Abstract
BACKGROUND:
Approximately 30% of fine-needle aspiration (FNA) biopsies of thyroid nodules are indeterminate or nondiagnostic. Recent studies suggest microRNA (miRNA, miR) is differentially expressed in malignant tumors and may have a role in carcinogenesis, including thyroid cancer. The authors therefore tested the hypothesis that miRNA expression analysis would identify putative markers that could distinguish benign from malignant thyroid neoplasms that are often indeterminate on FNA biopsy.
METHODS:
A miRNA array was used to identify differentially expressed genes (5-fold higher or lower) in pooled normal, malignant, and benign thyroid tissue samples. Real-time quantitative polymerase chain reaction was used to confirm miRNA array expression data in 104 tissue samples (7 normal thyroid, 14 hyperplastic nodule, 12 follicular variant of papillary thyroid cancer, 8 papillary thyroid cancer, 15 follicular adenoma, 12 follicular carcinoma, 12 Hurthle cell adenoma, 20 Hurthle cell carcinoma, and 4 anaplastic carcinoma cases), and 125 indeterminate clinical FNA samples. The diagnostic accuracy of differentially expressed genes was determined by analyzing receiver operating characteristics.
RESULTS:
Ten miRNAs showed >5-fold expression difference between benign and malignant thyroid neoplasms on miRNA array analysis. Four of the 10 miRNAs were validated to be significantly differentially expressed between benign and malignant thyroid neoplasms by quantitative polymerase chain reaction (P < .002): miR-100, miR-125b, miR-138, and miR-768–3p were overexpressed in malignant samples of follicular origin (P < .001), and in Hurthle cell carcinoma samples alone (P < .01). Only miR-125b was significantly overexpressed in follicular carcinoma samples (P < .05). The accuracy for distinguishing benign from malignant thyroid neoplasms was 79% overall, 98% for Hurthle cell neoplasms, and 71% for follicular neoplasms. The miR-138 was overexpressed in the FNA samples (P = .04) that were malignant on final pathology with an accuracy of 75%.
CONCLUSIONS:
MicroRNA expression differs for normal, benign, and malignant thyroid tissue. Expression analysis of differentially expressed miRNA could help distinguish benign from malignant thyroid neoplasms that are indeterminate on thyroid FNA biopsy.
Keywords: microRNA, thyroid, cancer, biomarker, diagnostic
The incidence of thyroid cancer, the most common endocrine malignancy, has increased more than 2-fold in the United States over the last 3 decades, mostly as a result of improved detection of subclinical tumors.1 Approximately 95% of all thyroid cancers are differentiated thyroid cancer of follicular cell origin (papillary, follicular, and Hurthle cell cancer). Although 5-year and 10-year survival rates are excellent, except for patients with poorly differentiated thyroid cancer, a vexing diagnostic problem remains. Cytology from fine-needle aspiration (FNA) biopsy, the most accurate preoperative test for diagnosing thyroid cancer, yields indeterminate or nondiagnostic results approximately 30% of the time. For that reason, there is considerable interest in whether genotyping for the most common thyroid cancer-related mutations, or if high-throughput genomic and proteomic approaches could be used to improve the diagnostic accuracy of FNA cytology to avoid unnecessary diagnostic thyroidectomy.
MicroRNAs (miRNAs, miRs) are small (approximately 22 nucleotide lengths), noncoding single-stranded RNAs that constitute a novel class of gene regulators.2 The miRNAs are transcribed from endogenous DNA and processed from primary transcript (pri-miRNA) to a hairpin precursor (pre-miRNA) comprising 2 strands: the leading strand used to produce the mature miRNA and the passenger strand that is believed to be degraded. Mature miRNAs target and bind to transcripts and interfere with their translation into protein or cause messenger RNA (mRNA) degradation.3 Although the function of miRNAs has only recently been recognized, emerging evidence indicates that particular miRNAs may be closely involved with human cancer pathogenesis.4 In fact, they regulate complicated biological behaviors such as cell growth, adhesion, differentiation, and apoptosis. We hypothesized that miRNA expression profiling would identify putative markers for distinguishing benign from malignant thyroid neoplasms that are often indeterminate on FNA biopsy.
MATERIALS AND METHODS
Thyroid Tissue Samples
After obtaining approval from the Committee on Human Research at the University of California, San Francisco (UCSF), we analyzed 104 tissue samples of patients with thyroid lesions (7 normal thyroid, 14 multinodular goiter, 12 follicular variant of papillary thyroid cancer, 8 papillary thyroid cancer [PTC], 15 follicular thyroid adenomas, 12 follicular thyroid carcinomas [FTC], 12 Hurthle cell adenomas, 20 Hurthle cell thyroid carcinomas, and 4 anaplastic thyroid carcinomas [ATC]) as well as 125 indeterminate FNA samples of patients with thyroid nodules. All thyroid tissue diagnoses were confirmed histologically. Both tissue and FNA samples used in the experiments were carefully snap-frozen in liquid nitrogen at the time of thyroidectomy and stored at −80°C until total RNA was extracted. All FNA samples were either an extra passes or the remnant of an aspirate if enough cellular material remained. FNA samples were used to validate the tissue candidate markers identified.5 Demographic data and TNM stage of all patients were documented.
Real-Time Quantitative Polymerase Chain Reaction
First, several hundred (approximately 850) miRNAs were investigated by miRNA assay (miRCURY LNA; Exiqon, Vedbaek, Denmark). Genes that were upregulated or downregulated by ≥5-fold in malignant versus benign thyroid tissue were then validated by real-time quantitative polymerase chain reaction (RT-qPCR). A pooled miRNA analysis was used for marker identification, which does not provide individual miRNA profiling information. The pooled samples were the tissue samples in the discovery set of candidate markers that were randomly chosen from our biobank at UCSF to identify ≥5-fold differently expressed miRNAs in our array studies. Total RNA was extracted from thyroid tissue using the Trizol reagent (Invitrogen, Carlsbad, Calif). Quantification using TaqMan miRNA assay was a 2-step RT-PCR. First, complementary DNA (cDNA) was derived from mRNA using specific miRNA primers with a TaqMan miRNA reverse-transcriptase kit (Applied Biosystems, Foster City, Calif). Total RNA (5 ng/μL) was reverse-transcribed into cDNA in a 10-μL reaction, containing 1 μL of 10× PCR buffer, 0.1 μL of 25 mM deoxynucleotide triphosphate (dNTP) mix, 0.67 μL of RT enzyme, 0.13 μL of ribonuclease inhibitor, 2 μL 5× RT primer, and 4.1 μL of distilled water. The cycle parameters for the reverse transcription reaction were 16°C for 30 minutes, 42°C for 30 minutes, 85°C for 5 minutes, and a hold at 4°C. PCR products were amplified from cDNA in a 10-μL reaction containing 2 μL of 5× TaqMan buffer, 2.2 μL of 25 mM MgCl2, 0.08 μL of 25 mM dNTP, 3.27 μL of water, 0.005 μL of Taq polymerase, 0.4 μL of TaqMan primer, and 2 μL of cDNA. All PCR reactions were performed in a final volume of 10 μL on an ABI-PRISM 7900 Sequence Detection System (Applied Biosystems). The gene expression level was normalized to U18 mRNA expression as follows. Normalized gene expression = 2^- (Ct for miR of interest − Ct for U18) × 100%, in which Ct = the quantitative PCR cycle threshold. All experiments were performed in triplicate.
Cell Growth/Proliferation Studies
The FTC cell line FTC-133 was cultured to confluence. Cells were trypsinized and placed in 96-well plates at the density of 7500 cells per well with 150 μL of media in each well. After 24 hours, cells were transfected with miRNA. The 3 miRNA genes (hsa-miR-100 [Applied Biosystems/Ambion, ID# AM12598], hsa-miR-125b [Applied Biosystems/Ambion, ID# PM10148], and hsamiR-138 [Applied Biosystems/Ambion, ID# PM11727]) were used at the concentration of 30 nM, and 0.8 μL per well of X-tremeGENE siRNA transfection reagent (catalog no. 04-476-093-001; Roche Applied Science,Indianapolis, Ind) was used according to the manufacturer’s protocol. After 15 minutes of incubation, the mixture of miRNA inhibitor/precursor and transfection reagent was added to each well. The cell plates were then reincubated at 37°C. The cell plates were harvested at days 0, 1, 2, 3, and 6 of transfection. Six wells for each condition were harvested in Trizol for the RNA preparation, using the Trizol/chloroform (Acros Organics, Pittsburgh, Pa) extraction protocol. Equal concentrations of RNA were used to synthesize cDNAs using TaqMan microRNA RT kit (catalog no. 4366597). TaqMan reactions (TaqMan 7900CRI) were performed at the concentration of 1 ng of cDNA, using primer mix: miR-100 (TM2142; Applied Biosystems), miR-125b (TM449; Applied Biosystems), and miR-138 (TM2284; Applied Biosystems). U18 primer (TM1204; Applied Biosystems) was used in parallel as a control. Six replicates were used to perform the Cyquant Assay (Cyquant Cell Proliferation Assay Kit, C7026; Invitrogen). The fluorescence microplate reader (Spectra Max GeminiXS; Molecular Devices) was used to measure optical density at 485-nanometer (nm) excitation and 538 nm emission, with a cutoff 530 nm. A standard curve of cell numbers was generated to determine the cell number for each condition.
Statistical Analysis
Kruskal-Wallis and Mann-Whitney rank sum tests were used to determine differences in normalized mRNA expression levels between the benign and malignant histologic groups and extent of disease. A P value < .05 was considered statistically significant. Data are presented as the mean ± standard deviation unless otherwise stated. Receiver operating characteristic curves and the area under the curve were calculated to measure the accuracy of the candidate diagnostic markers.6 The positive predictive value of the potential markers was calculated as the true-positive value/(true-positive + false-positive value). The negative predictive value(NPV) was calculated as the true-negative/(true-negative + false-negative value). Predicted targets for specific miRNAs were analyzed using publically available databases: miRBase,7 TargetScanHuman,8 and PicTar.9
RESULTS
Tissue Samples
Analysis of miRNA arrays revealed 10 genes that showed at least a >5-fold difference in expression between benign and malignant tissue samples (Table 1). Expression profiling of these 10 genes was performed on 104 tissue samples obtained from patients who underwent surgery at UCSF. Of the 3 candidate housekeeping genes (U18, U47, and RNU-44), RT-PCR analysis showed U18 to be the gene with the least variability and highest expression in both benign and malignant samples.
Table 1.
MicroRNAs With a >5-Fold Difference in Expression Between Benign and Malignant Samples
| miRNA Name | Malignant | Benign | Fold Difference | Downregulated/Upregulated |
|---|---|---|---|---|
| hsa-miR-149 | 24.3 | 438.1 | 0.06 | Down |
| hsa-miR-100 | 47.8 | 280.7 | 0.17 | Down |
| hsa-miR-138 | 92.2 | 512.8 | 0.18 | Down |
| hsa-miR-125b | 255.6 | 1394.6 | 0.18 | Down |
| hsa-miR-768–3p | 36.3 | 181.8 | 0.20 | Down |
| hsa-miR-628–3p | 1318.0 | 332.1 | 3.97 | Up |
| hsa-miR-550* | 217.8 | 39.3 | 5.54 | Up |
| hsa-miR-564 | 287.3 | 46.4 | 6.19 | Up |
| hsa-miR-635 | 444.6 | 65.2 | 6.82 | Up |
| hsa-miR-584 | 953.3 | 122.7 | 7.77 | Up |
Abbreviation: miRNA, microRNA.
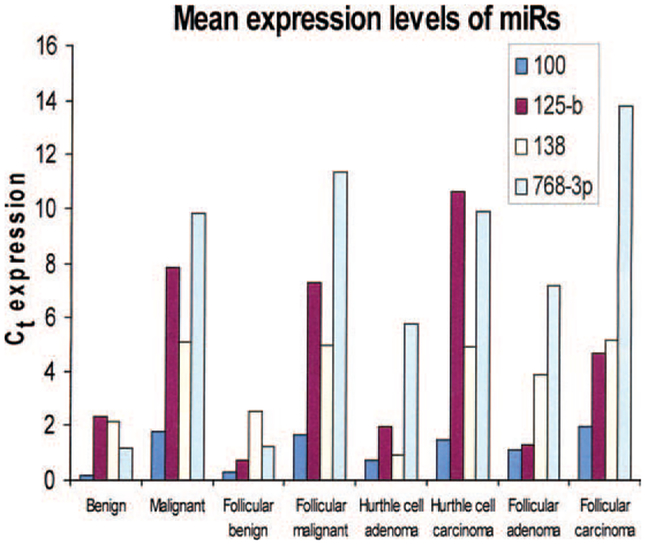
Of the 10 miRNAs, miR-100, miR-125b, miR-138, and miR-768–3p (all downregulated) were significantly differently expressed between all benign and malignant tissue samples (P < .002) by RT-qPCR. These 4 miRs were also significantly differently expressed between benign and malignant samples of follicular cell origin (P < .001) and between benign and malignant samples of Hurthle cell neoplasms (P < .01) (Figure 1). Only miR-125b was significantly overexpressed in FTCs compared with follicular adenomas (P < .05). None of the 10 miRNAs differed significantly between normal and benign tissue samples, or between PTC and ATC samples. Yet, miR-768–3p differed significantly between the follicular variant of PTC and benign samples (P < .001). All significant differences were noted in both miRNA Ct expressions only, as well as normalized miRNA expressions.
Figure 1.
Mean expression levels (normalized to delta Ct) of the 4 microRNAs are shown that are significantly differently expressed between benign and malignant tissue samples.
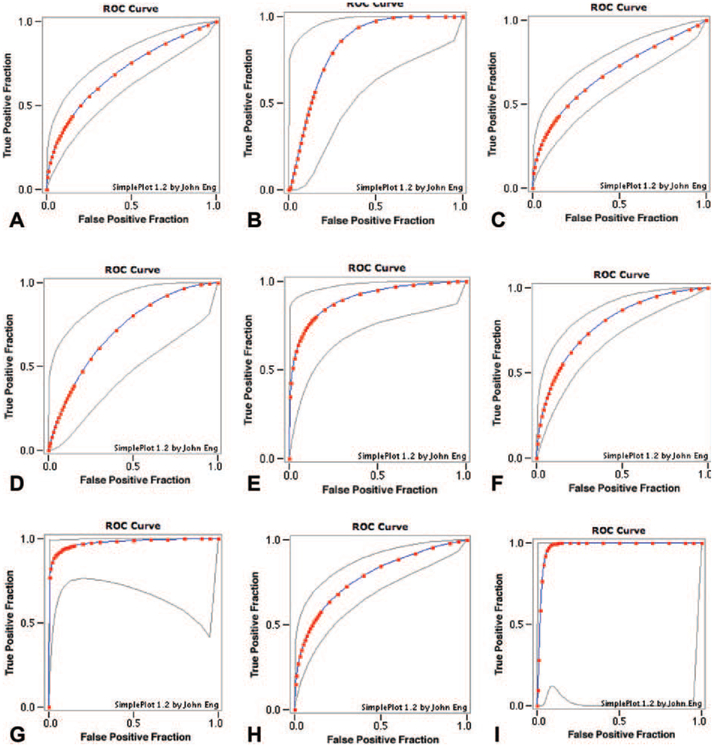
The highest diagnostic accuracy for distinguishing benign from malignant thyroid neoplasms was 79% (miR-138), 98% for Hurthle cell neoplasms (miR-138 and miR-768–3p), and 71% for follicular neoplasms (miR-125b) (Figure 2). Several searchable databases of published miRNA sequences and annotation provided target genes for the distinguishing miRNAs (Table 2).7–9
Figure 2.
(A) Mir-100, overall benign vs malignant samples, fitted ROC area: 0.703. (B) Mir-100, benign vs malignant Hürthle cell samples, fitted ROC area: 0.837. (C) Mir-125b, overall benign vs malignant samples, fitted ROC area: 0.689. (D) Mir-125b, benign vs malignant follicular samples, fitted ROC area: 0.717. (E) Mir-125b, benign vs malignant Hürthle cell samples, fitted ROC area: 0.903. (F) Mir-138, overall benign vs malignant samples, fitted ROC area: 0.791. (G) Mir-138, benign vs malignant Hürthle cell samples, fitted ROC area: 0.978. (H) Mir-768–3p, overall benign vs malignant samples, fitted ROC area: 0.785. (I) Mir-768–3p, benign vs malignant Hürthle cell samples, fitted ROC area: 0.978.
Table 2.
Target Genes Related to Each miRNA
| miRNA | Location | Abbreviation | Gene |
|---|---|---|---|
| miR-100 | 11q24.1 | ID1 | Inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
| EGR2 | Early growth response 2 | ||
| MMP13 | Matrix metallopeptidase 13 (collagenase 3) | ||
| FGFR3 | Fibroblast growth factor receptor 3 | ||
| miR-125b | 11q24.1 | KLF13 CXCL11 | Kruppel-like factor 13 Chemokine (C-X-C motif) ligand 11 |
| FOXA1 | Forkhead box A1 | ||
| miR-138 | 5p15.33 | hTERT | Telomerase reverse transcriptase |
| THRB | Thyroid hormone receptor, beta | ||
| miR-768–3p | 16q22.2 | AP1G1 | Adaptor-related protein complex 1, gamma 1 subunit |
Abbreviation: miRNA, microRNA.
FNA Samples
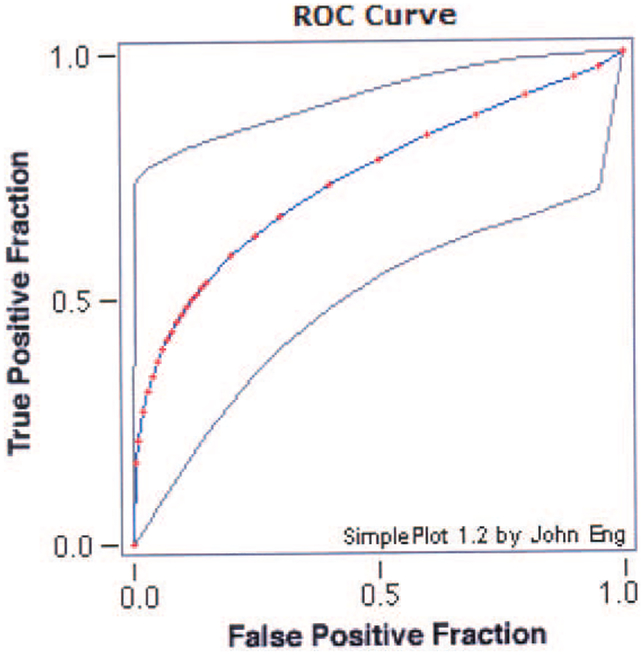
Of the 125 indeterminate FNA samples of thyroid tumors, 78 samples were eventually determined to be benign (of which 48 were follicular or Hurthle cell adenomas) and 37 were malignant (of which 10 were follicular or Hurthle cell carcinomas). Of the 4 miRNAs that could distinguish significantly between benign and malignant thyroid tissue samples, miR-100 showed a borderline significant difference between benign and malignant follicular FNA samples (P = 0.057). MiR-138 showed a highly significant difference between benign and malignant thyroid FNA samples (P < .001) with an accuracy rate of 75% (Figure 3). The NPV of miR-138 for distinguishing between benign and malignant samples was only 81%; however, it was 100% for distinguishing the follicular subtype.
Figure 3.
Receiver operating characteristic (ROC) curve of microRNA (miR)-138 is shown demonstrating its accuracy rate (75%) in distinguishing benign from malignant follicular (including Hurthle cell) fine-needle aspiration samples.
Studies of Cell Proliferation
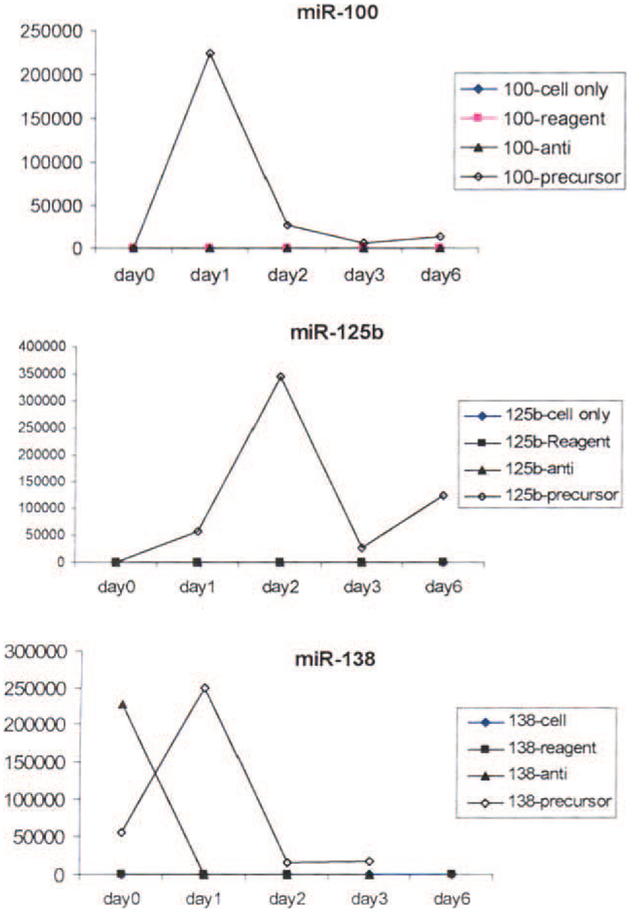
When we determined the in vitro inhibition and overexpression by transient transfection of miR-100, miR-125b, and miR-138 in a FTC cell line (FTC-133), we found that inhibition of the miRNAs by their specific anti-miRNA led to growth arrest. In contrast, overexpression by pre-miRNAs (pre–miR-100, pre–miR-125b, and pre–miR-138) dramatically increased proliferation. Proliferation was especially significant at 24 hours after miR-100 and miR-138 transfection, and expression levels peaked at 48 hours after miR-125b transfection (Figure 4).
Figure 4.
The effect of anti-microRNA (miR) and precursor-miR on miR TaqMan expression of (Top) miR-100, (Middle) miR-125b, and (Bottom) miR-138 is shown. Proliferation was especially significant at 24 hours after miR-100 and miR-138 transfection. Expression levels peaked 48 hours after miR-125b transfection.
Association Between TNM Stage and miRNA Expression
We compared TNM stages of all patients with the miRNA expression data from their tumor samples and observed that only miR-100 showed a significantly different expression (delta-Ct) between patients with T1 tumors and patients with T4 tumors (mean, 1.07 vs 2.78, P = .045).
DISCUSSION
Our study demonstrates that 4 miRs (miR-100, miR-125b, miR-138, and miR-768–3p) distinguished between benign and malignant thyroid tissue samples, and between benign and malignant Hurthle cell samples. Only miR-125b distinguished between follicular adenoma and carcinoma samples, and miR-768–3p distinguished between the benign and follicular variants of PTC samples. For FNA samples that had indeterminate results, only miR-138 distinguished between benign and malignant follicular lesions. We used a pooled miRNA analysis, which is good for marker identification but does not give individual miRNA profiling information. Housekeeping gene U18 was used as a control, because it had the least variable expression. Because it is still unknown in the literature which housekeeping gene is the best control, we calculated both differences in miRNA Ct expression as well as normalized miRNA expression relative to U18. Functional analysis studies confirmed the decrease in miRNA expression by its specific anti-miRNA (growth arrest) and dramatic increase of expression by its specific pre-miRNA (proliferation), which would suggest that these particular miRNAs could have an important role in thyroid carcinoma cell proliferation. These findings, together with the diagnostic accuracy of the 4 miRNAs for distinguishing benign from malignant thyroid neoplasms, suggest that these miRNAs might be markers for diagnosing thyroid cancer in both FNA and final histopathological samples. Of course, the clinical utility of such markers should first be determined, as well as the costs involved in developing a new diagnostic tool.
A variety of miRs are down- and upregulated in PTC, FTC, and ATC.10 Expression of miR-100, which we found to be downregulated, has not been evaluated previously in thyroid cancer, but it is known to be downregulated in ovarian cancer and oral squamous cell carcinoma, and upregulated in hepatocellular cancer and prostate cancer.11–15 MiR-125b, which we found to be downregulated, is reportedly upregulated in PTC compared with normal thyroid tissue, but is downregulated in ATC.16 MiR-125b expression is downregulated in ovarian cancer, oral squamous cell carcinoma, breast cancer, lung cancer, and bladder cancer, and is upregulated in urothelial cancer and prostate cancer.11–14,17–21 MiR-138, which we found to be downregulated, is reportedly downregulated in PTC and in ATC cell lines as compared with PTC cell lines.22–24 MiR-138 was also reported to be downregulated in colon cancer and upregulated both in lung cancer and squamous cell carcinoma of the tongue.14,17 The NPV of miR-138 for distinguishing between benign and malignant was only 81%; however, it was 100% for distinguishing the follicular subtype. Thus, for this subtype, it would be reasonable to consider close follow-up based on testing for miR-138 and forgo a diagnostic thyroidectomy.
MiR-768–3p, which we found to be downregulated, is reportedly downregulated in gastric cancer.4 Several studies have reported miR-221 and miR-222 to be overexpressed in PTC compared with normal thyroid tissue.22,25,26 Finally, a recent study that used a high-density miRNA chip array found 4 miRNAs (miR-192, miR-197, miR-328, and miR-346) to be overexpressed in FTC compared with follicular adenomas.27 This study also provided in vitro evidence for induction of significant growth arrest by anti–miR-197 and anti–miR-346 in cell line FTC-133, the cell line we used in our study as well. In the current study, we observed a difference in miR-100 expression in T1 and T4 tumors. A limitation, however, was the lack of enough prospective malignant FNA samples to validate these data.
PTCs feature commonly mutually exclusive somatic mutations (RET/PTC, BRAF, and RAS), all of which can activate the mitogen-activated protein kinase signaling pathway.28 BRAF mutation is associated with an aggressive tumor phenotype and higher risk of recurrent and persistent disease in patients with conventional PTC.29 Differential miRNA expression has been associated with RET/PTC-, BRAF-, or RAS-positive mutations, although BRAF mutation status was recently found to have no regulatory influence on the expression of the examined miRNAs in PTC.26,30 Geographical mapping of multifocal thyroid cancer by miRNA profiling found that miRNA expression might differ within the tumor.31
For miR-100 and miR-138, we identified some noteworthy target genes. In miR-100, inhibitor of differentiation 1, early growth response 2, and fibroblast growth factor receptor 3 play a part in cell growth and proliferation. In miR-138, the human telomerase reverse transcriptase (hTERT) has a role in cellular senescence, because it is normally repressed in postnatal somatic cells, resulting in progressive shortening of telomeres. Deregulation of telomerase expression in somatic cells may be involved in oncogenesis. The exact role of the other target genes of miR-125b and miR-768–3p is not currently known.
The results of the current study showed that 4 miRNAs (miR-100, miR-125b, miR-138, and miR-768–3p) distinguished between benign and malignant thyroid tissue samples, and that miR-138 even did so for thyroid FNA samples. These miRNAs might be putative markers and illustrate the potential for miRNA markers of thyroid cancer. Clinical applicability needs to be refined and validated in clinical trial, as does information on the related target genes. MicroRNA expression profiling could be a useful tool for optimizing diagnosis and treatment of patients with thyroid cancer.
FUNDING SOURCES
No specific funding was disclosed.
We thank Pamela Derish from the UCSF Department of Surgery Publication Office for her editorial assistance. M.R.V. was supported by the Dutch Cancer Society.
Footnotes
CONFLICT OF INTEREST DISCLOSURE
The authors made no disclosure.
REFERENCES
- 1.Davies L, Welch HG. Increasing incidence of thyroid cancer in the United States, 1973–2002. JAMA. 2006;295:2164–2167. [DOI] [PubMed] [Google Scholar]
- 2.Visone R, Pallante P, Vecchione A, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26:7590–7595. [DOI] [PubMed] [Google Scholar]
- 3.Jazdzewski K, Liyanarachchi S, Swierniak M, et al. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci U S A. 2009;106:1502–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo J, Miao Y, Xiao B, et al. Differential expression of microRNA species in human gastric cancer versus nontumorous tissues. J Gastroenterol Hepatol. 2009;24:652–657. [DOI] [PubMed] [Google Scholar]
- 5.Chudova D, Wilde JI, Wang ET, et al. Molecular classification of thyroid nodules using high-dimensionality genomic data. J Clin Endocrinol Metab. 2010;95:5296–5304. [DOI] [PubMed] [Google Scholar]
- 6.Kebebew E, Peng M, Reiff E, Duh Q-Y, Clark OH, McMillan A. Diagnostic and prognostic value of cell-cycle regulatory genes in malignant thyroid neoplasms. World J Surg. 2006;30:767–774. [DOI] [PubMed] [Google Scholar]
- 7.miRBase: the MicroRNA Database. Available at: http://microrna.sanger.ac.uk/sequences/index.shtml. Accessed September 15, 2011.
- 8.Whitehead Institute for Biomedical Research. TargetScan-Human 5.2. Available at: http://www.targetscan.org. Accessed September 15, 2011.
- 9.PicTar. Available at: http://pictar.mdc-berlin.de. Accessed September 15, 2011.
- 10.Nikiforova MN, Chiosea SI, Nikiforov YE. MicroRNA expression profiles in thyroid tumors. Endocr Pathol. 2009; 20:85–91. [DOI] [PubMed] [Google Scholar]
- 11.Nam EJ, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008; 1:14:2690–2695. [DOI] [PubMed] [Google Scholar]
- 12.Yang H, Kong W, He L, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. [DOI] [PubMed] [Google Scholar]
- 13.Henson BJ, Bhattacharjee S, O’Dee DM, Feingold E, Gollin SM. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer. 2009;48:569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14: 2588–2592. [DOI] [PubMed] [Google Scholar]
- 15.Varnholt H, Drebber U, Schulze F, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. [DOI] [PubMed] [Google Scholar]
- 16.Pallante P, Visone R, Ferracin M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. [DOI] [PubMed] [Google Scholar]
- 17.Baffa R, Fassan M, Volinia S, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–221. [DOI] [PubMed] [Google Scholar]
- 18.Yamada H, Yanagisawa K, Tokumaru S, et al. Detailed characterization of a homozygously deleted region corresponding to a candidate tumor suppressor locus at 21q11-21 in human lung cancer. Genes Chromosomes Cancer. 2008; 47:810–818. [DOI] [PubMed] [Google Scholar]
- 19.Ichimi T, Enokida H, Okuno Y, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–352. [DOI] [PubMed] [Google Scholar]
- 20.Veerla S, Lindgren D, Kvist A, et al. MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int J Cancer. 2009;124:2236–2242. [DOI] [PubMed] [Google Scholar]
- 21.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. [DOI] [PubMed] [Google Scholar]
- 22.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–19080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitomo S, Maesawa C, Ogasawara S, et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008;99:280–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leite KR, Sousa-Canavez JM, Reis ST, et al. Change in expression of miR-let7c, miR-100, and miR-218 from high grade localized prostate cancer to metastasis. Urol Oncol. 2011;29:265–269. [DOI] [PubMed] [Google Scholar]
- 25.Visone R, Russo L, Pallante P, et al. MicroRNAs (miR) −221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. [DOI] [PubMed] [Google Scholar]
- 26.Sheu SY, Grabellus F, Schwertheim S, Handke S, Worm K, Schmid KW. Lack of correlation between BRAF V600E mutational status and the expression profile of a distinct set of miRNAs in papillary thyroid carcinoma. Horm Metab Res. 2009;41:482–487. [DOI] [PubMed] [Google Scholar]
- 27.Weber F, Teresi RE, Broelsch CE, Frilling A, Eng C. A limited set of human microRNA is deregulated in follicur thyroid cancer. J Clin Endocrinol Metab. 2006;91:3584–3591. [DOI] [PubMed] [Google Scholar]
- 28.Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63:1454–1457. [PubMed] [Google Scholar]
- 29.Kebebew E, Weng J, Bauer J, et al. The prevalence and prognostic value of BRAF mutation in thyroid cancer. Ann Surg. 2007;246:466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aherne ST, Smyth PC, Flavin RJ, et al. Geographical mapping of a multifocal thyroid tumour using genetic alteration analysis & miRNA profiling. Mol Cancer. 2008;7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]