Abstract
Intraoperative diagnosis is essential for providing safe and effective care during cancer surgery1. The existing workflow for intraoperative diagnosis based on hematoxylin and eosin-staining of processed tissue is time-, resource-, and labor-intensive2,3. Moreover, interpretation of intraoperative histologic images is dependent on a contracting, unevenly distributed pathology workforce4. Here, we report a parallel workflow that combines stimulated Raman histology (SRH)5–7, a label-free optical imaging method, and deep convolutional neural networks (CNN) to predict diagnosis at the bedside in near real-time in an automated fashion. Specifically, our CNN, trained on over 2.5 million SRH images, predicts brain tumor diagnosis in the operating room in under 150 seconds, an order of magnitude faster than conventional techniques (e.g., 20–30 minutes)2. In a multicenter, prospective clinical trial (n = 278) we demonstrated that CNN-based diagnosis of SRH images was non-inferior to pathologist-based interpretation of conventional histologic images (overall accuracy, 94.6% vs. 93.9%). Our CNN learned a hierarchy of recognizable histologic feature representations to classify the major histopathologic classes of brain tumors. Additionally, we implemented a semantic segmentation method to identify tumor infiltrated, diagnostic regions within SRH images. These results demonstrate how intraoperative cancer diagnosis can be streamlined, creating a complimentary pathway for tissue diagnosis that is independent of a traditional pathology laboratory.
Approximately 15.2 million people are diagnosed with cancer across the world yearly and greater than 80% will undergo surgery1. In many cases, a portion of the excised tumor is analyzed during surgery to provide preliminary diagnosis, ensure the specimen is adequate for rendering final diagnosis and guide operative management. In the US, there are over 1.1 million biopsy specimens annually8, all of which must be interpreted by a contracting pathology workforce9. The conventional workflow for intraoperative histology, dating back over a century3, necessitates tissue transport to a laboratory, specimen processing, slide preparation by highly-trained technicians, and interpretation by a pathologist, with each step representing a potential barrier to delivering timely and effective surgical care.
Harnessing advances in optics5 and artificial intelligence (AI) we developed a streamlined workflow for microscopic imaging and diagnosis that ameliorates each of these barriers. Stimulated Raman histology (SRH) is an optical imaging method that provides rapid, label-free, sub-micron resolution, images of unprocessed biological tissues5. SRH utilizes the intrinsic vibrational properties of lipids, proteins, nucleic acids to generate image contrast, revealing diagnostic microscopic features and histologic findings poorly visualized with hematoxylin and eosin-stain (H&E) images, such as axons and lipid droplets7, while eliminating the artifacts inherent in frozen or smear tissue preparations6.
Advances in fiber-laser technology have enabled the development of an FDA-registered system for generating SRH images that can be used in the operating room. We have demonstrated that SRH images reveal microscopic architectural features comparable to conventional H&E images6. Given this finding, we recently deployed clinical SRH imagers in our operating rooms, making histologic data readily available during surgery6,10.
Whether histologic images are obtained via SRH or frozen sectioning, diagnostic interpretation has required the expertise of a trained pathologist. Both globally and within the US, there is an uneven distribution of expert pathologists available to provide intraoperative diagnosis. For example, many centers performing brain tumor surgery do not employ a neuropathologist and further shortages are expected given the 42% vacancy rate in neuropathology fellowships4. Moreover, while final pathologic diagnosis is increasingly driven by molecular rather than morphological criteria11, intraoperative diagnosis relies heavily on interpretation of cytologic and histo-architectural features. We hypothesized that the application of AI could be used to expand access to expert-level intraoperative diagnosis in the ten most commonly encountered brain tumors and augment the ability of pathologists to interpret histologic images.
We have previously demonstrated that SRH images are particularly well-suited for computer-aided diagnosis using hand-engineered feature extractors with random forest and multilayer perceptron classifiers6,10,12. However, manual feature engineering inherent in these methods is challenging, requires domain-specific knowledge, and poses a major bottleneck towards achieving human-level accuracy and clinical implementation13. In contrast, deep neural networks utilize trainable feature extractors, which provide a learned and optimized hierarchy of image features for classification. Human-level accuracy for image classification tasks has been achieved through deep learning in the fields of ophthalmology14, radiology15, dermatology16, and pathology17–19.
Consequently, we designed a three-step intraoperative tissue-to-diagnosis pipeline (Figure 1) consisting of: (1) image acquisition, (2) image processing, and (3) diagnostic prediction via a CNN. (Supplementary Video 1). A fresh, unprocessed surgical specimen is passed off the operative field and a small sample (e.g., 3 mm3) is compressed into a custom microscope slide. After inserting the slide into the SRH imager, images are acquired at two Raman shifts, 2845 cm−1 and 2930 cm−1. SRH images are then processed via a dense sliding window algorithm to generate overlapping, single-scale, high-resolution, and high-magnification patches used for CNN training and inference. In the prediction stage, individual patches are passed through the trained Inception-ResNet-v2 network, a benchmarked neural network that combines inception modules and residual connections in a deep CNN architecture for image classification20.
Figure 1. Intraoperative diagnostic pipeline using SRH and deep learning.
The intraoperative workflows for both conventional hematoxylin and eosin-staining (H&E) histology and stimulated Raman histology (SRH) plus convolutional neural networks (CNN) are shown in parallel. (1) Freshly excised specimens are loaded directly into an SRH imager for image acquisition. Operation of the SRH imager is performed by a single user, who loads tissue into a carrier and interacts with a simple touch-screen interface to initate imaging. Images are sequentially acquired at two Raman shifts, 2845 cm−1 and 2930 cm−1, as strips. After strip stitching, the two image channels are registered and virtual H&E provides SRH mosaics for intraoperative review by surgeons and pathologists. Time to acquire a 1×1-mm SRH image is approximately 2 minutes. (2) Image processing starts by using a dense sliding window algorithm with valid padding over the 2845 cm−1 and 2930 cm−1 images concurrently. Registered 2845 cm−1 and 2930 cm−1 image patches are subtracted pixelwise to generate a third image channel (2930 cm−1-2845 cm−1) that highlights nuclear contrast and cellular density. Each image channel is postprocessed to enhance image contrast and concatenated to produce a single three-channel RGB image for CNN input. (3) To provide an intraoperative prediction of brain tumor diagnosis, each patch undergoes a feedforward pass through the trained CNN and takes approximately 15 secs using a single GPU for 1×1-mm SRH image. Our inference algorithm (Extended Data Figure 3) for patient-level diagnosis acts by retaining the high probability tumor regions within the image based on patch-level predictions, and filtering the nondiagnostic and normal areas. Patch-level predictions from tumor regions are then summed and renormalized to generate a patient-level probability distribution over the diagnostic classes. Our pipeline is able to provide a diagnosis in less than 2.5 minutes using a 1×1-mm image, which corresponds to more than a 10x speedup in time-to-diagnosis compared to conventional intraoperative histology2. Scale bar, 50 μm.
Over 2.5 million labelled patches from 415 patients were used for CNN training (Extended Data Figure 1). The CNN was trained to classify tissue into 13 histologic categories organized into a taxonomy that includes output and inference nodes focusing on commonly encountered brain tumors (Extended Data Figure 2). To provide a final patient-level diagnostic prediction, an inference algorithm was developed to map all patches from a specimen to a single probability distribution over the diagnostic classes (Extended Data Figure 3).
Noting the commentary on the importance of rigorous clinical evaluations of deep-learning-based algorithms21, we executed a two-arm, prospective, multicenter, non-inferiority clinical trial comparing the diagnostic accuracy of pathologists interpreting conventional histologic images (control arm) to the accuracy of SRH image classification by the CNN (experimental arm) (Extended Data Figure 4 and Supplementary Table 1). Fresh brain tumor specimens were collected, split intraoperatively into sister specimens and randomly assigned to the control or experimental arm. Sister specimens in the control arm were processed via conventional frozen-section and smear preparation techniques and interpreted by board-certified pathologists. Sister specimens in the experimental arm were imaged with SRH and diagnosis was predicted by the CNN. Two hundred seventy-eight patients were included and the primary endpoint was overall multiclass diagnostic accuracy using final clinical diagnosis as the ground truth. Overall diagnostic accuracy was 93.9% (261/278) for the conventional H&E histology arm and 94.6% (264/278) for SRH plus CNN arm, exceeding our primary endpoint threshold for noninferiority (>91%) (Figure 2).
Figure 2. Prospective clinical trial of SRH plus CNN versus conventional H&E histology.
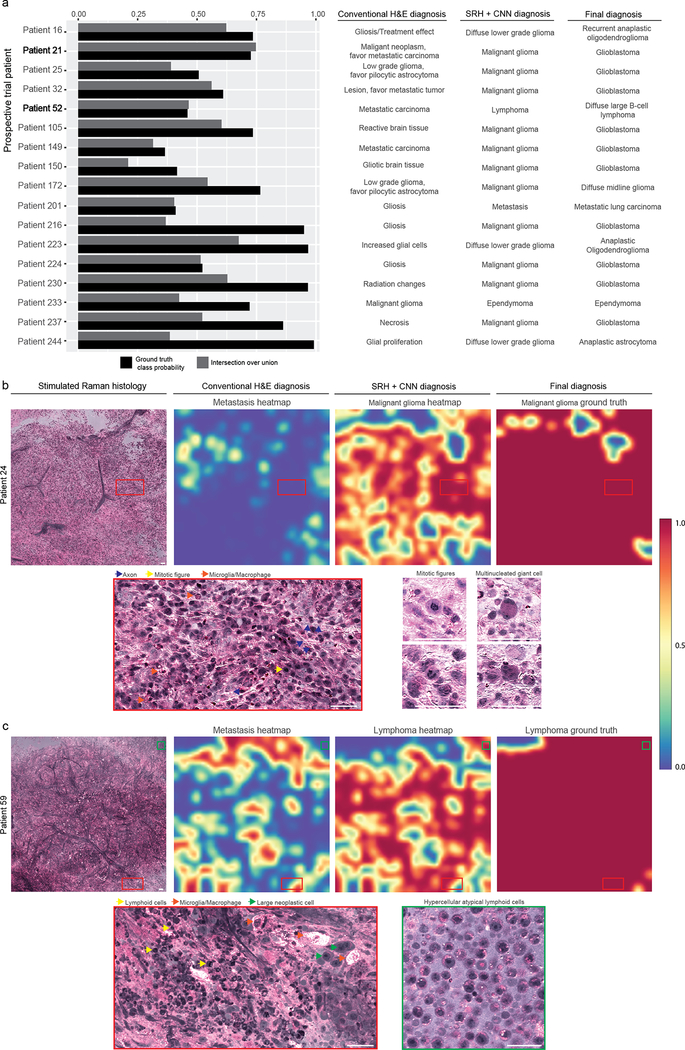
a, The prediction probabilities for the ground truth classes are plotted in descending order by medical center with indication of correct (green) or incorrect (red) classification. b, Multiclass confusion matrices for both the control arm and experimental arm. Mistakes in the control arm, traditional H&E histology with pathologist, were mostly misclassification of malignant gliomas (10/17). The glial tumors had the highest error rate in the SRH plus CNN arm (9/14). Less common tumors, including ependymoma, medulloblastoma, and pilocytic astrocytomas were also misclassified, likely due to insufficient number of cases for model training, resulting in lower mean class accuracy compared to the control arm. These errors are likely to improve with additional SRH training data. Model performance on cases misclassified using conventional H&E histology can be found in Extended Data Figure 6. The glioma inference class was used for the clinical trial in the setting where the control arm pathologist did not specify glioma grade at the time of surgery, thereby allowing for one-to-one comparison between study arms.
*No gliosis/treatment effect cases were enrolled during the clinical trial. This row is included because gliosis was a predicted label and to maintain the convention of square confusion matrices.
Notably, the CNN was designed to predict diagnosis independent of clinical or radiographic findings, which were reviewed by study pathologists and are often of central importance in diagnosis. Nine of the 14 errors in the SRH plus CNN arm were glial tumors, which often have overlapping morphologic characteristics but highly divergent clinical presentations and radiographic appearances. Ten of the 17 errors in the conventional H&E arm were malignant gliomas incorrectly classified by pathologists as metastatic tumors, gliosis/treatment effect, or pilocytic astrocytoma. In addition, the CNN correctly classified all 17 of the cases in which the pathologist’s diagnosis was incorrect (Extended Data Figure 5). Moreover, pathologists correctly diagnosed all 14 cases misdiagnosed in the CNN/SRH arm. These results indicate that CNN-based classification of SRH images could aid pathologists in the classification of challenging specimens.
While the CNN output classes in our study would cover greater than 90% of all brain tumors diagnosed in the US22, the diversity and scarcity of rare tumors precludes training of a fully universal CNN for brain tumor diagnosis. Understanding the limitations of our CNN, we developed and implemented a Mahalanobis distance-based confidence scoring system to detect rare tumors23. Thirteen of the patients enrolled in our trial were diagnosed with nine rare tumor types. Our method for rare tumor detection identified all 13 tumors as entities distinct from the output diagnostic classes (Extended Data Figure 6).
To gain insight into the learned representations utilized by the CNN for image classification, we used activation maximization, which generates an image that maximally activates a neuron in any neural network layer using iterations of gradient ascent in the input space (Figure 3 and Extended Data Figure 7)24. Deep hidden layers detected nuclear and chromatin morphology, axonal density, and histoarchitecture, indicating that our network learned recognizable domain-specific feature representations. We sampled 1000 SRH patches from normal brain tissue and two tumor classes to investigate class-specific, hidden-layer neuron activation. Neurons from a deep hidden layer (convolutional layer 159) with maximal mean activation for each class were recorded and the distribution of mean rectified linear unit activations was plotted.
Figure 3. Activation maximization reveals a hierarchy of learned SRH feature representations.
a, Images that maximize the activation of select filters from layers 5, 10, and 159 are shown. (Activation maximization images for each layer’s filter bank can be found in Extended Data Figure 7.) A hierarchy of increasingly complex and recognizable histologic feature representations can be observed. b, The activation maximization images for the 148th, 12th, and 101st filter in the 159th layer are shown as column headings. These filters were selected because they are maximally active for the grey matter, malignant glioma, and metastatic brain tumor class, respectively, with example images from each class shown as row labels. A spatial map of the rectified linear unit (ReLU) values for the class example images and corresponding mean ReLU value (± standard deviation) is shown in each cell of the grid. Each cell also contains the distribution of mean activation values for 1000 images randomly sampled from each diagnostic class. High magnification crops from the example images which maximally activate each neuron are shown. Activation maximization images show interpretable image features for each diagnostic class, such as axons (neuron 148), hypercellularity with lipid droplets and high nuclear:cytoplastic ratios (neuron 12), and large cells with prominent nucleoli and cytoplasmic vesicles (neuron 101). Example image scale bar, 50 μm; maximum ReLU activation area image scale bar, 20 μm.
The images generated through activation maximization reveal recognizable features for each histologic class. For example, green linear structures (neuron 148) represent lipid-rich axons found in grey matter. Neuron 12 was maximally active for malignant glioma and responds to high nuclear density and lipid droplets, features associated with higher grade gliomas25,26. Neuron 101 was maximally activated by patches containing large nuclei with prominent nucleoli and cytoplasmic vesicles commonly seen in metastatic tumor cells and pyramidal neurons. These results indicate that the CNN has learned the importance of specific histomorphologic, cytologic, and nuclear features for image classification, including some features classically used by pathologists to diagnose cancer. In addition, we used t-distributed stochastic neighbor embedding (t-SNE) to show that our histologic categories have similar internal CNN feature representations and form clusters based on diagnostic classes (Extended Data Figure 8).
We also implemented a semantic segmentation technique to provide pixel-level classification and demonstrate how CNN-based analysis could be used to highlight diagnostic regions within an SRH image (Extended Data Figure 9). By utilizing a dense sliding window algorithm, every pixel in an SRH image has an associated probability distribution over the diagnostic classes that is a function of the local overlapping patch-level predictions. Class probabilities, can be mapped to a pixel intensity scale. A three-channel RGB overlay indicating tumor tissue, normal/non-neoplastic tissue, and nondiagnostic regions allows for image overlay of pixel-level CNN predictions. Our segmentation method achieved a mean intersection over union (IOU) value of 61.6 ± 28.6 for the ground truth diagnostic class and 86.0 ± 19.2 for the tumor inference class for patients in our prospective cohort. Analysis of specimens collected at the tumor-brain interface in primary (Figure 4) and metastatic brain tumors (Extended Data Figure 10) demonstrates how the CNN can differentiate tumor from non-infiltrated brain and nondiagnostic regions.
Figure 4. Semantic segmentation of SRH images identifies tumor-infiltrated and diagnostic regions.
a, Full SRH mosaic of a specimen collected at the brain-tumor interface of a patient diagnosed with glioblastoma, WHO IV. b, Dense hypercellular glial tumor with nuclear atypia is seen diffusely on the left and peritumoral gliotic brain with reactive astrocytes on the right of the specimen. SRH imaging of fresh specimens without tissue processing preserves both the cytologic and histoarchitectural features allowing for visualization of the brain-tumor margin. c, Three-channel RGB CNN-prediction transparency is overlaid on the SRH image for surgeon and pathologist review intraoperatively with associated (d) patient-level diagnostic class probabilities. e, Inference class probability heatmap for tumor (IOU 0.869), nontumor (IOU 0.738), and nondiagnostic (IOU 0.400) regions within the SRH image are shown with ground truth segmentation. The brain-tumor interface is well delineated using CNN semantic segmentation and can be used in operating room to identify diagnostic regions, residual tumor burden and tumor margins. Scale bar, 50 μm.
Our semantic segmentation technique parallels that of Chen and colleagues who reported the development of an augmented reality microscope with real-time AI-based prostate and breast cancer diagnosis using conventional light microscopy27. Both methods superimpose diagnostic predictions of an AI algorithm on a microscopic image, calling the clinician’s attention to areas containing diagnostic information and providing insight into how AI could ultimately streamline tissue diagnosis.
In conclusion, we have demonstrated how combining SRH with deep learning can be employed to rapidly predict intraoperative brain tumor diagnosis. Our workflow provides a transparent means of delivering expert-level intraoperative diagnosis where neuropathology resources are scarce and improving diagnostic accuracy in resource-rich centers. The workflow also allows surgeons to access histologic data in near-real time, enabling more seamless use of histology to inform surgical decision-making based on microscopic tissue features.
In the future we anticipate that AI algorithms can be developed to predict key molecular alterations in brain tumors such as MGMT methylation, IDH, and ATRX status. In addition, it is possible that SRH will ultimately incorporate spectroscopic detection of the metabolic effects of diagnostic genetic mutations, such as accumulation of 2-hydroxyglutarate in IDH mutated gliomas. In the interim, however, we note that SRH preserves the integrity of imaged tissue for downstream analytic testing and integrates well within the modern practice of molecular diagnosis.
While our workflow was developed and validated in the context of neurosurgical oncology, many histologic features used to diagnose brain tumors are found in the tumors of other organs. Consequently, we predict a similar workflow incorporating optical histology and deep learning could apply to dermatology28, head and neck surgery29, breast surgery30, and gynecology31, where intraoperative histology is equally central to clinical care. Importantly, our AI-based workflow provides unparalleled access to microscopic tissue diagnosis at the bedside during surgery, facilitating detection of residual tumor, reducing the risk of removing histologically normal tissue adjacent to a lesion, enabling the study of regional histologic and molecular heterogeneity and minimizing the chance of non-diagnostic biopsy or misdiagnosis due to sampling error32,33.
Study design
The main objectives of the study were to 1) develop an intraoperative diagnostic computer vision system that combines clinical stimulated Raman histology (SRH) and a deep learning-based method to augment the interpretation of fresh surgical specimens in near real-time, and 2) perform a multicenter, prospective clinical trial to test the diagnostic accuracy of our clinical SRH system combined with trained convolutional neural networks (CNN). “Near real-time” diagnosis was defined as a nonclinically significant delay from the time of tissue removal from the resection cavity to tissue diagnosis (i.e., 2–3 minutes). Patient enrollment for intraoperative SRH imaging began June 1, 2015. Inclusion criteria for intraoperative imaging included: 1) male or female, 2) subjects undergoing central nervous system tumor resection or epilepsy surgery at Michigan Medicine, New York-Presbyterian/Columbia University Medical Center, or the University of Miami Health System, 3) subject or durable power of attorney able to give informed consent, and 4) subjects in which there was additional specimen beyond what was needed for routine clinical diagnosis. We then trained and validated a benchmarked CNN architecture on an image classification task to provide rapid and automated evaluation of fresh surgical specimens imaged with SRH. CNN performance was then tested using a two-arm, prospective, noninferiority trial conducted at three tertiary medical centers with dedicated brain tumor programs. A semantic segmentation method was developed to allow for surgeon and pathologist review of SRH images with integrated CNN predictions.
Stimulated Raman histology
All images used in our study were obtained using a clinical stimulated Raman scattering (SRS) microscope5. Biomedical tissue is excited with a dual-wavelength fiber laser with a fixed wavelength pump beam at 790 nm and a Stokes beam tunable from 1015 nm to 1050 nm. This configuration allows for spectral access to Raman shifts in the range from 2800 cm−1 to 3130 cm−1.35 Images are acquired via beam-scanning with a spatial sampling of 450 nm/pixel, 1000 pixels per strip and an imaging speed for 0.4 MPixel/s/Raman shift. The NIO Laser Imaging System (Invenio Imaging, Inc., Santa Clara, California), a clinical fiber-laser-based SRS microscope, was used to acquire all images in the prospective clinical trial. For SRH, samples were imaged sequentially at the two Raman shifts: 2850 cm−1 and 2950 cm−1. Lipid-rich brain regions (e.g., myelinated white matter) demonstrate high SRS signal at 2845 cm−1 due to CH2 symmetric stretching in fatty acids. Cellular regions produce high 2930 cm−1 intensity and large S2930/S2845 ratios to high protein and DNA content. A virtual H&E look-up table is applied to transform the raw SRS images to SRH images for intraoperative use and pathologic review. A video of intraoperative SRH imaging with automated CNN-based prediction can be found in Supplementary Video 1. The NIO Imaging System (Invenio Imaging, Inc., Santa Clara, California) is delivered ready-to-use for image acquisition. SRH images can be reviewed locally using the integrated high-definition monitor, remotely via the health systems picture archiving and communication system (PACS) or a cloud-based image viewer that allows images to be reviewed anywhere with a high-speed internet connection in less than 30 seconds.
Image preprocessing and data augmentation
The 2845 cm−1 image was subtracted from the 2930 cm−1 image, and the resultant image was concatenated to generate a three-channel image (2930 cm−1 minus 2845 cm−1, red; 2845 cm−1, green; 2930 cm−1, blue). A 300×300-pixel sliding window algorithm with 100-pixel step size (both horizontal and vertical directions) and valid padding was used to generate image patches. This single-scale sliding window method over high-resolution, high-magnification images has the following advantages: 1) accommodates the image input size of most CNN architectures without downsampling, 2) allows for efficient graphical processing unit-based model implementation, 3) boosts the number of training and inference images by approximately an order of magnitude, 4) allows for better learning of high-frequency image features, and 5) is faster and 6) easier to implement compared to multi-scale networks. Previous multi-scale CNN implementations have not yielded better performance for image classification tasks involving histologic images36. Additionally, the use of larger, lower magnification images complicates image label assignment in the setting of multiple class labels applying to separate regions within a single image (i.e., white matter, tumor tissue, nondiagnostic, gliotic tissue, etc.), which introduces an additional tunable hyperparameter to identify an optimal class labelling strategy. This problem is effectively avoided using high-magnification patches, where multiple class labels for a single image are rare. To optimize image contrast, the bottom and top 3% of pixels by intensity from each channel were clipped and images rescaled. All image patches in the training, validation, and testing datasets were reviewed and labelled by study authors (T.C.H, S.S.K., S.L, A.R.A., E.U.). To accommodate class imbalance due to variable incidence rates between the CNS tumors included in our study, oversampling was used for the underrepresented classes. We used multiple label-preserving affine transformations for data augmentation, including any uniformly distributed random combination of rotation, shift, and reflection. All images were mean zero centered by subtracting the channel mean of the training set.
Image datasets
Our study included 4 image datasets obtained from 4 SRH imagers: 1) University of Michigan (UM) images from a prototype clinical SRH microscope6, 2) UM images from one NIO Imaging System, 3) Columbia University images from a second NIO Imaging System and 4) University of Miami images from a third NIO Imaging System. Distribution of tumor classes by both number of patches and patients used for CNN training and validation can be found in Extended Data Figure 1. A total of 296 patients were imaged using the prototype SRH microscope and 339 using the NIO Imaging System. Final tissue diagnosis was provided by each institution’s board-certified neuropathologists. Only UM images were used for model training and validation. Images acquired at Columbia University and the University of Miami were only used in the prospective clinical trial to test model performance on SRH images acquired at other medical centers and optimize assessment of CNN generalizability within our study.
Convolutional neural network training
A total of 13 diagnostic classes were selected that 1) represent the most common central nervous system tumors11,22 and 2) optimally inform intraoperative decisions that effect surgical goals. Classes included malignant glioma (glioblastoma and diffuse midline glioma, WHO grade IV), diffuse lower grade gliomas (oligodendrogliomas and diffuse astrocytomas, WHO grade II and III), pilocytic astrocytoma, ependymoma, lymphoma, metastatic tumors, medulloblastoma, meningioma, pituitary adenoma, gliosis/reactive astrocytosis/treatment effect, white matter, grey matter, and nondiagnostic tissue. We implemented the Google (Google LLC, Mountain View, CA) Inception-ResNet-v2 architecture with 55.8 million trainable parameters randomly initialized. Similar to previous studies, our preliminary experiments using pretrained weights from the ImageNet challenge did not improve model performance, likely due to the large domain difference and limited feature transferability between histologic images and natural scenes (Extended Data Figure 6)36,37. The network was trained on approximately 2.5 million unique patches from 415 patients using a categorical cross-entropy loss function weighted using inverse class frequency. A randomly selected 16-patient validation set imaged using the NIO Imaging System at UM was used for hyperparameter tuning and model selection based on patch-level classification accuracy. We used the Adam optimizer with an initial learn rate of 0.001, β1 of 0.9, and β2 of 0.999, ε of 10E-8 and a 32-image batch size. An early stopping callback was used with a minimum validation accuracy increase of 0.05 and 5 epoch patience (Extended Data Figure 1). Training, validation and testing was done using the high-level Python-based neural network API, Keras (version 2.2.0), with a TensorFlow (version 1.8.0)38 backend running on two NVIDIA GeForce 1080 Ti graphical processing units.
Patient-level diagnosis and inference algorithm
Patch-level predictions from each patient need to be mapped to a mosaic-, specimen-, or patient-level diagnosis in order to provide a final intraoperative classification (Extended Data Figure 3). The set of diagnostic patch softmax output vectors from a specimen or patient are summed elementwise and renormalized to produce specimen-level or patient-level probability distribution. To account for normal brain and pathologic tissue contained within the same specimen, a thresholding procedure was used, such that if the probability of a normal specimen was greater than 90%, a normal label was assigned. Otherwise, the normal class probabilities were set to zero, the probability distribution renormalized, and the final diagnosis was the expected value of the renormalized distribution. Our inference algorithm leverages the fact that normal brain tissue and nondiagnostic regions have similar histologic features among all patients, resulting in high patch-level classification accuracy for normal brain and eliminating the need to train an additional classifier based on the patch-level probability histograms39. Similar to previous publications using deep learning for medical diagnosis15,16, a taxonomy of inference classes was used to allow for classification at various clinically relevant levels of granularity (Extended Data Figure 2). The probability of any parent/inference class is the sum of its child node probabilities.
Mahalanobis distance-based confidence score
The most common brain tumors types were used for model training and includes greater than 90% of all CNS tumors diagnosed in the United States22; however, rare tumor types will be encountered in the clinical setting. Therefore, in addition to a posterior probability distribution over the CNN output classes, we aimed to provide a confidence score to detect tumor samples that are far away from the training distribution to detect rare tumor types not included during training. We induce class conditional Gaussian distributions with respect to mid- and upper-level features (i.e. layer outputs) of our CNN under Gaussian discriminant analysis that results in a confidence score based on the Mahalanobis distance23. Without any modification to our pertained network, we obtain a generative model by converting the penultimate layer, for example, to a class conditional distribution which follows a multivariate Gaussian distribution. Specifically, we compute 13 class conditional Gaussian distributions, one for each histologic class, with a tied covariance matrix using our training set. Using these induced class conditional Gaussian distributions, we calculate a confidence score, M(x), using the Mahalanobis distance between the test specimen x and the closest class conditional Gaussian distribution,
where is the class mean, f(x) is the output from the penultimate layer, and is the tied covariance matrix. The specimen-level confidence is the mean patch-level confidence score. To improve performance and increase separability of common and rare tumor classes as previously described23, we implemented an ensemble method that included output from 7 layers: convolutional layers 159, 195, 199, 203, final average pooling layer, final dense layer, and softmax output layer. Mahalanobis distance-based confidence scores for each layer were then used as features to train a linear discriminant classifier on our training set and rare tumor specimens imaged prior to starting prospective trial enrollment.
Prospective clinical trial design
A noninferiority trial was designed to rigorously validate our proposed intraoperative diagnostic pipeline. An expected accuracy of 96%, a delta of 5%, alpha 0.05, and power of 0.9 were used to calculate a minimum patient sample size of 264 with the primary endpoint of overall multiclass diagnostic accuracy (Extended Data Figure 4). Prospective enrollment began on April 6, 2018 and closed on February 26, 2019 with a total of 302 patients enrolled. Clinical trial inclusion criteria were the same for intraoperative SRH imaging. Exclusion criteria were 1) poor quality of specimen on visual gross examination due to excessive blood, coagulation artifact, necrosis, or ultrasonic damage or 2) specimen classified as out-of-distribution by LDA classifier using the Mahalanobis distance-based confidence score. A total of 278 patients were included in the trial. The conventional intraoperative H&E diagnosis was used in the control arm and the SRH imaging plus CNN was used for the experimental arm. The final histopathologic diagnosis was used to label patients into the appropriate patient-level ground truth class. For example, a patient with final WHO classification of glioblastoma, WHO IV, is classified into the malignant glioma class, or a final diagnosis of diffuse astrocytoma, WHO II, is classified into the diffuse lower grade glioma class. The strategy does not bias either study arm and allows for a multiclass accuracy value to be calculated for each study arm. Three instances arose where the control arm diagnosis was limited to “glioma” without further specification. To allow for a one-to-one comparison between the two study arms, the “glial tumor” inference class was used in the experimental arm for these cases. To eliminate the possibility of sampling error in the control arm, all incorrectly classified specimens underwent secondary review by two board-certified neuropathologists (S.C.P., P.D.C.) to ensure the specimen was of sufficient quality to make a diagnosis and to ensure that tumor tissue was present in the specimen. Following completion of the trial, we prospectively imaged 8 stereotactic needle brain biopsies to validate our workflow in operations were sampling was based on stereotactic navigation rather than gross inspection of the tissue (Supplementary Figure 1). SRH with automated CNN diagnosis can play in essential role in these cases to confirm diagnostic tissue sampling, provide intraoperative histologic data, and cut total surgical time in half (i.e., 60–90 minutes to 20–30 mins).
Activation maximization
Activation maximization allows for qualitative evaluation of learned representations in deep neural network architectures24. The objective is to generate an image that maximally activates a neuron or filter in a CNN hidden layer given a set of fixed, trained weights, such that:
where x is the input image, θ denotes the neural network weights, is the activation of a jth neuron in hidden layer l, and R(x) is a regularization term. An image, x*, can be generated by computing the gradient of and updating the pixel values of x using iterations of gradient ascent. Our regularization term included weight decay, gaussian blur, and dark pixel clipping to improve image clarity and interpretability40. We used 500 iterations of gradient ascent for each of the images shown in Figure 3 and Extended Data Figure 7. We choose convolutional layer 159, a deep hidden layer with sufficient spatial information to identify regions of low and high activation within a single image, to evaluate class-specific activation.
Probability heatmaps and semantic segmentation of SRH
Class probability heatmaps can localize diagnostic tissue and spatially identify areas with different predicted class labels (e.g., normal versus tumor infiltrated tissue). Our single-scale sliding window approach allows for an intuitive image patch-to-heatmap pixel mapping that 1) is computationally efficient, 2) yields a 9-fold increase in heatmap pixel spatial resolution relative to patch size, and 3) integrates a local neighborhood of overlapping patch predictions for semantic segmentation. For example, a 1000×1000-pixel SRS image is divided into a 10×10-pixel grid. The image area contained within each heatmap pixel will overlap with 1 (grid corners) to 9 (inner 6×6 grid) neighboring patches due to valid padding and 100-pixel step size (Extended Data Figure 9). The softmax output vector from each overlapping patch is summed and renormalized to give a probability distribution for each heatmap pixel. This procedure yields a prediction heatmap for each output class to produce a 10×10xk array, where k is the number of output classes. This method can be repeated to produce heatmaps for arbitrarily large SRH images. Intersection over union (IOU) metric was used to evaluate segmentation performance. To produce effective prediction overlays for pathologist review, probabilities were uniformly mapped to a 0–255 scale for three diagnostic classes (i.e., nondiagnostic, nontumor inference class, and tumor inference class) to generate a three-channel RGB transparency overlay (alpha = 40%).
Statistics and reproducibility
All measures on central tendency were reported as mean and standard deviation. CNN training was replicated 10 times and the model with the highest validation accuracy was selected for use in the prospective clinical trial. Pearson correlation coefficient was used to measure linear correlations. Full R code for calculated trial sample size can be found in our code repository (See below). Please see the Life Sciences Reporting Summary for more details.
Data availability
A University of Michigan IRB protocol (HUM00083059) was approved for the use of human brain tumor specimens in this study. To obtain these samples or SRH images, contact D.A.O. Code repository for network training, evaluation, and visualizations is publicly available at https://github.com/toddhollon/srh_cnn.
Extended Data
Extended Data Figure 1: SRH image dataset and CNN training.
The class distribution of (a) training and (b) validation set images are shown as number of patches and patients. Class imbalance results from different incidence rates among human central nervous system tumors. The training set contains over 50 patients for each of the five most common tumor types (malignant gliomas, meningioma, metastasis, pituitary adenoma, and diffuse lower grade gliomas). In order to maximize the number of training images, no cases from medulloblastoma or pilocytic astrocytoma were included in the validation set and oversampling was used to augment the underrepresented class during CNN training. c, Training and validation categorical cross entropy loss and patch-level accuracy is plotted for the training session that yielded the model used for our prospective clinical trial. Training accuracy converges to near-perfect with a peak validation accuracy of 86.4% following epoch 8. Training procedure was repeated 10 times with similar accuracy and cross entropy convergence. Additional training did not result in better validation accuracy and early stopping criteria were reached.
Extended Data Figure 2: A taxonomy of intraoperative SRH diagnostic classes to inform intraoperative decision making.
a, Representative example SRH images from each of the 13 diagnostic class are shown. Both diffuse astrocytoma and oligodendroglioma are shown as examples of diffuse lower grade gliomas. Classic histologic features (i.e., piloid process in pilocytic astrocytomas, whorls in meningioma, and microvascular proliferation in glioblastoma) can be appreciated, in addition to features unique to SRH images (e.g., axons in gliomas and normal brain tissue). Scale bar, 50 μm. b, A taxonomy of diagnostic classes was selected specifically to inform intraoperative decision making, rather than to match WHO classification. Essential intraoperative distinctions, such as tumoral versus nontumoral tissue or surgical versus nonsurgical tumors, allow for safer and more effective surgical treatment. Inference node probabilities inform intraoperative distinctions by providing coarse classification with potentially higher accuracy due to summation of daughter node probabilities16. The probability of any inference node is the sum of all of its daughter node probabilities.
Extended Data Figure 3: Inference algorithm for patient-level brain tumor diagnosis.
A patch-based classifier that uses high-magnification, high-resolution images for diagnosis requires a method to aggregate patch-level predictions into a single intraoperative diagnosis. Our inference algorithm performs a feedforward pass on each patch from a patient, filters the nondiagnostic patches (line 12), and stores the output softmax vectors in an RN x 13 array. Each column of the array, corresponding to each class, is summed and renormalized (line 22) to produce a probability distribution. We then used a thresholding procedure such that if greater than 90% of the probability density is nontumor/normal, that probability distribution is returned. Otherwise, the normal/nontumor class (grey matter, white matter, gliosis) probabilities are set to zero (line 31), the distribution renormalized, and returned. This algorithm leverages the observation that normal brain and nondiagnostic tissue imaged using SRH have similar features across patients resulting in high patch-level classification accuracy. Using the expected value of the renormalized patient-level probability distribution for the intraoperative diagnosis eliminates the need to train an additional classifier based on patch predictions.
Extended Data Figure 4. Prospective clinical trial design and recruitment.
a, Minimum sample size was calculated under the assumption that pathologists’ multiclass diagnostic accuracy ranges from 93% to 97% based on our previous experiments6 and that a clinically significant lower accuracy bound was less than 91%. We, therefore, selected an expected accuracy of 96% and equivalence/non-inferiority limit, or delta, of 5%, yielding a non-inferiority threshold accuracy of 91% or greater. Minimum sample size was 264 (black point) patients using an alpha of 0.05 and a power of 0.9 (beta = 0.1). b, Flowchart of specimen processing in both the control and experimental arms is shown. c, A total of 302 patients met inclusion criteria and were enrolled for intraoperative SRH imaging. Eleven patients were excluded at the time of surgery due to specimens that were below the necessary quality for SRH imaging. A total of 291 patients were imaged intraoperatively and 13 patients were subsequently excluded due to a Mahalanobis distance-based confidence score (See Extended Data Figure 5), resulting in a total of 278 patients included. d, Meningioma, pituitary adenomas, and malignant gliomas were the most common diagnoses in our prospective cohort. University of Michigan, University of Miami, and Columbia University recruited 55.0%, 26.6%, 18.4% of the total patients, respectively.
Extended Data Figure 5. Mahalanobis distance-based confidence score.
a, Pairwise comparison and b, principal component analysis of class conditional Mahalanobis distance-based confidence score for each layer output included in the ensemble. The confidence score from the mid- and high-level hidden features are correlated, which demonstrate that out-of-distribution samples result in greater Mahalanobis distances throughout the network. As previously described and observed in our results, out-of-distribution (i.e. rare tumors) are better detected in the representation space of deep neural networks, rather than the “label-overfitted” output space of the softmax layer23. c, Specimen-level predictions (black hashes, n = 478) and kernel density estimate from the trained LDA classifier for all specimens imaged during the trial period projected onto the linear discriminant axis. Trial and rare tumor cases were linearly separable resulting in all 13 rare tumor cases imaged during the trial period correctly identified. d, SRH mosaics of rare tumors imaged during the trial period are shown. Germinomas show classic large round neoplastic cells with abundant cytoplasm and fibrovascular septae with mature lymphocytic infiltrate. Choroid plexus papilloma shows fibrovascular cores lined with columnar cuboidal epithelium. Papillary craniopharyngioma have fibrovascular cores with well-differentiated monotonous squamous epithelium. Clival chordoma has unique bubbly cytoplasm (i.e., physaliferous cells). Scale bar, 50 μm.
Extended Data Figure 6. Error analysis of pathologist-based classification of brain tumors.
a, The true class probability and intersection over union values for each of the prospective clinical trial patients incorrectly classified by the pathologists. All 17 were correctly classified using SRH plus CNN. All incorrect cases underwent secondary review by two board-certified neuropathologists (S.C.P., P.C.) to ensure the specimens were 1) of sufficient quality to make a diagnosis and 2) contained tumor tissue. b, SRH mosaic from patient 21 (glioblastoma, WHO IV) is shown. Pathologist classification was metastatic carcinoma; however, CNN metastasis heatmap does not show high probability. Malignant glioma probability heatmap shows high probability over the majority of the SRH mosaic, with a 73.4% probability of patient-level malignant glioma diagnosis. High-magnification views show regions of hypercellularity due to tumor infiltration of brain parenchyma with damaged axons, activated lipid-laden microglia, mitotic figures, and multinucleated cells. c, SRH mosaic from patient 52 diagnosed with diffuse large B-cell lymphoma predicted to be metastatic carcinoma by pathologist. While CNN identified patchy areas of metastatic features within the specimen, the majority of the image was correctly classified as lymphoma. High-magnification views show atypical lymphoid cells with macrophage infiltration. Regions with large neoplastic cells share cytologic features with metastatic brain tumors, as shown in Figure 3. Scale bar, 50 μm.
Extended Data Figure 7: Activation maximization to elucidate SRH feature extraction using Inception-ResNet-v2.
a, Schematic diagram of Inception-ResNet-v2 shown with repeated residual blocks compressed. Residual connections and increased depth resulted in better overall performance compared to previous Inception architectures. b, To elucidate the learned feature representations produced by training the CNN using SRH images, we used activation maximization24. Images that maximally activate the specified filters from the 159th convolutional layer are shown as a time series of iterations of gradient ascent. A stable and qualitatively interpretable image results after 500 iterations, both for the CNN trained on SRH images and for ImageNet images. The same set of filters from the CNN trained on ImageNet are shown in order to provide direct comparison of the trained feature extractor for SRH versus natural image classification. c, Activation maximization images are shown for filters from the 5th, 10th, and 159th convolutional layers for CNN trained using SRH images only, SRH images after pretraining on ImageNet images, and ImageNet images only. The resulting activation maximization images for the ImageNet dataset are qualitatively similar to those found in previous publications using similar methods34. CNN trained using only SRH images produced similar classification accuracy compared to pretraining and activation maximization images that are more interpretable compared to those generated using a network pretrained on ImageNet weights.
Extended Data Figure 8. t-SNE plot of internal CNN feature representations for clinical trial patients.
We used the 1536-dimensional feature vector from the final hidden layer of the Inception-ResNet-v2 network to determine how individual patches and patients are represented by the CNN using t-distributed stochastic neighbor embedding (t-SNE), an unsupervised clustering method to visualize high-dimensional data. a, One hundred representative patches from each trial patient (n = 278) were sampled for t-SNE and are shown in the above plot as small, semi-transparent points. Each trial patient is plotted as a large point located at their respective mean patch position. Recognizable clusters form that correspond to individual diagnostic classes, indicating that tumor types have similar internal CNN representations. b, Grey and white matter form separable clusters from tumoral tissue, but also from each other. lipid-laden myelin in white matter has significantly different SRH features compared to grey matter with axons and glial cells in a neuropil background. c, Diagnostic classes that share cytologic and histoarchitectural features form neighboring clusters, such as malignant glioma, pilocytic astrocytoma, and diffuse lower grade glioma (i.e., glial tumors). Lymphoma and medulloblastoma are adjacent and share similar features of hypercellularity, high nuclear:cytoplasmic ratios, and little to no glial background in dense tumor.
Extended Data Figure 9. Methods and results of SRH segmentation.
a, A 1000×1000-pixel SRH image is shown with the corresponding grid of probability heatmap pixels that results from using a 300×300-pixel sliding window with 100-pixel step size in both horizontal and vertical directions. Scale bar, 50 μm. b, An advantage of this method is that the majority of the heatmap pixels are contained within multiple image patches and the probability distribution assigned to each heatmap pixel results from a renormalized sum of overlapping patch predictions. This has the effect of pooling the local prediction probabilities and generates a smoother prediction heatmap. c, For our example, each pixel of the inner 6×6 grid has 9 overlapping patches from which the probability distribution is determined. d, An SRH image of a meningioma, WHO grade I, from our prospective trial is shown as an example. Scale bar, 50 μm. e, The meningioma probability heatmap is shown after bicubic interpolation to scale image to the original size. Nondiagnostic prediction and ground truth is for the same SRH mosaic and is shown. f, The SRH semantic segmentation results of the full prospective cohort (n = 278) are plotted. The upper plot shows the mean IOU and standard deviation (i.e., averaged over SRH mosaics from each patient) for ground truth class (i.e., output classes). Note that the more homogenous or monotonous histologic classes (e.g., pituitary adenoma, white matter, diffuse lower grade gliomas) had higher IOU values compared to heterogeneous classes (e.g., malignant glioma, pilocytic astrocytoma). The lower plot shows the mean inference class IOU and standard deviation (i.e., either tumor or normal inference class) for each trial patient. Mean normal inference class IOU for the full prospective cohort was 91.1 ± 10.8 and mean tumor inference class IOU was 86.4 ± 19.0. g, As expected, mean ground truth class IOU values for the prospective patient cohort (n = 278) were correlated with patient-level true class probability (Pearson correlation coefficient, 0.811).
Extended Data Figure 10. Localization of metastatic brain tumor infiltration in SRH images.
a, Full SRH mosaic of a specimen collected at the brain-tumor margin of a patient with a metastatic brain tumor (non-small cell lung adenocarcinoma). b, Metastatic rests with glandular formation are dispersed among gliotic brain with normal neuropil. c, Three-channel RGB CNN-prediction transparency is overlaid on the SRH image for pathologist review intraoperatively with associated (d) patient-level diagnostic class probabilities. e, Class probability heatmap for metastatic brain tumor (IOU 0.51), nontumor (IOU 0.86), and nondiagnostic (IOU 0.93) regions within the SRH image are shown with ground truth segmentation. Scale bar, 50 μm.
Supplementary Material
Extended Data Video 1: Intraoperative video of clinical stimulated Raman histology and automated diagnosis using CNN
https://youtu.be/eDsdtIruJUs or https://umich.box.com/s/w627nnxmnuaokkc6pvbkzm3vk1ow03gr
The video shows the automated tissue-to-diagnosis pipeline described in Figure 1 and used in the clinical trial. Each of the three steps, 1) image acquisition, 2) image processing, and 3) diagnostic prediction, are labelled onscreen. At the time of surgery, the CNN-predicted diagnosis was diffuse lower grade glioma (unnormalized probability, 30.6% (shown in video); renormalized probability, 83.0%). Conventional intraoperative H&E diagnosis was “atypical glial cells, favor glioma” and final histopathologic diagnosis was diffuse glioma, WHO grade II.
Acknowledgments
The authors would like to thank Tom Cichonski for manuscript editing.
Funding Sources: This work was supported by the NIH National Cancer Institute (R01CA226527-02), Neurosurgery Research Education Fund, University of Michigan MTRAC, and The Cook Family Foundation.
Footnotes
Competing interests: D.A.O. is an advisor and shareholder of Invenio Imaging, Inc., a company developing SRH microscopy systems. C.W.F., Z.U.F., and J.T. are employees and shareholders of Invenio Imaging, Inc.
Additional information
Extended data are available for this paper at https://doi.org/….
Reprints and permissions information is available at http://www.nature.com/reprints.
References
- 1.Sullivan R et al. Global cancer surgery: delivering safe, affordable, and timely cancer surgery. Lancet Oncol. 16, 1193–1224 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Novis DA & Zarbo RJ Interinstitutional comparison of frozen section turnaround time. A College of American Pathologists Q-Probes study of 32868 frozen sections in 700 hospitals. Arch. Pathol. Lab. Med 121, 559–567 (1997). [PubMed] [Google Scholar]
- 3.Gal AA & Cagle PT The 100-year anniversary of the description of the frozen section procedure. JAMA 294, 3135–3137 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Robboy SJ et al. Pathologist workforce in the United States: I. Development of a predictive model to examine factors influencing supply. Arch. Pathol. Lab. Med 137, 1723–1732 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Freudiger CW et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 322, 1857–1861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orringer DA et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat Biomed Eng 1, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji M et al. Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci. Transl. Med 5, 201ra119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Top 100 Lab Procedures Ranked by Service. www.cms.gov (2017). Available at: www.cms.gov/Research-Statistics-Data-and-Systems/Statistics-Trends-and-Reports. (Accessed: 4th November 2019)
- 9.Metter DM, Colgan TJ, Leung ST, Timmons CF & Park JY Trends in the US and Canadian Pathologist Workforces From 2007 to 2017. JAMA Netw Open 2, e194337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollon TC et al. Rapid Intraoperative Diagnosis of Pediatric Brain Tumors Using Stimulated Raman Histology. Cancer Res. 78, 278–289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louis DN et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 131, 803–820 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Ji M et al. Detection of human brain tumor infiltration with quantitative stimulated Raman scattering microscopy. Sci. Transl. Med 7, 309ra163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krizhevsky A, Sutskever I & Hinton GE ImageNet Classification with Deep Convolutional Neural Networks in Advances in Neural Information Processing Systems 25 (eds. Pereira F, Burges CJC, Bottou L & Weinberger KQ) 1097–1105 (Curran Associates, Inc., 2012). [Google Scholar]
- 14.Gulshan V et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 316, 2402–2410 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Titano JJ et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat. Med 24, 1337–1341 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Esteva A et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litjens G et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep 6, 26286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coudray N et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med 24, 1559–1567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He K, Zhang X, Ren S & Sun J Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. arXiv [cs.CV] (2015). [Google Scholar]
- 20.Szegedy C, Ioffe S, Vanhoucke V & Alemi AA Inception-v4, inception-resnet and the impact of residual connections on learning. in AAAI 4, 12 (2017). [Google Scholar]
- 21.Topol EJ High-performance medicine: the convergence of human and artificial intelligence. Nat. Med 25, 44–56 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Ostrom QT et al. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro. Oncol 19, v1–v88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K, Lee K, Lee H & Shin J A Simple Unified Framework for Detecting Out-of-Distribution Samples and Adversarial Attacks. arXiv [stat.ML] (2018). [Google Scholar]
- 24.Erhan D, Bengio Y, Courville A & Vincent P Visualizing higher-layer features of a deep network. University of Montreal 1341, 1 (2009). [Google Scholar]
- 25.Lu F-K et al. Label-Free Neurosurgical Pathology with Stimulated Raman Imaging. Cancer Res. 76, 3451–3462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohe S, Colmenero I, McConville C & Peet A Immunohistochemical staining of lipid droplets with adipophilin in paraffin-embedded glioma tissue identifies an association between lipid droplets and tumour grade. J Histol Histopathol 4, 4 (2017). [Google Scholar]
- 27.Chen P-HC et al. An augmented reality microscope with real-time artificial intelligence integration for cancer diagnosis. Nat. Med 25, 1453–1457 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Viola KV et al. Mohs micrographic surgery and surgical excision for nonmelanoma skin cancer treatment in the Medicare population. Arch. Dermatol 148, 473–477 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Hoesli RC, Orringer DA, McHugh JB & Spector ME Coherent Raman Scattering Microscopy for Evaluation of Head and Neck Carcinoma. Otolaryngol. Head Neck Surg 157, 448–453 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter CL, Allen C & Henson DE Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer 63, 181–187 (1989). [DOI] [PubMed] [Google Scholar]
- 31.Ratnavelu NDG et al. Intraoperative frozen section analysis for the diagnosis of early stage ovarian cancer in suspicious pelvic masses. Cochrane Database Syst. Rev 3, CD010360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sottoriva A et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. U. S. A 110, 4009–4014 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dammers R et al. Towards improving the safety and diagnostic yield of stereotactic biopsy in a single centre. Acta Neurochir. 152, 1915–1921 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeiler MD & Fergus R Visualizing and Understanding Convolutional Networks in Computer Vision – ECCV 2014 818–833 (Springer International Publishing, 2014). [Google Scholar]
References
- 35.Freudiger CW et al. Stimulated Raman Scattering Microscopy with a Robust Fibre Laser Source. Nat. Photonics 8, 153–159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y et al. Detecting Cancer Metastases on Gigapixel Pathology Images. arXiv [cs.CV] (2017). [Google Scholar]
- 37.Yosinski J, Clune J, Bengio Y & Lipson H How transferable are features in deep neural networks? arXiv [cs.LG] (2014). [Google Scholar]
- 38.Abadi M et al. Tensorflow: a system for large-scale machine learning. in OSDI 16, 265–283 (2016). [Google Scholar]
- 39.Hou L et al. Patch-based Convolutional Neural Network for Whole Slide Tissue Image Classification. arXiv [cs.CV] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin Z et al. How convolutional neural networks see the world --- A survey of convolutional neural network visualization methods. Mathematical Foundations of Computing 1, 149–180 (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Video 1: Intraoperative video of clinical stimulated Raman histology and automated diagnosis using CNN
https://youtu.be/eDsdtIruJUs or https://umich.box.com/s/w627nnxmnuaokkc6pvbkzm3vk1ow03gr
The video shows the automated tissue-to-diagnosis pipeline described in Figure 1 and used in the clinical trial. Each of the three steps, 1) image acquisition, 2) image processing, and 3) diagnostic prediction, are labelled onscreen. At the time of surgery, the CNN-predicted diagnosis was diffuse lower grade glioma (unnormalized probability, 30.6% (shown in video); renormalized probability, 83.0%). Conventional intraoperative H&E diagnosis was “atypical glial cells, favor glioma” and final histopathologic diagnosis was diffuse glioma, WHO grade II.
Data Availability Statement
A University of Michigan IRB protocol (HUM00083059) was approved for the use of human brain tumor specimens in this study. To obtain these samples or SRH images, contact D.A.O. Code repository for network training, evaluation, and visualizations is publicly available at https://github.com/toddhollon/srh_cnn.