Abstract
The compaction of DNA and the continuous action of DNA transactions, including transcription and DNA replication, create complex DNA topologies that require Type IIA Topoisomerases, which resolve DNA topological strain and control genome dynamics. The human TOP2 enzymes catalyze their reactions via formation of a reversible covalent enzyme DNA–protein crosslink, the TOP2 cleavage complex (TOP2cc). Spurious interactions of TOP2 with DNA damage, environmental toxicants and chemotherapeutic “poisons” perturbs the TOP2 reaction cycle, leading to an accumulation of DNA–protein crosslinks, and ultimately, genomic instability and cell death. Emerging evidence shows that TOP2-DNA protein crosslink (DPC) repair entails multiple strand break repair activities, such as removal of the poisoned TOP2 protein and rejoining of the DNA ends through homologous recombination (HR) or non-homologous end joining (NHEJ). Herein, we discuss the molecular mechanisms of TOP2-DPC resolution, with specific emphasis on the recently uncovered ZATTZnf451-licensed TDP2-catalyzed TOP2-DPC reversal mechanism.
Keywords: DNA strand breaks, Topoisomerase 2, Structural biology, Tyrosyl DNA phosphodiesterase 2, Tdp2, ZATT, ZNF451, TOP2, Chemotherapeutics
Introduction
Chromatin architecture and the collective action of DNA replication and RNA transcription creates positive and negative DNA supercoiling by over- and under-winding DNA strands, and DNA catenation—the linking together of DNA strands [1]. Topoisomerase 2 (TOP2) regulation of DNA topological strain is critical for transcription, DNA replication, and recombination [2–4]. The mammalian Type IIA topoisomerases, TOP2α and TOP2β, resolve DNA topology by creating paired DNA strand breaks in a DNA duplex, and gating passage of a second DNA duplex. A critical intermediate in this reaction is a protein-linked DNA double-strand break or a TOP2 protein-DNA crosslink (TOP2-DPC), also known as the TOP2 cleavage complex (TOP2cc). Despite being a requisite reaction state central to regulating DNA topology in the cell [5–8], TOP2-DPCs pose unique threats to genomic integrity. Clinically important and frontline chemotherapeutic TOP2 inhibitors, such as etoposide, exploit this critical intermediate and stabilize the TOP2cc, or “poison” TOP2 leading to accumulation of TOP2-DPCs double-strand breaks (DSBs) and cellular death [9]. When left unrepaired, TOP2-DPCs block progression of RNA and DNA polymerases, and are recombinogenic intermediates that can catalyze the generation of DNA deletions and translocations (reviewed in [10]).
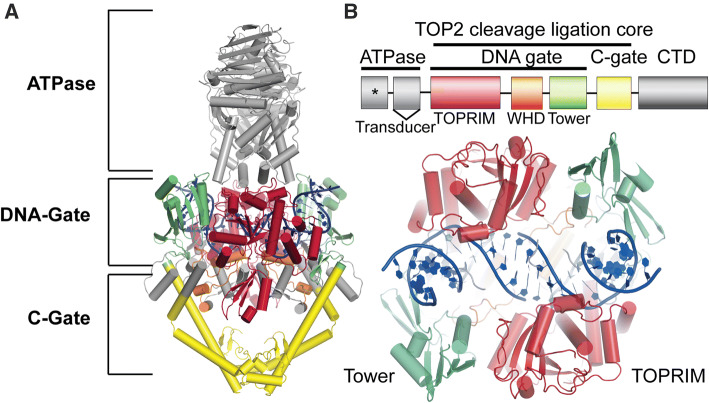
The two mammalian isoforms of TOP2, TOP2α (170 kDa) and TOP2β (183 kDa), share ~ 70% identity, despite originating from distinct genes [11–15]. TOP2α is expressed predominantly in proliferating cells, and TOP2β is expressed in all cells but is the only isoform found in non-dividing and terminally differentiated cells [16, 17]. TOP2 enzyme architecture can be delineated into three functional regions: the N-terminal ATPase, the DNA gate, and the C-gate (Fig. 1a) [18]. Both TOP2 isoforms also contain a C-terminal domain that is largely unstructured but plays a regulatory role in tuning TOP2 activity [19–22]. The DNA-gate is comprised of the TOPRIM domain, Winged-helix domain, the tower domain (Fig. 1a, b). The DNA gate and C-gate are collectively referred to as the cleavage ligation core, which is the minimum region required for DNA cleavage (Fig. 1b) [23–25]. Both TOP2α and TOP2β proceed via the same reaction mechanism to alter DNA topology. In this sequence, TOP2 binds to a DNA duplex (termed the G-segment) and creates paired nucleotide incisions in the phosphate backbone of both DNA strands with a four base-pair staggered spacing to create transient, reversible DNA double-strand breaks (DSBs) (refer to [5, 26] for detailed descriptions). A transesterification reaction and the formation of covalent enzyme-phosphotyrosyl linkages between the 5′-phosphate ends of the incised duplex and an active site TOP2 tyrosine facilitate the reversibility of TOP2-DNA cleavage reactions (Fig. 2a). Following the transit of a second DNA duplex (T-segment DNA), the DSB is reversed with re-ligation of the DNA ends, and the enzyme subsequently is released from DNA.
Fig. 1.
Schematic of topoisomerase 2 domain structure. a Four main protein regions: the N-terminal ATPase, a DNA gate, a C-gate, and a disordered C-terminal domain (CTD). PDB: 4GFH S. cerevisiae. b TOP2 is defined by ATPase, TOPRIM domain, Winged-helix domain, the tower domain, a C-gate, and a disordered C-terminal domain (CTD). The DNA gate and C-gate or the cleavage ligation core is the minimum region required for catalytic DNA cleavage. * indicates ATPase domain active site. Bottom: Rotation of panel a 90 degrees
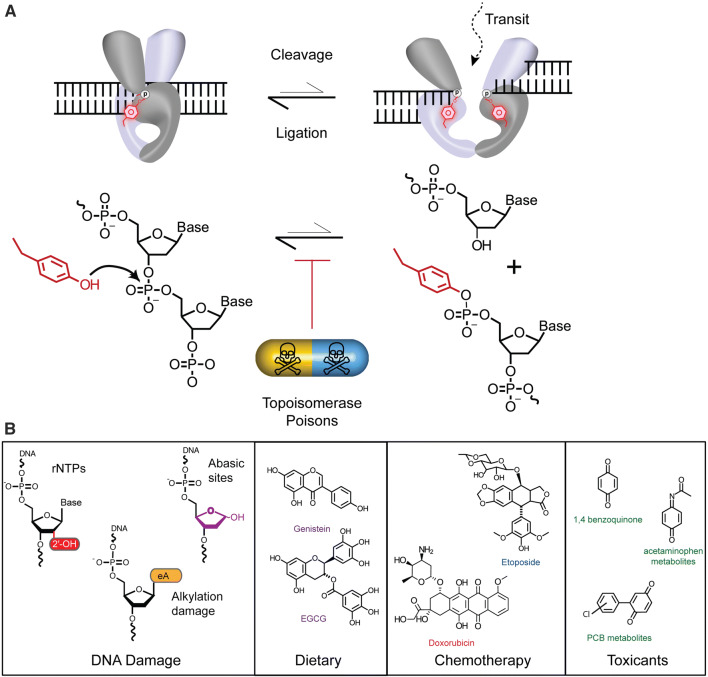
Fig. 2.
Topoisomerase-2 Cleavage Ligation Equilibrium and Topoisomerase Poisons. a TOP2 Catalytic cycle. TOP2 binding G-segment DNA (Left). TOP2 cleaves DNA generating a double-strand break of the G-segment DNA and the reaction intermediate is a TOP2-DPC (right). The presence of TOP2 “poisons” shifts the catalytic cycle equilibrium to favor a trapped DNA complex. b Examples of the chemical structures of Topoisomerases “poisons” include: DNA damage substrates (e.g. genome embedded ribonucleotides—red, alkylated DNA damage including 1,N6-ethenoadenine—orange, and abasic sites—purple), dietary supplements (e.g. Genistein and Epigallocatechin gallate, EGCG, chemotherapeutics (e.g. Doxorubicin and Etoposide), and various environmental toxicants
This complex mechanistic framework presents vulnerabilities through which the TOP2 reaction can be interrupted. Indeed, TOP2 reactions are variably sensitive to DNA damage, chemotherapeutics, and environmental perturbations that can either accelerate the TOP2-DNA cleavage reaction, or stall DNA re-ligation to poison the reaction cycle, and shift the cleavage/ligation equilibrium to favor the accumulation of TOP2-DPCs (Fig. 2a). TOP2 poisons can be both endogenous or exogenous factors (Fig. 2b) [27, 28]. Endogenous DNA damage poisons include, but are not limited to: abasic (also known as apurinic and apyrimidinic) sites formed spontaneously or during base excision repair reactions [29], ribonucleotides introduced during DNA replication and repair [30–34], and bulky alkylated bases [34–38] (Fig. 2b). Exogenous TOP2 poisons are compounds found in our food or environmental toxicants (Fig. 2b). This mechanistic vulnerability of TOP2 is exploited by potent chemotherapeutic TOP2 poisons such as etoposide or doxorubicin that stall the TOP2 reaction by binding directly to the TOP2cc intermediate and stabilizing TOP2-DPCs, causing genomic instability and cell death in cancer cells [25, 39–41].
TOP2-DPCs are typified by 5′-phosphotyrosyl (5′-Y) covalent linkages with the TOP2 protein adducted at the ends of a DSB. Failure to resolve and accurately repair TOP2-DPCs can lead to mutagenesis, translocations, neurological disease, and carcinogenesis [26, 42–45]. Furthermore, translocations caused by TOP2-DPCs [29] lead to leukemias with a poor prognosis [46]. It is estimated that 2–3% of patients that receive TOP2 inhibitors, as a chemotherapeutic agent, will develop acute myeloid leukemia (AML), which, are frequently accompanied by translocations within the mixed-lineage leukemia (MLL) gene [29, 47–49]. Given the consequences of unrepaired or incorrectly repaired TOP2-DPCs to genome integrity, the cell has evolved redundant pathways to safeguard the genome and facilitate the removal of TOP2-DPCs. Repair of TOP2–DPCs reverses damage caused by chemotherapeutics and underlies tumor drug resistance that causes treatment to fail. Therefore, defining the DNA damage response to chemotherapy-induced DPCs is vital to ensure the best possible treatment outcomes, and for the development of new therapies or cotherapies. In this review, we provide an overview of the direct reversal of TOP2-DPCs mediated by TDP2 and ZATT and discuss the alternative cellular strategies for TOP2-DPC resolution.
TDP2 directly resolves Phosphotyrosyl-DNA linkages
TDP2 (Tyrosyl-DNA Phosphodiesterase 2) is a highly specific enzyme that specifically reverses 5′-Y linkages characteristic of TOP2-DPCs, and is important for cellular survival following treatment with TOP2 poisons including etoposide [50]. Loss of TDP2 results in hypersensitivity to TOP2 poisons and impairs transcription and neuronal development [44]. Emerging evidence has revealed that the cellular response to TOP2-DPCs is multifaceted, and vertebrate cells have evolved redundant biochemical solutions to resolving aberrant topoisomerase reactions. TDP2 and nuclease complexes that resolve the TOP2-DNA linkage are blocked from acting directly on the TOP2-DNA bond by the proteinaceous TOP2 shell. This obstacle must be overcome to initiate the repair process and is an import regulatory step in TOP2-DPC repair. Cellular strategies that accomplish this task include remodeling of the TOP2-DPC with a chaperone-like function by the ZATTZNF451 (Zinc-Finger Associated with TOP2 and TDP2) SUMO2 (Small Ubiquitin-like modifier 2) E3 ligase [51] to license their removal by TDP2.
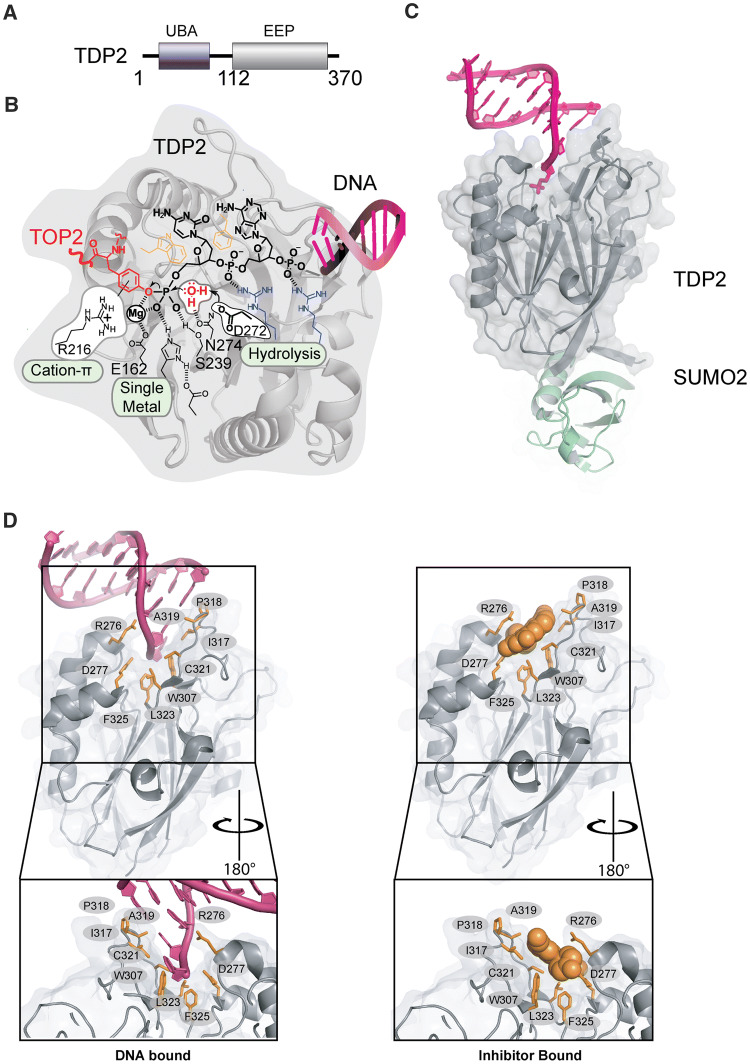
Our understanding of the molecular mechanisms for the specific resolution of TOP2-DPCs was propelled a decade ago with the discovery of the 5′ Tyrosyl- DNA phosphodiesterase TDP2 [50]. The 41 kDa TDP2 protein is structurally defined by two domains: an amino-terminal Ubiquitin Associated (UBA) domain that binds to Ubiquitin (Ub) [52] and a C-terminal catalytic domain sharing homology to the Exonuclease-Endonuclease-Phosphatase (EEP) nuclease superfamily (Fig. 3a). The UBA domain is connected to the catalytic domain via a flexible linker region, as determined through limited proteolysis and small angle X-ray scattering (SAXS), whereas the EEP domain is a globular folded catalytic domain [53, 54]. The TDP2 EEP domain contains a mixed α–β fold with 12 stranded anti-parallel β-sandwich enveloped by several helical elements and an active site that catalyzes phosphotyrosyl-DNA hydrolysis (Fig. 3b). Detailed work employing structural biology and biochemical analyses [53, 55] have shown that TDP2 engagement and processing of DPCs involves: (1) a two-sided active site pocket tailored to recognize both the DNA and protein components of a DNA–protein crosslink, (2) substrate-induced fit conformational change that regulates assembly of the TDP2 active site, and (3) a single Mg2+-mediated catalytic mechanism promoting hydrolysis of the phosphotyrosyl linkage DPC linkage.
Fig. 3.
Tyrosyl–DNA Phosphodiesterases-2 (TDP2). a Domain organization of TDP2. b Chemical reactions facilitating direct hydrolysis of a 5′ phosphotyrosyl bond, such as will exist in TOP2-DPC reaction intermediates. c Mouse TDP2 catalytic domain bound to SUMO PDB ID: 5TVQ. d Upper left panel, DNA bound TDP2 C-terminal Endonuclease-Exonuclease-Phosphate (EEP) (PDB ID: 5INL) Upper right panel: Inhibitor bound TDP2 EEP (PDB ID: 5J42). Green residues in stick and cartoon show conformation of motif 7 in inhibitor bound state. Residues important for engaging inhibitor or DNA binding are shown in orange sticks. Bottom: 180 degree rotation of upper panels
TDP2 is a phosphodiesterase that efficiently hydrolyzes 5′-Y DNA, even amongst the presence of a vast excess of undamaged DNA. This activity demands an active site specifically tailored to recognize a 5′-Y. Interestingly, the genetic screens that identified TDP2 were originally designed to detect alternative activities that repair Camptothecin induced TOP1 adducts typified by 3′-Y-linked DPCs. Indeed, TDP2 can process 3′-Y [50], albeit much less efficiently than 5′-Y adducts [50, 55]. Although apurinic site analogs (tetrahydrofuran) or 5′–alkylamine adducts (e.g. 6-amino hexanol) are hydrophobic and small enough to be accommodated in the 5′–adduct binding site of TDP2, they are resistant to hydrolysis. The specificity of TDP2 for an aromatic tyrosine leaving group is likely conferred by a conserved arginine which forms a cation–π interaction that stabilizes the negatively charged phenolate transition state [53] (Fig. 3b). Therefore, efficient TDP2 hydrolase activity is defined by three features of the 5′–adduct—hydrophobicity, size, and aromaticity that restrict it to 5′-Y DNA ends. The TDP2 DNA-binding trench recognizes the DNA 5′-end through sequence-independent interactions, and also serve an important regulatory function for the enzyme. Structures of TDP2 obtained in the absence of DNA substrate show that the β2Hβ helix “grasp” DNA-binding loop adopts multiple conformations, including an extended state that is stabilized by a cis-peptide [53]. In this “apo” state metal-coordinating residues in the active site are misaligned, which disrupts stable binding of metal in the active site. Binding the 5′–end of a DNA stabilizes a closed state (Fig. 5c) and relays a conformational shift through W307 and D350 (human TDP2 numbering) to shift Mg2+–coordinating residues and the Mg2+ ion into the correct position for catalysis. Thus, these DNA-induced fit conformational changes link DNA 5′–end binding to enzyme activation. In this way, and analogous to modern secure computing environments, TDP2 uses “two-factor authentication” to enforce hydrolysis of only the correct substrates containing an aromatic 5′ leaving group on a DNA end.
Fig. 5.
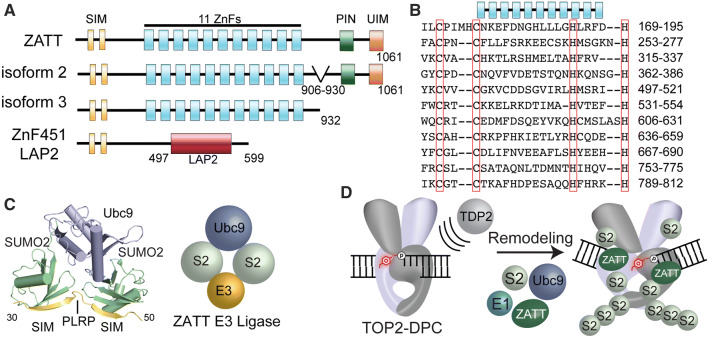
ZATT domain architecture and SUMOylation. a Four splicing isoforms ZnF451: ZATT (isoform1), isoform2 and 3 and ZnF451 Lap2a. ZATT domain structure consists of 2 N-terminal SUMO interacting motifs (SIMs), 11 Zinc Finger domains (ZnFs), PilT N-terminal domain (PIN) NYN domain, and a Ubiquitin Interacting Motif (UIM). b Uniprot alignment of predicted central ZnF domains of ZATT. c X-ray Crystal structure PDB ID: 5D2 M. ZNF451 in complex with SUMOylation machinery, SUMO2 and the E2 Ligase, Ubc9. Znf451 dual SIM domain and PLRP is composed of amino acids 30-50 bound to SUMO2. d Model of ZATT interaction in TOP2-DPC resolution. ZATT, an E3 ligase, along with other SUMOylation machinery (E1 ligase, E2 ligase (human Ubc9/yeast Sae2) and SUMO2 (S2) allows for SUMOylation of TOP2-DPCs. ZATT binds to TOP2-DPC allowing for remodeling and TDP2 accessibility to the 5′Y
TOP2-DPCs require multistep processing for resolution
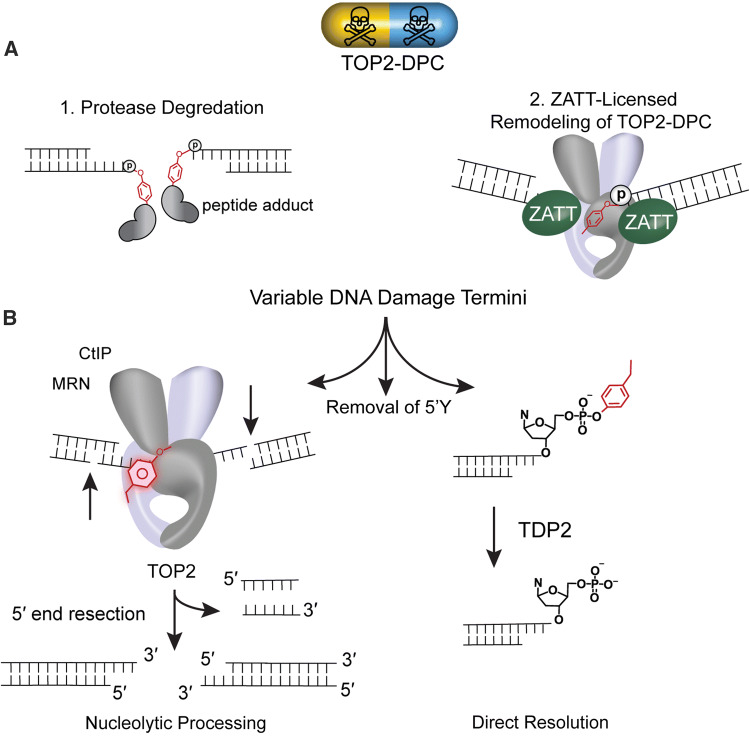
TOP2-DPCs bulky adduct architecture is a hindrance to the direct reversal phosphotyrosyl phosphodiesterase activity of TDP2. It is clear that TDP2 activity is accelerated on free phosphotyrosyl modified ends as compared to intact TOP2-DPCs [53, 56]. Biochemical and cellular biological evidence suggests that several strategies have evolved to license TOP2-DPCs for direct resolution. These strategies include proteolytic processing or remodeling of the TOP2-DPC (Fig. 4a). Both the proteasome and metalloproteases proteolytically degrade TOP2 protein [51, 57] in a process that presumably generates DNA ends adducted with variable sized peptide adducts. Proteosomal degradation of TOP2-DPCs has been observed in the presence of high concentrations of etoposide [58, 59]. Recently, metalloproteases that are capable of cleaving both TOP1 and TOP2 and include the human SPARTN, yeast WSS1 and the drosophila Maternal Haploid (MH) proteins [60–62]. SUMO and/or ubiquitination play a role in the metalloprotease-dependent resolution of TOP2-DPC for the human, yeast, and X. laevis [60, 61, 63] adding additional levels of regulation. It is not known if proteolytic activities are coordinated with TDP2 tyrosyl DNA phosphodiesterase activities at this time.
Fig. 4.
Overview of TOP2-DPC resolution. a Variable 5′ DNA termini produced subsequent to (1) protease degradation (2) ZATT-licensed Remodeling of TOP2-DPC. Protease digestion of TOP2 removes TOP2 protein leaving behind a variable length peptide of TOP2 and a 5-phosphotyrosol linkage. Accessibility of the 5′ phosphotyrosyl linkage can also be regulated by remodeling. ZATT binds to and remodels TOP2-DPC promoting 5′ phosphotyrosyl linkage accessibility. b Removal of 5′Y. (Left) Nucleolytic processing mediated by a large complex of DNA repair proteins importantly CtIP, MRN. (Right) Direct Resolution of the 5′ phosphotyrosyl by Tyrosyl–DNA Phosphodiesterases 2 (TDP 2) producing a ligation competent 5′P on DNA
Alternatively, remodeling of TOP2-DPC conformational states facilitates direct removal of the DPC [51]. The subsequent removal of variable 5′ phosphotyrosine DPCs ends then can occur either by direct hydrolysis of the phosphotyrosyl linkage by TDP2 or by nucleolytic processing, such as through the action of MRX Mre11/Rad50/Nbs1/CtIP nuclease complex which also excises the flanking DNA duplex region (Fig. 4b) [64]. In the former and simplest case, the DNA termini will be ligation competent (5′ P and 3′ OH) and DNA ligase will religate the DNA termini. If the termini of DNA does not contain ligation competent ends, the ends of DNA must then be repaired by the non-homologous end joining (NHEJ) and additional repair enzymes are required prior to ligation [31, 51, 65, 66]. The cellular decision to choose one pathway of removal over another i.e. proteolytic removal vs remodeling utilizes an undetermined mechanism.
Regulation of TDP2 activity by post-translational modifications
Similar to other cellular processes, posttranslational modifications have been implicated in the removal and remodeling of TOP2-DPCs [51, 67, 68]. Ubiquitination and SUMOylation may enhance the efficiency or even be critical for the recruitment of the proteasome and additional end-processing repair factors [69, 70]. Likewise, posttranslational modifications may also dictate pathway decisions and organization of repair. Various biochemical and structural observations highlight the importance of posttranslational modifiers in regulating TDP2 activity and recruitment during the resolution of TOP2-DPC [51, 52, 71]. It has been postulated that Ub could regulate intrinsic activities of TDP2 or bring about a conformation change which may remodel TDP2 active site [52]. C. elegans TDP2 binds to both K48-linked and K63-linked di-Ub via its N-terminal UBA domain [52]. Complementation with TDP2 WT, but not an N-terminal deletion of residues 1–100 reduces the hypersensitivity of etoposide in TDP2 deficient DT40 cells, indicating a role for Ub in the resolution of TOP2-DPCs [52]. However, the exact role of Ub in the resolution of TOP2-DPC is still unclear.
Phosphorylation also regulates TDP2, and TDP2 and extracellular signal-regulated kinase 3 (ERK3) were shown to interact in a yeast-two hybrid screen [72]. TDP2 is phosphorylated in an ERK3-dependent manner and ERK3 is upregulated in lung cancer cells and human lung tumors. Mutations that remove a phosphorylation site of TDP2 (S60A) in A549 cells yield cells that are hypersensitive to etoposide. However, S60A does not abrogate TDP2 phosphorylation in vitro, potentially indicating that ERK3 can possibly phosphorylate other sites of TDP2. S60A also substantially decreased 5′ phosphodiesterase activity and phosphomimic mutant S60D slightly increased activity in vitro as compared to WT activity [72], suggesting phosphorylation regulates catalytic activity. Phosphorylation may also serve as a mechanism for recruitment of additional repair factors to provide an additional level of TDP2 regulation.
Licensing of TOP2-DPCs for direct reversal by ZATT
Although intact TOP2-DPCs are resistant to TDP2 hydrolyase activity in vitro, heat denaturation of TOP2-DPCs facilitates TDP2 mediated phosphotyrosyl bond cleavage of denatured TOP2-DPCs [33]. This biochemical observation hinted that alternative mechanisms might facilitate remodeling of intact TOP2-DPCs to facilitate TDP2 processing. Remodeling of TOP2-DPCs might be advantageous and more efficient than energetically consuming processes of proteasome-mediated proteolytic degradation, which occurs on the order of hours following exposure TOP2 drug treatment. Moreover, in a liquid chromatography–tandem mass spectrometry screen using TDP2 as the bait protein, TDP2 associated with SUMO2/3-modified full-length TOP2α and TOP2β proteins, rather than proteolyzed remnants of the topoisomerase proteins [51]. Tandem affinity purification (TAP) of SUMOylated TDP2 associated proteins yielded a highly purified complex containing four components: (1) TOP2α, (2) TOP2β, (3) SUMO2 and (4) the ZATTZNF451 (poly Znf protein Associated with TDP2 and TOP2) SUMO2 E3 ligase. However, these SUMOylated TOP2 molecules did not purify with a catalytically inactive TDP2 mutant or when ZATT was knocked down, which suggests that these complexes are soluble TDP2 reactions products liberated from chromatin by TDP2, and that ZATTZNF451 was a previously unidentified regulator of TDP2 activity. Consistent with a DNA repair role, ZATT CRISPR knockout cells are hypersensitive to etoposide, and purified recombinant ZATT markedly stimulates TDP2-catalyzed direct reversal of reconstituted, intact TOP2-DPCs. Furthermore, probing of the global conformational states of purified TOP2-DPCs using limited tryptic proteolysis showed that ZATT binding to TOP2-DPCs regulates a conformational change in the TOP2 molecule. Together these results suggest that ZATT remodels the TOP2-DPC to license TDP2 processing of the DNA–protein crosslink [51].
ZATT (aka ZNF451 or COASTER, Coactivator for steroid receptors) is a transcriptional corepressor of androgen receptor [73–75]. ZATT is a 120 kDa protein comprised of 11 central Zinc finger domains (ZnFs) (Fig. 4a, b). In addition to the central ZnFs, other known domains or motifs of ZATT include: (1) two N-terminal SUMO-interacting motifs (SIMs) separated by a “PLRP” motif which comprise an E3 SUMO2 ligase domain, (2) an N-terminal coil–coil (cc) motif, (3) a PilT N-terminal “PIN” domain and (4) a ubiquitin-interacting motif (UIM) (Fig. 5a) [73, 74, 76, 77]. Interestingly, ZNF451 exists in four splicing isoforms. Isoform 1 encodes a 1061 amino acid protein, ZATT. Isoform 2 lacks residues 870–917, termed COASTER, and isoform 3 encodes a C-terminal deletion of 933–1061 with alternate amino acids in positions 919–932 [76]. An additional isoform of unknown function is generated from alternative splicing that fuses the N-terminal SUMO2 ligase domain with a lamina-associated polypeptide (LAP2) domain [75] (Fig. 5a).
The structure and function of the central ZNFs, PIN/NYN is largely unexplored. ZnF repeat domains often act as transcription factors that bind DNA and/or RNA among other functions. Other proteins containing similarly clustered C2H2 zinc fingers often bind to longer DNA sequences and recognize a target DNA sequence [78]. It has yet to be determined if the multiple consecutive ZNFs perform one or more of these functions. PIN/NYN domains are Rossman fold domains found frequently in nucleases and can be involved in a wide variety of cellular processes including DNA repair [77], but the function of ZATT–PIN/NYN domain is unknown. ZATT binding to Ubiquitin (Ub) occurs through the C-terminal UIM residues 1050–1061, and deletion of the UIM drastically reduced binding to GST-Ub [76]. However, it is not known if ZATT binds to a single mono-Ubiquitinated proteins, polyUb, or specific Ubiquitin chain linkages. Ubiquitin binding may function to recruit ZATT to ubiquitinated cellular proteins as part of the DNA damage response perhaps directing repair decisions, such as has been observed for RNF168 catalyzed the ubiquitination of H2A, which recruits 53BP1 [79].
A well-characterized function of ZATT is its E3 SUMO2 ligase activity. Both ZATT and TOP2 are amongst the most highly SUMOylated proteins in human cells [80]. ZATT binds to both SUMO1 and SUMO2/3, but with a strong preference for SUMO2/3 (Fig. 5c, PDB ID: 5D2 M) [73]. The dual-SIM domains (SIM-PLRP-SIM; residues 31–49) direct SUMO2 binding SUMO2 E3 ligase activity [73, 81] (Fig. 5c). Mutations in this region, L48/V49A, lead to a reduction in SUMO2 binding and impaired auto-SUMOylation of ZATT [76]. In addition to the dual SIM motif, E3 ligase activity requires the first predicted ZnF domain [81]. ZATT nuclear enrichment in promyelocytic leukemia (PML) [74] bodies may be driven by ZATT-mediated autoSUMOylation, whereby SENP1-driven deconjugation of SUMO disrupts cellular localization of ZATT to PML bodies [76]. Many sites of ZATT auto-SUMOylation have been identified but attempts to mutate known SUMO consensus site lysine mutations of these residues fail to abrogate SUMO conjugation of ZATT [81–83]. SUMO conjugation regulates cellular function, as was shown for the SUMO E2 Ubc9. AutoSUMOylation of Ubc9 alters the selection of its targets for SUMO conjugation. Additionally, Ubc9 autoSUMOylation negatively regulates Ubc9-dependent SUMOylation of septin, which in turn, alters cellular morphology [84]. It is possible that ZATT binding SUMO2/3 and autoSUMOylation may regulate targets of ZATT-dependent SUMOylation and TOP2-DPC repair.
SUMOylation of TOP2 and TOP2-DPCs
TOP2 is highly SUMOylated during mitosis [67, 85] and in response to TOP2 inhibitors (Fig. 4d) [51, 86, 87]. SUMOylation may also act to recruit other repair factors or act as a scaffold, regulating protein interactions, which may ultimately dictate the cellular decision to choose one pathway of repair verses another. It is posited that SUMOylation marks TOP2-DPCs to distinguish them from normal reaction state intermediates of TOP2. SUMOylation is known to play a role in the recruitment of other DNA repair factors as well as altering catalytic activity or other biological functions such as DNA binding. In X. laevis, PIASy-dependent SUMOylation of TOP2 on residue K660 (near the DNA binding cleft), inhibits decatenation [88] and impaired SUMOylation of TOP2. ZATT E3 SUMO ligase catalyzes the majority of TOP2 SUMOylation in mammalian HEK293 cells, and TOP2 SUMOylation is significantly impaired in ZATT knockout cells. Consistent with these observations, ZATT displays a marked preference for SUMOylating TOP2-DPCs compared to free TOP2. In HEK293 cells, the drugs ICRF-193 and etoposide both bind to TOP2 and trigger ZATT-dependent TOP2 SUMOylation. While the precise interplay of ZATT-dependent SUMOylation with other repair pathway remains to be explored, it is clear that ZATT-mediated SUMOylation regulates recruitment of TDP2 [51]. TDP2 engagement of SUMO2 occurs through a unique split-SUMO interacting motif (split-SIM) [51] (Fig. 3c). The split SIM interface uses five surface loops from the EEP domain to expand the canoncial SUMO2/3 binding SIM interface. TDP2 is likely targeted to TOP2-DPCs by ZATT mediated SUMO2/3 E3 ligase activity.
Clinical implications
While current chemotherapeutic TOP2 “poisons” stabilize TOP2-DPCs, it is likely that enhanced 5′-Y hydrolysis and TOP2-DPC resolution would confer etoposide resistance. Indeed, TDP2 which catalyzes 5′-Y linkage reversal is a key modulator of drug resistance to TOP2-targeted chemotherapeutic drugs [44, 50, 65] through a NHEJ DSB repair pathway [66, 89]. Etoposide treated TDP2 RNAi knockdown cells have increased the formation of nuclear γH2AX foci, a marker of DSBs [50], and TDP2 KO cells are hypersensitive to TOP2 poisons that are used in the clinic such as etoposide, doxorubicin, and amsacrine (m-AMSA) [90]. Additionally, TDP2 has been shown to be overexpressed in lung cancer and knockdown of TDP2 increases lung cancer cells sensitivity to TOP2 inhibitors [91, 92]. However, TDP2 deletion also increases genome rearrangements triggered by etoposide, underlining the importance of TDP2 in treatment outcomes [45, 65].
TDP2 remains a viable small molecule drug target and several small molecules that inhibit TDP2 in vitro have been generated (Fig. 5d) [93, 94]. Both deazaflavin derivatives and furoquinolinediones have shown potential for clinical applications. Deazaflavin derivatives show potent synergy in combination with etoposide and furoquinolinediones therapeutic benefit has been observed at non-toxic concentrations [95], but this effect may be complicated by impacts of the compounds on altering etoposide efflux [96].
Given the interplay of ZATT-TDP2 in TOP2-DPC reversal, as well as ZATT-catalyzed SUMOylation of TOP2-DPCs, uncovering effective ZATT or TDP2 inhibitors in combination with TOP2 inhibition may serve as a powerful chemotherapeutic regimen to overcome intrinsic or acquired tumor drug resistance. ZATT KO cells display etoposide sensitivity and overexpression of ZATT in a dose-dependent manner provides etoposide resistance [51]. Therefore, ZATT expression levels may be a good predictor of a tumor’s sensitivity to TOP2 drugs and a good predictor of treatment outcome.
Conclusion and outlook
DNA strand break repair is driven by DNA damage response (DDR) proteins bearing DNA damage-specific molecular recognition and reversal activities. DDR protein-nucleic acid interfaces, enzymatic activities, DNA scaffolding activities, and dynamic protein conformational states are impacted by heritable mutations linked to human cancer predisposition. DNA repair machinery is a viable therapeutic target to modulate cellular resistance to DNA damage for improved disease treatment. In order to best design therapeutics, it is essential to address the overarching challenge of understanding how DNA damage surveillance apparatus promotes DNA repair to maintain genome stability. Given the danger of a covalent DNA–protein crosslink, the cell has complex overlapping and multifaceted pathways for TOP2-DPC resolution. What is unclear is how the cell makes the decision to choose a proteasomal-dependent or remodeling of TOP2 to gain access to the 5′-Y. Future studies that explore the interplay between the known TOP2-DPC DNA damage response proteins, and yet to be discovered factors are needed to address these questions.
Acknowledgements
This research was supported by the intramural research program of the US National Institutes of Health (NIH), National Institute of Environmental Health Sciences (NIEHS) Grant 1Z01ES102765 to R.S.W.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Witz G, Stasiak A. DNA supercoiling and its role in DNA decatenation and unknotting. Nucleic Acids Res. 2009;38(7):2119–2133. doi: 10.1093/nar/gkp1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King IF, et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501(7465):58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tammaro M, Barr P, Ricci B, Yan H. Replication-dependent and transcription-dependent mechanisms of DNA double-strand break induction by the topoisomerase 2-targeting drug etoposide. PLoS One. 2013;8(11):e79202. doi: 10.1371/journal.pone.0079202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, Wang S, Yin S, Hong S, Kim KP, Kleckner N. Topoisomerase II mediates meiotic crossover interference. Nature. 2014;511(7511):551–556. doi: 10.1038/nature13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9(5):327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41(1):41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 7.Nitiss JL, Soans E, Berk J, Seth A, Mishina M, Nitiss KC. Repair of topoisomerase II-mediated DNA damage: fixing dna damage arising from a protein covalently trapped on DNA. In: Pommier Y, editor. Topoisomerases and cancer. Cancer drug discovery and development. New York, NY: Springer; 2012. [Google Scholar]
- 8.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 9.Chen GL, Yang L, Rowe TC, Halligan BD, Tewey KM, Liu LF. Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984;259:13560–13566. [PubMed] [Google Scholar]
- 10.Ashour ME, Atteya R, El-Khamisy SF. Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nat Rev Cancer. 2015;15(3):137–151. doi: 10.1038/nrc3892. [DOI] [PubMed] [Google Scholar]
- 11.McClendon AK, Osheroff N. DNA topoisomerase II, genotoxicity, and cancer. Mutat Res. 2007;623(1–2):83–97. doi: 10.1016/j.mrfmmm.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drake FH, et al. Purification of topoisomerase II from amsacrine-resistant P388 leukemia cells. Evidence for two forms of the enzyme. J Biol Chem. 1987;262:16739–16747. [PubMed] [Google Scholar]
- 13.Drake FH, Hofmann GA, Bartus HF, Mattern MR, Crooke ST, Mirabelli CK. Biochemical and pharmacological properties of p170 and p180 forms of topoisomerase II. Biochemistry. 1989;28:8154–8160. doi: 10.1021/bi00446a029. [DOI] [PubMed] [Google Scholar]
- 14.Mirabelli CK, Crooke ST, Mao Ji. Topoisomerase Ila and topoisomerase 11/3 genes: characterization and mapping to human chromosomes 17 and 3, repectively. Cancer Res. 1992;52(14):3831–3837. [PubMed] [Google Scholar]
- 15.Tsai-Pflugfelder M, et al. Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21-22. Proc Natl Acad Sci USA. 1988;85:7177–7181. doi: 10.1073/pnas.85.19.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capranico G, Tinelli S, Austin CA, Fisher ML, Zunino F. Different patterns of gene expression of topoisomerase II isoforms in differentiated tissues during murine development. Biochim Biophys Acta - Gene Struct Expr. 1992;1132(1):43–48. doi: 10.1016/0167-4781(92)90050-a. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe M, Tsutsui K, Tsutsui K, Inoue Y. Differential expressions of the topoisomerase IIα and IIβ mRNAs in developing rat brain. Neurosci Res. 1994;19(1):51–57. doi: 10.1016/0168-0102(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt BH, Osheroff N, Berger JM. Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity. Nat Struct Mol Biol. 2012;19(11):1147–1154. doi: 10.1038/nsmb.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryu H, et al. SUMOylation of the C-terminal domain of DNA topoisomerase IIα regulates the centromeric localization of Claspin. Cell Cycle. 2015;14(17):2777–2784. doi: 10.1080/15384101.2015.1066537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozuki T, et al. Roles of the C-terminal domains of topoisomerase IIα’ and topoisomerase IIβ in regulation of the decatenation checkpoint. Nucleic Acids Res. 2017;45(10):5995–6010. doi: 10.1093/nar/gkx325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickey JS, Osheroff N. Impact of the C-terminal domain of topoisomerase IIα on the DNA cleavage activity of the human enzyme. Biochemistry. 2005;44(34):11546–11554. doi: 10.1021/bi050811l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Austin CA, Sng JH, Patel S, Fisher LM. Novel HeLa topoisomerase II is the IIβ isoform: complete coding sequence and homology with other type II topoisomerases. BBA - Gene Struct Expr. 1993;1172(3):283–291. doi: 10.1016/0167-4781(93)90215-y. [DOI] [PubMed] [Google Scholar]
- 23.Chen S-F, et al. Structural insights into the gating of DNA passage by the topoisomerase II DNA-gate. Nat Commun. 2018;9(1):3085. doi: 10.1038/s41467-018-05406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wendorff TJ, Schmidt BH, Heslop P, Austin CA, Berger JM. The structure of DNA-bound human topoisomerase II alpha: conformational mechanisms for coordinating inter-subunit interactions with DNA cleavage. J Mol Biol. 2012;424(3–4):109–124. doi: 10.1016/j.jmb.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu CC, et al. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science (80–) 2011;333(6041):459–462. doi: 10.1126/science.1204117. [DOI] [PubMed] [Google Scholar]
- 26.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9(5):338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem Biol. 2013;8(1):82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karimi Goftar M, Alizadeh Rayeni N, Rasouli S. Topoisomerase inhibitors and types of them. Int J Adv Biol Biomed Res. 2014;2(8):2431–2436. [Google Scholar]
- 29.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res. 2009;37(3):738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams JS, Kunkel TA. Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst) 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andres SN, Schellenberg MJ, Wallace BD, Tumbale P, Williams RS. Recognition and repair of chemically heterogeneous structures at DNA ends. Environ Mol Mutagen. 2015;56(1):1–21. doi: 10.1002/em.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace BD, Williams RS. Ribonucleotide triggered DNA damage and RNA-DNA damage responses. RNA Biol. 2014;11(11):1340–1346. doi: 10.4161/15476286.2014.992283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao R, et al. Proteolytic degradation of topoisomerase II (Top2) enables the processing of Top2 DNA and Top2 RNA covalent complexes by tyrosyl-DNA-phosphodiesterase 2 (TDP2) J Biol Chem. 2014;289(26):17960–17969. doi: 10.1074/jbc.M114.565374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Knudsen BR, Bjergbæk L, Westergaard O, Andersen AH. Stimulated activity of human topoisomerases IIα and IIβ on RNA-containing substrates. J Biol Chem. 1999;274:22839–22846. doi: 10.1074/jbc.274.32.22839. [DOI] [PubMed] [Google Scholar]
- 35.Cline SD, Jones WR, Stone MP, Osheroff N. DNA abasic lesions in a different light: solution structure of an endogenous topoisomerase II poison. Biochemistry. 1999;38(47):15500–15507. doi: 10.1021/bi991750s. [DOI] [PubMed] [Google Scholar]
- 36.Khan QA, et al. Position-specific trapping of topoisomerase II by benzo[a]pyrene diol epoxide adducts: implications for interactions with intercalating anticancer agents. Proc Natl Acad Sci. 2003;100(21):12498–12503. doi: 10.1073/pnas.2032456100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabourin M, Osheroff N. Sensitivity of human type II topoisomerases to DNA damage: stimulation of enzyme-mediated DNA cleavage by abasic, oxidized and alkylated lesions. Nucleic Acids Res. 2000;28(9):1947–1954. doi: 10.1093/nar/28.9.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kingma PS, Osheroff N. Apurinic sites are position-specific topoisomerase II poisons. J Biol Chem. 1997;272(2):1148–1155. doi: 10.1074/jbc.272.2.1148. [DOI] [PubMed] [Google Scholar]
- 39.Hande KR. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur J Cancer. 1998;34(10):1514–1521. doi: 10.1016/s0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 40.Baldwin EL, Osheroff N. Etoposide, topoisomerase II and cancer. Curr Med Chem Anticancer Agents. 2005;5(4):363–372. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- 41.Wu C-C, Li Y-C, Wang Y-R, Li T-K, Chan N-L. On the structural basis and design guidelines for type II topoisomerase-targeting anticancer drugs. Nucleic Acids Res. 2013;41(22):10630–10640. doi: 10.1093/nar/gkt828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zagnoli-Vieira G, et al. Confirming TDP2 mutation in spinocerebellar ataxia autosomal recessive 23 (SCAR23) Neurol Genet. 2018;4(4):e262. doi: 10.1212/NXG.0000000000000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciaccio C, et al. Consolidating the role of TDP2 mutations in recessive spinocerebellar ataxia associated with pediatric onset drug resistant epilepsy and intellectual disability (SCAR23) The Cerebellum. 2019;18(5):972–975. doi: 10.1007/s12311-019-01069-7. [DOI] [PubMed] [Google Scholar]
- 44.Gómez-Herreros F, et al. TDP2 protects transcription from abortive topoisomerase activity and is required for normal neural function. Nat Genet. 2014;46:516. doi: 10.1038/ng.2929. [DOI] [PubMed] [Google Scholar]
- 45.Gómez-Herreros F, et al. TDP2 suppresses chromosomal translocations induced by DNA topoisomerase II during gene transcription. Nat Commun. 2017;8(1):233. doi: 10.1038/s41467-017-00307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Felix CA, Kolaris CP, Osheroff N. Topoisomerase II and the etiology of chromosomal translocations. DNA Repair (Amst) 2006;5(9):1093–1108. doi: 10.1016/j.dnarep.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 47.Strick R, Strissel PL, Borgers S, Smith SL, Rowley JD. Dietary bioflavonoids induce cleavage in the MLL gene and may contribute to infant leukemia. Proc Natl Acad Sci USA. 2000;97(9):4790–4795. doi: 10.1073/pnas.070061297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cowell IG, et al. Model for MLL translocations in therapy-related leukemia involving topoisomerase IIβ-mediated DNA strand breaks and gene proximity. Proc Natl Acad Sci USA. 2012;109(23):8989–8994. doi: 10.1073/pnas.1204406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strissel PL, Strick R, Rowley JD, Zeleznik-Le NJ. An in vivo topoisomerase II cleavage site and a DNase I hypersensitive site colocalize near exon 9 in the MLL breakpoint cluster region. Blood. 1998;92(10):3793–3803. [PubMed] [Google Scholar]
- 50.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461(7264):674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 51.Schellenberg MJ, et al. ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links. Science. 2017;357(6358):1412–1416. doi: 10.1126/science.aam6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao T, et al. Novel TDP2-ubiquitin interactions and their importance for the repair of topoisomerase II-mediated DNA damage. Nucleic Acids Res. 2016;44(21):10201–10215. doi: 10.1093/nar/gkw719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schellenberg MJ, et al. Reversal of DNA damage induced topoisomerase 2 DNA-protein crosslinks by Tdp2. Nucleic Acids Res. 2016;44(8):3829–3844. doi: 10.1093/nar/gkw228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi K, et al. Structural basis for recognition of 5′-phosphotyrosine adducts by TDP2. Nat Struct Mol Biol. 2012;19(12):1372–1377. doi: 10.1038/nsmb.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schellenberg MJ, Appel CD, Adhikari S, Robertson PD, Ramsden DA, Williams RS. Mechanism of repair of 5′-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nat Struct Mol Biol. 2012;19(12):1363–1371. doi: 10.1038/nsmb.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao R, Huang SN, Marchand C, Pommier Y. Biochemical characterization of human tyrosyl-DNA phosphodiesterase 2 (TDP2/TTRAP): a Mg(2+)/Mn(2+)-dependent phosphodiesterase specific for the repair of topoisomerase cleavage complexes. J Biol Chem. 2012;287(36):30842–30852. doi: 10.1074/jbc.M112.393983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J Biol Chem. 2001;276(44):40652–40658. doi: 10.1074/jbc.M104009200. [DOI] [PubMed] [Google Scholar]
- 58.Lee KC, Swan RL, Sondka Z, Padget K, Cowell IG, Austin CA. Effect of TDP2 on the level of TOP2-DNA complexes and SUMOylated TOP2-DNA complexes. Int J Mol Sci. 2018;19(7):2056. doi: 10.3390/ijms19072056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang A, et al. A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J Biol Chem. 2006;281(47):35997–36003. doi: 10.1074/jbc.M604149200. [DOI] [PubMed] [Google Scholar]
- 60.Stingele J, Jentsch S. DNA-protein crosslink repair. Nat Rev Mol Cell Biol. 2015;16(8):455–460. doi: 10.1038/nrm4015. [DOI] [PubMed] [Google Scholar]
- 61.Stingele J, Schwarz MS, Bloemeke N, Wolf PG, Jentsch S. A DNA-dependent protease involved in DNA-protein crosslink repair. Cell. 2014;158(2):327–338. doi: 10.1016/j.cell.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 62.Tang X, Cao J, Zhang L, Huang Y, Zhang Q, Rong YS. Maternal Haploid, a Metalloprotease enriched at the largest satellite repeat and essential for genome integrity in Drosophila embryos. Genetics. 2017;206(4):1829–1839. doi: 10.1534/genetics.117.200949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duxin JP, Dewar JM, Yardimci H, Walter JC. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell. 2014;159(2):349–357. doi: 10.1016/j.cell.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deshpande RA, Lee JH, Arora S, Paull TT. Nbs1 converts the human Mre11/Rad50 nuclease complex into an endo/exonuclease machine specific for protein-DNA adducts. Mol Cell. 2016;64(3):593–606. doi: 10.1016/j.molcel.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 65.Gómez-Herreros F, et al. TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo. PLoS Genet. 2013;9(3):e1003226. doi: 10.1371/journal.pgen.1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams GJ, Hammel M, Radhakrishnan SK, Ramsden D, Lees-Miller SP, Tainer JA. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst) 2014;17:110–120. doi: 10.1016/j.dnarep.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Azuma Y, Arnaoutov A, Anan T, Dasso M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 2005;24(12):2172–2182. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H, Wang Y, Liu X. Plk1-dependent phosphorylation regulates functions of DNA topoisomerase IIα in cell cycle progression. J Biol Chem. 2008;283(10):6209–6221. doi: 10.1074/jbc.M709007200. [DOI] [PubMed] [Google Scholar]
- 69.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49(5):795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 70.Pichler A, Fatouros C, Lee H, Eisenhardt N. SUMO conjugation—a mechanistic view. Biomol Concepts. 2017;8(1):13–36. doi: 10.1515/bmc-2016-0030. [DOI] [PubMed] [Google Scholar]
- 71.Kawale AS, Povirk LF. Tyrosyl–DNA phosphodiesterases: rescuing the genome from the risks of relaxation. Nucleic Acids Res. 2018;46(2):520–537. doi: 10.1093/nar/gkx1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bian K, et al. ERK3 regulates TDP2-mediated DNA damage response and chemoresistance in lung cancer cells. Oncotarget. 2016;7(6):6665–6675. doi: 10.18632/oncotarget.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cappadocia L, Pichler A, Lima CD. Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nat Struct Mol Biol. 2015;22(12):968–975. doi: 10.1038/nsmb.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koidl S, Eisenhardt N, Fatouros C, Droescher M, Chaugule VK, Pichler A. The SUMO2/3 specific E3 ligase ZNF451-1 regulates PML stability. Int J Biochem Cell Biol. 2016;79:478–487. doi: 10.1016/j.biocel.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 75.Abascal F, Tress ML, Valencia A. Alternative splicing and co-option of transposable elements: the case of TMPO/LAP2α and ZNF451 in mammals. Bioinformatics. 2015;31(14):2257–2261. doi: 10.1093/bioinformatics/btv132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karvonen U, Jääskeläinen T, Rytinki M, Kaikkonen S, Palvimo JJ. ZNF451 is a novel PML body- and SUMO-associated transcriptional coregulator. J Mol Biol. 2008;382(3):585–600. doi: 10.1016/j.jmb.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 77.Matelska D, Steczkiewicz K, Ginalski K. Comprehensive classification of the PIN domain-like superfamily. Nucleic Acids Res. 2017;45(12):6995–7020. doi: 10.1093/nar/gkx494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fedotova AA, Bonchuk AN, Mogila VA, Georgiev PG. C2H2 zinc finger proteins: the largest but poorly explored family of higher eukaryotic transcription factors. Acta Naturae. 2017;9(2):47–58. [PMC free article] [PubMed] [Google Scholar]
- 79.Fradet-Turcotte A, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499(7456):50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal ACO, Nielsen ML. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol. 2017;24:325. doi: 10.1038/nsmb.3366. [DOI] [PubMed] [Google Scholar]
- 81.Eisenhardt N, et al. A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly. Nat Struct Mol Biol. 2015;22(12):959–967. doi: 10.1038/nsmb.3114. [DOI] [PubMed] [Google Scholar]
- 82.Hendriks IA, D’Souza RCJ, Yang B, Verlaan-de Vries M, Mann M, Vertegaal ACO. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol. 2014;21(10):927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hendriks IA, Treffers LW, Verlaan-de Vries M, Olsen JV, Vertegaal ACO. SUMO-2 orchestrates chromatin modifiers in response to DNA damage. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ho C-W, Chen H-T, Hwang J. UBC9 autosumoylation negatively regulates sumoylation of septins in Saccharomyces cerevisiae. J Biol Chem. 2011;286(24):21826–21834. doi: 10.1074/jbc.M111.234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoshida MM, Azuma Y. Mechanisms behind topoisomerase II SUMOylation in chromosome segregation. Cell Cycle. 2016;15(23):3151–3152. doi: 10.1080/15384101.2016.1216928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mao Y, Desai SD, Liu LF. SUMO-1 conjugation to human DNA topoisomerase II isozymes. J Biol Chem. 2000;275(34):26066–26073. doi: 10.1074/jbc.M001831200. [DOI] [PubMed] [Google Scholar]
- 87.Agostinho M, et al. Conjugation of human topoisomerase 2α with small ubiquitin-like modifiers 2/3 in response to topoisomerase inhibitors: cell cycle stage and chromosome domain specificity. Cancer Res. 2008;68(7):2409–2418. doi: 10.1158/0008-5472.CAN-07-2092. [DOI] [PubMed] [Google Scholar]
- 88.Ryu H, Furuta M, Kirkpatrick D, Gygi SP, Azuma Y. PIASy-dependent SUMOylation regulates DNA topoisomerase IIalpha activity. J Cell Biol. 2010;191(4):783–794. doi: 10.1083/jcb.201004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Radhakrishnan SK, Jette N, Lees-Miller SP. Non-homologous end joining: emerging themes and unanswered questions. DNA Repair (Amst) 2014;17:2–8. doi: 10.1016/j.dnarep.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kont YS, et al. Depletion of tyrosyl DNA phosphodiesterase 2 activity enhances etoposide-mediated double-strand break formation and cell killing. DNA Repair (Amst) 2016;43:38–47. doi: 10.1016/j.dnarep.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 91.Do PM, et al. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 2012;26(8):830–845. doi: 10.1101/gad.181685.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li C, Fan S, Owonikoko TK, Khuri FR, Sun S-Y, Li R. Oncogenic role of EAPII in lung cancer development and its activation of the MAPK–ERK pathway. Oncogene. 2011;30(35):3802–3812. doi: 10.1038/onc.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hornyak P, et al. Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2. Biochem J. 2016;473(13):1869–1879. doi: 10.1042/BCJ20160180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kossmann BR, et al. Discovery of selective inhibitors of tyrosyl-DNA phosphodiesterase 2 by targeting the enzyme DNA-binding cleft. Bioorg Med Chem Lett. 2016;26(14):3232–3236. doi: 10.1016/j.bmcl.2016.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu LM, et al. Synthesis and structure-activity relationship of furoquinolinediones as inhibitors of Tyrosyl-DNA phosphodiesterase 2 (TDP2) Eur J Med Chem. 2018;151:777–796. doi: 10.1016/j.ejmech.2018.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Komulainen E, Pennicott L, Le Grand D, Caldecott KW. Deazaflavin inhibitors of TDP2 with cellular activity can affect etoposide influx and/or efflux. ACS Chem Biol. 2019;14(6):1110–1114. doi: 10.1021/acschembio.9b00144. [DOI] [PMC free article] [PubMed] [Google Scholar]