Abstract
The large isoform of the transmembrane protein angiomotin (AMOT130) controls cell proliferation and migration of many cell types. AMOT130 associates to the actin cytoskeleton and regulates tight-junction maintenance and signaling often via endosomal uptake of polarity proteins at tight junctions. AMOT130 is highly polarized and present only at the apical side of polarized cells. Here we show that bone morphogenetic protein (BMP) growth factor signaling and AMOT function are interlinked in apical-basal polarized cells. BMP6 controls AMOT internalization and endosomal trafficking in epithelial cells. AMOT130 interacts with the BMP receptor BMPR2 and facilitates SMAD activation and target gene expression. We further demonstrate that this effect of AMOT on BMP-SMAD signaling is dependent on endocytosis and specific to the apical side of polarized epithelial and endothelial cells. Knockdown of AMOT reduces SMAD signaling only from the apical side of polarized cells, while basolateral BMP-SMAD signaling is unaffected. This allows for the first time interference with BMP signaling in a polarized manner and identifies AMOT130 as a novel BMP signaling regulator.
INTRODUCTION
Bone morphogenetic proteins (BMPs) belong to the transforming growth factor-β (TGF-β) family of secreted growth factors and function as pleiotropic cytokines guiding various cellular processes ranging from mesenchymal cell differentiation to cancer cell migration (Sieber et al., 2009). BMPs signal via heterotetrameric receptor complexes containing type I and type II serine/threonine kinase receptors. Ligand binding to the receptor complex induces phosphorylation and thereby activation of receptor-regulated SMADs (R-SMADs) 1/5/8 through the activated type I receptor. Activated R-SMADs then oligomerize with SMAD4, the common mediator of BMP and TGF-β signals, and accumulate in the nucleus where they function as transcriptional regulators of BMP-SMAD target genes (Massague et al., 2005). These encompass inhibitor of differentiation (ID) genes, which for example potently inhibit myogenic differentiation in mesenchymal precursor cells and are associated with epithelial-to-mesenchymal transition and increased invasive potential of breast cancer (Katagiri et al., 2002; Tobin et al., 2011). BMPs also trigger non-SMAD signaling including phosphorylation of mitogen-activated protein kinases (MAPK) such as p38 or ERK. These effectors in turn initiate nontranscriptional responses by remodeling the actin cytoskeleton and influencing migration or transcriptional responses leading to the expression of non-SMAD transcription factors such as JunB (Derynck and Zhang, 2003). Both signaling routes need to be tightly controlled and fine-tuned at multiple levels, as aberrant signaling can lead to several diseases, including musculoskeletal disorders or cancer (Wang et al., 2014). Receptor internalization can adapt the intensity and duration of signaling responses and is often the first mechanism to shut down further signaling. In previous studies, our lab showed that BMP receptors are internalized by both clathrin- and caveolin-dependent endocytosis (Nohe et al., 2005; Hartung et al., 2006). It was furthermore postulated that endosomal regulation fine-tunes SMAD1/5 signaling, while initiation of the signaling cascade has been demonstrated to even occur in endocytosis-defective cells and in cells exposed to gold surface immobilized ligand, which cannot be internalized (Pohl et al., 2012; Schwab et al., 2015; Paarmann et al., 2016). This suggests that BMP signaling is initiated either directly at the plasma membrane (PM) or in endosomes. Besides shutting down the cascade, endosomes also positively affect signaling responses by stabilizing the endosomal receptor–ligand interaction and by compartmentalizing the respective cascade (Sorkin and von Zastrow, 2009). The latter increases signaling efficiency or velocity specifically in one cell compartment depending on the lipid composition of the PM or membrane-anchored proteins (Disanza et al., 2009).
The adaptor protein angiomotin (AMOT) is part of the Motin family of proteins and exists in two isoforms, AMOT80 (∼80 kDa) and AMOT130 (∼130 kDa), which are generated by alternative splicing (Moreau et al., 2005; Ernkvist et al., 2006). All family members share a coiled-coil domain and a conserved C-terminal PDZ motif, which allows them to interact with tight junction-associated proteins (Bratt et al., 2002). AMOT is expressed as a transmembrane protein and exhibits angiostatin binding in the endothelium (Troyanovsky et al., 2001; Bratt et al., 2005). It has been recently shown that both isoforms also localize in the cytoplasm and the nucleus (Moleirinho et al., 2017). However, the precise mechanism of translocation and the essential functional domains have yet to be described. AMOT130 contains a unique, extended N-terminus with XPXY motifs and a phosphorylation site at Ser-175 (human; Moleirinho et al., 2014). Phosphorylation at this site by large tumor suppressor kinases 1/2 (LATS1/2) was shown to target AMOT130 to the membrane and masks the F-actin binding site, thereby preventing interaction with the actin cytoskeleton (Mana-Capelli et al., 2014; Moleirinho et al., 2017). LATS1/2 kinases in turn are activated by mammalian sterile-20-type kinases (MST1/2) both upstream of the transcriptional coactivator Yes-associated protein 1 (YAP) (Adler et al., 2013; Chan et al., 2013; Dai et al., 2013; Mana-Capelli et al., 2014). Hence, the extended N-terminus directly interacts with the actin cytoskeleton, YAP and LATS1/2, endorsing AMOT130 to act as an upstream regulator of YAP. In confluent cells, AMOT130 is associated with tight junctions and induces YAP to localize in endosomes where it is inactive (Cox et al., 2015). Very likely, this is mediated by the AMOT CC/BAR domain, which enables high-affinity binding to curved, cholesterol-rich membrane structures and can induce tubulation and enlargement of endosomal compartments (Heller et al., 2010). Additionally, endosomal localization of AMOT was shown to regulate trafficking of tight junction-associated proteins (Wells et al., 2006). AMOT interacts directly with PATJ and is thereby targeted to a protein complex containing Pals1 and Crumbs, necessary for the formation and maintenance of apical-basal polarity in epithelial cells (Sugihara-Mizuno et al., 2007; Ernkvist et al., 2009; Campbell et al., 2016). It is furthermore associated to other tight-junction protein complexes containing MAGI-1 or Par-3 (Wells et al., 2006; Ebnet, 2008).
So far, most of the molecular analyses to unravel AMOT function have been performed in epithelial cells, specifically mammary epithelial cells. Therefore, we also made use of this model in our study using Michigan Cancer Foundation-7 (MCF7) cells. Furthermore, we extended our research to endothelial cells, also exhibiting distinct apical-basal polarity and being the cell type in which AMOT was discovered (Troyanovsky et al., 2001). Until now, the effects of AMOT130 on cellular processes have been attributed to its modulation of the Hippo/YAP pathway, GTPase signaling, or MAPK signaling. However, no upstream triggers besides actin rearrangements, cell–cell contacts, or serum stimulation have been investigated so far. We identified AMOT in a proteomics-based BMPR-interactome study. Here, we aimed to elucidate the molecular function of AMOT in BMP signaling. We show that AMOT130 specifically interacts with the BMP receptor BMPR2 and thereby facilitates BMP-SMAD signaling specifically at the apical side of polarized cells. BMP-stimulation leads to dephosphorylation of AMOT at Ser-175 and subsequent internalization of AMOT. With AMOT we identified the first component, which acts on BMP-SMAD signaling in a polarized manner. With this, polarized interference of BMP-SMAD signaling is made possible for the first time.
RESULTS
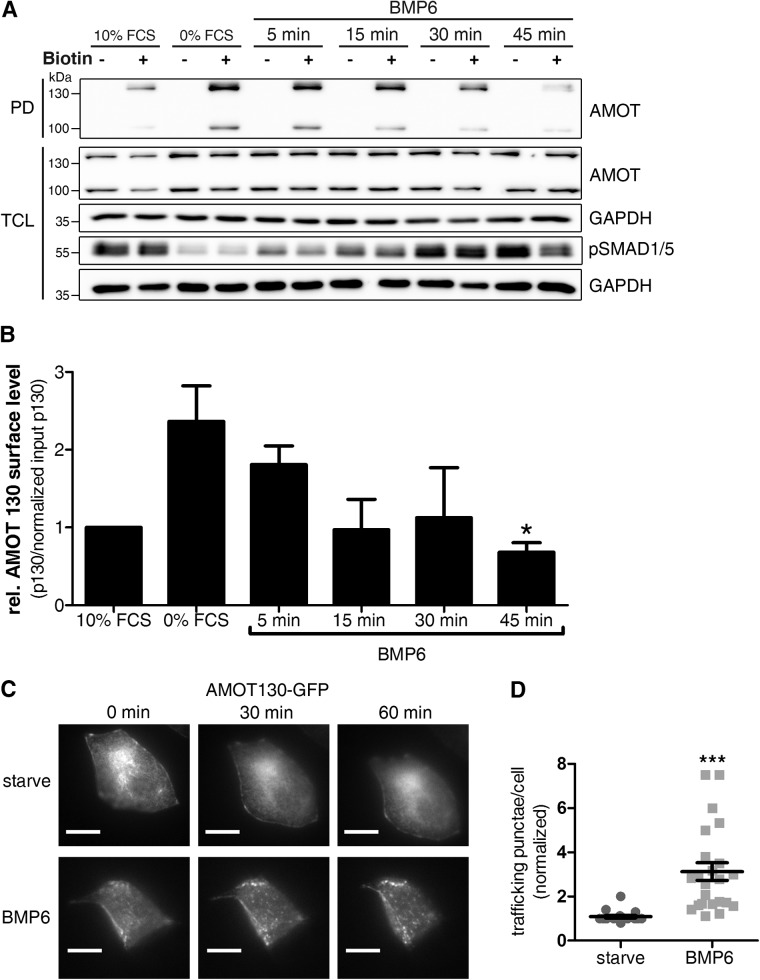
BMP stimulation induces internalization and trafficking of AMOT
AMOT acts as a receptor for circulating angiostatin in the endothelium but is not a classical transmembrane protein, since it also translocates to the nucleus where it modulates YAP target gene transcription (Troyanovsky et al., 2001; Moleirinho et al., 2017). Destabilization of the actin cytoskeleton or serum starvation is the only upstream regulator of AMOT phosphorylation investigated so far (Adler et al., 2013; Mana-Capelli et al., 2014). However, AMOT surface levels and potential internalization on growth factor stimulation were never analyzed. Although it has been postulated that serum itself leads to AMOT dephosphorylation via GPCR signaling (Dai et al., 2013), there is no experimental proof specifically showing GPCR activation prior to dephosphorylation and whether this induces relocalization of AMOT130. We aimed to investigate whether the internalization of AMOT could be induced by specific growth factors in the serum, particularly BMPs. Therefore, we measured the endogenous surface levels of AMOT130 and AMOT80 during BMP stimulation using a surface biotinylation assay in HEK293T cells. In this assay, the cell-impermeable biotinylation compound selectively labels proteins exposed to the extracellular space to be subsequently pulled down with Streptavidin beads. We found that surface levels of both isoforms of AMOT (AMOT80 and AMOT130) are reduced on BMP6 stimulation (Figure 1A). Both AMOT80 and more pronounced AMOT130 are increased at the cell surface after starvation but decrease significantly on stimulation with BMP6; similar dynamics were found for AMOT80 (Figure 1B, Supplemental Figure S1B). Interestingly, AMOT surface levels in cells stimulated with BMP6 for 45 min were even less than in the 10% serum control, in which cells were stimulated with 10% FCS for 45 min. As total levels of AMOT remained unchanged (see total cell lysate [TCL], Figure 1A), AMOT seems not to be degraded but rather changes trafficking under those conditions (Supplemental Figure S1C). Additionally, we detected that indeed phosphorylated AMOT130 (Ser-175) was most predominantly present at the cell surface and dephosphorylated on 1 h serum stimulation, which again led to internalization of AMOT130 (Supplemental Figure S1A). To assess this further, we monitored AMOT130 localization via live cell imaging. MCF7 cells expressing GFP-tagged AMOT130 were subjected to video analysis during the course of BMP stimulation. Initially, AMOT is equally distributed at the PM, while 30 min of BMP6 stimulation led to spot-like patterns, reflecting endosomes (Figure 1, C and D; Supplemental Videos S4 [starved conditions] and S5 [BMP6-stimulated conditions]).
FIGURE 1:
Trafficking of AMOT130 is increased after BMP6 stimulation. (A) HEK293T cells were stimulated with 10 nM BMP6 for the indicated time points, subjected to surface biotinylation, and lysed, and biotinylated proteins were precipitated with streptavidin beads. Eluted proteins were immunoblotted using the indicated antibodies. Pull down (PD) with streptavidin beads only represents proteins expressing an extracellular motif. TCL is the protein input before streptavidin pull down. The blot is representative of three independent experiments. (B) Densitometric quantification of AMOT130 surface levels as blotted in A. Band intensity of biotinylated AMOT130 was normalized to AMOT TCL levels (relative to GAPDH). Data are presented as mean ± SEM relative to 10% FCS of three independent experiments; *p < 0.05, one-way ANOVA with Bonferroni post-hoc test compared with 0% FCS control. (C) MCF7 cells, expressing GFP-tagged AMOT130, were analyzed microscopically during BMP6 stimulation. Cells were incubated in a live cell incubation chamber and stimulated for 1 h. Images of the GFP signal were taken every 30 s. Scale bar represents 10 µm. Representative cells are also depicted as movie files. (D) Quantification of GFP-positive punctae after 1 h of BMP6 stimulation for at least 20 cells per condition of three independent experiments. Data are presented as mean fold induction 60 min/0 min ± SEM signals per cell; ***p < 0.001, unpaired Student’s t test.
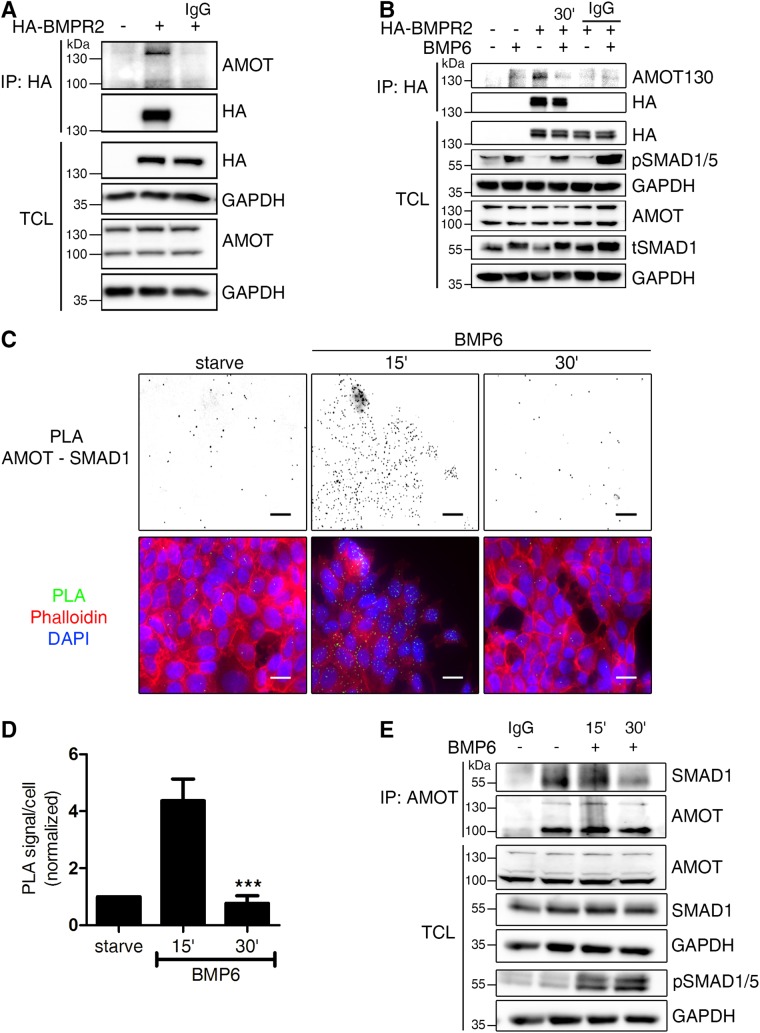
AMOT interacts with the BMP type II receptor (BMPR2) and SMAD1
On the basis of our observation that BMP triggers AMOT internalization, we hypothesized that there is a direct interaction between AMOT and BMP signaling components, which facilitates this effect. Therefore, we first used a semiendogenous coimmunoprecipitation (Co-IP) approach, in which we expressed HA-tagged BMP receptors in HEK293T cells and investigated whether endogenous AMOT associates to BMP receptors. Here, we show that only AMOT130, but not AMOT80, interacts with HA-tagged BMPR2 (Figure 2A; Supplemental Figure S2A). Interestingly, this interaction was lost after 30 min of BMP6 stimulation (Figure 2B). It is noteworthy that we did not observe any interaction between AMOT130 and BMP type I receptors (BMPR1) in HEK293T cells (Supplemental Figure S2B). When we analyzed the different BMPR2 isoforms for interaction with AMOT130, we found that only BMPR2 long form (LF), and not BMPR2 short form (SF), interacts with AMOT130 (Supplemental Figure S2C). Next, we investigated whether and where AMOT might interact with other BMP pathway components. Using proximity ligation assays (PLA) in MCF7 cells, we show that AMOT localized in close proximity to SMAD1 (Figure 2C; controls in Supplemental Figure S2D). Of note, this association was increased after 15 min of BMP6 stimulation and decreased again to starving levels after 30 min (Figure 2D). This interaction was further validated using coimmunoprecipitation analyses, demonstrating that AMOT formed a complex with SMAD1 under serum starvation and short-term BMP6 stimulation conditions (Figure 2E). Prolonged stimulation reduced the interaction markedly, which coincides with the internalization dynamics of AMOT (Figure 1A). This suggests that the interaction of AMOT130 with both BMPR2 and SMAD1 is transient and limited to the PM. Taken together, our data provide evidence for a novel, highly dynamic interaction between the adaptor protein AMOT130 and SMADs. BMP6 stimulates AMOT internalization and a concomitant loss of interaction with BMPR2 and SMAD1.
FIGURE 2:
AMOT130 but not AMOT80 dynamically associates with the BMPR2 and SMAD1. (A, B) Transfected HEK293T cells were subjected to immunoprecipitation using α-HA tag antibody. Before, cells were left in full medium (A) or starved and stimulated for 30 min with 10 nM BMP6 (B). Immunoprecipitates (IP) and TCL were analyzed by Western blotting using the indicated antibodies. Incubation with mouse immunoglobulin G (IgG) served as control. (C) In situ PLA of AMOT and SMAD1. MCF7 cells were subjected to in situ PLA (green signal) to visualize the endogenous association of AMOT and SMAD1 after the respective indicated treatments. Nuclei were stained with DAPI (blue) and F-actin with Phalloidin594 (red). PLA signal images were inverted to visualize the signal. Scale bar represents 20 µm. Relevant controls are depicted in Supplemental Figure S2. (D) Quantification of AMOT/SMAD1 heteromers shown in C. The bar chart represents mean ± SD from three independent experiments; ***p < 0.001, one-way ANOVA with Bonferroni post-hoc test compared with 15′ condition. (E) Endogenous interaction of AMOT with SMAD1. MCF7 cells were starved and stimulated for the indicated time points with 10 nM BMP6 before lysis and immunoprecipitation using α-AMOT (BL) antibody. Precipitates and TCL were analyzed by Western blotting using the indicated antibodies. Rabbit IgG served as control.
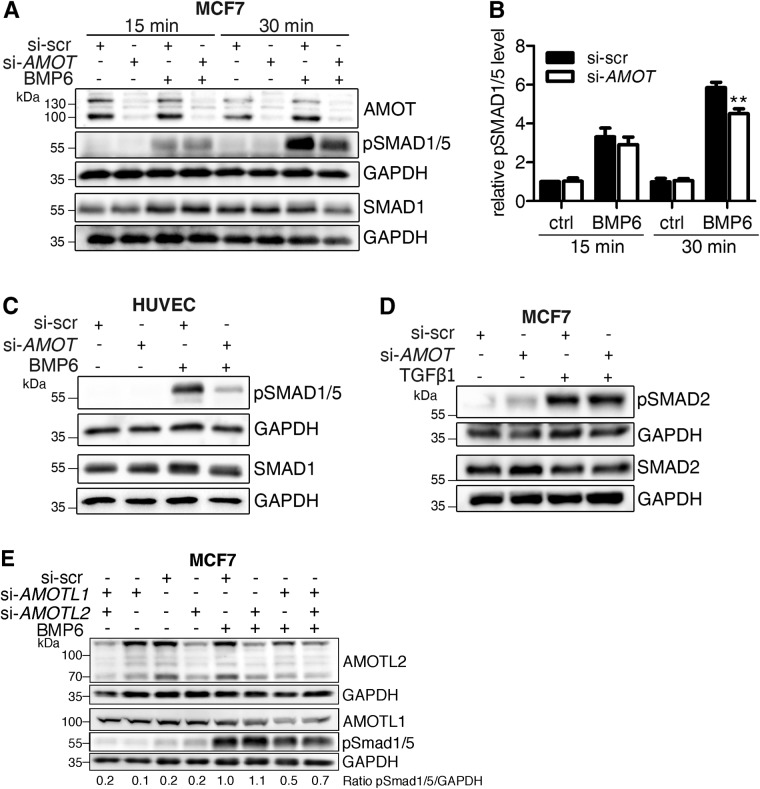
AMOT knockdown specifically inhibits BMP signaling
Next, we analyzed the impact of BMP6-induced endocytosis of AMOT and the association between AMOT and BMP pathway components on downstream signaling events. For this, we depleted AMOT in established cell lines and primary cells derived from different tissues and subsequently analyzed each step of the BMP signaling cascade at different time points during stimulation with BMP6. Of note, the small interfering RNAs (siRNAs) used target both isoforms of AMOT. Knockdown efficiency in each experiment was 80% as assessed by Western blot analyses (Supplemental Figure S3, D–G). First, we measured the impact of AMOT knockdown on BMP6-mediated phosphorylation of BMP-specific R-SMAD1/5 in MCF7 cells. AMOT depletion reduced SMAD1/5 phosphorylation significantly after 30 min of stimulation but not after 15 min (Figure 3, A and B). Similar results were obtained in primary endothelial cells (HUVECs; Figure 3C) and in three different mesenchymal precursor cell lines (mouse myoblast precursor cells [C2C12]), human fetal osteoblasts [hFOBs], human myoblasts; Supplemental Figure S3, A–C). To explore further whether this effect of AMOT was specific for SMAD1, we performed similar experiments in TGF-β1 stimulated MCF7 cells and assessed phosphorylation of SMAD2. Here, no difference in SMAD2 phosphorylation on AMOT knockdown was observed (Figure 3D). This finding suggests that the effect of AMOT on SMAD phosphorylation is BMP-specific. We also investigated whether the other two proteins of the Motin family, AMOT-like protein 1 (AMOTL1) and AMOT-like protein 2 (AMOTL2), have a similar effect on BMP-induced pSMAD1/5 levels. Therefore, AMOTL1 and AMOTL2 were depleted either separately or combined before stimulating MCF7 cells with BMP6. While AMOTL2 depletion did not affect SMAD1/5 phosphorylation, the knockdown of AMOTL1 as well as the combined knockdown inhibited BMP6-induced phosphorylation (Figure 3E).
FIGURE 3:
Targeted AMOT depletion reduces phosphorylation of BMP-SMAD proteins specifically. (A–D) MCF7 cells or HUVECs were transfected with siRNA targeting either nonspecific sequences (si-scr) or human AMOT (si-AMOT) and stimulated for the indicated time with 10 nM BMP6 (A–C) or 200 pM TGFβ1 (D). Protein lysates were subjected to Western blotting using the indicated antibodies. Blots represent at least three independent experiments. (B) Quantification depicts pSMAD1/5 level normalized to GAPDH relative to si-scr control. Bar charts represent mean ± SEM from six independent experiments of Western blots depicted in A; **p < 0.01, two-way ANOVA with Bonferroni post-hoc test compared with si-scr stimulated with BMP6 for 30 min. (E) MCF7 cells were transfected with siRNA targeting AMOTL1 (si-AMOTL1) or AMOTL2 (si-AMOTL2) and stimulated with 10 nM BMP6 for 30 min. Lysates were analyzed by Western blotting with the indicated antibodies. The blot is representative for three independent experiments.
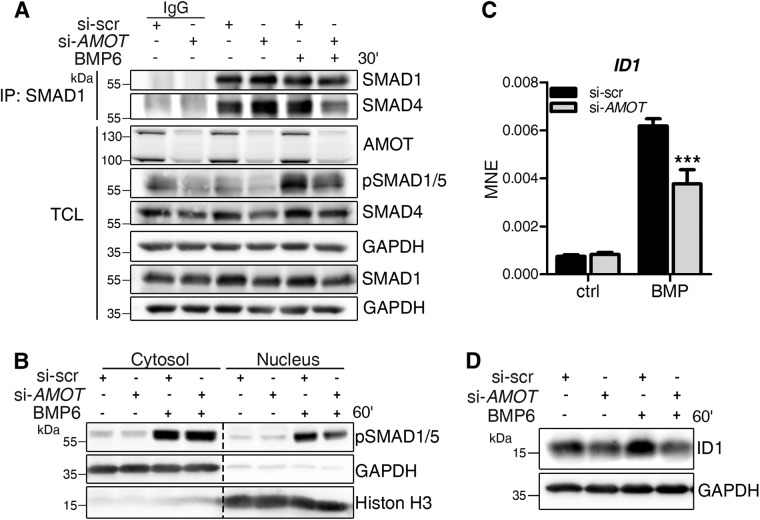
We further investigated whether decreased R-SMAD phosphorylation also affects downstream signaling steps, such as complex formation between activated R-SMADs and co-SMAD4. To measure this, we depleted AMOT in MCF7 cells and subsequently subjected them to coimmunoprecipitation analyses to investigate the interaction between SMAD1 and SMAD4. On AMOT knockdown, SMAD1–SMAD4 complex formation in BMP6 stimulated cells was decreased (Figure 4A). This is in line with the general decrease in pSMAD1/5 levels, visible in the TCL (Figure 4A). Next, the translocation of pSMADs to the nucleus was assessed by cellular fractionations and consecutive protein analyses. As expected from the decrease in SMAD complex formation, BMP6-mediated pSMAD1/5 translocation to the nucleus was reduced in AMOT-depleted cells (Figure 4B). Last, we analyzed the induction of ID1, a direct SMAD target gene. AMOT knockdown led to decreased mRNA expression of ID1 after 1 h of BMP6 stimulation compared with cells transfected with control siRNA (Figure 4C; Supplemental Figure S3H). This effect was also reflected on protein expression, as ID1 protein levels were decreased after BMP6 stimulation in AMOT-depleted cells (Figure 4D).
FIGURE 4:
AMOT depletion causes decreased BMP-SMAD signaling. (A–D) MCF7 cells were transfected with siRNA targeting either nonspecific sequences (si-scr) or human AMOT (si-AMOT) and stimulated for the indicated time points with 10 nM BMP6. (A) Endogenous complex formation of SMAD1 and SMAD4. After treatment, MCF7 cells were lysed and immunoprecipitation using αSMAD1 antibody was performed. Precipitates and TCL were analyzed by Western blotting using the indicated antibodies. Rabbit IgG served as control. (B) After 1 h of stimulation, MCF7 cells were lysed and their cytosolic and nuclear proteins fractionated and blotted with the indicated antibodies. The blot represents three independent experiments. (C) After 1 h of stimulation, cells were lysed and RNA was extracted, reverse transcribed, and used for gene expression analysis. qRT-PCR analysis of ID1 mRNA. Data are presented as mean ± SEM of three independent experiments; ***p < 0.001, two-way ANOVA with Bonferroni post-hoc test, compared with si-scr stimulated with BMP6 for 1 h. (D) Representative Western blot of MCF7 protein lysates after AMOT depletion and 1 h BMP6 stimulation shows ID1 and GAPDH level.
Together, these results show that AMOT depletion modulates BMP target gene expression by decreasing SMAD1/5 phosphorylation, SMAD complex formation, as well as its nuclear translocation.
AMOT is predominantly expressed apically in polarized cells
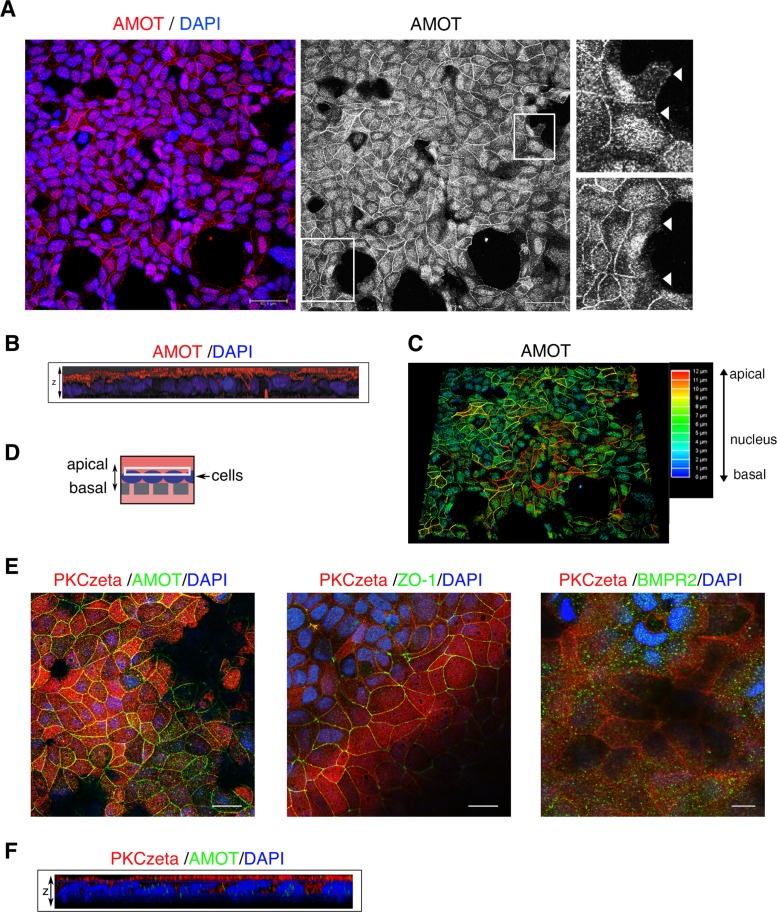
As described in the literature, AMOT function is associated with the maintenance and formation of apical-basal polarity and cell–cell contacts in polarized cells (Bratt et al., 2005; Ebnet, 2008). This is dependent on its predominant apical expression, which was demonstrated in Madin–Darby canine kidney (MDCK) cells (Wells et al., 2006). To verify this compartmentalization in MCF7 cells, we performed confocal z-stack analyses of AMOT immunofluorescence images. Figure 5A depicts the maximal projection of all planes ranging from apical to basal of a semiconfluent MCF7 cell layer. Endogenous AMOT predominantly localized at cell–cell junctions and in the nucleus. When cells lacked cell–cell contacts, AMOT was not concentrated at the outer rim of the PM (Figure 5A, right zoom-in images). Three-dimensional (3D) analysis shows that AMOT was almost exclusively located at the apical side of polarized cells (Figure 5B). Depth analysis of the AMOT signal confirmed this finding (Figure 5C). To verify that cells were indeed polarized, specifically in the transwell assay used later on (Figures 5D and 6C), the typical polarity marker protein kinase C zeta (PKCzeta) was investigated (Yanger et al., 2013; Yonemura, 2014). As expected, PKCzeta was exclusively present on the apical side, thereby indicating complete apical-basal polarization of cells (Figure 5, E and F). Further, ZO-1 was present at cell–cell contacts, demonstrating mature and functional tight junctions. The BMPR2, however, localized ubiquitously to the PM, both at the apical as well as at the basolateral sides (Figure 5E). These findings are further supported by z-stack movies of the shown IF-stainings (Supplemental Videos S1–S3).
FIGURE 5:
AMOT predominantly localizes at apical cell–cell junctions and the nucleus in polarized MCF7 cells. (A) MCF7 cells were subjected to immunofluorescence staining using α-AMOT (BL) antibody (red). Nuclei were stained with DAPI (blue). Scale bar represents 20 µm. Images were taken at a confocal microscope by applying a z-stack and depicting maximal projection. Right panel shows only AMOT. Zoom-in images depict AMOT staining on sites without cell–cell contacts, marked by white arrows. (B) 3D analysis of the picture shown in A. Z-stack merge is depicted from side view. (C) Depth analysis of the AMOT signal. (E) MCF7 cells were seeded in transwells and subjected to immunofluorescence staining using the indicated antibodies. Z-stacks were taken at a confocal microscope and representative images depict the apical membrane as indicated by the white rectangle in D. Cells were confluent; dark spots represent nonfocused areas, because transwell membranes were not flat. Scale bar represents 20 µm. (F) Z-stack merge is depicted from side view for PKCzeta/AMOT. Supplemental videos of z-stacks: Supplemental Video S1 depicts AMOT (green) and PKCzeta (red) from basal to apical; Supplemental Video S2 depicts ZO-1 (green) and PKCzeta (red) from basal to apical; Supplemental Video S3 depicts BMPR2 (green) and PKCzeta (red) from apical to basal.
FIGURE 6:
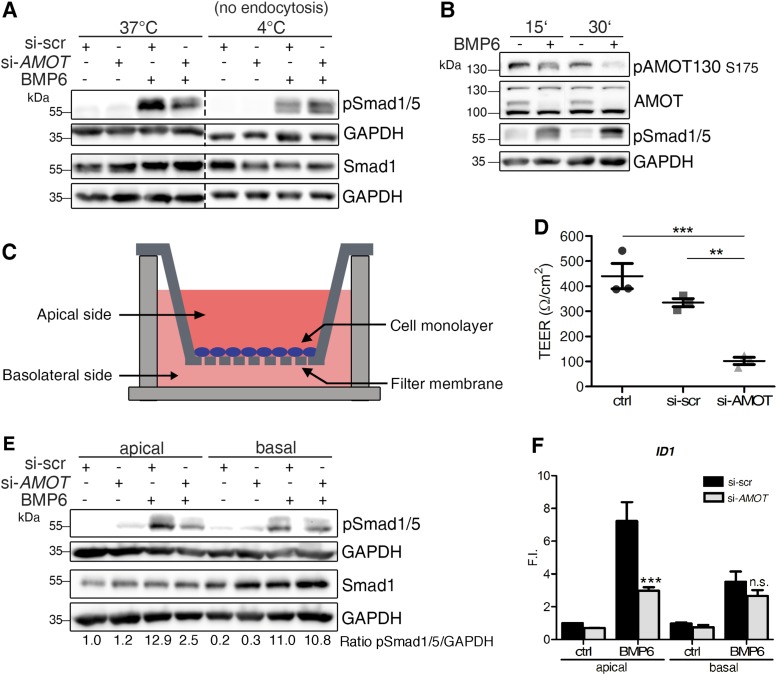
AMOT drives apical SMAD signaling via endocytosis. (A) MCF7 cells were depleted of AMOT and stimulated with BMP6 at 37°C or at 4°C. Lysates were analyzed by Western blotting using the indicated antibodies. (B) MCF7 cells were starved and stimulated with BMP6 for the indicated time points. Lysates were analyzed by Western blotting using the indicated antibodies. (C) Experimental setup for apical–basal-specific BMP stimulation of MCF7 cells in specific transwell inserts. (D) MCF7 cells transfected with siRNA targeting AMOT (si-AMOT) or scr control (si-scr) were seeded into transwells and their TEER was measured. Data are presented as mean ± SEM of three independent experiments; **p < 0.01, ***p < 0.001, one-way ANOVA with Bonferroni post-hoc test. (E) MCF7 cells transfected with siRNA targeting AMOT (si-AMOT) or scr control (si-scr) were seeded until confluency in transwells depicted in C and stimulated specifically from the apical or basal side. Lysates were analyzed by Western blotting with the indicated antibodies. All blots are representive for each four independent experiments. (F) Target gene expression measured by qRT-PCR after 1 h of BMP6 stimulation in transwell experiment. After 1 h of stimulation, cells were lysed and RNA was extracted, reverse transcribed, and used for gene expression analysis of ID1 mRNA. Data are presented as mean fold induction (F.I.) ± SEM of four independent experiments; ***p < 0.001, two-way ANOVA with Bonferroni post-hoc test compared with si-scr stimulated apically or basally with BMP6.
Video S1.
Video depicts a z-stack of MCF7 cells subjected to immunofluorescence staining of AMOT (green) and PKCzeta (red). Nuclei are stained with DAPI (blue). Z-stack is shown from basal to apical. For representative picture see Figure 5.
Video S2.
Video depicts a z-stack of MCF7 cells subjected to immunofluorescence staining of ZO-1 (green) and PKCzeta (red). Nuclei are stained with DAPI (blue). Z-stack is shown from basal to apical. For representative picture see Figure 5.
Video S3.
Video depicts a z-stack of MCF7 cells subjected to immunofluorescence staining of BMPR2 (green) and PKCzeta (red). Nuclei are stained with DAPI (blue). Z-stack is shown from apical to basal. For representative picture see Figure 5.
Video S4.
Video depicts a representative MCF7 cell, expressing GFP tagged AMOT130 and incubated in a live cell incubation chamber. Cells were starved and images of the GFP signal were taken every 30 seconds for 1 h. For representative pictures and quantification see Figure 1.
Video S5.
Video depicts a representative MCF7 cell, expressing GFP tagged AMOT130 and incubated in a live cell incubation chamber. Cells were starved and then stimulated with BMP6 for 1 h. images of the GFP signal were taken every 30 seconds for 1 h. For representative pictures and quantification see Figure 1.
Endocytosis of AMOT130 is crucial for apical BMP-SMAD signaling
We then assessed whether trafficking of AMOT130, as shown in Figure 1, is a prerequisite for its effect on pSMAD1/5 levels. Thus, AMOT-depleted MCF7 cells were stimulated with BMP either at 37°C or at 4°C. At this lower temperature, endocytosis is blocked because of the physical properties of the PM. Intriguingly, AMOT knockdown did not affect SMAD1/5 phosphorylation when stimulated at 4°C, while at 37°C pSMAD1/5 levels were clearly decreased (Figure 6A). This suggests that the trafficking of AMOT130 is crucial for its modulatory effect on BMP-SMAD signaling.
As mentioned, AMOT130 predominantly functions as a transmembrane protein, when it is phosphorylated at Ser-175 (Moleirinho et al., 2017). Hence, we investigated whether this phosphorylation status is affected by BMP stimulation. Using different stimulation times, we detected decreased pAMOT130 signal after 15 min of BMP6 stimulation (Figure 6B). The phosphorylation was further reduced after 30 min of BMP6 stimulation.
Since we showed that AMOT is mostly expressed at apical cell–cell junctions, we hypothesized that this is the only compartment in which it regulates BMP-SMAD signaling. We first analyzed whether AMOT has any effect on the integrity of this junctional compartment by measuring the transepithelial electrical resistance (TEER), which represents the permeability of a confluent cell layer. AMOT-depleted MCF7 cells were seeded as a confluent monolayer into transwell inserts (Figure 6C). Cells were then analyzed for their TEER. They exhibited a basal TEER of roughly 400 Ω/cm2, which was significantly decreased on AMOT depletion (Figure 6D). Next, we measured whether application of BMPs either apically or basolaterally would be similarly affected by AMOT knockdown (Figure 6E). Generally, pSMAD1/5 was induced by BMP6 stimulations from both sides suggesting BMP-receptors to be present unilaterally. Interestingly, AMOT-depleted cells showed decreased BMP-induced SMAD1/5 phosphorylation only when stimulated from the apical side, but not from the basolateral side (Figure 6E). Accordingly, when analyzing the expression of BMP target genes, we observed a significant reduction of ID1, ID2 and ID3 expression in AMOT depleted cells only after BMP6 stimulation from the apical side (Figure 6F; Supplemental Figure S3, I and J). This indeed suggests a compartment-specific function for AMOT in the BMP-SMAD pathway.
Taken together, our data provide evidence that AMOT130 is a novel modulator of BMP-SMAD signaling and likely exerts its function based on its polarized localization within cells and increased trafficking after BMP stimulation. As AMOT compartmentalizes at the apical side of polarized cells, it also modulates BMP signaling only in this compartment and therefore constitutes another layer of subcellular regulation to this crucial growth factor signaling network.
DISCUSSION
Polarization of epithelial and endothelial cells forms physiological barriers separating complex structures from the environment, allowing transport of nutrients across and directional secretion of biomolecules from these cell sheets. The PM of these cells is divided into an apical lumen-facing and basolateral membrane. The latter consists of cell–cell contact sites and the membrane facing the underlying extracellular membrane. The borders between these membrane regions are composed of tight and adherens junction complexes. Apical-basal polarity is essential for epithelial and endothelial function and is maintained by cell polarity and junction proteins. AMOT has been identified as an important regulator for cell–cell contacts and polarity in epithelial and endothelial cells. The role of AMOT is most intensively studied in the context of the Hippo/YAP pathway or small GTPase signaling (Ernkvist et al., 2009; Chan et al., 2011). Since its discovery, biochemical characteristics of AMOT have been only poorly understood. Both descriptions of AMOT as a transmembrane protein and receptor for angiostatin in the apical compartment and its role as transcription factor-binding nuclear protein have been published (Troyanovsky et al., 2001; Levchenko et al., 2008; Hong, 2013). Additionally, it has been proposed that AMOT130 is a mechanosensor, connecting the actin cytoskeleton with cell–cell contacts and the Hippo/YAP pathway (Paramasivam et al., 2011; Dai et al., 2013; Mana-Capelli et al., 2014). The BMP growth factor signaling cascade is known to integrate mechanical cues and shows compelling cross-talk with Hippo/YAP signaling (Alarcon et al., 2009; Kopf et al., 2012). After we identified AMOT from a proteomics-based BMP receptor-interactome study, we aimed to elucidate the molecular function of AMOT in the context of BMP signaling.
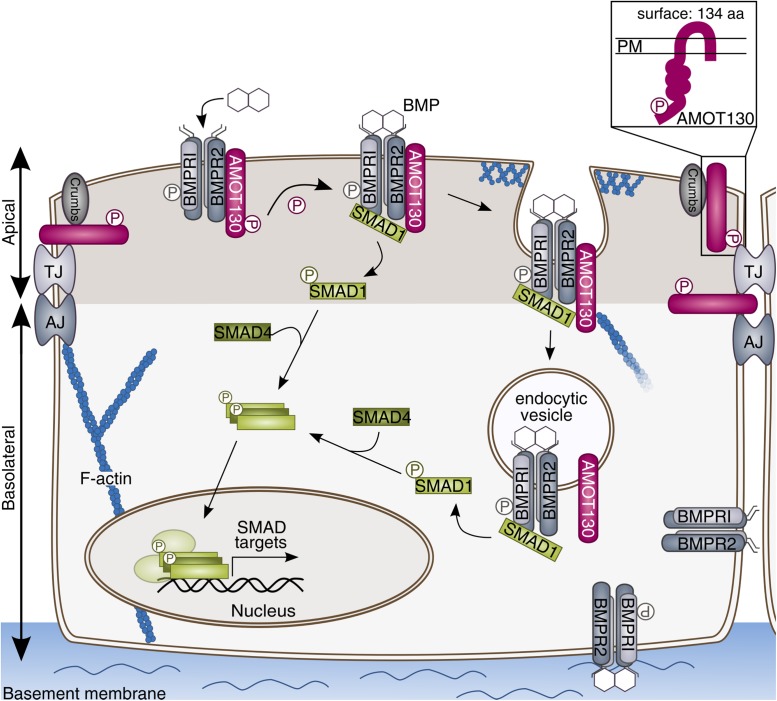
We examined the impact of AMOT on BMP signaling and vice versa BMP in AMOT function in polarized mammary epithelial cells. With this, we provide first evidence that AMOT130 is a novel regulator of the BMP-SMAD pathway, exclusively at the apical side of polarized cells. We show that AMOT130 interacts with the BMPR2 and the downstream signaling component SMAD1. On BMP stimulation, AMOT is dephosphorylated at Ser-175 and subsequently internalized. AMOT knockdown decreases SMAD1/5 phosphorylation and target gene expression derived exclusively from the apical side of polarized cells. We hypothesize that this effect depends on apical endocytosis as it can be reversed by blocking internalization. SMAD signaling derived from the basal side is not influenced by AMOT depletion (Figure 7).
FIGURE 7:
AMOT130 modulates BMP-SMAD signaling by enhanced trafficking at the apical membrane of polarized cells. AMOT130 associates to the BMP receptor complex and SMAD1. On BMP stimulation, AMOT130 is dephosphorylated and subsequently internalized. Increased trafficking of AMOT130 leads to more SMAD1/5 phosphorylation and subsequently more SMAD1/SMAD4 complex formation. These complexes translocate to the nucleus and activate target gene transcription, e.g., ID1. As AMOT is predominantly expressed apically, it facilitates only apical BMP-SMAD signaling and thereby only a subpopulation of receptor complexes. The proposed transmembrane topology was proven by surface biotinylation assays (box) (Bratt et al., 2005).
To measure cell surface levels of AMOT in the presence and absence of BMP, we applied surface biotinylation on polarized epithelial cells. Notably, biotinylation occurs preferentially at lysines or arginines and only one lysine was described in the extracellular angiostatin-binding domain (amino acids 871–1005 in AMOT130) (Bratt et al., 2005). Nevertheless, both AMOT isoforms were detected at the surface and were internalized on BMP stimulation after 45 min. The verification that AMOT is a transmembrane was a prerequisite for our study since the specific localization of AMOT varies in different cell contexts. It has, for example, been shown that AMOT only localizes to the PM at mature cell–cell contacts in dense cell cultures and dephosphorylated AMOT130 localizes in the nucleus (this study; Mana-Capelli et al., 2014; Moleirinho et al., 2017). Furthermore, AMOT is recruited to the PM on starvation (this study; Adler et al., 2013; Dai et al., 2013). However, so far no specific factors in the serum could be attributed to these effects and besides density, serum, and manipulations of the actin cytoskeleton, no upstream triggers were investigated. Here, we show that BMP6, a member of the BMP family of ligands regulates AMOT localization in polarized cells. BMP6 inhibits migration and proliferation of mammary cancer epithelial cells (Lian et al., 2013; Liu et al., 2014; Hu et al., 2016). Of note, AMOT is not degraded after BMP stimulation but rather is subjected to increased trafficking, potentially mediating recycling. This was further validated by live cell imaging studies, revealing that AMOT130-positive trafficking punctae increased after 30–60 min of BMP6 stimulation. The small isoform AMOT80 was internalized as well, although we only identified the large isoform to interact with the BMPR2, potentially facilitating the subsequent BMP-induced internalization. It has been described that the large and small isoform can build heterodimers and therefore we cannot exclude that AMOT130 in a complex with AMOT80 or even with other Motin family members binds to the BMPR2 (Ernkvist et al., 2008). Interaction of AMOT130 specifically with the BMPR2 long isoform (BMPR2-LF) suggests a binding motif for AMOT130 in the cytoplasmic tail of the BMPR2-LF and vice versa for the BMPR2-LF in the N-terminal tail of AMOT130, absent in AMOT80 (Ernkvist et al., 2006). AMOT130 could not be immunoprecipitated with BMPR1s, which is in line with other proteins modulating BMP signaling, such as PI3K, c-Src, and IRS4 (Hiepen et al., 2014; Benn et al., 2015; Dorpholz et al., 2017). However, BMP6 exhibits high affinity for ALK2 binding, making it tempting to speculate that AMOT130 is associated to single BMPR2 molecules or BMPR2-ALK2 complexes only (Yadin et al., 2016).
We furthermore report colocalization and interaction of AMOT with SMAD1 (Figure 2, C–E). Here, we cannot assure any AMOT isoform specificity, as the utilized antibody recognizes both isoforms. Interactions of AMOT with either BMPR2 or SMAD1 were abolished by ligand addition. Considering the internalization dynamics of AMOT, this suggests that the interactions are decreased in the course of AMOT internalization. In contrast, association of AMOT to SMAD1 was increased after 15 min of BMP stimulation, coinciding with the BMP-induced dephosphorylation of AMOT130 at Ser-175. So far, only the specific AMOT130 upstream kinases LATS1/2 were investigated for Ser-175 phosphorylation and no dephosphorylation mechanisms are known (Dai et al., 2013). Within the BMP pathway, multiple phosphatases are described to counterbalance critical phosphorylation events. These include PP2A, which directly interacts with the BMP receptor complex and causes dephosphorylation of R-SMADs (Bengtsson et al., 2009). It is not known in detail whether AMOT has to be phosphorylated at Ser-175 to reach its transmembrane topology. Although it has been published that pAMOT130 predominantly localizes at the PM, only ectopically expressed phosphomimetic or phosphoresistant AMOT130 mutants were used to show this (Moleirinho et al., 2017). Experiments investigating the lipid binding and tubulation function of AMOT did not consider the influence of putative PTMs (Heller et al., 2010). It is also not clear whether AMOT needs to penetrate the PM to induce tubulation and subsequent endocytosis or whether PM-association alone is sufficient. Hence, the question of cytoplasmic or transmembrane localization of AMOT still remains to be answered.
Beyond this, we also determined that AMOT depletion reduces SMAD1/5 phosphorylation in all cell systems used ranging from epithelial and endothelial to mesenchymal precursor cells. This suggests a general mechanism and confers AMOT an even more important function in BMP signaling. Further, the SMAD modulation was specific for BMP signaling, as we did not detect an effect of AMOT depletion on TGF–β-mediated SMAD2 phosphorylation. The observed decrease in SMAD1/5 phosphorylation affected all downstream signaling events, as it interfered with subsequent SMAD1–SMAD4 complex formation, translocation of SMADs to the nucleus, and target gene transcription.
In addition, we investigated whether AMOTL1 or AMOTL2 had similar effects on BMP-SMAD activation. Both proteins are structurally similar to AMOT130 and localize in a comparable manner, although they lack the Angiostatin-binding domain (Huang et al., 2018). Only AMOTL1 depletion affected pSMAD1/5 levels in MCF7 cells and thereby resembled the effect of AMOT knockdown. This can possibly be explained by the identical existence of XPXY motifs within AMOTL1 and AMOT, while AMOTL2 differs slightly (Huang et al., 2018). Further, AMOTL1 has similar effects on the paracellular permeability as AMOT, whereas AMOTL2 is apparently more involved in the link between tight junctions and the actin cytoskeleton (Zheng et al., 2009; Hultin et al., 2014). Here, further experiments are necessary to explore whether AMOTL1 has a similar role as AMOT130 in BMP signaling.
These findings suggest the following model (Figure 7): AMOT130 is expressed at the apical cell surface associating to BMPR2 in serum-starved cells, potentially phosphorylated at Ser-175. During the first 15 min of BMP6 stimulation, AMOT130 becomes dephosphorylated at Ser-175 and concomitantly SMAD1 is recruited to the BMP receptor complex. At this stage, AMOT and SMAD1 still interact and this connection is lost within the next 15 min. After 30 min of BMP6 stimulation, dephosphorylated AMOT130 is internalized, thereby losing its association to BMPR2 and SMAD1. We have shown previously that roughly 40% of BMPR2 surface levels are internalized after 30 min of BMP stimulation, similar to those of TGF-β type II receptor endocytosis dynamics (Horbelt et al., 2010; Amsalem et al., 2016). This suggests that BMPR2 is potentially internalized together with AMOT130 in this period. SMAD1 is already phosphorylated after 30 min and translocates to the nucleus to activate target gene transcription.
As mentioned above, AMOT is predominantly expressed at the apical side of epithelial cells, demonstrated by confocal z-stacks. Since MCF7 cells are not a widely used model for epithelial polarity, the established apical-basal polarity was verified by the expression and apical localization of PKCzeta and tight junction integrity by ZO-1, both accepted markers (Yonemura, 2014). We further measured the basal TEER of MCF7 cells within the transwell setup to check whether tight junctions were functional. The cells exhibited a basal TEER of roughly 400 Ω/cm2, which represents an intermediate level of resistance and demonstrates a sealed epithelial sheet according to the literature (Srinivasan et al., 2015; Etoc et al., 2016). Spatial compartmentalization is crucial to understand the manifold signaling outcome induced by activated BMP receptors. BMPs are secreted as precursors into the extracellular space, where they become cleaved and activated but at the same time bound by soluble antagonists to fine-tune their action. Once engaged into receptor complexes, it is the distinct localization of these BMP receptors, the constitution of their complexes, and their stability that have striking effects on downstream signaling outcome (Ramel and Hill, 2012). By these mechanisms BMP signaling can, for example, be elicited only at the leading edge of a migrating cell or specifically induce only SMAD or non-SMAD signaling depending on the PM side and composition it was activated in Guzman et al. (2012). Indeed, we have proven that AMOT regulates SMAD1/5 signaling in a spatially restricted mode, only when the ligand was added to the apical side of polarized cells. This establishes AMOT as a very specific apical fine-tuner of BMP-SMAD signaling and adds another layer of regulation to the complex signaling network. Interestingly, BMP stimulation elicited SMAD signaling from both apical and basolateral sides in MCF7 cells. This is in contrast to MDCK cells and human embryonic stem cells, in which it was shown SMAD1/5 activity is not induced when cells are stimulated from the apical side (Saitoh et al., 2013; Etoc et al., 2016). However, we demonstrate that the BMPR2 is not expressed exclusively basally but also in the apical cell compartment in polarized MCF7 cells. This demonstrates that polarized expression of BMP receptors is highly context dependent.
Additionally, we showed that endocytosis was crucial for AMOT regulation, as SMAD1/5 phosphorylation was not affected by AMOT knockdown when endocytosis was blocked. A previous study by us already revealed that endocytosis is dispensable to initiate SMAD1/5 phosphorylation but can enhance the signaling outcome (Paarmann et al., 2016). The same study determined that dynamin-dependent endocytosis affected steady-state levels of both type I and type II receptors but influenced the surface expression of only the type I receptors and not of BMPR2. This suggests that there are independent regulatory mechanisms modulating surface levels for each receptor type, probably including dynamin-independent mechanisms for BMPR2, such as actin-driven endocytosis (Johannes et al., 2015). It is tempting to speculate that actin-binding AMOT130 enhances trafficking of the BMPR2 and thereby promotes the subsequent signaling events. AMOT130 can induce strong F-actin bundling when overexpressed and only if Ser-175 is not phosphorylated (Mana-Capelli et al., 2014). This would be in line with our data showing that AMOT130 is first dephosphorylated at Ser-175 and then internalized. With these experiments, we only begin to understand the molecular mechanism by which AMOT influences SMAD1/5 phosphorylation. Yet, AMOT has been described as a trafficking regulator for YAP and tight junction-associated proteins, a function that very likely could be assigned to BMP signaling components as well (Wells et al., 2006; Cox et al., 2015).
Taken together, our results describe AMOT130 as a novel, spatially restricted BMP signaling regulator. We clearly demonstrate that AMOT130 specifically affects SMAD1/5 signaling at the apical side of polarized cells by enhanced trafficking (Figure 7). This provides new, valuable insights to the complex regulation of the BMP pathway, which will be beneficial to elucidate the role of AMOT in BMP-mediated processes such as cell differentiation, motility, and proliferation.
MATERIALS AND METHODS
Cell culture
HEK293T, MCF7, and C2C12 cells were grown in DMEM (Biochrom AG) supplemented with 10% fetal calf serum (FCS; Biochrom AG), 2 mM l-glutamine, and penicillin (100 U/ml)/streptomycin (10 µg/ml) (PAA Laboratories) at 37°C and 10% (C2C12) or 5% CO2. Immortalized human myoblasts were cultured in skeletal muscle growth medium (Provitro) supplemented with supplement mix (Provitro), 50 ng/ml amphotericin, 50 µg/ml gentamicin, 10% FCS, 2 mM l-glutamine, and penicillin (100 U/ml)/streptomycin (10 µg/ml) at 37°C and 5% CO2. hFOBs (1.19) were cultured in a 1:1 mixture of DMEM and Ham’s F12 medium supplemented with 10% FCS, 2 mM l-glutamine, penicillin (100 U/ml)/streptomycin (10 µg/ml), and 0.3 mg/ml G418 (Biochrom AG) at 34°C with 5% CO2 to keep them in a proliferative state. HUVECs were a kind gift from M. Lorenz and V. Stangl (Charité Universitätsmedizin, Berlin, Germany) and cultured on gelatin-coated tissue culture ware in M199 medium supplemented with 20% FCS, 50 µg/ml endothelial cell growth supplement (Corning), 25 µg/ml heparin, 2 mM l-glutamine, and penicillin (100 U/ml)/streptomycin (10 µg/ml) at 37°C and 5% CO2. HUVECs were used at passage 3 in all experiments. Unless stated otherwise, all cells were starved for 5 h prior to stimulation with their respective growth medium, without FCS supplement, containing 2 mM l-glutamine and penicillin/streptomycin.
Cell transfection
HEK293T cells were transiently transfected with plasmids using polyethylenimine (Sigma-Aldrich). MCF7 cells were transiently transfected using Lipofectamine2000 (Invitrogen) according to manufacturer’s instructions. Cells were cultured for 24 h before the experimental procedure. For siRNA-mediated knockdown of AMOT, Lipofectamine RNAiMAX (Invitrogen) or Lipofectamine2000 (HUVEC) was used according to manufacturer’s instructions. Cells were cultured in antibiotic-free medium during transfection and incubated for 48 h prior to subsequent assays. All siRNAs were obtained from Dharmacon-Healthcare. ON-TARGET plus nontargeting siRNA (si-scr) was used as control (5′-UGGUUUACAUGUCGACUAA-3′). Human AMOT siRNA (5′-GAAACAAGCUAGAGGGCGA-3′) and murine AMOT SMARTpool siRNA (L-058986-02) were used to deplete both isoforms of AMOT. Human AMOTL1 siRNA (5′-GGAAUGAUUUGAACUGAUA-3′) and human AMOTL2 siRNA (5′-GGUUCAUGUGCAUUGUUUA-3′) were used to deplete AMOTL1 and AMOTL2, respectively.
Plasmids
The full-length coding sequences of human AMOT130 and AMOT80 were amplified from HEK293T cell cDNA and cloned into the pcDNA3.1 V5/His-TOPO vector (Life Technologies) according to manufacturer’s instructions with an N-terminal FLAG-tag. The AMOT130 insert was furthermore subjected to site-directed mutagenesis and fused to an N-terminal GFP-Tag. Plasmids encoding human HA-BMPR2-LF, HA-ALK3, and HA-ALK6 were described previously (Gilboa et al., 2000).
Western blotting
Protein lysates were separated by SDS–PAGE and subsequently transferred on polyvinylidene difluoride (PVDF) membranes. After blocking for 1 h with 3% nonfat dry milk in TBS-T, membranes were incubated with primary antibodies overnight at 4°C. Membranes were then incubated with respective HRP-conjugated secondary antibody and analyzed using WesternBright Quantum ECL HRP reagents (Advansta) and a Fusion-FX7 detection system (Vilber-Lourmat). Primary antibodies used were rabbit anti-GAPDH (2118S), rabbit anti-pSMAD1/5 (9516S), rabbit anti-SMAD1 (D59D7), rabbit anti–histone H3 (9715S), rabbit anti-pSMAD2 (3108S), and rabbit anti-SMAD2 (3122) obtained from Cell Signaling Technologies. Mouse anti-HA tag antibody (HA7) was purchased from Sigma-Aldrich. Goat anti-AMOT (C18) and rabbit anti-ID1 (sc488) were purchased from Santa Cruz Biotechnology. Rabbit anti-AMOT (A303-305) was purchased from Bethyl Labs. Rabbit anti-pAMOT130 (Ser174, ABS1045) was purchased from Merck Millipore. Rabbit anti-AMOTL1 (HPA001196) was purchased from Atlas Antibodies, and rabbit anti-AMOTL2 (23351-1-AP) was purchased from Proteintech.
Coimmunoprecipitation studies
Immunoprecipitation of overexpressed proteins from HEK293T cells (24 h posttransfection) and endogenous proteins from MCF7 cells (48 h postseeding) was performed using a modified radio-immunoprecipitation assay buffer (RIPA) freshly supplemented with inhibitors (1 mM PMSF, 2 mM Na3VO4, 20 mM Na4P2O7, 50 mM NaF, complete protease inhibitor cocktails [Roche]) as previously described (Benn et al., 2015). TCLs were taken before incubating the lysates with the indicated primary antibody (1–4 µg/lysate) overnight. Rabbit anti-AMOT (TLE) antibody, used for AMOT-BMPR2 interactions, was kindly provided by Lars Holmgren (Karolinska Institutet, Stockholm, Sweden). Furthermore, mouse anti-HA tag (Sigma-Aldrich), rabbit anti-SMAD1 (Cell Signaling Technologies), and rabbit anti-AMOT (BethylLabs) antibodies were used for pull down. Control samples were incubated with either isotype control antibodies or recombinant protein A-Sepharose beads (GE Healthcare). Immunocomplexes were precipitated at 4°C for 2 h with recombinant protein A-Sepharose beads and subsequently washed 3–5× with fresh lysis buffer including inhibitors. Proteins were eluted with 2× Laemmli sample buffer and boiled for 10 min at 95°C prior to Western blotting.
Cell surface biotinylation
Twenty-four hours posttransfection, HEK293T cells were starved and stimulated with BMP6 for the indicated time points. Cells were then incubated with 0.5 mg/ml EZ-Link Sulfo-NHS-SS-Biotin solution (Thermo Fisher Scientific) at 4°C followed by incubation with 50 mM Tris, pH 8.0. Lysis was performed with modified RIPA buffer and biotinylated surface proteins were pulled down using streptavidin-coupled Sepharose beads (GE Healthcare). Beads were washed with lysis buffer, eluted with 2× Laemmli buffer, and subjected to Western blotting. Control cells (–) were not incubated with Biotin but otherwise treated the same.
Cell fractionation
For cell fractionation, MCF7 cells were first scraped in ice-cold phosphate-buffered saline and harvested by centrifugation posttreatment. The NE-PER nuclear and cytoplasmic extraction reagents kit (Thermo Fisher Scientific) was used according to manufacturer’s instructions. Fractions were then analyzed by Western blotting.
In situ PLA
MCF7 cells were seeded in Nunc Lab-Tec II 16-well glass chamber slides (Thermo Fisher Scientific) and incubated for 48 h before serum starvation followed by BMP stimulation for the indicated time points. Cells were then fixed using 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. Subsequently, PLA was performed using Duolink in situ proximity ligation (Sigma-Aldrich), as previously described (Thymiakou and Episkopou, 2011), using mouse anti-SMAD1 (sc7965; Santa Cruz Biotechnology), mouse anti-YAP/TAZ (sc101199; Santa Cruz Biotechnology), and rabbit anti-AMOT (Bethyl Labs) at a dilution of 1:200. Imaging occurred with an inverted epifluorescence Axiovert 200M microscope (Zeiss). The number of heteromers was quantified using BlobFinder image analysis software as previously described (Allalou and Wahlby, 2009). At least 500 cells per experimental condition were quantified of each replicate and normalized to starved cells.
Quantitative Real-Time PCR
Total RNA extraction was performed using NucleoSpin RNA II isolation kit (MACHEREY-NAGEL) according to manufacturer’s instructions; 0.5–1 μg of RNA was subjected to reverse transcription using MMLV reverse transcriptase (Promega) and random primers (Invitrogen). Gene expression was assessed by quantitative PCR utilizing StepOne Plus and SYBR Green PCR Master Mix (Applied Biosystems) or Luna PCR Master Mix (New England Biolabs). Transcript expression levels were calculated as mean normalized expression ratios referred to housekeeping gene using the ΔΔCT method considering primer efficiency correction (Livak and Schmittgen, 2001; Pfaffl, 2001). All measurements were done in triplicate and CT values were determined with the StepOne Software version 2.2 (Applied Biosystems). The following human primer pairs were used for amplification: GAPDH (fwd: 5′-GAAGGTGAAGGTCGGAGTC-3′, rev: 5′-GAAGATGGTGATGGGATTTC-3′), ID1 (fwd: 5′-GCTGCTCTACGACATGAACG-3′, rev: 5′-CCAACTGAAGGTCCCTGATG-3′), JunB (fwd: 5′-TGGAACAGCCCTTCTACCAC-3′, rev: 5′-GGTTTCAGGAGTTTGTAGTC-3′), both isoforms of AMOT (fwd: 5′-GATTCTGGCTCTGGAAGCTG-3′, rev: 5′-CTGTTGTGTCCCTCTGAGCA-3′).
Immunofluorescence
MCF7 cells were fixed with 4% PFA, permeabilized with 0.5% Triton X-100, and subsequently blocked with 3% bovine serum albumin/2% normal goat serum solution before incubation with specific primary antibodies at 4°C overnight followed by Alexa Fluor 594–conjugated secondary antibody incubation. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Images were taken with an inverted confocal SP8 microscope (Leica) and analyzed with the LAS X software (Leica).
Live cell imaging of AMOT130-GFP
Living MCF7 cells were imaged in a Zeiss Axiovert 200M microscope with a heating module with CO2 supply at 37°C and 5% CO2. To visualize internalization of AMOT130-GFP, cells were starved 5 h in medium without FCS supplement and then stimulated during image acquisition with 10 nM BMP6. Videos were all taken with the same exposure time. Images at 0 and 60 min were quantified using ImageJ.
Apical-basal transwell assay
MCF7 cells were seeded into six-well plates, transfected for 24 h with respective siRNAs and then reseeded into 12-mm Transwell 0.4-µm inserts (Corning). On confluency, cells were starved for 5 h with medium not containing FCS and subsequently stimulated with 10 nM BMP6 only applied from either the apical or the basal side. Cells were lysed for either protein or RNA analysis. Protein lysates were subjected to Western blotting and analyzed for SMAD1/5 phosphorylation. RNA lysates were subjected to quantitative real-time PCR (qRT-PCR).
TEER measurement with ECIS
MCF7 cells were seeded into six-well plates, and AMOT was depleted for 48 h. Cells were then seeded into 6.5-mm Transwell 0.4-µm inserts (Corning). After 48 h, inserts were introduced in the ECIS trans-Filter Adapter and for data acquisition, a multiple frequency time course program was chosen, and data were recorded for 48 h. Analysis of TEER data was evaluated with the ECIS software, and peak values for each corresponding condition were recorded for the graph.
Statistical analysis
Statistical analysis was performed with Prism Software (GraphPad) using either unpaired Student’s t test or one-way or two-way analysis of variance (ANOVA) with post-hoc Bonferroni test depending on the experiment; p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
Financial support was provided by the Deutsche Forschungsgemeinschaft (DFG) to P.K., J.J., and P.B. (Berlin School of Integrative Oncology; Berlin-Brandenburg School for Regenerative Therapies). Further support was provided by the SFB958 and by the Bundesministerium fuer Bildung und Forschung (BMBF) (PrevOP/OVERLOAD) to P.K. We thank Slobodan Vukicevic (University of Zagreb, Croatia) for providing recombinant BMP6, Walter Sebald (Julius-Maximilans-Universität, Würzburg, Germany) for providing recombinant BMP, Simone Spuler (Charité University Medical School, Berlin, Germany) for providing human myoblasts, and Lars Holmgren (Karolinska Institutet, Stockholm, Sweden) for the AMOT TLE antibody. We acknowledge the assistance of the Core Facility BioSupraMol supported by the DFG. We also thank Gina Dörpholz for valuable input.
Abbreviations used:
- AMOT
angiomotin
- AMOTL1
AMOT-like protein 1
- AMOTL2
AMOT-like protein 2
- BMP
bone morphogenetic protein
- BMPR1
BMP type I receptor
- BMPR2
BMP type II receptor
- BMPR2-LF
BMPR2 long isoform
- FCS
fetal calf serum
- hFOB
human fetal osteoblast
- HUVEC
human umbilical vein endothelial cells
- ID
inhibitor of differentiation
- LATS1/2
large tumor suppressor kinases 1/2
- LF
long form
- MAPK
mitogen-activated protein kinase
- MCF7
Michigan Cancer Foundation-7
- PKCzeta
protein kinase C zeta
- PLA
proximity ligation assay
- PM
plasma membrane
- RIPA
radio-immunoprecipitation assay
- SF
short form
- TCL
total cell lysate
- TEER
transepithelial electrical resistance
- TGF-β
fransforming growth factor β.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-03-0179) on December 4, 2019.
REFERENCES
- Adler JJ, Johnson DE, Heller BL, Bringman LR, Ranahan WP, Conwell MD, Sun Y, Hudmon A, Wells CD. (2013). Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc Natl Acad Sci USA , 17368–17373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. (2009). Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell , 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allalou A, Wahlby C. (2009). BlobFinder, a tool for fluorescence microscopy image cytometry. Comput Methods Programs Biomed , 58–65. [DOI] [PubMed] [Google Scholar]
- Amsalem AR, Marom B, Shapira KE, Hirschhorn T, Preisler L, Paarmann P, Knaus P, Henis YI, Ehrlich M. (2016). Differential regulation of translation and endocytosis of alternatively spliced forms of the type II bone morphogenetic protein (BMP) receptor. Mol Biol Cell , 716–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson L, Schwappacher R, Roth M, Boergermann JH, Hassel S, Knaus P. (2009). PP2A regulates BMP signalling by interacting with BMP receptor complexes and by dephosphorylating both the C-terminus and the linker region of Smad1. J Cell Sci , 1248–1257. [DOI] [PubMed] [Google Scholar]
- Benn A, Bredow C, Casanova I, Vukicevic S, Knaus P. (2015). VE-cadherin facilitates BMP-induced endothelial cell permeability and signaling. J Cell Sci , 206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratt A, Birot O, Sinha I, Veitonmaki N, Aase K, Ernkvist M, Holmgren L. (2005). Angiomotin regulates endothelial cell-cell junctions and cell motility. J Biol Chem , 34859–34869. [DOI] [PubMed] [Google Scholar]
- Bratt A, Wilson WJ, Troyanovsky B, Aase K, Kessler R, Van Meir EG, Holmgren L. (2002). Angiomotin belongs to a novel protein family with conserved coiled-coil and PDZ binding domains. Gene , 69–77. [DOI] [PubMed] [Google Scholar]
- Campbell CI, Samavarchi-Tehrani P, Barrios-Rodiles M, Datti A, Gingras AC, Wrana JL. (2016). The RNF146 and tankyrase pathway maintains the junctional Crumbs complex through regulation of angiomotin. J Cell Sci , 3396–3411. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. (2011). Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem , 7018–7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Guo F, Tan I, Leung T, Hong W. (2013). Actin-binding and cell proliferation activities of angiomotin family members are regulated by Hippo pathway-mediated phosphorylation. J Biol Chem , 37296–37307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CM, Mandell EK, Stewart L, Lu R, Johnson DL, McCarter SD, Tavares A, Runyan R, Ghosh S, Wilson JM. (2015). Endosomal regulation of contact inhibition through the AMOT:YAP pathway. Mol Biol Cell , 2673–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, She P, Chi F, Feng Y, Liu H, Jin D, Zhao Y, Guo X, Jiang D, Guan KL, et al. (2013). Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J Biol Chem , 34041–34051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. (2003). Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature , 577–584. [DOI] [PubMed] [Google Scholar]
- Disanza A, Frittoli E, Palamidessi A, Scita G. (2009). Endocytosis and spatial restriction of cell signaling. Mol Oncol , 280–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorpholz G, Murgai A, Jatzlau J, Horbelt D, Belverdi MP, Heroven C, Schreiber I, Wendel G, Ruschke K, Stricker S, Knaus P. (2017). IRS4, a novel modulator of BMP/Smad and Akt signalling during early muscle differentiation. Sci Rep , 8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebnet K. (2008). Organization of multiprotein complexes at cell-cell junctions. Histochem Cell Biol , 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernkvist M, Aase K, Ukomadu C, Wohlschlegel J, Blackman R, Veitonmaki N, Bratt A, Dutta A, Holmgren L. (2006). p130-angiomotin associates to actin and controls endothelial cell shape. FEBS J , 2000–2011. [DOI] [PubMed] [Google Scholar]
- Ernkvist M, Birot O, Sinha I, Veitonmaki N, Nystrom S, Aase K, Holmgren L. (2008). Differential roles of p80- and p130-angiomotin in the switch between migration and stabilization of endothelial cells. Biochim Biophys Acta , 429–437. [DOI] [PubMed] [Google Scholar]
- Ernkvist M, Persson NL, Audebert S, Lecine P, Sinha I, Liu M, Schlueter M, Horowitz A, Aase K, Weide T, et al. (2009). The Amot/Patj/Syx signalling complex spatially controls Rho A GTPase activity in migating endothelial cells. Blood , 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etoc F, Metzger J, Ruzo A, Kirst C, Yoney A, Ozair MZ, Brivanlou AH, Siggia ED. (2016). A balance between secreted inhibitors and edge sensing controls gastruloid self-organization. Dev Cell , 302–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa L, Nohe A, Geissendorfer T, Sebald W, Henis YI, Knaus P. (2000). Bone morphogenetic protein receptor complexes on the surface of live cells: a new oligomerization mode for serine/threonine kinase receptors. Mol Biol Cell , 1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman A, Zelman-Femiak M, Boergermann JH, Paschkowsky S, Kreuzaler PA, Fratzl P, Harms GS, Knaus P. (2012). SMAD versus non-SMAD signaling is determined by lateral mobility of bone morphogenetic protein (BMP) receptors. J Biol Chem , 39492–39504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung A, Bitton-Worms K, Rechtman MM, Wenzel V, Boergermann JH, Hassel S, Henis YI, Knaus P. (2006). Different routes of bone morphogenic protein (BMP) receptor endocytosis influence BMP signaling. Mol Cell Biol , 7791–7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller B, Adu-Gyamfi E, Smith-Kinnaman W, Babbey C, Vora M, Xue Y, Bittman R, Stahelin RV, Wells CD. (2010). Amot recognizes a juxtanuclear endocytic recycling compartment via a novel lipid binding domain. J Biol Chem , 12308–12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiepen C, Benn A, Denkis A, Lukonin I, Weise C, Boergermann JH, Knaus P. (2014). BMP2-induced chemotaxis requires PI3K p55gamma/p110alpha-dependent phosphatidylinositol (3,4,5)-triphosphate production and LL5beta recruitment at the cytocortex. BMC Biol , 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W. (2013). Angiomotin’g YAP into the nucleus for cell proliferation and cancer development. Sci Signal , pe27. [DOI] [PubMed] [Google Scholar]
- Horbelt D, Guo G, Robinson PN, Knaus P. (2010). Quantitative analysis of TGFBR2 mutations in Marfan-syndrome-related disorders suggests a correlation between phenotypic severity and Smad signaling activity. J Cell Sci , 4340–4350. [DOI] [PubMed] [Google Scholar]
- Hu F, Zhang Y, Li M, Zhao L, Chen J, Yang S, Zhang X. (2016). BMP-6 inhibits the metastasis of MDA-MB-231 breast cancer cells by regulating MMP-1 expression. Oncol Rep , 1823–1830. [DOI] [PubMed] [Google Scholar]
- Huang T, Zhou Y, Zhang J, Cheng ASL, Yu J, To KF, Kang W. (2018). The physiological role of Motin family and its dysregulation in tumorigenesis. J Transl Med , 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultin S, Zheng Y, Mojallal M, Vertuani S, Gentili C, Balland M, Milloud R, Belting HG, Affolter M, Helker CS, et al. (2014). AmotL2 links VE-cadherin to contractile actin fibres necessary for aortic lumen expansion. Nat Commun , 3743. [DOI] [PubMed] [Google Scholar]
- Johannes L, Parton RG, Bassereau P, Mayor S. (2015). Building endocytic pits without clathrin. Nat Rev Mol Cell Biol , 311–321. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Imada M, Yanai T, Suda T, Takahashi N, Kamijo R. (2002). Identification of a BMP-responsive element in Id1, the gene for inhibition of myogenesis. Genes Cells , 949–960. [DOI] [PubMed] [Google Scholar]
- Kopf J, Petersen A, Duda GN, Knaus P. (2012). BMP2 and mechanical loading cooperatively regulate immediate early signalling events in the BMP pathway. BMC Biol , 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko T, Veitonmaki N, Lundkvist A, Gerhardt H, Ming Y, Berggren K, Kvanta A, Carlsson R, Holmgren L. (2008). Therapeutic antibodies targeting angiomotin inhibit angiogenesis in vivo. FASEB J , 880–889. [DOI] [PubMed] [Google Scholar]
- Lian WJ, Liu G, Liu YJ, Zhao ZW, Yi T, Zhou HY. (2013). Downregulation of BMP6 enhances cell proliferation and chemoresistance via activation of the ERK signaling pathway in breast cancer. Oncol Rep , 193–200. [DOI] [PubMed] [Google Scholar]
- Liu G, Liu YJ, Lian WJ, Zhao ZW, Yi T, Zhou HY. (2014). Reduced BMP6 expression by DNA methylation contributes to EMT and drug resistance in breast cancer cells. Oncol Rep , 581–588. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods , 402–408. [DOI] [PubMed] [Google Scholar]
- Mana-Capelli S, Paramasivam M, Dutta S, McCollum D. (2014). Angiomotins link F-actin architecture to Hippo pathway signaling. Mol Biol Cell , 1676–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. (2005). Smad transcription factors. Genes Dev , 2783–2810. [DOI] [PubMed] [Google Scholar]
- Moleirinho S, Guerrant W, Kissil JL. (2014). The Angiomotins–from discovery to function. FEBS Lett , 2693–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moleirinho S, Hoxha S, Mandati V, Curtale G, Troutman S, Ehmer U, Kissil JL. (2017). Regulation of localization and function of the transcriptional co-activator YAP by angiomotin. Elife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau J, Lord M, Boucher M, Belleau P, Fernandes MJ. (2005). Protein diversity is generated within the motin family of proteins by alternative pre-mRNA splicing. Gene , 137–148. [DOI] [PubMed] [Google Scholar]
- Nohe A, Keating E, Underhill TM, Knaus P, Petersen NO. (2005). Dynamics and interaction of caveolin-1 isoforms with BMP-receptors. J Cell Sci , 643–650. [DOI] [PubMed] [Google Scholar]
- Paarmann P, Dorpholz G, Fiebig J, Amsalem AR, Ehrlich M, Henis YI, Muller T, Knaus P. (2016). Dynamin-dependent endocytosis of bone morphogenetic protein2 (BMP2) and its receptors is dispensable for the initiation of Smad signaling. Int J Biochem Cell Biol , 51–63. [DOI] [PubMed] [Google Scholar]
- Paramasivam M, Sarkeshik A, Yates JR, 3rd, Fernandes MJ, McCollum D. (2011). Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol Biol Cell , 3725–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res , e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl TL, Boergermann JH, Schwaerzer GK, Knaus P, Cavalcanti-Adam EA. (2012). Surface immobilization of bone morphogenetic protein 2 via a self-assembled monolayer formation induces cell differentiation. Acta Biomater , 772–780. [DOI] [PubMed] [Google Scholar]
- Ramel MC, Hill CS. (2012). Spatial regulation of BMP activity. FEBS Lett , 1929–1941. [DOI] [PubMed] [Google Scholar]
- Saitoh M, Shirakihara T, Fukasawa A, Horiguchi K, Sakamoto K, Sugiya H, Beppu H, Fujita Y, Morita I, Miyazono K, Miyazawa K. (2013). Basolateral BMP signaling in polarized epithelial cells. PLoS One , e62659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab EH, Pohl TL, Haraszti T, Schwaerzer GK, Hiepen C, Spatz JP, Knaus P, Cavalcanti-Adam EA. (2015). Nanoscale control of surface immobilized BMP-2: toward a quantitative assessment of BMP-mediated signaling events. Nano Lett , 1526–1534. [DOI] [PubMed] [Google Scholar]
- Sieber C, Kopf J, Hiepen C, Knaus P. (2009). Recent advances in BMP receptor signaling. Cytokine Growth Factor Rev , 343–355. [DOI] [PubMed] [Google Scholar]
- Sorkin A, von Zastrow M. (2009). Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol , 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan B, Kolli AR, Esch MB, Abaci HE, Shuler ML, Hickman JJ. (2015). TEER measurement techniques for in vitro barrier model systems. J Lab Autom , 107–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara-Mizuno Y, Adachi M, Kobayashi Y, Hamazaki Y, Nishimura M, Imai T, Furuse M, Tsukita S. (2007). Molecular characterization of angiomotin/JEAP family proteins: interaction with MUPP1/Patj and their endogenous properties. Genes Cells , 473–486. [DOI] [PubMed] [Google Scholar]
- Thymiakou E, Episkopou V. (2011). Detection of signaling effector-complexes downstream of bmp4 using PLA, a proximity ligation assay. J Vis Exp , 2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin NP, Sims AH, Lundgren KL, Lehn S, Landberg G. (2011). Cyclin D1, Id1 and EMT in breast cancer. BMC Cancer , 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky B, Levchenko T, Mansson G, Matvijenko O, Holmgren L. (2001). Angiomotin: an angiostatin binding protein that regulates endothelial cell migration and tube formation. J Cell Biol , 1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RN, Green J, Wang Z, Deng Y, Qiao M, Peabody M, Zhang Q, Ye J, Yan Z, Denduluri S, et al. (2014). Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis , 87–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CD, Fawcett JP, Traweger A, Yamanaka Y, Goudreault M, Elder K, Kulkarni S, Gish G, Virag C, Lim C, et al. (2006). A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell , 535–548. [DOI] [PubMed] [Google Scholar]
- Yadin D, Knaus P, Mueller TD. (2016). Structural insights into BMP receptors: Specificity, activation and inhibition. Cytokine Growth Factor Rev , 13–34. [DOI] [PubMed] [Google Scholar]
- Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, Wells RG, Greenbaum LE, Stanger BZ. (2013). Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev , 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S. (2014). Differential sensitivity of epithelial cells to extracellular matrix in polarity establishment. PLoS one , e112922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Vertuani S, Nystrom S, Audebert S, Meijer I, Tegnebratt T, Borg JP, Uhlen P, Majumdar A, Holmgren L. (2009). Angiomotin-like protein 1 controls endothelial polarity and junction stability during sprouting angiogenesis. Circ Res , 260–270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.