Abstract
Cell wall biosynthesis plays essential roles in cell division and expansion and thus is fundamental to plant growth and development. In this work, we show that an Arabidopsis mutant dpr3, isolated by a forward genetic screen, displays embryo defects and short, swelling primary root with the failure of maintenance of root apical meristem reminiscent to several cell wall–deficient mutants. Map-based cloning identified dpr3 is a mutant allele of RIBOSE PHOSPHATE ISOMERSASE 1 (RPI1), an enzyme involved in cellulose synthesis. Cellulose content in the mutant was dramatically decreased. Moreover, dpr3 (rpi1 from hereon) caused aberrant auxin distribution, as well as defective accumulation of root master regulators PLETHORA (PLT1 and PLT2) and misexpression of auxin response factor 5 (MONOPTEROS, MP). The abnormal auxin distribution is likely due to the reduced accumulation of auxin efflux transporters PIN-FORMED (PIN1 and PIN3). Surprisingly, we found that the orientation of actin microfilaments was severely altered in rpi1 root cells, whereas the cortical microtubules stay normal. Our study provides evidence that the defects in cellulose synthesis in rpi1 affect polar auxin transport possibly connected with altered F-actin organization, which is critically important for vesicle trafficking, thus exerting effects on auxin distribution, signaling, and auxin-mediated plant development.
Keywords: auxin, cell wall, embryo development, root development, RIBOSE PHOSPHATE ISOMERSASE 1 (RPI1)
Introduction
Root growth depends on quiescent center (QC) and the surrounding stem cells which form the root stem cell niche (SCN) (Heidstra and Sabatini, 2014). All types of root cells originate from the SCN; thus, the maintenance of root SCN is critical for root growth (Scheres, 2007). There are two parallel pathways that had been identified for root SCN maintenance. The first one is involved in AP2 transcription factors PLETHORA (PLT), and the other one is associated with GRAS transcription factors SHORT-ROOT (SHR) and SCARECROW (SCR) (Nakajima et al., 2001; Sabatini et al., 2003; Aida et al., 2004; Galinha et al., 2007). The phytohormone auxin plays an important role in maintaining root SCN. Auxin response maximum in the distal stem cell region is required for QC function (Blilou et al., 2005). The transcription of PLT genes is dependent on auxin, and the auxin–PLT pathway acts as a core module in root stem cell maintenance and cell division for developing root (Galinha et al., 2007; Mahonen et al., 2014). plt1plt2 double mutant showed strongly defective root SCN organization, giving rise to short root meristem phenotypes (Aida et al., 2004; Galinha et al., 2007).
Auxin distribution pattern acts as the developmental clue for growing plant, and the certain auxin pattern is mainly determined by polar auxin transport (PAT), which is mediated by polar located PIN-FORMED (PIN) efflux proteins on the plasma membrane (PM) to a great extent (Adamowski and Friml, 2015). Various regulators had been identified for the abundance and polarities maintenance of PINs, including the ARF-GTPase activator ARF-GEF (Kleine-Vehn et al., 2008a), AGCIII-type protein kinase PINOID (Friml et al., 2004), phosphatase 2A (Michniewicz et al., 2007), and D6 protein kinase and its family members (Zourelidou et al., 2014). Moreover, PINs undergo trafficking to the lytic vacuole for degradation, which is an important mechanism for maintaining the abundance of PIN proteins (Kleine-Vehn et al., 2008b; Nodzynski et al., 2013).
Other players are also involved in modulating PIN proteins. Firstly, studies have demonstrated the close correlation between cytoskeleton and PAT. Pharmacological investigations showed that treatments with microtubule (MT)-targeted drug oryzalin to depolymerize MTs reduced the basal distribution of PIN1 and PIN2 in root cells (Boutte et al., 2006; Kleine-Vehn et al., 2008b). The CLIP-ASSOCIATED PROTEIN (CLASP) mediates an association between PINs cycling and MTs by interacting with the retromer component sorting nexin 1. clasp mutants display a range of auxin-related phenotypes, including a reduction in root apical meristem size and increased lateral root abundance (Ambrose et al., 2007; Kirik et al., 2007; Ambrose et al., 2013). Several investigations confirmed actin cytoskeleton also links to the PAT. In Arabidopsis, actin-targeted drug latrunculin B inhibited intracellular PIN1 accumulation in brefeldin A (BFA) compartments and recycling of PIN1 to the PM after washout of BFA (Geldner et al., 2001). Latrunculin B treatment also caused PIN3 internalization in smaller compartments without any regular positioning in columella cells (Friml et al., 2002) and increased the [3H]indole-3-acetic acid (IAA) accumulation in Fucus distichus embryos (Sun et al., 2004). A study showed enhanced accumulation of the cortical fine actin of leaf epidermal cells inhibits clathrin-dependent PIN1 endocytosis, leading to enhanced PIN1 accumulation on the PM (Nagawa et al., 2012). In rice, defective F-actin arrays in rmd (ROOT MORPHOLOGY DETERMINANT) mutants disrupt expression of OsPIN1b and OsPIN2, auxin distribution, and auxin-mediated cell growth during root development (Li et al., 2014). Secondly, evidence was accumulating that the cell wall may function in the PAT. Treatment with cellulose synthesis inhibitor isoxaben led to hyperlocalization of PIN1 on the PM in shoot apical meristem (SAM) cells (Heisler et al., 2010). Genetic and pharmacological interference with cellulose synthesis led to enhanced lateral diffusion and reduced polarity of PIN2, indicating there is a connection between the cell wall and PIN polarity on the PM (Feraru et al., 2011). This connection was further confirmed by the finding that plant cell wall limits lateral diffusion of PM proteins (Martiniere et al., 2012). Hence, the tight link between the cell wall and PAT and, ultimately, plant SAM and root development has been proposed. Nevertheless, the molecular mechanism underlying, for now, remains largely elusive.
Ribose 5-phosphate isomerase (RPI) is a small group of enzymes which function in the oxidative pentose phosphate pathway (oxPPP), catalyzing the interconversion between ribose 5-phosphate (R5P) and ribulose 5-phosphate (Ru5P). There are four members in RPI gene family in Arabidopsis thaliana, RPI1–RPI4, with RPI1 and RPI2 showing cytosolic localization while the other two are located in plastids (Xiong et al., 2009). Genetic studies in Arabidopsis have revealed the role of RPI1 and RPI2 during plant development. rsw10, carrying a point mutation in RPI1 gene, showed a lower level of cellulose and swelling root phenotypes under 31°C temperature condition (Howles et al., 2006). RPI2 knockout plants appeared with defective chloroplast structure and reduced photosynthetic capacity. When grown at a relative high temperature, the mutants presented premature cell death in the leaves (Xiong et al., 2009).
In this report, we isolated a dpr3 mutant which is a novel allele of RPI1 gene. dpr3 displays short, swelling roots and abnormal cell divisions in the basal region of embryos in Arabidopsis, indicating RPI1 is required for proper embryo and root development. Further studies revealed the mutation in RPI1 gives rise to altered auxin distribution and defective auxin-dependent PLT1 and PLT2 accumulation as well as MP expression. By analyzing auxin transport markers, we found the aberrant auxin distribution in the mutant and the reduced abundance of auxin efflux proteins PIN1 and PIN3 on the PM. Moreover, the rpi1 presented a more transverse F-actin array rather than longitudinal aligned in root cells. Our results suggest a role of RPI1 for actin cytoskeleton may link cell wall and PAT in the coordination of auxin-dependent root cell growth and patterning.
Methods
Plant Materials and Growth Conditions
A. thaliana Columbia-0 (Col-0) accession was used in this research. The DR5rev:GFP and PIN1pro:PIN1-GFP (Benkova et al., 2003), PIN2pro:PIN2-GFP (Xu and Scheres, 2005), PIN3pro:PIN3-GFP (Dello Ioio et al., 2008), PLT1pro:PLT1-YFP, PLT2pro:PLT2-YFP (Grieneisen et al., 2007), WOX5pro:GFP (Sarkar et al., 2007), MPpro:n3xGFP (Rademacher et al., 2012), SHRpro: SHR-GFP (Nakajima et al., 2001), SCRpro : GFP (Wysocka-Diller et al., 2000), 35Spro : Actin-binding Domain 2 (ABD2)–GFP (Wang et al., 2008), and 35Spro : GFP-tubulin (Bannigan et al., 2006) marker lines have been described before. Surface-sterilized seeds were sowed on 1/2 Murashige and Skoog (MS) medium (1% sucrose, 0.8% agar) and then followed by cold-treated at 4°C for 3 days in darkness and transferring to a phytotron set (light:dark = 16:8 h, 70% humidity, 22 ℃) The marker lines were individually crossed into rpi1 mutant. Homozygous hybrid lines were obtained in F2 populations and analyzed in F3 or F4 generations.
Root Phenotypic Analysis
Seedlings were grown on 1/2 MS standard medium for 2–10 days. The primary root length and meristem cell length were measured using Image J software. The meristem length, which is determined by the number of cortical cells from the stem cell to the first elongated cell, was investigated after soaking in HCG solution (Dello Ioio et al., 2007). Data presented were means with SD of 30 to 40 seedlings. For genetic complementation tests, the primary root and meristem length were analyzed after 10 days. For chemical complementation, seedlings of Col-0 and dpr3 were grown on 1/2 MS standard medium with or without 2.5 mM uridine supplemented for 9 days, and the primary root and meristem length were measured. For 2,6-dichlorobenzonitrile (DCB; Sigma-Aldrich) treatment, seedlings of wild type bearing PIN1pro:PIN1-GFP, PIN3pro:PIN3-GFP, and 35Spro : ABD2-GFP markers, respectively, were grown on 1/2 MS standard medium with or without 0.1 μM of DCB (20 mM DCB stock solution, dissolved in DMSO).
Map-Based Cloning of DPR3
The EMS-mutagenized Arabidopsis plant dpr3 (Col-0) was crossed with Landsberg erecta (Ler) accession. And 40 dpr3 × Ler F2 plants of the dpr3 (short root) phenotype were used for rough mapping. Positional mapping showed that the site of dpr3 mutation was flanked by two simple sequence length polymorphism (SSLP) markers (within BACs F10D13 and F10A5 respectively) on the lower arm of chromosome 1. Additional 98 short root F2 plants were analyzed to be narrowed down to a 670 kbp region (26.60 to 27.36 Mbp). Candidate genes in this region were PCR-amplified and sequenced, and a C-to-T transition resulting in the predicted Ala113Val amino acid change in the locus At1g71100 (RPI1) was found to be the casual single nucleotide polymorphism (SNP).
Vector Construction and Plant Transformation
To construct rpi1/RPI1pro:RPI1-GFP, the RPI1 promoter and coding sequence (CDS) were amplified by PCR from Col-0 genomic DNA or cDNA templates respectively and then inserted into the binary vector pCAMBIA1300, together with in-frame fused GFP. Agrobacterium tumefaciens (C58) with the resulting plasmid was introduced into rpi1 mutant background using the floral dipping method (Clough and Bent, 1998). The relevant primer sequences were listed in Table S1 . PCR primers were designed with the Primer Premier 5.0 software.
Expression Pattern and Subcellular Localization Analysis of RPI1
The embryos and 4-day-old seedlings of transgenic rpi1/RPI1pro:RPI1-GFP plants were used for gene expression pattern analysis and subcellular localization analysis by confocal imaging.
qRT-PCR Analysis
Whole roots of 6-day-old seedlings were used for total RNA preparation by using the plant RNA extraction kit (Huayueyang, Beijing, China). cDNA was synthesized by PrimeScript reverse transcriptase (TakaRa, Kusatsu, Shiga, Japan) from 3 μg of total RNA. PCR reaction was carried out using Illumina Eco system (Illumina, San Diego, California, USA) with a SYBR green probe (Vazyme, Nanjing, China). Expression of PIN1 and PIN3 in rpi1 and wild type plants was normalized to the internal control ACTIN2, respectively. Three biological replicates were performed, and data presented are means with SD. Related primers used are listed in Table S1 .
Cellulose Measurement
Cellulose content measurement was performed according the method described by Bailey (1958) with minor modification. Briefly, 200 mg of 8-day-old dried seedlings was boiled in acetic acid and nitric acid for 25 min. The sample was then filtered, and 5 ml of this filtrate was mixed with 95 ml ddH2O for 1:20 dilution. Two milliliters of this dilution was used to perform colorimetric determination by adding 0.5 ml anthrone solution (2 g of anthrone in 100 ml of ethyl acetate) and 5 ml sulfuric acid was added to mix well and sit at room temperature for 12 min. The mixture was then subjected to a spectrophotometer (MAPADA® V1300, Shanghai, China) assay at the wavelength of 625 nm. The content of cellulose was calculated according to the glucose stand curve.
Microscopy
Ovules were cleared in Hoyer’s solution and roots in HCG solution as previously described (Chen et al., 2011). Differential interference contrast (DIC) pictures were taken on a Olympus BX51 microscope (Tokyo, Japan) connected with a Qimaging Ritiga 2000R digital camera (Surrey, BC, Canada). FM4-64 (5 μM) or propidium iodide (100 μg/ml) was used to visualize cell contour. Samples were imaged under ZEISS confocal microscopy (LSM 780) with excitation/emission (Ex/Em) wavelengths: GFP (488/505~530 nm), YFP (514/530~560 nm), FM4-64 (543/600~ nm), propidium iodide (561/591~635 nm), and kFluor615 (560/645~ nm). Twenty to 30 samples were examined for each group, and 15 to 20 samples were used for confocal imaging afterward. Similar results were obtained in three independent experiments.
Accession Numbers
ACT2 (At3g18780), PIN1 (At1g73590), PIN2 (At5g57090), PIN3 (At1g70940), PLT1 (At3g20840), PLT2 (At1g51190), RPI1 (AT1G71100), SCR (At3g54220), SHR (At4g37650), WOX5 (At3g11260).
Results
DPR3 Is Required for Maintenance of Root Meristem Development
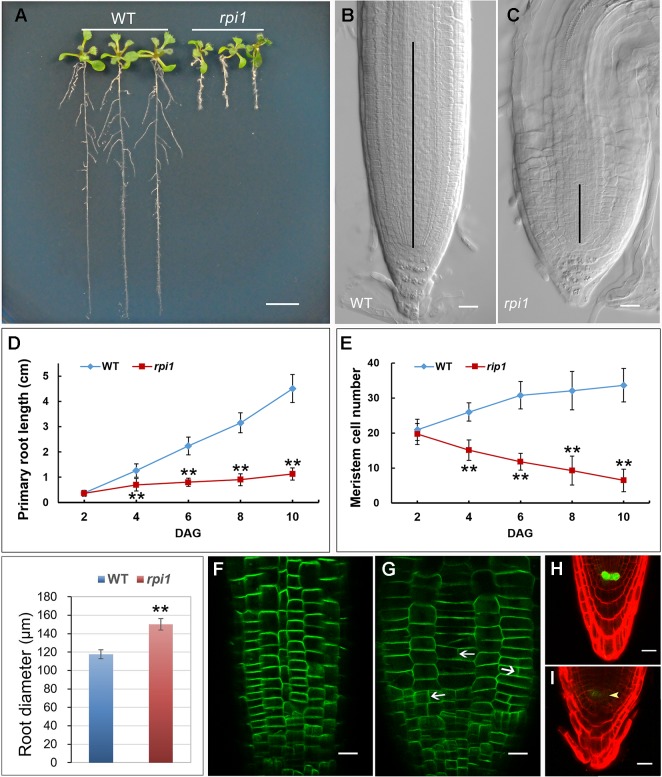
One mutant showing significantly short primary root was isolated from an ethyl methane sulfonate (EMS)–mutagenized population of Arabidopsis. The mutant, hereafter referred to as defective primary root 3 (dpr3), showed stunted root growth ( Figure 1A ) and reduced meristem length ( Figures 1B, C ). Primary root growth was markedly reduced in dpr3 from as early as 4 days after germination (DAG), compared with wild type ( Figure 1D ). And the number of meristematic cells significantly reduced over time in the root of dpr3 ( Figure 1E ). Besides, dpr3 mutant had a large diameter of roots ( Figure 1F ) and show swelling phenotype grown under normal condition ( Figure 1C ). Moreover, some meristematic epidermal cells showed vertical cell division plane (12%, n = 15) in dpr3 roots, in comparison to the horizontal division plane in wild type ( Figures 1G, H ). These results indicate that DPR3 plays an important role in root meristem pattern formation in Arabidopsis.
Figure 1.
Phenotypes of dpr3 mutant roots. (A) Phenotype of the wild type (WT, left) and dpr3 (right) seedlings at 10 days after germination (DAG). (B, C) Root tips of the wild type (b) and dpr3 (c) at 10 DAG. The black vertical lines indicate the root meristem region. (D) Primary root length of wild type and dpr3 seedlings from 2 to 10 DAG. Data shown are average and SD (n = 30). Asterisks indicate Student’s t-test significant difference (**P < 0.01). (E) Root meristem cell number of the wild type and dpr3 on 2 to 10 DAG. The root meristem cell number is designated as the number of cortex cells in the cortex file extending from the quiescent center (QC) to the transition zone. Data shown are average and SD (n = 30). Asterisks indicate Student’s t-test significant difference (**P < 0.01). (F) Diameters of primary root meristem. The diameters were measured where the transition zone appeared. Data shown are average and SD (n = 30). Asterisks indicate Student’s t-test significant difference (**P < 0.01). (G, H) The expression of PIN2pro:PIN2-GFP in wild type (g) and dpr3 (h) root tips at 6 DAG. White arrowheads indicate the abnormal cell plates. (I, J) The expression of WOX5pro:GFP in roots of 4-day-old wild type (I) and dpr3 mutant seedlings (J). Bars = 5 mm in (A); 20 μm in (B,C,G–J).
WUSCHEL-RELATED HOMEOBOX 5 (WOX5) is mainly required for root columella stem cell activity (Sarkar et al., 2007). We investigated whether WOX5 is affected in dpr3 through examining the expression of the WOX5pro:GFP (Blilou et al., 2005; Sarkar et al., 2007). The intensity of WOX5pro:GFP was significantly reduced in the QC of dpr3 roots, compared with wild type roots ( Figures 1I, J ). We also examined the WOX5 expression in the embryos and similar results were observed ( Figure S1 ).
DPR3 Shows Aberrant Cell Divisions in Embryo Development
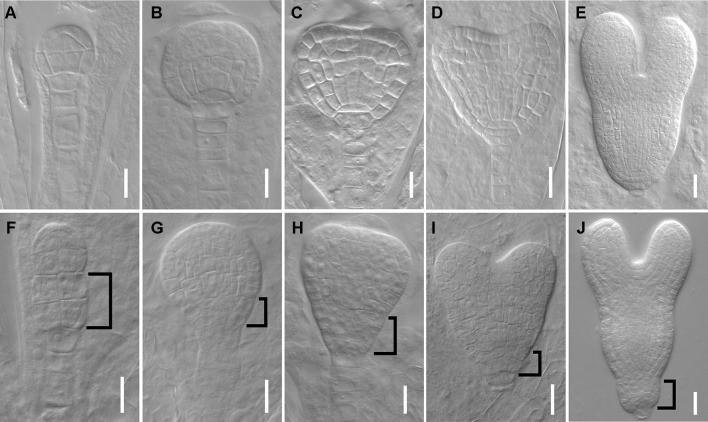
We next tested whether the mutant has defects in embryos since the root SCN is established at embryo stage. At 16-cell stage, the hypophysis cell was specified, generating a normal basal pole in wild type embryo ( Figure 2A ), whereas the dpr3 embryo displayed unclear boundary of the apical and basal pole ( Figure 2F ). From the globular stage onward, dpr3 embryos showed frequent cell division defects in embryonic root pole ( Figures 2G–J compared with Figures 2B–E ; also see Table S2 ). Besides, excessively dividing suspensor cells were observed in some individuals ( Figure 2G ), in contrast to wild type ( Figure 2B ). Taken together, these data demonstrate that DPR3 is important for promoting the normal pattern formation during embryonic root development.
Figure 2.
dpr3 mutants have defective cell divisions in the root pole of embryos. (A–E) Wild type embryos at 16-cell (A), globular (B), triangle (C), heart (D), and later heart (E) stages. (F–J) Embryos of dpr3 mutants at 16-cell (F), globular (G), triangle (H), heart (I), and later heart (J) stages. Bracketed area displays cell division defects in basal embryo region. Bars = 20 μm in (D, E, I, J) and 10 μm for the rest of the images.
Map-Based Cloning of DPR3 Gene
Using positional cloning, we first mapped the dpr3 locus on the short arm of chromosome 1, flanking by markers F10D13 and F10A5 in a physical region of ~3 Mb. Fine mapping was performed and localized the mutation in a region between 26.69 and 27.36 Mb. Sequencing analysis showed a point mutation occurring in a gene coding for RIBOSE-5-PHOSPHATE ISOMERASE 1 (RPI1) in Arabidopsis ( Figure S2A ). This point mutation involves substitution of C to T at 338 bp, leading to amino acid transition of Ala113Val ( Figure S2B ), which is different from the previously reported rsw10 (Glu115Lys) mutant of RPI1, which showed the temperature-dependent swelling root and a significant reduction in cellulose content (Howles et al., 2006).
To verify whether the dpr3 phenotypes are caused by RPI1 deficiency, a vector carrying RPI1pro:RPI1-GFP was introduced into dpr3 mutant and the phenotypes of transgenic plants were analyzed. The recovered root and meristem length of these dpr3/RPI1pro:RPI1-GFP plants indicate that introduction of RPI1 into the mutant background can rescue the mutant phenotypes ( Figures S3A , D – G , L , M ), including their embryo defects ( Figure S4 and Table S2 ). RPI1 catalyzes ribose interconversion, which is required for the synthesis of ribonucleotides. Furthermore, the mutant phenotype was complemented by exogenous application of uridine, which was one component of cellulose biosynthesis substrate, uridine 5’-diphosphate–glucose (UDP–glucose), as suggested in the study of rsw10 (Howles et al., 2006; McFarlane et al., 2014). The dpr3 mutant showed significantly shorter root and meristem lengths, compared with wild type ( Figures S3B , H , I , N , O ), and these defects were rescued while growing on the medium supplemented with 2.5 mM uridine ( Figures S3C , J , K , N , O ). Together, these data show that the developmental aberrations of dpr3 can be well restored via genetic and chemical complementation, revealing the role of RPI1 in maintaining normal root development.
To further understand the RPI1 function, we investigated its expression pattern and subcellular localization using rpi1/RPI1pro:RPI1-GFP transgenic plants. We found that RPI1 was expressed in embryos and multiple organs of seedling, root, and also other organs like hypocotyl and leaf ( Figure S5 ). The cytosolic subcellular localization of RPI1 was clearly observed by getting closer look to the GFP signal in root meristem cells ( Figure S5I ).
Mutation in RPI1 Affects the Expression of Auxin Reporter Gene
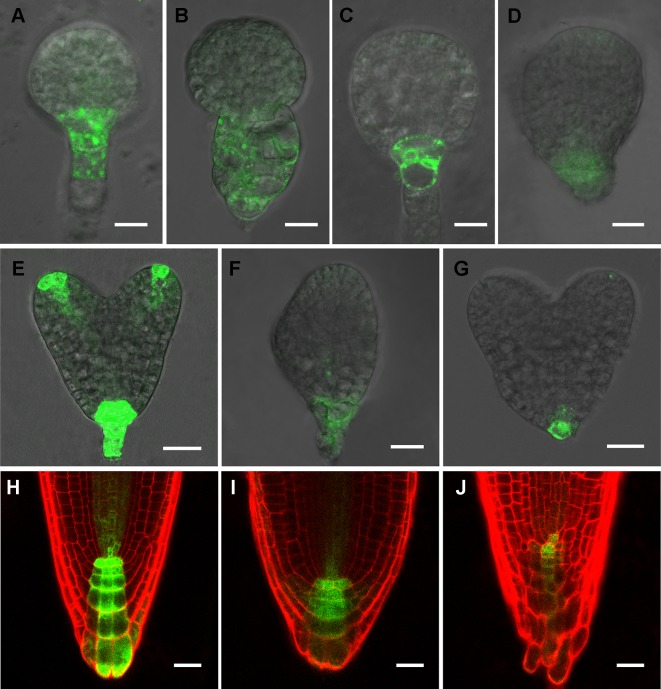
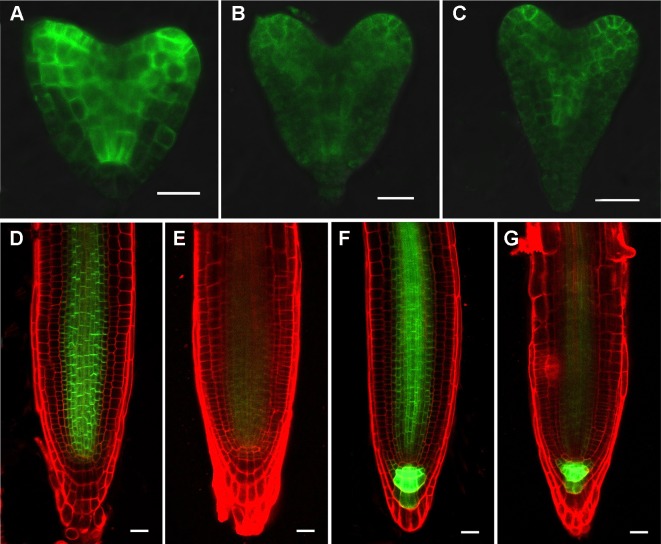
Phenotypic analyses suggest that auxin-regulated processes could be affected in rpi1 mutants ( Figures 1 and 2 ). As indicated by the auxin reporter DR5rev:GFP, an auxin maximum was observed at the embryonic root pole of wild type (100%, n = 30; Figures 3A, C, E ). In contrast, altered auxin maximum was detected in a majority of (77%, n = 31) rpi1 embryos ( Figures 3B, D, F, G ). We then checked DR5 activity in the roots of rpi1 seedlings. Comparing with the wild type ( Figure 3H ), the expression of DR5rev:GFP, auxin reporter was suppressed in rpi1 mutant ( Figures 3I, J ). These data suggested that auxin maximum was perturbed in rpi1 embryos and roots, which correlated with the observed abnormal cell divisions in the embryonic root pole and retarded root meristem development.
Figure 3.
RIBOSE PHOSPHATE ISOMERSASE 1 (RPI1) is required for auxin maximum maintenance. (A–G) DR5rev:GFP expression in embryos at early globular, later globular, and heart stages of the wild type (A, C, E) and rpi1 mutant (B, D, F, G). (H–J) DR5rev:GFP expression in roots of 4-day-old wild type (H) and rpi1 (I, J) seedlings. Bars = 10 μm in (A–D) and 20 μm for the rest of the images.
rpi1 Exhibited Altered Expression Pattern of PLT1/PLT2 and MP
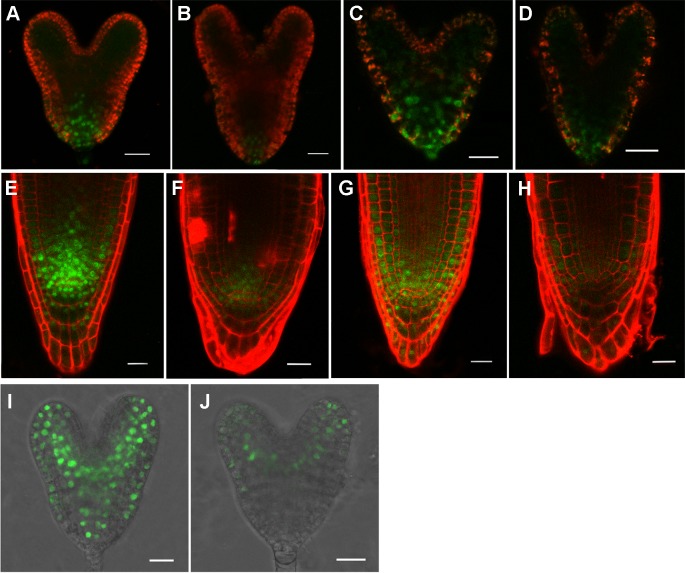
The above data suggested that auxin was involved in causing the defects in rpi1. We then asked whether the expression of auxin-induced regulators PLT1/PLT2, which were well known for controlling cell fate specification, were affected in rpi1. We found both PLT1 and PLT2 were down-regulated in rpi1 embryos and roots ( Figures 4B, D, F, H compared with Figures 4A, C, E, G ). MONOPTEROS (MP/ARF5) is a key transcription factor in auxin signaling in the embryo (Weijers et al., 2006). We next aimed to ascertain whether MP expression was affected. Strong MPpro:n3XGFP signal was detected in the pro-vascular tissue of heart stage embryos ( Figure 4I ), the expression of MP was largely altered in the rpi1 mutant embryos ( Figure 4J ). The SHR-SCR pathway acts parallel with PLTs for the specification and maintenance of the root SCN and root growth (Helariutta et al., 2000; Sabatini et al., 2003; Aida et al., 2004). In contrast to PLT1/PLT2, the localization patterns of SHR and SCR were not altered in rpi1 mutants ( Figure S6 ). These results suggest that the defects observed in rpi1 embryos and roots might result from aberrant expression pattern of PLTs and MP.
Figure 4.
rpI1 mutation affects the expression of PLETHORA (PLT1/PLT2) and MONOPTEROS (MP) in embryos and roots. (A, B) The expression of PLT1pro:PLT1-YFP in embryos of the wild type (A) and rpi1 mutant (B). (C, D) The expression of PLT2pro:PLT2-YFP in embryos of the wild type (c) and rpi1 mutant (D). (E, F) The expression of PLT1pro:PLT1-YFP in 4-day-old roots of the wild type (E) and rpi1 mutant (F). (G, H) The expression of PLT2pro:PLT2-YFP in 4-day-old roots of the wild type (G) and rpi1 mutant (H). (I, J) MPpro:3XGFP is expressed in the embryos of wild type (I) and rpi1 (J) at heart stages. Bars = 20 μm.
Mutation of RPI1 Affects the Accumulation of PIN1 and PIN3
We next set out to determine whether the reduced DR5 activity in embryos and root tips in rpi1 mutants is associated with an alteration in auxin transport. We first examined the level of PIN1 in the mutant. At heart stage of embryo development, strong and basal localization of PIN1-GFP was observed in wild type ( Figure 5A ), whereas there were 80% (n = 30) of rpi1 embryos, which displayed reduced PIN1-GFP accumulation in the corresponding regions ( Figures 5B, C ). In wild type seedling root tips, PIN1-GFP was expressed in the stele and endodermis ( Figure 5D ). In contrast, PIN1-GFP was strongly suppressed in the rpi1 roots ( Figure 5E ). Then we analyzed the expression of other auxin transporters and found that PIN3-GFP signal was faint in rpi1 root stele cells ( Figure 5G ) compared to its amount in wild type roots ( Figure 5F ). These observations suggest that perturbed auxin distribution in rpi1 embryos ( Figures 3B, D, F, G ) and roots ( Figures 3I, J ) could result from the altered accumulation of PIN1 and PIN3. However, we confirmed that mutation in RPI1 did not result in changes in PIN1 and PIN3 at the transcriptional level in roots ( Figure S9 ).
Figure 5.
RPI1 mutation reduces the expression of PIN1 and PIN3. (A–C) PIN1pro:PIN1-GFP is expressed in embryos of the wild type (B) and rpi1 mutant (B, C) at heart stages. (D, E) PIN1pro:PIN1-GFP expression pattern in 4-day-old wild type (D) and rpi1 (E) root tips. (F, G) PIN3pro:PIN3-GFP expression pattern in 4-day-old wild type (F) and rpi1 (G) root tips. Bars = 20 μm.
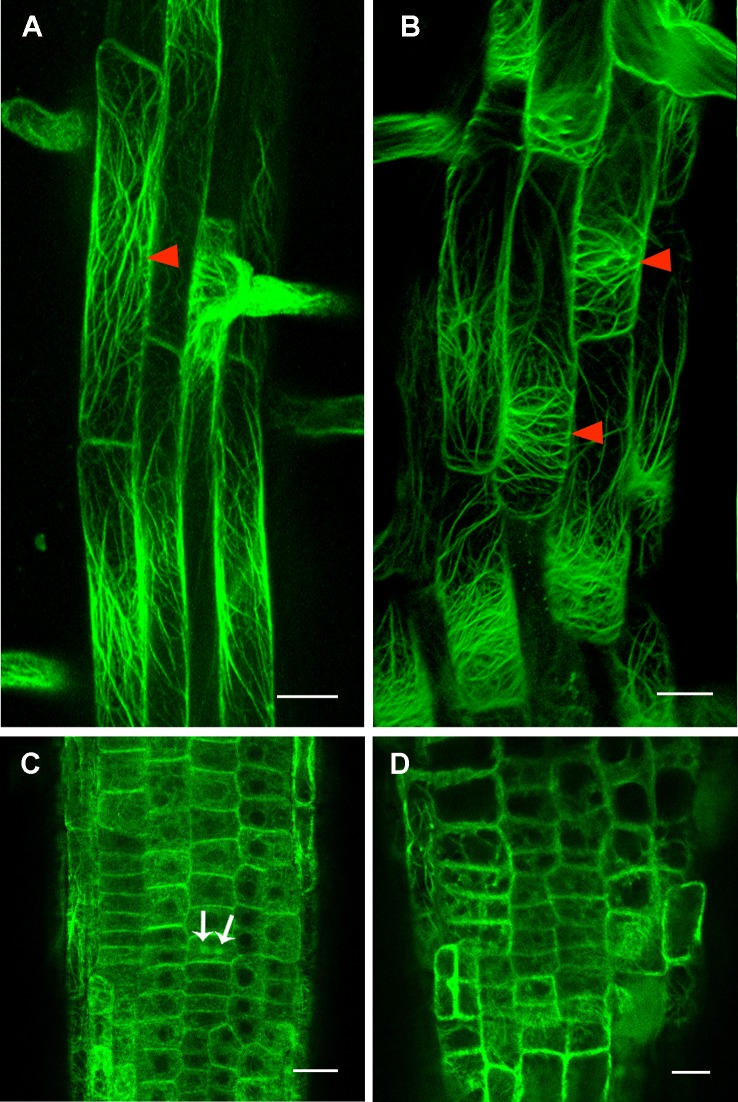
rpi1 Showed Abnormal Cellulose Synthesis and Altered Cortical Actin Filament Orientation
Our investigation demonstrated a severely decreased cellulose content in rpi1 mutant ( Figure S7 ), implying the cell wall architecture was largely affected. Previous studies have shown the cellulose deficiency could lead to the altered polarity of PIN proteins in root cells (Feraru et al., 2011) and disturbed cytoskeleton organization (Panteris et al., 2013; Paredez et al., 2008; Peng et al., 2013). To investigate the role of the cytoskeleton, we first explored whether the mutation in RPI1 had an effect on the MT cytoskeleton using a maker line expressing 35Spro : GFP-tubulin, and we found that the MT arrays were not affected in root mature zone and meristem region of the mutant ( Figure S8 ). Then we tested the actin cytoskeleton. As revealed by an actin marker, 35Spro : ABD2-GFP, actin filaments were mainly longitudinal oriented in wild type root cells in the elongation and differentiation zone ( Figure 6A ). By contrast, they showed transversely oriented pattern in the cells of the same zone in rpi1 root ( Figure 6B ). In the meristematic region of the root, actin filaments orient randomly both in wild type and rpi1 ( Figures 6C, D ), reflecting an isotropic manner of cell enlargement. And in wild type roots, we could observe ABD2-GFP–labeled phragmoplasts, indicating active cell division events ( Figure 6C ). These results demonstrate that RPI1 is required for the normal F-actin organization in root but not for the regulation of MTs.
Figure 6.
Cortical actin filament orientation is affected in rpi1 mutant. (A) Cortical actin filaments are mainly longitudinal along the direction of elongation in 4-day-old control seedling root cells (arrowheads). Each figure is a maximum projection of five slices z-stack. (B) Cortical actin filaments are transversely oriented in root cells of 4-day-old rpi1 seedling (arrowheads). Each figure is a maximum projection of five slices z-stack. (C, D) Cortical actin filaments are randomly oriented in the meristem zone in (C) wild type and (D) rpi1. Arrows indicate phragmoplast-enriched actin filaments. Bars = 50 μm in (A, B) and 20 μm in (C, D).
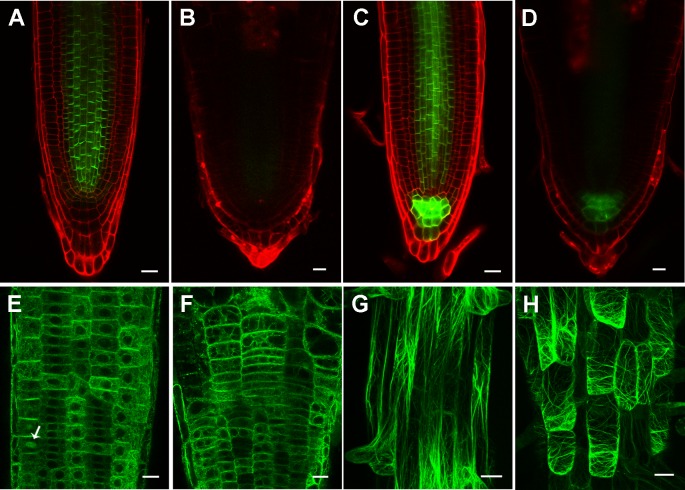
To understand how the altered expression of PIN1 and PIN3 and altered actin filament orientation were caused by defective cellulose biosynthesis in rpi1, we first tested whether they could be rescued by uridine supplemented into the media. Indeed, we found both PIN expression ( Figure S10 ) and actin orientation ( Figure S11 ) were clearly restored by uridine treatment. We further treated wild type seedlings bearing these markers with 0.1 μM of cellulose synthesis inhibitor DCB and observed the expected swelling root tips, significantly reduced PIN levels, and transversely oriented actin filaments ( Figure 7 ), which was the same case as in rpi1. Treatment with DCB also inhibits primary root growth of these wild type marker lines ( Figure S12 ). These results demonstrate the key role of cellulose biosynthesis in maintaining normal expression levels of PIN1 and PIN3, and normal acting actin orientation in the root tips.
Figure 7.
Inhibition of cellulose synthesis by 2,6-dichlorobenzonitrile (DCB) leads to significantly reduced PIN levels, and transversely oriented actin filaments. (A, B) PIN1pro:PIN1-GFP expression pattern in 4-day-old wild type root tips treated without (A) and with (B) 0.1 μM DCB. (c, d) PIN3pro:PIN3-GFP expression pattern in 4-day-old wild type root tips treated without (C) and with (D) by 0.1 μM DCB. (E–H) Actin filament organization indicated by 35Spro : ABD2-GFP in 4-day-old wild type root tips (E, F) in the meristem zone and (G, H) mature zone treated without (E, G) and with (F, H) 0.1 μM DCB. Arrow indicates phragmoplast-enriched actin filaments. Each figure in (A, B) is a maximum projection of five slices z-stack. Bars = 20 μm in (a-f) and 50 μm in (g, h).
Discussion
Mutation in RPI1 Leads to Defective Cellulose Synthesis and Influences Auxin-Dependent Root Development
Several studies showed that the plant cell wall played a key role in auxin-related root development (Zhang et al., 2011; Yang et al., 2014). However, the molecular mechanism underlying remained far from clear. In this work, we further revealed a molecular link between the cell wall and primary root development in Arabidopsis. The single-recessive mutant rpi1 was isolated from our forward genetic screening. The mutant showed significant short and swelling of the primary root and meristem ( Figure 1 ). Abnormal cell divisions were observed in the basal region of the embryo from eight-cell to later heart stages ( Figure 2 ). These embryonic phenotypes in rpi1 are similar to those PAT mutants pin1 and pin7 (Friml et al., 2003), auguring that auxin-related pathway may function in root development affected by RPI1 mutation. Indeed, auxin reporter DR5rev:GFP displayed altered distribution pattern in the rpi1 roots and embryos ( Figure 3 ). Moreover, the mutant showed reduced accumulation of auxin-induced PLT1/PLT2 ( Figure 4 ) and misexpression of MP ( Figure 4 ), which is critical for embryonic root development. In fact, RPI1 is also one of the many PLT-activated genes (Santuari et al., 2016). Besides, the greatly decreased WOX5proGFP ( Figure 1 ) suggested that the QC and columella stem cell identity might be affected. Further investigation substantiated that the phenotypes of rpi1 embryos and seedlings can be attributed to dramatically decreased abundance of auxin efflux carriers PIN1 and PIN3 ( Figure 5 ). Interestingly, the transcription level of PIN1 and PIN3 were not obviously affected ( Figure S9 ), implying that the reduced abundance of PIN1 and PIN3 could be associated with post-transcription regulated mechanism, such as 26S proteasome (Abas et al., 2006; Kleine-Vehn et al., 2008b; Laxmi et al., 2008) or vacuole-targeted (Abas et al., 2006; Kleine-Vehn et al., 2008b; Laxmi et al., 2008) pathways of protein degradation. Together, our results suggest that RPI1 influences root growth and development through auxin-related pathway.
Impairment of the RPI1 could result in the decrease in the synthesis of UDP–glucose, a substrate for cellulose synthesis, which eventually led to abnormal cell wall composition in the mutant. Consistent with this, the mutant contains notably lower levels of cellulose compared to the control ( Figure S7 ). These phenotypes, combined with the swelling roots, were similar to those observed in rsw10, a conditional mutant of RPI1, when growing under relatively high temperature (Howles et al., 2006). Besides, the other two radial swelling (rsw) mutants, rsw1 and rsw2, which mutated in the genes encoding cellulose synthase subunit or glycosyltransferase respectively, also showed changes in cellulose levels and short root phenotypes (Arioli et al., 1998; Peng et al., 2000; Lane et al., 2001). Moreover, seedlings grown on the plates containing cellulose synthesis inhibitor, DCB, are almost identical to the root phenotype of rpi1 seedlings (Peng et al., 2013). These genetic and pharmacological data support the suggestion that the RPI1 gene does not directly function in the auxin-related pathway but exerts its effects by affecting cellulose synthesis. Interestingly, compared with the rsw10, the rpi1 displayed short and swelling root phenotypes under normal growth conditions, due to their different mutant sites with Glu115Lys in rsw10 and Ala113Val in rpi1 ( Figure S2 ). The different sensitivity to temperature between these two mutants was probably associated with the different structural changes in RPI1. But the precise mechanism underlying needs to be further addressed.
The Role of the Cytoskeleton in PAT and Plant Cell Morphogenesis Regulated by Cell Wall
A previous study has revealed that cell wall functions in regulating PINs localization (Feraru et al., 2011). However, so far, few studies focused on the detailed mechanism underlying interaction between the cell wall and PAT. Several investigations demonstrated an important role of actin cytoskeleton in regulating PIN endocytosis, thus affecting their abundance on the PM (Geldner et al., 2001; Dhonukshe et al., 2008; Nagawa et al., 2012). Moreover, there is evidence showing that defects in cell wall led to abnormal actin arrangement (Zhang et al., 2011; Peng et al., 2013). Given that cell wall defects caused both abnormal actin orientations and reduced PIN protein levels in rpi1, it is of high interest to investigate the relationship between actin cytoskeleton reorganization and changes in PIN-dependent polar auxin in future studies.
Given that MTs is the trajectory for cellulose synthesis (McFarlane et al., 2014), most studies on the relationship between cell wall and cytoskeleton showed that defects in wall compositions affect MTs or both MTs and actin filaments organization. Disturbed cortical MT stability and orientation were revealed by genetic and inhibitor analysis (Himmelspach et al., 2003; Chu et al., 2007; Paredez et al., 2008; Peng et al., 2013). Mutation in the FORMIN HOMOLOGY 5 (FH5), an actin-nucleating protein which functions in rice morphology determination, led to abnormal MTs and actin filaments organization (Zhang et al., 2011). Seedlings growing on DCB containing plates also exhibited impairments in both MTs and actin filaments organization, with the actin cytoskeleton only showing changes when treated with high DCB concentration (Peng et al., 2013). In addition to cytoskeleton changes, we also observed reduction in PIN protein accumulation with DCB treatment in wild type plants ( Figure 7 ). Compared to previous findings, rpi1 showed obviously aberrant F-actin orientation ( Figure 6 ) whereas quite normal MTs arrangement in root cells ( Figure S8 ), suggesting a unique mechanism underlying cell wall regulating cytoskeleton in this mutant.
Besides, it has been believed that actin filaments play fundamental roles in designating a specific division plane during cell division (Rasmussen et al., 2013). Application of actin depolymerizing drugs caused abnormal orientation of division planes in tobacco BY-2 cells (Sano et al., 2005). Loss-of-function mutants of ACT7 showed defects in division plane orientation (Gilliland et al., 2003). Moreover, the orientation of actin filaments is consistent with the direction of cell elongation in root cells. Collectively, these findings raise a possibility that the transversal F-actin array could be a cause of the abnormal cell division planes observed in rpi1 mutant root cells ( Figure 1 ), which eventually led to the swelling root phenotype.
The influence on actin cytoskeleton from cell wall may be involved in signal transduction. It is reported that mutation in FEI1 and FEI2, two LRR type receptor-like kinase, gives rise to root swelling phenotype which is very similar to rpi1 (Xu et al., 2008). The fei1 fei2 showed significantly impaired cellulose synthesis under nonpermissive conditions compared to the wild type. They thought the FEI genes may sense the cell wall integrity and then induce the signal transduction to provide a feedback signal for cell wall synthesis (Xu et al., 2008).
In this study, we provide cellular evidence showing the cell wall may act on PAT by modulating actin cytoskeleton. However, the detailed mechanism underlying, for now, is poorly understood. Some researchers argued that the actin-associated formin family proteins might mediate the interplay between the cell wall and actin microfilaments (Liu et al., 2015). This notion is supported by the study of rice type II Formin homology 5 (OsFH5) (Zhang et al., 2011). A report demonstrated that defects in OsFH5 caused abnormal cell wall structure and disorganized cytoskeleton, with actin microfilaments showed more transverse than longitudinal in the root cells (Zhang et al., 2011), suggesting plant formins might play an essential role in cell wall and actin cytoskeleton interaction. Although the actin filament phenotype in osfh5 is very similar to that in rpi1, like most of the studies on cell wall and cytoskeleton, the MTs also severely altered in osfh5 (Zhang et al., 2011). Investigation for the function of formin proteins in rpi1 remains to be an attractive subject in the future which will probably clarify the unique mechanism underlying cell wall and actin cytoskeleton interaction.
Data Availability Statement
All datasets generated for this study are included in the article/ Supplementary Material .
Author Contributions
J-BH, YZ, MW, and L-ZT designed the experiments. J-BH, YZ, XZ, MW, and QD performed the experiments. J-BH, YZ, MW, and L-ZT analyzed the data. J-BH and YZ wrote the manuscript. L-ZT revised the manuscript.
Funding
This research was supported by the grants from National Natural Science Foundation of China (91417316 and 31870280).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Alice Y. Cheung at the University of Massachusetts, Ben Scheres at Wageningen University & Research, Chuanyou Li at the Institute of Genetics and Developmental Biology (CAS), and Philip N. Benfey at Duke University for providing seeds used in this study. We thank Prof. Yihua Zhou at CAS for providing the DCB reagent as a kind gift.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01641/full#supplementary-material
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wisniewska J., Moulinier-Anzola J. C., et al. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8, 249–256. 10.1038/ncb1369 [DOI] [PubMed] [Google Scholar]
- Adamowski M., Friml J. (2015). PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27, 20–32. 10.1105/tpc.114.134874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., et al. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120. 10.1016/j.cell.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Ambrose J. C., Shoji T., Kotzer A. M., Pighin J. A., Wasteneys G. O. (2007). The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19, 2763–2775. 10.1105/tpc.107.053777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C., Ruan Y., Gardiner J., Tamblyn L. M., Catching A., Kirik V., et al. (2013). CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana . Dev. Cell 24, 649–659. 10.1016/j.devcel.2013.02.007 [DOI] [PubMed] [Google Scholar]
- Arioli T., Peng L., Betzner A. S., Burn J., Wittke W., Herth W., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis . Science 279, 717–720. 10.1126/science.279.5351.717 [DOI] [PubMed] [Google Scholar]
- Bailey R. W. (1958). The reaction of pentoses with anthrone. Biochem. J. 68, 669–672. 10.1042/bj0680669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannigan A., Wiedemeier A. M., Williamson R. E., Overall R. L., Baskin T. I. (2006). Cortical microtubule arrays lose uniform alignment between cells and are oryzalin resistant in the Arabidopsis mutant, radially swollen 6. Plant Cell Physiol. 47, 949–958. 10.1093/pcp/pcj067 [DOI] [PubMed] [Google Scholar]
- Benkova E., Michniewicz M., Sauer M., Teichmann T., Seifertova D., Jurgens G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. 10.1038/nature03184 [DOI] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., et al. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. 10.1242/jcs.02847 [DOI] [PubMed] [Google Scholar]
- Boutte Y., Crosnier M. T., Carraro N., Traas J., Satiat-Jeunemaitre B. (2006). The plasma membrane recycling pathway and cell polarity in plants: studies on PIN proteins. J. Cell Sci. 119, 1255–1265. 10.1105/tpc.111.085514 [DOI] [PubMed] [Google Scholar]
- Chen M., Liu H., Kong J., Yang Y., Zhang N., Li R., et al. (2011). RopGEF7 regulates PLETHORA-dependent maintenance of the root stem cell niche in Arabidopsis . Plant Cell 23, 2880–2894. 10.1104/pp.106.088393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z., Chen H., Zhang Y., Zhang Z., Zheng N., Yin B., et al. (2007). Knockout of the AtCESA2 gene affects microtubule orientation and causes abnormal cell expansion in Arabidopsis . Plant Physiol. 143, 213–224. 10.1046/j.1365-313x.1998.00343.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. Cell Mol. Biol. 16, 735–743. 10.1073/pnas.0711414105 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F. S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., et al. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. CB 17, 678–682. 10.1016/j.cub.2011.01.036 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M. T., et al. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384. 10.1126/science.1164147 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Grigoriev I., Fischer R., Tominaga M., Robinson D. G., Hasek J., et al. (2008). Auxin transport inhibitors impair vesicle motility and actin cytoskeleton dynamics in diverse eukaryotes. Proc. Nat. Acad. Sci. U. S. A. 105, 4489–4494. 10.1038/nature02085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E., Feraru M. I., Kleine-Vehn J., Martiniere A., Mouille G., Vanneste S., et al. (2011). PIN polarity maintenance by the cell wall in Arabidopsis . Curr. Biol. CB 21, 338–343. 10.1038/415806a [DOI] [PubMed] [Google Scholar]
- Friml J., Wisniewska J., Benkova E., Mendgen K., Palme K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis . Nature 415, 806–809. 10.1038/nature06206 [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., et al. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis . Nature 426, 147–153. 10.1126/science.1100618 [DOI] [PubMed] [Google Scholar]
- Friml J., Yang X., Michniewicz M., Weijers D., Quint A., Tietz O., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306, 862–865. 10.1038/35096571 [DOI] [PubMed] [Google Scholar]
- Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., et al. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057. 10.1046/j.1365-313X.2003.01626.x [DOI] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y. D., Jurgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428. 10.1038/nature06215 [DOI] [PubMed] [Google Scholar]
- Gilliland L. U., Pawloski L. C., Kandasamy M. K., Meagher R. B. (2003). Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J. Cell Mol. Biol. 33, 319–328. 10.1038/nrm3790 [DOI] [PubMed] [Google Scholar]
- Grieneisen V. A., Xu J., Maree A. F., Hogeweg P., Scheres B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449, 1008–1013. 10.1371/journal.pbio.1000516 [DOI] [PubMed] [Google Scholar]
- Heidstra R., Sabatini S. (2014). Plant and animal stem cells: similar yet different. Nat. Rev. Mol. Cell Biol. 15, 301–312. 10.1046/j.1365-313X.2003.01906.x [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., et al. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567. 10.1016/S0092-8674(00)80865-X [DOI] [PubMed] [Google Scholar]
- Heisler M. G., Hamant O., Krupinski P., Uyttewaal M., Ohno C., Jonsson H., et al. (2010). Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PloS Biol. 8, e1000516. 10.1111/j.1365-313X.2006.02902.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelspach R., Williamson R. E., Wasteneys G. O. (2003). Cellulose microfibril alignment recovers from DCB-induced disruption despite microtubule disorganization. Plant J. Cell Mol. Biol. 36, 565–575. 10.1242/jcs.024950 [DOI] [PubMed] [Google Scholar]
- Howles P. A., Birch R. J., Collings D. A., Gebbie L. K., Hurley U. A., Hocart C. H., et al. (2006). A mutation in an Arabidopsis ribose 5-phosphate isomerase reduces cellulose synthesis and is rescued by exogenous uridine. Plant J. Cell Mol. Biol. 48, 606–618. 10.1016/j.cub.2008.03.021 [DOI] [PubMed] [Google Scholar]
- Kirik V., Herrmann U., Parupalli C., Sedbrook J. C., Ehrhardt D. W., Hulskamp M. (2007). CLASP localizes in two discrete patterns on cortical microtubules and is required for cell morphogenesis and cell division in Arabidopsis . J. Cell Sci. 120, 4416–4425. 10.1073/pnas.0808073105 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Sauer M., Brewer P. B., Wisniewska J., Paciorek T., et al. (2008. a). ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis . Curr. Biol. CB 18, 526–531. 10.1104/pp.126.1.278 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., et al. (2008. b). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. U. S. A. 105, 17812–17817. 10.1371/journal.pone.0001510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. R., Wiedemeier A., Peng L., Hofte H., Vernhettes S., Desprez T., et al. (2001). Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis . Plant Physiol. 126, 278–288. 10.1073/pnas.1401680111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxmi A., Pan J., Morsy M., Chen R. (2008). Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana . PloS One 3, e1510. 10.1111/jipb.12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Liang W., Zhang X., Ren H., Hu J., Bennett M. J., et al. (2014). Rice actin-binding protein RMD is a key link in the auxin-actin regulatory loop that controls cell growth. Proc. Natl. Acad. Sci. U. S. A. 111, 10377–10382. 10.1038/nature13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Persson S., Zhang Y. (2015). The connection of cytoskeletal network with plasma membrane and the cell wall. J. Integr. Plant Biol. 57, 330–340. 10.1073/pnas.1202040109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahonen A. P., Ten Tusscher K., Siligato R., Smetana O., Diaz-Trivino S., Salojarvi J., et al. (2014). PLETHORA gradient formation mechanism separates auxin responses. Nature 515, 125–129. 10.1146/annurev-arplant-050213-040240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiniere A., Lavagi I., Nageswaran G., Rolfe D. J., Maneta-Peyret L., Luu D. T., et al. (2012). Cell wall constrains lateral diffusion of plant plasma-membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 109, 12805–12810. 10.1016/j.cell.2007.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane H. E., Doring A., Persson S. (2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65, 69–94. 10.1371/journal.pbio.1001299 [DOI] [PubMed] [Google Scholar]
- Michniewicz M., Zago M. K., Abas L., Weijers D., Schweighofer A., Meskiene I., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130, 1044–1056. 10.1038/35095061 [DOI] [PubMed] [Google Scholar]
- Nagawa S., Xu T., Lin D., Dhonukshe P., Zhang X., Friml J., et al. (2012). ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PloS Biol. 10, e1001299. 10.1093/mp/sst044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P. N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307–311. 10.1371/journal.pone.0082442 [DOI] [PubMed] [Google Scholar]
- Nodzynski T., Feraru M. I., Hirsch S., De Rycke R., Niculaes C., Boerjan W., et al. (2013). Retromer subunits VPS35A and VPS29 mediate prevacuolar compartment (PVC) function in Arabidopsis . Mol. Plant 6, 1849–1862. 10.1104/pp.108.120196 [DOI] [PubMed] [Google Scholar]
- Panteris E., Adamakis I. D., Daras G., Hatzopoulos P., Rigas S. (2013). Differential responsiveness of cortical microtubule orientation to suppression of cell expansion among the developmental zones of Arabidopsis thaliana root apex. PloS One 8, e82442. 10.1186/s13007-015-0094-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A. R., Persson S., Ehrhardt D. W., Somerville C. R. (2008). Genetic evidence that cellulose synthase activity influences microtubule cortical array organization. Plant Physiol. 147, 1723–1734. 10.1007/s004250000301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Hocart C. H., Redmond J. W., Williamson R. E. (2000). Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta 211, 406–414. 10.1016/j.devcel.2011.10.026 [DOI] [PubMed] [Google Scholar]
- Peng L., Zhang L., Cheng X., Fan L. S., Hao H. Q. (2013). Disruption of cellulose synthesis by 2,6-dichlorobenzonitrile affects the structure of the cytoskeleton and cell wall construction in Arabidopsis . Plant Biol. 15, 405–414. 10.1111/tpj.12177 [DOI] [PubMed] [Google Scholar]
- Rademacher E. H., Lokerse A. S., Schlereth A., Llavata-Peris C. I., Bayer M., Kientz M., et al. (2012). Different auxin response machineries control distinct cell fates in the early plant embryo. Dev. Cell 22, 211–222. 10.1111/j.1365-313X.2005.02558.x [DOI] [PubMed] [Google Scholar]
- Rasmussen C. G., Wright A. J., Muller S. (2013). The role of the cytoskeleton and associated proteins in determination of the plant cell division plane. Plant J. Cell Mol. Biol. 75, 258–269. 10.1038/nature05703 [DOI] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes & Dev. 17, 354–358. 10.1101/gad.252503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T., Higaki T., Oda Y., Hayashi T., Hasezawa S. (2005). Appearance of actin microfilament ‘twin peaks’ in mitosis and their function in cell plate formation, as visualized in tobacco BY-2 cells expressing GFP-fimbrin. Plant J. Cell Mol. Biol. 44, 595–605. 10.1038/nrm2164 [DOI] [PubMed] [Google Scholar]
- Santuari L., Sanchez-Perez G. F., Luijten M., Rutjens B., Terpstra I., Berke L., et al. (2016). The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis Roots. Plant Cell 28, 2937–2951. 10.1105/tpc.16.00656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A. K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., et al. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. 10.1104/pp.103.034900 [DOI] [PubMed] [Google Scholar]
- Scheres B. (2007). Stem-cell niches: nursery rhymes across kingdoms. Nature reviews. Mol. Cell Biol. 8, 345–354. 10.1038/nrm2164 [DOI] [PubMed] [Google Scholar]
- Sun H., Basu S., Brady S. R., Luciano R. L., Muday G. K. (2004). Interactions between auxin transport and the actin cytoskeleton in developmental polarity of Fucus distichus embryos in response to light and gravity. Plant Physiol. 135, 266–278. 10.1111/j.1399-3054.2009.01276.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. S., Yoo C. M., Blancaflor E. B. (2008). Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol. 177, 525–536. 10.1105/tpc.108.063354 [DOI] [PubMed] [Google Scholar]
- Weijers D., Schlereth A., Ehrismann J. S., Schwank G., Kientz M., Jürgens G., et al. (2006). Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell. 10, 265–270. 10.1016/j.devcel.2005.12.001 [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller J. W., Helariutta Y., Fukaki H., Malamy J. E., Benfey P. N. (2000). Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127, 595–603. 10.1105/tpc.114.127993 [DOI] [PubMed] [Google Scholar]
- Xiong Y., DeFraia C., Williams D., Zhang X., Mou Z. (2009). Deficiency in a cytosolic ribose-5-phosphate isomerase causes chloroplast dysfunction, late flowering and premature cell death in Arabidopsis . Physiol. Plant. 137, 249–263. 10.1105/tpc.110.081349 [DOI] [PubMed] [Google Scholar]
- Xu J., Scheres B. (2005). Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17, 525–536. 10.1105/tpc.104.028449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. L., Rahman A., Baskin T. I., Kieber J. J. (2008). Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis . Plant Cell 20, 3065–3079. 10.7554/eLife.02860.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. B., Geng X., He C., Zhang F., Wang R., Horst W. J., et al. (2014). TAA1-regulated local auxin biosynthesis in the root-apex transition zone mediates the aluminum-induced inhibition of root growth in Arabidopsis . Plant Cell 26, 2889–2904. 10.1105/tpc.114.127993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang Y., Tan H., Wang Y., Li G., Liang W., et al. (2011). RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. Plant Cell 23, 681–700. 10.1105/tpc.110.081349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M., Absmanner B., Weller B., Barbosa I. C., Willige B. C., Fastner A., et al. (2014). Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/ Supplementary Material .