Abstract
UVB radiation results in the formation of potentially mutagenic photoproducts in the DNA of epidermal skin cells. In vitro approaches have demonstrated that the nucleotide excision repair (NER) machinery removes UV photoproducts from DNA in the form of small (~30-nt-long), excised, damage-containing DNA oligonucleotides (sedDNAs). Though this process presumably takes place in human skin exposed to UVB radiation, sedDNAs have not previously been detected in human skin. Using surgically discarded human skin, we have optimized the detection of the sedDNA products of NER from small amounts of human epidermal tissue ex vivo within minutes of UVB exposure and after UVB doses that normally lead to minimal erythema. Moreover, sedDNA generation was inhibited by treatment of skin explants with spironolactone, which depletes the epidermis of the essential NER protein XPB to mimic the skin of xeroderma pigmentosum patients. Time course experiments revealed that a partially degraded from of the sedDNAs could be readily detected even 12 hours following UVB exposure, which indicates that these repair products are relatively stable in human skin epidermis. Together, these data suggest that sedDNA detection may be a useful assay for determining how genetic, environmental, and other factors influence NER activity in human skin epidermis and whether abnormal sedDNA processing contributes to photosensitive skin disorders.
Keywords: Nucleotide excision repair, UV radiation, human skin, DNA repair activity, DNA oligonucleotides
1. INTRODUCTION
The epidermis of human skin is routinely exposed to UVB wavelengths of sunlight that induce the formation of cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6-4) pyrimidone photoproducts [(6-4)PPs] in genomic DNA [1]. These UV photoproducts interfere with DNA and RNA polymerase movement and can result in cell death and/or mutagenesis, which contribute to erythema and skin carcinogenesis, respectively. Nucleotide excision repair (NER) is the sole system in humans for removing UV photoproducts from genomic DNA [2–4], and mutations in NER genes are found in patients with the disease xeroderma pigmentosum (XP) [5], which predisposes individuals to photosensitivity, premature aging, and skin cancer.
Determining how various genetic, environmental, dietary, and other factors contribute to inter-individual differences in DNA repair activity is important for understanding cancer susceptibility [6]. Taking advantage of the fact that the NER system generates small, excised, damage-containing DNA oligonucleotides (sedDNAs) as part of its mechanism of action to remove UV photoproducts form the genome (Figure 1A) [2,7], methods have been developed to detect sedDNAs in cultured cells in vitro following exposure to UV radiation and other DNA damaging agents [8–10]. Though damage removal/sedDNA generation via NER is important for preventing mutagenesis and skin carcinogenesis, the observation that defects in nucleic acid degradation contribute to autoimmunity and inflammation [11,12] suggests that defects in sedDNA degradation could contribute to various pathologies associated with UV-exposed skin. In this report, sedDNAs are shown to be efficiently isolated and sensitively detected in small amounts of human skin epidermis even after exposure to sub-erythemal doses of UVB radiation. Moreover, sedDNAs can be detected within minutes of UVB exposure but remain in the skin epidermis for hours following UVB exposure. Thus, the detection of the sedDNA products of NER in human skin epidermis may therefore provide a new approach for measuring inter-individual differences in NER activity and help to identify factors that affect repair activity, inflammation, and autoimmunity in the context of human skin epidermis in vivo.
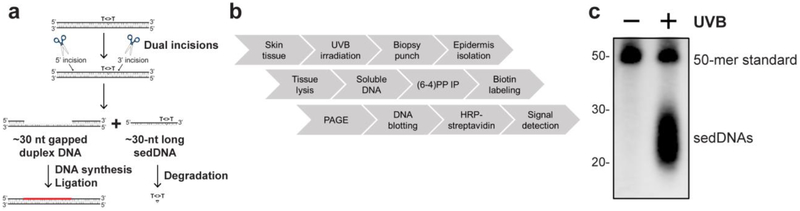
Figure 1. Method for detecting sedDNAs in human skin epidermis.
(a) Schematic of the nucleotide excision repair mechanism, in which UVB-damaged bases (indicated by thymine dimer, T<>T) are removed by via dual incisions to generate a sedDNA. (b) Sample workflow for skin processing and detection of the sedDNA products of NER. Small, 6 mm punch biopsies of discarded human skin were obtained from UVB-irradiated skin and snap frozen. Epidermal tissue was removed from the biopsy by heat shock and lysed. Soluble DNAs obtained from the epidermal lysates were then immunoprecipitated with (6-4)PP antibody, 3’ end-labeled with a biotinylated nucleotide, electrophoresed on a polyacrylamide gel, transferred to a nylon membrane, and then detected with HRP-coupled streptavidin and chemiluminescence. (c) The image shows a typical experimental result from skin harvested 1 hr after exposure to 800 J/m2 of UVB radiation. sedDNA labeling reactions were supplemented with a 50-mer ssDNA oligonucleotide standard. Location of DNA size markers (nt) is indicated.
2. MATERIALS AND METHODS
2.1. Skin treatments
Discarded, de-identified human skin from routine panniculectomies was used in these studies. Patient consent for experiments was not required because de-identified, leftover surgical human tissue is considered discarded material by our institution, and thus the studies were considered exempt. Small, approximately 2.5 x 2.5 cm sections of skin were exposed to the indicated fluences of UVB radiation using a Philips F20T12 broadband UVB light source at a dose rate of 5 J/m2/sec and then placed in a pyrex containiner in a 37°C water bath for the indicated periods of time. Small, 6 mm punch biopsies were obtained, frozen in liquid nitrogen, and stored at −80°C until ready for further processing. Skin explants (8 mm punch biopsies) were incubated above medium containing DMSO or 20 μM spironolactone to deplete XPB for 3 days as previously described [13].
2.2. sedDNA detection and quantification
To isolate and detect the sedDNA products of NER in human skin, the skin punch biopsies were briefly subjected to a heat shock (6 sec at 60°C and 9 sec in ice cold water) to facilitate release of the epidermis, which was scraped off the biopsy with a curette. The epidermal tissue was then disrupted with several strokes of a pellet pestle in 200 μl of cold Triton X-100 lysis buffer (20 mM Tris-HCl, pH7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 1% Triton X-100) and incubated on ice for 15-20 min with occasional vortexing. Following centrifugation for 60 min at maximum speed in a microcentrifuge at 4°C to pellet genomic DNA, the soluble lysates were digested with RNase A (5 μl of a 25 mg/ml stock) for 5 min at room-temperature and then treated with SDS (8 μl of a 10% solution) and Proteinase K (4 μl of a 20 U/μl stock) for 15 min at 50-60°C. The samples were then extracted twice with an equal volume of 25:24:1 phenol:chloroform:isoamylalcohol, once with chloroform, and precipitated overnight in ethanol in the presence of 20 μg of glycogen. Following a 15 min centrifugation at maximum speed in a microcentrifuge at 4°C, the DNA pellets were washed with 1 ml of cold 70% ethanol, centrifuged again for 5 min, and then air dried. The subsequent methods of sedDNA labeling and detection from skin epidermis were similar to that reported for cultured cells [9,10,14,15]. Briefly, the soluble DNAs were resuspended in 50 μl of TE (10 mM Tris-HCl pH 8.0, 1 mM EDTA), immunoprecipitated with 5 μl of Dynabead slurry bound to anti-(6-4)PP antibody, washed with Wash Buffer I (20 Mm Tris-HCl pH 8.0, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, and 0.1% SDS), Wash Buffer II (20 mM Tris-HCl pH 8.0, 2 mM EDTA, 500 mM NaCl, 1% Triton X-100, 0.1% SDS), Wash Buffer III (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 150 mM LiCl, 1% Nonidet P-40, 1% sodium deoxycholate), Wash Buffer IV (100 mM Tris-HCl pH 8.0, 1 mM EDTA, 500 mM LiCl, 1% Nonidet-P40, 1% sodium deoxycholate), and twice with TE buffer. The DNAs were eluted with 100 μl of elution buffer (50 mM NaHCO3, 1% SDS, and 20 μg.ml glycogen) at 65°C for 15 min, extracted with phenol-chloroform, precipitated in ethanol, and dried. DNAs were then labeled with 20 units of terminal deoxynucleotidyl transferase (New England Biolabs) and 10 μM of biotin-11-dUTP in 50 μl reactions containing Terminal Transferase Reaction Buffer (New England Biolabs) and 2 fmol of a 50-mer ssDNA control oligonucleotide. Reactions were stopped with 10 mM EDTA, and DNAs were purified following treatment with RNase A (10-20 μg/ml) and RNase T1 (30-50 units/ml) for 20 min at 37°C, treatment with proteinase K (0.4 mg/ml) in the presence of 0.4% SDS, extraction with phenol-chloroform, and precipitation in ethanol. Purified DNAs were separated on 10-12% TBE-urea gels in 1X TBE at 150 V, transferred to a modified nylon Biodyne B membrane (Pierce) in 0.5X TBE buffer at 300 mA for 1 hr, and then fixed by UV crosslinking. The membrane was incubated with PBS containing 2% SDS for 30 min and then incubated for 30 min with HRP-conjugated streptavidin in the same buffer. Membranes were next washed three times with PBS containing 0.5% SDS and then incubated with 200 mM Tris-HCl (pH 9.0) containing 10 mM MgCl2 for 5 min. Chemiluminescence was visualized with ECL reagents (GE Healthcare) on an ImageQuant LAS 4000 Mini apparatus (GE Healthcare) and quantified with ImageQuantTL software (GE Healthcare). For the data presented in Figures 2–4, the chemiluminescent sedDNA signals for each sample were normalized to the 50-mer ssDNA control oligonucleotide signal and then to the highest sedDNA/50-mer signal on each blot (which was set to an arbitrary value of 100).
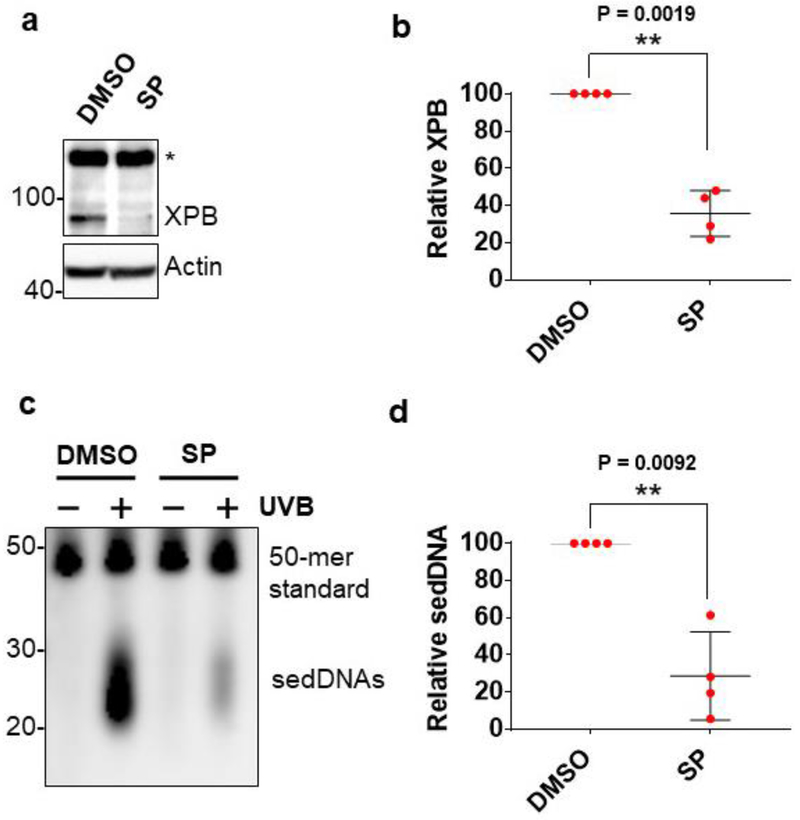
Figure 2. sedDNA generation in human skin is dependent on the NER protein XPB.
(a) Medium below skin biopsy explants were supplemented with vehicle (DMSO) or spironolactone (SP, 20 μM) for 3 days before immunoblot analysis of skin epidermal protein. (b) The graph shows the level of XPB protein (average and standard deviation, normalized to actin) in SP-treated skin epidermis relative to DMSO-treated skin epidermis (n=4). (c) The image shows a representative experiment detecting sedDNAs from skin biopsies treated as in (a) and harvested 1 hr after exposure to 800 J/m2 of UVB radiation. (d) The graph quantifies the relative level of sedDNAs in DMSO- and SP-treated skin epidermis (n=4).
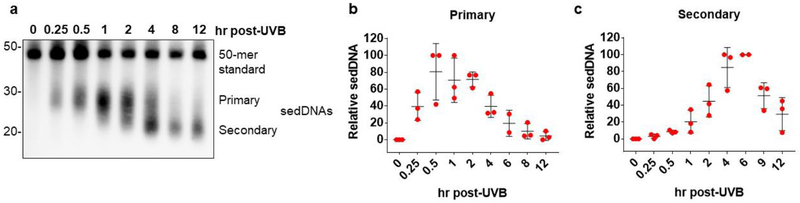
Figure 4. Kinetics of sedDNA generation in UVB-irradiated human skin.
(a) Human skin was exposed to 800 J/m2 of UVB radiation at a dose rate of 5 J/m2/sec and then biopsies were taken at the indicated time points for detection of (6-4)PP-containing sedDNAs. The primary, full-length and secondary, partially degraded sedDNA species are indicated. (b) Relative quantification (average and standard deviation) of the primary and secondary (6-4)PP-containing-sedDNAs from different skin samples treated as in (a) (n=3).
2.3. Protein Immunoblotting
Epidermal tissues were lysed in RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCI, 2 mM EDTA, 1% triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing a 1:200 dilution of protease inhibitor cocktail (Sigma), 1 mM DTT, 0.1 mM PMSF, 10 mM NaF, 1 mM Na2VO3, and 10 mM glycerophosphate and analyzed by immunoblotting as previously described [13].
2.4. Statistical analyses
For each experimental approach, the number of independent skin samples that were used is provided in the appropriate figure legend. Two-tailed, paired or unpaired Student’s t-tests were carried out where indicated to determine whether differences between treatment samples reached statistical significance (p<0.05).
3. RESULTS
3.1. Method for detecting sedDNAs in human skin
UVB photoproducts are removed from DNA through a dual incision mechanism, in which the damaged nucleotide bases are cut out from DNA in the form of a sedDNA approximately 24-32 nt in length (Figure 1A) [2,3,7]. The generation of UVC-induced sedDNAs has been extensively studied using in vitro systems and recently observed in cultured human cells [8–10]. Therefore, to determine whether sedDNAs can be isolated and detected in human skin epidermis, discarded human skin from routine panniculectomies was exposed to UVB radiation and then incubated for a short period of time to allow for NER to take place. Isolated epidermal tissue from small punch biopsies of control- or UVB-irradiated skin was then subjected to tissue lysis to purify soluble DNAs. Though UVB radiation induces the formation of both pyrimidine (6-4) pyrimidone photoproducts [(6-4)PPs] and cyclobutane pyrimidine dimers (CPDs) in genomic DNA, (6-4)PPs are repaired faster than CPDs, and thus an anti-(6-4)PP antibody was used here to enrich for (6-4)PP-containing sedDNAs from the total population of soluble DNAs. The immunoprecipitated sedDNAs were next subjected to 3’-end labeling with terminal transferase and a biotinylated nucleotide, transferred to a nylon membrane, probed with HRP-streptavidin, and finally detected with a chemiluminescent substrate [15]. A schematic of this approach is provided in Figure 1B, and a sample experimental result demonstrating the UVB-dependent generation of small DNAs approximately 20 to 30 nt in length is shown in Figure 1C. Importantly, these small DNAs were not observed in non-irradiated skin, which indicates that the assay is highly specific for the detection of sedDNAs generated following UVB exposure.
3.2. sedDNA generation in human skin requires the NER factor XPB
To demonstrate that sedDNA generation in UVB-irradiated skin is indeed dependent on the NER machinery, skin punch biopsies were incubated ex vivo for 3 days above a basal medium containing spironolactone (SP), which has been shown to lead to the proteolytic degradation of the essential NER protein XPB in human cells in vitro [16] and skin epidermis ex vivo [13]. As shown in Figure 2A, spironolactone treatment resulted in a nearly 70% reduction in XPB protein levels in human skin epidermis in several different skin samples. Importantly, and consistent with the role of XPB in NER, this reduction in XPB protein was closely associated with a significant reduction in sedDNA levels following UVB exposure of the same skin samples (Figure 2B). These results clearly demonstrate that sedDNA production in UVB-irradiated skin requires an intact NER system.
3.3. sedDNAs are generated in a UVB dose-dependent manner
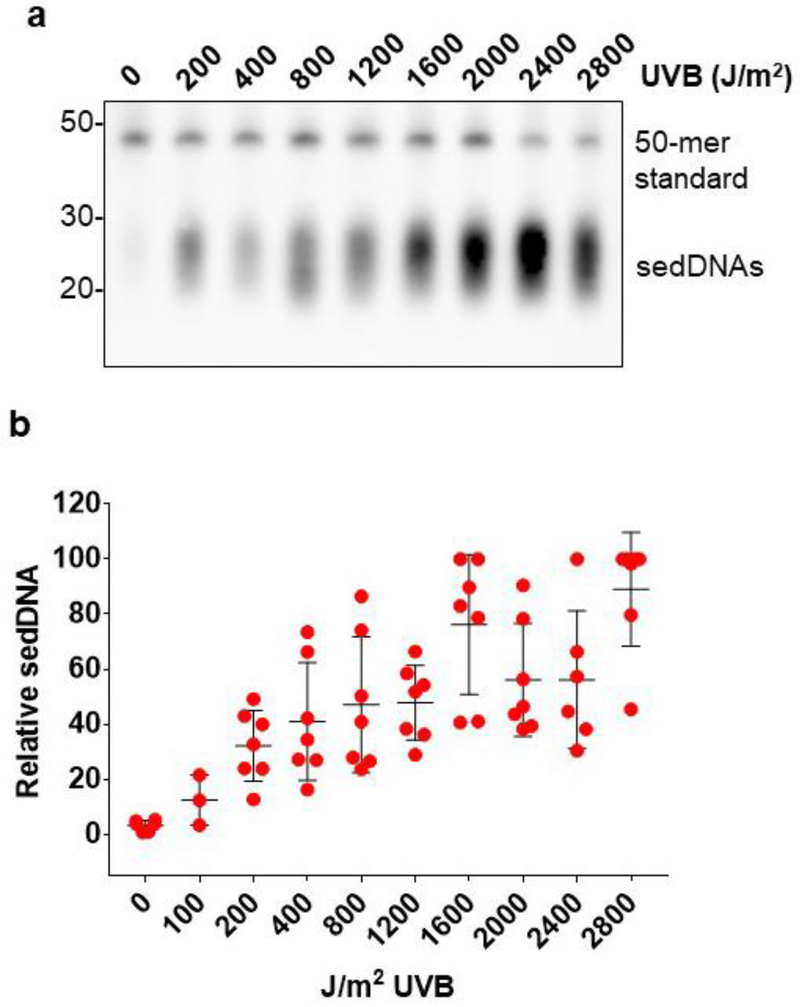
To examine the sensitivity of the sedDNA detection assay for monitoring NER activity in human skin epidermis, skin samples were exposed to different doses of UVB and then incubated for 1 hr. As shown in Figure 3, sedDNA abundance was strongly correlated with the UVB dose. Importantly, even sub-erythemal doses of UVB (100-200 J/m2) led to detectable sedDNA signals above the background level found in non-irradiated skin. We observed that the amount of sedDNAs generated upon UVB irradiation in human skin epidermis was largely dose-dependent up to approximately 1,600 J/m2, above which NER activity appeared to begin to become saturated. Thus, these data indicate that the sedDNA detection assay allows for the sensitive detection of NER activity even in response to very low doses of UVB that are physiologically relevant to UVB exposures in daily life.
Figure 3. UVB induces sedDNA generation in a dose-dependent manner.
(a) Representative experiment detecting sedDNAs generated 1 hr after exposure to the indicated doses of UVB radiation. (b) Relative quantification (average and standard deviation) of epidermal sedDNAs from several different skin samples (n=3-7 for each dose) treated as in (a). Densitometry was used to quantify sedDNA (and 50-mer) signal for each sample, and the highest sedDNA/50-mer signal on each blot was set to an arbitrary value of 100. All other sedDNA signals in each experiment were compared to this value.
3.4. Kinetics of sedDNA generation in human skin
In cultured cells in vitro, sedDNAs have been detected within minutes of exposure to UVC radiation [9], which shows that the NER machinery responds immediately to the presence of DNA damage by beginning to excise damaged nucleotides. Using cell-free extracts with defined DNA substrates [17] and UV-irradiated cultured cells [8,9,18], two major species of sedDNAs have been reported to be detected. These include a larger species of sedDNAs nearly 30 nt in length and a partially degraded species approximately 20 nt in length. These two sedDNA species have been referred to as the primary and secondary products of NER [18,19], and have been shown to be primarily bound to the NER factors TFIIH (transcription factor II-H) and RPA (replication protein A), respectively [8,17]. Thus, current models suggest that sedDNAs are initially excised from DNA in association with TFIIH before being released in an ATP-dependent manner to become bound by RPA and undergo limited nucleolytic degradation to form the secondary, partially degraded species of sedDNAs [17,19].
To determine whether a similar phenomenon occurs in human skin ex vivo, small pieces of human skin were exposed to UVB radiation and then incubated for various periods of time. As shown in Figure 4A, the primary sedDNA products of NER were clearly detectable in skin epidermis within 15 minutes of exposure, and the level of the primary products peaked approximately 30 min to 1 hr after UVB exposure. Interestingly, nearly identical results have been reported in cultured cells in vitro [8,9]. Moreover, in both cultured cells in vitro and human skin epidermis ex vivo (Figure 4A), (6-4)PP-containing sedDNAs were primarily of a smaller size close to 20 nt in length at later time points after UV exposure. However, in contrast to observations in cultured cells in vitro, in which (6-4)PP-containing sedDNAs are almost completely degraded by 6 hr following UV exposure, (6-4)PP-containing sedDNAs could still be readily detected even 12 hr after UVB exposure in skin epidermis ex vivo. These results suggest that once excised from genomic DNA, (6-4)PP-containing sedDNAs are only slowly and partially degraded by cellular nucleases in human skin epidermis.
4. DISCUSSION
Understanding how genetic, environmental, dietary, pharmacological, and other factors impact UV photoproduct removal from human skin epidermis is important for predicting skin cancer risk. Though in vitro studies with cultured cells are critical for carrying out this work, it is also important to understand the interplay of these factors within the context of human skin. However, many approaches aimed at understanding inter-individual differences in DNA repair capacity [6] are not readily applicable for use in skin. Currently, the most widely used technique for monitoring changes in UV photoproduct levels in human skin is immunohistochemical staining of CPDs. However, because CPDs are only slowly removed from the genome, this method frequently requires extended periods of time following irradiation to detect clear reductions in staining intensity. Moreover, CPD dilution due to DNA replication and cell division [21] or apoptosis in the skin may lead to artifactual reductions in apparent CPD levels. Lastly, procedures that utilize formaldehyde fixation of skin tissue may crosslink sedDNAs to genomic DNA [18], and thus excised UV photoproducts may appear as unrepaired damage.
The results shown here demonstrate that the NER machinery begins to remove (6-4)PPs photoproducts from human skin epidermis within minutes of UVB exposure (Figure 4), and thus, (6-4)PP-sedDNA detection may offer a more sensitive approach for quantifying the basal enzymatic activity of the NER system in human skin exposed to physiological doses of UVB radiation within a single sample of skin epidermis (Figure 3). However, it should be noted that sedDNAs provide a measure of the activity of the NER system and do not necessarily take into account the total amount of photoproduct damage that is induced by UVB exposure. Thus differences in damage formation between individual skin samples or throughout the different layers of the epidermis due to melanin content [22] or epidermal thickness may influence the accuracy of sedDNA quantification as a measure of NER activity and should be considered in future studies. Moreover, differences in sedDNA degradation by as-yet-unknown nucleases may influence the accuracy of using sedDNA detection as a measure of NER activity. Nonetheless, as demonstrated in Figure 2, the method described here could be useful in testing the how pharmaceuticals and other factors specifically impact NER activity in human skin explants ex vivo. Through DNA sequencing of the sedDNAs with eXcision Repair (XR-seq) [23], it should also be possible to measure NER activity at nucleotide resolution throughout the epidermal cell genome. Though punch biopsies were used in the work here to isolate defined areas of epidermal tissue, less invasive methods involved the scraping of skin epidermis should also be possible to allow for the detection of epidermal sedDNAs in human subjects exposed to natural or artificial sources of UV radiation.
Little information is currently known regarding the ultimate fate of the sedDNA products of NER in either cultured cells or human skin [19,23]. Thus, an additional future application of skin sedDNA detection will be to understand how epidermal skin cells process and degrade sedDNAs following excision from the genome. A recent study using cultured cells indicated that sedDNA degradation occurs independent of apoptotic signaling [14], and thus important work remains to be done in this area. Efficient degradation of sedDNAs may be necessary to avoid aberrant stimulation of innate immune signaling pathways [11,12], and therefore defective clearance of sedDNAs may contribute to autoimmunity or other photosensitive skin reactions.
Highlights.
UV photoproducts are removed from DNA in the form of sedDNAs
sedDNAs have been detected for the first time in UVB-irradiated human skin ex vivo
sedDNA detection may allow for quantification of DNA repair activity in human skin
Acknowledgements
The authors thank the WSU Proteome Analysis Laboratory for the use of equipment to carry out this work.
Funding
This work was supported in part by grants from the National Institute of General Medical Sciences (GM130583 to M.G.K.), the Development of New Chemical Medial Measurement Standard Technology Program funded by the Korea Research Institute of Standards and Science (KRISS-2019-GP2019-0009), and the Nano Material Technology Development Program (NRF-2014M3A7B6020163) of the National Research Foundation (NRF) funded by the Ministry of Science and ICT.
Abbreviations
- NER
nucleotide excision repair
- sedDNA
small, excised, damage-containing DNA oligonucleotide
- SP
spironolactone
- XP
xeroderma pigmentosum
- XPB
XP group B
- (6-4)PP
pyrimidine (6-4) pyrimidone photoproduct
- CPD
cyclobutane pyrimidine dimer
- TFIIH
transcription factor II-H
- RPA
replication protein A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing financial interest
The authors declare that they have no conflict of interest.
References
- [1].Cadet J, Grand A, Douki T. Solar UV radiation-induced DNA bipyrimidine photoproducts: Formation and mechanistic insights, Top. Curr. Chem 356 (2015) 249–275. [DOI] [PubMed] [Google Scholar]
- [2].Sancar A. Mechanisms of DNA repair by photolyase and excision nuclease (nobel lecture), Angew. Chem. Int. Ed Engl 55 (2016) 8502–8527. [DOI] [PubMed] [Google Scholar]
- [3].Reardon JT, Sancar A. Nucleotide excision repair, Prog. Nucleic Acid Res. Mol. Biol 79 (2005) 183–235. [DOI] [PubMed] [Google Scholar]
- [4].Wood RD. Nucleotide excision repair in mammalian cells, J. Biol. Chem 272 (1997) 23465–23468. [DOI] [PubMed] [Google Scholar]
- [5].DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum, J. Invest. Dermatol 132 (2012) 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nagel ZD, Chaim IA, Samson LD. Inter-individual variation in DNA repair capacity: A need for multi-pathway functional assays to promote translational DNA repair research, DNA Repair (Amst). 19 (2014) 199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Huang JC, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5’ and the 6th phosphodiester bond 3’ to the photodimer, Proc. Natl. Acad. Sci. U. S. A 89 (1992) 3664–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hu J, Choi JH, Gaddameedhi S, Kemp MG, Reardon JT, Sancar A. Nucleotide excision repair in human cells: Fate of the excised oligonucleotide carrying DNA damage in vivo, J. Biol. Chem 288 (2013) 20918–20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Choi JH, Gaddameedhi S, Kim SY, Hu J, Kemp MG, Sancar A. Highly specific and sensitive method for measuring nucleotide excision repair kinetics of ultraviolet photoproducts in human cells, Nucleic Acids Res. 42 (2014) e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Choi JH, Kim SY, Kim SK, Kemp MG, Sancar A. An integrated approach for analysis of the DNA damage response in mammalian cells: Nucleotide excision repair, DNA damage checkpoint, and apoptosis, J. Biol. Chem 290 (2015) 28812–28821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Elkon KB. Review: Cell death, nucleic acids, and immunity: Inflammation beyond the grave, Arthritis Rheumatol. 70 (2018) 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Unterholzner L. The interferon response to intracellular DNA: Why so many receptors? Immunobiology. 218 (2013) 1312–1321. [DOI] [PubMed] [Google Scholar]
- [13].Kemp MG, Krishnamurthy S, Kent MN, Schumacher DL, Sharma P, Excoffon KJDA, Travers JB. Spironolactone depletes the XPB protein and inhibits DNA damage responses in UVB-irradiated human skin, J. Invest. Dermatol 139 (2019) 448–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Baek S, Han S, Kang D, Kemp MG, Choi JH. Simultaneous detection of nucleotide excision repair events and apoptosis-induced DNA fragmentation in genotoxin-treated cells, Sci. Rep 8 (2018) 2265-018-20527-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Song J, Kemp MG, Choi JH. Detection of the excised, damage-containing oligonucleotide products of nucleotide excision repair in human cells, Photochem. Photobiol 93 (2017) 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Alekseev S, Ayadi M, Brino L, Egly JM, Larsen AK, Coin F. A small molecule screen identifies an inhibitor of DNA repair inducing the degradation of TFIIH and the chemosensitization of tumor cells to platinum, Chem. Biol 21 (2014) 398–407. [DOI] [PubMed] [Google Scholar]
- [17].Kemp MG, Reardon JT, Lindsey-Boltz LA, Sancar A. Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair, J. Biol. Chem 287 (2012) 22889–22899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kemp MG, Gaddameedhi S, Choi JH, Hu J, Sancar A. DNA repair synthesis and ligation affect the processing of excised oligonucleotides generated by human nucleotide excision repair, J. Biol. Chem 289 (2014) 26574–26583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kemp MG, Hu J. PostExcision events in human nucleotide excision repair, Photochem. Photobiol 93 (2017) 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Berube R, Drigeard Desgarnier MC, Douki T, Lechasseur A, Rochette PJ. Persistence and tolerance of DNA damage induced by chronic UVB irradiation of the human genome, J. Invest. Dermatol 138 (2018) 405–412. [DOI] [PubMed] [Google Scholar]
- [21].Shih BB, Farrar MD, Cooke MS, Osman J, Langton AK, Kift R, Webb AR, Berry JL, Watson REB, Vail A, de Gruijl FR, Rhodes LE. Fractional sunburn threshold UVR doses generate equivalent vitamin D and DNA damage in skin types I-VI but with epidermal DNA damage gradient correlated to skin darkness, J. Invest. Dermatol 138 (2018) 2244–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hu J, Li W, Adebali O, Yang Y, Oztas O, Selby CP, Sancar A. Genome-wide mapping of nucleotide excision repair with XR-seq, Nat. Protoc 14 (2019) 248–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kemp MG, Sancar A. DNA excision repair: Where do all the dimers go? Cell. Cycle 11 (2012) 2997–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]