Abstract
Monophosphoryl lipid A (MPLA) species, including MPL (a trade name of GlaxoSmithKline) and GLA (a trade name of Immune Design, a subsidiary of Merck), are widely used as an adjuvant in vaccines, allergy drugs, and immunotherapy to boost the immune response. Even though MPLA is a derivative of lipopolysaccharide (LPS), a component of the outer membrane of Gram-negative bacteria, bacterial strains producing MPLA have not been found in nature nor engineered. In fact, MPLA generation involves expensive and laborious procedures based on synthetic routes or chemical transformation of precursors isolated from Gram-negative bacteria. Here, we report the engineering of an Escherichia coli strain for in situ production and accumulation of MPLA. Furthermore, we establish a succinct method for purifying MPLA from the engineered E. coli strain. We show that the purified MPLA (named EcML) stimulates the mouse immune system to generate antigen-specific IgG antibodies similarly to commercially available MPLA, but with a dramatically reduced manufacturing time and cost. Our system, employing the first engineered E. coli strain that directly produces the adjuvant EcML, could transform the current standard of industrial MPLA production.
Keywords: Adjuvant, monophosphoryl lipid A, lipopolysaccharide biosynthesis, Gram-negative bacterial outer membrane, lipid A 1-phosphatase, vaccine adjuvant
1. Introduction
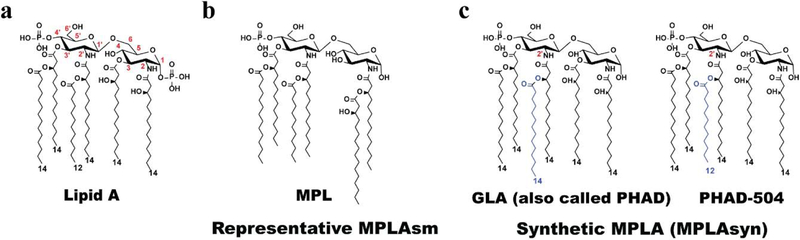
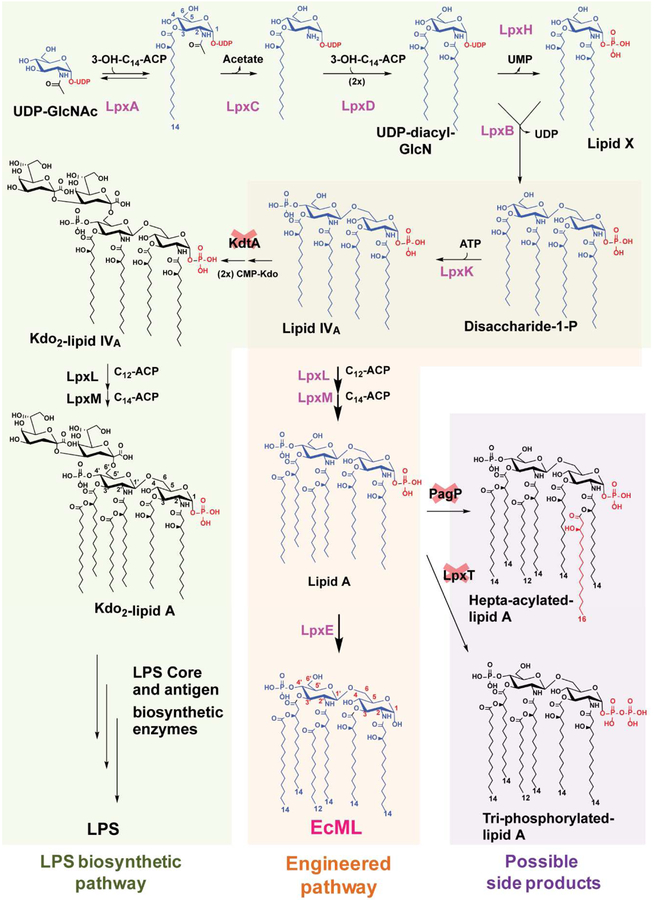
Lipid A (Fig. 1a) is a component of bacterial LPS, and it is a potent ligand for Toll-like receptor 4/myeloid differentiation 2 (TLR4/MD2) [1] as well as caspase-4, −5, and −11 [2–4]. Lipid A (Fig. 1a), also known as the toxic component of endotoxin, can trigger excessive immune responses that result in septic shock [5]; however, MPLAsm, which is chemically derived from Salmonella LPS (Fig. 1b and Supplementary Fig. 1), e.g., 3-O-desacyl-4’-monophosphoryl lipid A (MPL), and synthetic hexa-acylated MPLAs (MPLAsyn), e.g., GLA (also called PHAD) (Fig. 1c), PHAD-504, etc, have toxicity levels of only ~0.1% of that of LPS, but with comparable immune-stimulating activity [6]. Owing to this property, MPL has been used as an adjuvant in human vaccines, including Cervarix, Fendrix, Mosquirix, and Shingrix [7], which are marketed by GSK. MPL was also proposed as a potential therapeutic option for Alzheimer’s disease [8]. A synthetic hexa-acylated MPLA, GLA [9] (Fig. 1c), showed anticancer activity [10] and was developed by Immune Design as an orphan drug for follicular non-Hodgkin’s lymphoma and soft tissue sarcoma. GLA and MPL have similar modes of action, targeting the TLR4/MD2 signaling pathway [11]. MPL is typically prepared via acid/base hydrolysis of a precursor compound, Kdo2-lipid A species, isolated from S. minnesota R549 [12] (Supplementary Fig. 1b). Since the resulting MPL has diverse congener compositions including hexa-, penta-, and tetra-MPL (Fig. 1b and Supplementary Fig. 1a), it is difficult to control the quality of the product. By contrast, synthetic GLA (Fig. 1c) is homogeneous, but its production requires multiple synthetic steps [13]. Given that MPLA is derived from lipid A, bacteria that could directly produce MPLA would offer tremendous value as a renewable resource along with simplified production steps to reduce production time and cost. Engineering a biosynthetic pathway for MPLA production in E. coli (Fig. 2, orange-colored box) is conceptually straightforward, but technically challenging, because the native LPS biosynthetic pathway has evolved to transfer Kdo sugars via the kdo transferase KdtA before incorporation of the secondary acyl chains by acyltransferases, i.e., lauroyltransferase (LpxL) and myristoyltransferase (LpxM) (Fig. 2, green-colored box). In wild-type E. coli cells, Kdo2-lipid IVA is first synthesized followed by Kdo2-lipid A [34], and MPLA is not produced. Therefore, to obtain MPLA, incorporation of the secondary acyl chains must occur in the absence of the Kdo2 unit (Fig. 2, orange-colored box). Lipid A 1-phosphate phosphatase LpxE dephosphorylates the 1-phosphate of Kdo2-lipid A or Kdo2-lipid IVA most efficiently (Supplementary Fig. 2) [21]; however, it is not known whether any LpxE enzymes from Gram-negative bacteria can dephosphorylate lipid A substrates lacking the Kdo2 sugar unit during outer membrane biogenesis. Moreover, prior to this study, it was thought that production of MPLA would be lethal for E. coli [14] since the minimum structure of LPS to support E. coli growth was reported as lipid IVA that contains two phosphate groups [35]. This report describes an unprecedented E. coli strain that is bioengineered to produce MPLA directly without the need for additional hydrolysis and related purification processes.
Figure 1.
Chemical structures of lipid A (the toxic component of endotoxin), MPLAsm, and MPLAsyn. (a) Lipid A structure. (b) A representative structure of MPLAsm (hexa-acylated MPL). Other MPLAsm structures are shown in Supplementary Figure 1a. (c) Synthetic MPLA (GLA and PHAD-504) structures. PHAD-504 contains a lauroyl group (C12) at the secondary acyl chain (blue colored fatty acid) of the 2’-position instead of a myristoyl group (C14) compared with the GLA (PHAD) structure.
Figure 2.
The proposed de novo biosynthetic pathway for MPLA production. LPS biosynthesis and the proposed de novo biosynthetic pathway for MPLA. E. coli evolved to incorporate Kdo sugar units via Kdo transferase KdtA before addition of the lauroyl and myristoyl groups by LpxL and LpxM, respectively, during LPS biogenesis (green-colored box). A proposed MPLA biosynthetic pathway in E. coli is shown in the orange-colored box (blue-colored pathway). Red crosses indicate the necessary gene knockouts for generating the MPLA-producing strains. LpxT and PagP potentially compete with LpxE for lipid A as a substrate (purple-colored box).
2. Material and Methods
The strains, plasmids, and primers used in this study are listed in Table 1 and Supplementary Table 1.
Table 1.
Strains and plasmids used in this study
| Strain | Relevant properties | Source or reference |
|---|---|---|
| W3110 | Wild type F− λ− rph-1 IN(rmD, rmE) | E. coli Genetic Stock Center, Yale |
| C41(DE3) | F− ompT hsdSB (rB−mB−) gal dcm (DE3) F- φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 | [42] |
| DH5α | endA1 hsdR17(rk−, mk+) phoA supE44 thi-1 gyrA96 relA1 λ− | [18] |
| JW2162 | lpxT::kan BW25113 | [26] |
| JW0617 | pagP::kan BW25113 | [26] |
| HSC1/pEcKdtA | kdtA::kan DY330/pEcKdtA | [22] |
| KIHSC001 | lpxT::kan W3110 | This work |
| KIHSC002 | ΔlpxT W3110 | This work |
| KIHSC003 | ΔlpxT pgpP::kan W3110 | This work |
| KHSC0001 | ΔlpxT ApagP W3110 | This work |
| KHSC0001–1 | KHSC0001/pWSK29-lpxLEClpxMEC | This work |
| KHSC0003 | kdtA::kan KHSC0001–1 | This work |
| KHSC0003–1 | KHSC0003/pBAD33.1 | This work |
| KHSC0003–2 | KHSC0003/pBAD33.1-lpxEAA | This work |
| KHSC0003–3 | KHSC0003/pBAD33.1-lpxEHP | This work |
| KHSC0003–4 | KHSC0003/pBAD33.1-lpxEFN | This work |
| KHSC0003–5 | KHSC0003/pBAD33.1-lpxERL | This work |
| Plasmid | Relevant properties | Reference |
| pWSK29 | Low copy vector, lac promoter, AmpR | [23] |
| pWSK29-lpxLEclpxMEc | pWSK29 harboring E. coli lpxL and lpxM | This work |
| pET21a-lpxEAA | pET21a harboring A. aeolicus lpxE (aq_1706) | [19] |
| pBAD33.1 | Medium copy vector, CamR | [22] |
| pBAD33.1-lpxEAA | pBAD33.1 harboring A. aeolicus lpxE (aq_1706) | This work |
| pBAD33.1-lpxEHP | pBAD33.1 harboring H. pyrolilpxEHP(hp0021) | This work |
HPLC-grade chloroform and methanol and reagent-grade hydrochloric acid, sulfuric acid, aqueous ammonia, kanamycin, chloramphenicol, ammonium acetate, LB broth, ampicillin, and LB broth were purchased from Merck (USA) or Sigma-Aldrich (USA), and Bacto agar was purchased from BD (USA). GLA (catalog no.: tlrl-mpls, lot: MPS-40–02; GLA and PHAD have the same chemical structure) and MPLAsm (catalog no.: tlrl-mpla, lot: MPL-38–02) were purchased from Invivogen (USA). Purified lipid A detoxified (from Salmonella minnesota R595, catalog no.: 699200P, lot: 699200P-1MG-A-031, MPLAsm), PHAD (catalog no.: 699800P, lot: 699800P-1MG-A-029), and PHAD-504 (catalog no.: 699810P, lot: 699810P-1MG-A-010) were obtained from Avanti Polar Lipids (USA). 1,2-dipalmitoyl-sn-glycero-3-phosphocholine was purchased from Sigma-Aldrich (USA, catalog no.: P0763) or Avanti Polar Lipids (USA, catalog no.: 850355). HRP-conjugated goat anti-mouse IgG (catalog no.: 1030–05), IgG1 (catalog no.: 1070–05), or IgG2a (catalog no.: 1080–05) antibodies were purchased from Southern Biotech (USA).
2.1. Plasmid construction
All primers were purchased from Macrogen (ROK). All polymerase chain reactions (PCRs) were performed using KOD Hot Start DNA Polymerase (Novagen, USA), PrimeSTAR HS (Takara, ROK), or Pfu polymerase (Elpisbio, ROK) in a T3000 Thermocycler (Biometra) or T100 Thermal Cycler (Bio-Rad). PCR products were purified using the DokDo-Prep PCR purification kit (Elpisbio, ROK) or the QlAquick PCR purification kit (Qiagen, USA). Plasmids were amplified in DH5α [18] cells and purified using the GeneJET plasmid Miniprep kit (Thermo Fisher Scientific, USA). The inserted genes in the plasmids constructed in this study were sequenced by Macrogen (ROK).
The lpxEHP (hp0021) gene was synthesized by IDT (USA) with a silent mutation at Ser17 (from AGC to TCG) to remove a HindIII recognition site and amplified with the LpxEHP-5’ and LpxEHP-3’ primer pair. The lpxEAA (aq_1706) gene was amplified using the LpxEAA-5’ and LpxEAA-3’ primer pair with pET21-AaLpxE [19] as the template. The lpxEFN and lpxERL genes were amplified using Primer pairs: LpxEFN-5’ and LpxEFN-3’ and LpxERL-5’ and LpxERL-3’ with pET28b-FnLpxE [20] and pLpxE-4 [21] as templates, respectively. The lpxE genes were inserted individually into pBAD33.1 [22] using XbaI and HindIII restriction enzyme recognition sites. To construct the pWSK29-lpxLEClpxMEC plasmid, two sequential PCRs were performed. First, the lpxLEC and lpxMEC genes were amplified with two primer pairs, LpxL-5’ and LpxL-3’ and LpxM-5’ and LpxM-3’, respectively, and the fragments were then fused to generate the lpxLEClpxMEC fragment via extended PCR using the LpxL-5’ and LpxM-3’ primers and the PCR products from the first PCR reactions as the template. The resulting lpxLEClpxMEC fragment was inserted into pWSK29 [23] using the XbaI and HindIII restriction enzyme recognition sites.
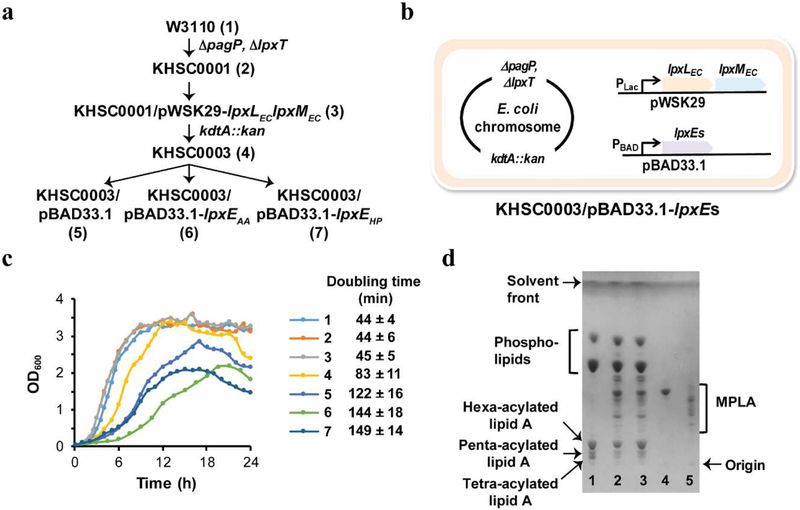
2.2. Construction of KHSC0003/pBAD33.1, KHSC0003/pBAD33.1-lpxEAA, and KHSC0003/pBAD33.1-lpxEHP (Fig. 3a and 3b)
Figure 3.
Engineered E. coli strains that directly produce MPLA. (a) Schematic illustration of the construction of the MPLA-producing E. coli strain KHSC0003/pBAD33.1-lpxEs. (b) Schematic diagram of the MPLA-producing strain. (c) Growth curves for all engineered strains and the mother strain W3110. The number 1–7 correspond to the strains shown on panel (a). (d) TLC profiles of the lipids extracted from the engineered E. coli strains compared to those of GLA (lane 4) and MPLAsm (lane 5). Total lipids extracted from KHSC0003/pBAD33.1 (lane 1), KHSC0003/pBAD33.1-lpxEAA (lane 2), and KHSC0003/pBAD33.1-lpxEHP (lane 3).
P1vir generation and P1 transduction were performed following a previously described procedure [24]. The kanamycin-resistance (kanR) cassette was removed from the W3110 chromosome using pCP20 according to the previously published procedure [25].
A lpxT::kan knockout mutation was transduced into W3110 via P1vir grown on JW2162 (lpxT::kan) [26], and the kanR cassette was removed via pCP20. The pagP::kan mutation was transduced into the ΔlpxT W3110 strain with P1vir grown on JW0617, and the kanR cassette was then removed. The resulting strain was named KHSC0001 (ΔlpxT ΔpagP W3110). To construct KHSC0003, pWSK29-lpxLEClpxMEC was transformed into KHSC0001 and kdtA was then knocked out via P1vir grown on HSC1/pEckdtA[22] in the presence of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG).
The pBAD33.1, pBAD33.1-lpxEAA, and pBAD33.1-lpxEHP plasmids were transformed singly into KHSC0003 via electroporation [27], and the transformants were selected on LB agar plates containing 1 mM IPTG, 100 μg/mL ampicillin, and 30 μg/mL chloramphenicol. The chromosomal knockouts of lpxT, pagP, and kdtA were confirmed by PCR using the primer pairs lpxT-up/lpxT-down, pagP-up/pagP-down, and kdtA-up/kdtA-down, respectively (Supplementary Fig. 3b). Genomic DNA was isolated from KHSC0003/pBAD33.1-lpxEAA using QIAamp genomic DNA kits (Qiagen, USA). De novo sequencing of KHSC0003/pBAD33.1-lpxEAA genome and analyses for single nucleotide polymorphism (SNP)s and indels in comparison with the published W3110 genome (Genebank accession number AP009048) were done by Macrogen (ROK). The identified SNPs and indels were compared to the sequence of PCR products amplified from W3110 genome used for KHSC0003/pBAD33.1-lpxEAA construction.
2.3. Growth conditions of strains and lipid extraction from cell pellets
Growth curve: W3110 and KHSC0001 in LB broth (Miller) (Merck 1.10285.5000) medium, KHSC0001–1 in LB broth medium supplemented with ampicillin (100 μg/mL), KHSC0003 in LB broth medium supplemented with ampicillin (100 μg/mL) and 1 mM IPTG, and KHSC0003/pBAD33.1, KHSC0003/pBAD33.1-lpxEAA, and KHSC0003/pBAD33.1-lpxEHP in LB broth medium supplemented with ampicillin (100 μg/mL) and chloramphenicol (30 μg/mL), and 1 mM IPTG were grown at 30 °C with shaking (180 rpm) for 24 h. Then cultures were diluted (OD600 ~ 0.05) into 300–500 mL of LB medium with the same supplements. After 12 h of growth, KHSC0003, KHSC0003/pBAD33.1, KHSC0003/pBAD33.1-lpxEAA, and KHSC0003/pBAD33.1-lpxEHP were supplemented with 1 mM IPTG and incubated at 30 °C with shaking (180 rpm). Doubling time was calculated using the equation (doubling time = ln2/r where r = (ln OD2 – ln OD1)/(t2-t1)) based on the growth curve (OD600 range from ~ 0.2 to 1.2 for each strain).
IPTG effect on the growth and the MPLA production of KHSC0003/pBAD33.1-lpxEAA: The seeding culture was diluted into LB medium in the presence of ampicillin (100 μg/mL) and chloramphenicol (30 μg/mL) with 0 or 1 mM IPTG. After 12 h of growth, 0 or 1 mM IPTG was added into the culture.
l-arabinose (Ara) effect on the growth and the MPLA production of KHSC0003/pBAD33.1-lpxEAA: The seeding culture was diluted into the LB broth medium containing ampicillin (100 μg/mL), chloramphenicol (30 μg/mL) and 1 mM IPTG in the presence of 0, 0.01, 0.1, or 1 mM arabinose and 1 mM IPTG was added into the culture at 12 hours after inoculation.
Lipid extraction: After 22–28 hours growth, cells were harvested via centrifugation at 2900 RPM (1,777 g, Hanil s500 4B rotor, Centrifuge COMBI-514R, Hanil Science Industrial Co., Ltd.) for 1 h or centrifugation at 8,000 g for 15 minutes at 4 °C, and the resulting pellets were washed with deionized water or phosphate-buffered saline (PBS). Lipids were extracted from the pellets via the Bligh-Dyer system [28] and analyzed via thin-layer chromatography (TLC) in a solvent system consisting of a chloroform/methanol/water/ammonium hydroxide solution (40:25:4:2 v/v).
2.4. Purification and characterization of EcML
The extracted lipids were fractionated via ion-exchange chromatography using a DEAE cellulose column (IonSep DE52 Cellulose Preswollen, Biophoretics) [29] with increasing ammonium acetate concentrations following the procedure described for phosphatidylglycerol phosphate fractionation [30]. Fractions containing EcML were converted to a two-phase Bligh-Dyer system by adding appropriate amounts of water and chloroform. The lower organic phase was evaporated under a stream of N2 (g). EcML was analyzed in comparison with GLA and MPLAsm via TLC. The TLC plates were visualized by spraying 10% (v/v) sulfuric acid (in ethanol) followed by charring on a hot plate at 300 °C. The purity of EcML was assessed via the TLC and ESI-MS methods and determined based on TLC images using the ImageJ software [31]. Quantification of EcML was determined by weighing samples using an analytical balance with a precision of 0.1 mg.
2.5. MALDI-TOF, ESI-MS, and HPLC analysis of lipids
Total lipids were dissolved in chloroform and methanol at a ratio of 4:1 (v/v). The resuspended lipid samples were loaded on a matrix consisting of 10 mg/mL 2,5-dihydroxybenzoic acid solution (Sigma-Aldrich) and acetonitrile (Sigma-Aldrich) at a ratio of 1:4 and then analyzed using a MALDI-TOF mass spectrometer (Shimadzu Biotech Axima Resonance) at Korea Basic Science Institute. The mass spectra were analyzed via the mMass software (http://www.mmass.org/).
EcML was dissolved in chloroform and methanol at a ratio of 4:1 (v/v) at a concentration of 1 mg/mL and then diluted with 4:1 (v/v) chloroform:methanol solution at a 1:400 ratio. The sample was analyzed via ESI-MS (TSQ Quantum Ultra EMR) at Korea Basic Science Institute. For the congener analysis, EcML, GLA, MPLAsm, MPLAsyn, and PHAD-504 were derivatized with dinitrobenzyloxyamine (DNBA, Sigma-Aldrich) and analyzed via HPLC chromatography (Prominence, Shimadzu) as described previously [32].
2.6. Mouse immunization
All animal experiments were performed via protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Chuncheon Bioindustry Foundation. Aqueous formulations of EcML, GLA, and MPLAsm were produced following the procedure described by Orr, M. T., et al. [33].
Ovalbumin (OVA, 5 μg/dose) was used as an antigen. Female BALB/cAnNCri (Orientbio, ROK) mice (7 weeks old) were immunized via two intramuscular injections (0 and 14 days) with PBS alone, OVA (5 μg/dose) alone, EcML (5 μg/dose) alone, EcML (5 μg/dose) and OVA (5 μg/dose), GLA (5 μg/dose) and OVA (5 μg/dose), or MPLAsm (5 μg/dose) and OVA (5 μg/dose). Serum samples were collected 14 days after the second immunization. Each test group consisted of seven mice.
2.7. Analysis of antigen-specific antibodies
ELISA plates (SPL, ROK) were coated with OVA (Sigma-Aldrich) dissolved in carbonate buffer (BioLegend) overnight at 4 °C. The plates were then washed with PBST (0.05% Tween20 in PBS) and blocked with 1% bovine serum albumin dissolved in PBS buffer for 1 h at room temperature (RT). After washing the plates, 100 μL of 2-fold serial dilutions of serum were added to the wells, followed by incubation for 2 h at RT. After washing, the plates were incubated with HRP-conjugated goat anti-mouse IgG, IgG1, or IgG2a antibodies (Southern Biotech) for 2 h at RT. 3,3’,5,5’-tetramethylbenzidine (BioLegend) substrate (100 μL) was added to each well, and the plates were incubated for 10 min at RT. The reaction was terminated via addition of 2 N H2SO4 (50 μL per well). The absorbance values were read at 450 nm and were corrected for optical imperfections in the plates by subtracting absorbance values at 540 nm. Antibody titers were obtained by plotting the maximum serum dilution that gave an optical density > 5× the background. The first serum dilution was started at 27 to analyze the IgG, IgG1, and IgG2a levels; therefore, the lowest titer for antibody was arbitrarily determined as 26.
2.8. Statistical analysis
All of the data are presented as mean ± standard deviation (S.D.) for control and experimental samples. The sample size was chosen based on the most commonly used sizes in this area. Significance of differences comparing two groups was analyzed by two tailed unpaired student t-test. Statistical significances were set at p < 0.05; individual P values are indicated in figure legends.
3. Results and Discussion
3.1. Metabolic engineering of E. coli for MPLA production
To develop a de novo biosynthetic pathway for MPLA production in E. coli, we engineered the strain according to the scheme shown in Figure 3a. First, the genes encoding lipid A palmitoyltransferase and lipid A 1-diphosphate synthase (pagP [36] and lpxT [37], respectively) were deleted from the E. coli chromosome to minimize potential side products (Fig. 2, purple-colored box, Fig. 3a). Second, a lipid A-producing strain was obtained by introducing a plasmid that co-overexpresses LpxLEC and LpxMEC followed by knocking out the gene encoding Kdo transferase (kdtAEC from the E. coli chromosome (Fig. 2, orange-colored box, Fig. 3a). While we were not able to obtain a colony by P1vir transduction carrying a kdtA::kan cassette into KHSC0001/pWSK29 within 48 hours on a LB-agar plate at 30 °C, co-overexpression of LpxLEC and LpxMEC effectively suppresses the lethality of kdtAEC knockout, and the strain KHSC0003 survived and supported the efficient production of hexa-acylated lipid A. Thus, the resulting E. coli strain, KHSC0003 transformed with an empty pBAD33.1 vector (Table 1 and Fig. 3a), generated hexa-acylated lipid A species as one of its primary LPS molecules without the core oligosaccharide, as detected via thin-layer chromatography (TLC) (Fig. 3d, lane 1). Finally, to identify the appropriate LpxE enzymes that effectively dephosphorylate lipid A in E. coli, we cloned lpxE genes, including Aquifex aeolicus lpxEAA (locus tag: aq_1706) [19], Helicobacter pylori lpxEHP[39] (locus tag: hp0021), Francisella novicida lpxEFN (locus tag: ftn_0416) [20], and Rhizobium leguminosarum lpxERL (locus tag: rl4708) [40] in pBAD33.1 [22] and transformed the constructs into KHSC0003 (Fig. 3a and 3b). Prior to this study, none of LpxE enzymes is known for the dephosphorylating activity of lipid A 1-phosphate in the absence of the Kdo2 unit during LPS biosynthesis. Among the tested KHSC0003/pBAD33.1-lpxEs, KHSC0003/pBAD33.1-lpxEAA and KHSC0003/pBAD33.1-lpxEHP effectively produced MPLA assessed by TLC (Supplementary Fig. 3a).
3.2. Characterization of a MPLA production strain, KHSC0003/pBAD33.1-lpxEAA
In order to characterize strains along with the steps of metabolic engineering, we measured growth curves and calculated doubling times of the strains (Fig. 3c). As shown in Figure 3c, pagP and lpxT double knockouts did not affect E. coli growth at 30 °C in LB medium compared to that of wild type W3110 (doubling time is about 44 minutes). However, the growth rate of KHSC0003 having a chromosomal kdtA knockout decreased to about 50% of that of W3110 (Fig. 3c), with a longer initial lag phase when the strain was cultured in LB medium supplemented with the addition of 1 mM IPTG at time 0 and 12 hours. A longer initial lag phase was also observed in the ClearColi strain grown in LB, which lacks Kdo sugar biosynthetic pathway and has a suppressive mutation in msbA gene [35], resulting in lipid IVA production instead of LPS (https://www.lucigen.com/docs/literature/eLucidations/ClearColi-Media-App-Note-April-2016.pdf). Growth rates of KHSC0003/pBAD33.1, KHSC0003/pBAD33.1-lpxEAA, and KHSC0003/pBAD33.1-lpxEHP (doubling time: 122 ± 16, 144 ± 18, and 149 ± 14 minutes, respectively, Fig. 3c) decreased compared to that of KHSC0003 (doubling time: 83 ± 11 minutes, Fig. 3c), probably due to the additional plasmid pBAD33.1 and leaky expression of LpxEs. OD600 values of KHSC0003 were gradually decreased at stationary phase indicating possible cell lysis.
Surprisingly, leaky LpxEAA or LpxEHP expression from pBAD33.1 led to dephosphorylation of the 1-phosphate group from hexa-, penta-, and tetra-acylated lipid A species and resulted in MPLA production, which was confirmed by performing TLC (Fig. 3d, lane 2 and 3, respectively) along with commercially available GLA (Fig. 3d, lane 4) and MPLAsm (Fig. 3d, lane 5) as standards. Since LpxEAA and LpxEHP produced similar amounts of MPLA in KHSC0003 and had similar growth rates, further characterization of the strain and MPLA production was done using the KHSC0003/pBAD33.1-lpxEAA strain. To analyze the genome sequence of KHSC0003/pBAD33.1-lpxEAA, de novo sequencing was executed and compared to the published W3110 genome (Genbank accession number: AP009048) [41], Table 2). The identified SNPs and indels were compared to the corresponding sequence of W3110 used for the construction of KHSC0003/pBAD33.1-lpxEAA (Table 2). As seen in Table 2, the KHSC0003/pBAD33.1-lpxEAA strain has only one SNP, which is not originated from the mother strain at location 151,154 based on the nucleotide numbering of AP009048 genome. The C to G change at location 151,154 makes a 149th amino acid mutation of YadK (putative fimbrial-like protein) from glycine to alanine. Considering that the genomic location of yadK is too far to be transduced by P1vir during pagP (located at 656,979 – 657,539), lpxT (located at 2,272,188 – 2,272,901), or kdtA (located at 3,830,597 – 3,831,875) knockout process, this mutation is likely acquired spontaneously.
Table 2.
Mutation analyses of KHSC0003/pBAD33.1-lpxEAA genome via de novo sequencing in comparison with the published W3110 genome (Genbank accession number: AP009048) and that of the mother strain W3110 used for the strain construction.
| Nucleotide position based on AP009048 | W3110 (AP009048) | W3110 (mother strain) | KHSC0003/pBAD33.1-lpxEAA |
|---|---|---|---|
| 151,154 | C | C | G (gly149ala mutation YadK) |
| 547,694 | A | G | G |
| 547,831 | AGGGG | AGGGGG | AGGGGG |
| 556,858 | A | T | T |
| 1,065,700 | C | T | T |
| 1,093,686 | T | C | C |
| 1,097,580 | C | T | T |
| 1,117,836 | C | C | C |
| 1,303,712 | A | G | G |
| 2,019,202 | C | T | T |
| 2,906,975 | C | G | G |
| 4,371,270 | GAAAAA | GAAA | GAAA |
| 653,806 – 656,391 | IS elements inserted in dcuC | Intact ducC | Intact ducC |
| 656,979 – 657,539 | pagP | pagP | Deleted |
| 2,272,188 – 2,272,901 | lpxT | lpxT | Deleted |
| 3,830,597 – 3,831,875 | kdtA | kdtA | Replaced with a kanamycin resistance cassette |
3.3. Evaluate the effects of LpxL, LpxM, and LpxE overexpression on the growth rate and MPLA production of KHSC0003/pBAD33.1-lpxEAA
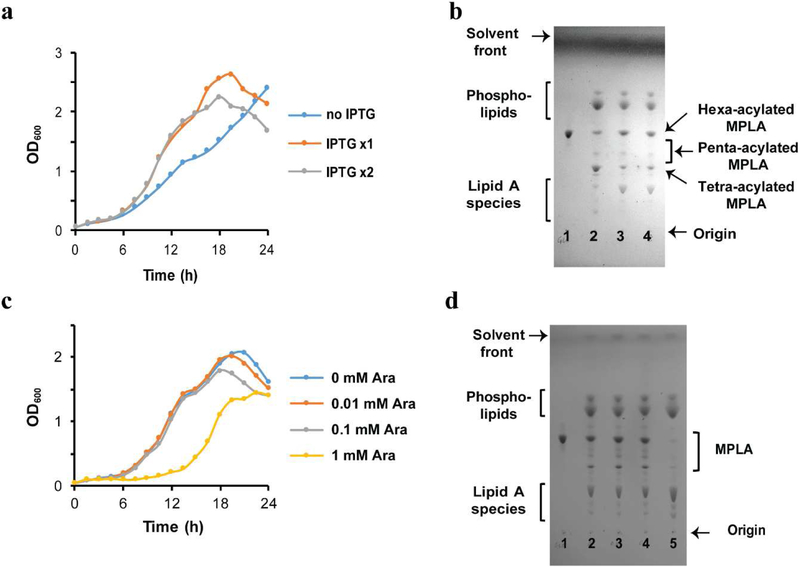
In order to optimize the strain growth and production of hexa-acylated MPLA that is suggested as the most potent adjuvant among MPLA congeners in human [9]. We first checked the effect of LpxL and LpxM overexpression on the strain growth and MPLA production profile. In the absence of IPTG, we observed obvious decrease in the growth rate compared to that of the cultures supplemented with 1 mM IPTG at time 0 or time 0 and 12 hours after dilution (Fig. 4a). As a note, while we did not add IPTG at the dilution point, there was IPTG carried over from the seeding culture. In addition, we observed the shift in the major congener composition of the MPLA from hexa-acylated to tetra-acylated MPLA depending on the LpxL and LpxM overexpression (Fig. 4b, lane 2 vs lane 3 and 4). Even though further experiments should be required to determine the optimum IPTG amounts, no obvious difference was observed in the growth rates (Fig. 3a) and MPLA composition assessed via TLC analysis (Fig. 4b, lane 3 vs lane 4) between the cultures supplemented with 1 mM IPTG at time 0 or time 0 and 12 hours after dilution. Therefore 1 mM IPTG would be sufficient for hexa-acylated MPLA production. In order to increase MPLA formation relative to lipid A species, we overexpressed LpxE with different concentrations of arabinose. Unexpectedly, the culture grown with 1 mM arabinose had a long lagging phase (about 10 hours) before starting growth (Fig. 4c) and dramatically reduced the amount of MPLA production (Fig. 4d, lane 5) compared to those of the cultures supplemented with 0, 0.01, or 0.1 mM arabinose (Fig, 4d, lane 2, 3, and 4, respectively). Such a result indicated that overexpression of LpxE with 1 mM arabinose might be toxic to the strain, generating spontaneous mutant strains, in which LpxE could not function properly. The cultures supplemented with 0, 0.01, or 0.1 mM arabinose showed similar growth rates, and the MPLA production profiles assessed by TLC analysis indicated that leaky expression of LpxE from the pBAD33.1 plasmid is the best growth condition for MPLA production to mitigate the risk of mutants’ selection. At this point, it is not clear why the kdtA knockout with overexpression of LpxL and LpxM showed gradual lysis phenotype at the stationary phase (Fig. 3c, 4a, and 4c) and it would be an interesting research topic to understand the outer membrane biogenesis of the final strain to enhance biomass.
Figure 4.
Efficient production of hexa-acylated MPLA is achieved by overexpression of LpxL and LpxM along with leaky expression of LpxE. (a) Growth curves for KHSC0003/pBAD33.1-lpxEAA supplemented with or without IPTG. The strain was grown in LB medium containing ampicillin and chloramphenicol supplemented with 0 (no IPTG), 1 mM IPTG (IPTG 1x) at time 0 and 1 mM IPTG at time 0 and 12 hours (IPTG 2x) after dilution. Overexpression of LpxL and LpxM increases the growth rate of the strain. (b) TLC profiles of the lipids extracted from the engineered E. coli strains compared with those of GLA (lane 1). Total lipids extracted from KHSC0003/pBAD33.1-lpxEAA grew supplemented with 0 (lane 2), IPTG 1x (lane 3), and IPTG 2x (lane 4). Overexpression of LpxL and LpxM is required for efficient hexa-acylated MPLA production. (c) Growth curves for KHSC0003/pBAD33.1-lpxEAA supplemented with different concentrations of l-arabinose (Ara). The strain was grown in LB medium containing ampicillin, chloramphenicol, and IPTG supplemented with 0, 0.01, 0.1, 1 mM Ara. Overexpression of LpxE by 1 mM Ara caused a long lagging phase. (d) TLC profiles of the lipids extracted from the engineered E. coli strains compared with those of GLA (lane 1). Total lipids extracted from KHSC0003/pBAD33.1-lpxEAA grew supplemented with 0 (lane 2), 0.01 mM Ara (lane 3), 0.1 mM Ara (lane 4), and 1 mM Ara (lane 5) at time 0. The presence of 1 mM Ara caused a long lagging phase of the strain growth, and the strain grown in the presence of 1 mM IPTG barely produced MPLA. The presence of 0, 0.01, or 0.1 mM Ara does not affect MPLA production.
3.4. Characterization of total lipids and purified MPLA, EcML
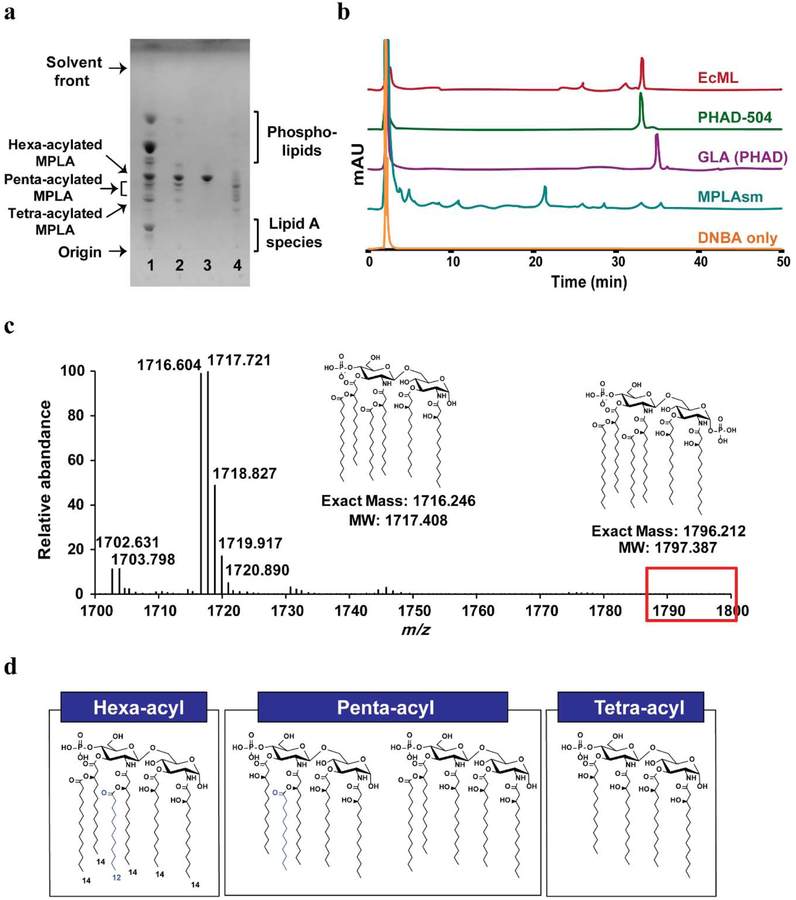
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis of the total lipids extracted from KHSC0003/pBAD33.1-lpxEAA cells confirmed the presence of hexa-acylated MPLA ([M-H]− of hexa-acylated MPLA: calculated = 1716.246 and observed = 1716.305, Supplementary Fig. 4). Based on the TLC (Fig. 5a, lane 1) and MALDI-TOF (Supplementary Fig. 4) data, the KHSC0003/pBAD33.1-lpxEAA total lipid extract contained lipid A (calculated [M-H]− = 1796.212 and observed [M-H]− = 1796.270) (~40–50% of the total LPS species), and MPLA. To allow the MPLA produced by the engineered E. coli strains to be safely used as an adjuvant, we removed lipid A by taking advantage of the negative charge difference between MPLA (mono-phosphorylated species) and lipid A (bis-phosphorylated species).
Figure 5.
Characterization of EcML purified from engineered E. coli KHSC0003/pBAD33.1-lpxEAA cells. (a) TLC profile of EcML (lane 2) purified from the total lipids of KHSC0003/pBAD33.1-lpxEAA (lane 1) compared to those of GLA (lane 3) and MPLAsm (lane 4). (b) The HPLC profile of EcML compared to those of DNBA, MPLAsm, GLA (PHAD), and PHAD-504. (c) The ESI-MS spectrum of EcML in the m/z range of 1700–1800 amu. Lipid A (calculated [M-H]−: 1796.212), was not detected, while hexa-acylated MPLA (calculated [M-H]−: 1716.246) was observed at 1716.604 by ESI-MS. (d) Suggested chemical structures of hexa-, penta-, and tetra-acylated EcML.
After purification of the MPLA via anion-exchange chromatography (DE52 resin, Biophoretics) (Fig. 5a, lane 2), we named the purified MPLA “E. coli-derived monophosphoryl lipid A (EcML)”. EcML was purified at a yield of 8–12 mg/1 L of LB and 13–19 mg/g dry cell weight (DCW) (Table 3). The purity and lipid A contamination of EcML were also assessed via TLC with increasing amounts of EcML (Supplementary Fig. 5) and electrospray ionization-mass spectrometry (ESI-MS) (Fig. 5c and Supplementary Fig. 6), both of which showed no lipid A carry-over (Fig. 5c, red box). Using the ImageJ program [31], we determined the purities of EcML, GLA, and MPLAsm as 80–89% (depending on the different batches), 90%, and 88%, respectively based on the TLC images shown in Figures 3d and 5a, and Supplementary Fig. 5. Majority of impurities are phospholipids (Fig. 5a). Considering that each Cervarix dose contains 50 μg of MPL, about 10 mg of EcML would be sufficient to generate 200 doses. Although EcML contained more of a complex mixture of hexa-, penta-, and tetra-acylated MPLA (Fig. 5a, lane 2) than GLA (Fig 5a, lane 3), its composition was simpler than that of MPLAsm (Fig. 5a, lane 4). To determine the relative distribution of hexa-, penta-, and tetra-acylated MPLA and batch variation of EcML, three different batches of EcML, MPLAsyn (GLA and PHAD-504), and MPLAsm were modified with dinitrobenzyloxyamine hydrochloride (DNBA) according to a previously described procedure [32] and analyzed via high-performance liquid chromatography (HPLC) (Fig. 5b and Supplementary Fig. 7). Since LpxLEC preferentially transfers a lauroyl group during LPS and MPLA biosynthesis in E. coli (Fig. 2), the major hexa-acylated EcML (Fig. 5d) component has the same chemical structure as PHAD-504 (Fig. 1c, right), resulting in the same retention time as shown in HPLC profiles (Fig. 5b). EcML contained about 69.7% ± 3.8% of hexa-, 22.2% ± 3.3% of penta-, and 8.2% ± 0.7% of tetra-acylated MPLA (Table 3, the predicted chemical structures of hexa-, penta-, tetra-acylated MPLA are shown in Fig. 5d). Furthermore, our results demonstrated that the congener compositions of EcML from different batches are fairly consistent (Table 3 and Supplementary Fig. 7). It is worth noting that the HPLC analyses of MPLAsm from two different vendors, Invivogen and Avanti Polar Lipids, showed large variations in their congener compositions due to uncontrollable aspects of the acid/base hydrolysis process (Supplementary Fig. 8), which significantly complicates the MPLAsm quality control. Accordingly, the simplicity and low batch-to-batch variations in congener composition intrinsic to our EcML preparation method offer particular advantages for its use in chemistry, manufacturing, and controls for clinical trials or marketing applications.
Table 3.
Batch-to-batch variations in EcML yield and congener composition.
| Batch | Yield mg/25g LB (mg/g DCWa) | Composition (%): number of acyl chain | |||
|---|---|---|---|---|---|
| Hexa | Penta | Tetra | |||
| EcML | 1 | 8.4 (12.7) | 68.5 | 23.7 | 7.9 |
| 2 | 12.4 (18.6) | 66.6 | 24.5 | 9.0 | |
| 3 | 8.4 (13.9) | 73.9 | 18.5 | 7.6 | |
| Average ± STD | 9.7 ± 2.3 (14.7 ± 3.3) | 69.7 ± 3.8 | 22.2 ± 3.3 | 8.2 ± 0.7 | |
DCW of KHSC0003/pBAD33.1-lpxEAA is 0.29 g for 1L of OD600 = 1.0
3.5. EcML effectively enhances antigen-specific antibody formation in mice model
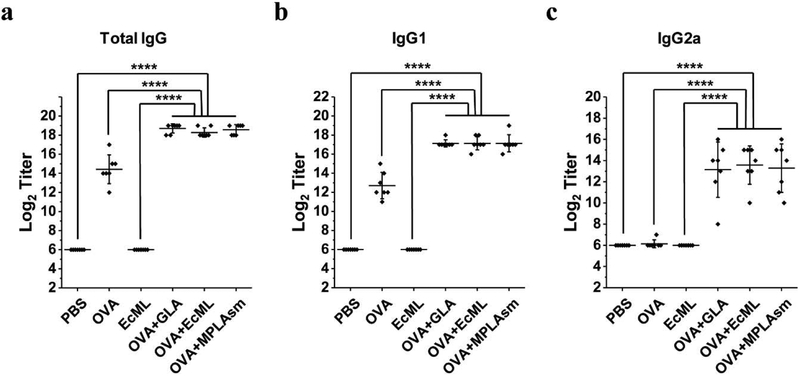
After characterization of the prepared EcML, we determined its adjuvanticity. Aqueous formulations of EcML, along with MPLAsm and GLA as positive controls, were produced according to a published procedure [33]. As shown in Figure 6, EcML potentiated antigen-specific IgG, IgG1, and IgG2a production to a similar extent as the positive controls in BALB/c mice when ovalbumin was used as an antigen. Taken together, these results strongly suggest that the adjuvanticity of EcML purified from the engineered E. coli strain is equal to those of commercially available GLA and MPLAsm for antigen-specific antibody formation after vaccination.
Figure 6.
Evaluation of antigen (ovalbumin, OVA)-specific antibody responses show that EcML has equal adjuvanticity to those of GLA and MPLAsm (Invivogen). Total IgG, IgG1, and IgG2a antibody titers measured 14 days after the second immunization with PBS only, OVA only, EcML only, OVA together with GLA, OVA together with EcML, or OVA together with MPLAsm. (a) Total IgG endpoint titers, (b) IgG1 endpoint titers, and (c) IgG2a endpoint titers. Data are presented as mean ± S.D. (n = 7; ****p < 0.0001; two-tailed unpaired student t-test). The lowest antibody titer was arbitrarily determined as 26, as the first sample tested was a 27 dilution of the serum. Error bars represent the standard deviations.
4. Conclusion
Over the course of the rapid growth of the vaccine and immunotherapy markets, MPLA has played pivotal roles in the development of vaccines and immunotherapy for allergies, cancer, and Alzheimer’s disease. However, the production methods for GLA and MPLAsm suffer from complexity in synthetic steps and quality control, respectively, and the production costs are high for both methods. The MPLA-producing E. coli strain would be a succinct, cost-effective, and sustainable resource to meet the increasing demand of MPLA not only as an adjuvant but also as emerging therapeutic options for humans and animals.
Supplementary Material
Highlights.
Monophosphoryl lipid A (MPLA) is a safe and effective adjuvant
Engineering lipopolysaccharide biosynthetic pathway enables to produce MPLA
Employing lipid A 1-phosphatase from Aquifex dephosphorylates lipid A in vivo.
The succinct purification method of MPLA (EcML) is developed
EcML purified from the engineered strain effectively functions as an adjuvant.
Acknowledgements
We thank Robert A. Gillespie for providing pET28b-lpxEAA.
Funding information
This work was supported by KIST intramural grants and by the Pioneer Research Center Program (2014M3C1A3054141) through the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning, and by the National Institute of General Medical Sciences, USA (GM115355 to PZ). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B, Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene, Science 282(5396) (1998) 2085–8. [DOI] [PubMed] [Google Scholar]
- [2].Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F, Inflammatory caspases are innate immune receptors for intracellular LPS, Nature 514(7521) (2014) 187–92. [DOI] [PubMed] [Google Scholar]
- [3].Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA, Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock, Science 341(6151) (2013) 1250–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszynski A, Forsberg LS, Carlson RW, Dixit VM, Noncanonical inflammasome activation by intracellular LPS independent of TLR4, Science 341(6151) (2013) 1246–9. [DOI] [PubMed] [Google Scholar]
- [5].Lu YC, Yeh WC, Ohashi PS, LPS/TLR4 signal transduction pathway, Cytokine 42(2) (2008) 145–51. [DOI] [PubMed] [Google Scholar]
- [6].Casella CR, Mitchell TC, Putting endotoxin to work for us: Monophosphoryl lipid A as a safe and effective vaccine adjuvant, Cellular and Molecular Life Sciences 65(20) (2008) 3231–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Reed SG, Carter D, Casper C, Duthie MS, Fox CB, Correlates of GLA family adjuvants’ activities, Semin Immunol 39 (2018) 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Michaud JP, Halle M, Lampron A, Theriault P, Prefontaine P, Filali M, Tribout-Jover P, Lanteigne AM, Jodoin R, Cluff C, Brichard V, Palmantier R, Pilorget A, Larocque D, Rivest S, Toll-like receptor 4 stimulation with the detoxified ligand monophosphoryl lipid A improves Alzheimer’s disease-related pathology, Proceedings of the National Academy of Sciences of the United States of America 110(5) (2013) 1941–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Coler RN, Bertholet S, Moutaftsi M, Guderian JA, Windish HP, Baldwin SL, Laughlin EM, Duthie MS, Fox CB, Carter D, Friede M, Vedvick TS, Reed SG, Development and Characterization of Synthetic Glucopyranosyl Lipid Adjuvant System as a Vaccine Adjuvant, Plos One 6(1) (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].C.V.S.F. Paya Cuenca, CA, US), Ter Meulen, Jan Henrik (Mercer Island, WA, US), GLA monotherapy for use in cancer treatment, United States Patent, Immune Design Corp (Seattle, WA, US), United States, 2015. [Google Scholar]
- [11].Reed SG, Hsu FC, Carter D, Orr MT, The science of vaccine adjuvants: advances in TLR4 ligand adjuvants, Curr Opin Immunol 41 (2016) 85–90. [DOI] [PubMed] [Google Scholar]
- [12].Qureshi N, Takayama K, Ribi E, Purification and structural determination of nontoxic lipid A obtained from the lipopolysaccharide of Salmonella typhimurium, The Journal of biological chemistry 257(19) (1982) 11808–15. [PubMed] [Google Scholar]
- [13].Reed SG, Carter D, Synthetic Glucopyranosyl lipid adjuvants, in: T.U.S.P.a.T. Office (Ed.) United States Patent, Infectious Disease Research Institute, US, 2014. [Google Scholar]
- [14].Wang B, Han Y, Li Y, Li Y, Wang X, Immuno-Stimulatory Activity of Escherichia coli Mutants Producing Kdo2-Monophosphoryl-Lipid A or Kdo2-Pentaacyl-Monophosphoryl-Lipid A, PLoS One 10(12) (2015) e0144714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chen J, Tao G, Wang X, Construction of an Escherichia coli mutant producing monophosphoryl lipid A, Biotechnology letters 33(5) (2011) 1013–9. [DOI] [PubMed] [Google Scholar]
- [16].Needham BD, Carroll SM, Giles DK, Georgiou G, Whiteley M, Trent MS, Modulating the innate immune response by combinatorial engineering of endotoxin, Proceedings of the National Academy of Sciences of the United States of America 110(4) (2013) 1464–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gregg KA, Harberts E, Gardner FM, Pelletier MR, Cayatte C, Yu L, McCarthy MP, Marshall JD, Ernst RK, Rationally Designed TLR4 Ligands for Vaccine Adjuvant Discovery, MBio 8(3) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Grant SG, Jessee J, Bloom FR, Hanahan D, Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants, Proceedings of the National Academy of Sciences of the United States of America 87(12) (1990) 4645–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhao J, An J, Hwang DH, Wu Q, Wang S, Gillespie RA, Yang EG, Guan Z, Zhou P, Chung HS, The Lipid A 1-phosphatase, LpxE, Functionally Connects Multiple Layers of Bacterial Envelope Biogenesis, MBio 10(3) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang X, Karbarz MJ, McGrath SC, Cotter RJ, Raetz CR, MsbA transporter-dependent lipid A 1-dephosphorylation on the periplasmic surface of the inner membrane: topography of francisella novicida LpxE expressed in Escherichia coli, The Journal of biological chemistry 279(47) (2004) 49470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Karbarz MJ, Six DA, Raetz CR, Purification and characterization of the lipid A 1-phosphatase LpxE of Rhizobium leguminosarum, The Journal of biological chemistry 284(1) (2009) 414–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chung HS, Raetz CR, Interchangeable domains in the Kdo transferases of Escherichia coli and Haemophilus influenzae, Biochemistry 49(19) (2010) 4126–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang RF, Kushner SR, Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli, Gene 100 (1991) 195–9. [PubMed] [Google Scholar]
- [24].Thomason LC, Costantino N, Court DL, E. coli Genome Maniplation by P1 Transduction, Current Protocols in Molecular Biology (2007) 1.17.1–1.17.8. [DOI] [PubMed] [Google Scholar]
- [25].Datsenko KA, Wanner BL, One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products, Proceedings of the National Academy of Sciences of the United States of America 97(12) (2000) 6640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H, Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection, Mol Syst Biol 2 (2006) 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Seidman CE, Struhl K, Sheen J, Jessen J, Introduction of Plasmid DNA into Cells, Current Protocols in Molecular Biology (1997) 1.8.1–1.8.10. [DOI] [PubMed] [Google Scholar]
- [28].Bligh EG, Dyer WJ, A rapid method of total lipid extraction and purification, Can J Biochem Physiol 37(8) (1959) 911–7. [DOI] [PubMed] [Google Scholar]
- [29].Raetz CR, Purcell S, Meyer MV, Qureshi N, Takayama K, Isolation and characterization of eight lipid A precursors from a 3-deoxy-D-manno-octylosonic acid-deficient mutant of Salmonella typhimurium, The Journal of biological chemistry 260(30) (1985) 16080–8. [PubMed] [Google Scholar]
- [30].Lu YH, Guan Z, Zhao J, Raetz CR, Three phosphatidylglycerol-phosphate phosphatases in the inner membrane of Escherichia coli, The Journal of biological chemistry 286(7) (2011) 5506–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis, Nat Methods 9(7) (2012) 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hagen SR, Thompson JD, Snyder DS, Myers KR, Analysis of a monophosphoryl lipid A immunostimulant preparation from Salmonella minnesota R595 by high-performance liquid chromatography, J Chromatogr A 767(1–2) (1997) 53–61. [DOI] [PubMed] [Google Scholar]
- [33].Orr MT, Fox CB, Baldwin SL, Sivananthan SJ, Lucas E, Lin S, Phan T, Moon JJ, Vedvick TS, Reed SG, Coler RN, Adjuvant formulation structure and composition are critical for the development of an effective vaccine against tuberculosis, J Control Release 172(1) (2013) 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Raetz CR, Whitfield C, Lipopolysaccharide endotoxins, Annual review of biochemistry 71 (2002) 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mamat U, Meredith TC, Aggarwal P, Kuhl A, Kirchhoff P, Lindner B, Hanuszkiewicz A, Sun J, Holst O, Woodard RW, Single amino acid substitutions in either YhjD or MsbA confer viability to 3-deoxy-d-manno-oct-2-ulosonic acid-depleted Escherichia coli, Molecular microbiology 67(3) (2008) 633–48. [DOI] [PubMed] [Google Scholar]
- [36].Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR, Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria, The EMBO journal 19(19) (2000) 5071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Touze T, Tran AX, Hankins JV, Mengin-Lecreulx D, Trent MS, Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate, Molecular microbiology 67(2) (2008) 264–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Goodall ECA, Robinson A, Johnston IG, Jabbari S, Turner KA, Cunningham AF, Lund PA, Cole JA, Henderson IR, The Essential Genome of Escherichia coli K-12, MBio 9(1) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tran AX, Whittimore JD, Wyrick PB, McGrath SC, Cotter RJ, Trent MS, The lipid A 1-phosphatase of Helicobacter pylori is required for resistance to the antimicrobial peptide polymyxin, J Bacteriol 188(12) (2006) 4531–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Karbarz MJ, Kalb SR, Cotter RJ, Raetz CR, Expression cloning and biochemical characterization of a Rhizobium leguminosarum lipid A 1-phosphatase, The Journal of biological chemistry 278(41) (2003) 39269–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T, Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110, Mol Syst Biol 2 (2006) 2006 0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Miroux B, Walker JE, Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels, Journal of molecular biology 260(3) (1996) 289–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.