fig. 2C). Additionally, IL-2 increased the transcriptional activity of the constructs irrespective of the orientation of the regulatory DNA (Fig. 2C). These results indicated that an IL-2–inducible enhancer located within the 500-bp region from −18,302 to −17,802 bp functions in the context of a heterologous SV40 promoter irrespective of its transcriptional orientation.
Mutation of the STAT5 binding site abrogates granulysin enhancer activity
The results above indicated that there is an IL-2–responsive enhancer upstream of the granulysin promoter in the region of −18,302 to −17,802 bp. To characterize further the specific region, sequential deletions of pGL3−18,302/−17,802 were generated (Fig. 3). The granulysin promoter activity was decreased significantly by deletion of the region between −18,177 and −17,802 (Fig. 3), confirming that an enhancer regulatory element may exist in the 125-bp region from −18,302 to −18,177. The TRANSFAC database revealed a putative STAT5 binding site, TTCCAGGAA, within this region (30), which was consistent with our previous result that STAT5 was required for the late expression of granulysin by microbicidal CD4+ T cells (23). As expected, the putative STAT5 sequence alone could enhance the granulysin promoter activity in both primary CD4+ T cells and CRL-2105 T cells (Fig. 3). To demonstrate further whether the binding site could function as an enhancer for the granulysin promoter, the putative STAT5 binding site sequence, TTCCAGGAA, was mutated to TGCCAGCAA using site-directed mutagenesis. It was previously shown that this mutation completely abrogates STAT5 binding activity (31). A reporter plasmid pGL −18,302/−18,177Mut was constructed and confirmed by sequencing to evaluate the functional relevance of the STAT5 binding site. Mutating the STAT5 binding site resulted in a dramatic reduction of enhancer activity to the level of the core promoter −329/+62 (Fig. 3). A construct containing the STAT binding site was also transfected into primary CD4+ T cells, which demonstrated a similar level of enhanced activity. This result demonstrates the presence of an IL-2–inducible enhancer element in the region between −18,302 and −18,177 bp and suggests that a STAT5 binding site is responsible for the granulysin enhancer activity in CD4+ T cells.
FIGURE 3.
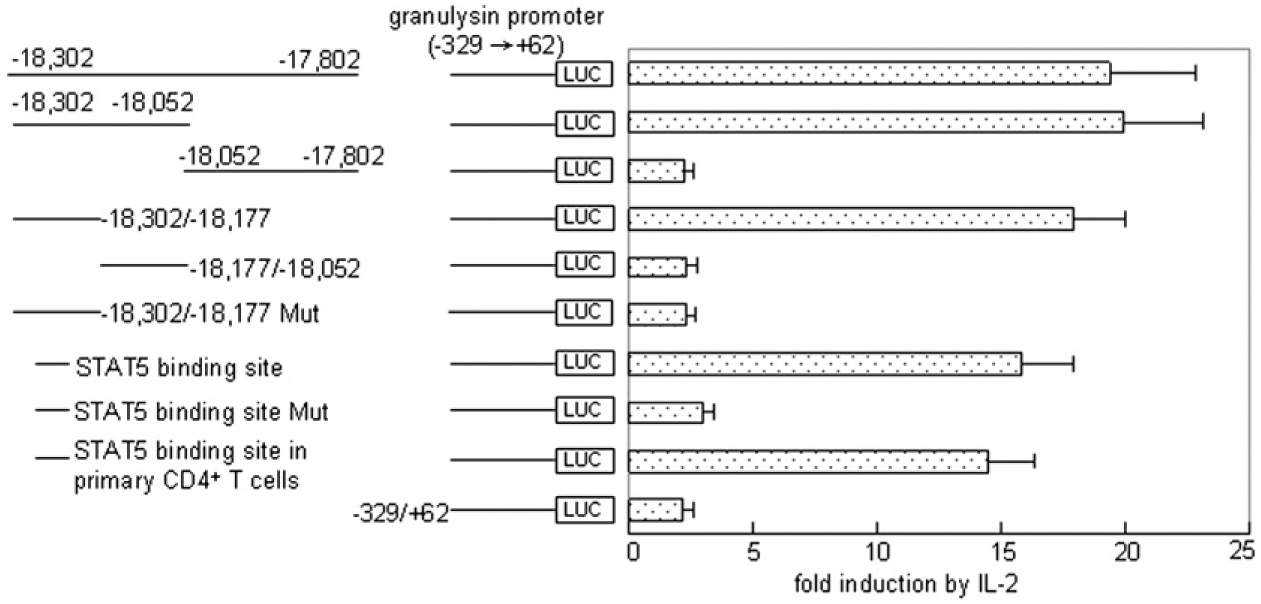
Mutation of STAT5 binding site abrogates the granulysin enhancer activity. The sequence of the STAT5 binding site construct is TTCCAGGAA. The potential STAT5 binding site TTCCAGGAA at the 59 end of the construct pGL−18,302/−18,177Mut was mutated to TGCCAGCAA using site-directed mutagenesis. After transient transfection and IL-2 stimulation, the granulysin enhancer activities of the depicted DNAs in the context of the granulysin promoter (−329 to +62) in CRL-2105 T cells or primary CD4+ T cells (as indicated) were detected as described in Fig. 2. Results are expressed as mean ± SEM. Data are representative of three independent experiments.
Dominant negative STAT5 blocked the activation of the enhancer
To determine whether STAT5 was required for IL-2–stimulated enhancer activity, a dominantnegative (DN) construct of STAT5a was generated. Coexpression of a DN STAT5 (27) abolished the activation of the enhancer activity by IL-2R signals in a dose-dependent manner (Fig. 4A), in contrast with the cotransfection with the parental wild-type STAT5a expression vector, which slightly increased the enhancer activity (data not shown). Furthermore, experiments were performed to investigate the effect of STAT5 enhancer on anticryptococcal activity of primary CD4+ T cells or CRL-2105 T cells. When C. neoformans was cultured in medium with or without untreated CRL-2105 cells, there was a 3- to 6-fold increase in the number of organisms after incubation (Fig. 4B). When C. neoformans was incubated with IL-2- stimulated primary CD4+ T cells or CRL-2105 T cells transfected with DN STAT5a, their CFU at 24 and 48 h were similar to those of the group containing C. neoformans alone (Fig. 4B). In contrast, the primary CD4+ T cells stimulated with IL-2 at 24 and 48 h had similar levels of anticryptococcal activity to those of IL-2- stimulated CRL-2105 T cells (Fig. 4B). Western blot analysis revealed that granulysin expression of IL-2–stimulated primary CD4+ T cells and CRL-2105 T cells were profoundly increased, whereas granulysin expression of T cells transfected with DN STAT5a were dramatically reduced (Fig. 4C). These data indicated that the activation of STAT5 plays a key role in the activation of the granulysin enhancers by IL-2R signals and is critical for the IL-2–stimulated primary CD4+ T cells and CRL-2105 T cells to acquire anticryptococcal activity and kill C. neoformans.
FIGURE 4.
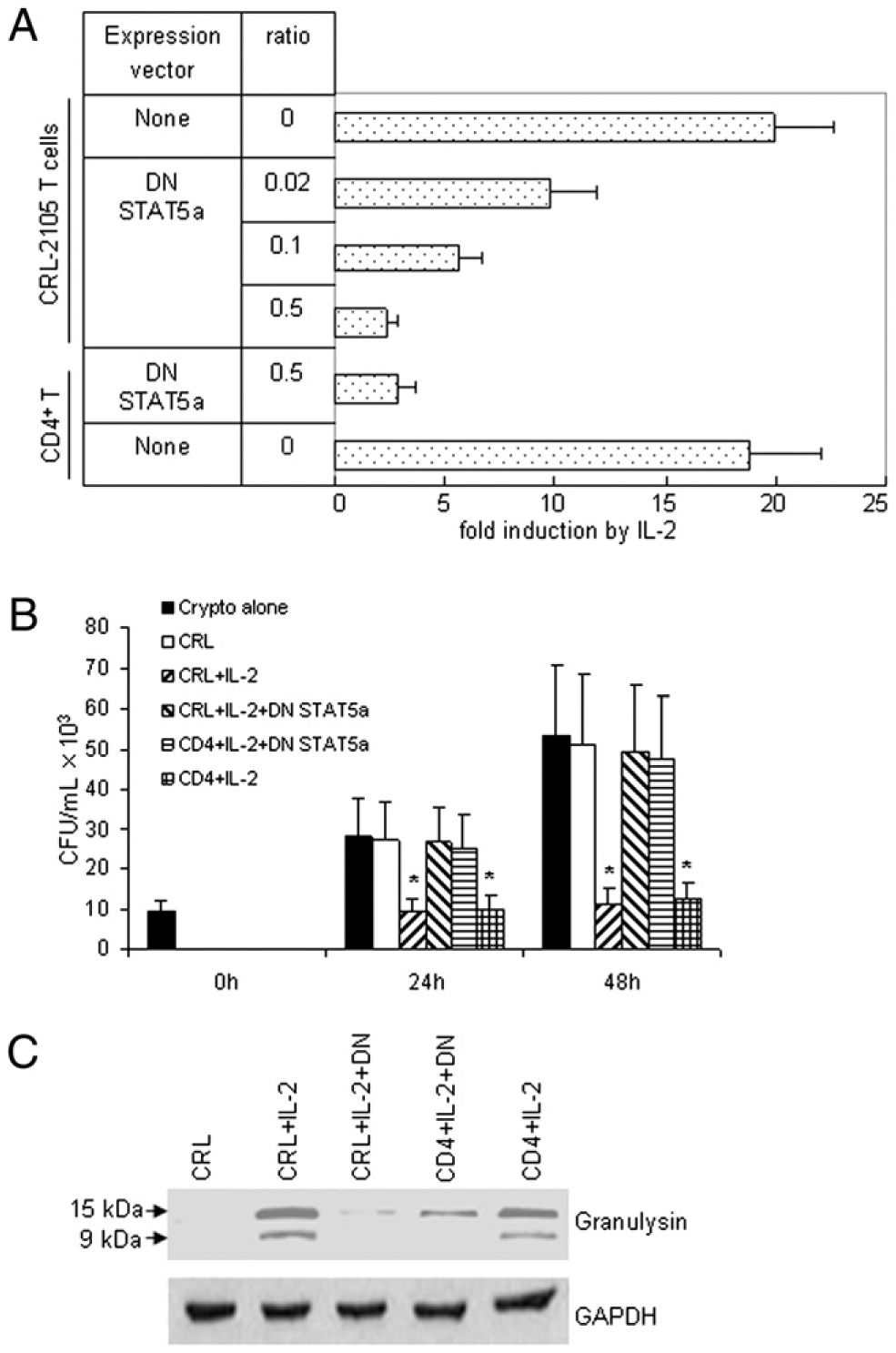
DN STAT5a blocked the activation of the enhancer. A, Primary CD4+ T cells or CRL-2105 T cells were activated in the presence or absence of 100 U/ml IL-2. The data summarize three independent transient transfections of CRL-2105 T cells or primary CD4+ T cells using 2 mg DNA comprising the tabulated molar ratios of the indicated expression vectors to the minimal enhancer SV40 plasmids. The CMV promoter-driven expression construct DN STAT5a was constructed by mutating aa 750 of murine STAT5a into a stop codon as described (27). Transfected cells were split and cultured in the absence or presence of IL-2. Reporter expression was quantitated and corrected for the protein content of the samples. B, Primary CD4+ T cells or CRL-2105 T cells were transfected with DN STAT5a expression vector as indicated, followed by treatment with IL-2 and incubation with C. neoformans. The number of C. neoformans (CFU) was determined in each group as indicated. Results are expressed as mean ± SEM. Data are representative of three independent experiments. *p < 0.01 compared with the number of C. neoformans alone or resting CRL-2105 T cells incubated with C. neoformans. C, Primary CD4+ T cells or CRL-2105 T cells were transfected with DN STAT5a as indicated, followed by treatment with IL-2. Expression of granulysin was assessed by Western blot.
Mutation within the STAT5 binding site eliminates its ability to bind to STAT5
To demonstrate further specific protein binding to the putative STAT5 binding site, an EMSA was performed. A 20-bp DNA oligonucleotide encompassing the STAT binding site from the region between −18,302 and −18,177 bp was used as a probe to detect specific protein binding in the nuclear extract from CRL-2105 T cells stimulated with IL-2 at concentration of 100 U/ml or 400 U/ml, as indicated. When the nuclear extracts were incubated with the 32P-labeled STAT5 consensus probe, a retarded band was observed (Fig. 5, lane 6) that was not detectable with a STAT5 wild-type probe alone (without nuclear extract) (Fig. 5, lane 7). In a competition assay, the presence of 100-fold molar excess unlabeled STAT5 wild-type competitor resulted in a loss of the retarded band (Fig. 5, lane 4), whereas an unlabeled STAT5 mutant competitor failed to do so (Fig. 5, lane 5). To determine whether STAT5 was binding, rather than another STAT protein, the nuclear extract from stimulated CRL-2105 T cells was preincubated with anti-STAT5 Ab before addition of the radiolabeled probe. The retarded band was supershifted in the presence of anti-STAT5 Ab (Fig. 5, lane 1). However, in the presence of NRS, the specific retarded band was not supershifted (Fig. 5, lanes 2 and 3). Additionally, a stronger band was detected from stimulated CRL-2105 T cells at higher IL-2 concentration 400 U/ml (Fig. 5, lane 2 compared with lane 3). These results show that the transcription factor STAT5 specifically binds to the STAT binding site in the granulysin promoter, and mutation within the STAT5 binding site abolishes its ability to bind to STAT5.
FIGURE 5.
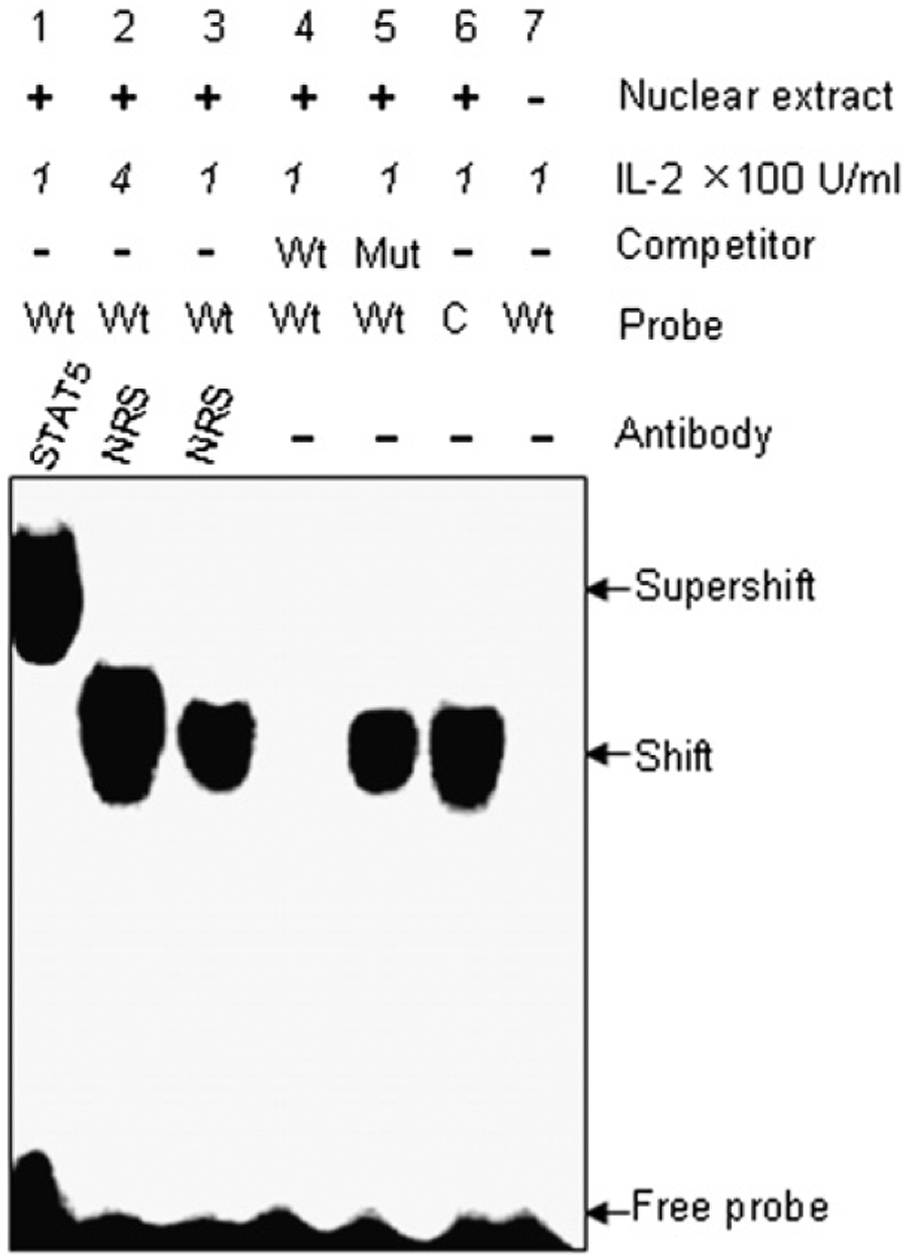
Mutation within the STAT5 binding site abolishes its ability to bind to STAT5. EMSA with the putative STAT5 binding site sequence of the granulysin enhancer was performed. 32P-labeled STAT5 consensus (C) probe (lane 6) and STAT5 wild-type (Wt) probes based on granulysin enhancer sequence (lanes 1–5), were incubated with nuclear extracts from CRL-2105 T cells stimulated with IL-2 at concentration of 100 U/ml or 400 U/ml as indicated. For competition assays, a 100-fold molar excess of unlabeled STAT5 Wt (lane 4), STAT5 mutant (Mut; lane 5) oligonucleotides were incubated with CRL-2105 nuclear extracts prior to incubation with 32P-labeled STAT5 Wt probe. Lane 7, Labeled STAT5 Wt probe without nuclear extract. The labeled STAT5 Wt probe incubated with CRL-2105 nuclear extracts after incubating with 400 U/ml IL-2 (lane 2) or 100 U/ml IL-2 (lane 3). Ab-mediated supershift assays were performed with 2 mg rabbit anti-STAT5 Ab and NRS as a control.
Confirmation of the in vivo binding of STAT5 protein to the human granulysin enhancer
To verify that the human granulysin enhancer has the capability directly to bind STAT5 protein in living cells, a ChIP assay was applied to precipitate the STAT5–DNA complexes with an anti-STAT5 Ab. After cross-linking the DNA with bound STAT5 proteins in situ in IL-2–stimulated versus mock-stimulated human primary CD4+ T cells or CRL-2105 T cells, the DNA fragments containing the STAT5 binding sequences in the granulysin promoter were precipitated with anti-STAT5 Ab and measured by PCR amplification. PCR with primers flanking the identified STAT5 binding sequences generated a band from DNA that had been coprecipitated with anti-STAT5 in both primary CD4+ T cells and CRL-2105 T cells (Fig. 6, lanes 2 and 5). As a negative control, immunoprecipitation with normal IgG did not generate a corresponding PCR product (Fig. 6, lanes 3 and 6). As expected, ChIP assays failed to detect DNA bands from either cell type in the absence of IL-2 stimulation (Fig. 6, lanes 1 and 2). These data strongly suggest that STAT5 interacts with the identified enhancer upstream of the human granulysin promoter after IL-2 stimulation in both human primary CD4+ T cells and CRL-2105 T cells.
FIGURE 6.
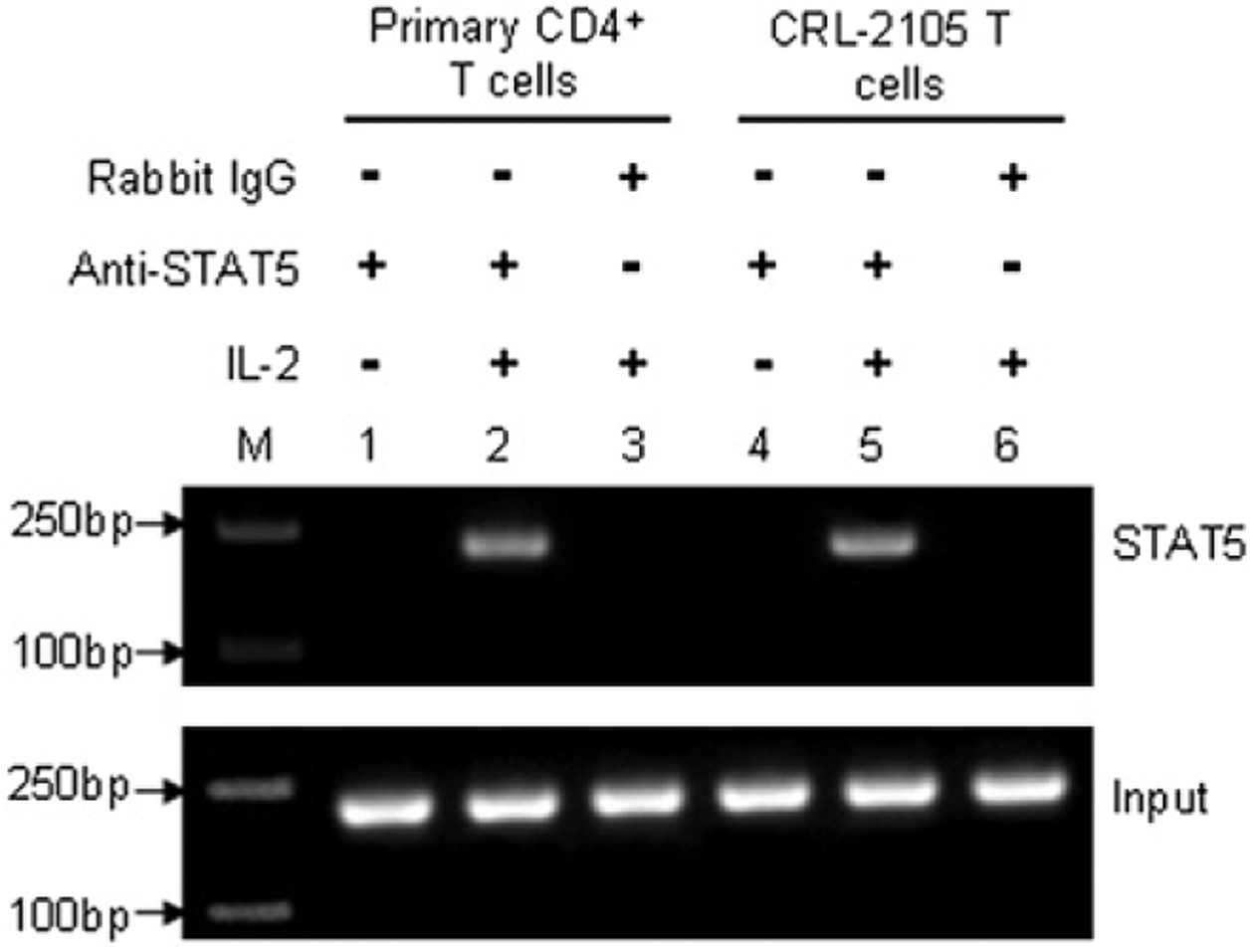
Detection of the in vivo binding of STAT5 protein to the human granulysin enhancer element in a ChIP assay. Formaldehyde cross-linked chromatin was prepared from both mock-treated and IL-2–treated human primary CD4+ T cells (lanes 1–3) or CRL-2105 T cells (lanes 4–6) as described in Materials and Methods. ChIP assays were performed using an anti-STAT5 Ab (lanes 1, 2, 4, and 5) or isotype-matched IgG (lanes 3 and 6) as a negative control, and M indicates the m.w. markers of nucleic acids. Immunoprecipitated chromatin was subjected to PCR analysis using STAT5 enhancer element specific primers. The equivalent amount of chromatin in the immunoprecipitations was monitored by PCR amplification of input chromatin as an internal control. ChIP assay was performed at least three times.
Discussion
An antimicrobial protein, granulysin, is expressed selectively in CTL and NK cells (3, 32) and plays a critical role in defense against microbial invasion (33), including that by C. neoformans (11, 12). Previously, we have demonstrated that cytotoxic CD4+ T cells use granulysin to kill C. neoformans (12), and both STAT5 and PI3K are required for increased IL-2Rβ and granulysin expression in CD4+ T cells (23). However, little is known about the precise regulatory mechanisms underlying granulysin gene expression. T cell lymphoma lines of murine and human origin are often used as versatile tools for immunologic research (34). Additionally, in cell lines, IL-2 stimulation activates Jak/STAT cascades (35). Therefore, CRL-2105, a human CD4+ T cell line derived from peripheral blood of a patient with aggressive cutaneous T cell leukemia/lymphoma, was used in this study to characterize the expression of granulysin. We have demonstrated herein that the granulysin expression at both the level of mRNA and protein is augmented in CRL-2105 T cells stimulated with IL-2. Additionally, IL-2–stimulated CRL-2105 T cells acquired anticryptococcal activity and killed C. neoformans similar to primary CD4+ T cells. It was previously reported that A. laidlawii caused increased granulysin gene expression via the transcription factor AP-1 in the THP-1 human monocytic cell line (24). However, microbial lysates containing a wide array of components may induce multiple factors that stimulate transduction pathways and result in complicated and indirect genetic regulation.
Studies examining the regulation of granulysin are limited compared with those of perforin and granzymes, in part due to the lack of a murine gene homolog (26). Our results demonstrated that IL-2 stimulated and enhanced the expression of granulysin via transcription factor binding sites upstream of the granulysin promoter. Indeed, strong enhancer activity was detected in construct pGL3−19,802/−17,802 that increased promoter activity ~17-fold in CRL-2105 T cells, suggesting that cell-specific transcription factors were binding to this region upstream of the promoter. Furthermore, high-level inducible transcriptional activity of pGL3−19,802/+62 was also detected in human primary CD4+ T cells, further demonstrating that the data from CRL-2105 T cells correctly reflected those of human primary CD4+ T cells. Additionally, IL-2 increased the transcriptional activity of the construct compared with the SV40 promoter alone irrespective of the orientation of the regulatory DNA. This region, −19,802 to −17,802 bp, contains a potential STAT binding site (TTCCAGGAA) at 218,215. The same palindromic core motif, TTCN2–4GAA, has been found in sequences recognized by all STATs (36), which further confirmed that the enhancer element of the granulysin gene identified in this study was a genuine STAT5 binding site. Mutation of the STAT5 binding element further demonstrated the essential role of the element in driving granulysin promoter activity in T cells. Indeed, a search for single-nucleotide polymorphisms as an indication of natural human variants in the identified STAT5 binding site failed to reveal any known single-nucleotide polymorphisms in this region, suggesting that this identified enhancer element is highly conserved.
STAT proteins were discovered in the course of the analysis of IFN signaling pathways, and seven mammalian genes coding for members of this family of intracellular signaling proteins have been found (37). Signaling by a large number of cytokine, growth factor, and hormone receptors leads to activation of one or more STATs (38). STAT5 is a transcription factor important for normal and malignant physiology of hematopoietic cells (39). Interestingly, the granulysin enhancer identified as STAT5 binding site located far-upstream of the transcription start site, which was similar to the perforin enhancers (40). We have demonstrated previously that STAT5 and PI3K are required for granulysin expression in the setting of increased expression of IL-2Rb in primary CD4+ T cells (23). It is of interest to note that DN STAT5 blocked the activation of the enhancer in response to IL-2R signals in a dose-dependent manner. Moreover, IL-2–stimulated human primary CD4+ T cells or CRL-2105 T cells transfected with DN STAT5a all abolished the anticryptococcal activity indicating that the STAT5 enhancer we identified herein was critical for the IL-2- stimulated human primary CD4+ T cells and CRL-2105 T cells to acquire anticryptococcal activity and kill C. neoformans. The ChIP and EMSA assays further demonstrated that STAT5 specifically bound to the STAT5 binding site upstream of the granulysin promoter both in vivo and in vitro.
The identification of a functional STAT5 binding element upstream of the human granulysin promoter provides a molecular explanation for the ability of cytokines to modulate granulysin expression. Our study provides strong evidence that transcriptional activation of the granulysin gene in human CD4+ T cells involves STAT5 protein activation that acts far upstream of the promoter. Additionally, the effect of STAT5 enhancer we identified on granulysin-mediated anticryptococcal activity of human primary CD4+ T cells and CRL-2105 CD4+ T cells is an important element of microbicidal T cell activity.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (30972670) and the Start-up Fund of the Hundred Talents Program of the Chinese Academy of Sciences (20071010-141) (to C.Z.), the Canadian Institutes for Health Research (to C.H.M.), and the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (to A.M.K.).
Abbreviations used in this paper:
- C
32P-labeled STAT5 consensus
- γc
common γ chain
- ChIP
chromatin immunoprecipitation
- DN
dominant negative
- Mut
STAT5 mutant
- NRS
normal rabbit serum
- RT
reverse transcription
- Wt
wild-type
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Krensky AM, and Clayberger C. 2005. Granulysin: a novel host defense molecule. Am. J. Transplant 5: 1789–1792. [DOI] [PubMed] [Google Scholar]
- 2.Jongstra J, Schall TJ, Dyer BJ, Clayberger C, Jorgensen J, Davis MM, and Krensky AM. 1987. The isolation and sequence of a novel gene from a human functional T cell line. J. Exp. Med 165: 601–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peña SV, Hanson DA, Carr BA, Goralski TJ, and Krensky AM. 1997. Processing, subcellular localization, and function of 519 (granulysin), a human late T cell activation molecule with homology to small, lytic, granule proteins. J. Immunol 158: 2680–2688. [PubMed] [Google Scholar]
- 4.Ernst WA, Thoma-Uszynski S, Teitelbaum R, Ko C, Hanson DA,Clayberger C, Krensky AM, Leippe M, Bloom BR, Ganz T, and Modlin RL. 2000. Granulysin, a T cell product, kills bacteria by altering membrane permeability. J. Immunol 165: 7102–7108. [DOI] [PubMed] [Google Scholar]
- 5.Gamen S, Hanson DA, Kaspar A, Naval J, Krensky AM, and Anel A. 1998. Granulysin-induced apoptosis. I. Involvement of at least two distinct pathways. J. Immunol 161: 1758–1764. [PubMed] [Google Scholar]
- 6.Kaspar AA, Okada S, Kumar J, Poulain FR, Drouvalakis KA, Kelekar A,Hanson DA, Kluck RM, Hitoshi Y, Johnson DE, et al. 2001. A distinct pathway of cell-mediated apoptosis initiated by granulysin. J. Immunol 167: 350–356. [DOI] [PubMed] [Google Scholar]
- 7.Okada S, Li Q, Whitin JC, Clayberger C, and Krensky AM. 2003. Intracellular mediators of granulysin-induced cell death. J. Immunol 171: 2556–2562. [DOI] [PubMed] [Google Scholar]
- 8.Kishi A, Takamori Y, Ogawa K, Takano S, Tomita S, Tanigawa M, Niman M,Kishida T, and Fujita S. 2002. Differential expression of granulysin and perforin by NK cells in cancer patients and correlation of impaired granulysin expression with progression of cancer. Cancer Immunol. Immunother 50: 604–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Choice E, Kaspar A, Hanson D, Okada S, Lyu SC, Krensky AM, and Clayberger C. 2000. Bactericidal and tumoricidal activities of synthetic peptides derived from granulysin. J. Immunol 165: 1486–1490. [DOI] [PubMed] [Google Scholar]
- 10.Kumar J, Okada S, Clayberger C, and Krensky AM. 2001. Granulysin: a novel antimicrobial. Expert Opin. Investig. Drugs 10: 321–329. [DOI] [PubMed] [Google Scholar]
- 11.Ma LL, Wang CL, Neely GG, Epelman S, Krensky AM, and Mody CH. 2004. NK cells use perforin rather than granulysin for anticryptococcal activity.J. Immunol 173: 3357–3365. [DOI] [PubMed] [Google Scholar]
- 12.Zheng CF, Ma LL, Jones GJ, Gill MJ, Krensky AM, Kubes P, and Mody CH. 2007. Cytotoxic CD4+ T cells use granulysin to kill Cryptococcus neoformans, and activation of this pathway is defective in HIV patients. Blood 109: 2049–2057. [DOI] [PubMed] [Google Scholar]
- 13.Stenger S, Hanson DA, Teitelbaum R, Dewan P, Niazi KR, Froelich CJ,Ganz T, Thoma-Uszynski S, Melián A, Bogdan C, et al. 1998. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science 282: 121–125. [DOI] [PubMed] [Google Scholar]
- 14.Farouk SE, Mincheva-Nilsson L, Krensky AM, Dieli F, and Troye-Blomberg M. 2004. Gamma delta T cells inhibit in vitro growth of the asexual blood stages of Plasmodium falciparum by a granule exocytosis-dependent cytotoxic pathway that requires granulysin. Eur. J. Immunol 34: 2248–2256. [DOI] [PubMed] [Google Scholar]
- 15.Mori S, Jewett A, Cavalcanti M, Murakami-Mori K, Nakamura S, and Bonavida B. 1998. Differential regulation of human NK cell-associated gene expression following activation by IL-2, IFN-alpha and PMA/ionomycin. Int. J. Oncol 12: 1165–1170. [DOI] [PubMed] [Google Scholar]
- 16.Samten B, Wizel B, Shams H, Weis SE, Klucar P, Wu S, Vankayalapati R,Thomas EK, Okada S, Krensky AM, and Barnes PF. 2003. CD40 ligand trimer enhances the response of CD8+ T cells to Mycobacterium tuberculosis. J. Immunol 170: 3180–3186. [DOI] [PubMed] [Google Scholar]
- 17.Stegelmann F, Bastian M, Swoboda K, Bhat R, Kiessler V, Krensky AM,Roellinghoff M, Modlin RL, and Stenger S. 2005. Coordinate expression of CC chemokine ligand 5, granulysin, and perforin in CD8+ T cells provides a host defense mechanism against Mycobacterium tuberculosis. J. Immunol 175: 7474–7483. [DOI] [PubMed] [Google Scholar]
- 18.Ma LL, Spurrell JC, Wang JF, Neely GG, Epelman S, Krensky AM, and Mody CH. 2002. CD8 T cell-mediated killing of Cryptococcus neoformans requires granulysin and is dependent on CD4 T cells and IL-15. J. Immunol 169: 5787–5795. [DOI] [PubMed] [Google Scholar]
- 19.Leonard WJ, Zeng R, and Spolski R. 2008. Interleukin 21: a cytokine/cytokine receptor system that has come of age. J. Leukoc. Biol 84: 348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waldmann TA 2006. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat. Rev. Immunol 6: 595–601. [DOI] [PubMed] [Google Scholar]
- 21.Hogg AE, Bowick GC, Herzog NK, Cloyd MW, and Endsley JJ. 2009. Induction of granulysin in CD8+ T cells by IL-21 and IL-15 is suppressed by human immunodeficiency virus-1. J. Leukoc. Biol 86: 1191–1203. [DOI] [PubMed] [Google Scholar]
- 22.Boyman O, Purton JF, Surh CD, and Sprent J. 2007. Cytokines and T-cell homeostasis. Curr. Opin. Immunol 19: 320–326. [DOI] [PubMed] [Google Scholar]
- 23.Zheng CF, Jones GJ, Shi M, Wiseman JC, Marr KJ, Berenger BM,Huston SM, Gill MJ, Krensky AM, Kubes P, and Mody CH. 2008. Late expression of granulysin by microbicidal CD4+ T cells requires PI3K- and STAT5-dependent expression of IL-2Rbeta that is defective in HIV-infected patients. J. Immunol 180: 7221–7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kida Y, Kuwano K, Zhang Y, and Arai S. 2001. Acholeplasma laidlawii up-regulates granulysin gene expression via transcription factor activator protein-1 in a human monocytic cell line, THP-1. Immunology 104: 324–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mody CH, Toews GB, and Lipscomb MF. 1988. Cyclosporin A inhibits the growth of Cryptococcus neoformans in a murine model. Infect. Immun 56: 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang LP, Lyu SC, Clayberger C, and Krensky AM. 2007. Granulysin-mediated tumor rejection in transgenic mice. J. Immunol 178: 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moriggl R, Gouilleux-Gruart V, Jähne R, Berchtold S, Gartmann C, Liu X,Hennighausen L, Sotiropoulos A, Groner B, and Gouilleux F. 1996. Deletion of the carboxyl-terminal transactivation domain of MGF-Stat5 results in sustained DNA binding and a dominant negative phenotype. Mol. Cell. Biol 16: 5691–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreiber E, Matthias P, Müller MM, and Schaffner W. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17: 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva M, Benito A, Sanz C, Prosper F, Ekhterae D, Nuñez G, and Fernandez-Luna JL. 1999. Erythropoietin can induce the expression of bcl-x(L) through Stat5 in erythropoietin-dependent progenitor cell lines. J. Biol. Chem 274: 22165–22169. [DOI] [PubMed] [Google Scholar]
- 30.Quandt K, Frech K, Karas H, Wingender E, and Werner T. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23: 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu CR, Lin JX, Fink DW, Akira S, Bloom ET, and Yamauchi A. 1996. Differential utilization of Janus kinase-signal transducer activator of transcription signaling pathways in the stimulation of human natural killer cells by IL-2, IL-12, and IFN-alpha. J. Immunol 157: 126–137. [PubMed] [Google Scholar]
- 32.Yabe T, McSherry C, Bach FH, and Houchins JP. 1990. A cDNA clone expressed in natural killer and T cells that likely encodes a secreted protein. J. Exp. Med 172: 1159–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krensky AM, and Clayberger C. 2009. Biology and clinical relevance of granulysin. Tissue Antigens 73: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abraham RT, and Weiss A. 2004. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat. Rev. Immunol 4: 301–308. [DOI] [PubMed] [Google Scholar]
- 35.Ellery JM, and Nicholls PJ. 2002. Alternate signalling pathways from the interleukin-2 receptor. Cytokine Growth Factor Rev. 13: 27–40. [DOI] [PubMed] [Google Scholar]
- 36.Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, and Bucher P. 2001. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J. Biol. Chem 276: 6675–6688. [DOI] [PubMed] [Google Scholar]
- 37.Yu H, Pardoll D, and Jove R. 2009. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9: 798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darnell JE Jr. 1997. STATs and gene regulation. Science 277: 1630–1635. [DOI] [PubMed] [Google Scholar]
- 39.Calò V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, and Russo A. 2003. STAT proteins: from normal control of cellular events to tumorigenesis. J. Cell. Physiol 197: 157–168. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Scordi I, Smyth MJ, and Lichtenheld MG. 1999. Interleukin 2 receptor signaling regulates the perforin gene through signal transducer and activator of transcription (Stat)5 activation of two enhancers. J. Exp. Med 190: 1297–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]


