Abstract
We aimed to evaluate the clinical significance of bacterial coexistence and the coinfection dynamics between bacteria and respiratory viruses among young children. We retrospectively analyzed clinical data from children aged < 5 years hospitalized with a community-acquired single respiratory viral infection of influenza, adenovirus, or RSV during 2 recent consecutive influenza seasons. Remnant respiratory specimens were used for bacterial PCR targeting Moraxella catarrhalis, Haemophilus influenzae, Streptococcus pneumoniae, and Staphylococcus aureus.
A total of 102 children were included; median age was 0.8 years and 44.1% had underlying comorbidities. Overall, 6.8% (7/102) of cases were classified as severe diseases requiring intensive care unit admission and/or mechanical ventilation and ranged from 8.8% for a patient with RSV and 7.6% for those with adenovirus to 0% for those with influenza viruses. The overall viral–bacterial codetection rate was 59.8% (61/102); M catarrhalis was the most frequent (33.3%), followed by H influenzae (31.4%). Influenza cases showed higher bacterial codetection rates (80.0%; 8/10) compared with those with adenoviruses (69.2%; 9/13) and RSV (55.7%; 44/79). S pneumoniae and H influenzae codetections were associated with reduced severity (aOR, 0.24; 95% CI, 0.07–0.89), and reduced risk of wheezing (aOR, 0.36; 95% CI, 0.13–0.98), respectively.
We observed the interactions between respiratory viruses and bacteria and the clinical significance of viral–bacterial coexistence in upper airway on disease severity. Future study will be necessary to elucidate the active interactions between different viruses and bacteria and give clues to risk stratified strategy in the management of respiratory infections among young children.
Keywords: children, colonization, respiratory tract, severity, viral–bacterial coinfection
KEY POINTS
Among young children with solo-infections of influenza viruses, adenoviruses, and RSV, the overall viral–bacterial codetection rate was 60%. The most frequently codetected bacteria were H influenzae in adenoviral infection (46.2%), M catarrhalis in RSV infection (34.2%), and S pneumoniae in influenza (50.0%; 5/10). Although the presence of viral–bacterial coinfection was not a risk factor for severe LRTIs, the codetection of H influenzae was the protective factor for wheezing, and pneumococcal codetection was associated with reduced severity of viral LRTIs.
1. Introduction
Respiratory tract infections cause a significant morbidity and mortality in children, especially during infancy to preschool age. The majority of acute respiratory infections can be attributed to viral infection, possessing a self-limiting clinical course and often confined to upper respiratory tract involvement. Among diverse respiratory viruses, respiratory syncytial virus (RSV) and influenza virus are the most common pathogens during the winter season in temperate countries. RSV is one of the most important causes of lower respiratory tract infections in infants leading to respiratory failure; internationally 200,000 deaths and 3,000,000 hospitalizations each year are attributed to RSV infection by either direct or nondirect impacts.[1–3] Although influenza viruses are far less common than RSV infection, except for seasonal outbreaks, severe complications can occur when the elderly, children, and patients with underlying disease become infected. Adenovirus infection can manifest with diverse clinical presentations based on serotype all year round and is the causative organism in 5% to 10% of lower respiratory tract infections in children.[4,5]
Viral–bacterial coinfection frequently occurs, and a number of studies emphasize the potential risk of synergistic presentation during coinfection with respiratory viruses and bacteria, showing longer hospital stays and higher morbidity with variations in which bacterial strains were invaders being modified by personal or population immunity.[2,6–8] Secondary bacterial infection serves as an aggravating factor for disease severity as formerly shown during the influenza pandemics in which mortality cases were vastly attributed to secondary bacterial infection from Streptococcus pneumoniae (S pneumoniae), or Staphylococcus aureus (S aureus).[6,9,10] Although most coinfection research has focused on influenza viruses, other viruses also participate in viral–bacterial coinfections, including RSV and adenoviruses.[10]
In healthy children, the upper respiratory tract is colonized by several microorganisms which can become potential pathogens in certain circumstances. S pneumoniae is detected as a colonizer of 8% to15% of healthy, asymptomatic adults and adolescents, and 30% to 70% of children, which is responsible for most cases of community-acquired pneumonia, sinusitis, otitis media, and meningitis.[11] Evidence is accumulating that bacterial colonization or infection of the respiratory tract might modulate the viral infection by disturbing various steps of defense mechanism spanning to signal pathway and viral infection might also modulate the subsequent bacterial infection in similar context.[8,12–14] However, the exact epidemiology and degree of impact on clinical course of community acquired respiratory infections are still being gathered.
In this study, we analyzed viral–bacterial codetection in the upper respiratory tract among young children with a single respiratory viral infection in distinct severity categories. We aimed to assess the coinfection dynamics between bacteria and respiratory viruses especially focused on influenza viruses, adenoviruses, and RSV and the clinical significance of viral–bacterial codetection.
2. Methods
2.1. Study population
Data from the clinical virology laboratory at Asan Medical Center was used to determine the relative frequency of respiratory viruses detected at Asan Medical Center Children‘s Hospital during 2 recent consecutive influenza seasons (November 2015 to April 2016 and November 2016 to April 2017). During the study period, clinical and demographic data from hospitalized pediatric patients aged under 5 years with community-acquired single viral infections of influenza A/B, RSV A/B, or adenovirus detected by real-time multiplex polymerase chain reaction (PCR) (Seeplex: Seegene Inc, Seoul, Korea) were abstracted from electronic medical records including clinical diagnosis, duration of hospital stay, underlying medical conditions, antibiotics use, need for intensive care unit stay, and/or mechanical ventilation. The analysis only included the first virus detected during a single clinical episode occurring within a 4-week period, and duplicates from the same patient were excluded. The following cases were excluded: healthcare-associated infection, coexistence of ≥ 2 different respiratory viruses, influenza cases confirmed only by rapid influenza antigen test, and respiratory viral infections other than adenovirus, influenza, or RSV.
Patients with a respiratory infection were grouped into 3 severity subgroups, as mild, moderate, and severe infections; “mild” infection referred to no need for respiratory support, absence of tachypnea (respiratory rate > 50/min at age of 2–11 mo and > 40/min at age of 1–5 yrs),[15] and no definite chest wall indrawing; “moderate” infection meant the presence of oxygen need (oxygen saturation < 93% on room air), tachypnea or chest wall indrawing; “severe” infection was defined as need for advanced respiratory support due to respiratory failure (i.e., invasive or noninvasive mechanical ventilation), or need for pediatric intensive care unit (PICU) admission for reasons other than respiratory failure (i.e., hemodynamic instability, lethargy, reduced level of consciousness, convulsions), or cases of death.
This study was approved by the Institutional Review Board of Asan Medical Center with a waiver of informed consent for a retrospective, deidentified data collection, and analysis (IRB No. 2018-0117).
2.2. Definition
Acute respiratory infection was defined as an illness of < 7 days duration with ≥ 2 of the following symptoms: a cough, fever, nasal congestion, and sore throat. Lower respiratory tract infection (LRTI) including acute bronchitis, acute bronchiolitis, and pneumonia was defined as the presence of wheezing, rales, hypoxia, and/or chest radiographic opacity/pleural effusion. Respiratory symptoms not satisfying those of LRTI were diagnosed as upper respiratory tract infections (URTI). Community-acquired infection was defined as an infection excluding healthcare-associated infections, which referred to infections newly occurring after 48 hours of admission, or within 30 days of discharge from any healthcare facility.[16]
2.3. Viral identification
With the exception of respiratory specimens only for rapid influenza antigen testing which were obtained by nasopharyngeal swabs, respiratory specimens of nasopharyngeal aspirates submitted to the clinical virology laboratory for detection of respiratory viruses were tested using real-time multiplex PCR (Seeplex), which differentiated 15 species of common respiratory viruses including influenza A and B, RSV A and B, and adenovirus. Nasopharyngeal aspirates were collected within 3 days of symptom onset and no later than 7 days from the patients who might have respiratory diseases with/or without fever, if possible prior to the initiation of antimicrobial therapy at our institute.
2.4. Bacterial identification
Remnants of nasopharyngeal aspirates after viral confirmation using multiplex PCR, which were kept at −70°C were used for the bacterial PCR as follows. Nasopharyngeal aspirates stored at −70°C, were thawed and centrifuged. We suspended a pellet in 180 μL of the enzyme solution (20 mg/mL lysozyme, 20 mM Tris-HCl [pH 8.0], 2 mM EDTA, 1% Triton X-100) and incubated at 37°C for 30 minutes. Extraction and purification of DNA from this sample was done according to the manufacturer's instructions for the QIAmp DNA Mini Kit (QIAGEN GmbH, Hilden, Germany).
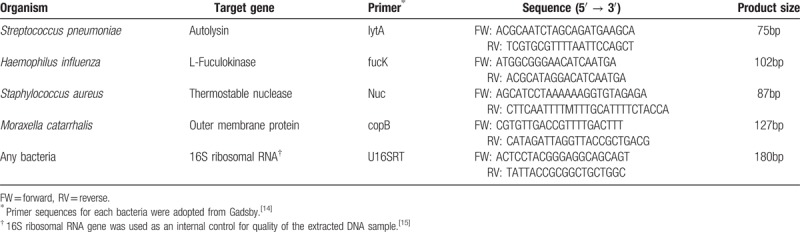
Bacterial PCR detecting Moraxella catarrhalis (M catarhallis), Haemophilus influenzae (H influenzae), S pneumoniae, and S aureus was performed using primer sequences adopted from the 2015 Publication by Gadsby,[17] and the 16S ribosomal RNA gene was used as an internal control[18] (Table 1). Bacterial PCR amplifications were performed using 20 μL reaction mixtures containing distilled water 10.8 μL, 10× PCR buffer 2 μL, 25 mN MgCl2 1.6 μL, primer set 0.4 μL each, 2.5 mM dNTP mixture 1.6 μL, 5U/μL Taq DNA polymerase 0.2 μL (TaKaRa Bio Inc, Shiga, Japan), and 3 μL of DNA extract. After placing reaction tubes within the GeneAmp PCR System 9700 (Applied Biosystems, Foster, CA), we maintained the temperature at 95°C for 5 minutes and then immediately went through 35 cycles, each consisting of 95°C for 1 minute, 55°C for 30 seconds, and 72°C for 1 minute. The final cycle had a prolonged extension time of 5 minutes. Distilled water was used as a negative control.
Table 1.
Primers for bacterial PCR.
2.5. Statistical analysis
Categorical data were analyzed with χ2 test and Fischer exact test, and continuous variables with independent t test, or 1-way ANOVA. A multivariate logistic regression analysis was used to assess the potential impact of variables on clinical severity, with bootstrapping to obtain confidence intervals. A P value of <.05 was considered statistically significant for the comparisons. Statistical Program for Social Science release 11 was used for all statistical calculations.
3. Results
3.1. Study population and clinical manifestation
Among the 796 nasopharyngeal aspirates from hospitalized patients who showed positive results for any respiratory viruses using real-time multiplex PCR during 2 consecutive influenza seasons, 51.5% (410/796) comprised of adenovirus (n = 114), influenza viruses (n = 53), or RSV (n = 271); 50.2% (206/410) of these infections occurred in children < 5 years of age (Fig. 1). Excluding coinfection with ≥ 2 different viruses (n = 67) and healthcare-associated infections (n = 37), a total of 102 children aged under 5 years with community-acquired single viral infections of adenovirus (n = 13), influenza A (n = 7), influenza B (n = 3), and RSV (n = 79) were included in this analysis.
Figure 1.
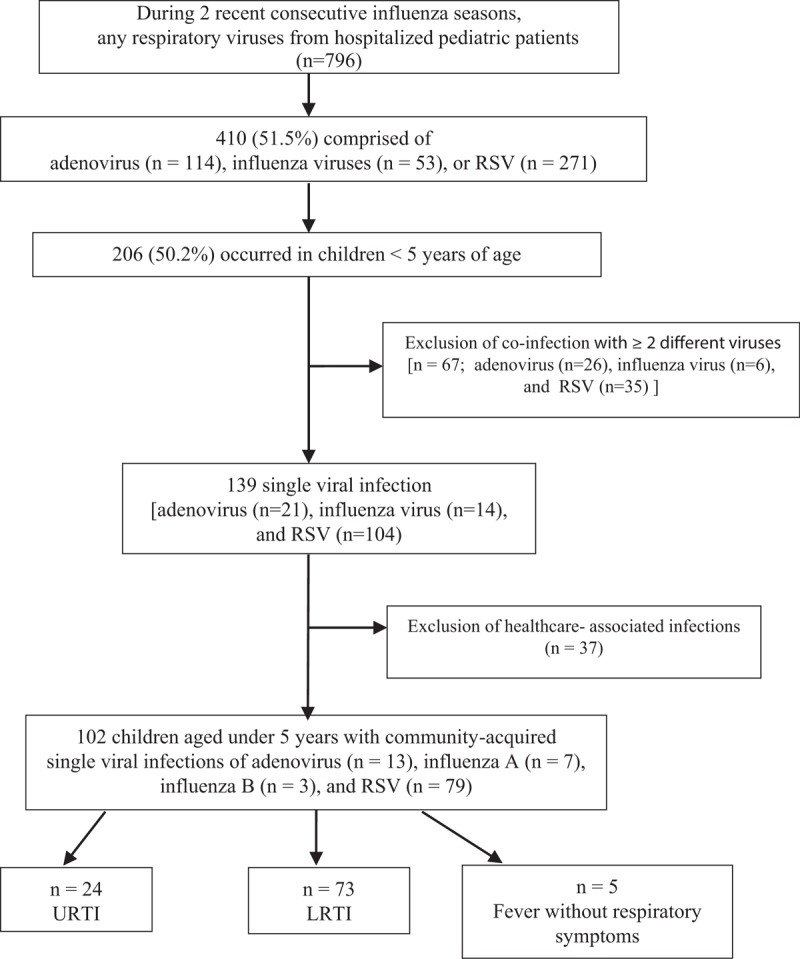
Study flow of recruitment of children hospitalized under age 5, with proven single viral infections of adenovirus, influenza A and B viruses, and respiratory syncytial virus (RSV). LRTI = lower respiratory tract infection, RSV = respiratory syncytial virus, URTI = upper respiratory tract infection.
The demographic and clinical characteristics of 102 patients are shown in Table 2. Median age was 0.83 years [interquartile range (IQR), 0.3–2.0 yrs] and 45 (44.1%) patients had underlying diseases including chronic respiratory conditions (n = 15) and congenital heart diseases (n = 13). The most common diagnosis was LRTI (71.5%), followed by URTI (23.5%) and fever without localizing signs (4.9%). Fever ≥ 38°C as an initial manifestation was present more frequently with adenovirus (81.8%) than with influenza (60.0%) or RSV (53.1%). Overall median length of hospital stay was 4 days (IQR 3.0–6.0 d); 4 days (IQR 3.0–6.0 d) in adenovirus and RSV, and 3 days (IQR 2.0–5.0 d) in influenza viruses, respectively.
Table 2.
Demographic data and clinical characteristics of hospitalized children < 5 years old with community-acquired single viral infections during 2 consecutive influenza seasons.
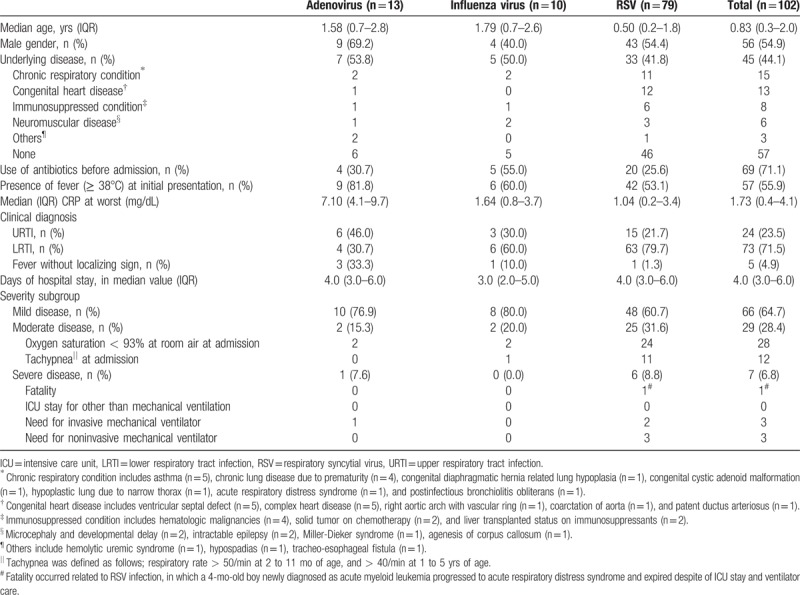
The occurrence of severe clinical outcome requiring PICU admission and mechanical ventilation ranged from 8.8% for patients with RSV and 7.6% for those with adenovirus to 0% for those with influenza viruses. Overall, 6.8% (7/102) of cases were classified as severe with 1 fatal case associated with RSV infection, in which a 4-month-old boy newly diagnosed with acute myeloid leukemia progressed to acute respiratory distress syndrome and expired despite ICU stay and ventilator care.
3.2. Prevalence of viral–bacterial codetection
Overall viral–bacterial codetection rate was 59.8% (61/102); M catarrhalis was the most frequent bacteria in the nasopharyngeal aspirates (33.3%), followed by H influenzae (31.4%), S pneumoniae (21.6%), and S aureus (7.8%) (Table 3). There was no significant difference in the distribution of 4 different bacteria in the upper respiratory tract according to the clinical diagnosis of LRTI and non-LRTI. Patients with influenza viruses showed higher bacterial coinfection rates (80.0%; 8/10) compared with those with adenoviruses (69.2%; 9/13) and RSV (55.7%; 44/79) even though there was no statistically significant difference. The most frequently observed pairs of viral and bacterial combination in each viral infection were as follows: adenovirus and H influenzae (46.2%; 6/13), RSV and M catarrhalis (34.2%; 27/79), and influenza viruses and S pneumoniae (50.0%; 5/10). Pneumococcal coinfection rate was 50% in influenza viral infection, which was higher than that of adenovirus (15.3%) or RSV (18.9%).
Table 3.
Distribution of viral–bacterial coinfection among children infected with any single respiratory viruses.
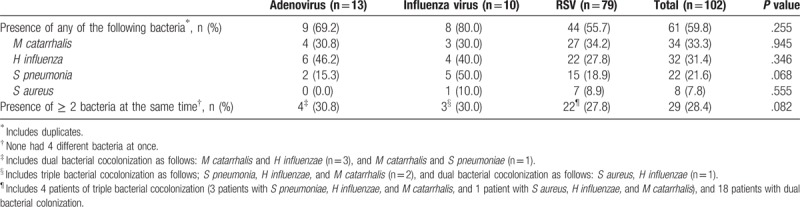
Overall, 29 pairs of viral–bacterial codetections had ≥ 2 different bacteria simultaneously; dual or triple bacterial coinfection rate was not statistically different according to each virus, ranging from 27.8% to 30.8% (Table 3). The percent with ≥ 2 bacterial species was not statistically different between LRTI and non-LRTI [24.7% (18/73) and 37.9% (11/29), respectively; P = .180]. Although simultaneous detection of ≥ 3 different bacteria in the nasopharyngeal aspirates was not observed in adenoviral infection, 5 of the children infected with RSV or influenza virus were colonized with 3 different bacteria. However, none of the aspirates exhibited 4 different bacteria at once.
3.3. Clinical significance of viral–bacterial codetection
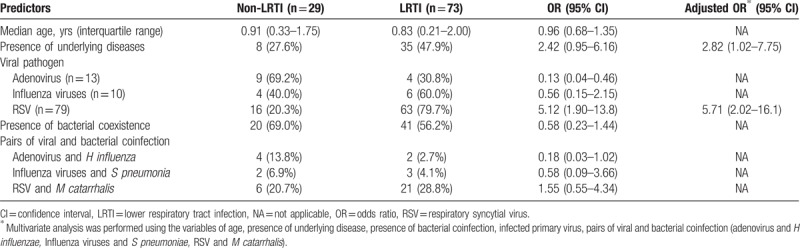
Statistically significant risk factors associated with LRTIs were as follows: the presence of underlying diseases [adjusted odds ratio (aOR), 2.82; 95% confidence interval (CI), 1.02–7.75] and viral pathogen of RSV (aOR, 5.71; 95% CI, 2.02–16.1) (Table 4). However, age was not an additional risk factor for progression to LRTIs among young children aged < 5 years with respiratory viral infections (OR, 0.96; 95% CI, 0.68–1.35). In addition, the presence of viral–bacterial coinfection and frequently observed pairs of viral–bacterial combinations did not have any significant influence on progression to LRTI.
Table 4.
Risk factors for progression to LRTIs.
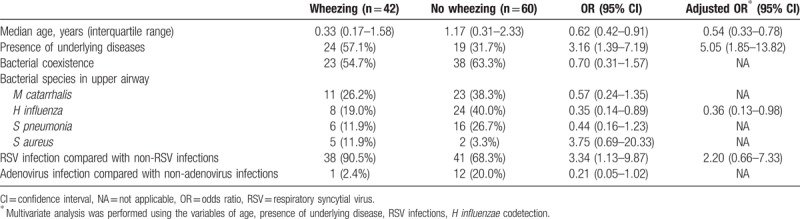
Younger age and presence of underlying diseases were associated with the increased risk of accompanying wheezing (aOR, 1.85 and 5.05, respectively) (Table 5). Although RSV itself was a risk factor for wheezing in univariate analysis (OR, 3.34; 95% CI, 1.13–9.87), there was no statistical significance in multivariate analysis. The codetection of H influenzae was the protective factor for wheezing (aOR, 0.36; 95% CI, 0.13–0.98).
Table 5.
Factors associated with presentation of wheezing.
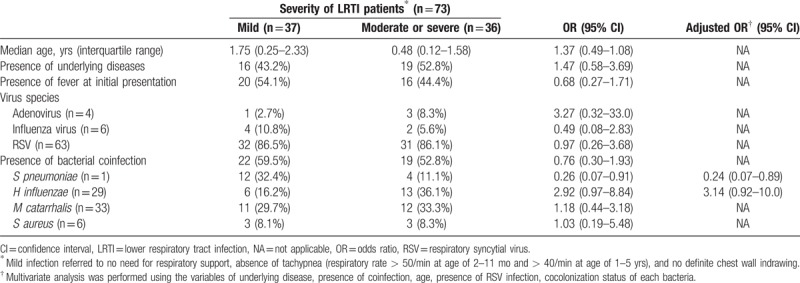
Among the 73 patients with LRTIs, age, underlying disease, and viral species did not affect the clinical severity (Table 6); RSV infection itself had no association with the severity of LRTIs compared with adenoviral or influenza viral infections (OR 0.97; 95% CI 0.26–3.68). Although codetection of H influenzae had a tendency to be associated with increased severity (aOR 3.14, 95% CI 0.92–10.0), the presence of viral–bacterial coinfection was not a risk factor for severe LRTIs (OR, 0.76; 95% CI, 0.30–1.93). However, pneumococcal codetection itself was associated with reduced severity (aOR, 0.24; 95% CI, 0.07–0.89).
Table 6.
Comparison among 73 patients presented with LRTI according to the clinical severity.
4. Discussion
This study over 2 consecutive influenza seasons showed that 60% of single respiratory viral infections including adenovirus, influenza viruses, and RSV occurred in the setting of coexistence of potentially pathogenic respiratory bacteria in the upper respiratory tract of young children. Frequently detected bacteria in the nasopharyngeal aspirates were different according to each viral infection regardless of upper or lower respiratory tract involvement. Although clinical severity of lower respiratory tract involvement from a single respiratory viral infection in young children was not affected by the presence of bacterial coinfection in the upper respiratory tract, codetection of any specific bacteria including H influenzae and S pneumoniae might affect the presence of wheezing and the clinical severity, respectively.
In this study, codetection of bacteria in the nasopharyngeal aspirates by PCR was referred to as coinfection or colonization implying no difference in the meaning. The detection rate of any single viral pathogen of interest with any bacterial pathogen was 60%, in line with the previous studies of children with acute viral respiratory infections showing 23% to 83% of bacterial codetection.[19–21] Colonization might directly influence transmission rate by the evolution of virulence factors, facilitated host adherence, and evasion from host immunity.[12,13,22] Coinfections with viral pathogens enhance bacterial colonization of the airway, due to increased bacterial adherence facilitated by denuded airway epithelium during acute respiratory tract infections,[12,23,24] and a small percentage of children colonized with bacteria will develop invasive disease immediately or later following colonization, especially on an inflamed nasopharynx.[25,26] Although it is well known that the secondary bacterial infection serves as an aggravating factor of disease severity as formerly shown during the influenza pandemics,[9,10,27] pneumococcal coinfection in the upper respiratory tract was associated with reduced severity of viral LRTIs in this study, which might conflict with the known synergism between viral–bacterial coinfection. It can be partly explained by the small number of influenza cases; influenza cases diagnosed only by rapid influenza antigen test were not included in this study because no remnant frozen specimens were available for bacterial PCR assays. In addition, only seasonal influenza epidemics were observed during this study period, and none of the patients infected with influenza ended up with a severe clinical outcome.
Dynamics of the polymicrobial carriage were previously reported, showing increased polymicrobial colonization especially during acute respiratory infection, and cooperative relationship of S pneumoniae and H influenzae.[28,29] Our study also showed corresponding data supporting the patterns of polymicrobial dynamics, implying the complex competitive interactions during an acute respiratory infection might shift the association between bacteria from negative to positive. Except for 1 patient infected with RSV who showed cocolonization of both S pneumoniae and S aureus at the same time, those 2 bacteria were not detected together elsewhere. These findings were also concordant with the previous knowledge of negative associations between carriage of S pneumoniae and S aureus, which may be associated with immune-mediated interspecies inferences.[29–32]
Emerging evidence reveals that the airway microbiome changes were associated with postviral wheeze; especially, higher risk of developing persistent wheezing and asthma in the presence of M catarrhalis in viral infection.[33,34] In this study, however, there was no increased risk of wheeze in young children experiencing viral infections, irrespective of coexistence of M catarrhalis. Our study suggested that codetection of H influenzae was significantly associated with lower odds of presentation with wheezing. Although this association does not necessarily mean causation, and the effect of previous or concomitant use of antibiotics cannot be excluded, the airway microbiome during viral infections is a relevant determinant of wheeze in young children.
Our study has some limitations. First, the sample size was relatively small and had low power to evaluate the significance of viral–bacterial coinfection. Even though the study period included 2 consecutive influenza seasons in Korea, the number of influenza cases was too small to analyze the synergism of influenza virus and pneumococci or S aureus. Secondly, respiratory specimens obtained from children without viral infections such as influenza, adenovirus, or RSV were not included as negative controls. So, it is uncertain whether the codetection of certain bacteria in the upper respiratory tract is under the influence of each virus. And third, from the study design, we only included patients with solo—infections of adenovirus, influenza, and RSV which are known to be associated with severe LRTIs. Rhinovirus, for example, is another frequently detected respiratory virus associated with both upper and lower respiratory tract infections. It can possibly cause synergistic bacterial coinfection as shown from previous studies revealing rhinovirus with S pneumoniae as the most common combination.[35–37] By restricting the viruses of interest, the overall comprehension of distribution and dynamics of the pathogens of respiratory tract might have been precluded. In addition, the presence of bacteria in the nasopharyngeal aspirates was determined not by semiquantitative real-time PCR but conventional PCR, which leaves some uncertainty of significant bacterial abundance on viral–bacterial interaction. Furthermore, the presence of bacteria in the upper respiratory tract might not reflect the viral–bacterial interaction in lower respiratory tract infections. Finally, we did not review the individual histories of immunization including pneumococcal and H influenza type b (Hib) vaccines. However, in Korea, Hib and pneumococcal protein-conjugate vaccine had been included in the national immunization programs since 2013 and May 2014, respectively, and as of 2017, the coverage rate for both vaccines is estimated to have risen up to 97% to 98%.[38] So, the incidence of viral–bacterial codetection might not reflect the differences between vaccinated or nonvaccinated patients.
5. Conclusions
Although coexistence of bacteria does not have an apparent impact on progression to viral LRTI, the codetection of H influenzae was the protective factor for wheezing, and pneumococcal codetection was associated with reduced severity of viral LRTIs. Despite the small study population, influenza virus most frequently detected with S pneumoniae, RSV with M catarrhalis, and adenovirus with H influenzae. Dual or triple bacterial coinfection rate was 28%, which was not statistically different according to each virus or progression to LRTIs. A future study with a larger study population and perhaps including lower respiratory tract aspirates might help to elucidate the active interaction mechanism between viral–bacterial interactions and provide insight to the risk-stratified management of respiratory infections among young children.
Acknowledgment
The authors are very grateful to Jung-Hwa Kim for assistance with an experiment during the course of this research.
Author contributions
Data curation: Jiwon Jung.
Formal analysis: Jiwon Jung.
Investigation: Jiwon Jung, Euri Seo, Ree Nar Yoo, Hungseop Sung, Jina Lee.
Methodology: Jiwon Jung, Ree Nar Yoo, Jina Lee.
Project administration: Jina Lee.
Resources: Euri Seo, Ree Nar Yoo, Hungseop Sung.
Supervision: Jina Lee.
Writing – original draft: Jiwon Jung.
Jina Lee orcid: 0000-0002-3435-251X.
Footnotes
Abbreviations: CI = confidential interval, IQR = interquartile range, LRTI = lower respiratory tract infection, OR = odds ratio, PCR = polymerase chain reaction, PICU = pediatric intensive care unit, RSV = respiratory syncytial virus, URTI = upper respiratory tract infections.
How to cite this article: Jung J, Seo E, Yoo RN, Sung H, Lee J. Clinical significance of viral–bacterial codetection among young children with respiratory tract infections: Findings of RSV, influenza, adenoviral infections. Medicine. 2020;99:2(e18504).
This work was supported by a grant from the Korean Society of Pediatric Infectious Diseases and Boryung Pharmaceuticals.
The authors have no conflicts of interest to disclose.
References
- [1].Nair H, Nokes DJ, Gessner BD, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 2010;375:1545–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Thorburn K, Harigopal S, Reddy V, et al. High incidence of pulmonary bacterial co-infection in children with severe respiratory syncytial virus (RSV) bronchiolitis. Thorax 2006;61:611–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Weinberger DM, Klugman KP, Steiner CA, et al. Association between respiratory syncytial virus activity and pneumococcal disease in infants: a time series analysis of US hospitalization data. PLoS Med 2015;12:e1001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hong JY, Lee HJ, Piedra PA, et al. Lower respiratory tract infections due to adenovirus in hospitalized Korean children: epidemiology, clinical features, and prognosis. Clin Infect Dis 2001;32:1423–9. [DOI] [PubMed] [Google Scholar]
- [5].Jin Y, Zhang RF, Xie ZP, et al. Prevalence of adenovirus in children with acute respiratory tract infection in Lanzhou, China. Virol J 2013;10:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Brealey JC, Sly PD, Young PR, et al. Viral bacterial co-infection of the respiratory tract during early childhood. FEMS Microbiol Lett 2015;362: fnv062. [DOI] [PubMed] [Google Scholar]
- [7].Michelow IC, Olsen K, Lozano J, et al. Epidemiology and clinical characteristics of community-acquired pneumonia in hospitalized children. Pediatrics 2004;113:701–7. [DOI] [PubMed] [Google Scholar]
- [8].Smith CM, Sandrini S, Datta S, et al. Respiratory syncytial virus increases the virulence of Streptococcus pneumoniae by binding to penicillin binding protein 1a. A new paradigm in respiratory infection. Am J Respir Crit Care Med 2014;190:196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Joseph C, Togawa Y, Shindo N. Bacterial and viral infections associated with influenza. Influenza Other Respir Viruses 2013;7: suppl 2: 105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].McCullers JA. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol 2014;12:252–62. [DOI] [PubMed] [Google Scholar]
- [11].Regev-Yochay G, Raz M, Dagan R, et al. Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin Infect Dis 2004;38:632–9. [DOI] [PubMed] [Google Scholar]
- [12].Bellinghausen C, Rohde GGU, Savelkoul PHM, et al. Viral-bacterial interactions in the respiratory tract. J Gen Virol 2016;97:3089–102. [DOI] [PubMed] [Google Scholar]
- [13].Siegel SJ, Weiser JN. Mechanisms of bacterial colonization of the respiratory tract. Annu Rev Microbiol 2015;69:425–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].de Steenhuijsen Piters WA, Heinonen S, Hasrat R, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med 2016;194:1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hyde TB, Gay K, Stephens DS, et al. Macrolide resistance among invasive Streptococcus pneumoniae isolates. JAMA 2001;286:1857–62. [DOI] [PubMed] [Google Scholar]
- [16].Inweregbu K, Dave J, Pittard A. Nosocomial infections. BJA Educ 2005;5:14–7. [Google Scholar]
- [17].Gadsby NJ, McHugh MP, Russell CD, et al. Development of two real-time multiplex PCR assays for the detection and quantification of eight key bacterial pathogens in lower respiratory tract infections. Clin Microbiol Infect 2015;21:781.e1–8.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Clifford RJ, Milillo M, Prestwood J, et al. Detection of bacterial 16S rRNA and identification of four clinically important bacteria by real-time PCR. PLoS One 2012;7:e48558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Skevaki CL, Tsialta P, Trochoutsou AI, et al. Associations between viral and bacterial potential pathogens in the nasopharynx of children with and without respiratory symptoms. Pediatr Infect Dis J 2015;34:1296–301. [DOI] [PubMed] [Google Scholar]
- [20].Teo SM, Mok D, Pham K, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015;17:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cebey-Lopez M, Herberg J, Pardo-Seco J, et al. Does viral co-infection influence the severity of acute respiratory infection in children? PLoS One 2016;11:e0152481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Givon-Lavi N, Fraser D, Porat N, et al. Spread of Streptococcus pneumoniae and antibiotic-resistant S. pneumoniae from day-care center attendees to their younger siblings. J Infect Dis 2002;186:1608–14. [DOI] [PubMed] [Google Scholar]
- [23].Hirano T, Kurono Y, Ichimiya I, et al. Effects of influenza A virus on lectin-binding patterns in murine nasopharyngeal mucosa and on bacterial colonization. Otolaryngol Head Neck Surg 1999;121:616–21. [DOI] [PubMed] [Google Scholar]
- [24].Plotkowski MC, Puchelle E, Beck G, et al. Adherence of type I Streptococcus pneumoniae to tracheal epithelium of mice infected with influenza A/PR8 virus. Am Rev Respir Dis 1986;134:1040–4. [DOI] [PubMed] [Google Scholar]
- [25].Ghaffar F, Friedland IR, McCracken GH., Jr Dynamics of nasopharyngeal colonization by Streptococcus pneumoniae. Pediatr Infect Dis J 1999;18:638–46. [DOI] [PubMed] [Google Scholar]
- [26].Robinson J. Colonization and infection of the respiratory tract: what do we know? Paediatr Child Health 2004;9:21–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev 2006;19:571–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].DeMuri GP, Gern JE, Eickhoff JC, et al. Dynamics of bacterial colonization with Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis during symptomatic and asymptomatic viral upper respiratory tract infection. Clin Infect Dis 2018;66:1045–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lewnard JA, Givon-Lavi N, Huppert A, et al. Epidemiological markers for interactions among Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus in upper respiratory tract carriage. J Infect Dis 2016;213:1596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lijek RS, Luque SL, Liu Q, et al. Protection from the acquisition of Staphylococcus aureus nasal carriage by cross-reactive antibody to a pneumococcal dehydrogenase. Proc Natl Acad Sci U S A 2012;109:13823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Regev-Yochay G, Dagan R, Raz M, et al. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in Children. JAMA 2004;292:716–20. [DOI] [PubMed] [Google Scholar]
- [32].Shak JR, Vidal JE, Klugman KP. Influence of bacterial interactions on pneumococcal colonization of the nasopharynx. Trends Microbiol 2013;21:129–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bisgaard H, Hermansen MN, Buchvald F, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med 2007;357:1487–95. [DOI] [PubMed] [Google Scholar]
- [34].Zhou Y, Bacharier LB, Isaacson-Schmid M, et al. Azithromycin therapy during respiratory syncytial virus bronchiolitis: upper airway microbiome alterations and subsequent recurrent wheeze. J Allergy Clin Immunol 2016;138:1215.e5–9.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Honkinen M, Lahti E, Österback R, et al. Viruses and bacteria in sputum samples of children with community-acquired pneumonia. Clin Microbiol Infect 2012;18:300–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bhuiyan MU, Snelling TL, West R, et al. Role of viral and bacterial pathogens in causing pneumonia among Western Australian children: a case-control study protocol. BMJ Open 2018;8:e020646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Toivonen L, Schuez-Havupalo L, Karppinen S, et al. Rhinovirus infections in the first 2 years of life. Pediatrics 2016;138: e20161309. [DOI] [PubMed] [Google Scholar]
- [38].Republic of Korea: WHO and UNICEF estimates of immunization coverage: 2018 revision [Available at: https://www.who.int/immunization/monitoring_surveillance/data/kor.pdf Accessed July 2, 2019. [Google Scholar]


