Abstract
The plant secretome is usually considered in the frame of proteomics, aiming at characterizing extracellular proteins, their biological roles and the mechanisms accounting for their secretion in the extracellular space. In this review, we aim to highlight recent results pertaining to secretion through the conventional and unconventional protein secretion pathways notably those involving plant exosomes or extracellular vesicles. Furthermore, plants are well known to actively secrete a large array of different molecules from polymers (e.g. extracellular RNA and DNA) to small compounds (e.g. ATP, phytochemicals, secondary metabolites, phytohormones). All of these play pivotal roles in plant-fungi (or oomycetes) interactions, both for beneficial (mycorrhizal fungi) and deleterious outcomes (pathogens) for the plant. For instance, recent work reveals that such secretion of small molecules by roots is of paramount importance to sculpt the rhizospheric microbiota. Our aim in this review is to extend the definition of the plant and fungal secretomes to a broader sense to better understand the functioning of the plant/microorganisms holobiont. Fundamental perspectives will be brought to light along with the novel tools that should support establishing an environment-friendly and sustainable agriculture.
Keywords: agriculture, apoplast, extracellular vesicles, fungal effectors, plant–fungi interactions, root exudates, whole and integrated secretomics
Introduction
Plants and fungi secrete a wide range of molecules into the extracellular space, where they play crucial roles in signaling, development and stress responses (Delaunois et al., 2014). Proteins constitute the most intensively studied group of these molecules. Until recently, current paradigm in plant-microbe interactions suggested that secreted proteins are synthesized and delivered through the conventional endoplasmic reticulum (ER) secretory pathway, which is based on the specific recognition of N-terminally located transit peptides (Agrawal et al., 2010; Giraldo et al., 2013). However, recent work disclosed the secretion of a new type of secreted proteins, devoid of transit peptide, referred to as leaderless secretory proteins (LSPs) (Wang et al., 2017), supporting the existence of novel secretory mechanisms independent of the classical ER-Golgi pathway. These unconventional secretion pathways involve small extracellular vesicles (EVs) for the export of proteins to the extracellular milieu and have recently been shown to mediate plant defense against invasive fungal pathogens. Indeed, it has been observed that when an invasive fungal pathogen takes up plant EVs, its growth is inhibited as a direct consequence of plant EVs absorption (Regente et al., 2017).
EVs, including exosomes, are small, membrane-enclosed structures released from a cell into the surrounding environment. They play a pivotal role in cell-to-cell communications and host-pathogen interactions, through the transport and exchange of molecules. Ubiquitous, EVs are found in prokaryotes and eukaryotes, as well as unicellular and multicellular organisms (Yanez-Mo et al., 2015). To date, several studies have mainly focused on human EVs for their potential applications in medicine, underpinned by the development of EV biomarkers for diagnostic and therapeutic tools (Bielska et al., 2019). The idea that organisms possessing strong cellular physical barriers such as cell walls (CWs), like plants and fungi, could produce entities as large as EVs that make it past the CW has seemed questionable, albeit is now a fact (Casadevall et al., 2009; Brown et al., 2015; Bleackley et al., 2019). How this phenomenon eventuates remains to be elucidated.
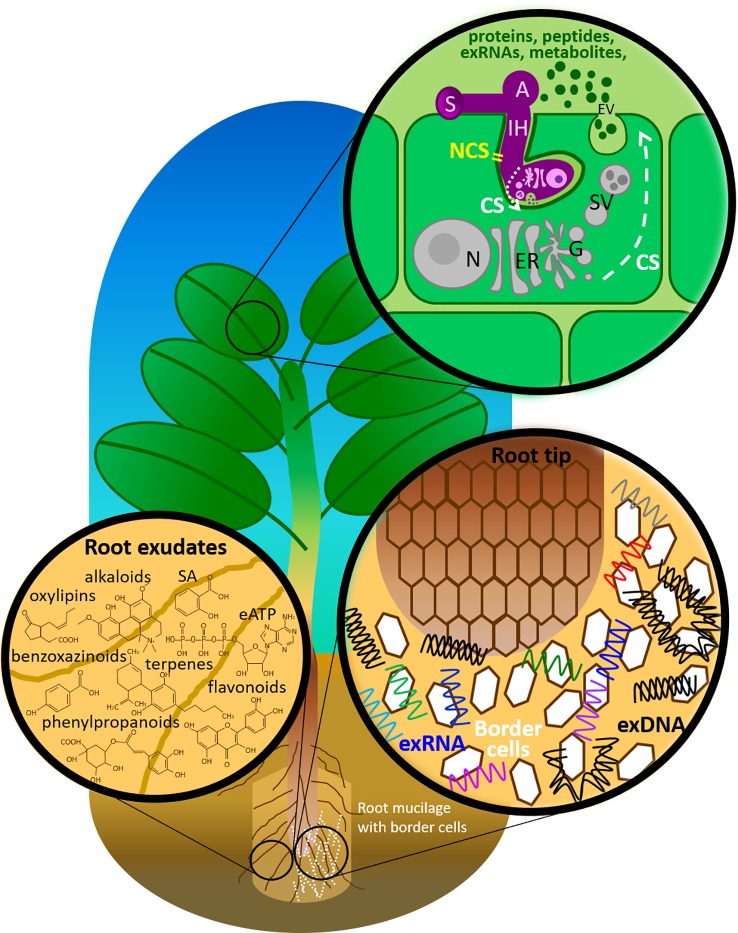
Besides proteins, plants and their interacting fungi secrete also a wide variety of molecules, such as peptides, metabolites, phytohormones and nucleic acids ( Figure 1 and Table 1 ). A number of studies have documented the secretion of metabolites by plant roots, but this is often referred to as exudation rather than a secretion process, even though there is compiling evidence that these small molecules can be secreted not only by passive diffusion but also by making use of specific transporters (Baetz and Martinoia, 2014). In addition, EVs are suggested to drive the universal evolution of ribonucleic acid (RNA) export. Microbial RNAs transported in EVs mediate intra- and inter-kingdom communications, including plants’, by regulating gene expression in target cells directly and indirectly via host immune receptor signaling (Tsatsaronis et al., 2018).
Figure 1.
Components of the plant immune system deployed in the extracellular milieu against fungal pathogens. A, appressorium; CS, conventional secretion; eATP, extracellular adenosine 5′-triphosphate; ER, endoplasmic reticulum; EV, extracellular vesicle; G, Golgi apparatus; IH, invasive hyphae; N, nucleus with nucleolus; NCS, non-conventional secretion; SA, salicylic acid; SV, secretary vesicle; S, spore.
Table 1.
List of studies demonstrating the secretion of molecules involved in plant immune response. Grey highlights molecules secreted by fungi.
| Plant | Mutant/Plant organ | Partner | Vehicle | Secreted molecule | Targets/modulator | Reference |
|---|---|---|---|---|---|---|
| Arabidopsis thaliana (Mouse-ear cress) and Solanum lycopersicum (Tomato) | leaf | Botrytis cinerea | EV | siRNA effectors | EVs of fungal pathogens in cross-kingdom interaction; fungal plant pathogens siRNA effectors | Weiberg et al., 2013 |
| Nicotiana benthamiana (Tjuntiwari and Muntju) | leaf | Phytophthora infestans | RXLR effectors | pathogen virulence | Wang et al., 2019 | |
| Zea mays (Maize) | root apoplast | Trichoderma virens | secreted proteins | suppression of plant immune responses | Nogueira-Lopez et al., 2018 | |
| in vitro fungal secretome | Phytophthora infestans | Leaderless secreted proteins (LSPs), effectors | pathogen virulence | Meijer et al., 2014 | ||
| predicted fungal secretome | Aspergillus fumigatus | Small secreted and effector-like proteins | pathogen virulence | Vivek-Ananth et al., 2018 | ||
| Arabidopsis thaliana | ein2 mutant insensitive to eNAD | Pseudomonas syringae pv maculicola ES4326 | exocytose, transporters | NAD | plant defense | An et al., 2016 |
| Oryza sativa (Rice) | cks1 mutant of Magnaporthe oryzae | Magnaporthe oryzae | cytokinins | fungal virulence | Chanclud and Morel, 2016 | |
| Tomato | leaves; mutants of Botrytis cinerea with reduced virulence | Botrytis cinerea | botrydial, botcinic acid | secondary metabolites (phytotoxins) in fungal virulence | Dalmais et al., 2011 | |
| Arabidopsis thaliana | apoplast; Arabidopsis thaliana mutants accumulating nucleosides in the extracellular space | Botrytis cinerea | transporters | ATP, NAD | susceptibility toward B. cinerea infection | Daumann et al., 2015 |
| Rice | suppression of pathogen-induced IAA accumulation | Magnaporthe oryzae, Xanthomonas oryzae pv oryzae and Xanthomonas oryzae pv oryzicola | transporters | auxin (IAA) | broad-spectrum disease resistance in rice | Fu and Wang, 2011 |
| Rice | Magnaporthe oryzae mutants deficient in siderophore biosynthesis | Magnaporthe oryzae | siderophore | fungal virulence | Hof et al., 2007 | |
| Maize | rhizosphere microbiote | Spodoptera frugiperda | secondary metabolites: benzoxazinoids | root exudate, soil microbiote | Hu et al., 2018 | |
| Rice | RNAi-knockdown of key regulators of the SA signaling pathway in rice | Magnaporthe oryzae | cytokinins | fungal virulence | Jiang et al., 2013 | |
| Arabidopsis thaliana | rhizosphere microbiome, Arabidopsis thaliana mutants in the SA pathway | root microbiote | salicylic acid | root exudate, soil microbiote | Lebeis et al., 2015 | |
| Tomato | rhizosphere microbiome | Trichoderma harzianum | peroxidases and oxylipins | root exudate, soil microbiote | Lombardi et al., 2018 | |
| Maize | Ustilago maydis | cytokinins | fungal virulence | Morrison et al., 2017 | ||
| Vitis vinifera L. (Grapevine) | grape berry, apoplast | Botrytis cinerea | transporters, extracellular vesicles | flavonoids | defense against phytopathogens; hypersensitive-like response in grapevine leaves | Petrussa et al., 2013 |
| Rice | leaves | Magnaporthe oryzae | tyrosine-derived cytochalasan compound | M. oryzae avirulence signalling compound mediated by the PKS-NRPS ACE1 | Song et al., 2015 | |
| Arabidopsis thaliana | roots | exudates | nucleosides, deoxynucleosides, aromatic AAs, anabolites and catabolites of glucosinolates, dipeptides, indolics, SA and JA catabolites, coumarins, mono-, di- and trilignols, hydroxycinnamic acid derivatives and oxylipins | Strehmel et al., 2014) | ||
| Arabidopsis thaliana | leaves | Botrytis cinerea | ATP | induction of plant defense responses | Tripathi et al., 2018 | |
| Medicago truncatula (Barrel medick) | root border cells | root exudates: 70 compounds identified by metabolomics include AA, OA, sugars, beta-alanine, urea, phenolics, saponins/sapogenins | plant-microbe signaling, defense, and interactions | Watson et al., 2015 | ||
| Helianthus annuus (Sunflower) | extracellular fluid of seedling | Sclerotinias clerotiorum | plant EV | proteins | Sunflower pathogenesis-related proteins and defense proteins | Regente et al., 2017 |
| Arabidopsis thaliana | apoplast | plant EV | exRNA | Arabidopsis EV-transported tiny RNAs (tyRNA) | Baldrich et al., 2019 | |
| Gossypium hirsutum (Cotton), Arabidopsis thaliana | leaf | Verticillium dahliae | exRNA | V. dahliae genes encoding a Ca2+-dependent cysteine protease (Clp-1) and an isotrichodermin C-15 hydroxylase (HiC-15) | Zhang et al., 2016 | |
| Arabidopsis thaliana | protoplast | Botrytis cinerea | exosomes | exRNA | transferred Arabidopsis sRNA TAS1c-siR483, TAS2- siR453 | Cai et al., 2018 |
| Arabidopsis thaliana | aerial part | Verticillium dahliae | exRNA | Arabidopsis RNA-silencing components AGO1, AGO7, DCL4, NRPD1a, RDR2, RDR6/SGS2, SDE1, HEN1, HST, SGS3, and SGS1 | Ellendorff et al., 2009 | |
| Arabidopsis thaliana | stem | Cuscuta campestris | exRNA | Arabidopsis miRNAs encoding the TIR1, AFB2, and AFB3 auxin receptors; the pathogen-induced BIK1 kinase; the injuruy-induced phloem protein SEOR1; and the predicted transcriptional repressor HSFB4 | Shahid et al., 2018 | |
| Nicotiana tabacum (Tobacco) | stem | Cuscuta pentagona | exRNA | C. pentagona SHOOT MERISTEMLESS-like Cp-STM | Alakonya et al., 2012 | |
| Arabidopsis thaliana, Tomato | leaf, fruit | Botrytis cinerea | Botrytis cinerea small RNAs (Bc-siR3.2, Bc-siR3.1, Bc-siR5) | mitogen activated protein kinase (MPK1, MPK2), Peroxiredoxin-2F (PRXIIF), cell wall-associated kinase (WAK) from host | Weiberg et al., 2013 | |
| Various plants | leaf, fruit, petal | Botrytis cinerea | plant exRNA | B. cinerea Dicer-like protein 1 (Bc-DCL1) and Bc-DCL2 | Wang et al., 2016 | |
| Pisum sativum (Pea), Tomato | root mucilage | border cell | exDNA | R solanacearum extracellular DNases Rs-nucA, Rs-nucB | Tran et al., 2016 | |
| Phaseolus vulgaris (Common bean) | root | itself, Phaseolus lunatus, Acacia farnesiana | self vs. non-self DNA; exDNA | exDNA as a damage‐associated molecular pattern (DAMP) | Duran-Flores and Heil, 2018 | |
| Pea | root tip | Nectria haematococca | border cell | exDNA | root tip resistance to fungal infection | Wen et al., 2009 |
| Pea, Maize | root mucilage | border cell | exDNA | root tip defense against pathogens | Wen et al., 2017 | |
| Arabidopsis thaliana | itself and various microbes | self vs. non-self DNA | autotoxicity | Mazzoleni et al., 2015a; Mazzoleni et al., 2015b | ||
| Maize | leaf | Cochliobolus heterostrophus | exDNase | C. heterostrophus TatD type deoxyribonuclease Ch-NUC1 | Park et al., 2019 |
The aim of the present review is to summarize the current findings on proteinic and nonproteinic secretion in plants and their interacting fungi, exploring both conventional and nonconventional secretion pathways.
Secretion of Proteins and Peptides by Plants and Fungi
Plants: Cargos Are Diverse and So Are Export Processes At the Apoplast Gates
Plants export a wide variety of proteins into the extracellular space, defined in plants as the zone beyond the plasma membrane, i.e. the CW, the apoplast and the rhizosphere. The CW is a dynamic structure constituting a barrier that pathogens need to breach in order to colonize host plant tissues. Plants have developed a sophisticated system for sensing pathogens and monitoring the CW integrity, upon which they activate defense responses that lead to dynamic CW remodeling that prevent disease development. Pathogens, on the other hand, may exploit the host CW metabolism to establish infection. Several studies have documented the role of different classes of secreted proteins in the strategies deployed by both plants and pathogens in this CW battleground. These proteins include CW degrading enzymes (CWDEs), CWDE-inhibitors, CW methylesterases (CWMEs), CWME-inhibitors and oligosacharide-oxidases in the strategies utilized by both plants and pathogens to prevail in this CW battleground. For example, Bellincampi et al. (2014) reported that when pathogens start degrading components of the plant CW, plants are capable of perceiving the loss of CW integrity, and subsequently activate defense signaling pathways. In turn, successful pathogens escape plant defense and take advantage of the host CW metabolism to facilitate their entry into the host tissues. An interesting area of research toward improving plant protection is to study the dynamics of the expression of endogenous and microbial CWDEs and their inhibitors (Bellincampi et al., 2014). Consistent with this, Benedetti et al. (2015) have shown that oligogalacturonides (OGs) released from the degradation of pectin, a major CW component, acted as a damage-associated molecular pattern (DAMP) signal to trigger plant immunity. OGs were generated in planta by partial inhibition of pathogen-encoded polygalacturonases (PGs). Under these conditions, plants were more resistant to the phytopathogens Botrytis cinerea, Pectobacterium carotovorum, and Pseudomonas syringae. This strongly supports the idea that controlled release of OGs upon infection may be a valuable tool to protect plants against infectious diseases.
Besides OGs, cellodextrins, which are by-products of cellulose breakdown, are also well-known DAMPs. Recently, Locci et al. (2019) showed enhanced resistance to B. cinerea in Arabidopsis plants overexpressing a berberine bridge enzyme-like (BBE-like) protein named CELLOX (cellodextrin oxidase), which is the only BBE‐like enzyme identified so far that oxidizes cellulose fragments but not glucose. Restriction of fungal growth might arise from the overexpression of BBE-like enzyme which may have prevented the accumulation of the cellodextrins-type DAMPS. Moreover, oxidized cellodextrins are a less valuable carbon source for the pathogenic fungus, further limiting fungal growth. Based on these results, the authors speculated that other members of the large BBE‐like family may control the homeostasis of CW fragments other than OGs and cellodextrins (Locci et al., 2019). The pectin matrix is the main CW target of B. cinerea. Lionetti et al. (2017) showed that pectin methylesterase (PME) activity and pectin methylesterification are dynamically modulated by endogenous PME inhibitors (PMEIs) during the infection, thereby pointing out the role of PMEIs in mediating the maintenance of CW integrity in plant immunity. Interestingly, proteomics analyses of EVs isolated from sunflower seedlings revealed that a large part (112 out of 237 proteins, 47%) of the identified EV proteins are CW-interacting proteins, including enzymes involved in the degradation and reorganization of polysaccharides (de la Canal and Pinedo, 2018). This implicates EVs in the unconventional secretion of CW-modifying enzymes and suggests a role of EVs in modifying the composition of plant CW. Finally, it is worth noting the work of Zhao et al. (2019) showing that yeast strains with deletions in genes involved in CW biosynthesis produce more EVs than the wild type, indicating a potential role for yeast EVs in CW remodeling.
Exported proteins are implicated in a variety of processes other than CW modification, including signaling, development and stress responses (Delaunois et al., 2014). The classical view of secretion of these proteins assumes that they are synthesized and delivered using the conventional ER secretory pathway. Several reviews on this topic have been published (Agrawal et al., 2010; Ghahremani et al., 2016; Martínez-González et al., 2018). A plant protein secreted via the canonical secretory pathway presents a N-terminal signal peptide typically 15–30 AAs long, which enables translocation of the protein across the ER in plants and which is cleaved upon secretion (Emanuelsson et al., 2007). The presence of a transit or signal peptide (SP) in secreted proteins can be predicted in silico using online algorithms such as signalP or targetP (http://www.cbs.dtu.dk/services) (Nielsen et al., 2019) and the localization of secreted proteins to the apoplast can be in silico validated using the online tool apoplastP (http://apoplastp.csiro.au) (Sperschneider et al., 2018b). Proteomics analyses of apoplast fluids have revealed that most pathosystems exhibit conserved biochemical responses involving enzymes acting in CW remodeling (e.g. xylosidases, arabinofuranosidases, fucosidases, pectin methylesterases, galactosidases) as described above, reactive oxidative species (ROS) detoxification (e.g. superoxide dismutases, catalases, peroxidases), as well as pathogenesis-related (PR) proteins (e.g. PR-1, 2, 3, 4, 5, 8, 10, 16 and 17) (Tanveer et al., 2014; Ghahremani et al., 2016; Martínez-González et al., 2018). We refer the reader to these comprehensive reviews for further information on this topic.
Leaderless Secreted Proteins: The Puzzle of Unconventional Secretion of Proteins
Over the past decade, one of the most intriguing findings in plant secretomics (as well as in fungi secretomics, as discussed below) has been the discovery of a new type of secreted proteins, devoid of a SP, referred to as LSPs (Agrawal et al., 2010; Albenne et al., 2013; Bellucci et al., 2017). Interestingly, LSPs account for more than 50% of the total identified secretome, supporting the existence of novel secretory mechanisms independent of the classical ER–Golgi secretion pathway in plants, which is similar to what has been reported in other eukaryotes. One way these LSPs could be exported to the extracellular milieu is through EVs. In this context, several recent studies led to the characterization of unconventional protein secretory (UPS) pathways that involve EVs for the export of proteins to the extracellular space (Pinedo et al., 2012; Davis et al., 2016; de la Canal and Pinedo, 2018).
Owing to their rigid CWs, the production of EVs in plants had first seemed improbable. We now know that plant cells do indeed secrete EVs, although little is known about the origin, composition and mode of function of EVs (Rutter and Innes, 2018). A protocol for isolating EVs from apoplastic fluids of Arabidopsis thaliana leaves, initially devised by Regente and colleagues (Regente et al., 2009) and later implemented with a quantitative fluorometric dye assay to assess total membrane content and screening of the exosome marker PEN1 by Rutter and colleagues (Rutter and Innes, 2017; Rutter et al., 2017), is available. The size of recovered vesicles range between 50 and 300 nm. By using a combination of filtration and differential centrifugation steps, Rutter and Innes (2017) were the first to isolate and purify EVs from Arabidopsis leaf apoplasts. The authors also used a proteomics approach to analyze the protein content of purified EVs. EVs are highly enriched in proteins involved in responses to biotic and abiotic stresses, suggesting their likely role in plant defense against pathogens. Consistent with this, EVs secretion was enhanced in plants infected with P. syringae and in response to treatment with the plant hormone salicylic acid (SA). Among the recovered defense-related proteins, several proteins involved in signal transmission were identified, many of which were highly induced in response to stress and/or contribute to immunity. Other defense-related proteins included members of the myrosinase–glucosinolate system, involved in ROS signaling, and various membrane-trafficking proteins (e.g. syntaxins, RAB GTPases, patellin family proteins) (Rutter and Innes, 2017). This method was further applied to analyze EVs from sunflower seedlings. In the presence of Sclerotinia sclerotiorum ascospores, isolated sunflower EVs were found to be incorporated by fungal cells, which subsequently led to the inhibition of spore germination, stunted mycelial growth, and loss of vitality. Proteomics analyses of EVs taken up by fungal cells have identified several defense proteins (Regente et al., 2017). The biological function of EVs in plant–pathogen interactions is just emerging (Boevink, 2017; Hansen and Nielsen, 2017). Their role as key mediators of such interactions needs to be further investigated.
In parallel to experimental studies, new bioinformatics tools are currently being developed for in silico characterization of LSPs. For example, ApoplastP can predict whether an effector candidate or a plant protein localizes to the apoplast and, in doing so, allows the identification of sequence features, common to both effectors and plant proteins, that are required for apoplastic localization. Such sequence properties include depletion in glutamic acid, acidic and charged amino acids as well as enrichment in small amino acids (AAs) (Sperschneider et al., 2018a). SecretomeP is an online tool that provides ab initio predictions of LSPs (Bendtsen et al., 2004). Because this algorithm has initially been designed for mammalian proteins it might prove unreliable for plant LSPs (Delaunois et al., 2014). To overcome this shortcoming, a plant secretome knowledgebase PlantSecKB has been developed (https://omictools.com/plantseckb-tool) (Lum et al., 2013). Besides proteins, plant cells synthesize and export an array of peptides originated from larger precursors (thus not different from protein export discussed above) or directly made from small Open Reading Frames (ORFs). For instance, antimicrobial peptides (AMPs) are peptides of up to 100 AAs that are structurally and biochemically highly diverse, are of ribosomal or non-ribosomal origin, and display activities against microbial pathogens. Plant AMPs have various roles, covering functions such as hormones like systemin and HypSys, defense signaling like Pep1 and GmPep914 and GmPep890, or DAMP elicitor like CAPE1, as well as defensins that are constitutively expressed in planta and carry antifungal activity [reviewed in (Breen et al., 2015)].
An intriguing question is whether LSPs exported via UPS are glycosylated. While it is rational to assume that glycosylation is the exclusive signature of the passage through the ER secretory pathway, very little is known about the subcellular localization of O‐ and N‐glycosylated enzymes in plants. A study by Poulsen et al. (2014) has presented data demonstrating that at least some of the O-glycosyltransferases are localized to unique subcellular compartments that are distinct from Golgi apparatus and that may be part of UPS pathway. Therefore, these data support the view that it is possible for glycosylated proteins to be exported through unconventional pathways. On the other hand, Stock et al. (2016) could show, through heterologous expression of a modified β-glucuronidase (GUS) reported gene in Ustilago maydis, that glycosylation only took place when the ER secretory pathway directed protein secretion. No glycosylation was observed in the UPS pathway. This was concluded based on monitoring the activity of secreted GUS enzyme, which is hampered when the protein is glycosylated. GUS activity could not be detected in the supernatant of U. maydis culture when GUS was fused to N-terminal SP. Instead, GUS activity was restored when GUS was fused to Chitinase Cts1, an enzyme known to be released via UPS in U. maydis.
Although it is possible that UPS pathways may be dissimilar in plants and fungi, the above-mentioned puzzling results do not allow to draw clear-cut conclusions regarding whether or not glycosylated proteins are being exported through the UPS pathway. In addition, it is as yet completely unknown to science how many UPS pathways exist in plants and fungi. Indeed, more empirical work is needed to address these questions.
Posttranslational modifications (PTMs) are likely to play an important role, including additional functions associated with protein localization and delivery, incorporation of other cargo proteins/RNAs, or exclusion from secretion (Claridge et al., 2019). More generally, the mechanisms that define the sorting of proteins into EVs and subsequent trafficking are the focus of recent work. Several other basic features regarding EVs’ biology, diversity and function are, to date, simply assumed or based on untested hypotheses that need to be experimentally tested for the field to advance (Margolis and Sadovsky, 2019).
In Fungi Size Matters: Plant Pathogens Possess the Largest Secretomes
Fungi export large amount of proteins and small peptides, including effectors, into the extracellular milieu. Accumulating genome sequence data has helped understand both the size and nature of fungal secretomes. Pathogenic fungi secrete larger numbers of proteins than symbionts. Among plant and animal pathogens, the largest secretomes are found in crop-infecting necrotrophs, while the smallest secretomes are released by biotrophs. Hemibiotrophs and wood-decaying necrotrophs produce secretomes with intermediate sizes. The small size of biotrophic secretomes is mostly attributed to the fact that this group of fungi lack many proteins with enzymatic activity (Kim et al., 2016).
A recent genome-wide survey of 250 fungal secretomes unveiled the presence of putative KEX2-processed repeat proteins (KEPs) in nearly all fungi, including mycorrhizal fungi and human and plant pathogens. This class of secreted peptides are processed in the Golgi apparatus upon their passage through the secretory pathway and are exported to the extracellular space by exocytosis (Le Marquer et al., 2019a; Le Marquer et al., 2019b). While some KEPs are predicted to be ɑ-type sexual pheromones that play a role in mating, others are predicted to act as mycotoxins and many other KEPs are of unknown function.
Many secreted fungal proteins with enzymatic activities have predicted biological functions, including the breakdown of the host CW, self-protection or nutrient acquisition, such as carbohydrate-active enzymes (CAZymes (Cantarel et al., 2009)), oxidoreductases, proteases, and lipases (Kim et al., 2016; Franceschetti et al., 2017). A net reduction in these enzymes in mycorrhizal fungal genomes has placed these fungi in a separate group (Tisserant et al., 2013; Pellegrin et al., 2015; Garcia and Ane, 2016). However, a large group of secreted putative fungal effectors carry no recognizable Pfam domain (Rafiqi et al., 2012; Vivek-Ananth et al., 2018). This group of unknown secreted proteins is thought to play a crucial role in enabling fungal colonization of plant tissue. Yet, this is the least characterized group of secreted proteins. While the exact biological function in the host is yet to be discovered for most effectors, knowledge is being accumulated on how their expression is regulated upon fungal invasion of plant tissue along with their final host subcellular destination. An interesting study by Zeng et al. (2018) has shown that while some secreted proteins conserve the same expression upon interaction of the arbuscular mycorrhizal fungus Rhizophagus irregularis with different plant species, a subset of these proteins are host-specific. A recent study has shown that long-distance retrograde motility of early endosomes is required for the expression of effector genes in U. maydis during host invasion (Bielska et al., 2018). A mitogen-activated kinase, Crk1, transported in early endosomes is involved in the retrograde signaling that coordinates effectors expression. Wang and colleagues have used fusion to fluorescent proteins and heterologous expression in the model host Nicotiana benthamiana to identify the subcellular targets of 52 Phytophthora infestans effectors (Wang et al., 2019). While most of these effectors are nucleocytoplasmic, many others target more specific localizations, including the plasma membrane, the ER, microtubules or host organelles such as the nucleus, indicating the vast diversity of host subcellular compartments targeted by the pathogen and, consequently, indicating where host molecular interactors might be located. The diversity of the final cellular targets of fungal effectors strengthens the hypothesis that adapted coevolved pathogens use far-reaching and diverse strategies that, when combined, have the potential to take control over molecular processes in infected host cells in favor of disease development.
At the Frontline: Plant and Fungal Secretomes Merge
Plants recognize molecules of pathogens or microbes that attempt infection. These molecules are referred to as invasion patterns (IPs) or invasion molecules (IMs) (Cook et al., 2015; Kanyuka and Rudd, 2019) and were previously known as microbe- or pathogen-associated molecular patterns (MAMPs or PAMPs) in the original Zigzag model proposed by Jones and Dangl (2006). Despite the importance of this early immune response, relatively little is known about the size and nature of the plant secretomes deployed to control microbial and pathogenic invasions. Upon perception of IMs in the apoplastic space, plants secrete an arsenal of proteins and peptides with the primary function of halting fungal growth (Breen et al., 2015; Gupta et al., 2015). When secreted plant proteins encounter the fungal secretome, intensive biochemical interactions are expected to take place at the fungal–plant interface. It is expected that all microbes must face this first layer of the plant immune system.
Necrotrophic pathogens, which kill the cells they infect, secrete a range of proteins with enzymatic activity (discussed above) or protein effectors that exploit host cell death mechanisms to promote pathogen growth and cause disease. One example is SnToxA effector from wheat-infecting Stagonospora nodorum (Vincent et al., 2012) In contrast, biotrophic and mutualistic fungi, which grow and reproduce on living plant cells, deploy more exquisite secretomes. Trichoderma virens is a biocontrol and opportunistic endophytic fungus (Harman et al., 2004). A recent study on the interaction of T. virens with maize roots (Nogueira-Lopez et al., 2018) has identified 95 secretory proteins of maize using a gel-free shotgun proteomics approach. In this study, maize secretome was found to be reduced by 36% upon colonization with T. virens (Nogueira-Lopez et al., 2018). Seven secreted Uaca_Ns effectors from the bean rust pathogen Uromyces appendiculatus were shown to suppress plant host innate immunity, by either dampening pathgogen-triggered immunity or preventing hypersensitive response (Qi et al., 2019).
Such proteomics analyses will be further needed when comparing secretomes of susceptible versus resistant plants, as it will help identify critical components that underlie resistance versus susceptibility in the host, and virulence versus avirulence in the pathogen. This in turn will help develop pathogen control strategies through enhancing resistance traits in the host (e.g. via introgression of resistance genes into commercial varieties or via GMO approach) and through suppression of virulence genes in the pathogen (e.g. via agrochemical development or via RNAi mechanisms discussed below).
Are We Missing Something? Non-Conventional Secretory Pathways in Fungi
Proteinic secretomes of fungi have so far been identified through mining fungal genomes using bioinformatics pipelines (Saunders et al., 2012) that filter out proteins harboring SP and consequently being predicted to be exported outside fungal cells through the classical ER secretory pathway. However, an increasing number of experimental studies recover proteins lacking SP within fungal and oomycetes secretomes. For example, Meijer and colleagues have recovered, through mass spectrometry, proteins lacking obvious SP when analyzing proteins exported by P. infestans in different growth media (Meijer et al., 2014). Similarly, Nogueira-Lopez and colleagues have found evidence for non-conventional secretion mechanisms when analyzing the secretomes of T. virens on its own and within infected maize cells (Nogueira-Lopez et al., 2018).
A recent analysis of the secretome of Aspergillus fumigatus and ten other related species has also concluded that 64 A. fumigatus proteins (0.65% of the proteome) are secreted through unconventional mechanisms, as opposed to 598 (6.1% of the proteome) that are secreted through the ER secretory pathway (Vivek-Ananth et al., 2018). Altogether, these findings point to the necessity of considering experimental approaches to characterize secretomes combined with in silico methods in order to ensure a comprehensive analysis of plant and fungal secretomes.
Several methods are now available for the identification of secreted proteins both in growth media as well as in the apoplastic space (Tanveer et al., 2014; Gupta et al., 2015; Miura and Ueda, 2018). Inclusion of empirical analyses, in addition to bioinformatics prediction, when identifying fungal extracellular proteins is of high importance. Empirical approaches are increasingly needed to identify the whole spectrum of extracellular proteins that play a role not only in fungal colonization of plant tissue, but also those deployed in fungal cell growth and modifications.
Beyond Proteins: What Else Do Plants and Fungi Secrete?
Secretome studies in plants and their interacting fungi most often address the role of proteins, including effectors that are secreted in the extracellular space (Kamoun, 2006; Agrawal et al., 2010; Ghahremani et al., 2016; Cologna et al., 2018). However, both plants and fungi secrete molecules other than proteins, notably small molecules such as metabolites, secondary metabolites and hormones ( Figure 1 and Table 1 ). In the following sections, we review the secretion of such nonproteinic molecules by plants and their interacting fungi.
Export of Metabolites by Plants
Root Exudates
Root exudation is an important source of organic carbon in the soil, which accounts for up to 2–10% of the total photosynthetic production (Kuzyakov and Domanski, 2000). This leads to a process called “soil priming”, whereby the microbial community becomes more active and liberates nutrients that are important for plants (Fontaine et al., 2013; Gargallo-Garriga et al., 2018). Plant roots release complex mixtures of bioactive molecules, including those affecting the activity and composition of the rhizosphere microbiota (Sasse et al., 2018). For example, tomato root exudates, which act as chemoattractants of the biocontrol fungus Trichoderma harzianum, were shown to contain peroxidases and oxylipins, both known to be released by roots in response to stress (Lombardi et al., 2018). Other examples are benzoxazinoids, a class of defensive secondary metabolites released by roots of cereals such as wheat and maize. Benzoxazinoids alter fungal and bacterial communities associated with roots and increase plant defenses (Hu et al., 2018). Similarly, studies conducted on different plant-metabolites concluded that secondary metabolites contained in root exudates, including alkaloids, flavonoids and phenolics, can potentially combat bacterial, viral and fungal infections (reviewed by (Zaynab et al., 2018)). Unexpectedly, SA, a major phytohormone usually involved in plant defense in the shoot system, proved also able to sculpt the microbiota of Arabidopsis roots (Lebeis et al., 2015).
The chemical composition of root exudates was explored through metabolomics studies (Strehmel et al., 2014; Watson et al., 2015; van Dam and Bouwmeester, 2016; Gargallo-Garriga et al., 2018), notably through the recent field of exometabolomics that aims at understanding how such exudates inhibit pathogen growth or, on the contrary, recruit mutualistic microbial species (Jacoby and Kopriva, 2018; Zhalnina et al., 2018). Metabolomics revealed the large chemical diversity of root exudates. Consequently, a number of compounds including nucleosides, deoxynucleosides, aromatic AAs, anabolites and catabolites of glucosinolates, dipeptides, indolics, SA and jasmonic acid (JA) catabolites, coumarins, mono-, di- and trilignols, hydroxycinnamic acid derivatives and oxylipins, were identified in Arabidopsis root exudates (Strehmel et al., 2014) and Medicago truncatula root border cells (Watson et al., 2015).
Vesicles are involved in secondary metabolites and flavonoids transport in plant cells (Petrussa et al., 2013). Membrane-bound proteins are implicated in metabolite release from plant cells. Some localize to the plasma membrane where they can directly export compounds from the cell. Others are associated with internal membranes where they may sequester compounds within subcellular compartments or perhaps load vesicles ready for exocytosis. These proteins include the ATP-binding cassette (ABC) family, the Multidrug and Toxic Compound Extrusion (MATE) family, the Major Facilitator Superfamily (MFS), and the Aluminum-Activated Malate Transporter (ALMT) family of transport proteins (Badri et al., 2008; Badri et al., 2009; Badri and Vivanco, 2009; Weston et al., 2012; Ziegler et al., 2017). Therefore, both conventional (transporters) and unconventional (extracellular trafficking) pathways ensure the secretion of plant secondary metabolites.
Similarly, fungal pathogens release volatile substances, such as the sesquiterpene-derived trichotecene toxins from Fusarium culmorum that are potent inhibitors of protein synthesis and inhibit the activation of plant-defense response genes prior to any physical contact with the pathogen (reviewed in (Zeilinger et al., 2016)). Transporters are involved in the secretion from fungal cells of several secondary metabolites and phytohormones (Yazaki, 2006; Badri and Vivanco, 2009; Lefevre and Boutry, 2018).
Extracellular Nucleosides and Nucleotides
One secreted metabolite, adenosine 5′-triphosphate (ATP), was the object of several studies in plants. While traditional studies emphasized the roles of nucleotides in intracellular energy metabolism, recent findings, first in animals then in plants, highlighted their potential roles outside the plasma membrane. Extracellular ATP (eATP) plays critical roles in plant stress responses, suggesting that it can act as a DAMP. A major finding arose from the characterization of the ATP-insensitive Arabidopsis mutant, dorn1 (Does not Respond to Nucleotides 1), which is defective in the lectin receptor kinase I.9. The data disclosed that DORN1 protein binds eATP with high affinity and is required for ATP-induced calcium response, mitogen-activated protein kinase (MAPK) activation, and gene expression. Thus, DORN1 is essential for the perception of eATP and as such plays a variety of roles in plant stress resistance. DORN1 is the founding member of a new plant-specific purinoceptor subfamily, P2K (P2 receptor kinase). It consists of an extracellular legume L-type lectin domain, a transmembrane domain, and an intracellular serine/threonine kinase domain. The predicted structure of the extracellular domain revealed putative key ATP binding residues (Choi et al., 2014a; Choi et al., 2014b; Tanaka et al., 2014; Cho et al., 2017). Another study on the susceptibility to B. cinerea infection in Arabidopsis mutants accumulating nucleosides in the extracellular space was addressed. These plants markedly accumulated adenosine and uridine in leaves, were highly susceptible toward B. cinerea and showed a reduced induction of pathogen-related genes PR1 and WRKY33 (Daumann et al., 2015; Feng et al., 2015).
In addition, eATP is also implicated in plant-fungi symbiotic interactions. For example, the beneficial root endophyte Serendipita indica secretes SiE5′NT, an enzymatically active ecto‐5′‐nucleotidase capable of hydrolyzing nucleotides in the apoplast. Importantly, Arabidopsis lines producing extracellular SiE5′NT were significantly more colonized, had reduced eATP levels, and altered responses to biotic stress, indicating that SiE5′NT functions as a compatibility factor (Nizam et al., 2019).
As in animals, ATP appears to be released by plant cells via vesicular exocytosis (Marzec, 2011; Tanaka et al., 2014), thereby highlighting the involvement of unconventional secretion mechanisms. However, in Arabidopsis, AtPGP1, an ABC transporter, and PM-ANT1, a plasma membrane-localized nucleotide transporter, were shown to export intracellular ATP into the extracellular milieu by conventional secretion (Tanaka et al., 2014). Therefore, the secretion of eATP by plant cells makes use of both unconventional and conventional mechanisms. No information is presently available on the existence of eATP in fungi.
Phytohormones
Several recent studies pointed out the central role of phytohormones, including SA, JA, auxins, cytokinins (CKs), abscisic acid (ABA), in regulating plant-microbe interactions (Lebeis et al., 2015; Boivin et al., 2016; Di et al., 2016; Carvalhais et al., 2017). Of importance is the discovery that SA sculpts the Arabidopsis root microbiome. Thus, isogenic Arabidopsis mutants lacking biosynthesis of SA, and/or SA-related signaling, display root microbiota that differ in the relative abundance of specific bacterial families relative to those of wild type (Lebeis et al., 2015). This study demonstrated that different bacterial strains could make use of SA in different ways, whether as a growth signal or as a carbon source. Together, these findings show that a central regulator of the plant immune system, largely uncharacterized in roots, directly influences the root microbiota composition (Lebeis et al., 2015). Consistent with this, exogenous SA and derivatives proved able to inhibit the growth of B. cinerea in vitro (Dieryckx et al., 2015). Similarly, several studies showed that ABA exhibits antifungal activity in plants (Khedr et al., 2018).
Export of Small Molecules by Phytopathogenic Fungi
Phytohormone-Mimicking Compounds
It is established that classical plant hormones, including auxins, CKs, gibberellins (GAs), ABA, ethylene, SA, JA, or metabolites mimicking phytohormones, are produced by plant-interacting fungi (Robert-Seilaniantz et al., 2007; Chanclud and Morel, 2016). Converging evidence suggests that these fungal-derived molecules can perturb plant processes, either positively or negatively, notably to favor fungal invasion (Chanclud and Morel, 2016; Ma and Ma, 2016; Vrabka et al., 2018; Zhang et al., 2018). For example, upon infection of rice by the blast fungus Magnaporthe oryzae, diverse CK species could be detected in the hyphae, conidia, and culture filtrates, indicating that the fungus is capable of producing and secreting CKs (Jiang et al., 2013). In agreement with the role of CKs in plant infection by fungal pathogens, CK-deletion strains of the smut fungus U. maydis elicited fewer and smaller tumors than the pathogenic strain SG200. Consistent with this, mining of the U. maydis genome identified genes encoding CK signaling- and biosynthesis-related proteins (Morrison et al., 2017). Also, diverse pathogens are able of synthesizing and secreting indole-3-acetic acid (IAA), the major form of auxin in plants. This secretion of IAA may have a direct virulence effect by loosening the plant CW, opening stomata, and inhibiting SA-dependent defense signaling (Fu and Wang, 2011).
Furthermore, several phytopathogenic fungi, such as Cercospora rosicola, B. cinerea and M. oryzae have the ability to produce ABA through a biosynthetic pathway that is distinct from that of plants (Izquierdo-Bueno et al., 2018). Impairing ABA biosynthesis in M. oryzae dramatically reduces virulence (Spence et al., 2015).
The endophytic fungus A. niger CSR3 produces IAA and GAs to promote growth of the GA-deficient rice mutant Waito-C (Lubna Asaf et al., 2018). Multifunctional plant growth-promoting fungi (PGPF) include the fungal genera Aspergillus, Fusarium, Penicillium, Piriformospora, Phoma, and Trichoderma. The associations between plants and multipurpose PGPF have been shown to control numerous foliar and root pathogens through induced systemic resistance (ISR) in host plants, notably through phytohormones production (Hossain et al., 2017).
Phytotoxic Fungal Metabolites
Necrotrophic fungi secrete phytotoxic secondary metabolites to kill the host tissues and suppress plant-defense responses (Osbourn, 2001; Keller et al., 2005; Muria-Gonzalez et al., 2015; Pusztahelyi et al., 2015; Collemare et al., 2019). These metabolites are secreted into infected plants and either act as virulence factors, i.e. intensify disease symptoms, or act as pathogenicity factors, i.e. are exclusively responsible for the development of disease symptoms. B. cinerea secretes several toxic compounds, including the secondary metabolites botrydial (a sesquiterpene) and botcinic acid (a polyketide), causing the death of infected plant cells (Dalmais et al., 2011). Other examples of secondary metabolites essential for fungal virulence are siderophores. M. oryzae produces the siderophore ferricrocin, which contributes to pathogenicity on rice by interfering with appressorium turgor pressure (Hof et al., 2007).
Of interest is the finding that M. oryzae ACE1, which encodes a polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) fusion protein, differs from other fungal avirulence (AVR) genes in that it is not a secreted protein, despite behaving like a classical AVR gene. When carrying functional AVR gene ACE1, M. oryzae is unable to infect rice cultivars carrying the corresponding Pi33 resistance gene, whereas isolates or mutants defective for ACE1 are virulent and bypass the rice Pi33-mediated resistance. ACE1 accumulates exclusively in appressoria during fungal penetration of host tissue. Mutation of its β-ketoacyl synthase domain abolishes recognition of the fungus by resistant rice, indicating that ACE1 enzyme activity is required for avirulence. These findings provide evidence that the avirulence signal recognized by Pi33 rice cultivars is not the ACE1 protein itself, but rather the secondary metabolite (most presumably a tyrosine-derived cytochalasan compound) synthesised by this enzyme and exported into infected plant cells (Collemare et al., 2008a; Collemare et al., 2008b; Song et al., 2015).
Export Mechanisms of Fungal Small Compounds
In animal systems, it is well established that vesicular trafficking plays a key role in fungal secondary metabolism and transport. For example, this has been documented for the filamentous fungus A. parasiticus in relation with the transport of aflatoxin, a secondary metabolite considered as one of the most potent naturally occurring carcinogens known to date (Chanda et al., 2009). These studies allowed the discovery of aflatoxisomes, which are specialised trafficking vesicles that are implicated in the export of the toxin outside the cell (Roze et al., 2011). Moreover, endosomal trafficking was shown to be critical for subcellular localization of melanin biosynthetic enzymes in the human fungal pathogen A. fumigatus (Upadhyay et al., 2016).
Information on extracellular vesicles in the transport of secondary metabolites from plant pathogenic fungi remains scarce. However, endosomes were suggested to be implicated in the synthesis and secretion of aflatoxins by A. parasiticus (Chanda et al., 2010). Thus, this vesicular‐mediated secretion could provide a route for fungal export of metabolites that is distinct from efflux by membrane transporters. It is anticipated that vesicular-mediated secretion in plant pathogenic fungi will become an active topic of research in the near future (Collemare et al., 2019). Conventional export/secretion of secondary metabolites seems to operate in plant pathogenic fungi as they possess several ABC and MFS transporters (Amnuaykanjanasin and Daub, 2009; Coleman and Mylonakis, 2009).
The New Nucleic Acid Kids on the Block: Secretion of Rna and Dna by Plants and Fungal Phytopathogens
In the previous sections, we highlighted how plant-fungal interactions are mediated by the exchange of diverse proteins, peptides and metabolites between both partners. Here, we address the export of nucleic acids (RNA and deoxyribonucleic acid (DNA)) as novel secreted molecules ( Figure 1 and Table 1 ).
Extracellular RNA
Extracellular RNA (exRNA) comprises RNA molecules located outside cells through active secretion mediated by EVs or membrane transporters, or through passive release from apoptotic cells (Tsatsaronis et al., 2018). ExRNAs include rRNAs, tRNAs, mRNAs, and small RNAs (sRNAs). sRNAs are short [21- to 24-nucleotides (nt)] non-coding regulatory RNAs resulting from cleavage of double-stranded RNA substrates by dicer-like (DCL) enzymes. Small RNAs silence genes whose sequences are complementary. Small RNAs are further subcategorised into microRNAs (miRNAs) and small interfering RNAs (siRNAs), based on the differences in their biogenesis and modes of action. They move between hosts and interacting organisms and induce gene silencing, known as cross-kingdom/organism RNA interference (RNAi) (Cai et al., 2018). sRNAs are transferred in a selective fashion across cells and tissues of individuals and across species, thus connecting animal, plant, and microbial kingdoms.
Such is the interest in this active field of research that the Extracellular RNA Communication Consortium (ERCC) was launched in 2013 (Ainsztein et al., 2015) with its first progress report released this year (Das et al., 2019). Aware of the role of exRNAs in the intercellular communications in human, the ERCC aimed at profiling exRNAs and their carriers from diverse biofluids from healthy individuals, identifying exRNA biomarkers for a broad range of diseases and optimizing methods for large-scale production of safe and effective exRNA- and EV-based therapeutics. Significant progress was achieved through refined sample preparation and next generation sequencing (NGS) methods. This has enabled the exploration of the entire RNA populations from various samples and treatments over a time-course. However, mining this large amount of data remains a challenge (Morgado and Johannes, 2017).
Compared to human sRNAs, research on plant and fungal sRNAs and EVs has been lagging behind. However, it is quickly catching up as illustrated by the number of reviews published on this topic (Weiberg et al., 2014; Han and Luan, 2015; Bhat and Ryu, 2016; Islam et al., 2017; Hua et al., 2018; Rutter and Innes, 2018; Tsatsaronis et al., 2018; Zhao et al., 2018; de Vries et al., 2019; Lee, 2019; Zeng et al., 2019).
Plant Extracellular sRNAs and Vesicles
In plants, sRNAs have key regulatory functions in development, physiology, response to biotic and abiotic stresses, genome stability and transposon control (Mirouze, 2012). Plants transport viral RNAs, mRNAs, miRNAs and siRNAs through the phloem (Kehr and Buhtz, 2008). Loading of RNA into the phloem is likely to be facilitated by plasmodesmata and associated RNA-binding proteins. EVs represent an alternative process for the export of RNAs into the phloem and possibly the transport of RNA through the phloem or apoplast (Rutter and Innes, 2018).
EVs were isolated from apoplastic fluids of Arabidopsis leaves (Baldrich et al., 2019) and analysed for sRNAs. Arabidopsis EVs contain a high number of sRNAs whose sises, classes and identities vary, including miRNAs, siRNAs, as well as so-called tinyRNAs (tyRNAs, 10–17 nt-long) of unknown function and originating from mRNA, rRNA and miRNA precursors. Conversely, apoplastic fluids devoid of EVs are enriched in sRNAs able of inducing silent expression in genes with matching sequences, which led to hypothesise on the existence of two independent pathways for sRNA export, with or without EVs as vehicles (Baldrich et al., 2019). Analysis of whole tissue sRNA public datasets in repositories revealed the over-representation of tyRNAs in plants compared to mammals. However, tyRNAs have been overlooked due to bioinformatic analysis pipelines that discard reads shorter than 18 nt. Adjusting pipelines parameters to include such tyRNAs should help find out whether plant tyRNAs are taken up by pathogens via uptake of EVs, and will help reveal their potential role in plant immunity (Bielska et al., 2019).
Extracellular sRNAs Associated With Plant Immunity Against Fungi
The involvement of extracellular miRNA in the plant immune system has become apparent in the seminal study of cotton plants infected by Verticillium dahliae (Zhang et al., 2016). Cotton plants increased expression of native cotton miRNAs Gh-miRNA166 and Gh-miR159 in response to V. dahlia infection. Both miRNAs were delivered into the fungus, via as yet an unknown process, where they targeted genes essential for pathogen virulence, namely a Ca2+ dependent cysteine protease (Clp-1) and an isotrichodermin C-15 hydroxylase (HiC-15) (Zhang et al., 2016). Future research should investigate the mechanisms of miRNA export from the host plant into fungal pathogens and translational applications.
A recent outstanding study of the Arabidopsis–B. cinerea pathosystem by Cai and colleagues (2018) developed a sequential protoplast purification method to isolate pure fungal cells from infected plant tissues. A number of Arabidopsis siRNAs were identified inside B. cinerea cells that would have been transferred not merely through concentration-dependent diffusion but possibly through a more selective process mediated by EVs. Conducting such comprehensive analyses in other pathosystems will highlight how prevalent this immune strategy is in the plant kingdom.
The combined facts that sRNAs contribute to plant immunity, transit through and between organisms, and act rapidly on multiple targets conserved across different species make them promising candidates to be considered in disease resistance breeding programs for valuable crops (Rose et al., 2019). The method of Host-Induced Gene Silencing (HIGS) exploits the silencing effect of sRNA signals in interacting organisms and involves host expression of sRNA-generating constructs directed against genes in associated pathogens to dampen their virulence. The efficacy and persistence of disease resistance reported in tightly-controlled laboratory conditions remains to be validated under field conditions (Knip et al., 2014).
Family Feud: Plant Virulence Against Other Plant Species
Plant crops fall not only victim to the molecular clutches of various microbial pathogens but also to those of weeds and parasitic plants. Connection with the host can take place by root invasion, as with Striga hermonthica, or by constricting and invading the host’s aerial photosynthetic parts and stem, as with Cuscuta (dodder) genus (Johnson and Axtell, 2019). Through physical connections between Cuscuta and its hosts, movements of water, nutrients and metabolites take place, carrying along macromolecules such as proteins and mRNAs (Kim and Westwood, 2015; Westwood and Kim, 2017). Due to its relative wide host range, Cuscuta can parasitize several species from diverse range of plant families, and therefore act as a sink for host mobile RNA from many different species. Furthermore, host mRNA sequences are highly divergent from those of Cuscuta, thus simplifying the bioinformatical process of filtering out host mRNA that has trafficked into the parasite (Leblanc et al., 2012). Consequently, an impressive body of knowledge has recently been accumulated on RNA trafficking in this system (for review, see afore-mentioned references).
Naturally occurring sRNAs can be exchanged across the parasite haustorium thereby affecting gene expression in the host plant. C. campestris haustoria accumulate high levels of many novel miRNAs, 22-nt long, while parasitizing Arabidopsis plants. miRNAs of 22-nt occur less frequently than 21-nt miRNAs in plants and are often associated with accumulation of secondary siRNA from their targets. C. campestris-derived miRNAs are active inside host cells and hijack the host’s own silencing machinery to produce secondary siRNAs. Together, these data suggest that C. campestris trans-species miRNAs function as virulence factors to remodel host gene expression to the parasite’s advantage (Shahid et al., 2018).
Cuscuta is susceptible to HIGS, as exemplified hereafter. Interspecific sRNA-mediated silencing of parasite genes is one of the strategies to produce crops resistant to dodder invasion. A likely candidate would be the KNOTTED-like homeobox transcription factors, including SHOOT MERISTEMLESS-like (STM), which is essential to the development and subsequent establishment of haustorial connections by C. pentagona on tobacco plants. Interspecific silencing of the STM gene in dodder driven by a vascular-specific promoter in transgenic host plants disrupted dodder growth (Alakonya et al., 2012). HIGS technology is therefore a potential effective method for control of infection by plant parasites.
Extracellular DNA
Intracellular or internal DNA (iDNA) is the DNA located within the cell membranes while extracellular, external or environmental DNA (exDNA) represents the DNA located outside cells originating from iDNA by active or passive extrusion mechanisms and/or by cell lysis (Pietramellara et al., 2009). A method to discriminate between iDNA and bound or free exDNA, as well as evaluate various DNA fractions and infer related ratios (ex:iDNA) was developed for microbes, including anaerobic fungi (Nagler et al., 2018b). A thorough overview on the main research areas dealing with exDNA, its origins and functions, and existing and emerging exDNA-based methods and applications was recently published (Nagler et al., 2018a).
Self Versus Non-Self DNA
Across the tree of life, from plants to mammals, immune and nonimmune cells express evolutionarily conserved pattern recognition receptors (PRRs) that sense and recognize DNA as a potential marker of self-damage and/or non-self-organisms, which in turn trigger responses to inflammation, immunity, or pathogen resistance. Self-DNA is an indicator of self-damage and acts as a DAMP. Non-self DNA indicate the presence of a foreign organism and act as a PAMP or a MAMP (Gallucci and Maffei, 2017).
As previously explained, we limit the scope of our review to plants and their interacting fungal partners. However, we acknowledge the breakthrough study by Tran and colleagues (Tran et al., 2016) on the tomato bacterial pathogen Ralstonia solanacearum. Deletion of two exDNase genes, nucA and nucB, trapped bacterial mutants in the exDNA matrix of tomato root border cells, resulting in dampening of the pathogen’s virulence. These findings demonstrate for the first time that exDNases are virulence factors deployed by plant pathogens in the counter defense strategy against host exDNA.
Plants not only depend on small molecules or ions to garner mineral nutrients for growth but are also able to absorb into their roots large organic molecules such as proteins used as an alternative source of nitrogen (Paungfoo-Lonhienne et al., 2008) and DNA as an alternative source of inorganic phosphorous (Paungfoo-Lonhienne et al., 2010a). Uptake of non-self exDNA by plant roots was also shown to have a role in signaling by supplementing hydroponic cultures of Arabidopsis plants with foreign DNA from herring sperm. The addition of foreign DNA induced changes in expression of CLAVATA3/ESR (CLE)-related gene, encoding a peptide hormone implicated in root morphogenesis, that can potentially enhance nutrient absorption by the plant, thus pointing to a DNA-elicited signal pathway (Paungfoo-Lonhienne et al., 2010b).
Once Upon a Slime: Root Border Cells Trap Pathogens
Soil-borne plant pathogens invade their plant hosts, attacking them at the roots. Root tips of plants usually manage to escape infection even when directly inoculated with spores of a fungal pathogen. This happens through a process in which the pathogen is prevented from forming intimate contact with the root surface (Gunawardena and Hawes, 2002). Plant roots harbor a mucilaginous matrix or “slime” around them that contains a population of living cells, referred to as “border cells”, which have separated from the root cap into the environment. Border cells bear intact CWs and are metabolically active, yet their metabolism is significantly different from that of the root cap cells they originate from. The export of newly synthesized DNA into the extracellular matrix among detaching cells at the root cap periphery from Convolvulus arvensis was first documented in 1971 (Phillips and Torrey, 1971), but at the time its biological significance remained unknown.
Analysis of the extracellular mucilage surrounding border cells confirmed the presence of host exDNA even in the absence of pathogens. Enzymatic digestion of this exDNA using DNase I abolished root tip resistance to infection (Wen et al., 2009). Recently, the delivery of plant exDNA in living border cell populations separated from the root caps of pea and corn plants was visualised for the first time (Wen et al., 2017). This study revealed that exDNA is secreted as new border cells disperse from the root cap periphery and that exDNA plays a critical scaffolding role in the structural integrity of the complex extracellular trap structures surrounding root border cells. Border cells may thus provide a protective role to the root tip meristem against infection and injury to maintain development and ensure survival (Hawes et al., 2011; Wen et al., 2017).
Taking Secretomics Seriously: Biotechnological Applications to Solve Food Security Constraints
Given the significant economical and societal impact of plant diseases caused by microbial pathogens, molecular components involved in the control of such devastating diseases have been the object of several biotechnological studies and applications, notably genetic improvement of plant resistance. Identification and introgression of resistance traits into desirable varieties through accelerated breeding and gene editing technologies is essential to global food security (Ghag, 2017; Silva et al., 2018; Pixley et al., 2019; Yin and Qiu, 2019).
Owing to their central role in plant-fungi interactions, fungal secreted effectors are being investigated to address their biotechnological potential in agriculture, using the knowledge gained from effector biology in recent years. In a recent review by Van de Wouw et al. (2019), one strategy is referred to as ‘effector-guided’ breeding and involves the use of purified or heterologously-expressed effectors as well as recombinant effector proteins in agroinfiltration experiments for pathogen-resistance screening purpose. Another approach is based on the characterization of effector-induced defense responses, also referred to as effector-triggered immunity, in particular those mediated by a core of conserved effectors that play key roles in disease development. This will allow the identification of interacting resistance genes in host plants, hence hopefully a more durable resistance. Therefore, the identification of candidate plant resistance genes, using effector-triggered immunity, provides high-throughput tools for screening germplasm and breeding material. Given that traditional breeding is a slow process requiring numerous generations of selection, resorting to biotechnology approaches such as genome editing and GM crops can accelerate the crop improvement process. (Van de Wouw and Idnurm, 2019).
The use of natural and engineered EVs in drug delivery has been used to address several human health problems such as cancer, hepatitis C, neurodegenerative diseases, inflammatory states etc. (Saenz-Cuesta et al., 2015; Raimondo et al., 2019; Wiklander et al., 2019). Furthermore, it is surmised that vesicle-mimetic delivery systems offering properties similar to those of natural EVs will bring new opportunities to deliver cargos to a target cell (Garcia-Manrique et al., 2018; Surman et al., 2019). In recent years, particular attention has been drawn to non-animal (especially plant) EVs and their potential use in human therapy. In particular, fruit-derived exosomes have been isolated, characterized and tested as beneficial products (Wang et al., 2014; Raimondo et al., 2015; Zhuang et al., 2016; Garcia-Manrique et al., 2018). Therefore, the possibility of using edible plant-derived vesicles for the loading of other compounds, whether of vegetal or synthetic origin, for therapeutic purposes appears very promising (Rome, 2019), especially considering that plant EVs can be produced in large quantities and at low cost (Raimondo et al., 2019).
Finally, the demonstration that plant EVs isolated from sunflower seedlings are incorporated by the highly invasive fungal pathogen S. sclerotiorum and cause fungal growth inhibition (Regente et al., 2017) pave the way to develop plant EVs as a delivery system of drugs (metabolites, proteins, nucleic acids) able to combat bacterial and fungal pathogens, as demonstrated for animal EVs in human therapy (Wiklander et al., 2019).
Conclusions
Although numerous plant and fungal secretome studies have predominantly focused on protein secretion, the data reported in the present review highlight that both plants and their interacting fungi secrete a cocktail of nonproteinic molecules including metabolites, phytohormones and nucleic acids, thereby unveiling the multiple facets of plant-fungal interactions. Accordingly, we propose to extend the definition of the plant and fungal secretomes to a broader sense that includes both proteinic and non-proteinic secretions, to comprehensively unravel the functioning of the plant/microorganisms holobiont. How all these secreted molecules are orchestrated and how they interact to sculpt and regulate communication mechanisms between plants and fungi, in both beneficial and pathogenic interactions, remain to be elucidated. It will certainly be the object of future studies aiming at improving crop protection and achieving sustainable agriculture, particularly in the context of anticipated climate changes and increasing global food demand.
Plant–microbe interactions have evolved over hundreds of millions of years, generating a diversity of associations covering a broad spectrum from pathogenic to mutualistic coexistence (Martin et al., 2017; Uhse and Djamei, 2018). Although these various lifestyles incur different needs, they all bear in common the use of secreted molecules, which enable interacting partners to communicate and have an impact on each other and on their environment, and vice versa. Given that secreted molecules, including secondary metabolites, are critical for intercellular communication, it is likely that both plants and their associated fungi have evolved to adopt identical secretion processes, namely conventional and unconventional mechanisms, allowing them to optimize their mutualistic interactions or to combat and counterattack during pathogenesis.
We anticipate that the development of comprehensive secretome studies in plants and fungi will offer new avenues for the identification of proteinic and nonproteinic molecules that can be exploited to develop novel crop protection strategies, notably through the use of plant EVs in drug delivery. Comprehensive secretome analysis will also allow the characterization of plant genes and metabolites central to plant root-rhizospheric microbiote interactions towards improving plant production and protection, while optimizing the use of agrochemical products.
Author Contributions
DV, MR, DJ: drafted, edited, and reviewed the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank our respective institutions for their continuous support.
Abbreviations
AA, amino acid; ABA, abscisic acid; ABC, ATP-binding cassette; ALMT, Aluminum-Activated Malate Transporter; AMP, antimicrobial peptide; ATP, adenosine 5′-triphosphate; AVR, avirulence; BBE, berberine bridge enzyme; CAZyme, carbohydrate-active enzyme; CELLOX, cellodextrin oxidase; CK, cytokinin; CLE, CLAVATA3/ESR; CW, cell wall; CWDE, CW degrading enzyme; CWME, CW methylesterase; DAMP, damage-associated molecular pattern; DCL, dicer-like; DNA, deoxyribonucleic acid; DORN1, Does not Respond to Nucleotides 1; eATP, extracellular ATP; ER, endoplasmic reticulum; ERCC, Extracellular RNA Communication Consortium; EV, extracellular vesicle; exDNA, external DNA; exDNase, extracellular DNases; exRNA, extracellular RNA; GA, gibberellin; GMO, genetically-modified (GM) organism; GUS, β-glucuronidase; HIGS, Host-Induced Gene Silencing; IAA, indole-3-acetic acid; iDNA, internal DNA; IM, invasion molecule; IP, invasion patterns; ISR, induced systemic resistance; JA, jasmonic acid; KEP, KEX2-processed repeat protein; LSP, leaderless secretory proteins; MAMP, microbe-associated molecular pattern; MAPK, mitogen-activated protein kinase; MATE, Multidrug and Toxic compound Extrusion; MFS, Major Facilitator Superfamily; miRNA, microRNA; NGS, next generation sequencing; nt, nucleotides; OG, oligogalacturonide; ORF, Open Reading Frame; P2K, P2 receptor kinase; PAMP, pathogen-associated molecular pattern; PG, polygalacturonase; PGPF, plant growth-promoting fungi; PRR, pattern recognition receptor; PKS-NRPS, polyketide synthase-nonribosomal peptide synthetase; PME, pectin methylesterase; PMEI, PME inhibitor; PR, pathogenesis-related; PRR, pattern recognition receptor; PTM, posttranslational modification; RNA, ribonucleic acid; RNAi, RNA interference; ROS, reactive oxygen species; SA, salicylic acid; siRNA, small interfering RNA; SP, signal peptide; sRNA, small RNA; STM, SHOOT MERISTEMLESS; tyRNA, tinyRNAs; UPS, unconventional protein secretory.
References
- Agrawal G. K., Jwa N. S., Lebrun M. H., Job D., Rakwal R. (2010). Plant secretome: unlocking secrets of the secreted proteins. Proteomics 10 (4), 799–827. 10.1002/pmic.200900514 [DOI] [PubMed] [Google Scholar]
- Ainsztein A. M., Brooks P. J., Dugan V. G., Ganguly A., Guo M., Howcroft T. K., et al. (2015). The NIH Extracellular RNA Communication Consortium. J. Extracell. Vesicles 4, 27493. 10.3402/jev.v4.27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alakonya A., Kumar R., Koenig D., Kimura S., Townsley B., Runo S., et al. (2012). Interspecific RNA interference of SHOOT MERISTEMLESS-like disrupts Cuscuta pentagona plant parasitism. Plant Cell 24 (7), 3153–3166. 10.1105/tpc.112.099994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albenne C., Canut H., Jamet E. (2013). Plant cell wall proteomics: the leadership of Arabidopsis thaliana. Front. Plant Sci. 4, 111. 10.3389/fpls.2013.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amnuaykanjanasin A., Daub M. E. (2009). The ABC transporter ATR1 is necessary for efflux of the toxin cercosporin in the fungus Cercospora nicotianae. Fungal Genet. Biol. 46 (2), 146–158. 10.1016/j.fgb.2008.11.007 [DOI] [PubMed] [Google Scholar]
- An C., Ding Y., Zhang X., Wang C., Mou Z. (2016). Elongator plays a positive role in exogenous NAD-induced defense responses in Arabidopsis. Mol. Plant-Microbe Interact. 29 (5), 396–404. 10.1094/MPMI-01-16-0005-R [DOI] [PubMed] [Google Scholar]
- Badri D. V., Vivanco J. M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32 (6), 666–681. 10.1111/j.1365-3040.2008.01926.x [DOI] [PubMed] [Google Scholar]
- Badri D. V., Loyola-Vargas V. M., Broeckling C. D., De-la-Pena C., Jasinski M., Santelia D., et al. (2008). Altered profile of secondary metabolites in the root exudates of Arabidopsis ATP-binding cassette transporter mutants. Plant Physiol. 146 (2), 762–771. 10.1104/pp.107.109587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri D. V., Quintana N., El Kassis E. G., Kim H. K., Choi Y. H., Sugiyama A., et al. (2009). An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 151 (4), 2006–2017. 10.1104/pp.109.147462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz U., Martinoia E. (2014). Root exudates: the hidden part of plant defense. Trends Plant Sci. 19 (2), 90–98. 10.1016/j.tplants.2013.11.006 [DOI] [PubMed] [Google Scholar]
- Baldrich P., Rutter B. D., Karimi H. Z., Podicheti R., Meyers B. C., Innes R. W. (2019). Plant extracellular vesicles contain diverse small RNA species and are enriched in 10- to 17-Nucleotide “Tiny” RNAs. Plant Cell 31 (2), 315–324. 10.1105/tpc.18.00872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellincampi D., Cervone F., Lionetti V. (2014). Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions. Front. Plant Sci. 5, 228. 10.3389/fpls.2014.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellucci M., De Marchis F., Pompa A. (2017). The endoplasmic reticulum is a hub to sort proteins toward unconventional traffic pathways and endosymbiotic organelles. J. Exp. Bot. 69 (1), 7–20. 10.1093/jxb/erx286 [DOI] [PubMed] [Google Scholar]
- Bendtsen J. D., Jensen L. J., Blom N., Von Heijne G., Brunak S. (2004). Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17 (4), 349–356. 10.1093/protein/gzh037 [DOI] [PubMed] [Google Scholar]
- Benedetti M., Pontiggia D., Raggi S., Cheng Z., Scaloni F., Ferrari S., et al. (2015). Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proc. Natl. Acad. Sci. U.S.A. 112 (17), 5533–5538. 10.1073/pnas.1504154112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat A., Ryu C. M. (2016). Plant perceptions of extracellular DNA and RNA. Mol. Plant 9 (7), 956–958. 10.1016/j.molp.2016.05.014 [DOI] [PubMed] [Google Scholar]
- Bielska E., Sisquella M. A., Aldeieg M., Birch C., O’Donoghue E. J., May R. C. (2018). Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nat. Commun. 9 (1), 1556. 10.1038/s41467-018-03991-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielska E., Birch P. R. J., Buck A. H., Abreu-Goodger C., Innes R. W., Jin H., et al. (2019). Highlights of the mini-symposium on extracellular vesicles in inter-organismal communication, held in Munich, Germany, August 2018. J. Extracell Vesicles 8 (1), 1590116. 10.1080/20013078.2019.1590116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleackley M. R., Dawson C. S., Anderson M. A. (2019). Fungal extracellular vesicles with a focus on proteomic analysis. Proteomics 19 (8), e1800232. 10.1002/pmic.201800232 [DOI] [PubMed] [Google Scholar]
- Boevink P. C. (2017). Exchanging missives and missiles: the roles of extracellular vesicles in plant-pathogen interactions. J. Exp. Bot. 68 (20), 5411–5414. 10.1093/jxb/erx369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin S., Fonouni-Farde C., Frugier F. (2016). How auxin and cytokinin phytohormones modulate root microbe interactions. Front. Plant Sci. 7, 1240. 10.3389/fpls.2016.01240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen S., Solomon P. S., Bedon F., Vincent D. (2015). Surveying the potential of secreted antimicrobial peptides to enhance plant disease resistance. Front. Plant Sci. 6, 900. 10.3389/fpls.2015.00900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L., Wolf J. M., Prados-Rosales R., Casadevall A. (2015). Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 13 (10), 620–630. 10.1038/nrmicro3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Qiao L., Wang M., He B., Lin F. M., Palmquist J., et al. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360 (6393), 1126–1129. 10.1126/science.aar4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37 (Database issue), D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalhais L. C., Schenk P. M., Dennis P. G. (2017). Jasmonic acid signalling and the plant holobiont. Curr. Opin. Microbiol. 37, 42–47. 10.1016/j.mib.2017.03.009 [DOI] [PubMed] [Google Scholar]
- Casadevall A., Nosanchuk J. D., Williamson P., Rodrigues M. L. (2009). Vesicular transport across the fungal cell wall. Trends Microbiol. 17 (4), 158–162. 10.1016/j.tim.2008.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanclud E., Morel J. B. (2016). Plant hormones: a fungal point of view. Mol. Plant Pathol. 17 (8), 1289–1297. 10.1111/mpp.12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda A., Roze L. V., Kang S., Artymovich K. A., Hicks G. R., Raikhel N. V., et al. (2009). A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 106 (46), 19533–19538. 10.1073/pnas.0907416106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda A., Roze L. V., Linz J. E. (2010). A possible role for exocytosis in aflatoxin export in Aspergillus parasiticus. Eukaryot. Cell 9 (11), 1724–1727. 10.1128/EC.00118-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. H., Nguyen C. T., Choi J., Stacey G. (2017). Molecular mechanism of plant recognition of extracellular ATP. Adv. Exp. Med. Biol. 1051, 233–253. 10.1007/5584_2017_110 [DOI] [PubMed] [Google Scholar]
- Choi J., Tanaka K., Cao Y., Qi Y., Qiu J., Liang Y., et al. (2014. a). Identification of a plant receptor for extracellular ATP. Science 343 (6168), 290–294. 10.1126/science.343.6168.290 [DOI] [PubMed] [Google Scholar]
- Choi J., Tanaka K., Liang Y., Cao Y., Lee S. Y., Stacey G. (2014. b). Extracellular ATP, a danger signal, is recognized by DORN1 in Arabidopsis. Biochem. J. 463 (3), 429–437. 10.1042/BJ20140666 [DOI] [PubMed] [Google Scholar]
- Claridge B., Kastaniegaard K., Stensballe A., Greening D. W. (2019). Post-translational and transcriptional dynamics - regulating extracellular vesicle biology. Expert Rev. Proteomics 16 (1), 17–31. 10.1080/14789450.2019.1551135 [DOI] [PubMed] [Google Scholar]
- Coleman J. J., Mylonakis E. (2009). Efflux in fungi: la piece de resistance. PloS Pathog. 5 (6), e1000486. 10.1371/journal.ppat.1000486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collemare J., Billard A., Bohnert H. U., Lebrun M. H. (2008. a). Biosynthesis of secondary metabolites in the rice blast fungus Magnaporthe grisea: the role of hybrid PKS-NRPS in pathogenicity. Mycol. Res. 112 (Pt 2), 207–215. 10.1016/j.mycres.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Collemare J., Pianfetti M., Houlle A. E., Morin D., Camborde L., Gagey M. J., et al. (2008. b). Magnaporthe grisea avirulence gene ACE1 belongs to an infection-specific gene cluster involved in secondary metabolism. New Phytol. 179 (1), 196–208. 10.1111/j.1469-8137.2008.02459.x [DOI] [PubMed] [Google Scholar]
- Collemare J., O’Connell R., Lebrun M. H. (2019). Nonproteinaceous effectors: the terra incognita of plant–fungal interactions. New Phytol. 223, 590–596. 10.1111/nph.15785 [DOI] [PubMed] [Google Scholar]
- Cologna N. M. D., Gomez-Mendoza D. P., Zanoelo F. F., Giannesi G. C., Guimaraes N. C. A., Moreira L. R. S., et al. (2018). Exploring Trichoderma and Aspergillus secretomes: proteomics approaches for the identification of enzymes of biotechnological interest. Enzyme Microb. Technol. 109, 1–10. 10.1016/j.enzmictec.2017.08.007 [DOI] [PubMed] [Google Scholar]
- Cook D. E., Mesarich C. H., Thomma B. P. (2015). Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 53, 541–563. 10.1146/annurev-phyto-080614-120114 [DOI] [PubMed] [Google Scholar]
- Dalmais B., Schumacher J., Moraga J., P L. E. P., Tudzynski B., Collado I. G., et al. (2011). The Botrytis cinerea phytotoxin botcinic acid requires two polyketide synthases for production and has a redundant role in virulence with botrydial. Mol. Plant Pathol. 12 (6), 564–579. 10.1111/j.1364-3703.2010.00692.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Extracellular R.N.A.C.C., Ansel K. M., Bitzer M., Breakefield X. O., Charest A., et al. (2019). The extracellular RNA communication consortium: establishing foundational knowledge and technologies for extracellular RNA research. Cell 177 (2), 231–242. 10.1016/j.cell.2019.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daumann M., Fischer M., Niopek-Witz S., Girke C., Mohlmann T. (2015). Apoplastic nucleoside accumulation in arabidopsis leads to reduced photosynthetic performance and increased susceptibility against botrytis cinerea. Front. Plant Sci. 6, 1158. 10.3389/fpls.2015.01158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D. J., Kang B. H., Heringer A. S., Wilkop T. E., Drakakaki G. (2016). Unconventional protein secretion in plants. Methods Mol. Biol. 1459, 47–63. 10.1007/978-1-4939-3804-9_3 [DOI] [PubMed] [Google Scholar]
- de la Canal L., Pinedo M. (2018). Extracellular vesicles: a missing component in plant cell wall remodeling. J. Exp. Bot. 69 (20), 4655–4658. 10.1093/jxb/ery255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S., de Vries J., Rose L. E. (2019). The elaboration of miRNA regulation and gene regulatory networks in plant(-)microbe interactions. Genes (Basel) 10 (4), 13. 10.3390/genes10040310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunois B., Jeandet P., Clement C., Baillieul F., Dorey S., Cordelier S. (2014). Uncovering plant-pathogen crosstalk through apoplastic proteomic studies. Front. Plant Sci. 5, 249. 10.3389/fpls.2014.00249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di X., Takken F. L., Tintor N. (2016). How phytohormones shape interactions between plants and the soil-borne fungus fusarium oxysporum. Front. Plant Sci. 7, 170. 10.3389/fpls.2016.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieryckx C., Gaudin V., Dupuy J. W., Bonneu M., Girard V., Job D. (2015). Beyond plant defense: insights on the potential of salicylic and methylsalicylic acid to contain growth of the phytopathogen Botrytis cinerea. Front. Plant Sci. 6, 859. 10.3389/fpls.2015.00859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Flores D., Heil M. (2018). Extracellular self-DNA as a damage-associated molecular pattern (DAMP) that triggers self-specific immunity induction in plants. Brain Behav. Immun. 72, 78–88. 10.1016/j.bbi.2017.10.010 [DOI] [PubMed] [Google Scholar]
- Ellendorff U., Fradin E. F., de Jonge R., Thomma B. P. (2009). RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 60 (2), 591–602. 10.1093/jxb/ern306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2 (4), 953–971. 10.1038/nprot.2007.131 [DOI] [PubMed] [Google Scholar]
- Feng H., Guan D., Bai J., Sun K., Jia L. (2015). Extracellular ATP: a potential regulator of plant cell death. Mol. Plant Pathol. 16 (6), 633–639. 10.1111/mpp.12219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine S., Mariotti A., Abbadie L. (2013). The priming effect of organic matter: a question of microbial competition? Soil Biol. Biochem. 35, 837–843. 10.1016/S0038-0717(03)00123-8 [DOI] [Google Scholar]
- Franceschetti M., Maqbool A., Jimenez-Dalmaroni M. J., Pennington H. G., Kamoun S., Banfield M. J. (2017). Effectors of filamentous plant pathogens: commonalities amid diversity. Microbiol. Mol. Biol. Rev. 81 (2), 17. 10.1128/MMBR.00066-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Wang S. (2011). Insights into auxin signaling in plant-pathogen interactions. Front. Plant Sci. 2, 74. 10.3389/fpls.2011.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallucci S., Maffei M. E. (2017). DNA sensing across the tree of life. Trends Immunol. 38 (10), 719–732. 10.1016/j.it.2017.07.012 [DOI] [PubMed] [Google Scholar]
- Garcia K., Ane J. M. (2016). Comparative analysis of secretomes from ectomycorrhizal fungi with an emphasis on small-secreted proteins. Front. Microbiol. 7, 1734. 10.3389/fmicb.2016.01734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Manrique P., Matos M., Gutierrez G., Pazos C., Blanco-Lopez M. C. (2018). Therapeutic biomaterials based on extracellular vesicles: classification of bio-engineering and mimetic preparation routes. J. Extracell. Vesicles 7 (1), 1422676. 10.1080/20013078.2017.1422676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargallo-Garriga A., Preece C., Sardans J., Oravec M., Urban O., Penuelas J. (2018). Root exudate metabolomes change under drought and show limited capacity for recovery. Sci. Rep. 8 (1), 12696. 10.1038/s41598-018-30150-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghag S. B. (2017). Host induced gene silencing, an emerging science to engineer crop resistance against harmful plant pathogens. Physiol. Mol. Plant Pathol. 100, 242–254. 10.1016/j.pmpp.2017.10.003 [DOI] [Google Scholar]
- Ghahremani M., Stigter K. A., Plaxton W. (2016). Extraction and characterization of extracellular proteins and their post-translational modifications from arabidopsis thaliana suspension cell cultures and seedlings: a critical review. Proteomes 4 (3), 26. 10.3390/proteomes4030025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo M. C., Dagdas Y. F., Gupta Y. K., Mentlak T. A., Yi M., Martinez-Rocha A. L., et al. (2013). Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 4, 1996. 10.1038/ncomms2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena U., Hawes M. C. (2002). Tissue specific localization of root infection by fungal pathogens: role of root border cells. Mol. Plant Microbe Interact. 15 (11), 1128–1136. 10.1094/MPMI.2002.15.11.1128 [DOI] [PubMed] [Google Scholar]
- Gupta R., Lee S. E., Agrawal G. K., Rakwal R., Park S., Wang Y., et al. (2015). Understanding the plant-pathogen interactions in the context of proteomics-generated apoplastic proteins inventory. Front. Plant Sci. 6, 352. 10.3389/fpls.2015.00352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Luan Y. S. (2015). Horizontal transfer of small RNAs to and from plants. Front. Plant Sci. 6, 1113. 10.3389/fpls.2015.01113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L. L., Nielsen M. E. (2017). Plant exosomes: using an unconventional exit to prevent pathogen entry? J. Exp. Bot. 69 (1), 59–68. 10.1093/jxb/erx319 [DOI] [PubMed] [Google Scholar]
- Harman G. E., Petzoldt R., Comis A., Chen J. (2004). Interactions between trichoderma harzianum strain T22 and maize inbred line Mo17 and effects of these interactions on diseases caused by pythium ultimum and colletotrichum graminicola. Phytopathology 94 (2), 147–153. 10.1094/PHYTO.2004.94.2.147 [DOI] [PubMed] [Google Scholar]
- Hawes M. C., Curlango-Rivera G., Wen F., White G. J., Vanetten H. D., Xiong Z. (2011). Extracellular DNA: the tip of root defenses? Plant Sci. 180 (6), 741–745. 10.1016/j.plantsci.2011.02.007 [DOI] [PubMed] [Google Scholar]
- Hof C., Eisfeld K., Welzel K., Antelo L., Foster A. J., Anke H. (2007). Ferricrocin synthesis in Magnaporthe grisea and its role in pathogenicity in rice. Mol. Plant Pathol. 8 (2), 163–172. 10.1111/j.1364-3703.2007.00380.x [DOI] [PubMed] [Google Scholar]
- Hossain M., Sultana F., Islam S. (2017). Plant growth-promoting fungi (PGPF): Phytostimulation and induced systemic resistance. Plant-Microbe Interact. In Agro-Ecol. Perspect. 2, 135–191. 10.1007/978-981-10-6593-4_6 [DOI] [Google Scholar]
- Hu L., Robert C. A. M., Cadot S., Zhang X., Ye M., Li B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9 (1), 2738. 10.1038/s41467-018-05122-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua C., Zhao J. H., Guo H. S. (2018). Trans-kingdom RNA silencing in plant-fungal pathogen interactions. Mol. Plant 11 (2), 235–244. 10.1016/j.molp.2017.12.001 [DOI] [PubMed] [Google Scholar]
- Islam W., Islam S. U., Qasim M., Wang L. (2017). Host-Pathogen interactions modulated by small RNAs. RNA Biol. 14 (7), 891–904. 10.1080/15476286.2017.1318009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-Bueno I., González-Rodríguez V. E., Simon A., Dalmais B., Pradier J.-M., Le Pêcheur P., et al. (2018). Biosynthesis of abscisic acid in fungi: identification of a sesquiterpene cyclase as the key enzyme in Botrytis cinerea. Environ. Microbiol. 20, 2469–2482. 10.1111/1462-2920.14258 [DOI] [PubMed] [Google Scholar]
- Jacoby R. P., Kopriva S. (2018). Metabolic niches in the rhizosphere microbiome: new tools and approaches to analyse metabolic mechanisms of plant-microbe nutrient exchange. J. Exp. Bot. 70 (4), 1087–1094. 10.1093/jxb/ery438 [DOI] [PubMed] [Google Scholar]
- Jiang C. J., Shimono M., Sugano S., Kojima M., Liu X., Inoue H., et al. (2013). Cytokinins act synergistically with salicylic acid to activate defense gene expression in rice. Mol. Plant Microbe Interact. 26 (3), 287–296. 10.1094/MPMI-06-12-0152-R [DOI] [PubMed] [Google Scholar]
- Johnson N. R., Axtell M. J. (2019). Small RNA warfare: exploring origins and function of trans-species microRNAs from the parasitic plant Cuscuta. Curr. Opin. Plant Biol. 50, 76–81. 10.1016/j.pbi.2019.03.014 [DOI] [PubMed] [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. Nature 444 (7117), 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kamoun S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60. 10.1146/annurev.phyto.44.070505.143436 [DOI] [PubMed] [Google Scholar]
- Kanyuka K., Rudd J. J. (2019). Cell surface immune receptors: the guardians of the plant’s extracellular spaces. Curr. Opin. Plant Biol. 50, 1–8. 10.1016/j.pbi.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehr J., Buhtz A. (2008). Long distance transport and movement of RNA through the phloem. J. Exp. Bot. 59 (1), 85–92. 10.1093/jxb/erm176 [DOI] [PubMed] [Google Scholar]
- Keller N. P., Turner G., Bennett J. W. (2005). Fungal secondary metabolism - from biochemistry to genomics. Nat. Rev. Microbiol. 3 (12), 937–947. 10.1038/nrmicro1286 [DOI] [PubMed] [Google Scholar]
- Khedr M. A., Massarotti A., Mohamed M. E. (2018). Rational discovery of (+) (S) abscisic acid as a potential antifungal agent: a repurposing approach. Sci. Rep. 8 (1), 8565. 10.1038/s41598-018-26998-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G., Westwood J. H. (2015). Macromolecule exchange in Cuscuta-host plant interactions. Curr. Opin. Plant Biol. 26, 20–25. 10.1016/j.pbi.2015.05.012 [DOI] [PubMed] [Google Scholar]
- Kim K. T., Jeon J., Choi J., Cheong K., Song H., Choi G., et al. (2016). Kingdom-wide analysis of fungal small secreted proteins (SSPs) reveals their potential role in host association. Front. Plant Sci. 7, 186. 10.3389/fpls.2016.00186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knip M., Constantin M. E., Thordal-Christensen H. (2014). Trans-kingdom cross-talk: small RNAs on the move. PloS Genet. 10 (9), e1004602. 10.1371/journal.pgen.1004602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyakov Y., Domanski G. (2000). Carbon input by plants into the soil. J. Plant Nutr. Soil Sci. 163, 421–431. [DOI] [Google Scholar]
- Le Marquer M., Clemente H. S., Roux C., Savelli B., Dit Frey N. F. (2019. a). Correction to: Identification of new signalling peptides through a genome-wide survey of 250 fungal secretomes. BMC Genomics 20 (1), 94. 10.1186/s12864-019-5466-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Marquer M., San Clemente H., Roux C., Savelli B., Frei Dit Frey N. (2019. b). Identification of new signalling peptides through a genome-wide survey of 250 fungal secretomes. BMC Genomics 20 (1), 64. 10.1186/s12864-018-5414-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeis S. L., Paredes S. H., Lundberg D. S., Breakfield N., Gehring J., McDonald M., et al. (2015). Plant microbiome. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349 (6250), 860–864. 10.1126/science.aaa8764 [DOI] [PubMed] [Google Scholar]
- Leblanc M., Kim G., Westwood J. H. (2012). RNA trafficking in parasitic plant systems. Front. Plant Sci. 3, 203. 10.3389/fpls.2012.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J. (2019). Microbe-host communication by small RNAs in extracellular vesicles: vehicles for transkingdom RNA transportation. Int. J. Mol. Sci. 20 (6). 10.3390/ijms20061487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre F., Boutry M. (2018). Towards identification of the substrates of ATP-binding cassette transporters. Plant Physiol. 178 (1), 18–39. 10.1104/pp.18.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V., Fabri E., De Caroli M., Hansen A. R., Willats W. G., Piro G., et al. (2017). Three pectin methylesterase inhibitors protect cell wall integrity for arabidopsis immunity to botrytis. Plant Physiol. 173 (3), 1844–1863. 10.1104/pp.16.01185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci F., Benedetti M., Pontiggia D., Citterico M., Caprari C., Mattei B., et al. (2019). An Arabidopsis berberine bridge enzyme-like protein specifically oxidizes cellulose oligomers and plays a role in immunity. Plant J. 98 (3), 540–554. 10.1111/tpj.14237 [DOI] [PubMed] [Google Scholar]
- Lombardi N., Vitale S., Turra D., Reverberi M., Fanelli C., Vinale F., et al. (2018). Root exudates of stressed plants stimulate and attract trichoderma soil fungi. Mol. Plant Microbe Interact. 31 (10), 982–994. 10.1094/MPMI-12-17-0310-R [DOI] [PubMed] [Google Scholar]
- Lubna Asaf S., Hamayun M., Gul H., Lee I. J., Hussain A. (2018). Aspergillus niger CSR3 regulates plant endogenous hormones and secondary metabolites by producing gibberellins and indoleacetic acid. J. Plant Interact. 13, 100–111. 10.1080/17429145.2018.1436199 [DOI] [Google Scholar]
- Lum G., Meinken J., Orr J., Frazier S., Min J. M. (2013). PlantSecKB: the plant secretome and subcellular proteome knowledgebase. Comput. Mol. Biol. 4 (1), 1–17. 10.5376/cmb.2014.040001 [DOI] [Google Scholar]
- Ma K. W., Ma W. (2016). Phytohormone pathways as targets of pathogens to facilitate infection. Plant Mol. Biol. 91 (6), 713–725. 10.1007/s11103-016-0452-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis L., Sadovsky Y. (2019). The biology of extracellular vesicles: The known unknowns. PloS Biol. 17 (7), e3000363. 10.1371/journal.pbio.3000363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-González A. P., Ardila H. D., Martínez-Peralta S. T., Melgarejo-Muñoz L. M., Castillejo-Sánchez M. A., Jorrín-Novo J. V. (2018). What proteomic analysis of the apoplast tells us about plant–pathogen interactions. Plant Pathol. 67 (8), 1647–1668. 10.1111/ppa.12893 [DOI] [Google Scholar]
- Martin F. M., Uroz S., Barker D. G. (2017). Ancestral alliances: Plant mutualistic symbioses with fungi and bacteria. Science 356 (6340), 9. 10.1126/science.aad4501 [DOI] [PubMed] [Google Scholar]
- Marzec M. (2011). Extracellular nucleotides in plants. Postepy Biologii. Komorki. 38, 3–18. [Google Scholar]
- Mazzoleni S., Cartenì F., Bonanomi G., Senatore M., Termolino P., Giannino F., et al. (2015. a). Inhibitory effects of extracellular self-DNA: a general biological process? New Phytol. 206 (1), 127–132. 10.1111/nph.13306 [DOI] [PubMed] [Google Scholar]
- Mazzoleni S., Bonanomi G., Incerti G., Chiusano M. L., Termolino P., Mingo A. (2015. b). Inhibitory and toxic effects of extracellular self-DNA in litter: a mechanism for negative plant-soil feedbacks? New Phytol. 205 (3), 1195–1210. 10.1111/nph.13121 [DOI] [PubMed] [Google Scholar]
- Meijer H. J., Mancuso F. M., Espadas G., Seidl M. F., Chiva C., Govers F., et al. (2014). Profiling the secretome and extracellular proteome of the potato late blight pathogen Phytophthora infestans. Mol. Cell. Proteomics 13 (8), 2101–2113. 10.1074/mcp.M113.035873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M. (2012). The small RNA-based odyssey of epigenetic information in plants: from cells to species. DNA Cell Biol. 31 (12), 1650–1656. 10.1089/dna.2012.1681 [DOI] [PubMed] [Google Scholar]
- Miura N., Ueda M. (2018). Evaluation of Unconventional Protein Secretion by Saccharomyces cerevisiae and other Fungi. Cells 7 (9), 14. 10.3390/cells7090128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgado L., Johannes F. (2017). Computational tools for plant small RNA detection and categorization. Brief Bioinform. 1–12. 10.1093/bib/bbx136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison E. N., Emery R. J. N., Saville B. J. (2017). Fungal derived cytokinins are necessary for normal Ustilago maydis infection of maize. Plant Pathol. 66, 726–742. 10.1111/ppa.12629 [DOI] [Google Scholar]
- Muria-Gonzalez M. J., Chooi Y. H., Breen S., Solomon P. S. (2015). The past, present and future of secondary metabolite research in the Dothideomycetes. Mol. Plant Pathol. 16 (1), 92–107. 10.1111/mpp.12162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagler M., Insam H., Pietramellara G., Ascher-Jenull J. (2018. a). Extracellular DNA in natural environments: features, relevance and applications. Appl. Microbiol. Biotechnol. 102 (15), 6343–6356. 10.1007/s00253-018-9120-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagler M., Podmirseg S. M., Griffith G. W., Insam H., Ascher-Jenull J. (2018. b). The use of extracellular DNA as a proxy for specific microbial activity. Appl. Microbiol. Biotechnol. 102 (6), 2885–2898. 10.1007/s00253-018-8786-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., Tsirigos K. D., Brunak S., von Heijne G. (2019). A Brief History of protein sorting prediction. Protein J. 38 (3), 200–216. 10.1007/s10930-019-09838-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizam S., Qiang X., Wawra S., Nostadt R., Getzke F., Schwanke F., et al. (2019). Serendipita indica E5’NT modulates extracellular nucleotide levels in the plant apoplast and affects fungal colonization. EMBO Rep. 20 (2), 17. 10.15252/embr.201847430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira-Lopez G., Greenwood D. R., Middleditch M., Winefield C., Eaton C., Steyaert J. M., et al. (2018). The apoplastic secretome of trichoderma virens during interaction with maize roots shows an inhibition of plant defence and scavenging oxidative stress secreted proteins. Front. Plant Sci. 9, 409. 10.3389/fpls.2018.00409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A. E. (2001). Tox-boxes, fungal secondary metabolites, and plant disease. Proc. Natl. Acad. Sci. U.S.A. 98 (25), 14187–14188. 10.1073/pnas.261573598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. J., Wang W., Curlango-Rivera G., Xiong Z., Lin Z., Huskey D. A., et al. (2019). A DNase from a fungal phytopathogen is a virulence factor likely deployed as counter defense against host secreted extracellular DNA. mBio 10, e02805–e02818. 10.1128/mBio.02805-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paungfoo-Lonhienne C., Lonhienne T. G., Rentsch D., Robinson N., Christie M., Webb R. I., et al. (2008). Plants can use protein as a nitrogen source without assistance from other organisms. Proc. Natl. Acad. Sci. U.S.A. 105 (11), 4524–4529. 10.1073/pnas.0712078105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paungfoo-Lonhienne C., Lonhienne T. G., Mudge S. R., Schenk P. M., Christie M., Carroll B. J., et al. (2010. a). DNA is taken up by root hairs and pollen, and stimulates root and pollen tube growth. Plant Physiol. 153 (2), 799–805. 10.1104/pp.110.154963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paungfoo-Lonhienne C., Lonhienne T. G., Schmidt S. (2010. b). DNA uptake by Arabidopsis induces changes in the expression of CLE peptides which control root morphology. Plant Signal. Behav. 5 (9), 1112–1114. 10.4161/psb.5.9.12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin C., Morin E., Martin F. M., Veneault-Fourrey C. (2015). Comparative analysis of secretomes from ectomycorrhizal fungi with an emphasis on small-secreted proteins. Front. Microbiol. 6, 1278. 10.3389/fmicb.2015.01278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrussa E., Braidot E., Zancani M., Peresson C., Bertolini A., Patui S., et al. (2013). Plant flavonoids–biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 14 (7), 14950–14973. 10.3390/ijms140714950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips H. L., Torrey J. G. (1971). Deoxyribonucleic Acid synthesis in root cap cells of cultured roots of convolvulus. Plant Physiol. 48 (2), 213–218. 10.1104/pp.48.2.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietramellara G., Ascher J., Borgogni F., Ceccherini M. T., Guerri G., Nannipieri P. (2009). Extracellular DNA in soil and sediment: fate and ecological relevance. Biol. Fert. Soils 45 (3), 219–235. 10.1007/s00374-008-0345-8 [DOI] [Google Scholar]
- Pinedo M., Regente M., Elizalde M., Quiroga I. Y., Pagnussat L. A., Jorrin-Novo J., et al. (2012). Extracellular sunflower proteins: evidence on non-classical secretion of a jacalin-related lectin. Protein Pept. Lett. 19 (3), 270–276. 10.2174/092986612799363163 [DOI] [PubMed] [Google Scholar]
- Pixley K. V., Falck-Zepeda J. B., Giller K. E., Glenna L. L., Gould F., Mallory-Smith C. A., et al. (2019). Genome editing, gene drives, and synthetic biology: will they contribute to disease-resistant crops, and who will benefit? Annu. Rev. Phytopathol. 57, 165–188. 10.1146/annurev-phyto-080417-045954 [DOI] [PubMed] [Google Scholar]
- Poulsen C. P., Dilokpimol A., Mouille G., Burow M., Geshi N. (2014). Arabinogalactan glycosyltransferases target to a unique subcellular compartment that may function in unconventional secretion in plants. Traffic 15 (11), 1219–1234. 10.1111/tra.12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusztahelyi T., Holb I. J., Pocsi I. (2015). Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 6, 573. 10.3389/fpls.2015.00573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M., Mei Y., Grayczyk J. P., Darben L. M., Rieker M. E. G., Seitz J. M., et al. (2019). Candidate effectors from uromyces appendiculatus, the causal agent of rust on common bean, can be discriminated based on suppression of immune responses. Front. Plant Sci. 10, 1182. 10.3389/fpls.2019.01182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi M., Ellis J. G., Ludowici V. A., Hardham A. R., Dodds P. N. (2012). Challenges and progress towards understanding the role of effectors in plant-fungal interactions. Curr. Opin. Plant Biol. 15 (4), 477–482. 10.1016/j.pbi.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Raimondo S., Naselli F., Fontana S., Monteleone F., Lo Dico A., Saieva L., et al. (2015). Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death. Oncotarget 6 (23), 19514–19527. 10.18632/oncotarget.4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondo S., Giavaresi G., Lorico A., Alessandro R. (2019). Extracellular vesicles as biological shuttles for targeted therapies. Int. J. Mol. Sci. 20 (8), E1848. 10.3390/ijms20081848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regente M., Corti-Monzon G., Maldonado A. M., Pinedo M., Jorrin J., de la Canal L. (2009). Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Lett. 583 (20), 3363–3366. 10.1016/j.febslet.2009.09.041 [DOI] [PubMed] [Google Scholar]
- Regente M., Pinedo M., San Clemente H., Balliau T., Jamet E., de la Canal L. (2017). Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J. Exp. Bot. 68 (20), 5485–5495. 10.1093/jxb/erx355 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Navarro L., Bari R., Jones J. D. (2007). Pathological hormone imbalances. Curr. Opin. Plant Biol. 10 (4), 372–379. 10.1016/j.pbi.2007.06.003 [DOI] [PubMed] [Google Scholar]
- Rome S. (2019). Biological properties of plant-derived extracellular vesicles. Food Funct. 10 (2), 529–538. 10.1039/c8fo02295j [DOI] [PubMed] [Google Scholar]
- Rose L. E., Overdijk E. J. R., van Damme M. (2019). Small RNA molecules and their role in plant disease. Eur. J. Plant Pathol. 153 (1), 1–14. 10.1007/s10658-018-01614-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze L. V., Chanda A., Linz J. E. (2011). Compartmentalization and molecular traffic in secondary metabolism: a new understanding of established cellular processes. Fungal Genet. Biol. 48 (1), 35–48. 10.1016/j.fgb.2010.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter B. D., Innes R. W. (2017). Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 173 (1), 728–741. 10.1104/pp.16.01253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter B. D., Innes R. W. (2018). Extracellular vesicles as key mediators of plant-microbe interactions. Curr. Opin. Plant Biol. 44, 16–22. 10.1016/j.pbi.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Rutter B. D., Rutter K. L., Innes R. W. (2017). Isolation and quantification of plant extracellular vesicles. Bio-protocol 7 (17), e2533. 10.21769/BioProtoc.2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz-Cuesta M., Mittelbrunn M., Otaegui D. (2015). Editorial: novel clinical applications of extracellular vesicles. Front. Immunol. 6, 381. 10.3389/fimmu.2015.00381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse J., Martinoia E., Northen T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23 (1), 25–41. 10.1016/j.tplants.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Saunders D. G., Win J., Cano L. M., Szabo L. J., Kamoun S., Raffaele S. (2012). Using hierarchical clustering of secreted protein families to classify and rank candidate effectors of rust fungi. PloS One 7 (1), e29847. 10.1371/journal.pone.0029847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid S., Kim G., Johnson N. R., Wafula E., Wang F., Coruh C., et al. (2018). MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553 (7686), 82–85. 10.1038/nature25027 [DOI] [PubMed] [Google Scholar]
- Silva M. S., Arraes F. B. M., Campos M. A., Grossi-de-Sa M., Fernandez D., Candido E. S., et al. (2018). Review: potential biotechnological assets related to plant immunity modulation applicable in engineering disease-resistant crops. Plant Sci. 270, 72–84. 10.1016/j.plantsci.2018.02.013 [DOI] [PubMed] [Google Scholar]
- Song Z., Bakeer W., Marshall J. W., Yakasai A. A., Khalid R. M., Collemare J., et al. (2015). Heterologous expression of the avirulence gene ACE1 from the fungal rice pathogen Magnaporthe oryzae. Chem. Sci. 6 (8), 4837–4845. 10.1039/c4sc03707c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence C. A., Lakshmanan V., Donofrio N., Bais H. P. (2015). Crucial roles of abscisic acid biogenesis in virulence of rice blast fungus magnaporthe oryzae. Front. Plant Sci. 6, 1082. 10.3389/fpls.2015.01082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J., Dodds P. N., Gardiner D. M., Singh K. B., Taylor J. M. (2018. a). Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 19 (9), 2094–2110. 10.1111/mpp.12682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J., Dodds P. N., Singh K. B., Taylor J. M. (2018. b). ApoplastP: prediction of effectors and plant proteins in the apoplast using machine learning. New Phytol. 217 (4), 1764–1778. 10.1111/nph.14946 [DOI] [PubMed] [Google Scholar]
- Stock J., Terfruchte M., Schipper K. (2016). A Reporter System to Study Unconventional Secretion of Proteins Avoiding N-Glycosylation in Ustilago maydis. Methods Mol. Biol. 1459, 149–160. 10.1007/978-1-4939-3804-9_10 [DOI] [PubMed] [Google Scholar]
- Strehmel N., Bottcher C., Schmidt S., Scheel D. (2014). Profiling of secondary metabolites in root exudates of Arabidopsis thaliana. Phytochemistry 108, 35–46. 10.1016/j.phytochem.2014.10.003 [DOI] [PubMed] [Google Scholar]
- Surman M., Drozdz A., Stepien E., Przybylo M. (2019). Extracellular vesicles as drug delivery systems - methods of production and potential therapeutic applications. Curr. Pharm. Des. 25 (2), 132–154. 10.2174/1381612825666190306153318 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Choi J., Cao Y., Stacey G. (2014). Extracellular ATP acts as a damage-associated molecular pattern (DAMP) signal in plants. Front. Plant Sci. 5, 446. 10.3389/fpls.2014.00446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanveer T., Shaheen K., Parveen S., Kazi A. G., Ahmad P. (2014). Plant secretomics: identification, isolation, and biological significance under environmental stress. Plant Signal. Behav. 9 (8), e29426. 10.4161/psb.29426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisserant E., Malbreil M., Kuo A., Kohler A., Symeonidi A., Balestrini R., et al. (2013). Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. U. S. A. 110 (50), 20117–20122. 10.1073/pnas.1313452110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T. M., MacIntyre A., Hawes M., Allen C. (2016). Escaping underground nets: extracellular DNases degrade plant extracellular traps and contribute to virulence of the plant pathogenic bacterium ralstonia solanacearum. PloS Pathog. 12 (6), e1005686. 10.1371/journal.ppat.1005686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi D., Zhang T., Koo A. J., Stacey G., Tanaka K. (2018). Extracellular ATP acts on jasmonate signaling to reinforce plant defense. Plant Physiol. 176 (1), 511–523. 10.1104/pp.17.01477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsatsaronis J. A., Franch-Arroyo S., Resch U., Charpentier E. (2018). Extracellular vesicle RNA: a universal mediator of microbial communication? Trends Microbiol. 26 (5), 401–410. 10.1016/j.tim.2018.02.009 [DOI] [PubMed] [Google Scholar]
- Uhse S., Djamei A. (2018). Effectors of plant-colonizing fungi and beyond. PloS Pathog. 14 (6), e1006992. 10.1371/journal.ppat.1006992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S., Xu X., Lowry D., Jackson J. C., Roberson R. W., Lin X. (2016). Subcellular compartmentalization and trafficking of the biosynthetic machinery for fungal melanin. Cell Rep. 14 (11), 2511–2518. 10.1016/j.celrep.2016.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam N. M., Bouwmeester H. J. (2016). Metabolomics in the rhizosphere: tapping into belowground chemical communication. Trends Plant Sci. 21 (3), 256–265. 10.1016/j.tplants.2016.01.008 [DOI] [PubMed] [Google Scholar]
- Van de Wouw A. P., Idnurm A. (2019). Biotechnological potential of engineering pathogen effector proteins for use in plant disease management. Biotechnol. Adv. 37 (6), 107387. 10.1016/j.biotechadv.2019.04.009 [DOI] [PubMed] [Google Scholar]
- Vincent D., Du Fall L. A., Livk A., Mathesius U., Lipscombe R. J., Oliver R. P., et al. (2012). A functional genomics approach to dissect the mode of action of the Stagonospora nodorum effector protein SnToxA in wheat. Mol. Plant Pathol. 13 (5), 467–482. 10.1111/j.1364-3703.2011.00763.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivek-Ananth R. P., Mohanraj K., Vandanashree M., Jhingran A., Craig J. P., Samal A. (2018). Comparative systems analysis of the secretome of the opportunistic pathogen Aspergillus fumigatus and other Aspergillus species. Sci. Rep. 8 (1), 6617. 10.1038/s41598-018-25016-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrabka J., Niehaus E. M., Munsterkotter M., Proctor R. H., Brown D. W., Novak O., et al. (2018). Production and role of hormones during interaction of fusarium species with maize (Zea mays L.) seedlings. Front. Plant Sci. 9, 1936. 10.3389/fpls.2018.01936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Weiberg A., Lin F. M., Thomma B. P., Huang H. D., Jin H. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2, 16151. 10.1038/nplants.2016.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Zhuang X., Deng Z. B., Jiang H., Mu J., Wang Q., et al. (2014). Targeted drug delivery to intestinal macrophages by bioactive nanovesicles released from grapefruit. Mol. Ther. 22 (3), 522–534. 10.1038/mt.2013.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chung K. P., Lin W., Jiang L. (2017). Protein secretion in plants: conventional and unconventional pathways and new techniques. J. Exp. Bot. 69 (1), 21–37. 10.1093/jxb/erx262 [DOI] [PubMed] [Google Scholar]
- Wang S., McLellan H., Bukharova T., He Q., Murphy F., Shi J., et al. (2019). Phytophthora infestans RXLR effectors act in concert at diverse subcellular locations to enhance host colonization. J. Exp. Bot. 70 (1), 343–356. 10.1093/jxb/ery360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson B. S., Bedair M. F., Urbanczyk-Wochniak E., Huhman D. V., Yang D. S., Allen S. N., et al. (2015). Integrated metabolomics and transcriptomics reveal enhanced specialized metabolism in Medicago truncatula root border cells. Plant Physiol. 167 (4), 1699–1716. 10.1104/pp.114.253054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Lin FM., Zhao H., Kaloshian I., Huang HD., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342 (6154), 118–123. 10.1126/science.1239705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Bellinger M., Jin H. (2014). Small RNAs: a new paradigm in plant-microbe interactions. Annu. Rev. Phytopathol. 52, 495–516. 10.1146/annurev-phyto-102313-045933 [DOI] [PubMed] [Google Scholar]
- Wen F., White G. J., VanEtten H. D., Xiong Z., Hawes M. C. (2009). Extracellular DNA is required for root tip resistance to fungal infection. Plant Physiol. 151 (2), 820–829. 10.1104/pp.109.142067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F., Curlango-Rivera G., Huskey D. A., Xiong Z., Hawes M. C. (2017). Visualization of extracellular DNA released during border cell separation from the root cap. Am. J. Bot. 104 (7), 970–978. 10.3732/ajb.1700142 [DOI] [PubMed] [Google Scholar]
- Weston L. A., Ryan P. R., Watt M. (2012). Mechanisms for cellular transport and release of allelochemicals from plant roots into the rhizosphere. J. Exp. Bot. 63 (9), 3445–3454. 10.1093/jxb/ers054 [DOI] [PubMed] [Google Scholar]
- Westwood J. H., Kim G. (2017). RNA mobility in parasitic plant - host interactions. RNA Biol. 14 (4), 450–455. 10.1080/15476286.2017.1291482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklander O. P. B., Brennan M. A., Lotvall J., Breakefield X. O., El Andaloussi S. (2019). Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 11 (492), 15. 10.1126/scitranslmed.aav8521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Mo M., Siljander P. R. M., Andreu Z., Zavec A. B., Borras F. E. (2015). Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066. 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K. (2006). ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett. 580 (4), 1183–1191. 10.1016/j.febslet.2005.12.009 [DOI] [PubMed] [Google Scholar]
- Yin K., Qiu J. L. (2019). Genome editing for plant disease resistance: applications and perspectives. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 374 (1767), 20180322. 10.1098/rstb.2018.0322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaynab M., Fatima M., Abbas S., Sharif Y., Umair M., Zafar M. H., et al. (2018). Role of secondary metabolites in plant defense against pathogens. Microb. Pathog. 124, 198–202. 10.1016/j.micpath.2018.08.034 [DOI] [PubMed] [Google Scholar]
- Zeilinger S., Gupta V. K., Dahms T. E., Silva R. N., Singh H. B., Upadhyay R. S., et al. (2016). Friends or foes? Emerging insights from fungal interactions with plants. FEMS Microbiol. Rev. 40 (2), 182–207. 10.1093/femsre/fuv045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng T., Holmer R., Hontelez J., Te Lintel-Hekkert B., Marufu L., de Zeeuw T., et al. (2018). Host- and stage-dependent secretome of the arbuscular mycorrhizal fungus Rhizophagus irregularis. Plant J. 94 (3), 411–425. 10.1111/tpj.13908 [DOI] [PubMed] [Google Scholar]
- Zeng J., Gupta V. K., Jiang Y., Yang B., Gong L., Zhu H. (2019). Cross-Kingdom Small RNAs Among Animals, Plants and Microbes. Cells 8 (4), 15. 10.3390/cells8040371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhalnina K., Louie K. B., Hao Z., Mansoori N., da Rocha U. N., Shi S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3 (4), 470–480. 10.1038/s41564-018-0129-3 [DOI] [PubMed] [Google Scholar]
- Zhang T., Zhao Y. L., Zhao J. H., Wang S., Jin Y., Chen Z. Q., et al. (2016). Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2 (10), 16153. 10.1038/nplants.2016.153 [DOI] [PubMed] [Google Scholar]
- Zhang S., Deng Y. Z., Zhang L. H. (2018). Phytohormones: the chemical language in Magnaporthe oryzae-rice pathosystem. Mycology 9 (3), 233–237. 10.1080/21501203.2018.1483441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Liang X., Zhou J. M. (2018). Small RNA trafficking at the forefront of plant-pathogen interactions. F1000Res 7, 8. 10.12688/f1000research.15761.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Bleackley M., Chisanga D., Gangoda L., Fonseka P., Liem M., et al. (2019). Extracellular vesicles secreted by Saccharomyces cerevisiae are involved in cell wall remodelling. Commun. Biol. 2, 305. 10.1038/s42003-019-0538-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Teng Y., Samykutty A., Mu J., Deng Z., Zhang L., et al. (2016). Grapefruit-derived nanovectors delivering therapeutic miR17 through an intranasal route inhibit brain tumor progression. Mol. Ther. 24 (1), 96–105. 10.1038/mt.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J., Schmidt S., Strehmel N., Scheel D., Abel S. (2017). Arabidopsis Transporter ABCG37/PDR9 contributes primarily highly oxygenated Coumarins to Root Exudation. Sci. Rep. 7 (1), 3704. 10.1038/s41598-017-03250-6 [DOI] [PMC free article] [PubMed] [Google Scholar]