Abstract
Traditionally, proteomic studies have been carried out on whole tissues or organs enabling the profiling of thousands of proteins within a single LC-MS analysis. A disadvantage of this approach is that proteomes generated from whole tissues are an “average” that represents a blend of cell types and distinct anatomical regions which can obscure important biological phenomena. Laser capture microdissection (LCM) is an elegant method that allows tissue features of interest, as small as a single cell, to be identified and isolated for downstream analysis. Herein we describe an approach that utilizes an immobilized enzyme reactor (IMER) coupled directly to nanoLC-MS/MS for highly sensitive, automated, quantitative proteomic analysis of the microscopic tissue specimens generated by LCM.
Keywords: Immobilized enzyme reactor, Laser capture microdissection, Mass spectrometry, NanoLC, Nanoproteomics, Proteomics
1. Introduction
Quantitative global proteomics analyses are now broadly applied across the field of biological research to study an organism (s) protein expression in response to perturbation or phenotype of interest [1]. In order to produce high quality quantitative data, efficient and reproducible sample preparation workflows are crucial, especially when employing label-free methods [2,3]. This becomes increasingly challenging as sample size decreases due to the protein losses incurred in the multiple processing steps required for producing peptides amenable to MS analysis [4]. This challenge has limited the effective application of proteomics to many sample types of great interest, including LCM dissected tissues, FACS sorted cells, circulating tumor cells, and developing embryos.
The potential significance of these measurements has driven the development of a wide variety of approaches to address the inherent limitations of proteomic sample handling. These methodologies employ three main approaches, or a combination thereof, to reducing sample losses: substitute denaturants, proteomic reactors, and online sample handling. Sample extraction and protein denaturation, a critical first step in all proteomic workflows, is achieved using chaotropes, most often high concentrations of urea or detergents. Both of these reagents are incompatible with LC-MS analysis and need to be removed resulting in significant protein losses. By substituting these reagents with volatile solvents or acid-cleavable detergents, all sample manipulations can be made within the same vessel greatly reducing losses [5–9]. This approach has the clear advantage of being the most straightforward to implement and has been used robustly with LCM tissue samples [7, 8, 10]. Neverthe-less, many sample isolations require the use of salts or detergents which make this approach incompatible. The proteomic reactor approach employs a solid phase support to immobilize proteins and/or peptides, allowing protein chemistry, digestion, and buffer exchange without sample transfers. This approach has been successfully applied in both manual and online configurations [11–13]. Proteomic reactor approaches offer greater reagent flexibility; however, they require many manual sample handling steps and long preparation times challenging reproducibility and requiring expert practitioners [14, 15]. Online sample handling borrows the technologies of the microscale separations community to carry out proteomics sample preparation within enclosed systems, i.e., fused silica capillaries, controlled by chromatography pumps and fluidic systems [16–18]. Most commonly, immobilized enzyme reactors (IMER) are used to achieve rapid protein digestions and eliminate the need for frequent additions of fresh protease [19–25]. These configurations allow for the miniaturization of reaction vessels and solid phase supports, reducing losses to nonspecific binding while facilitating reproducible handling of nanoliter sample volumes in an automated fashion. In theory, online handling represents the ideal solution for nanoproteomics, but in practice online handling systems are highly complex creating large numbers of potential fail points, resulting in significant downtimes and precluding their application to biological problems requiring large sample numbers to achieve sufficient statistical power.
To address this challenge, we have developed a simplified online platform capable of reproducibly handling small protein quantities and allowing for label-free proteomics analysis, termed the Simplified Nano-Proteomics Platform (SNaPP) [19, 26]. Prior to injection, proteins are extracted, denatured, and reduced by the addition of a small volume of extraction buffer. The SNaPP system utilizes immobilized trypsin beads packed into a fused silica capillary to create a high-efficiency enzyme reactor with minimal surface area [27, 28 ]. This reactor is directly interfaced with a capillary solid phase extractor (SPE) for online desalting of the resultant peptides. The SPE is then switched in line with a 50 μm I.D. analytical column for LC-MS/MS analysis. This system has been successfully applied for quantitative analysis of blastocysts, LCM dissected tissues, sorted cells, glomerular filtrate, and exosomes.
2. Materials
Prepare buffer solutions using Milli-Q purified water and analytical grade reagents. Mobile phase A and B should be premixed HPLC grade solvents. Dithiothreitol (DTT) can be purchased from Sigma in 7.7 mg preweighed individually sealed aliquots to ensure freshness and accuracy.
Tris buffer: 50 mM Tris, 1 mM CaCl2, pH 8.0. Weigh out 767 mg Tris–HCl and 925 mg Tris-Base, and then transfer to 250 mL graduated cylinder. Next, weigh out 27.5 mg CaCl2 and add to the cylinder. Add water to bring the total volume to 250 mL and check the pH to verify (see Note 1).
DTT stock: 500 mM dithiothreitol in Tris buffer (see Note 2). Empty the contents of a DTT preweigh tube into 0.6 mL micro-centrifuge tube (see Note 3). Add 100 μL of Tris buffer and vortex for 1 min to solubilize DTT.
IMER column wash: Mix 25 mL mobile phase A with 25 mL of mobile phase B.
3. Methods
3.1. Sample Preparation
Prepare the homogenization buffer by weighing out 480 mg of urea into a 1.5 mL microcentrifuge tube. Add 650 μL of Tris buffer and 20 μL of DTT stock to dissolve urea (see Note 4). Once dissolved bring the final volume to 1 mL with Tris buffer.
Deposit 10 μL of homogenization buffer on top of the dissected tissue mounted on the LCM cap. Allow the buffer to absorb into the tissue for 10 min.
After absorption, the tissue will be become very soft. Pipette the entire volume of homogenization buffer volume up and down 5–10 times to remove the tissue from the cap and transfer vial to LC-MS vial (see Note 5).
Rinse the LCM cap with a 20 μL aliquot of Tris buffer and transfer wash to LC-MS vial.
Incubate sample in orbital shaker at 60 °C for 30 min to denature and reduce the proteins.
Spin samples at 15,000 rcf for 5 min to pellet any remaining particulates and remove condensed solvent from the walls of the vial (see Note 6).
3.2. SNaPP Analysis
Load samples into the autosampler set at 4 °C. (See Note 7).
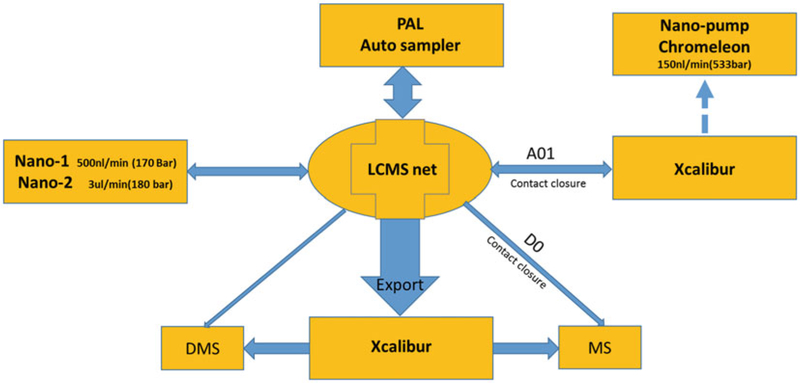
LCMSnet software is used to build the sample queue and initiate SNaPP analysis, as explained in Fig. 1. (See Note 8).
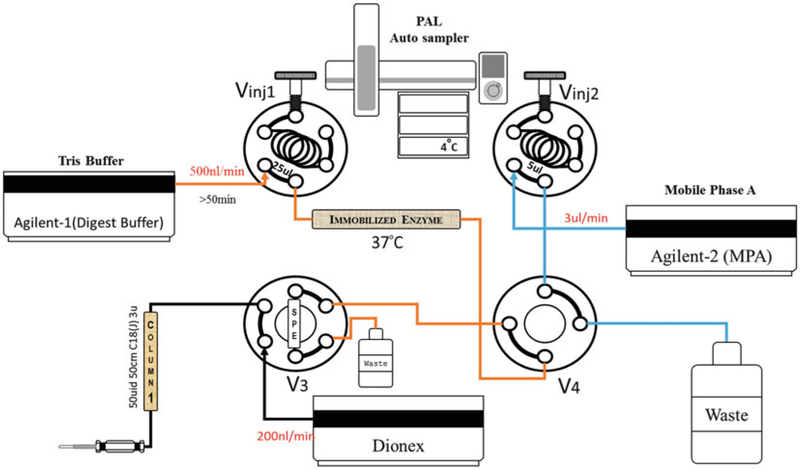
Sample is processed by SNaPP system, a diagram of the process is shown if Fig. 2.
Fig. 1.
Instrument control diagram. All sample retrieval, valve positioning, and method switching is controlled by LCMSnet software
Fig. 2.
The SNaPP system was constructed based on the previously reported configuration [19]. Two Agilent 1200 series nano pumps are used to deliver the digestion buffer (Agilent-1) and SPE wash buffer (Agilent-2). (See Note 9). A Dionex UltiMate 3000 RSLCnano pump is used to deliver the RPLC separation gradient. Two 6 ports injection valves were used for loading, SNaPP samples (Vinj1), and predigested peptide samples (Vinj2). The third 6 port valve (V3) is used to set up the SPE (5 μm C18,150 μm × 4 cm) and analytical column (1.7 μm AQ C18, 50 μm × 25 cm) and one 4 port valve was used for switching between SNaPP or peptide methods (See Note 10). The system is coupled to a Thermo QExactive Mass spectrometer by a replaceable custom ESI emitter (150 μm O.D. × 20 μm I.D.); (see Notes 11 and 12)
3.3. Data Analysis
Place all the Thermo “.raw” files and a fasta file containing the protein sequences for your organism(s) in a single folder.
Estimate the retention time shifts in your analyses by checking the retention times of multiple peptides using MS1 masses across the different datasets (e.g., using Xcalibur for viewing and exploring Thermo Raw files).
If necessary install Maxquant 1.5.2.8 or higher, open it and configure the fasta file(s) of interest in the Andromeda search engine.
Load your Raw files click on the “No fraction” button then set your experiment names.
In the “Group-specific parameters” and the “Global parameters” tabs, keep all the values to default except for the following: enable the LFQ quantification; use “Trypsin” as digestion enzyme in “Specific” mode use “Oxidation(M)” as “Variable modification” and no fixed modification; enable “re-quantify” and “Match between runs” and use your estimation of the time shifts (Step 2) as “match time window [min]”; for the identification set the “Min. peptide length” at 6, and the “Min. peptides,” “Min razor + unique peptide” and “Min unique peptides” at 2. (See Notes 13 and 14).
Click on the start button.
When the search is done, go to the folder containing your raw files. MaxQuant should have created the subfolders “combined/txt/.” In this folder, use the files “peptide.txt” and “proteinGroups.txt” for the peptide and protein information respectively. Use the columns starting with “Intensity” in the “peptide. txt” file and the column starting with “LFQ intensity” in the “proteinGroups.txt” file. (See Note 15).
For the peptide table, remove the rows containing a “Reverse” match or a “Potential contaminant” match if appropriate, for the protein table remove the rows containing a “Reverse” match a “Potential contaminant” match or a “Only identified by site” match.
Perform the normalization of the dataset and the statistics with appropriate statistical standards.
4. Notes
The pH of Tris buffer solutions is temperature dependent. This recipe is designed to give pH 8.0 at 37 °C. This is used because the digestion is carried out at this temperature. Tris buffer solutions are good for about 2 months if stored at 4 °C when not in use. Buffer should be kept no more than 2 weeks if stored at room temperature.
DTT is a reducing agent and its strength decreases over time when in solution. This solution should be made up fresh immediately prior to sample extraction.
DTT pellet can get stuck in the bottom of the preweigh tube and it is sometimes necessary to break up the pellet by pinching the bottom of tube to transfer. When this happens, it is essential that the Tris buffer used for solubilization is used to rinse preweigh tube to ensure quantitative transfer of the DTT.
Solvation of urea is an endothermic process and 8 M is a nearly saturated solution. Solubilization can be greatly sped up by shaking at 37 °C.
We have found that most tissues will lift off the cap easily; in some cases, it is necessary to use the pipette tip to dislodge the tissue slice by scraping the cap. Any transferred solids will be broken down further during incubation.
If there is a visible pellet, supernatant should be carefully transferred to a fresh LC-MS vial.
Samples should be spun upon thawing to check for protein precipitation. Avoid keeping undigested samples in autosampler at 4 °C for more than ~3 days as protein precipitation may occur.
LCMSnet software and tutorials are available at https://github.com/PNNL-Comp-Mass-Spec/LCMSNet.
We use Poroszyme™ Bulk for packing our columns because it is commercially available, highly consistent and low cost. IMER columns are slurry packed using a sixfold dilution in digestion buffer. For column prep we use a 10 cm long × 150 μm I. D. × 360 μm O.D. capillary section, prefitted with a 2 mm sol-gel frit. Slurry is loaded into a 500 μL syringe with 100 μL of slurry and connected to the capillary via metal union. Transfer approximately 2 cm of trypsin beads to the capillary. We have found that 2 cm length columns can handle protein loadings of up to 1.5 μg in the current cart configuration.
The second injection port is not critical to the operation of the SNaPP system but it is highly recommended for troubleshooting as it allows the digestion column to be easily isolated.
The SNaPP system is largely agnostic to the mass spectrometer it is coupled to and thus is compatible with all mass spectrometers capable of data-dependent MS/MS acquisition. Preliminary experiments suggest that the Thermo Lumos Orbitrap provides increased sensitivity compared with a QExactive.
Depending on the protein mass of the sample, proteomic data collection can be carried out with default settings. As the sample gets smaller, the MS2 maximum IT becomes important. Increasing this time to 100 ms can greatly increase the peptide identification rate. Longer ion times can decrease the instrument duty cycle, so when using 100 ms we recommend using a TOPN of no more than 12.
For the MaxQuant search, we have tested multiple options for the identification parameters. In our hands with the FDR set at 0.01 for the identification, with a “Min. peptide length” of 6 with 2 unique peptides and a maximum number of missed cleavages set at 2 provided the most identifications; however, depending on your specific setup and your trypsin digestion efficiency, you might want to test and adjust these parameters.
The method described in this paper resulted in a higher number of identifications, a better reproducibility and a lower number of contaminant peptides compared to the FASP method [12], both at the peptide and protein level [26]. However, if you have a specific interest in cysteine containing peptides, be aware that the method described here will provide a reduced cysteine containing peptide coverage compared to FASP as the method does not block the DTT reduced cysteines with a carbamidomethyl group.
Using MaxQuant we have compared label-free quantification using iBAQ, LFQ (ref: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4159666/), and spectral counting methods. For sample-limited analyses performed by the SNaPP approach, the LFQ quantification approach performed the best.
Acknowledgment
Portions of this research were supported by grants from the National Heart Lung Blood Institute of NIH (U01 HL122703), National Institute of General Medical Sciences of NIH (P41 GM103493) and the PNNL Laboratory-Directed Research and Development (LDRD) program. Work was performed in W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a Department Of Energy (DOE) office of Biological and Environmental Research (BER) national user facility located at Pacific Northwest National Laboratory (PNNL).
References
- 1.Nilsson T, Mann M, Aebersold R, Yates JR III, Bairoch A, Bergeron JJ (2010) Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods 7:681–685 [DOI] [PubMed] [Google Scholar]
- 2.Piehowski PD, Petyuk VA, Orton DJ, Xie F, Moore RJ, Ramirez-Restrepo M, Engel A, Lie-berman AP , Albin RL, Camp DG (2013) Sources of technical variability in quantitative LC–MS proteomics: human brain tissue sample analysis. J Proteome Res 12:2128–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domon B, Aebersold R (2010) Options and considerations when selecting a quantitative proteomics strategy. Nat Biotechnol 28:710–721 [DOI] [PubMed] [Google Scholar]
- 4.Altelaar AFM, Heck AJR (2012) Trends in ultrasensitive proteomics. Curr Opin Chem Biol 16:206–213 [DOI] [PubMed] [Google Scholar]
- 5.Wang N, MG X, Wang P, Li L (2010) Development of mass spectrometry-based shotgun method for proteome analysis of 500 to 5000 cancer cells. Anal Chem 82:2262–2271 [DOI] [PubMed] [Google Scholar]
- 6.Wang HX, Qian WJ, Mottaz HM, Clauss TRW, Anderson DJ, Moore RJ, Camp DG, Khan AH, Sforza DM, Pallavicini M, Smith DJ, Smith RD (2005) Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res 4:2397–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braakman RBH, Tilanus-Linthorst MMA, Liu NQ, Stingl C, Dekker LJM, Luider TM, Martens JWM, Foekens JA, Umar A (2012) Optimized nLC-MS workflow for laser capture microdissected breast cancer tissue. J Proteomics 75:2844–2854 [DOI] [PubMed] [Google Scholar]
- 8.Liu NQ, Braakman RBH, Stingl C, Luider TM, Martens JWM, Foekens JA, Umar A (2012) Proteomics pipeline for biomarker discovery of laser capture microdissected breast cancer tissue. J Mammary Gland Biol Neoplasia 17:155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11:319–324 [DOI] [PubMed] [Google Scholar]
- 10.De Marchi T, Braakman RBH, Stingl C, van Duijn MM, Smid M, Foekens JA, Luider TM, Martens JWM, Umar A (2016) The advantage of laser-capture microdissection over whole tissue analysis in proteomic profiling studies. Proteomics 16:1474–1485 [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Wang S, Adhikari S, Deng Z, Wang L, Chen L, Ke M, Yang P, Tian R (2016) Simple and integrated spintip-based technology applied for deep proteome profiling. Anal Chem 88:4864–4871 [DOI] [PubMed] [Google Scholar]
- 12.Wisniewski JR, Ostasiewicz P, Mann M (2011) High recovery FASP applied to the proteomic analysis of microdissected formalin fixed paraffin embedded cancer tissues retrieves known colon cancer markers. J Proteome Res 10:3040–3049 [DOI] [PubMed] [Google Scholar]
- 13.Ethier M, Hou WM, Duewel HS, Figeys D (2006) The proteomic reactor: a microfluidic device for processing minute amounts of protein prior to mass spectrometry analysis. J Proteome Res 5:2754–2759 [DOI] [PubMed] [Google Scholar]
- 14.Liebler DC, Ham AJL (2009) Spin filter-based sample preparation for shotgun proteomics. Nat Methods 6:785–785 [DOI] [PubMed] [Google Scholar]
- 15.Nel AJ, Garnett S, Blackburn JM, Soares NC (2015) Comparative re-evaluation of FADP and enhanced FASP methods by LC-MS/MS. J Proteome Res 14(3):1637–1642 [DOI] [PubMed] [Google Scholar]
- 16.Tian RJ, Alvarez-Saavedra M, Cheng HYM, Figeys D (2011) Uncovering the proteome response of the master circadian clock to light using an AutoProteome system. Mol Cell Proteomics 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Ferrer D, Petritis K, Robinson EW, Hixson KK, Tian ZX, Lee JH, Lee SW, Tolic N, Weitz KK, Belov ME, Smith RD, Pasa-Tolic L (2011) Pressurized pepsin digestion in proteomics. Mol Cell Proteomics 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Ferrer D, Petritis K, Lourette NM, Clowers B, Hixson KK, Heibeck T, Prior DC, Pasa-Tolic L, Camp DG, Belov ME, Smith RD (2008) On-line digestion system for protein characterization and proteome analysis. Anal Chem 80:8930–8936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang EL, Piehowski PD, Orton DJ, Moore RJ, Qian W-J, Casey CP, Sun X, Dey SK, Burnum-Johnson KE, Smith RD (2016) SNaPP: simplified nanoproteomics platform for reproducible global proteomic analysis of nanogram protein quantities. Endocrinology 157:1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hustoft HK, Brandtzaeg OK, Rogeberg M, Misaghian D, Torsetnes SB, Greibrokk T, Reubsaet L, Wilson SR, Lundanes E (2013) Integrated enzyme reactor and high resolving chromatography in “sub-chip” dimensions for sensitive protein mass spectrometry. Sci Rep 3:3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hustoft HK, Vehus T, Brandtzaeg OK, Krauss S, Greibrokk T, Wilson SR, Lundanes E (2014) Open tubular lab-on-column/mass spectrometry for targeted proteomics of nano-gram sample amounts. PLoS One 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang S, Zhang Z, Li L (2015) A one-step preparation method of monolithic enzyme reactor for highly efficient sample preparation coupled to mass spectrometry-based proteomics studies. J Chromatogr A 1412:75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi H, Miyazaki M (2013) Enzyme-immobilized reactors for rapid and efficient sample preparation in MS-based proteomic studies. Proteomics 13:457–466 [DOI] [PubMed] [Google Scholar]
- 24.Safdar M, Sproß J, Jänis J (2014) Microscale immobilized enzyme reactors in proteomics: latest developments. J Chromatogr A 1324:1–10 [DOI] [PubMed] [Google Scholar]
- 25.Colquhoun DR, Feild BJ (2015) Automated, online sample preparation for LC-MS analyses: affinity capture, digestion, and clean-up. State-of-the-art and emerging technologies for therapeutic monoclonal antibody characterization volume 3. Defining the next generation of analytical and biophysical techniques. Am Chem Soc 1202:335–356 [Google Scholar]
- 26.Clair G, Piehowski PD, Nicola T, Kitzmiller JA, Huang EL, Zink EM, Sontag RL, Orton DJ, Moore RJ, Carson JP (2016) Spatially-resolved proteomics: rapid quantitative analysis of laser capture microdissected alveolar tissue samples. Sci Rep 6:39223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slysz GW, Lewis DF, Schriemer DC (2006) Detection and identification of sub-nanogram levels of protein in a nanoLC-trypsin-MS system. J Proteome Res 5:1959–1966 [DOI] [PubMed] [Google Scholar]
- 28.Slysz GW, Schriemer DC (2009) Integrating accelerated tryptic digestion into proteomics workflows. Methods Mol Biol 492:241–254 [DOI] [PubMed] [Google Scholar]