Abstract
The UV‐B photoreceptor UVR8 mediates multiple UV‐B responses in plants, but the function of UVR8 in regulating root development has not previously been investigated. Here, we show that UV‐B light inhibits Arabidopsis lateral root growth in a UVR8‐dependent manner. Monomeric UVR8 inhibits auxin responses in a tissue‐autonomous manner and thereby regulates lateral root growth. Genome‐wide gene expression analysis demonstrated that auxin and UV‐B irradiation antagonistically regulate auxin‐regulated gene expression. We further show that UVR8 physically interacts with MYB73/MYB77 (MYB DOMAIN PROTEIN 73/77) in a UV‐B‐dependent manner. UVR8 inhibits lateral root development via regulation of MYB73/MYB77. When activated by UV‐B light, UVR8 localizes to the nucleus and inhibits the DNA‐binding activities of MYB73/MYB77 and directly represses the transcription of their target auxin‐responsive genes. Our results demonstrate that UV‐B light and UVR8 are critical for both shoot morphogenesis and root development. The UV‐B‐dependent interaction of UVR8 and MYB73/MYB77 serves as an important module that integrates light and auxin signaling and represents a new UVR8 signaling mechanism in plants.
Keywords: auxin responses, lateral root, MYB73/MYB77, UV‐B photomorphogenesis, UVR8
Subject Categories: Development & Differentiation, Plant Biology, Signal Transduction
The UV‐B light inhibits auxin‐induced root and shoot growth via direct interaction between the photoreceptor UVR8 and auxin‐responsive transcription factors MYB73/MYB77.

Introduction
Ultraviolet B (UV‐B) light is an inherent component of sunlight that markedly affects plant development (Heijde & Ulm, 2012). The UV‐B photoreceptor UVR8 (UV RESISTANCE LOCUS 8) is required for UV‐B responses (Rizzini et al, 2011; Tilbrook et al, 2013; Jenkins, 2014). While UVR8 protein is localized in both the cytoplasm and the nucleus, it is hypothesized to function mainly in the nucleus. UV‐B does not affect UVR8 protein abundance, but UV‐B irradiation induces the monomerization and nuclear accumulation of UVR8 (Kaiserli & Jenkins, 2007; Favory et al, 2009). UVR8 interacts with COP1 (CONSTITUTIVELY PHOTOMORPHOGENIC 1), the central regulator of light signaling (Yi & Deng, 2005), in a UV‐B‐dependent manner, and thereby mediates UV‐B signaling (Rizzini et al, 2011; Huang et al, 2014; Qian et al, 2016; Yin et al, 2016). The WD40‐repeat proteins REPRESSOR OF UV‐B PHOTOMORPHOGENESIS (RUP1) and RUP2 physically interact with UVR8 to disrupt the UVR8–COP1 interaction and mediate UVR8 re‐dimerization (Gruber et al, 2010). UVR8 also physically interacts with the transcription factors BES1, BIM1, and WRKY36 in the nucleus, directly regulating the transcription of their target genes and photomorphogenesis (Liang et al, 2018; Yang et al, 2018b). Whether or not UVR8 interacts with transcription factors in a UV‐B‐dependent manner and directly regulates transcription is unknown.
UV‐B light is also reported to function in root photomorphogenesis. RUS1 (ROOT UV‐B SENSITIVE 1) and RUS2 have been identified in forward genetic screens and play critical roles in UV‐B photomorphogenesis, including root growth. Root growth is affected when the Arabidopsis thaliana seedlings (including roots) are grown under UV‐B light (Tong et al, 2008; Leasure et al, 2009). It is proposed that light is sensed by the roots directly under natural soil conditions. Specifically, light may pass through the soil to the root or may be conducted through vascular tissue to the root (Sun et al, 2005). Recently, it was reported that light was channeled through the stem to the roots, where it activated the red light photoreceptor phytochrome (Lee et al, 2016), consistent with the hypothesis that the aboveground light environment is monitored by the roots by stem‐piped light under natural conditions. UVR8 is expressed in roots, but its role in root development has not previously been studied.
The phytohormone auxin is fundamental to plant growth and development (Weijers & Wagner, 2016; Leyser, 2018). Auxin is recognized by nuclear‐localized TIR/AFB F‐box proteins that are responsible for the ubiquitination‐dependent protein degradation of AUX/IAA proteins. AUX/IAA proteins inhibit the transcription activities of auxin‐responsive factors (ARFs) (Gray et al, 2001; Zenser et al, 2001; Tiwari et al, 2004). Auxin plays an essential role in lateral root development. The MYB transcription factor MYB77 is reported to interact with ARFs to modulate auxin signal transduction and lateral root growth (Shin et al, 2007; Zhao et al, 2014). Whether UVR8 is directly involved in regulating auxin signaling is largely unknown. R2R3‐MYB transcription factors PFG1 (PRODUCTION OF FLAVONOL GLYCOSIDES)/MYB12, PFG2/MYB11, and PFG3/MYB111 are specifically involved in the regulation of flavonol biosynthesis. MYB12 controls flavonol biosynthesis mainly in the root, while MYB111 controls flavonol biosynthesis primarily in cotyledons (Stracke et al, 2007). UVR8 and HY5 induce the transcription of PFG MYBs in response to UV‐B light (Favory et al, 2009). Whether other MYB transcription factors are involved in UV‐B signaling or are regulated by UV‐B is unknown.
Here, we show that UV‐B inhibits lateral root growth via UVR8, and that nuclear‐localized, UV‐B‐activated monomeric UVR8 inhibits auxin responses, thus repressing lateral root growth. We further demonstrate that UVR8 interacts with MYB73/MYB77 in a UV‐B‐dependent manner in vivo. MYB73/MYB77 acts downstream of UVR8 to regulate lateral root growth. UV‐B‐activated UVR8 represses lateral root growth and the transcription of MYB73/MYB77 targets by repressing MYB73/MYB77 DNA‐binding activity. These results demonstrate that UV‐B and UVR8 are important for both shoot development and root development. UVR8 interacts with transcription factors in a UV‐B‐dependent manner to regulate gene expression in response to UV‐B light, and MYB73/MYB77 is integrators of light and auxin signaling. The UV‐B light‐dependent UVR8–MYB73/MYB77 interaction enables UV‐B light and auxin signaling to coordinately regulate shoot and root development.
Results
UV‐B inhibits lateral root growth in a UVR8‐dependent manner
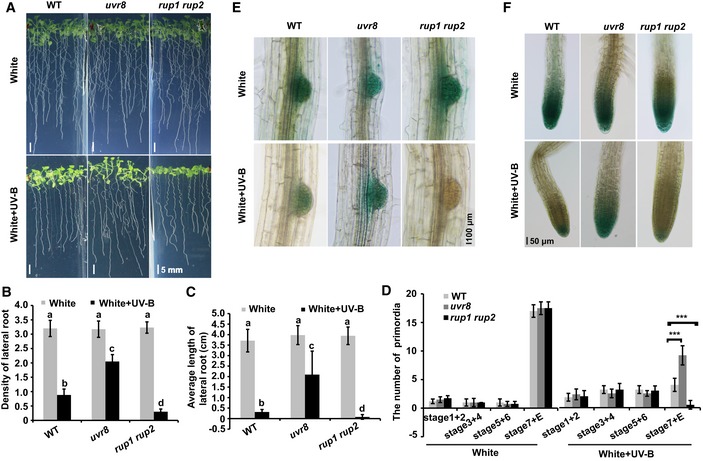
UV‐B has striking effects on plant growth and developmental processes, such as hypocotyl elongation. To examine whether UV‐B‐activated UVR8 affects root growth, we grew wild‐type (WT) Arabidopsis plants on 1/2 Murashige and Skoog (MS) plates under continuous white light and under white light plus UV‐B conditions and analyzed root development. UV‐B treatment not only inhibited hypocotyl elongation (Liang et al, 2018; Yang et al, 2018b), but also negatively regulated the growth of lateral roots (Fig 1A). The lateral root density of the WT was dramatically (about 3.6 times) lower under the UV‐B treatment than without (Fig 1B), and in the presence of UV‐B, the lateral root density of the WT was much lower than that of uvr8 (deficient in UVR8) but much higher than that of rup1 rup2 (deficient in RUP1 and RUP2 that mediate UVR8 re‐dimerization; Fig 1B).
Figure 1. UV‐B inhibits the growth of lateral roots in a UVR8‐dependent manner.
-
A–CPhenotypic analysis. Wild‐type, uvr8, and rup1 rup2 seedlings were grown in 1/2 MS with or without UV‐B (1 W/m2) for 2 weeks. Images are shown in (A); scale bars = 5 mm. The lateral root density (number of lateral roots/length of primary root) (B) and average length of lateral roots (each plant) (C) of the indicated genotypes were measured. SDs (n > 8 independent seedlings) are indicated. Letters “a” to “d” indicate statistically significant differences for the indicated values, as determined by a one‐way analysis of variance (ANOVA), followed by Tukey's least significant difference (LSD) test (P < 0.05). There was statistically significant difference between values marked with different letters (for example “a” and “b”), while there was no statistically significant difference between values marked with the same letters (for example “a” and “a”).
-
DDistribution of lateral root primordia stages. Seedlings of the indicated genotypes were grown in 1/2 MS treatment with or without UV‐B (1 W/m2) for 2 weeks. SDs (n > 8) are indicated. “***” indicates statistically significant differences (P < 0.001), as determined by Student's t‐test.
-
E, FGUS staining of seedlings expressing DR5p::GUS transgene in the WT, uvr8, and rup1 rup2 background. Seedlings were grown in 1/2 MS with or without UV‐B for 2 weeks. Images of lateral root primordia (E) and lateral roots (F) are shown. Scale bars = 100 and 50 μm, respectively.
Furthermore, the lateral root length of the WT was longer than that of rup1 rup2 and shorter than that of uvr8 in the presence of UV‐B, but did not differ among these three genotypes in the absence of UV‐B (Fig 1C), indicating that UVR8 is involved in UV‐B‐regulated root growth. The number of LR primordia of different stages with or without UV‐B treatment was also quantified. Numbers of lateral root primordia of WT, uvr8, and rup1 rup2 were similar under white light; however, the numbers of stage 7 and emerged primordia of the WT were lower than those of uvr8 but higher than those of rup1 rup2 under UV‐B (Fig 1D).
When WT, uvr8, and rup1 rup2 plants grown in soil were treated with or without UV‐B, UV‐B treatment again inhibited the elongation of lateral roots, and the lateral roots of WT plants were longer than those of rup1 rup2 plants, but shorter than those of uvr8 (Fig EV1A).
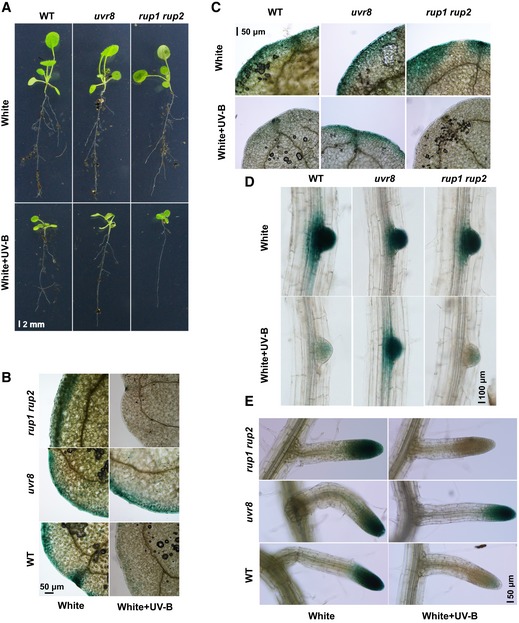
Figure EV1. UV‐B inhibits lateral root growth and auxin signaling.
-
APhenotypic analysis. Wild‐type, uvr8, and rup1 rup2 seedlings were grown in soil in LD condition (16‐h light/8‐h dark) for 5 days and then transferred to continuous white light or white light plus UV‐B (1 W/m2) for 10 days. Scale bar = 2 mm.
-
BGUS staining of seedlings expressing the DR5p::GUS transgene in the WT, uvr8, and rup1 rup2 background. Seedlings were grown in 1/2 MS with or without UV‐B for 2 weeks. Images show leaves; scale bar = 50 μm.
-
C–EGUS staining of seedlings expressing the DR5p::GUS transgene in the WT, uvr8, and rup1 rup2 background. Seedlings of the indicated genotypes were grown in soil under LD conditions for 5 days and then transferred to continuous white light or white light plus UV‐B (1 W/m2) for 10 days. Images show leaves (C), lateral root primordia (D), and lateral roots (E). Scale bars = 50 μm, 100 μm, and 50 μm, respectively.
It is well known that auxin plays a key role in regulating lateral root growth and development. Because ARF7 and ARF19 are reported to play critical roles in lateral root growth and development (Tatematsu et al, 2004; Weijers et al, 2005), we examined the root growth of arf7, arf19, and arf7 arf19 mutants with or without UV‐B. The arf7 and arf19 single mutants were more sensitive than the WT to UV‐B treatment, exhibiting an even greater decrease than the WT in lateral root length following UV‐B relative to white light treatment (the lateral root length ratio of WT White+UV‐B/White was 57.13%, that of arf7 was 0%, and that of arf19 was 25.24%), indicating that UV‐B may regulate lateral root growth by inhibiting ARF7 and ARF19. Indeed, arf7 arf19 double mutants formed no lateral roots, regardless of UV‐B treatment (Appendix Fig S1A–C).
The auxin‐responsive DR5p::GUS reporter construct was introduced into the uvr8 and rup1 rup2 mutant backgrounds so that we could analyze auxin‐induced gene expression in planta. GUS activity was analyzed in seedlings grown on 1/2 MS plates (Figs 1E and F, and EV1B) and in soil (Fig EV1C–E), and treated or not with UV‐B. Decreased GUS activity was observed in the leaves, lateral root primordia, and lateral roots of WT plants subjected to UV‐B treatment, and the decrease in GUS activity was more severe in the rup1 rup2 mutant. By contrast, no dramatic decrease in GUS activity was recorded in the uvr8 mutant background under UV‐B treatment. These results suggest that UV‐B and UVR8 inhibit lateral root growth by inhibiting auxin responses.
UV‐B inhibits auxin responses via UVR8
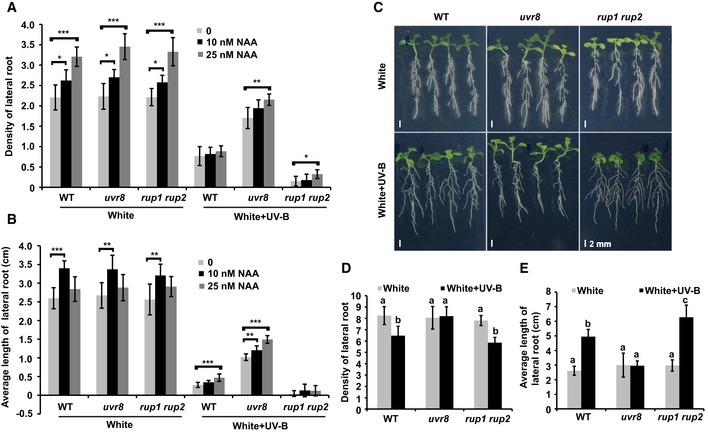
We further tested whether UV‐B regulates lateral root growth by regulating auxin responses. As auxin homeostasis could be affected by UV‐B via photooxidative damage, degradation, biosynthesis, and conjugation (Vanhaelewyn et al, 2016), we first assessed whether UV‐B affected the stability of IAA and NAA. IAA and NAA plates were pre‐irradiated with or without UV‐B for 7 days and then used for root experiments. IAA plates pre‐irradiated with UV‐B for 7 days had a reduced effect on root growth compared to plates exposed to white light without UV‐B, whereas NAA plates pre‐irradiated with UV‐B had the same effect on root growth as plates exposed to white light without UV‐B (Appendix Fig S1D). These results indicate that UV‐B affects IAA stability but not NAA stability. Therefore, we used mainly NAA in our experiments. Medium supplementation with low concentrations of NAA (0, 10, and 25 nM) promoted lateral root growth, while UV‐B inhibited lateral root growth and initiation in a UVR8‐dependent manner (Figs 2A and B, and EV2A). Supplementation with a high concentration of auxin repressed the growth of main roots and lateral roots in the WT, while promoting the generation of lateral roots (Shin et al, 2007; Appendix Fig S1E–G). The repression of lateral root growth and the promotion of lateral root generation by a high concentration of auxin (NAA) were suppressed by UV‐B treatment in the WT (Fig 2C–E). Furthermore, a high concentration of auxin inhibited the lateral root growth of uvr8, but did not inhibit that of rup1 rup2 in the presence of UV‐B (Fig 2C–E). In nature, roots are usually covered by soil, so they normally receive less light than shoots. Roots were covered with foil and a black box to mimic growth under natural conditions (soil) and reduce the light signal. Under these conditions, the repressed lateral root growth and increased lateral root generation induced by a high concentration of auxin were still reversed by UV‐B treatment in the WT but not in uvr8 (Appendix Fig S2A and B).
Figure 2. UV‐B inhibits auxin responses through UVR8.
-
A, BPhenotypic analysis. Seedlings of the indicated genotypes were grown in 1/2 MS with the addition of a series of concentrations of NAA under continuous white light or white plus UV‐B (1 W/m2) light for 2 weeks. The lateral root density (number of lateral roots/length of primary root) (A) and average length of lateral roots (B) of the indicated genotypes were measured. SDs (n > 8) are indicated. “*” indicates statistically significant differences (*P < 0.05, **P < 0.01, ***P < 0.001), as determined by Student's t‐test.
-
C–EUV‐B inhibits the response of Arabidopsis seedlings to exogenously added NAA in a UVR8‐dependent manner, since the response to NAA was not repressed in uvr8. Seedlings of the indicated genotypes were grown in LD (16‐h light/8‐h dark) conditions for 5 days, then transplanted to new plates containing 0.4 μM NAA, and kept in continuous white light or white light plus UV‐B (1 W/m2) for 7 days. Images are shown in (C); scale bars = 2 mm. The lateral root density (number of lateral roots/length of primary root) (D) and average length of lateral roots (E) of the indicated genotypes were measured. SDs (n > 8) are indicated. Letters “a” to “c” indicate statistically significant differences for the indicated values, as determined by a one‐way analysis of variance (ANOVA), followed by Tukey's least significant difference (LSD) test (P < 0.05). There was statistically significant difference between values marked with different letters (for example “a” and “b”), while there was no statistically significant difference between values marked with the same letters (for example “a” and “a”).
Figure EV2. UV‐B represses the auxin responses and inhibits lateral root growth in a UVR8‐dependent manner.
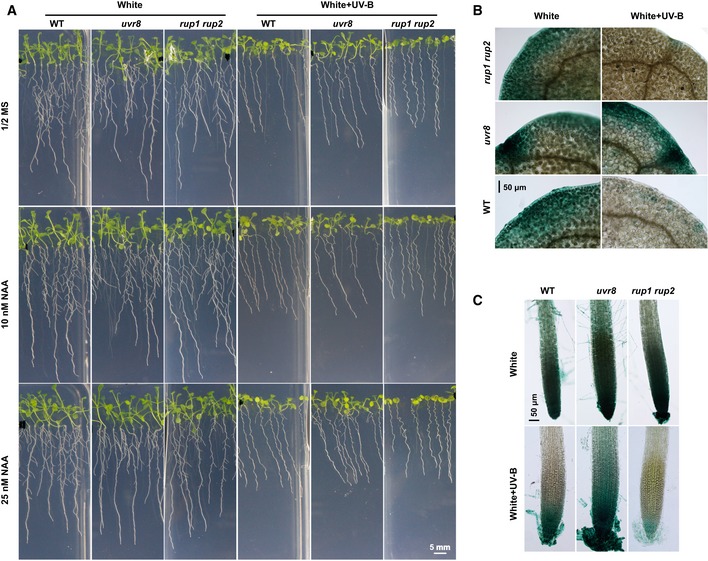
-
APhenotypic analysis. Seedlings of the indicated genotypes were grown in 1/2 MS with the addition of a series of concentrations of NAA under either continuous white light or white plus UV‐B (1 W/m2) for 2 weeks. Images of representative seedlings are shown; scale bar = 5 mm.
-
B, CGUS staining of seedlings expressing the DR5p::GUS transgene in the WT, uvr8, and rup1 rup2 mutant background. Seedlings of each indicated genotype were grown under LD conditions (16‐h light/8‐h dark) for 5 days, then transplanted to new medium containing 0.4 μM NAA, and kept in continuous white light or white light plus UV‐B (1 W/m2) for 7 days. Images of leaves (B) and lateral roots (C); scale bars = 50 μm.
Next, we examined the GUS activity of seedlings expressing DR5p::GUS and grown on 1/2 MS with 0.4 μM NAA, in the presence and absence of UV‐B treatment. UV‐B still inhibited auxin responses in a UVR8‐dependent manner (Fig EV2B and C). L‐kyn, an inhibitor of auxin biosynthesis, and PIC, an auxin analog, were applied to examine their effects on hypocotyl elongation. L‐kyn treatment abolished the long‐hypocotyl phenotype of uvr8 under UV‐B light but did not affect the hypocotyl elongation of rup1 rup2 (Appendix Fig S2C–E), suggesting that the long‐hypocotyl phenotype of uvr8 is at least partially dependent on the auxin response. Indeed, uvr8 was extremely sensitive to exogenous PIC after its long‐hypocotyl phenotype was abolished with L‐kyn treatment (Appendix Fig S2F–H). These results indicate that UV‐B represses auxin responses and inhibits hypocotyl elongation in a UVR8‐dependent manner.
Monomeric UVR8 inhibits auxin responses in a tissue‐autonomous manner
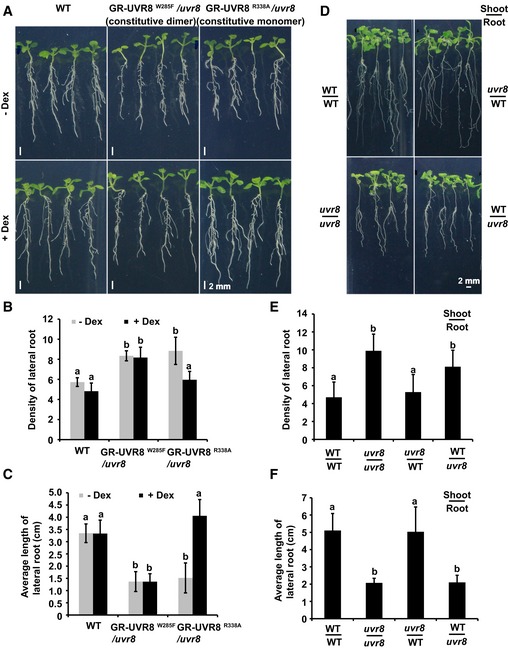
We hypothesized that only the UV‐B‐activated nuclear‐localized monomeric form of UVR8 could repress auxin responses. To test this hypothesis, transgenic uvr8 Arabidopsis plants containing GR‐UVR8W285F (Qian et al, 2016) (glucocorticoid receptor [GR] fused to a constitutively dimeric UVR8 mutant form) or GR‐UVR8R338A (Qian et al, 2016; GR fused to a constitutively monomeric UVR8 mutant form), each fused to YFP under the native UVR8 promoter, were used to conditionally localize UVR8W285F and UVR8R338A into distinct subcellular fractions (Qian et al, 2016). The application of dexamethasone (DEX) drove root cytosolic UVR8W285F or UVR8R338A fusion proteins into the nucleus (Fig EV3A). The constitutive monomeric UVR8R338A interacted with COP1 even in darkness (Fig EV3B). Accordingly, GR‐UVR8R338A complemented the lateral root phenotype of uvr8 in the presence of DEX and UV‐B (Figs 3A–C and EV3C), indicating that UV‐B represses auxin responses in transgenic seedlings expressing GR‐UVR8R338A and treated with DEX. By contrast, UV‐B did not repress the auxin responses in GR‐UVR8W285F, even with DEX and UV‐B treatment (Figs 3A–C and EV3C). These results demonstrate that only nucleus‐localized, UV‐B‐activated monomeric UVR8 inhibits auxin responses.
Figure EV3. Monomeric UVR8 inhibits lateral root growth and development.
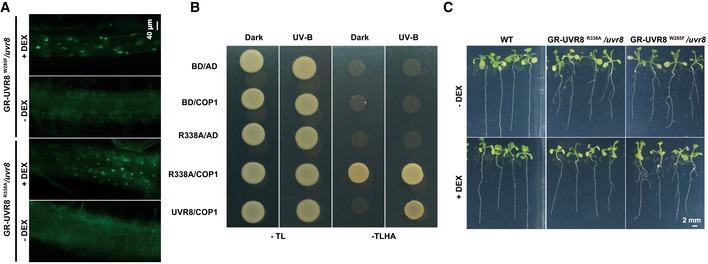
- DEX treatment induces the nuclear accumulation of GR‐UVR8R338A and GR‐UVR8W285F in roots. Transgenic plants expressing GR‐UVR8R338A or GR‐UVR8W285F in the uvr8 background were grown in LD (16‐h light/8‐h dark) for 5 days and then transplanted to new medium containing 1 μM IAA and with or without 20 μM DEX. Plants were photographed after 7 days. Scale bar = 40 μm.
- Yeast two‐hybrid analysis showed that UVR8R338A could interact with COP1 with or without UV‐B.
- Phenotypic analysis. Seedlings of the indicated genotypes were grown in 1/2 MS with or without 20 μM DEX for 2 weeks under UV‐B. Scale bar = 2 mm.
Figure 3. Monomeric UVR8 inhibits auxin responses in a tissue‐autonomous manner.
-
A–CPhenotypic analysis. WT, GR‐UVR8R338A/uvr8, and GR‐UVR8W285F/uvr8 seedlings were grown in LD (16‐h light/8‐h dark) for 5 days, then transplanted to new medium with 0.4 μM NAA and with or without 20 μM DEX, and kept in white light plus UV‐B (1 W/m2) for 7 days. Images are shown in (A); scale bars = 2 mm. The lateral root density (number of lateral roots/length of primary root) (B) and average length of lateral roots (C) of indicated genotypes were measured. SDs (n > 8) are indicated. Letters “a” and “b” indicate statistically significant differences for the indicated values, as determined by a one‐way analysis of variance (ANOVA), followed by Tukey's least significant difference (LSD) test (P < 0.05). There was statistically significant difference between values marked with different letters (for example “a” and “b”), while there was no statistically significant difference between values marked with the same letters (for example “a” and “a”).
-
D–FUVR8 inhibited auxin responses under UV‐B in a tissue‐autonomous way. WT and uvr8 seedlings grown in LD for 5 days were used for reciprocal grafting. Seedlings were kept in LD for 7 days after grafting, then transplanted to new medium containing 0.4 μM NAA, and kept in UV‐B light (1 W/m2) for 10 days. Images are shown in (D); scale bar = 2 mm. The lateral root density (number of lateral roots/length of primary root) (E) and average length of lateral roots (F) of indicated genotypes were measured. SDs (n > 8) are indicated. Letters “a” and “b” indicate statistically significant differences for the indicated values, as determined by a one‐way analysis of variance (ANOVA), followed by Tukey's least significant difference (LSD) test (P < 0.05). There was statistically significant difference between values marked with different letters (for example “a” and “b”), while there was no statistically significant difference between values marked with the same letters (for example “a” and “a”).
Reciprocal grafting experiments were used to determine whether root or shoot UVR8 is responsible for regulating lateral root growth and auxin responses in roots. Lateral root phenotypes of grafted plants with a uvr8 scion and a WT root were similar to those of nongrafted and self‐grafted WT plants, whereas grafted plants with a WT scion and uvr8 root had a similar lateral root growth phenotype to nongrafted or self‐grafted uvr8 mutant plants (Fig 3D–F). When roots of grafted seedlings were covered with foil and a black box to mimic growth under natural conditions (soil) with reduced light, the result was the same: Lateral root phenotypes of grafted plants with a uvr8 scion and WT root were similar to those of nongrafted and self‐grafted WT plants (Appendix Fig S3A). These observations indicate that lateral root growth is driven by root UVR8, and not shoot UVR8.
UV‐B represses the expression of auxin‐responsive genes through UVR8
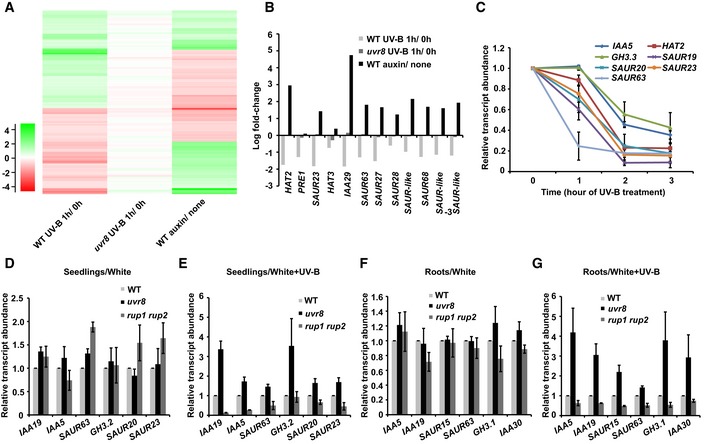
If UV‐B regulates lateral root development by repressing auxin responses, UV‐B would be expected to affect the expression of auxin‐responsive genes. The detection of UV‐B by UVR8 has been reported to antagonize auxin‐responsive genes that regulate shade avoidance and bending toward UV‐B (Hayes et al, 2014; Vandenbussche et al, 2014). Indeed, previously reported microarray datasets of genes affected by UV‐B, UVR8, and auxin significantly overlap (ArrayExpress, E‐MEXP‐1957, E‐GEOD‐627; Fig 4A). For 60.13% of these genes, the effects of auxin were reversed by UV‐B (as evidenced by comparing WT under UV‐B vs. no UV‐B and auxin vs. no auxin; Fig 4A, Table EV1). Many auxin‐responsive genes were repressed by UV‐B in the WT but not in uvr8 (Fig 4B). Previously reported high‐throughput sequencing data of root genes affected by UV‐B (Wan et al, 2018) and microarray datasets of genes affected by auxin also significantly overlap. For 23.26% of these genes, the effects of auxin were reversed by UV‐B (as evidenced by comparing UV‐B vs no UV‐B and auxin vs. no auxin; Appendix Fig S3B, Table EV2).
Figure 4. UV‐B represses the expression of auxin signaling genes.
-
A, BTranscriptome analysis of gene expression regulated by auxin, UV‐B, and UVR8. (A) Heat map of UV‐B‐, UVR8‐, and auxin‐regulated genes. The parameter measured by color key shows the log‐fold change. (B) Auxin signaling genes are up‐regulated by auxin treatment but down‐regulated by UV‐B in a UVR8‐dependent manner. Three biological replicates were analyzed, and final log‐fold change is shown.
-
CQuantitative RT–PCR results showing that UV‐B represses the expression of auxin signaling genes. Six‐day‐old WT seedlings grown in continuous white light were transferred to UV‐B light (2 W/m2) for the indicated period. Error bars are SD of three biological replicates.
-
D, EQuantitative RT–PCR analysis of auxin signaling gene expression in wild‐type (Col‐0), uvr8, and rup1 rup2 seedlings. Seedlings grown in continuous white light for 5 days were kept in white light (D) or transferred to white light plus UV‐B (E) for 1 day with the addition of 1 μM IAA. Error bars are SD of three biological replicates.
-
F, GqPCR analysis of auxin signaling gene expression in roots of WT, uvr8, and rup1 rup2 seedlings. Seedlings grown in continuous white light for 12 days were kept in white light (F) or transferred to white light plus UV‐B (G) for 1 day with the addition of 0.4 μM NAA. Error bars are SD of three biological replicates.
We verified the transcriptomic data using qPCR; the transcript abundances of auxin‐responsive genes such as IAA19 (INDOLE‐3‐ACETIC ACID INDUCIBLE 19) were all down‐regulated by UV‐B treatment (Fig 4C, Appendix Fig S3C). Furthermore, the expression of these genes was similar in the whole seedlings (Fig 4D) and roots (Fig 4F) of WT, uvr8, and rup1 rup2 mutants without UV‐B treatment. However, their expression was higher in uvr8 than in the WT but lower in rup1 rup2 with UV‐B treatment in both seedlings (Fig 4E) and roots (Fig 4G), because UV‐B repressed the expression of these genes in the WT, strongly repressed their expression in rup1 rup2, and weakly repressed their expression in uvr8. These results indicate that UV‐B represses the expression of auxin‐responsive genes through UVR8.
UVR8 interacts with MYB73/MYB77 in a UV‐B‐dependent manner
To decipher the mechanism by which UVR8 inhibits auxin responses, we performed a yeast two‐hybrid screen with a library of A. thaliana transcription factor ORFs (Castrillo et al, 2011) with and without UV‐B treatment to identify transcription factors that interact with UVR8 and are also involved in auxin responses. MYB73 was identified in this screen in the presence of UV‐B treatment. Unlike BIM1, BES1, and WRKY36, MYB73 interacted with UVR8 in a UV‐B‐dependent manner. In yeast cells, MYB73 interacted with UVR8 in the presence of UV‐B, but the interaction was hardly detected in darkness (Fig EV4A). A bimolecular luminescence complementation (BiLC) assay also indicated that UVR8 interacted with MYB73 in plant cells and that the interaction was enhanced by UV‐B treatment. Stronger luminescence was detected in leaves co‐transformed with cLUC‐UVR8 and MYB73‐nLUC plasmids with UV‐B treatment than without UV‐B (Fig EV4B and C). We also tested for interactions between UVR8 and three proteins that are highly similar to MYB73 (i.e., MYB77, MYB70, and MYB44; Appendix Fig S4A), using a BiLC assay. Only MYB77 interacted with UVR8, and the interaction was enhanced by UV‐B (Fig EV4D and E).
Figure EV4. UVR8 physically interacts with MYB73/MYB77 in a UV‐B‐enhanced manner.
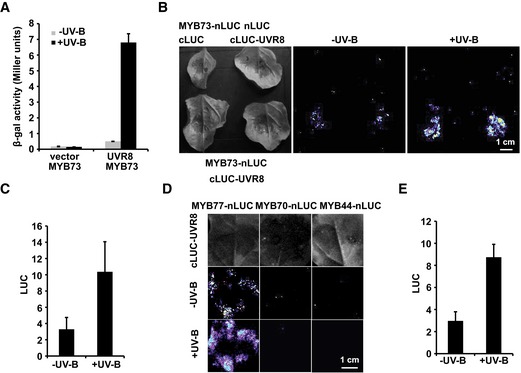
- β‐gal assay showing the UV‐B‐dependent interaction between UVR8 and MYB73. β‐gal activity was present in yeast cells expressing UVR8 and MYB73 together in the presence of UV‐B and almost absent in yeast cells in the absence of UV‐B or expressing MYB73 alone. Error bars, SDs of three biological replicates.
- BiLC assay showing that UV‐B treatment promotes the formation of the UVR8–MYB73 complex. Leaf epidermal cells of N. benthamiana were co‐transformed with MYB73‐nLUC and cLUC‐UVR8 and treated with or without UV‐B (2 W/m2) for 30 min before being photographed. Scale bar = 1 cm.
- Quantification of luciferase (LUC) activities of MYB73‐nLUC + cLUC‐UVR8 in (B) with or without UV‐B treatment (UV‐B + white light versus white light only). Error bars, SDs of three biological replicates.
- BiLC assay showing that UV‐B treatment promotes the formation of the UVR8–MYB77 complex. Scale bar = 1 cm.
- Quantification of LUC activities of MYB77‐nLUC + cLUC‐UVR8 in (D) with or without UV‐B treatment. Error bars, SDs of three biological replicates.
Previous studies showed that MYB77 regulated the growth and development of lateral roots by regulating auxin signaling (Shin et al, 2007; Zhao et al, 2014). An in vitro pull‐down assay was also applied to test the interaction between UVR8 and MYB73/MYB77. UVR8 and MYB73/MYB77 were expressed and purified as reported previously (Yang et al, 2018b). More MYB73/MYB77 was pulled down with UVR8 in the presence of UV‐B light than without it (Fig 5A and B), indicating that UVR8 interacts with MYB73/MYB77 in a UV‐B‐enhanced manner in vitro. A BiFC assay confirmed that UVR8 interacted with MYB73/MYB77 in plant cells (Fig 5C).
Figure 5. UVR8 physically interacts with MYB73/MYB77 in a UV‐B‐dependent manner.
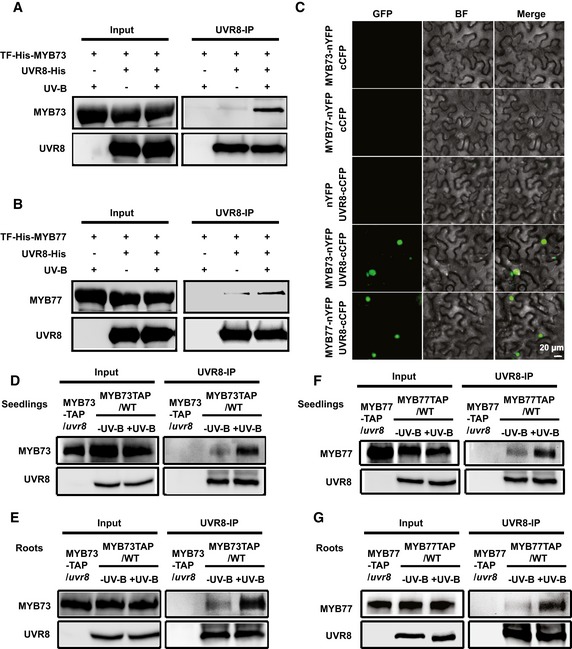
-
A, BIn vitro pull‐down assay showing that UV‐B treatment promotes the interaction between MYB73 (A) or MYB77 (B) and UVR8. The bound proteins were eluted and analyzed by probing immunoblots with an anti‐His antibody.
-
CBiFC assay showing that UVR8 interacts with MYB73/MYB77 in white light condition. N. benthamiana was co‐transformed with MYB73‐nYFP/MYB77‐nYFP and cCFP, nYFP, and UVR8‐cCFP, or MYB73‐nYFP/MYB77‐nYFP and UVR8‐cCFP. BF, bright field. Merge, overlay of the YFP and bright‐field images. Scale bar = 20 μm.
-
D–GCo‐IP assays showing that MYB73 and MYB77 interact with UVR8 in a UV‐B‐dependent manner in whole seedlings (D, F) and roots (E, G). Input: immunoblots showing the levels of UVR8, MYB73‐TAP, and MYB77‐TAP in the total protein extract. UVR8 IP: the IP products precipitated by the UVR8 antibody. Total proteins (Input) or IP products were probed, on immunoblots, with an anti‐UVR8 or anti‐Myc antibody, which detects the TAP tag.
Source data are available online for this figure.
To investigate whether the UVR8–MYB73/MYB77 interaction in plant cells is regulated by UV‐B light, we performed co‐immunoprecipitation (co‐IP) analyses in transgenic Arabidopsis plants expressing TAP‐tagged MYB73/MYB77. Seedlings were either kept in white light or exposed to UV‐B light for 20 min, and then, whole seedlings or roots were subjected to co‐IP analyses. More MYB73/MYB77 was coprecipitated with UVR8 from seedlings or roots irradiated with UV‐B than without UV‐B (Fig 5D–G). These results strongly suggest that UV‐B light promotes the accumulation of UVR8–MYB73 and UVR8–MYB77 protein complexes in plant cells, including root cells. Thus, MYB73/MYB77 undergo UV‐B‐dependent physical interactions with UVR8 in vivo.
Furthermore, co‐IP analyses were performed using transgenic Arabidopsis plants expressing MYB73‐ or MYB77‐TAP/YFP‐UVR8W285F (containing the constitutively dimeric UVR8 form) and MYB73‐ or MYB77‐TAP/YFP‐UVR8R338A (containing the constitutively monomeric UVR8 form). More MYB73/MYB77 was coprecipitated with UVR8R338A than with UVR8W285F (Appendix Fig S4B and C), indicating that MYB73/MYB77 preferentially interacted with monomeric UVR8. BiFC and in vitro pull‐down assays showed that the N‐terminus of MYB73/MYB77, which includes an R2R3 DNA‐binding domain (MYB73N: amino acids [aa] 1–120; MYB73C: aa 121–320; MYB77N: aa 1–120; MYB77C: amino 121–301), interacted with both the N‐ and C‐termini of UVR8 (UVR8N: aa 1–396; UVR8C: aa 397–440; Appendix Fig S4D–I).
MYB73/MYB77 acts downstream of UVR8 to regulate lateral root growth in response to UV‐B light
MYB77 is reported to function in auxin signal transduction, and interacts with ARFs to promote auxin‐responsive gene expression and lateral root development (Shin et al, 2007). MYB73/MYB77 is expressed in both shoot and root (Shin et al, 2007; Fig EV5A). To determine the biological roles of MYB73/MYB77 under UV‐B, we first examined whether UV‐B affected MYB73/MYB77 expression. The photomorphogenic UV‐B light (narrowband UV‐B) slightly induced the transcription of MYB73/MYB77 in an UVR8‐independent manner (Appendix Fig S5A and B). Transgenic plants constitutively expressing epitope‐tagged MYB73/MYB77 (35S:MYB73/MYB77‐TAP) were used to analyze MYB73/MYB77 post‐transcriptional protein expression. UV‐B treatment did not affect the protein accumulation of MYB73/MYB77 (Appendix Fig S5C and D). We obtained myb73 and myb77 T‐DNA insertion mutants from the Arabidopsis Biological Resource Centre (Appendix Fig S6A and B). The lateral root growth of myb73 and myb77 was similar to that of the WT with or without auxin treatment (Fig EV5B–D). However, the average length of myb73 myb77 lateral roots was longer than that of the WT when grown in the presence of a high concentration of auxin under UV‐B light (Fig 6A–C).
Figure EV5. MYB73/MYB77 works redundantly to mediate UVR8‐controlled auxin responses.
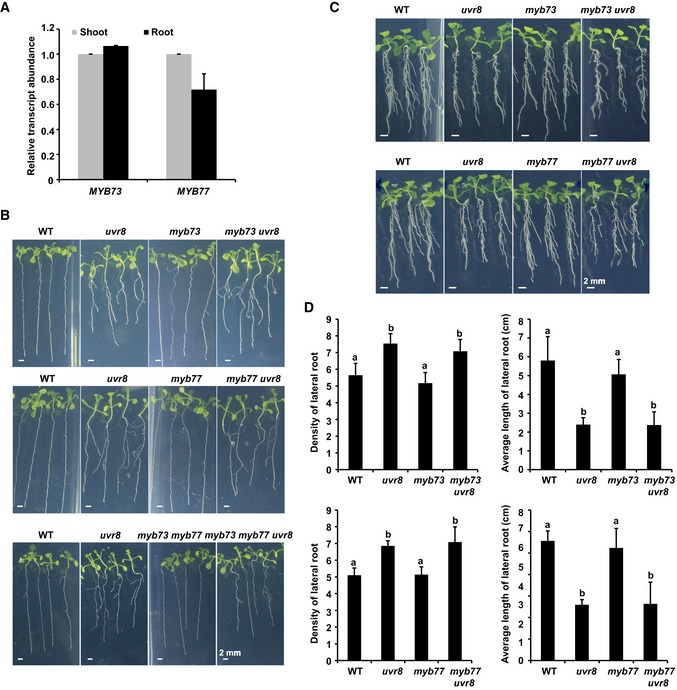
-
AqPCR assay showing that MYB73/MYB77 is expressed in both the shoot and root. Wild‐type seedlings were grown in 1/2 MS for 12 days. ACT7 expression was analyzed as an internal control. Error bars are SD of three biological replicates.
-
BPhenotypic analysis. Seedlings of the indicated genotypes were grown in 1/2 MS with UV‐B treatment for 2 weeks. Scale bars = 2 mm.
-
C, DPhenotypic analysis. Seedlings of the indicated genotypes were grown in LD (16‐h light/8‐h dark) conditions for 5 days and then transplanted to new plates containing 0.4 μM NAA with UV‐B treatment for 7 days. Images are shown in (C); scale bars = 2 mm. The lateral root density (number of lateral roots/length of primary root) and average length of lateral roots of the indicated genotypes were measured (D). SDs (n > 8) are indicated. Letters “a” and “b” indicate statistically significant differences for the indicated values, as determined by analysis of variance (ANOVA), followed by Tukey's least significant difference (LSD) test (P < 0.05). There was statistically significant difference between values marked with different letters (for example “a” and “b”), while there was no statistically significant difference between values marked with the same letters (for example “a” and “a”).
Figure 6. MYB73/MYB77 acts downstream of UVR8 in controlling the auxin response.
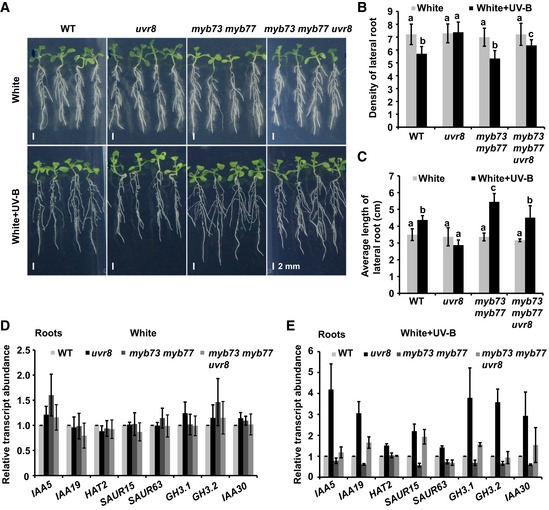
-
A–CPhenotypic analysis. Seedlings of the indicated genotypes were grown in LD conditions (16‐h light/8‐h dark) for 5 days, then transplanted to new medium containing 0.4 μM NAA, and kept in continuous white light or white plus UV‐B light (1 W/m2) for 7 days. Images are shown in (A); scale bars = 2 mm. Lateral root density (number of lateral roots/length of primary root) (B) and average length of lateral roots (C) of the indicated genotypes were measured. SDs (n > 8) are indicated. Letters “a” to “c” indicate statistically significant differences for the indicated values, as determined by a one‐way analysis of variance (ANOVA), followed by Tukey's least significant difference (LSD) test (P < 0.05). There was statistically significant difference between values marked with different letters (for example “a” and “b”), while there was no statistically significant difference between values marked with the same letters (for example “a” and “a”).
-
D, EQuantitative RT–PCR analysis of auxin signaling gene expression in roots of wild type (Col‐0), uvr8, myb73 myb77, and myb73 myb77 uvr8. (D) Results under white light and (E) under white plus UV‐B light. Seedlings grown in continuous white light for 12 days were kept in white light or transferred to white light plus UV‐B for 1 day with the addition of 0.4 μM NAA, and roots were collected to analyze auxin signaling gene expression. Error bars represent SD of three biological replicates.
We next investigated genetic interactions between UVR8 and MYB73/MYB77 genes. The uvr8 mutant was crossed with myb73, myb77, and myb73 myb77, resulting in myb73 uvr8, myb77 uvr8, and myb73 myb77 uvr8 mutant plants (Appendix Fig S6C–F). The lateral root phenotypes of myb73 uvr8 and myb77 uvr8 were similar to those of uvr8 grown with or without auxin under UV‐B light (Fig EV5B–D). However, the lateral root phenotype of uvr8 was partially suppressed in the myb73 myb77 uvr8 triple mutant both in the presence (i.e., it was less sensitive to high concentrations of auxin than uvr8 under UV‐B light) and absence (i.e., it had less and shorter lateral roots than uvr8 under UV‐B light) of a high concentration of auxin under UV‐B light (Figs 6A–C, and EV5B), even when the roots were covered (Appendix Fig S7A), suggesting that MYB73/MYB77 acts downstream of UVR8. The long‐hypocotyl phenotype of uvr8 was also partially rescued in myb73 myb77 uvr8 under UV‐B light (Appendix Fig S7B and C).
Next, we examined the abundance of auxin‐regulated transcripts and found that their levels were decreased in myb73 myb77 compared with the WT under UV‐B, suggesting that MYB73/MYB77 acts as positive regulators in auxin signaling (Fig 6D and E, Appendix Fig S7D and E), consistent with previous reports (Shin et al, 2007; Zhao et al, 2014). Furthermore, consistent with the observation that the lateral root and long‐hypocotyl phenotypes of uvr8 were partially suppressed in myb73 myb77 uvr8 mutants treated with UV‐B, the expression levels of auxin signaling genes were lower in the myb73 myb77 uvr8 mutants than in uvr8 under UV‐B (Fig 6D and E). However, the expression levels of those genes in myb73 uvr8 and myb77 uvr8 double mutants were similar to those in uvr8 (Appendix Fig S7F). These results indicate that MYB73/MYB77 acts downstream of UVR8 to regulate lateral root growth and development in response to UV‐B light, through the regulation of auxin responses.
UVR8 inhibits the DNA‐binding activity of MYB73/MYB77 under UV‐B
MYB73/MYB77 is R2R3 transcription factors that preferentially bind to the MBSI motif (Shin et al, 2007; Zhao et al, 2014). Many genes involved in auxin signaling such as IAA19, IAA7, and GH3.2 (AUXIN UP‐REGULATED 3) contain MBSI motifs. Electrophoretic mobility‐shift assays (EMSAs) were performed using recombinant MYB73, MYB77, and UVR8 proteins expressed in E. coli. MYB73/MYB77 bound to the MBSI motif, and to the promoter of IAA19 (IAA19p: −409 bp to −349 bp), which contains two MBSI motifs, in vitro. MYB73 and MYB77 had much lower binding activity when assayed with a mutated MBSI motif (Fig 7A and B, Appendix Fig S8A). Interestingly, UVR8W285A did not bind to the MBSI motif by itself (Fig 7C and D, Appendix Fig S8B–D), but both UVR8 and UVR8W285A inhibited the DNA‐binding activity of MYB73/MYB77 (Fig 7C and D, Appendix Fig S8B–D).
Figure 7. UVR8 inhibits the DNA‐binding activity of MYB73/MYB77.
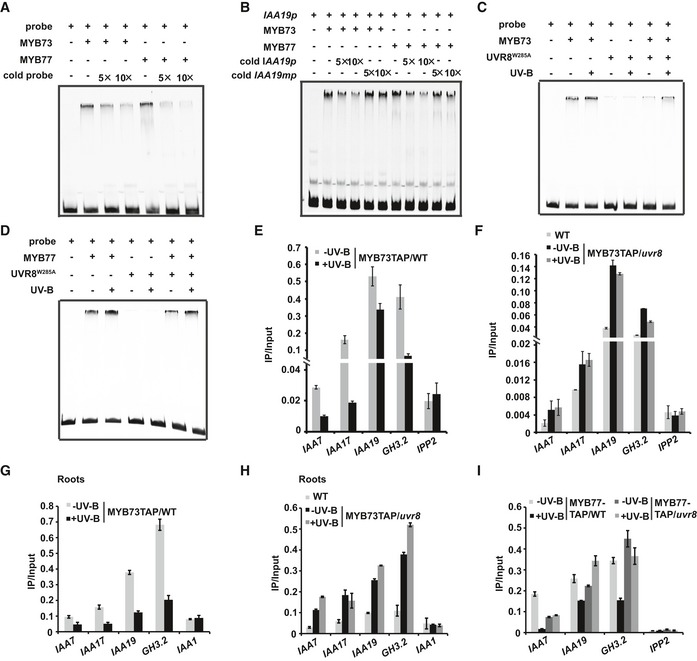
-
AElectrophoretic mobility‐shift assay (EMSA) showing that MYB73/MYB77 binds to the MBSI motif in vitro. Cold probe was added as a competitor.
-
BEMSA showing that MYB73/MYB77 binds to the promoter of IAA19. Cold IAA19p or IAA19mp (MBSI motif mutated) was added as a competitor.
-
C, DEMSA results showing that monomeric UVR8 inhibits the DNA‐binding activity of MYB73(C) and MYB77(D).
-
EChIP‐qPCR analysis showed that UV‐B inhibits the DNA‐binding activity of MYB73. ChIP‐qPCR assays were performed using transgenic seedlings expressing 35S::MYB73‐TAP, treated with or without UV‐B for 5 h before harvesting samples. Chromatin fragments (˜500 bp) were immunoprecipitated by anti‐Myc antibody, and the precipitated DNA was analyzed by qPCR using the primer pairs indicated. The IP/Input ratios are shown with SDs (n = 3).
-
FChIP‐qPCR showing that UV‐B inhibits the DNA‐binding activity of MYB73 in a UVR8‐dependent manner. The IP/Input ratios are shown with SDs (n = 3).
-
G, HChIP‐qPCR showing that UV‐B inhibits the DNA‐binding activity of MYB73 in roots (G) in a UVR8‐dependent manner (H). The IP/Input ratios are shown with SDs (n = 3).
-
IChIP‐qPCR showing that UV‐B inhibits MYB77 from binding to the indicated promoters in a UVR8‐dependent manner. The IP/Input ratios are shown with SDs (n = 3).
Source data are available online for this figure.
Next, we performed chromatin immunoprecipitation (ChIP)‐qPCR to determine whether MYB73/MYB77 bound to the promoters of auxin signaling genes in vivo. MYB73/MYB77 indeed bound to promoters of several auxin signaling genes in both seedlings and roots. The DNA‐binding activity of MYB73/MYB77 was inhibited by UV‐B in a UVR8‐dependent manner, as evidenced by the fact that UV‐B inhibition of the DNA‐binding activity of MYB73/MYB77 was abolished in uvr8 (Fig 7E–I, Appendix Fig S8E and F). However, UVR8 did not bind to the promoters of auxin signaling genes by itself (Appendix Fig S8G). Furthermore, constitutively monomeric UVR8 strongly repressed the DNA‐binding activity of MYB73/MYB77, since UVR8R338A (but not UVR8W285F) strongly repressed the DNA‐binding activity of MYB73/MYB77 in transgenic lines co‐expressing MYB73/MYB77‐TAP and YFP‐UVR8R338A or MYB73/MYB77‐TAP and YFP‐UVR8W285F (Appendix Fig S8H and I). Together, these data indicate that UVR8 interacts with MYB73/MYB77 and inhibits their DNA‐binding activity in a UV‐B‐dependent manner.
Discussion
There are many reports of light affecting root growth when roots are directly exposed to light under experimental conditions. Phytochromes (PHYs), which are red/far‐red light photoreceptors, are expressed in roots and mediate primary root development, the jasmonic acid response, and gravitropism (Costigan et al, 2011). Blue‐light photoreceptors, namely crytochromes (CRYs) and phototropins (PHOTs), mediate blue‐light‐regulated primary root growth and phototropism (Briggs & Christie, 2002). In addition, RUS1 and RUS2 play critical roles in UV‐B‐regulated root growth (Tong et al, 2008; Leasure et al, 2009). Recently, it was reported that light is channeled to the roots through the stem, activating phytochrome and triggering light responses in the root (Lee et al, 2016). Auxin is essential for lateral root development, and ARF7 and ARF19 are reported to play critical roles in lateral root growth and development (Tatematsu et al, 2004; Weijers et al, 2005). The MYB transcription factor MYB77 interacts with ARFs to modulate auxin signal transduction and lateral root growth (Shin et al, 2007; Zhao et al, 2014). Here, we show that UV‐B negatively regulates lateral root growth and development and the auxin response in a UVR8‐dependent manner. UVR8 physically interacts with MYB73/MYB77 to regulate the auxin response and lateral root development. Furthermore, monomeric UVR8 inhibits lateral root growth and auxin responses in a root‐autonomous manner, indicating that root UVR8 is responsible for the UV‐B‐regulated auxin responses and lateral root growth. Although UV‐B light might be conducted through the soil to the root, it is also possible that UV‐B light is conducted through the plant stem. Coordination is required between shoot and root growth for normal plant growth; when shoot growth is repressed by UV‐B, root growth is also inhibited. The root could also respond to light environmental changes.
A fundamental mechanism underlying photoreceptor signal transduction is direct interaction between photoreceptors and their respective target proteins. Plant photoreceptors may interact with a target protein by two alternative modes of action. A photoreceptor may interact with its target protein in response to light, leading to light‐dependent physiological responses. Alternatively, a photoreceptor may constitutively interact with a target protein, but only trigger changes in biochemical activities associated with the target protein when photoexcited. UVR8 interacts with COP1 in a UV‐B‐dependent manner to mediate UV‐B signaling (Rizzini et al, 2011; Huang et al, 2014; Qian et al, 2016; Yin et al, 2016). UVR8 interacts with RUP1 and RUP2 in a UV‐B‐independent manner to mediate UVR8 re‐dimerization (Gruber et al, 2010).
UVR8 also physically interacts with transcription factors BES1, BIM1, and WRKY36 in the nucleus in a UV‐B‐independent manner, while only the UV‐B‐activated monomeric UVR8 inhibits their DNA‐binding activity to regulate transcription and photomorphogenesis (Liang et al, 2018; Yang et al, 2018b). It was previously unknown, however, whether UVR8 interacted with some transcription factors in a UV‐B‐dependent manner. Here, we show that UVR8 interacts with the MYB73/MYB77 transcription factors in a UV‐B‐dependent manner and regulates their DNA‐binding activity and gene expression.
UVR8 can function by physically interacting with different transcription factors to regulate gene expression via multiple mechanisms. HY5 and its homolog HYH mediate UV‐B‐induced gene expression changes downstream of UVR8 (Ulm et al, 2004; Brown et al, 2005; Oravecz et al, 2006; Brown & Jenkins, 2008; Stracke et al, 2010; Feher et al, 2011; Huang et al, 2012). UV‐B treatment induces the transcription and translation of HY5 in a UVR8‐ and COP1‐dependent manner (Brown et al, 2005, 2009; Oravecz et al, 2006; Kaiserli & Jenkins, 2007; Favory et al, 2009). As UVR8 can interact directly with several transcription factors (e.g., BES1, BIM1, WRKY36, MYB73, and MYB77) and thus mediate UV‐B‐regulated gene expression, UVR8 could also regulate the expression of the transcription factor gene HY5 to mediate UV‐B‐regulated gene expression.
Liang et al (2018) and Wang et al (2018) established the molecular basis of cross‐talk between UV‐B light or blue light and brassinosteroid (BR) signaling. Nucleus‐localized UVR8 interacts with BR‐activated dephosphorylated BES1 and BIM1 in a UV‐B‐independent manner, while CRYs interact with dephosphorylated BES1 and BIM1 in a blue‐light‐dependent manner, repressing their DNA‐binding activity and also the expression of elongation genes (Liang et al, 2018; Wang et al, 2018). CRY1 interacts with PIF4 to regulate its transcriptional activity and auxin biosynthesis (Ma et al, 2016). Furthermore, CRYs and PHYs interact directly with AUX/IAA proteins, stabilizing them and thus inhibiting auxin signaling (Xu et al, 2018; Yang et al, 2018a). Auxin homeostasis is affected by UV‐B via photooxidative damage, degradation, biosynthesis, and conjugation (Vanhaelewyn et al, 2016). UV‐B‐activated UVR8 strongly attenuates thermomorphogenesis and shade avoidance through modulating PIF4 transcription and protein stability. UV‐B also stabilizes inhibitors of PIF4 including DELLA and HFR1 (LONG HYPOCOTYL IN FAR RED) proteins, thus affecting auxin biosynthesis (Hayes et al, 2014, 2017). UV‐B regulation on auxin also occurs at the level of redistribution via transport, influx, or efflux (Wargent et al, 2009; Ge et al, 2010; Yu et al, 2013). Furthermore, the detection of UV‐B by UVR8 antagonized auxin‐responsive genes that regulate shade avoidance and bending toward UV‐B (Hayes et al, 2014; Vandenbussche et al, 2014).
Auxin has been identified as a key long‐distance signal in light‐regulated root development (Sassi et al, 2012). COP1 controls the transcription of the auxin efflux carrier gene PIN‐FORMED1 (PIN1), modulating shoot‐derived auxin levels in the root (Sassi et al, 2012). HY5 mediates far‐red light‐regulated lateral root development by decreasing the plasma membrane abundance of PIN‐FORMED3 and LIKE‐AUX1 3 auxin transporters (van Gelderen et al, 2018). UV‐B and UVR8 might regulate auxin transport via COP1 and HY5. Here, we report that UVR8 interacts with MYB73/MYB77 in a UV‐B‐dependent manner to repress their DNA‐binding activity and the expression of their targets, so as to modulate auxin signaling. UV‐B represses growth through modulating different endogenous hormone signaling pathways (such as BR, auxin, and gibberellin pathways).
In summary, we show here that UV‐B regulates shoot and root growth by activating shoot and root UVR8 and that UV‐B‐activated UVR8 physically interacts with MYB73/MYB77 and thereby modulates auxin signaling and hypocotyl elongation and lateral root growth. The UV‐B‐dependent UVR8–MYB73/MYB77 interaction makes it possible for light and auxin signaling to coordinately regulate shoot and root growth (Fig 8).
Figure 8. A hypothetical model depicting how UVR8 acts with MYB73/MYB77 to regulate auxin signaling and UV‐B photomorphogenesis in Arabidopsis .
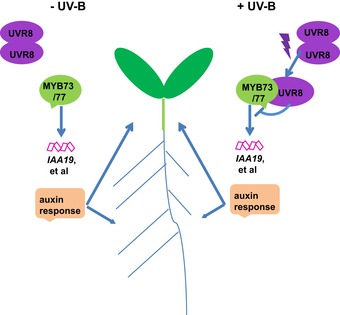
UVR8 is localized in both the shoot and root, and is responsible for both shoot morphogenesis and root development. In the absence of UV‐B light, MYB73/MYB77 positively regulates the expression of auxin‐responsive genes and promotes hypocotyl elongation and lateral root development and growth. However, in the presence of UV‐B light, UV‐B induces the monomerization and nuclear accumulation of UVR8, which then interacts with MYB73/MYB77 in the nuclei. This interaction inhibits the DNA‐binding activity of MYB73/MYB77, thus repressing the expression of auxin‐responsive genes and auxin responses.
Materials and Methods
Plant materials and growth conditions
The Columbia ecotype of Arabidopsis thaliana was used. GR‐UVR8R338A (YFP‐GR‐UVR8R338A/uvr8‐6) and GR‐UVR8W285F (YFP‐GR‐UVR8W285F/uvr8) have previously been described (Qian et al, 2016). T‐DNA insertion mutants uvr8‐6 (SALK_033468), myb73 (SALK_023478), and myb77 (SALK_067655) were obtained from ABRC. The myb73 myb77, myb73 uvr8, myb77 uvr8, and myb73 myb77 uvr8 mutants were prepared by genetic crossing, and their identities were verified by genotyping. The full‐length MYB73/MYB77 coding sequences were cloned into pCambia1300 (Cambia), bearing a TAP (Myc‐His‐Flag) tag (Pro35S::MYB73/MYB77‐TAP). WT(Col‐0) and uvr8 were transformed with Pro35S::MYB73/MYB77‐TAP; for every transformation, more than 10 independent transgenic lines with a single copy of the transgene were generated. Immunoblots were performed to verify overexpression of the transgenes. Plants expressing MYB73/MYB77‐TAP/GR‐UVR8R338A and MYB73/MYB77‐TAP/GR‐UVR8W285F were prepared by genetic crossing.
Seeds were sterilized in 10% (v/v) bleach, placed on ½‐strength MS medium containing 0.8% (w/v) agar and 1% (w/v) sucrose, and stratified for 4 days at 4°C in darkness before being transferred to white light (Philips TLD18W/54‐765, 6 μmol/m2/s, measured by an ILT1400 Radiometer Photometer) or white light plus UV‐B (Philips TL20W/01RS narrowband UV‐B tubes, 1 or 2 W/m2, measured by a LUYOR‐340 UV Light Meter).
Arabidopsis thaliana grafting
Reciprocal grafting was performed as previously described (Gao et al, 2017). After the graft unions were established, the grafted plants were examined under a stereoscopic microscope before being transferred into new MS plates. Healthy grafted plants without adventitious roots were transferred to new MS plates grown in the controlled environment described above.
GUS staining
The expression of GUS (beta‐glucuronidase) was analyzed as described (Ma et al, 2016). Five milligrams of 5‐bromo‐4‐chloro‐3‐indolyl glucuronide (X‐Gluc) was dissolved in 1 ml dimethylformamide, and then, 50 mM NaPO4 (pH 7.0) was added to a final volume of 10 ml. Samples were placed in this solution for staining overnight at 37°C and then rinsed in 70% ethanol for 8 h.
Root covering
Roots were covered with foil pieces (12 cm × 13 cm and 11 cm × 13 cm) and black boxes (Appendix Fig S3A). The 12 cm × 13 cm foil pieces were folded about 0.5 cm along one of the long edges, and then, all of the foil pieces were autoclave‐sterilized. First, 1/2 MS medium containing 0.8% (w/v) agar and 1% (w/v) sucrose was poured into the dishes to just cover the bottom of the dishes (13 cm × 13 cm). Second, the foil pieces (12 cm × 13 cm) were put into the dishes with the folded foil edges (0.5 cm) bent upward, perpendicular to the bottoms of the dishes. Third, about 50 ml of 1/2 MS was poured into each dish to a depth of about 0.5 cm (the same as the height of the folded foil edge). After the medium had solidified, the seedlings were transferred into the dishes, with the bottoms of the shoots at the position of the folded foil so that roots could be covered by it later. Finally, the 11 cm × 13 cm foil pieces were used to cover the roots of seedlings (Appendix Fig S3A). Dishes were put into a black box with the shoots at the top, extending out of the black box.
Transcriptome analysis
Microarray analyses were based on publicly available data. UV‐B‐regulated genes were compiled from the ArrayExpress database: accession numbers E‐MEXP‐1957 and E‐GEOD‐627. The data were analyzed by RMA. For differential expression analysis, a moderated t‐statistic was computed using the Limma package in R, and fold‐change values were also calculated. Significance for the differentially expressed genes was determined based on a P‐value ≤ 0.05 and log‐fold change 0.5. RNA‐seq data for root were obtained from the National Center for Biotechnology Information's Short Read Archive (SRA) under accession number SRP094914. The raw sequences were transformed into clean tags after certain steps of data processing were performed, including the removal of the adaptor sequence, empty reads, and low‐quality tags. All of the clean tags were mapped to the Arabidopsis reference sequence (TAIR 10) using RSEM (Li & Newey, 2011). Differentially expressed genes were identified using EBSeq (Leng et al, 2013). The significance of the differentially expressed genes was determined by a P‐value 0.05 and log‐fold change 0.5.
mRNA expression analyses
Total RNAs were isolated using an RNAiso Plus Kit (Takara). cDNA was synthesized from 500 ng of total RNA using a PrimeScript RT Reagent Kit with gDNA Eraser (Takara). SYBR Premix Ex Tag (Takara) was used for qPCRs, using the MX3000 System (Stratagene). The level of ACTIN7 mRNA expression (AT5G09810) was used as the internal control. qRT–PCR data for each sample were normalized to the respective ACT7 expression level. The cDNAs were amplified following denaturation, using a 40‐cycle program (95°C, 5 s; 60°C, 20 s per cycle). Biological replicates represent three independent experiments involving about 30 seedlings per experiment. Three technical replicates were performed for each experiment. Primers used are listed in Appendix Table S1.
Yeast two‐hybrid analysis
The coding sequence of UVR8 was fused in‐frame with the GAL4 DNA‐binding domain (BD) of the bait vector pDest32 (Clontech) and transformed into Y187. The library of 1362 transcription factors (in vector pDest22) was in yeast strain YM4271 (Clontech). The Y187 yeast strains were mated with the YM4271 yeast strains (6‐h YPD media), suspended in SD‐Trp‐Leu medium, and incubated overnight. Yeast cells were then grown on SD‐Trp‐Leu plates in darkness and treated with or without UV‐B light (2 W/m2) for 2–3 h per day for 4 days. The interactions were tested by galactosidase assays.
BiFC and BiLC assays
UVR8 or MYB73/MYB77 was fused to the C‐ or N‐terminus of firefly luciferase and transformed into Agrobacterium strain GV3101. Nicotiana benthamiana plants were left under long‐day (LD; 16‐h light/8‐h dark), white light conditions for 3 days after infiltration. The leaves were infiltrated with luciferin solution, and images were captured using a CCD camera 5 min later. The leaves were then treated with UV‐B for 30 min and photographed again.
UVR8 or MYB73/MYB77 was fused to the C‐ or N‐terminus of YFP and transformed into Agrobacterium strain GV3101. N. benthamiana plants were left under LD white light for 3 days after infiltration with Agrobacterium.
In vitro pull‐down
The in vitro pull‐down protein–protein interaction assay was modified from a previously described method (Liu et al, 2008, 2013; Ma et al, 2016; Liang et al, 2018; Yang et al, 2018b). The full‐length coding sequences of MYB73/MYB77, N‐terminal sequences of MYB73N (aa 1–120)/MYB77N (aa 1–120), or C‐terminal sequences of MYB73C (aa 121–320)/MYB77C (aa 121–301) were cloned into pCold‐TF. pET29b‐UVR8 (UVR8‐His) was reported previously (Wu et al, 2012; Yang et al, 2018b). UVR8N and UVR8C were described before. These proteins were expressed and purified from E. coli BL21. UVR8‐His was incubated with His‐TF‐MYB73/MYB77 under white light (Philips TLD18W/54‐765, 6 μmol/m2/s, measured by an ILT1400 Radiometer Photometer) or white light plus UV‐B (Philips TL20W/01RS narrowband UV‐B tubes, 2 W/m2, measured by a LUYOR‐340 UV Light Meter) for 30 min. Anti‐UVR8 (Youke Biotech) was used to pull down the protein complexes, and unbound proteins were removed via washing. The bound proteins were eluted and analyzed by probing immunoblots with anti‐His antibody. Samples were boiled before SDS–PAGE (UVR8 dimers became monomeric after heat denaturation). GST‐UVR8N or GST‐UVR8C was incubated with His‐TF‐MYB73/MYB77N or His‐TF‐MYB73/MYB77C. His beads were used to pull down the protein complexes. UVR8 antibody was a polyclonal antibody made by Youke Company (Shanghai, China) using the reported peptide.
Co‐immunoprecipitation
The co‐immunoprecipitation (co‐IP) procedure was described previously (Liu et al, 2008, 2013; Ma et al, 2016; Liang et al, 2018; Yang et al, 2018b). For co‐IP, 14‐days‐old WT and MYB73/MYB77‐TAP/WT seedlings grown under LD conditions were kept in white light or moved to white light plus UV‐B (1 W/m2) for 20 min, and 14‐days‐old MYB73/MYB77‐TAP/GR‐UVR8R338A and MYB73/MYB77‐TAP/GR‐UVR8W285F seedlings grown under LD conditions were moved to UV‐B (1 W/m2) for 20 min. After treatment, seedlings were ground in liquid nitrogen, homogenized in binding buffer [20 mM HEPES (pH 7.5), 40 mM KCl, 1 mM EDTA, 1% (v/v) Triton X‐100, 1 mM PMSF], incubated at 4°C for 5 min, pushed twice through a 1‐ml syringe and metal needle to promote nuclear lysis, and centrifuged at 14,000 g for 10 min at 4°C. The supernatant was mixed with 35 μl of anti‐UVR8 beads, incubated at 4°C for 30 min, and washed twice with washing buffer [20 mM HEPES (pH 7.5), 40 mM KCl, 1 mM EDTA, 0.1% Triton X‐100]. The bound proteins were eluted from the affinity beads with 4× SDS–PAGE sample buffer and analyzed by immunoblotting.
Electrophoretic mobility‐shift assays (EMSAs)
To make the probe, synthetic oligonucleotides complementary to the MBSI motif (CNGTTA/G) or the IAA19 promoter were annealed and cloned into the T‐vector. The probe was then PCR‐amplified using Cy5‐labeled M13 primer pairs. Cy5‐labeled DNA on the gel was then detected with a Starion FLA‐9000 scanner (Fujifilm, Japan).
Chromatin immunoprecipitation (ChIP) assays
ChIP experiments were performed as described previously (Liu et al, 2008, 2013; Ma et al, 2016; Liang et al, 2018; Yang et al, 2018b), using 14‐day‐old WT, MYB73/MYB77‐TAP/WT, MYB73/MYB77‐TAP/uvr8, MYB73/MYB77‐TAP/GR‐UVR8R338A, or MYB73/MYB77‐TAP/GR‐UVR8W285F transgenic plants grown under LD conditions and treated or not with 5 h of UV‐B before harvesting the samples. Chromatin fragments (~500 bp) were immunoprecipitated with anti‐Myc or anti‐UVR8 antibody, and the precipitated DNA was analyzed by qPCR using the primer pairs indicated or IPP2 as a negative control (Appendix Table S1). The level of binding was calculated as the ratio between IP and Input. Two grams of starting material was harvested and cross‐linked with 1% (v/v) formaldehyde (Sigma) for 15 min under vacuum. Cross‐linking was stopped by adding glycine to a final concentration of 0.125 M. The seedlings were rinsed with water, frozen in liquid nitrogen, and ground to a fine powder.
Accession numbers
Sequence data from this work can be found in the Arabidopsis Information Resource or GenBank databases under the following accession numbers: AT5G63860 (UVR8), AT4G37260 (MYB73), AT3G50060 (MYB77), AT2G23290 (MYB70), AT5G67300 (MYB44), AT3G15540 (IAA19), AT1G15580 (IAA5), AT5G18020 (SAUR20), AT5G18060 (SAUR23), AT1G29440 (SAUR63), AT4G37390 (GH3.2), AT2G23170 (GH3.3), AT4G14560 (IAA1), AT3G23050 (IAA7), and AT1G04250 (IAA17).
Author contributions
YY and HL conceived the project. YY performed most of the experiments, and PC and XL performed the genomic expression analysis. YY, LZ, and TL made some of the constructs. YY and HL analyzed the data and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Source Data for Figure 5
Source Data for Figure 7
Acknowledgements
The authors thank Drs. Tongda Xu, Yunde Zhao, Ronald Pierik, Xi Huang, Zhaojun Ding, and Yang Zhao for materials and technical assistance. This work is supported in part by the National Natural Science Foundation of China (31825004, 31721001, 31730009, and 31670282), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27030000), and the Program of Shanghai Academic Research Leader.
The EMBO Journal (2020) 39: e101928
References
- Briggs WR, Christie JM (2002) Phototropins 1 and 2: versatile plant blue‐light receptors. Trends Plant Sci 7: 204–210 [DOI] [PubMed] [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI (2005) A UV‐B‐specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 102: 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Jenkins GI (2008) UV‐B signaling pathways with different fluence‐rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol 146: 576–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Headland LR, Jenkins GI (2009) UV‐B action spectrum for UVR8‐mediated HY5 transcript accumulation in Arabidopsis . Photochem Photobiol 85: 1147–1155 [DOI] [PubMed] [Google Scholar]
- Castrillo G, Turck F, Leveugle M, Lecharny A, Carbonero P, Coupland G, Paz‐Ares J, Onate‐Sanchez L (2011) Speeding cis‐trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PLoS One 6: e21524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan SE, Warnasooriya SN, Humphries BA, Montgomery BL (2011) Root‐localized phytochrome chromophore synthesis is required for photoregulation of root elongation and impacts root sensitivity to jasmonic acid in Arabidopsis . Plant Physiol 157: 1138–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ et al (2009) Interaction of COP1 and UVR8 regulates UV‐B‐induced photomorphogenesis and stress acclimation in Arabidopsis . EMBO J 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feher B, Kozma‐Bognar L, Kevei E, Hajdu A, Binkert M, Davis SJ, Schafer E, Ulm R, Nagy F (2011) Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8‐controlled UV‐B signaling pathways in Arabidopsis thaliana . Plant J 67: 37–48 [DOI] [PubMed] [Google Scholar]
- Gao YQ, Chen JG, Chen ZR, An D, Lv QY, Han ML, Wang YL, Salt DE, Chao DY (2017) A new vesicle trafficking regulator CTL1 plays a crucial role in ion homeostasis. PLoS Biol 15: e2002978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Peer W, Robert S, Swarup R, Ye S, Prigge M, Cohen JD, Friml J, Murphy A, Tang D et al (2010) Arabidopsis ROOT UVB SENSITIVE2/WEAK AUXIN RESPONSE1 is required for polar auxin transport. Plant Cell 22: 1749–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen K, Kang C, Paalman R, Keuskamp D, Hayes S, Pierik R (2018) Far‐red light detection in the shoot regulates lateral root development through the HY5 transcription factor. Plant Cell 30: 101–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)‐dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R (2010) Negative feedback regulation of UV‐B‐induced photomorphogenesis and stress acclimation in Arabidopsis . Proc Natl Acad Sci USA 107: 20132–20137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Velanis CN, Jenkins GI, Franklin KA (2014) UV‐B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc Natl Acad Sci USA 111: 11894–11899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Sharma A, Fraser DP, Trevisan M, Cragg‐Barber CK, Tavridou E, Fankhauser C, Jenkins GI, Franklin KA (2017) UV‐B perceived by the UVR8 photoreceptor inhibits plant thermomorphogenesis. Curr Biol 27: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M, Ulm R (2012) UV‐B photoreceptor‐mediated signalling in plants. Trends Plant Sci 17: 230–237 [DOI] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Yang P, Lau OS, Li G, Li J, Chen H, Deng XW (2012) Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV‐B light. Plant Cell 24: 4590–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Yang P, Ouyang X, Chen L, Deng XW (2014) Photoactivated UVR8‐COP1 module determines photomorphogenic UV‐B signaling output in Arabidopsis . PLoS Genet 10: e1004218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI (2014) Structure and function of the UV‐B photoreceptor UVR8. Curr Opin Struct Biol 29: 52–57 [DOI] [PubMed] [Google Scholar]
- Kaiserli E, Jenkins GI (2007) UV‐B promotes rapid nuclear translocation of the Arabidopsis UV‐B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leasure CD, Tong H, Yuen G, Hou X, Sun X, He ZH (2009) ROOT UV‐B SENSITIVE2 acts with ROOT UV‐B SENSITIVE1 in a root ultraviolet B‐sensing pathway. Plant Physiol 150: 1902–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Ha JH, Kim SG, Choi HK, Kim ZH, Han YJ, Kim JI, Oh Y, Fragoso V, Shin K et al (2016) Stem‐piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots. Sci Signal 9: ra106 [DOI] [PubMed] [Google Scholar]
- Leng N, Dawson J, Thomson J, Ruotti V, Rissman A, Smits B, Haag J, Gould M, Stewart R, Kendziorski C (2013) EBSeq: an empirical Bayes hierarchical model for inference in RNA‐seq experiments. Bioinformatics 29: 1035–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O (2018) Auxin signaling. Plant Physiol 176: 465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Newey C (2011) RESM: accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinformatics 12: 323–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Mei S, Shi C, Yang Y, Peng Y, Ma L, Wang F, Li X, Huang X, Yin Y et al (2018) UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis . Dev Cell 44: 512–523.e5 [DOI] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis . Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Liu Y, Li X, Li K, Liu H, Lin C (2013) Multiple bHLH proteins form heterodimers to mediate CRY2‐dependent regulation of flowering‐time in Arabidopsis . PLoS Genet 9: e1003861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H (2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature‐mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci USA 113: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Mate Z, Brzezinska A, Molinier J, Oakeley EJ, Adam E, Schafer E, Nagy F, Ulm R (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV‐B response in Arabidopsis . Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Mao W, Liu Y, Ren H, Lau OS, Ouyang X, Huang X (2016) Dual‐source nuclear monomers of UV‐B light receptor direct photomorphogenesis in Arabidopsis . Mol Plant 9: 1671–1674 [DOI] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O'Hara A, Kaiserli E, Baumeister R, Schafer E, Nagy F, Jenkins GI et al (2011) Perception of UV‐B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Sassi M, Lu Y, Zhang Y, Wang J, Dhonukshe P, Blilou I, Dai M, Li J, Gong X, Jaillais Y et al (2012) COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1‐ and PIN2‐dependent auxin transport in Arabidopsis . Development 139: 3402–3412 [DOI] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP (2007) The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3‐MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R (2010) The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet‐B radiation. Plant, Cell Environ 33: 88–103 [DOI] [PubMed] [Google Scholar]
- Sun Q, Yoda K, Suzuki H (2005) Internal axial light conduction in the stems and roots of herbaceous plants. J Exp Bot 56: 191–203 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004) MASSUGU2 encodes Aux/IAA19, an auxin‐regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana . Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook K, Arongaus AB, Binkert M, Heijde M, Yin R, Ulm R (2013) The UVR8 UV‐B photoreceptor: perception, signaling and response. Arabidopsis Book 11: e0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Leasure CD, Hou X, Yuen G, Briggs W, He ZH (2008) Role of root UV‐B sensing in Arabidopsis early seedling development. Proc Natl Acad Sci USA 105: 21039–21044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schafer E, Nagy F (2004) Genome‐wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV‐B response of Arabidopsis . Proc Natl Acad Sci USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Tilbrook K, Fierro AC, Marchal K, Poelman D, Van Der Straeten D, Ulm R (2014) Photoreceptor‐mediated bending towards UV‐B in Arabidopsis . Mol Plant 7: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Vanhaelewyn L, Prinsen E, Van Der Straeten D, Vandenbussche F (2016) Hormone‐controlled UV‐B responses in plants. J Exp Bot 67: 4469–4482 [DOI] [PubMed] [Google Scholar]
- Wan J, Zhang P, Wang R, Sun L, Wang W, Zhou H, Xu J (2018) UV‐B radiation induces root bending through the flavonoid‐mediated auxin pathway in Arabidopsis . Front Plant Sci 9: 618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lu X, Li L, Lian HL, Mao ZL, Xu PB, Guo T, Xu F, Du SS, Cao XL et al (2018) Photoexcited CRYPTOCHROME 1 interacts with dephosphorylated BES1 to regulate brassinosteroid signaling and photomorphogenesis in Arabidopsis . Plant Cell 30: 1989–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargent JJ, Gegas VC, Jenkins GI, Doonan JH, Paul ND (2009) UVR8 in Arabidopsis thaliana regulates multiple aspects of cellular differentiation during leaf development in response to ultraviolet B radiation. New Phytol 183: 315–326 [DOI] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jager KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jurgens G (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24: 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Wagner D (2016) Transcriptional responses to the Auxin hormone. Annu Rev Plant Biol 67: 539–574 [DOI] [PubMed] [Google Scholar]
- Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Zhang J, Yang P, Deng H, Wang J et al (2012) Structural basis of ultraviolet‐B perception by UVR8. Nature 484: 214–219 [DOI] [PubMed] [Google Scholar]
- Xu F, He S, Zhang J, Mao Z, Wang W, Li T, Hua J, Du S, Xu P, Li L et al (2018) Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit Auxin signaling in Arabidopsis . Mol Plant 11: 523–541 [DOI] [PubMed] [Google Scholar]
- Yang C, Xie F, Jiang Y, Li Z, Huang X, Li L (2018a) Phytochrome a negatively regulates the shade avoidance response by increasing Auxin/Indole acidic acid protein stability. Dev Cell 44: 29–41.e4 [DOI] [PubMed] [Google Scholar]
- Yang Y, Liang T, Zhang L, Shao K, Gu X, Shang R, Shi N, Li X, Zhang P, Liu H (2018b) UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis . Nat Plants 4: 98–107 [DOI] [PubMed] [Google Scholar]
- Yi C, Deng XW (2005) COP1 – from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Yin R, Skvortsova MY, Loubery S, Ulm R (2016) COP1 is required for UV‐B‐induced nuclear accumulation of the UVR8 photoreceptor. Proc Natl Acad Sci USA 113: E4415–E4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Karampelias M, Robert S, Peer WA, Swarup R, Ye S, Ge L, Cohen J, Murphy A, Friml J et al (2013) ROOT ULTRAVIOLET B‐SENSITIVE1/weak auxin response3 is essential for polar auxin transport in Arabidopsis . Plant Physiol 162: 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J (2001) Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci USA 98: 11795–11800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xing L, Wang X, Hou YJ, Gao J, Wang P, Duan CG, Zhu X, Zhu JK (2014) The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77‐dependent transcription of auxin‐responsive genes. Sci Signal 7: ra53 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Source Data for Figure 5
Source Data for Figure 7


