Abstract
RPEL proteins, which contain the G-actin binding RPEL motif, coordinate cytoskeletal processes with actin dynamics. We show that the ArhGAP12- and ArhGAP32-family GTPase activating proteins are RPEL proteins. We determine the structure of the ArhGAP12/G-actin complex, and show that G-actin contacts the RPEL motif and GAP domain sequences. G-actin inhibits ArhGAP12 GAP activity, and this requires the G-actin contacts identified in the structure. In B16 melanoma cells, ArhGAP12 suppresses basal Rac and Cdc42 activity, F-actin assembly, invadopodia formation, and experimental metastasis. In this setting, ArhGAP12 mutants defective for G-actin binding exhibit more effective downregulation of Rac GTP loading following HGF stimulation, and enhanced inhibition of Rac-dependent processes, including invadopodia formation. Potentiation or disruption of G-actin/ArhGAP12 interaction, by treatment with the actin-binding drugs latrunculin B or cytochalasin D, has corresponding effects on Rac GTP loading. G-actin interaction with RPEL family rhoGAPs thus provides a negative feedback loop that couples Rac activity to actin dynamics.
Keywords: RhoGAP, ArhGAP12, RPEL, MRTF, actin, GAP, melanoma, Rac, Cdc42
Introduction
Spatial and temporal control of the actin cytoskeleton in response to local signalling or mechanical cues plays a critical role in development and disease1–3. Underpinning it is the regulation of actin treadmilling, the dynamic transition between actin's monomeric (G-actin) and polymerised (F-actin) forms1,4, which is controlled by rho family small GTPases5,6. Rho GTPase activity is potentiated by multiple rho GEF proteins, which catalyse GTP loading and effector protein binding7,8, and antagonised by inhibitory rho GAPs, which catalyse GTP hydrolysis9,10. Both are regulated by specific subcellular targeting, and by biological and mechanical signals, but relatively little is known about how their activity responds to the status of the actin cytoskeleton.
One connection between cytoskeletal dynamics and control of protein function is provided by the RPEL proteins, which act as G-actin sensors11,12. Their regulatory domains contain RPEL motifs (Pfam PF02755), short polypeptide sequences that bind G-actin13. Two RPEL protein families, the MRTFs and the Phactrs, have been characterised11,12,14,15. The MRTFs are coactivators for the SRF transcription factor, regulating expression of dozens of cytoskeletal structural and regulatory proteins11,16,17, while the Phactrs regulate cytoskeletal dynamics by poorly understood mechanisms14,15,18,19. G-actin controls the subcellular localisation and activity of the MRTFs and Phactrs, at least in part by binding competitively with their regulatory and effector proteins, such as importin αβ and PP1, to sites within their regulatory RPEL domains14,15,20,21.
Here we characterise two hitherto unidentified RPEL protein families, the ArhGAP12 and ArhGAP32 subfamilies of Rac1/Cdc42-specific GTPase activating proteins (GAPs)22–25. The ArhGAP12 family is associated with actin-dependent cell surface structures and processes, including adherens junctions23,26–28, plasma membrane blebs29, phagocytosis30, and dendritic spines31, while the ArhGAP32 family proteins have been implicated in protein trafficking and neuronal development25,32. However, little is known about their regulation. We show that binding of G-actin to an atypical RPEL motif adjoining the ArhGAP12 GAP domain inhibits its GAP activity for Rac1 in vitro. Disruption of the G-actin/ArhGAP12 interaction potentiates GAP activity in vivo. Our findings demonstrate that G-actin/ArhGAP12 interaction constitutes a feedback loop that couples Rac/Cdc42 GTP loading to the state of cytoskeletal dynamics.
Results
Two subfamilies of ArhGAP proteins contain a G-actin binding motif
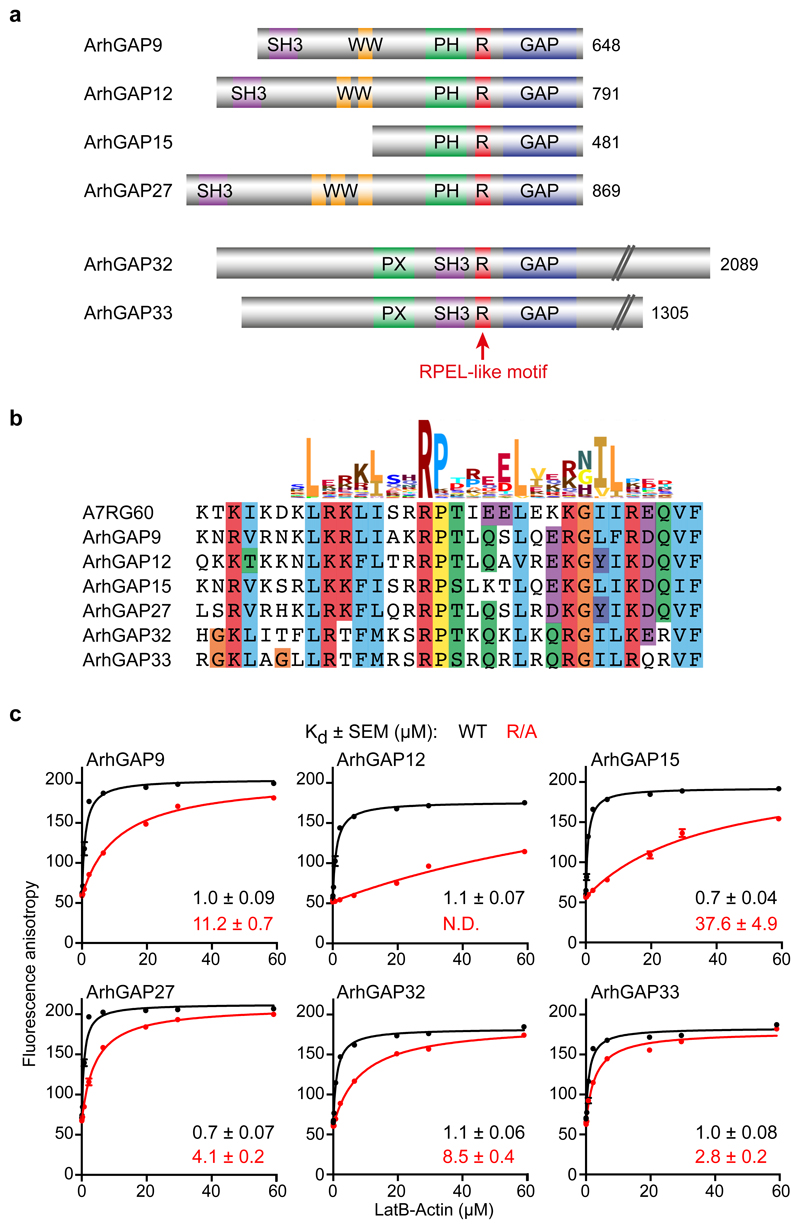
The Starlet sea anemone Nematostella vectensis A7RG60 protein contains a single canonical RPEL motif between its PH and GAP domains (http://pfam.xfam.org/family/rpel; Fig. S1a,b). The A7RG60 RPEL-GAP region is closely related to two subfamilies of vertebrate ArhGAP proteins, ArhGAP9/12/15/27 and ArhGAP32/33, although the RPEL motif in these proteins lacks the conserved RPEL glutamate residue (Fig. 1a,b; Fig. S1c,d). This residue does not directly contact the bound actin, however, but contacts a second G-actin/RPEL unit in proteins containing multiple RPEL motifs33.
Figure 1. Two families of rhoGAPs contain an RPEL motif.
(a) Domain structure of ArhGAP12 and ArhGAP32 rhoGAP subfamilies. The RPEL-like motif is indicated in red. (b) Clustal X sequence alignment of the RPEL-like motifs of ArhGAP12/32 family GAPs with the RPEL motif of Nematostella vectensis A7RG60, aligned with the Pfam PF02755 HMM logo. (c) Fluorescence anisotropy analysis of LatB-actin binding to the FAM-conjugated RPEL peptides shown in (b), or derivatives in which the core RPEL arginine is replaced by alanine. Data were fitted by non-linear regression; data are means ± SEM, n=6 independent experiments. N.D., not determined. See Supplementary Figure 1 for related data. Source data for c are shown in Supplementary Table 1.
To test whether the ArhGAP non-consensus RPEL motif indeed binds G-actin, we performed fluorescence anisotropy assays34(Fig. 1b, c). Peptides encompassing the A7RG60, ArhGAP12- and ArhGAP32-family RPEL motifs bound G-actin with micromolar affinities, comparable to those of the MRTFs and Phactrs13,14,34, and binding was impaired to varying extents by alanine substitution of the core RPEL arginine (Fig. 1c, Fig. S1e). Biolayer interferometry analysis of a GST-RPELArhGAP12 fusion protein gave a comparable result (2.85 μM; Fig. S1f).
Intact ArhGAP12 binds G-actin
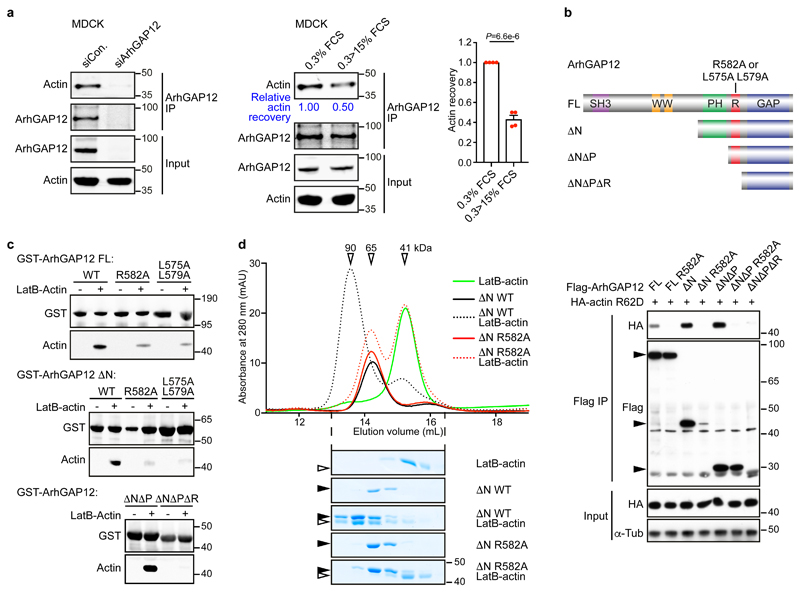
In MDCK epithelial cells, where ArhGAP12 promotes cell scattering23, actin was readily detectable in ArhGAP12 immunoprecipitates, and its recovery decreased following serum stimulation, as seen with other RPEL proteins (Fig. 2a)11,14. Similarly, ArhGAP12 coimmunoprecipitated with the non-polymerisable actin derivative R62D35 upon transient expression in NIH3T3 cells (Fig. 2b). ArhGAP12 did not colocalise with the F-actin cytoskeleton in either cell type (Fig. S1g). Actin binding was not affected by deletion of the SH3, WW or PH domain but was substantially reduced by the mutation or deletion of the RPEL motif (Fig. 2b). Immobilised recombinant GST-ArhGAP12 and GST-ArhGAP32 derivatives could recover purified rabbit skeletal muscle LatB-actin from solution, provided the RPEL motif was intact (Fig. 2c; Fig. S1h). Size exclusion chromatography of complexes formed between LatB-actin and ArhGAP12 ΔN resolved an apparent 1:1 complex of Mr 90 kDa, whose formation was abolished by the R582A RPEL mutation (Fig. 2d); in contrast, ArhGAP1, which does not contain an RPEL motif, did not bind actin in this assay (Fig. S1i).
Figure 2. ArhGAP12 interaction with G-actin requires the RPEL motif.
(a) Endogenous ArhGAP12 was immunoprecipitated and actin recovery was analysed by immunoblot in starved MDCK cells. Cells were transfected with control or ArhGAP12 siRNA (left; data shown represent 4 independent experiments), or serum-stimulated as indicated (middle and right). Bar graph data are mean ± SEM, n=4 independent experiments, two-tailed unpaired t-test. (b) Top, ArhGAP12 derivatives: full-length (FL), amino acids 1-791; ΔN 410-791; ΔNΔP 568-791; ΔNΔPΔR, 582-791. RPEL point mutants were R582A and L575A L579A. Bottom, nonpolymerisable actin mutant R62D was coexpressed with wildtype ArhGAP12 or RPEL mutant R582A in NIH3T3 cells and their interaction analysed by immunoprecipitation and immunoblotting. (c) Immobilised recombinant GST-ArhGAP12 proteins were used to recover purified LatB-actin from solution; actin recovery was analysed by immunoblot. (d) Analytical gel filtration. Elution profiles of recombinant ArhGAP12 ΔN (4 μM) and purified LatB-actin (5 μM) either alone (solid lines) or in a mixture (dotted lines), analysed by absorbance (top) or SDS-PAGE/Coomassie blue staining (bottom). Apparent Mr are indicated. Black and open horizontal arrowheads point to ArhGAP12 and actin respectively. Data shown in (b-d) are representative of 3 independent experiments, respectively. See Supplementary Figure 1 for related data. Source data for a and d are shown in Supplementary Table 1. Unprocessed blots and Coomassie gels are shown in Supplementary Figure 8.
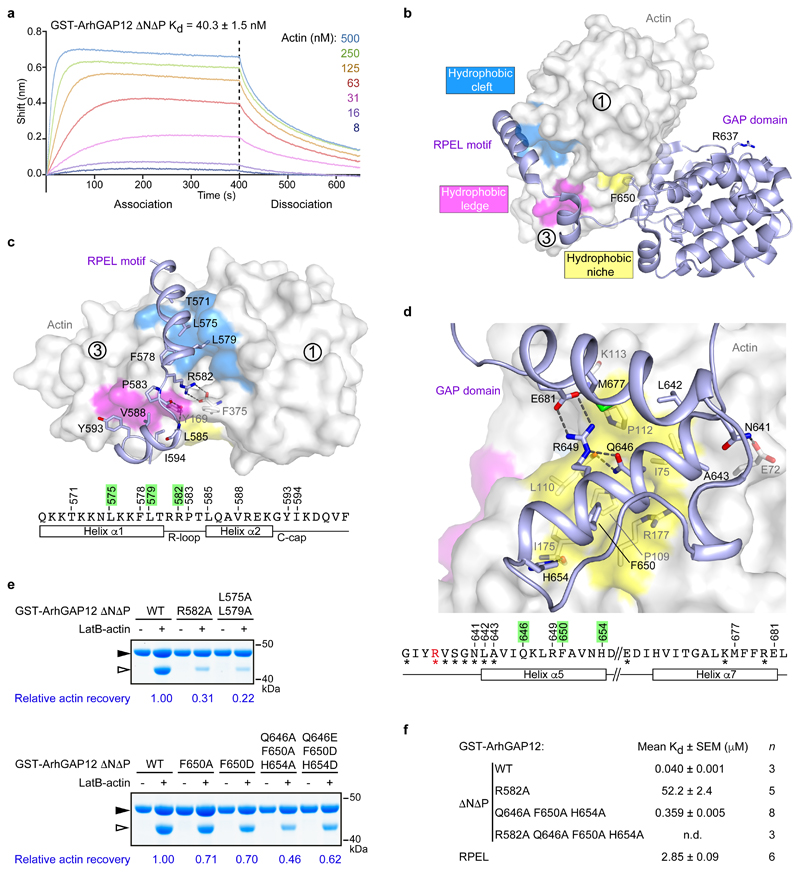
ArhGAP12/G-actin interaction involves both the RPEL motif and sequences within the GAP domain
Using biolayer interferometry, we determined the affinity of LatB-actin for ArhGAP12 ΔNΔP as 40.3 ± 1.5 nM (Fig. 3a). This is significantly greater than the ~micromolar affinity of the G-actin/RPEL motif-peptide interaction, and suggests that G-actin might contact additional sequences in the RPEL-GAP domain. To investigate interaction between G-actin and ArhGAP12 in detail, we determined the structure of ArhGAP12 ΔNΔP bound to LatB-actin (hereafter ArhGAP12•G-actin) at a resolution of 2.6Å (Fig. 3b; Table 1). The asymmetric unit contains four virtually identical copies of the complex, which superimpose with RMSDs ranging from 0.18 Å to 0.35 Å (over 500 Cα) (Fig. S2a). In the ArhGAP12•G-actin complex, ArhGAP12 forms a striking U-shape, clasping G-actin with its RPEL motif and GAP domain, which wrap around subdomains 1 and 3 (Fig. 3b). The extended ArhGAP12/G-actin interaction surface (1700 Å2) includes close contacts between the RPEL motif and the G-actin hydrophobic cleft and ledge (Fig. 3c, Fig. S2b), and between the GAP domain and a hydrophobic niche at the subdomain 1/3 interface, composed of actin I75, I175, and R177, and P109, L110, and P112 from the actin Pro-rich loop (Fig. 3d, Fig. S2c).
Figure 3. Structural analysis of the ArhGAP12•G-actin complex.
(a) Octet biolayer interferometry assay. Biosensors loaded with GST-ArhGAP12 ΔNΔP were incubated with different concentrations of G-actin, which was washed out at 400s. Kd is the mean ± SEM; a representative of 3 independent experiments is shown. (b) The ArhGAP12 ΔNΔP•LatB-actin complex. ArhGAP12 ΔNΔP is shown as blue ribbon, and LatB-actin in white surface representation, with subdomains indicated and the hydrophobic cleft, ledge and niche surfaces coloured in blue, pink and yellow, respectively. The GAP domain catalytic arginine finger is indicated. (c) RPEL-actin interactions. RPEL residues interacting with actin are shown as sticks; RPEL sequence, secondary structures, and interacting residues (mutated residues highlighted) are summarised below. (d) GAP domain interactions with the actin hydrophobic niche. ArhGAP12 residues interacting with the actin niche, or stabilising the orientation of the helices, are shown as sticks. GAP domain helix interaction residues and secondary structures are summarised as in (c), with asterisks indicating residues implicated in interaction with rho-family GTPases10 (catalytic arginine finger R637 in red). (e) Effect of RPEL and GAP domain mutations on G-actin binding, assessed by pulldown assay as in Fig. 2c and detected by Coomassie blue staining. LatB-actin recovery, quantified relative to GST-ArhGAP12 ΔNΔP WT, is indicated below the gels. Black and open arrowheads point to ArhGAP12 and actin respectively. Representative data of three experiments. (f) Summary of Octet biolayer interferometry assays for GST-ArhGAP12 ΔNΔP and its mutant derivatives, and GST-RPEL. Kd is the mean ± SEM, n, independent experiments as indicated; n.d., no binding detectable under the assay conditions. See Supplementary Figure 2 for related data. Source data for a and f are shown in Supplementary Table 1. Unprocessed Coomassie gels are shown in Supplementary Figure 8.
Table 1. Crystallographic data collection and structure refinement statistics.
| ArhGAP12•G-actin (6GVC) | |
|---|---|
| Data collection | |
| Space group | P 21 |
| Cell dimensions | |
| a, b, c (Å) | 101.6, 130.2, 109.3 |
| α, β, γ (°) | 90, 111.1, 90 |
| Resolution (Å) | 54.13 - 2.6 |
| (2.69 - 2.60)a | |
| Rmerge | 0.17 (0.77)a |
| I / σI | 10.82 (1.66)a |
| Completeness (%) | 99.43 (95.83)a |
| Redundancy | 6.2 (3.4)a |
| Refinement | |
| Resolution (Å) | 54.13 - 2.6 |
| (2.69 - 2.60)a | |
| No. reflections | 81 098 |
| (7 746) | |
| Rwork | 0.208 (0.317) |
| Rfree | 0.252 (0.359) |
| No. atoms | |
| Protein | 17,894 |
| Ligand/ion | 268 |
| Water | 182 |
| B-factors | |
| Actin | 48.5/47.5/53.0/52.8 |
| ArhGAP12 | 55.3/54.6/56.0/55.0 |
| Ligand/ion | 53.2 |
| Water | 42.5 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.52 |
| Ramachandran | |
| Favored (%) | 97.45 |
| Allowed (%) | 2.51 |
| Outliers (%) | 0.04 |
One crystal was used for the structure
Values in parentheses are for highest-resolution shell
The ArhGAP12 RPEL motif interacts in a manner largely indistinguishable from that seen in canonical RPEL motif•G-actin complexes13,33,36, but makes two additional contacts through T571 and F578 in helix α1 (Fig. 3c, Fig. S2d). Alanine substitution of the conserved RPEL motif core residue R582, or helix α1 hydrophobic residues L575 and L579, significantly reduced recovery of LatB-actin by ArhGAP12 ΔNΔP in the pull-down assay (Fig. 3e).
The GAP domain structure is essentially identical (RMSD 0.58 Å, 164 Cα) to a previously determined structure of the ArhGAP15 GAP domain (PDB 3BYI; Fig. S2e). The aromatic sidechain of F650, from its helix α5/α7 unit, is deeply buried in the hydrophobic niche, while L642, A643 and M677 interact with the niche edges. These interactions are further stabilised by a network of hydrogen bonds formed between N641, Q646, H654, E681 from the ArhGAP12 helix α5/α7 unit and actin E72, L110, I175 and K113 respectively (Figure 3d, Fig. S2c). Alanine- or charge-substitution of F650 reduced the recovery of G-actin in the pulldown assay, with or without additional alanine substitutions at Q646 and H654 (Fig. 3e). Biolayer interferometry analysis demonstrated that mutation of the GAP helix α5/α7 unit reduced binding affinity ~9-fold, while the RPEL R582A mutation reduced it 1300-fold; binding of actin to a protein containing both mutations was undetectable (Fig. 3f; Fig. S2f). Thus, the high binding affinity of G-actin for ArhGAP12 arises from contacts with both the RPEL motif and GAP domain.
G-actin inhibits ArhGAP12 GAP activity in vitro
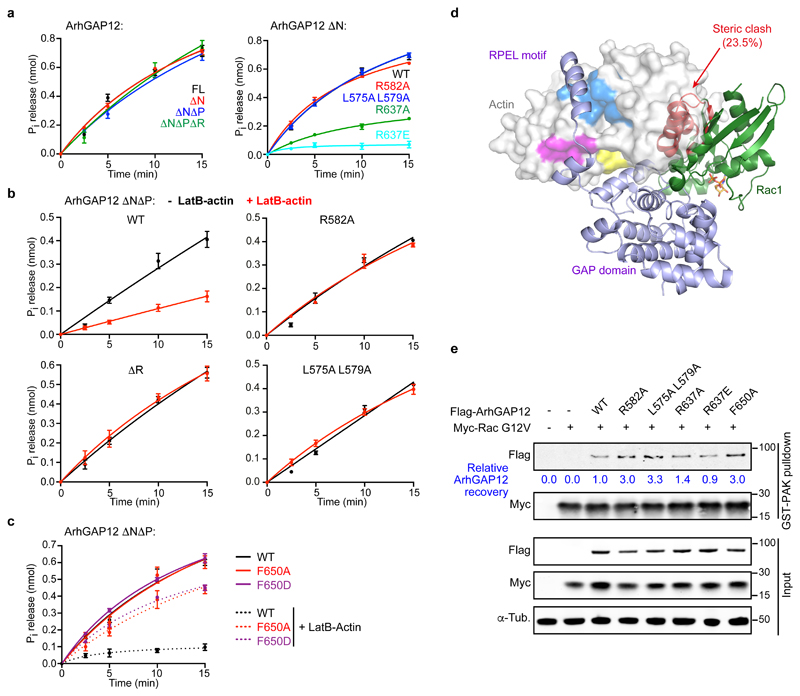
We next investigated the effect of ArhGAP12/G-actin interaction on GAP activity. In a colorimetric GAP assay measuring phosphate release, ArhGAP12 potentiated GTPase activity of Rac1 and Cdc42, but not RhoA (Fig. S3a-c), in agreement with previous studies of ArhGAP12/32 family members22–25, but it remains possible that other rho-family GTPases are ArhGAP12 targets. GAP activity towards Rac1 was unaffected by the presence of ArhGAP12 N-terminal sequences including the RPEL motif, or by RPEL point mutations that reduce G-actin binding (Fig. 4a), but impaired by mutation of R637, the catalytic "arginine finger" (Fig. 4a). Thus, in the absence of actin, the RPEL motif does not affect the in vitro catalytic activity of ArhGAP12.
Figure 4. G-actin inhibits ArhGAP12 GAP activity by occluding rho protein binding.
GAP activity towards Rac1 was assessed using a colorimetric assay for Pi release. Data were fitted by non-linear regression; data are means ± SEM, n=3 (a left, b), n=4 (a right, c) independent experiments. (a) Effect of ArhGAP12 truncations and point mutations of the RPEL motif or catalytic R637. (b) GAP activity is suppressed by 10 μM LatB-actin, and this requires the RPEL motif. (c) Alanine or aspartate substitutions at niche contact residue F650 do not affect GAP activity, but relieve the inhibitory effect of LatB-actin. (d) Model of Rac1 bound to ArhGAP12. The GAP domain of the MgcRacGAP:Cdc42.GDP structure (PDB ID 5C2J) was superimposed onto the GAP domain of the ArhGAP12 ΔNΔP•actin structure. The Rac1 structure (PDB 5N6O) was then superimposed onto the Cdc42 model (RMSD 0.50Å, 148 Cα). Exposed and occluded Rac1 residues are shown as green and red ribbons, GDP in orange. The degree of occlusion is similar for Cdc42 (23.7%) and Rac1 (23.5%). (e) Flag-ArhGAP12 derivatives and constitutively active Myc-RacG12V were co-expressed in NIH 3T3 cells; and recovery of ArhGAP12 and Myc-RacG12V in GST-PAK CRIB pulldown assays assessed by immunoblotting. Representative immunoblots from 3 independent experiments are shown. See Supplementary Figure 3 for related data. Source data for a-c are shown in Supplementary Table 1. Unprocessed blots are shown in Supplementary Figure 8.
Inclusion of increasing concentrations of LatB-actin in the GAP reactions, however, effectively inhibited ArhGAP12 GAP activity in vitro (Fig. 4b, Fig. S3d). Strikingly the RPEL mutations R582A and L575A L579A, which reduce ArhGAP12/G-actin interaction, rendered the GAP activity insensitive to inhibition by LatB-actin (Fig. 4b) as did mutations F650A and F650D, which weaken the interaction between helix 5 of the GAP domain and the actin hydrophobic niche (Fig. 4c). Thus, inhibition of ArhGAP12 GAP activity requires contact between actin and both the GAP domain and the RPEL motif.
G-actin partially occludes the GTPase binding site in the complex
To understand further the molecular mechanism by which G-actin binding inhibits ArhGAP12 activity, we modelled the interaction of Rac and Cdc42 with ArhGAP12. Superposition of the structure of the MgcRacGAP•Cdc42 complex GAP domain (PDB ID 5C2J) onto that of ArhGAP12•G-actin (RMSD=0.63Å for 123 Cα) revealed a substantial steric clash – 23.7% of GTPase atoms – between G-actin and Cdc42, and superposition of the Rac1 structure (PDB 5N6O) onto this model (RMSD 0.50Å for 148 Cα), revealed a similar clash for Rac1 (23.5%; Fig. 4d, Fig. S3e). To test this we co-expressed ArhGAP12 derivatives with RacG12V, which is locked in the GTP-bound state, and assessed the ability of a GST-PAK CRIB domain fusion protein to recover ArhGAP12 from cell lysates in association with Rac. RPEL and GAP domain mutations that impair G-actin/ArhGAP12 interaction increased ArhGAP12 recovery in this assay (Fig. 4e). Taken together, these data suggest that GAP domain interaction with the actin hydrophobic niche constrains its position so as to inhibit interaction with its substrate GTPases.
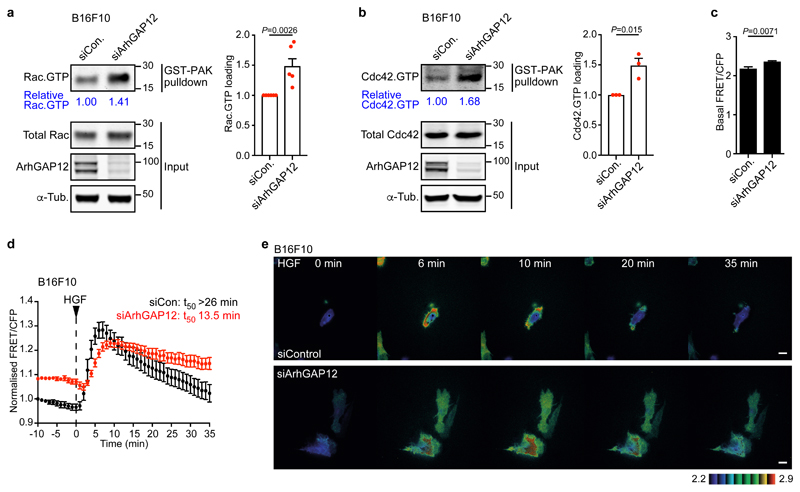
ArhGAP12 controls GTP loading on Rac and Cdc42 in melanoma cells
ArhGAP15, an ArhGAP12 family member, is implicated in maintenance of basal Rac.GTP levels in various cell types37–39. To investigate the role of ArhGAP12 in rho family GTPase regulation we studied melanoma cells, whose behaviour in invasiveness and experimental metastasis assays is Rac/Cdc42 dependent40–44. In B16F10 melanoma cells, siRNA-mediated ArhGAP12 depletion did not appreciably affect transcription of other ArhGAP12- and ArhGAP32-family members (Fig. S4a,b), but increased GTP loading on Rac and Cdc42, (Fig. 5a,b), as did depletion of ArhGAP32 (Fig. S4c). We used the RaichuEV-Rac FRET-based biosensor45 to test how ArhGAP12 affects the kinetics of GTP loading on Rac. In agreement with the pulldown experiments, the biosensor detected elevated basal GTP loading on Rac in ArhGAP12-depleted cells (Fig. 5c); following stimulation by HGF, downregulation was inhibited, taking at least twice as long to decrease to 50% of peak levels (Fig. 5d,e; Fig. S4d). Thus, ArhGAP12 antagonises Rac and Cdc42 activity in B16F10 melanoma cells.
Figure 5. ArhGAP12 controls GTP loading on Rac and Cdc42 in melanoma cells.
B16F10 melanoma cells were transfected with control or ArhGAP12 siRNA. (a) Rac.GTP and (b) Cdc42.GTP levels, as assessed by GST-PAK pulldown assays. Left, representative immunoblots. Right, data summary. Data are means ± SEM, n=6 (a) or n=3 (b) independent experiments, two-tailed unpaired t-test. (c) Increased basal Rac GTP loading in serum-starved B16F10 cells, measured using the RaichuEV-Rac FRET biosensor. FRET/CFP ratio was measured over 9 min in control (n=22) or ArhGAP12-depleted (n=32) cells. Data are means ± SEM, two-tailed Mann Whitney test. (d) Kinetics of Rac GTP loading in control (n=10) and ArhGAP12-depleted (n=9) B16F10 cells following HGF stimulation, measured as in (c). Data are expressed relative to control cell value at the start of the experiment. T50, time to recover to 50% peak Rac GTP loading. Data are means ± SEM. (e) Representative FRET/CFP ratio images displayed in 8-color, intensity modulated display mode. Representative images of three independent experiments. Scale bar 20 μm. See Supplementary Figures 4 and 5 for related data. Source data for a-d are shown in Supplementary Table 1. Unprocessed blots are shown in Supplementary Figure 8.
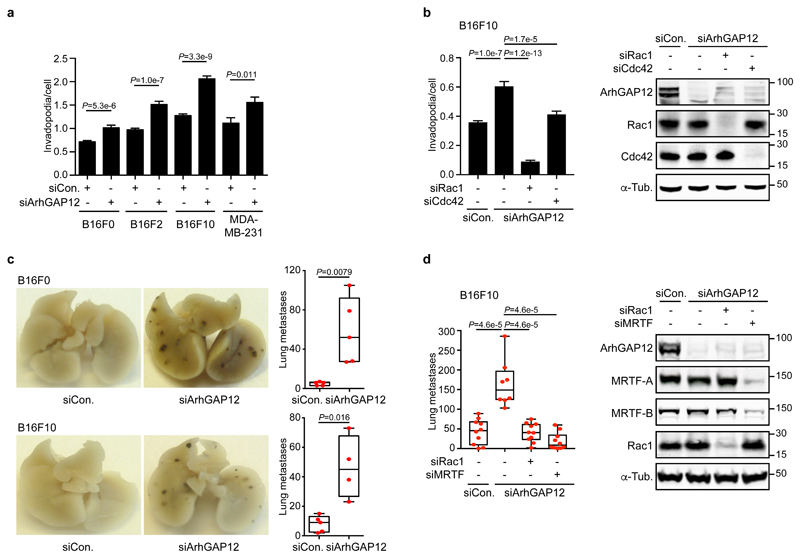
ArhGAP12 controls invadopodia and experimental metastasis in melanoma cells
Invadopodia are actin-rich membrane protrusions that degrade the extracellular matrix to drive tumour cell invasion46. Depletion of ArhGAP12 potentiated invadopodia formation by B16F10 and B16F2 melanoma cells and their less invasive parent B16F0, and by MDA-MB-231 mammary carcinoma cells (Fig. 6a; Fig. S4e,f); in B16F10 cells, invadopodia formation was strongly dependent on Rac and to a lesser extent on Cdc42 (Fig. 6b). ArhGAP12 depletion also significantly increased the ability of B16F10 and B16F0 cells to induce experimental metastasis in the mouse tail vein assay (Fig. 6c) without affecting cell proliferation (Fig. S4g,h). Increased metastasis was strongly dependent on Rac (Fig. 6d) and required MRTF/SRF signalling, as expected from our previous studies of B16F2 cells (Fig. 6d)47. Consistent with this, ArhGAP12-depleted B16F10 cells exhibited a Rac-dependent increase in F-actin (Fig. S5a,b), which was accompanied by increased nuclear accumulation of MRTF-A and increased expression of MRTF/SRF target genes (Fig. S5c,d). ArhGAP12 also contributes to the maintenance of basal Rac activity in NIH3T3 fibroblasts, where it is the only family member expressed (Fig. S5e): in resting cells, its depletion increased F-actin levels, and promoted MRTF-A nuclear accumulation and MRTF/SRF target gene expression11 (Fig. S5f-i). ArhGAP12 thus controls Rac-dependent processes in melanoma and fibroblast cells.
Figure 6. ArhGAP12 regulates Rac-dependent processes in cells.
Cells were transfected with control, ArhGAP12 or other siRNA as indicated. (a,b) Invadopodia formation by cells plated overnight on Oregon-green labelled gelatin was detected by loss of staining. (a) 7 fields per well have been imaged and averaged. Data shown represent n=16 independent wells pooled from three independent experiments. At least 8,507 cells were imaged per condition. (b) 4 fields per well have been imaged and averaged. Data shown represent n=24 independent wells pooled from three independent experiments. At least 30,675 cells were imaged per condition. Data in a and b are means ± SEM, two-tailed Mann Whitney test. (c,d) Experimental metastasis assay. B16F0 and F10 cells were injected in the tail vein of C57BL/6J mice. Images show lung colonisation after 12 days. Box-and-whiskers plots indicate the number of lung metastases, showing median, quartiles, and highest and lowest values. Representative results of three experiments are shown; n=5 (c) and n=10 (d) mice per group, except B16F10/siArhGAP12 for which n=4 (c) and n=8 (d), two-tailed Mann Whitney test. See Supplementary Figures 4 and 5 for related data. Source data for a-d are shown in Supplementary Table 1. Unprocessed blots are shown in Supplementary Figure 8.
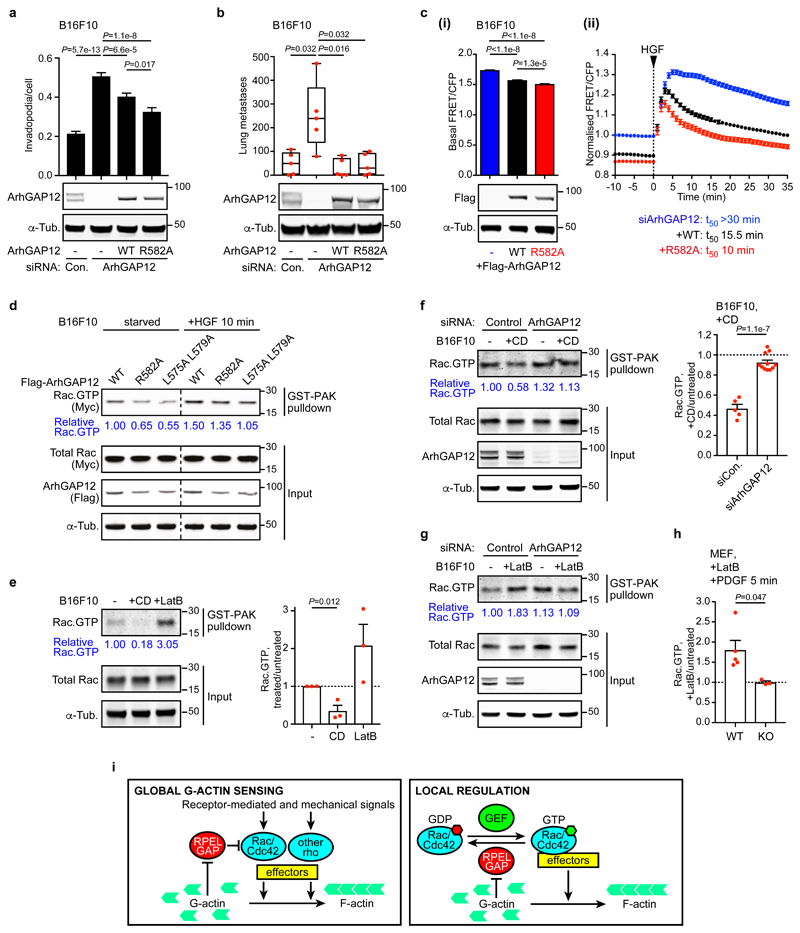
G-actin binding controls ArhGAP12 GAP activity in vivo
We next investigated the role of G-actin in ArhGAP12 regulation in vivo. B16F10-derived cell lines conditionally expressing siRNA-resistant ArhGAP12 derivatives were depleted of endogenous ArhGAP12, and their behaviour compared in the invadopodia and metastasis assays. Re-expression of both wildtype and R582A ArhGAP12 effectively suppressed the increased invadopodia formation, experimental metastasis, and F-actin formation associated with ArhGAP12 depletion; the R582A mutant was more effective than the wildtype protein at suppressing invadopodia formation, and was as effective as the wildtype protein in the other assays, even though it was expressed at lower level (Fig. 7a,b, Fig. S6a).
Figure 7. G-actin regulates Rac activity in melanoma cells.
(a,b) B16F10 conditional lines expressing control or siRNA-resistant Flag-ArhGAP12 derivatives were transfected with control or ArhGAP12 siRNA. (a) Invadopodia formation assessed as in Figure 6b. 4 fields per well have been imaged and averaged. Data shown represent n=24 independent wells pooled from three independent experiments. At least 22,437 cells were imaged per condition. Data are means ±SEM, two-tailed Mann-Whitney test. (b) Experimental metastasis assay, displayed as in Figure 6d. Representative results of three experiments are shown, n=5 mice, two-tailed Mann-Whitney test. (c) HGF-induced Rac GTP loading imaged using the RaichuEV-Rac biosensor. siRNA-resistant Flag-ArhGAP12 WT or R582A were transiently re-expressed in serum-starved ArhGAP12-depleted B16F10 cells. Images were acquired from control (n=96), +ArhGAP12 WT (n=58) and +ArhGAP12 R582A (n=37) cells. (i) Basal Rac GTP loading, measured by FRET/CFP ratio over 10 min before stimulation. Data are means ±SEM, two-tailed Mann-Whitney test. Note the lower expression level of ArhGAP12 R582A. (ii) Kinetics of Rac GTP loading following HGF stimulation, normalised taking the basal activity in control cells as 1.0. Data are means ±SEM. (d) Immunoblot analysis of GST-PAK Rac pulldown assays using lysates of B16F10 cells cotransfected with Flag-ArhGAP12 derivatives and Myc-Rac. Representative immunoblot of three independent experiments. (e) B16F10 cells, maintained in 0.3% FCS, following treatment with Cytochalasin D (CD) or Latrunculin B (LatB) for 30 min before Rac.GTP pulldown assay. Data are means ±SEM n=3, two-tailed unpaired t-test. (f) Cells transfected with control or ArhGAP12 siRNA were maintained in 10% FCS and treated with CD for 30 min before Rac.GTP pulldown assays. Data are means ±SEM, n=5 (control), n=11 (siArhGAP12) independent experiments, two-tailed unpaired t-test. (g) Cells as in (f) were treated with LatB for 30 min before Rac.GTP pulldown assay. Representative immunoblot of three independent experiments. (h) Wildtype and ArhGAP12-knockout MEFs were treated with LatB (30 min), and PDGF (5 min) before Rac.GTP pulldown assay. Data are means ±SEM, n=5 (WT), n=3 (KO) independent experiments, two-tailed unpaired t-test. (i) Global and local regulation by RPEL rhoGAP proteins. See Supplementary Figures 5 and 6 for related data. Source data for a-c, e, f and h are shown in Supplementary Table 1. Unprocessed blots are shown in Supplementary Figure 8.
We used the depletion-rescue approach in conjunction with the RaichuEV-Rac FRET biosensor to investigate the consequences of G-actin/ArhGAP12 interaction for Rac GTP loading in vivo. B16F10 melanoma cells transiently expressing siRNA-resistant wildtype or RPEL R582A mutant ArhGAP12 were depleted of endogenous ArhGAP12, and the kinetics of Rac GTP loading following HGF stimulation measured. In this setting, expression of ArhGAP12 R582A lowered the basal level of Rac GTP loading more than wildtype ArhGAP12, and altered the kinetics of Rac GTP loading such that Rac downregulation occurred more rapidly (Fig. 7c, Fig. S6b). Consistent with this, Rac.GTP pulldown assays from B16F10 cells transiently expressing ArhGAP12 derivatives, together with myc-tagged Rac, showed that Rac.GTP levels were lower in cells expressing the ArhGAP12 mutants R582A and L575A L579A, which cannot bind G-actin, than in cells expressing wildtype ArhGAP12, even though the mutant proteins were less efficiently expressed (Fig. 7d). Consistent with these results, expression of ArhGAP12 R582A in tetracycline-inducible NIH3T3 cell lines decreased F-actin levels to a greater extent than wildtype ArhGAP12 (Fig. S6c,d).
The preceding results show that the RPEL motif exerts an inhibitory effect on ArhGAP12 Rac GAP activity in vivo. To verify that this is a direct result of changes in ArhGAP12/G-actin interaction, we examined the effects of actin-binding drugs on ArhGAP12 activity. Cytochalasin D (CD) and Latrunculin B (LatB) both bind G-actin, promoting F-actin disassembly, but have opposing effects on RPEL proteins: CD binds G-actin competitively with the RPEL motif and disrupts G-actin binding, while LatB is compatible with G-actin/RPEL interaction11,12. Accordingly, treatment of B16F10 cells with CD decreased Rac.GTP levels, whereas LatB treatment potentiated it, as assessed using the GST-PAK pulldown assay (Fig. 7e). Strikingly, neither drug treatment affected Rac.GTP levels in ArhGAP12-depleted cells, indicating that their effects require ArhGAP12 (Fig. 7f,g; Fig. S6e). Similar results were obtained upon comparison of wildtype and ArhGAP12 knockout fibroblasts (Fig. 7h). Thus, G-actin controls cellular Rac GTP loading through its interaction with ArhGAP12-family GAPs.
Discussion
We have shown that the ArhGAP12- and ArhGAP32-family rhoGAPs are RPEL proteins, each containing a single atypical RPEL motif immediately N-terminal to the GAP domain. G-actin forms a 1:1 complex with ArhGAP12, inhibiting its GAP activity towards Rac1 and Cdc42. Actin makes canonical interactions with the RPEL motif, and also interacts with the GAP domain through a hydrophobic ‘niche’ on its subdomain 1/3 interface. Although the GAP domain contacts contribute only modestly to the overall G-actin binding affinity, they are critical for the repressive effect of actin binding on GAP activity. Inhibition of GAP activity by occlusion of the GTPase binding site is also seen in the inhibitory interaction between DLC1 and the SH3 domain of p120RasGAP48. Our results show that G-actin binding to ArhGAP12 downregulates its GAP activity in melanoma cells in vivo, sculpting the kinetics of Rac.GTP accumulation, and controlling Rac-dependent processes such as invadopodia formation and experimental metastasis.
Extracellular chemical or environmental signals, and changes in cell differentiation state are all associated with changes in actin dynamics. Since G-actin diffuses rapidly, ArhGAP12/32-family members would effectively link the downstream functions of their target GTPases, which include F-actin assembly, to the general state of actin cytoskeletal dynamics, thereby constituting a feedback loop (Fig. 7i, left). ArhGAP12-family members are also enriched at specific subcellular locations, such as epithelial cell junctions and macrophage phagocytic cups23,30. In such settings they could directly monitor local G-actin fluctuations induced by their target GTPases, thereby fine-tuning GTPase activity, as part of a local homeostatic feedback loop (Fig. 7i, right). Indeed, it has been proposed that ArhGAP12 fulfils such a function at the phagocytic cup30, and we are currently investigating this further.
The ArhGAP12- and ArhGAP32-family rhoGAPs contain only a single atypical RPEL motif lacking the conserved RPEL core glutamate (Pfam PF02755). Our previous studies of multivalent G-actin/RPEL complexes showed that the glutamate contacts a second RPEL/G-actin unit on its C-terminal side33,36, and it is therefore unsurprising that atypical RPEL motif peptides bind G-actin with comparable ~micromolar-range affinities13,14. We estimated the G-actin binding affinity of the intact ArhGAP12 RPEL-GAP fragment to be 40.3 ±1.5 nM, which is comparable to that estimated for the MRTF-A RPEL domain (~25nM35). We therefore think it likely that the ArhGAPs and MRTFs will be similarly responsive to changes in G-actin concentration, even though the ArhGAPs contain one rather than three RPEL motifs. The development of sensors that allow tracking of G-actin concentration and measurement of G-actin/RPEL interaction in vivo will be important to resolve this issue.
The actin hydrophobic niche identified here is conserved amongst different actin family members, and it is therefore unlikely that different actins have differential effects on ArhGAP12 activity. Although not previously implicated in interactions with other G-actin binding proteins, the niche region mediates actin-actin interactions within the ADP F-actin filament49,50. In this context, however, it displays a more open conformation, with the subdomain 1 Pro-rich loop interaction being disrupted to form the phosphate exit channel. ADP-ribosylation of residue R177, at the niche edge, by bacterial toxins disrupts filament formation (reviewed in ref51). The niche makes intimate contacts with the helix 5-7 unit of the rhoGAP domain, just C-terminal to the catalytic arginine, which is implicated in GTPase recognition10. ArhGAP12 F650, which docks in the niche, is conserved or substituted by tyrosine or histidine in the other RPEL GAPs, but is generally hydrophilic in other rhoGAPs10 (Fig. S7). This, and the lack of RPEL motifs in other rhoGAPs, suggests that only the ArhGAP12 and ArhGAP32 families are regulated by G-actin.
ArhGAP12 is present at high levels at adherens junctions, where it promotes cell-cell adhesion23,26–28, and at other actin-regulated cell surface structures such as plasma membrane blebs, phagocytic cups and dendritic spines29–31. Both ArhGAP12 and ArhGAP15 localisation is controlled by PI 3-kinase signalling30,39, and the PH domain of the ArhGAP12 family member ArhGAP9 binds the phospholipid products of PI 3-kinase52. The PH domain is just N-terminal to the RPEL motif, so we are currently investigating whether G-actin binding also affects its function. The two ArhGAP32 family GAPs, contain an SH3 domain N-terminal to the RPEL motif, and it will be interesting to see if G-actin influences its interactions.
ArhGAP12 suppresses basal levels of GTP loading on Rac and Cdc42 in mouse melanoma cells, as does the ArhGAP12 family protein ArhGAP15, in diverse settings, including brain, glioma, 293 kidney cells and myeloid lineages37–39. This could occur in two ways. First, depletion of ArhGAP12 from specific subcellular locations might increase Rac.GTP at these locations, which could rapidly exchange with Rac pools elsewhere in the cell. Alternatively, depletion of ubiquitously localised ArhGAP12 might impact global Rac.GTP level directly, although one might expect such effects would be small given most cells express multiple rho GAPs9,10. Either way, depletion of ArhGAP12 in B16F10 cells raises Rac GTP loading sufficiently to potentiate invadopodia formation and experimental metastasis40–42, the latter appearing to reflect Rac-dependent F-actin assembly and MRTF activation. Interestingly, low ArhGAP12-family expression levels are associated with poor survival in human melanoma in the TCGA database analysed using the OncoLnc tool (http://www.oncolnc.org).
We found that direct G-actin/ArhGAP12 interaction plays a significant role in control of ArhGAP12 GAP activity. In melanoma cells, ArhGAP12 RPEL mutants defective in actin binding exhibited greater GAP activity, were more effective at Rac downregulation following growth factor stimulation, and in at least some biological assays, such as invadopodia formation, were significantly more active than the wildtype protein. Moreover, the actin-binding drugs CD and LatB had opposing effects on Rac GTP loading consistent with their differential effects on G-actin/RPEL interaction, which were ArhGAP12-dependent. Interestingly, in macrophages, which exhibit ArhGAP12-dependent phagocytosis, CD and actin siRNA inhibit Rac and Cdc42 activation, and LPS-stimulated phagocytosis30,53, while in neutrophils, LatB treatment prevents ArhGAP15-dependent Rac down-regulation following PI3K activation39. Our findings suggest that these observations reflect the direct control of ArhGAP12 family proteins by G-actin in these contexts.
The RPEL motif present in the ArhGAP12 and ArhGAP32 proteins couples their activity to the surrounding availability of G-actin (Fig. 7i). We therefore consider it likely that these rhoGAPs will be involved in biological processes that are critically reliant on local control of F-actin assembly. Our future work will focus on elucidating how ArhGAP12 family members' activity relates to fluctuations in local G-actin concentration.
Methods
Plasmids
ArhGAP12 cDNA (short isoform encoding a 791 amino acid-protein, Uniprot S4R248) was amplified by PCR from an NIH 3T3 cDNA library using standard techniques. ArhGAP12 derivatives were expressed in mammalian cells with N-terminal Flag tag in pEF-plink11 or pcDNA4TO (Invitrogen) or as N-terminal GFP fusion in pcDNA6.2-N-EmGFP (Invitrogen), and in bacteria as GST fusion in pGex-6P-2 vector. Point mutants and siRNA-oligonucleotide 11-resistant ArhGAP12 derivatives were generated by site-directed mutagenesis (three mismatches: 5’-GAG CAT GTC-3’ to 5’-GAA CAC GTT-3’)(Quikchange, Agilent). Deletion mutants were created using the Phusion high-fidelity protocol (New England Biolabs). For lentiviral transduction, Flag-ArhGAP12 sequences were cloned into a modified pTripz vector (Dharmacon), where RFP and microRNA regulation sequences were replaced by a bGH Poly(A) sequence using In-fusion HD cloning (Takara). The ArhGAP32 RPEL-GAP domain (amino-acids 339-569) was expressed in bacteria as GST fusion using a pGex-6P-2 vector. Expression plasmids for Actin R62D and Rac have been described35. All plasmids were sequenced using Sanger sequencing.
Protein expression, purification and size-exclusion chromatography
Rabbit skeletal muscle LatB-actin was prepared as described34. ArhGAP12 and ArhGAP32 protein expression was induced at 37°C in Escherichia coli Rosetta (DE3) pLysS. Bacteria were harvested by centrifugation and lysed in 20 mM Tris, pH 6.8 and 8.5 respectively, 150 mM NaCl, 10 mM MgCl2, 1% Triton X-100, 1 mM DTT, 1 mM phenylmethylsulphonyl fluoride and protease inhibitors (Roche). The GST-fusion proteins were adsorbed onto a glutathione-sepharose resin (GE Healthcare), and ArhGAP12 derivatives were recovered by cleavage with 3C protease overnight at 4°C in 20 mM Tris pH 6.8, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT. Proteins were then purified by size exclusion chromatography using a Superdex 200 column (GE Healthcare) in 50 mM Tris, pH 6.8, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT. The purity of the proteins was examined by SDS-PAGE and Coomassie brilliant blue staining (Fig. S3b).
Analytical size exclusion chromatography and in vitro pulldown assays
For gel filtration analyses, 4 μM of purified ArhGAP12 derivatives or recombinant GST-ArhGAP1 (Cytoskeleton, GAS01) were incubated with 5 μM or 2 μM LatB-actin respectively and loaded on a calibrated Superdex 200 (10:300) column (GE Healthcare) in 50 mM Tris, pH 6.8 and 7.6 respectively, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT. Fractions collected were concentrated, and a tenth of each fraction was analysed by SDS-PAGE and Coomassie brilliant blue staining. For GST pulldown experiments, glutathione-sepharose beads (GE Healthcare) were saturated with GST-ArhGAP12 or GST-ArhGAP32 from E. coli lysates, and used as an affinity resin in a binding reaction with 10 μM LatB-actin in binding buffer (50 mM Tris, pH 7.0 and pH 8.5 respectively, 50 mM NaCl, 5 mM MgCl2) for 1h at 4°C. The resin was then washed 4 times in binding buffer and subjected to SDS-PAGE, Coomassie staining or Western blotting. Coomassie brilliant blue staining was performed according to standard techniques, and quantified using Image Studio after scanning with an Odyssey infrared scanner (Licor). Actin recovery was quantified relative to input GST-ArhGAP12. Unprocessed scans of blots and Coomassie gels are shown in Supplementary Figure 8.
Crystallisation, data collection and refinement of the ArhGAP12•G-actin complex
The protein complex was prepared by mixing purified ArhGAP12 ΔNΔP and LatB-actin in a 1:2 molar ratio and further purified by Superdex 200 size-exclusion chromatography equilibrated in 20 mM Tris, pH 8, 50 mM NaCl, 3 mM MgCl2, 0.2 mM EGTA, 0.2 mM ATP and 0.3 mM TCEP. To grow crystals, the protein solution was concentrated to 30 mg/mL. The complex was crystallised at 20°C using the sitting-drop vapour diffusion method. Drops of 0.5 μL consisted of a 1:1 (vol:vol) mixture of protein and a well solution containing 0.1 M Bis-Tris Propane, pH 6.5, 20% PEG 3350, 0.2 M sodium thiocyanate. Crystals appeared after five days and reached their maximum size after ten days. Crystals were cryoprotected in mother liquor supplemented with 20% glycerol and then flash-frozen in liquid nitrogen. X-ray data were collected at 100 K at the ID24 beamline (mx8015) of the Diamond Light Source synchrotron (DLS, Oxford, United Kingdom). Data collection and refinement statistics are summarised in Table 1. The data set was indexed, scaled and merged using xia254. Molecular replacement was achieved by using the high resolution atomic coordinates of G-actin extracted from the RPEL2•LatB-actin13 structure (PDB ID 2V52) and the GAP domain extracted from ArhGAP15 structure (PDB ID 3BYI) in PHASER55. Refinement was carried out by using Phenix56. Model building was carried out in COOT57. Model validation used PROCHECK58, and figures were prepared using the graphics program PYMOL 2.1.159. The asymmetric unit contains 4 copies of the complex. The ArhGAP12•G-actin structure has been deposited in PDB (ID 6GVC).
Protein affinity measurements
Fluorescence anisotropy assays were performed as described previously34. Dissociation constants were derived by nonlinear regression analysis of the data using Prism (GraphPad software). Biolayer interferometry analysis of G-actin binding to immobilised GST-ArhGAP12 was performed using the Octet Red96 (ForteBio); typical immobilisation levels were above 2.5 nm. GST-ArhGAP12 loaded anti-GST biosensors were incubated with various concentrations of G-actin in the kinetics buffer (25 mM Tris pH7.5, 100 mM NaCl, 0.1% Tween 20, 5 mM MgCl2, 0.5 mM TCEP, 1 mg/mL BSA). Binding experiments were performed in solid-black 96-well plates, at 25°C with an agitation speed of 1,000 rpm. Data analysis was done using the Octet software version 7.1 (ForteBio). Global fitting of the binding curves generated a best fit with a 1:1 model and the kinetic association and dissociation constants were calculated. The quality of the fit was assessed by evaluation of the χ2 and R2 values generated from all the fitting analyses. Experiments were repeated at least 3 times.
RhoGTPase activity assays
GAP activity was measured using a colorimetric rhoGAP assay kit (Cytoskeleton, BK105). The reactions were performed in 20 μL with 4.75 μM rho GTPase, 2 μM ArhGAP12 derivatives (Fig. S3b), in presence or absence of 10 μM LatB-actin, at 37°C for the indicated time after addition of 200 μM GTP. The release of inorganic phosphate (Pi) was detected at 650 nm using a SpectraMax Plus 384-well plate reader. A KH2PO4 solution was used to calibrate the quantity of Pi (nmol) released to the absorbance. A non-linear regression analysis was applied to the data using Prism (GraphPad software).
Cell lines
B16F0, B16F2, B16F10 melanoma cells, SV40 immortalised MEFs, NIH3T3 fibroblasts, MDCK II epithelial cells and MDA-MB-231 breast carcinoma cells were maintained in DMEM with 10% fetal calf serum (FCS). Where indicated, cells were serum starved (0.3% FCS) overnight, then treated for 30 min with 15% FCS, 100 ng/mL HGF (Millipore, GF414), 5 μM Cytochalasin D (Merck, 250255), or 1 μM Latrunculin B (Merck, 428020). NIH 3T3 monoclonal lines stably expressing Flag-tagged ArhGAP12 derivatives and the Tet repressor were generated using pcDNA4TO-Flag-ArhGAP12 and pcDNA6/TR (Invitrogen) plasmids, and selected for zeocin (200 μg/mL) and blasticidin (5 μg/mL) resistance. Expression was induced overnight with 2 μg/mL Tetracycline. B16F10 polyclonal lines conditionally expressing Flag-ArhGAP12 derivatives were generated by lentiviral transduction using pTripz-Flag-ArhGAP12 plasmids, and selected for puromycin (0.5 μg/mL) resistance. Expression was induced for 24h with 2 μg/mL Doxycycline. Cell growth was analysed following seeding and siRNA transfection of 50,000 cells in a well of a 6-well plate. Each day, cells of replicate wells were trypsinised, resuspended in media and counted using the Countess II instrument (Invitrogen). For cell cycle analysis cells were fixed after 2.5h of BrdU incorporation, counterstained with Propidium iodide, and analysed by flow cytometry using standard methods and FlowJo software as previously described47. All cell lines tested negative for mycoplasma and were authenticated by STR profiling by Crick Cell Services.
Transfection, immunoblotting and immunofluorescence
Cells were transfected with expression plasmids using Lipofectamine 2000 according to the manufacturer’s protocol (Invitrogen, 11668-019). Cells were reverse transfected with RNAi oligonucleotides using Lipofectamine RNAiMax (Invitrogen, 13778-150). siRNAs were: control UUCUCCGAACGUGUCACGU; MRTF-A/B UGGAGCUGGUGGAGAAGAA; ArhGAP32: L057176-01; Rac1: L041170-00; Cdc42: L043087-01; ArhGAP12: mouse L-040581-01 (a pool of oligonucleotides J-040581-9, -10, -11 and -12) and human L-008729-01 Dharmacon smartpools. In experiments where siRNA-resistant ArhGAP12 derivatives were re-expressed, either transiently or in ArhGAP12-expressing lines, ArhGAP12 siRNA oligonucleotide 11 (GCAUUGAGCAUGUCGAAGA) was used. In MDCK II cells, the oligonucleotide targeting ArhGAP12 was GAACAGAACUGCUAAUUCAUU. Assays were performed 72h after siRNA transfection, with the exception of the experimental metastasis assay (40h); where required, siRNA-depleted cells were transfected with ArhGAP12 plasmids 24h before analysis.
Whole cell extract preparation and immunoblotting were performed using standard techniques. Unprocessed scans of blots are shown in Supplementary Figure 8. For phenotypic experiments, samples were taken for analysis of protein expression at the time assays were commenced. Antibodies used were against β-actin (Santa-Cruz Biotechnology, clone C4, sc47778, 1:1,000 dilution), HA (Roche, 3F10, 11867431001, 1:1,000 dilution), Rac (Millipore, clone 23A8, 05-389, 1:500 dilution), Cdc42 (Millipore, 05-542, 1:250 dilution), MRTF-A (Santa-Cruz Biotechnology, C-19, sc21558, 1:1,000 dilution), MRTF-B (Bethyl Laboratories, A302-768, 1:1,000 dilution), Myc (Crick Biological Resources Facilities, clone 9E10, 1:1,000 dilution), ArhGAP12 (Sigma, HPA000412, 1:1,000 dilution), α-Tubulin (Sigma, clone B5-1-2, T5168, 1:6,000 dilution), GST (Sigma, G7781, 1:10,000 dilution) and Flag (Sigma, F7425, 1:1,000 dilution). Secondary antibodies were IRDye-680LT or -800CW conjugated (Licor, 925-68022, -68023, -32212, -32214, -32219, 1:10,000 dilution). For coimmunoprecipitation experiments in MDCK cells, Alexa Fluor 790 conjugated light chain specific IgG (Jackson Immunoresearch laboratories, 211-652-171 and 115-655-174, 1:5,000 dilution) were used. Immunoblots were scanned with an Odyssey infrared scanner (Licor) and quantified using Image Studio. Immunofluorescence assays were carried out as described previously11. F-actin was detected with Alexa Fluor 647- or Texas Red-Phalloidin (Invitrogen, A22287 and T7471) and DNA was counterstained using DAPI. F-actin staining was imaged in 96-well glass bottom plates on the automated Cellomics Arrayscan VTi and the intensity was measured using the Target Activation Bioapplication (Cellomics). Where indicated, cells were imaged using a confocal Laser Scanning Microscope LSM710 controlled by the Zen software (Zeiss), with a 63x/1.40 oil Plan Apochromat objective lens (Zeiss), utilising the 405 (DAPI), 488 (GFP) and 561 nm (Texas-Red) lasers for excitation, and a pinhole set at 1 Airy unit.
Immunoprecipitation and Rac/Cdc42 pulldown experiments
For Flag immunoprecipitation, cells were lysed in IP buffer (50 mM Tris, pH 7.8, 100 mM NaCl, 1% Triton X-100, 1 mM DTT and protease inhibitors (Roche)). The soluble fraction was precleared with Protein A-Sepharose beads (Sigma), and incubated with M2-Agarose beads (2h, 4°C, Sigma) with rotation. ArhGAP12 immunoprecipitation was performed with essentially the same protocol with the exception that it used 50 mM NaCl, Dynabeads Protein G (Invitrogen) and a pan-ArhGAP12 polyclonal antibody generated by Crick Biological Resources Facilities. Beads were washed four times in IP buffer and resuspended in SDS Laemmli buffer. Actin recovery was quantified relative to input. Rac/Cdc42 pulldown experiments were performed using 15 μL of GST-tagged human PAK1 p21-binding domain (residues 67-150, 1 μg/μL) bound to glutathione magnetic beads (Millipore, 17-10394), carried out according to the manufacturer’s instructions. Rac/Cdc42 GTP loading was quantified relative to total Rac/Cdc42. ArhGAP12 recovery was quantified relative to input.
Gene expression
Total RNA was isolated and cDNA was synthesised as described previously16. Amounts of cDNA corresponding to 10 ng of RNA were analysed in SYBR Green based real-time quantitative PCR (Invitrogen) using ABI Prism 7900HT and QuantStudio 5 detection systems (Applied Biosystems). Absolute quantification of cDNA abundance was determined using a mouse genomic DNA standard. Data were normalised to the abundance of Gapdh cDNA. Gene-specific exonic primers were as follows: Arhgap9 (CAGAGGGCACTGACCAGAAGA and TTGGCGATTAGCCGCTTTAA), Arhgap12 (ACAACCCAGGAGCGAACCT and TCGGCTTGTGCTCACATCTC), Arhgap15 (CTACAGGAGCTGTGCAAATGAGA and TTGGCTCTGCCTGTCTTGGT), Arhgap27 (GAGGCCTGGAAAGCGACTT and GGGTCGTCTCTGTAGGAATTTACG), Arhgap32 (CACCGCCTCCGAAAAATG and TGCAGACTCAGCTAACGCTAGTG), Arhgap33 (TGGCGATGATCTGGATTTCA and AAGTCAAGTCCCCGAAGTCCTT), Srf (GGTTGGAGGGAACCACTGT and CTGGGAGAAGGGGGAAGAC), Cyr61 (AATCGCAATTGGAAAAGGCA and TGAAAAGAACTCGCGGTTCG), Vcl (AGCCCAGATGCTTCAGTCAGA and GGTCAGATGTGCCAGAAAGGA), Gapdh (TCTTGTGCAGTGCCAGCCT and CAATACGGCCAAATCCGTTCA).
Intronic primers were as follows: Cyr61 (CGTAAACTGCCCTGAGCCTA and GACGCGATCGAGACACTTCT), Klf7 (CACTGGCTCCCTATACCGTG and GATCCAAAGCAGGGTTTGCC), Slc2a1 (CCGGATTTACGGAACCCCTC and GCAAAGGCGGGACAAGAAAG), Srf (TCAAGGCAGCAGCAGTTTCT and CAGGCAGGGTTAGGAACCAG), Vcl (CGTCACTTGCGTTGAGTACC and GAAACCACCCACAGGTTGGA), Zyx (CAACCTGGCTCGTTCTCACT and GACCATAACGAGGGGCTCAG).
Time-lapse FRET imaging
Cells were transiently transfected with the RaichuEV-Rac FRET biosensor45, 24h before imaging. Cells were imaged in phenol-red free DMEM using an inverted microscope with Perfect Focus System (Nikon Ti2), controlled by the Micro-Manager software60, with a 60x/1.4 NA Plan Apochromat objective lens (Nikon), an ASI XY stage with piezo Z, a scientific CMOS camera (Photometrics Prime), a SpectraX LED light engine (Lumencor) utilising the blue excitation light fitted with a 440/20 nm filter, an FF459/526/596-Di01 dichroic mirror (Semrock), and two emission filters (FF01-482/25 for CFP, FF01-544/24 for YFP). After background subtraction, FRET/CFP ratio images were generated using Metamorph (Molecular Devices) and represented in the intensity modulated display mode (8 colours). CFP and FRET intensities were averaged over the whole cell area using Fiji software61. For kinetics experiments, data were expressed relative to the start of the experiment, and normalised as indicated. The determination of t50 (time to downregulate to 50% of the maximum activity) for each condition is calculated using the formula: t50 = t(Min + 50% (Max - Min)) - t(Max) where t represents time, Min the minimum value, Max the maximum value (Fig. S4d, S6b).
Invadopodia assay
Invadopodia assays were carried out in 96-well glass bottom plates, coated with Poly-D-Lysine (50 μg/mL), functionalised with 0.5% glutaraldehyde, and coated for 30 min at 37°C with 33 μg/mL Oregon Green-Gelatin (Invitrogen, G13186) and 1% unlabelled gelatin in PBS. Cells (3000 per well) were seeded and incubated for 16h, fixed with 4% PFA, and stained for F-actin and DNA. Images were captured on the Cellomics Arrayscan VTi with a 5x objective and analysed using the automated Morphology Explorer Bioapplication (Cellomics). Quantitation was by loss of fluorescence, normalised to cell number.
Animals and experimental metastasis assay
The experimental metastasis assays were performed as described previously47. B16F0 (900,000 cells) and B16F10 cells (200,000 or 500,000 cells) were injected into the tail vein of 7-week old C57BL/6J females, and lungs were analysed 12 days after injection by counting surface metastatic foci macroscopically. For phenotypic rescue experiments, mice were given water supplemented with 2 mg/mL Doxycycline and 1% sucrose two days prior to injection and for the duration of the experiment, and replaced every two days. ArhGAP12 knockout embryos were obtained from the Jackson Laboratories, and knockout MEFs were generated and genotyped using standard techniques. Animal experimentation complied with all ethical regulations and was carried out under the UK Home Office Project licence P7C307997 in the Crick Biological Resources Facilities.
Multiple sequence alignment and phylogenetic tree
The protein sequences of all mouse rhoGAP domains and fifty amino acids N-terminally were taken from the RefSeq database. A multiple sequence alignment was generated using default parameters in Clustal Omega, and used to produce a phylogenetic tree by the neighbour-joining method. The alignment was edited and Clustal X-coloured in Jalview (blue, hydrophobic; red, positively charged; magenta, negatively charged; green, polar; pink, cysteine; orange, glycine; yellow, proline; cyan, aromatic; white, unconserved). The cladogram was drawn using Dendroscope.
Statistics and reproducibility
Each experiment was performed at least three times. Unless indicated otherwise, nonparametric two-tailed Mann-Whitney tests were used to determine statistical significance, where * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001; ns, not significant. Error bars represent SEM for n independent experiments, as indicated in the legends. Statistical analyses were performed using Prism (GraphPad software).
Supplementary Material
Acknowledgements
We thank the Crick Science Technology platforms for support and advice during this work, especially Matthew Renshaw and Kurt Anderson (Advanced Light Microscopy), Probir Chakravarty and Aengus Stewart (Bioinformatics and Biostatistics), Clare Watkins and Julie Bee (Biological Resources), Namita Patel and Alireza Alidoust (Fermentation Facility), Derek Davis (Flow Cytometry), Graham Clark (Genomics Equipment Park), Mike Howell (Highthroughput screening), Nicola O'Reilly (Peptide Chemistry), and Phil Walker (Structural biology). X-ray data were collected at the Diamond Light Source (ID24 beamline, mx8015). We thank Michiyuki Matsuda (Kyoto University) for the RaichuEV-Rac plasmid, and Michael Way and members of the RT and NM groups for helpful discussions. This work was supported by Cancer Research UK core funding until March 31st 2015. Since then support to RT and NQM has been by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001-190, FC001-115), the UK Medical Research Council (FC001-190, FC001-115) and the Wellcome Trust (FC001-190, FC001-115); and by ERC Advanced Grant 268690 to RT.
Footnotes
Data Availability
The ArhGAP12/G-actin structure has been deposited in the Protein Data Bank (PDB; https://www.rcsb.org) with the primary accession code 6GVC. Structures of MRTF-A RPEL2/G-actin and ArhGAP15 that were re-analysed in this study were obtained from PDB under the accession codes 2V52 and 3BYI, respectively.
Previously published RNA-seq data that were re-analysed here are available under accession code GSE45888.
The human melanoma survival data were derived from the TCGA Research Network: http://cancergenome.nih.gov. The data-set derived from this resource that supports the findings of this study is available in OncoLnc (http://www.oncolnc.org).
Source data for Fig. 1c, Fig. 2a,d, Fig. 3a,f, Fig. 4a,b,c, Fig. 5a,b,c,d, Fig. 6a,b,c,d, Fig. 7a,b,c,e,f,h and Supplementary Fig. S1e,f,i, Fig. S2f, Fig. S3c,d, Fig. S4b,c,d,f,g,h, Fig. S5a,b,c,d,g,h,i, Fig. S6a,b,c,e have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Author Contributions
All authors designed and interpreted experiments. JD conducted biochemical and cell biological studies; SM determined the structure of the actin/ArhGAP12 complex and conducted comparative structural analysis. JD and RT wrote the manuscript with input from SM and NQM.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–590. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- 2.Przybyla L, Muncie JM, Weaver VM. Mechanical Control of Epithelial-to-Mesenchymal Transitions in Development and Cancer. Annu Rev Cell Dev Biol. 2016;32:527–554. doi: 10.1146/annurev-cellbio-111315-125150. [DOI] [PubMed] [Google Scholar]
- 3.Skau CT, Waterman CM. Specification of Architecture and Function of Actin Structures by Actin Nucleation Factors. Annu Rev Biophys. 2015;44:285–310. doi: 10.1146/annurev-biophys-060414-034308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawson CD, Ridley AJ. Rho GTPase signaling complexes in cell migration and invasion. J Cell Biol. 2018;217:447–457. doi: 10.1083/jcb.201612069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodge RG, Ridley AJ. Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol. 2016;17:496–510. doi: 10.1038/nrm.2016.67. [DOI] [PubMed] [Google Scholar]
- 7.Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene. 2014;33:4021–4035. doi: 10.1038/onc.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laurin M, Cote JF. Insights into the biological functions of Dock family guanine nucleotide exchange factors. Genes & development. 2014;28:533–547. doi: 10.1101/gad.236349.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- 10.Amin E, et al. Deciphering the Molecular and Functional Basis of RHOGAP Family Proteins: A systematic approach toward selective inactivation of rho family proteins. J Biol Chem. 2016;291:20353–20371. doi: 10.1074/jbc.M116.736967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 12.Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- 13.Mouilleron S, Guettler S, Langer CA, Treisman R, McDonald NQ. Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL. The EMBO journal. 2008;27:3198–3208. doi: 10.1038/emboj.2008.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiezlak M, et al. G-actin regulates the shuttling and PP1 binding of the RPEL protein Phactr1 to control actomyosin assembly. Journal of cell science. 2012;125:5860–5872. doi: 10.1242/jcs.112078. [DOI] [PubMed] [Google Scholar]
- 15.Huet G, et al. Actin-regulated feedback loop based on Phactr4, PP1 and cofilin maintains the actin monomer pool. Journal of cell science. 2013;126:497–507. doi: 10.1242/jcs.113241. [DOI] [PubMed] [Google Scholar]
- 16.Esnault C, et al. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes & development. 2014;28:943–958. doi: 10.1101/gad.239327.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cen B, et al. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Molecular and cellular biology. 2003;23:6597–6608. doi: 10.1128/MCB.23.18.6597-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen PB, Greenfield AT, Svenningsson P, Haspeslagh DC, Greengard P. Phactrs 1-4: A family of protein phosphatase 1 and actin regulatory proteins. Proc Natl Acad Sci U S A. 2004;101:7187–7192. doi: 10.1073/pnas.0401673101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sagara J, et al. Scapinin, a putative protein phosphatase-1 regulatory subunit associated with the nuclear nonchromatin structure. J Biol Chem. 2003;278:45611–45619. doi: 10.1074/jbc.M305227200. [DOI] [PubMed] [Google Scholar]
- 20.Pawłowski R, Eeva Kaisa Rajakylä EK, Vartiainen MK, Treisman R. An actin-regulated importin α/β-dependent extended bipartite NLS directs nuclear import of MRTF-A. The EMBO journal. 2010;29:3448–3458. doi: 10.1038/emboj.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirano H, Matsuura Y. Sensing actin dynamics: structural basis for G-actin-sensitive nuclear import of MAL. Biochem Biophys Res Commun. 2011;414:373–378. doi: 10.1016/j.bbrc.2011.09.079. [DOI] [PubMed] [Google Scholar]
- 22.Furukawa Y, et al. Isolation of a novel human gene, ARHGAP9, encoding a rho-GTPase activating protein. Biochem Biophys Res Commun. 2001;284:643–649. doi: 10.1006/bbrc.2001.5022. [DOI] [PubMed] [Google Scholar]
- 23.Gentile A, et al. Met-driven invasive growth involves transcriptional regulation of Arhgap12. Oncogene. 2008;27:5590–5598. doi: 10.1038/onc.2008.173. [DOI] [PubMed] [Google Scholar]
- 24.Seoh ML, Ng CH, Yong J, Lim L, Leung T. ArhGAP15, a novel human RacGAP protein with GTPase binding property. FEBS Lett. 2003;539:131–137. doi: 10.1016/s0014-5793(03)00213-8. [DOI] [PubMed] [Google Scholar]
- 25.Zhao C, et al. GC-GAP, a Rho family GTPase-activating protein that interacts with signaling adapters Gab1 and Gab2. J Biol Chem. 2003;278:34641–34653. doi: 10.1074/jbc.M304594200. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda M, et al. Identification of adherens junction-associated GTPase activating proteins by the fluorescence localization-based expression cloning. Exp Cell Res. 2008;314:939–949. doi: 10.1016/j.yexcr.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Monastyrskaya K, et al. miR-199a-5p regulates urothelial permeability and may play a role in bladder pain syndrome. Am J Pathol. 2013;182:431–448. doi: 10.1016/j.ajpath.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 28.Rudnicki A, et al. Next-generation sequencing of small RNAs from inner ear sensory epithelium identifies microRNAs and defines regulatory pathways. BMC Genomics. 2014;15:484. doi: 10.1186/1471-2164-15-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lecat S, Matthes HW, Pepperkok R, Simpson JC, Galzi JL. A Fluorescent Live Imaging Screening Assay Based on Translocation Criteria Identifies Novel Cytoplasmic Proteins Implicated in G Protein-coupled Receptor Signaling Pathways. Mol Cell Proteomics. 2015;14:1385–1399. doi: 10.1074/mcp.M114.046698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlam D, et al. Phosphoinositide 3-kinase enables phagocytosis of large particles by terminating actin assembly through Rac/Cdc42 GTPase-activating proteins. Nat Commun. 2015;6 doi: 10.1038/ncomms9623. 8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ba W, et al. ARHGAP12 Functions as a Developmental Brake on Excitatory Synapse Function. Cell Rep. 2016;14:1355–1368. doi: 10.1016/j.celrep.2016.01.037. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura T, et al. PX-RICS mediates ER-to-Golgi transport of the N-cadherin/beta-catenin complex. Genes Dev. 2008;22:1244–1256. doi: 10.1101/gad.1632308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mouilleron S, Wiezlak M, O'Reilly N, Treisman R, McDonald NQ. Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity. Structure. 2012;20:1960–1970. doi: 10.1016/j.str.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 34.Guettler S, Vartiainen MK, Miralles F, Larijani B, Treisman R. RPEL motifs link the serum response factor cofactor MAL but not myocardin to Rho signaling via actin binding. Molecular and cellular biology. 2008;28:732–742. doi: 10.1128/MCB.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Posern G, Sotiropoulos A, Treisman R. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol Biol Cell. 2002;13:4167–4178. doi: 10.1091/mbc.02-05-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mouilleron S, Langer CA, Guettler S, McDonald NQ, Treisman R. Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator. Sci Signal. 2011;4:ra40. doi: 10.1126/scisignal.2001750. [DOI] [PubMed] [Google Scholar]
- 37.Radu M, et al. ArhGAP15, a Rac-specific GTPase-activating protein, plays a dual role in inhibiting small GTPase signaling. J Biol Chem. 2013;288:21117–21125. doi: 10.1074/jbc.M113.459719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zamboni V, et al. Disruption of ArhGAP15 results in hyperactive Rac1, affects the architecture and function of hippocampal inhibitory neurons and causes cognitive deficits. Sci Rep. 2016;6 doi: 10.1038/srep34877. 34877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graziano BR, et al. A module for Rac temporal signal integration revealed with optogenetics. J Cell Biol. 2017;216:2515–2531. doi: 10.1083/jcb.201604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurisu S, Suetsugu S, Yamazaki D, Yamaguchi H, Takenawa T. Rac-WAVE2 signaling is involved in the invasive and metastatic phenotypes of murine melanoma cells. Oncogene. 2005;24:1309–1319. doi: 10.1038/sj.onc.1208177. [DOI] [PubMed] [Google Scholar]
- 41.Nakahara H, et al. Involvement of Cdc42 and Rac small G proteins in invadopodia formation of RPMI7951 cells. Genes Cells. 2003;8:1019–1027. doi: 10.1111/j.1365-2443.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- 42.Yamaguchi H, et al. Sphingosine-1-phosphate receptor subtype-specific positive and negative regulation of Rac and haematogenous metastasis of melanoma cells. Biochem J. 2003;374:715–722. doi: 10.1042/BJ20030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stengel K, Zheng Y. Cdc42 in oncogenic transformation, invasion, and tumorigenesis. Cell Signal. 2011;23:1415–1423. doi: 10.1016/j.cellsig.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Revach OY, Winograd-Katz SE, Samuels Y, Geiger B. The involvement of mutant Rac1 in the formation of invadopodia in cultured melanoma cells. Exp Cell Res. 2016;343:82–88. doi: 10.1016/j.yexcr.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komatsu N, et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eddy RJ, Weidmann MD, Sharma VP, Condeelis JS. Tumor Cell Invadopodia: Invasive Protrusions that Orchestrate Metastasis. Trends Cell Biol. 2017;27:595–607. doi: 10.1016/j.tcb.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol. 2009;11:257–268. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaiswal M, et al. Functional cross-talk between ras and rho pathways: a Ras-specific GTPase-activating protein (p120RasGAP) competitively inhibits the RhoGAP activity of deleted in liver cancer (DLC) tumor suppressor by masking the catalytic arginine finger. J Biol Chem. 2014;289:6839–6849. doi: 10.1074/jbc.M113.527655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 50.Murakami K, et al. Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell. 2010;143:275–287. doi: 10.1016/j.cell.2010.09.034. [DOI] [PubMed] [Google Scholar]
- 51.Aktories K, Lang AE, Schwan C, Mannherz HG. Actin as target for modification by bacterial protein toxins. FEBS J. 2011;278:4526–4543. doi: 10.1111/j.1742-4658.2011.08113.x. [DOI] [PubMed] [Google Scholar]
- 52.Ceccarelli DF, et al. Non-canonical interaction of phosphoinositides with pleckstrin homology domains of Tiam1 and ArhGAP9. J Biol Chem. 2007;282:13864–13874. doi: 10.1074/jbc.M700505200. [DOI] [PubMed] [Google Scholar]
- 53.Kong L, Ge BX. MyD88-independent activation of a novel actin-Cdc42/Rac pathway is required for Toll-like receptor-stimulated phagocytosis. Cell Res. 2008;18:745–755. doi: 10.1038/cr.2008.65. [DOI] [PubMed] [Google Scholar]
- 54.Winter G, Lobley CM, Prince SM. Decision making in xia2. Acta Crystallogr D Biol Crystallogr. 2013;69:1260–1273. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D Biol Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 59.Schrodinger L. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- 60.Edelstein AD, et al. Advanced methods of microscope control using muManager software. J Biol Methods. 2014;1 doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.