Abstract
Plants establish mutualistic associations with beneficial microbes while deploying the immune system to defend against pathogenic ones. Little is known about the interplay between mutualism and immunity and the mediator molecules enabling such crosstalk. Here, we show that plants respond differentially to a volatile bacterial compound through integral modulation of the immune system and the phosphate‐starvation response (PSR) system, resulting in either mutualism or immunity. We found that exposure of Arabidopsis thaliana to a known plant growth‐promoting rhizobacterium can unexpectedly have either beneficial or deleterious effects to plants. The beneficial‐to‐deleterious transition is dependent on availability of phosphate to the plants and is mediated by diacetyl, a bacterial volatile compound. Under phosphate‐sufficient conditions, diacetyl partially suppresses plant production of reactive oxygen species (ROS) and enhances symbiont colonization without compromising disease resistance. Under phosphate‐deficient conditions, diacetyl enhances phytohormone‐mediated immunity and consequently causes plant hyper‐sensitivity to phosphate deficiency. Therefore, diacetyl affects the type of relation between plant hosts and certain rhizobacteria in a way that depends on the plant's phosphate‐starvation response system and phytohormone‐mediated immunity.
Keywords: diacetyl, immunity, mutualism, phosphate, plant–bacteria interactions
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Plant Biology
A volatile compound produced by a plant growth‐promoting rhizobacterium differentially affects plant immunity and plant response to symbiont colonization depending on phosphate availability.

Introduction
Plants naturally live with a diversity of microbes in the rhizosphere and the phyllosphere. Some microbes are considered as beneficial due to their capacities of increasing plant biomass and/or stress tolerance (Lugtenberg & Kamilova, 2009; Pieterse et al, 2014). In return, plants provide nutrient‐rich root exudates and niches for beneficial microbes, leading to mutualistic symbiosis (Sasse et al, 2008). Plants activate immune responses including production of reactive oxygen species (ROS) to combat pathogens (Yan & Dong, 2014; Couto & Zipfel, 2016; Wu et al, 2018). When encountering beneficial microbes, a balanced regulation of plant immunity is presumably required for mutualistic associations. Plant immune responses can be triggered by pathogen‐associated molecular patterns (PAMPs) such as bacterial flagellin; similarly, establishment of mutualistic symbiosis involves plant perception of microbial symbiotic signals, as demonstrated by symbiosis with arbuscular mycorrhizal fungi or nodule‐forming rhizobia (Cao et al, 2017; Martin et al, 2017; Zipfel & Oldroyd, 2017). Nonetheless, the interplay between mutualism and immunity is unclear. The establishment of Arabidopsis symbiosis with Colletotrichum tofieldiae, an endophytic fungus that can transfer phosphate to its host, requires phosphate (Pi) deficiency in the plant (Hacquard et al, 2016; Hiruma et al, 2016). In addition, the master regulators of phosphate‐starvation response (PSR) in Arabidopsis not only positively regulate PSR but also suppress plant immunity, and thereby influence root microbiome (Castrillo et al, 2017). It is thus intriguing whether plant mutualistic associations with beneficial soil microbes commonly prefer Pi deficiency.
Bacillus amyloliquefaciens strain GB03 and its microbial volatiles (hereafter referred to as GMVs) are recognized as beneficial to plants both in soil and in artificial medium. GMVs were shown to modulate plant hormone homeostasis and nutrient uptake (Ryu et al, 2003; Zhang et al, 2007, 2009; Paré et al, 2011; Beauregard et al, 2013); however, it remains unclear how plant biological processes were integrated by the microbial factors to produce multiple beneficial traits (Paré et al, 2011; Liu & Zhang, 2015). In this study, we show that Arabidopsis allows mutualistic association with B. amyloliquefaciens GB03 only under the Pi‐sufficient condition, whereas Pi‐deficient plants strongly activate immunity in response to the same bacterium. Our investigation further identified a bacterial volatile compound that influences the plant decision on mutualism or immunity. Our findings not only demonstrate that bacterial factor‐triggered modulation of the immune system and the PSR system in plants determines the relationship between the two organisms, but also provide an example where plants use different strategies for bacteria and fungi in determining mutualism or immunity.
Results
A plant abiotic stress condition disclosed a mutualism‐to‐pathogenicity transition
We were initially interested in studying whether GB03 would relieve plant stress caused by simultaneous deficiency of multiple nutrients. In order to do this, we grew seedlings of Arabidopsis thaliana in petri dishes containing 1/2‐strength and 1/20‐strength Murashige and Skoog medium as the nutrient‐sufficient and nutrient‐deficient medium, respectively. The petri dishes contained plastic partitions which separated different medium and also separated plants from the bacteria, so that the bacteria could influence plants only through volatile emissions (Fig 1A). In such conditions, we unexpectedly observed deleterious effects of GMVs on Arabidopsis grown in nutrient‐deficient medium, while the same GMVs promoted growth of plants supplemented with sufficient nutrients (Figs 1A and EV1A). Under nutrient‐deficient conditions, Arabidopsis not only lose GMV‐induced plant growth promotion (Figs 1B and EV1B), but also clearly displayed stress symptoms, including impaired photosynthesis (Fig 1C), increased leaf cell death (Fig 1D), strong accumulation of anthocyanin (Fig EV1C), and hyper‐induction or reduction of genes known to be up‐ or down‐regulated, respectively, by environmental stress (Fig EV1D). Thus, GMVs can be either beneficial or deleterious to plants, although GB03 has been recognized as a representative plant mutualistic bacterium (Paré et al, 2011; Choi et al, 2014).
Figure 1. A plant abiotic stress condition disclosed a mutualism‐to‐pathogenicity transition.
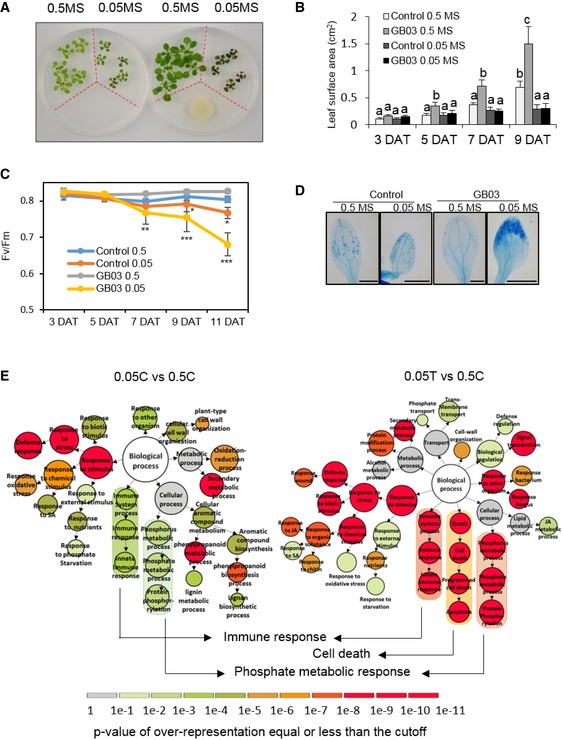
-
AThe same MVs from GB03 caused opposed impacts on plants grown in different medium. The petri dishes contain plastic partitions (red dotted lines) that separate different medium. 0.5 MS and 0.05 MS indicate 1/2‐strength (nutrient‐sufficient) and 1/20‐strength (nutrient‐deficient) Murashige and Skoog medium. Images were taken at 11 days after treatments (DAT).
-
BQuantification of total leaf area per plant (square centimeter, cm2). Values correspond to the means ± SE of three biological replicates. Different letters denote significant differences at P < 0.05, Tukey's multiple comparison test within each group of the same DAT.
-
CEffective quantum yield of photosystem II in plants. n = 3 biological replicates, mean ± SE. Asterisks denote significant differences at *P < 0.05, **P < 0.01, and ***P < 0.001, Dunnett's multiple comparison test (comparing with the control) within each group of the same DAT
-
DCell‐death visualization by trypan blue staining of 11 DAT leaves. Scale bar = 1 mm.
-
EGene Ontology (GO) comparative analysis of Arabidopsis genes that were induced at 5 DAT by nutrient deficiency alone (0.05C vs. 0.5C) and that were induced by the nutrient deficiency plus GMVs (0.05T vs. 0.5C). Diagrams are designed based on VirtualPlant platform. The size of circles represents the number of genes in each GO category. Scale color bar indicates the P‐value cutoff of over‐representation equal or less than the cutoff for each GO category. Darker color indicates higher possibility of each GO category. DEG lists for key terms are provided in Tables EV1 and EV2.
Figure EV1. Nutrient availability determines Arabidopsis thaliana responses to Bacillus amyloliquefaciens GB03 (related to Fig 1).
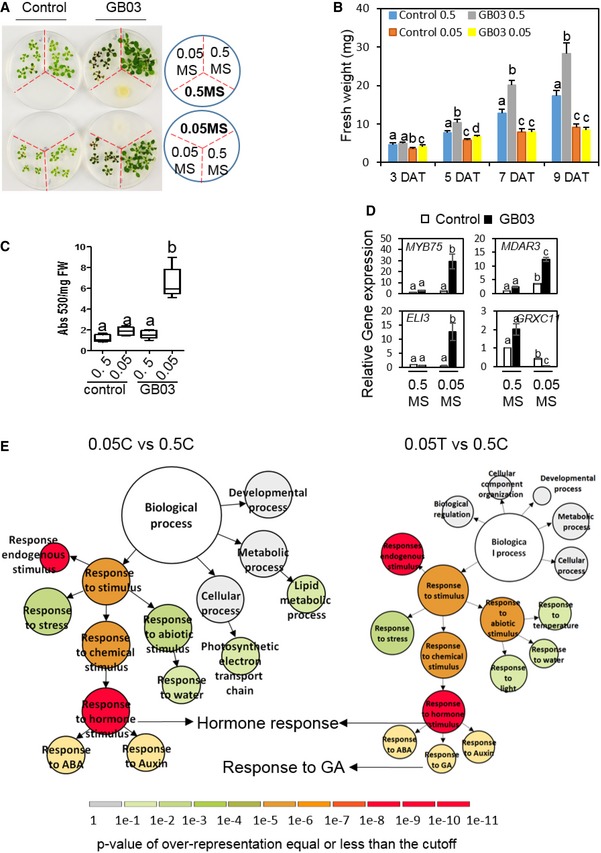
-
AExposure to MVs from GB03, which was either grown on the nutrient‐sufficient (0.5 MS) or grown on the nutrient‐deficient (0.05 MS) medium, caused opposed impacts on plants grown in different medium. The petri dishes contain plastic partitions (red dotted lines) that separate different medium.
-
BQuantification of fresh weight of seedlings grown under different treatment conditions. Values correspond to the means ± SE of three biological replicates. Different letters denote significant differences at P < 0.05, Tukey's multiple comparison test within each group of the same DAT.
-
CAnthocyanin accumulation levels in plants at 11 DAT. The boxplots show representative data from three independent experiments (n = 9). Whiskers represent the min to max data range, and the median is represented by the central horizontal line. The upper and lower limits of the box outline represent the first and third quartiles. Different letters denote significantly different means at P < 0.05, Tukey's multiple comparison test.
-
DRelative gene expression levels of Arabidopsis ELI3, MYB75, MDAR3, and GRXC11, which are indicative of environmental stress conditions (Somssich et al, 1996; Teng et al, 2005; Li et al, 2010; Mehterova et al, 2012), in 5 DAT seedlings grown in 0.5 and 0.05 MS medium with or without exposure to GMVs. Values are means ± SE of three biological replicates. Different letters denote significantly different means at P < 0.05, Tukey's multiple comparison test.
-
EGene Ontology (GO) comparative analysis of Arabidopsis genes that were repressed at 5 DAT by nutrient deficiency (0.05C vs. 0.5C) alone and that were repressed by the nutrient deficiency plus GMVs (0.05T vs. 0.5C). Diagrams are designed based on VirtualPlant platform. The size of circles represents the number of genes in each GO category. Scale color bar indicates the P‐value cutoff of over‐representation equal or less than the cutoff for each GO category. Darker color indicates higher possibility of each GO category. DEG lists for key terms are provided in Table EV3.
To understand how GMVs exacerbated the stress in plants that are under nutrient deficiency, we compared the transcriptomes of Arabidopsis with and without GMV treatment under nutrient‐deficient conditions. Gene Ontology (GO) analysis of RNAseq results revealed that genes induced by nutrient deficiency were enriched in immune response and phosphate metabolic response processes, and that these patterns were strongly intensified by GMVs (Fig 1E; Appendix Fig S1C; Tables EV1 and EV4). Compared with nutrient deficiency alone, nutrient deficiency with GMV treatment also additionally induced cell‐death genes in plants (Fig 1E; Table EV2). These results indicate that GMV‐induced stress in Arabidopsis is mediated through microbial regulation of plant immunity and phosphate homeostasis. Meanwhile, genes that were repressed by nutrient deficiency were enriched in hormone response processes, among which genes responsive to gibberellic acid (GA) were repressed only in nutrient‐deficient plants with GMV treatment (Fig EV1E; Table EV3), suggesting that GMVs inhibit GA‐mediated plant growth. Under nutrient‐sufficient conditions, GMVs induced genes related to cell wall organization and photosynthesis (Appendix Fig S1A; Table EV4), consistent with previous reports that GMVs induced leaf cell expansion and enhanced photosynthesis efficiency of Arabidopsis grown in artificial medium (Zhang et al, 2007, 2008). Interestingly, in contrast to GMV‐activated immunity in nutrient‐deficient plants, GMV treatment to nutrient‐sufficient plants repressed immunity‐related genes involved in plant responses to fungus, bacteria, chitin, jasmonic acid (JA), and salicylic acid (SA) (Appendix Fig S1B; Table EV4). In nutrient‐deficient condition, GMVs induced phosphate starvation, jasmonic acid signaling, and responses triggered by lipid metabolism (Appendix Fig S1C; Table EV4) and repress responses to reactive oxygen species and primary root development (Appendix Fig S1D; Table EV4). Altogether, the transcriptome results suggest that plant immunity and Pi homeostasis are strongly correlated with GMV‐induced plant vigor or stress.
Phosphate availability determines Arabidopsis responses to GMVs
Because Pi homeostasis was highlighted in the transcriptional analysis of nutrient‐deficient plants, we wondered whether GMVs affect the plant PSR. As shown in the RNAseq results, while nutrient deficiency induced most of the Pi homeostasis genes in Arabidopsis, these genes were further induced by GMVs (Fig EV2A; Table EV5), demonstrating that GMVs caused plant hyper‐activation of the PSR. This conclusion was confirmed by organ‐specific measurements of PSR gene expression, which showed that PSR gene hyper‐induction in both shoots and roots was generally observable starting at 5 days after treatments (Figs 2A, and EV2B and C). GMV‐induced hyper‐PSR was further supported by the observations that, in nutrient‐deficient plants, GMVs increased the activity of root acid phosphatases and the accumulation of the microRNA miR399 (Figs 2B and EV2D), which are also known to be induced by Pi deficiency in Arabidopsis (Fujii et al, 2005; Zhang et al, 2014b). Therefore, exposure to GMVs resulted in hyper‐stimulated PSR in the nutrient‐deficient plants.
Figure EV2. Phosphate availability determines Arabidopsis responses to GMVs (related to Fig 2).
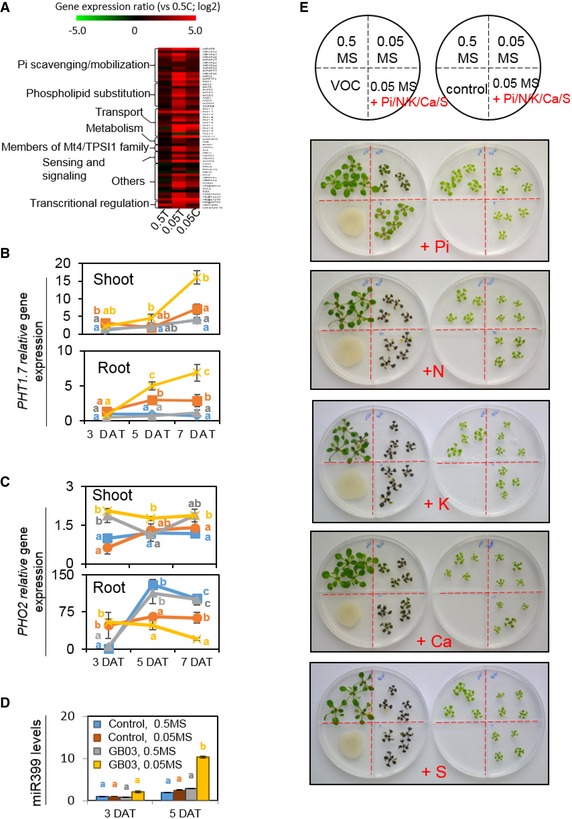
-
AA heatmap of RNAseq results showing the expression levels of Pi homeostasis genes in Arabidopsis grown under different conditions, including 0.5T (0.5 MS medium with GMV treatment), 0.05T (0.05 MS medium with GMV treatment), and 0.05C (0.05 MS medium without GMV treatment). Color scale indicates fold changes (log2) compared with gene expression in plants grown in 0.5C (0.5 MS medium without GMV treatment). DEG lists are provided in Table EV5.
-
B, CGMVs caused plant hyper‐sensitivity to Pi deficiency, as shown by GMV‐dependent hyper‐induction of PHT1.7 (B) and hyper‐suppression of PHO2. (C) Data points indicate mean ± SE (n = 3). Different letters denote significantly different means at P < 0.05, Tukey's multiple comparison test within each group of the same DAT.
-
DThe accumulation level of miR399 was strongly elevated by GMVs in nutrient‐deficient plants. Values are means ± SE of three biological replicates. Different letters denote significantly different means at P < 0.05, Tukey's multiple comparison test within each group of the same DAT.
-
EDifferent macronutrients including phosphorus (P), nitrogen (N), potassium (K), calcium (Ca), and sulfur (S) were supplemented individually to the 0.05 MS medium, in order to bring the corresponding nutrient content to a level that is equal to that in the 0.5 MS medium. The petri dishes contain plastic partitions (red dotted lines) that separate different medium. A scheme showing medium partitions is shown on the top right corner.
Figure 2. Phosphate availability determines Arabidopsis responses to GMVs.
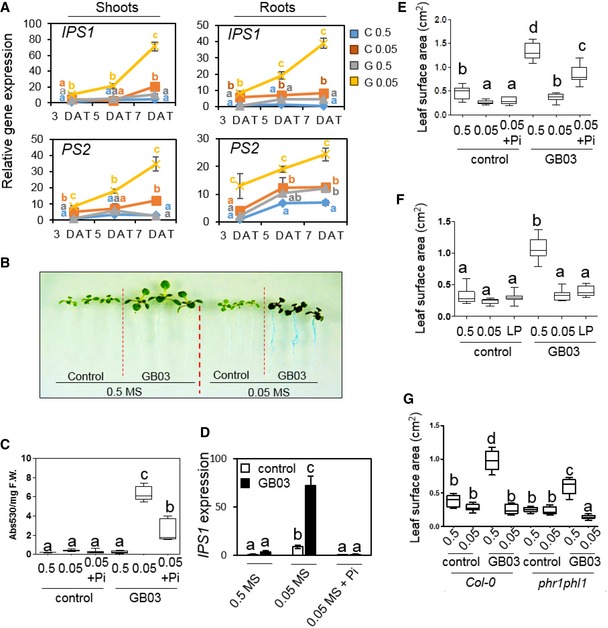
-
APlants grown with 0.05 MS and GMVs (G 0.05) showed hyper‐induction of IPS1 and PS2 gene expression in both shoots and roots, compared to plants grown with 0.05 MS alone (C 0.05). Values are means ± SE of three biological replicates.
-
BRoot acid phosphatase activity as detected by the blue color from the 5‐bromo‐4‐chloro‐3‐indolyl phosphate p‐toluidine salt (BCIP) treatment. Image contrast enhanced for improved visualization of the blue color in roots.
-
C, DSupplementation of Pi to the nutrient‐deficient plants (0.05 + Pi) significantly reduced hyper‐accumulation of anthocyanin (C) and hyper‐induction of IPS1 gene expression (D) triggered by GMVs.
-
EPi supplementation to the 0.05 MS medium partially restored GMV‐induced plant vigor, as indicated by increases in total leaf area per plant.
-
FA low Pi level (LP), which was equal to 1/20 of that in the 0.5 MS medium, in plant growth medium blocked GMV‐induced plant vigor.
-
GThe Arabidopsis phr1phl1 mutant showed substantially decreased growth promotion, compared with the wild‐type plants.
We next tested whether Pi was the main nutrient that determined the transition between GMV‐triggered plant vigor and stress in conditions of nutrient shortage. Phosphate supplementation to nutrient‐deficient plants blocked GMV‐dependent hyper‐activation of PSR genes and substantially reduced anthocyanin accumulation (Figs 2C and D, and EV2E), indicating that GMV‐induced stress in the nutrient‐deficient plants mainly resulted from Pi deficiency. In contrast, supplementation of several other nutrients including nitrogen (N), potassium (K), sulfur (S), and calcium (Ca) to the nutrient‐deficient medium did not decrease GMV‐induced stress symptoms in plants (Fig EV2E). Thus, in the medium with simultaneous deficiency of multiple nutrients, Pi deficiency is the major reason for GMV‐induced plant stress. Consistently, when plants were grown in the medium that was only deficient in Pi but not in the other nutrients, they showed GMV‐induced strong accumulation of anthocyanin and hyper‐activation of PSR genes (Appendix Fig S2A–C), demonstrating that plant Pi deficiency is sufficient to disclose the deleterious effects of GMVs. In addition, GMVs failed to trigger growth promotion in plants with Pi deficiency, whereas Pi supplementation to the nutrient‐deficient medium substantially restored GMV‐induced plant growth promotion (Fig 2E and F). Moreover, GMV‐induced plant growth promotion and stress were both impaired by mutation of PHR1 and PHL1 (Fig 2G; Appendix Fig S2D–F), which are important positive regulators of PSR in Arabidopsis (Castrillo et al, 2017). Together, these results demonstrate that the PSR system plays a key role in plant responses to GMVs. Importantly, these results reveal that phosphorus, instead of the other plant macronutrients, has a uniquely influential role in plant responses to microbes.
GMV‐triggered plant hyper‐sensitivity to Pi deficiency is mediated by diacetyl
GMVs include over 30 compounds as detected by chromatography coupled with mass spectrometry (GC‐MS), with varying individual abundance ranging from 0.6 ng per 24 h to 614 μg per 24 h (Farag et al, 2006). We examined plant responses to individual GMVs by using seven compounds, which are major GMV components and are commercially available. The compounds were applied by direct supplementation to the plant growth medium or by airborne transmission only (Appendix Table S1). Among the examined GMVs, only diacetyl (DA) induced anthocyanin hyper‐accumulation and PSR gene hyper‐activation in plants with Pi deficiency (Fig 3A–C; Appendix Fig S3A–C); in contrast, these stress patterns were not observed during treatments with the other examined GMVs, including acetoin and 2,3‐butanediol, which are structurally similar to DA and are produced by GB03 in greater abundance than DA (Fig 3A–C; Appendix Fig S3A and B). DA‐induced transcriptional activation of the anthocyanin biosynthesis pathway was observable at 5 DAT (Appendix Fig S3D), which is consistent with anthocyanin hyper‐accumulation quantitated later on at 10 DAT. Although PSR genes were hyper‐induced by DA at 5 DAT, DA does not further decrease plant soluble Pi contents in Pi‐deficient plants, as shown by plants collected at 3 and 5 DAT (Appendix Fig S3E). Thus, it appears that DA‐induced hyper‐PSR results from an interruption in the PSR system instead of from defective Pi uptake. Similar to the observations using the natural GMVs, DA‐induced anthocyanin hyper‐accumulation and PSR gene hyper‐induction were both impaired in the phr1phl1 mutant (Fig 3D and E; Appendix Fig S3F). Collectively, these results indicate that DA mediates GMV‐triggered plant hyper‐sensitivity to Pi deficiency.
Figure 3. GMV‐triggered plant hyper‐sensitivity to Pi deficiency is mediated by diacetyl.
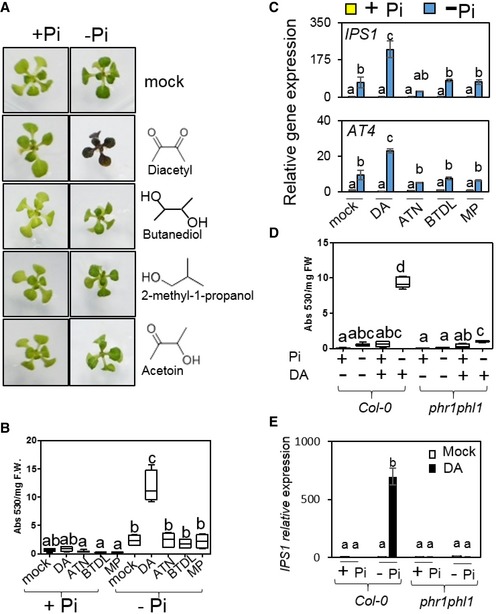
-
AImages of Arabidopsis plants exposed to four individual GMV components, including diacetyl (DA), 2,3‐butanediol (BTDL), 2‐methyl‐1‐propanol (MP), and acetoin (ATN). The compounds were applied at dosages that, when the compounds totally evaporate from the agar‐containing solid droplets, would yield in volatile concentrations of 9.7 μg (DA), 32.5 μg (BTDL), 7.9 μg (MP), and 28.5 μg (ATN) per ml free space in the petri dish, which resembled the 1:3.3:0.8:2.9 ratio among the four GMV components in natural GMVs.
-
B, CAnthocyanin hyper‐accumulation (B) and PSR gene hyper‐induction (C) were observed in Pi‐deficient plants exposed to DA but not to the other three GMV components.
-
D, ECompared with the wild‐type plants, phr1phl1 showed substantially decreased anthocyanin hyper‐accumulation (D) and PSR gene hyper‐induction (E) triggered by DA.
DA exacerbates plant sensitivity to Pi deficiency via activation of immunity
To understand why DA has a deleterious effect when the plants are under Pi deficiency, we investigated its influence on plant immunity, because transcriptome analyses of Pi‐deficient plants treated with GMVs had implied a potential link between up‐regulated immunity and GMV‐induced stress (Fig 1E). In plants with Pi deficiency, SA biosynthesis and response genes, such as ICS1 and PAD4, were strongly induced by DA, while this pattern was not observed in plants with sufficient Pi (Fig 4A). Consistently, Pi‐deficient plants with DA treatment showed 81% increase in SA levels compared with their untreated counterparts, whereas plants with sufficient Pi showed similar SA levels independent of DA treatment (Fig 4B). These results indicate that DA causes an enhancement of SA biosynthesis and related signaling in Pi‐deficient plants.
Figure 4. DA exacerbates plant sensitivity to Pi deficiency via activation of immunity.
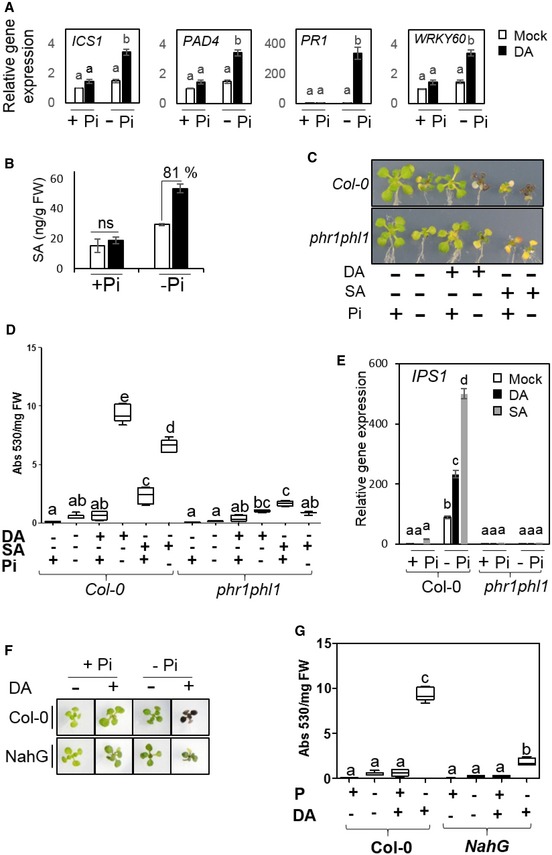
-
ADA induces genes involved in SA biosynthesis and signaling in Arabidopsis grown with Pi deficiency.
-
BDA elevates SA accumulation levels in Pi‐deficient Arabidopsis. Means ± SE of three biological replicates.
-
CImages of plants grown with different treatments using DA (9.7 μg/ml free space) or SA (100 μM in plant growth medium).
-
D, EExogenous application of SA mimics DA‐induced anthocyanin accumulation (D) and IPS1 gene induction (E) patterns in Pi‐deficient plants.
-
F, GCompared with the wild‐type plants, the NahG transgenic plants showed altered responses to DA under Pi‐deficiency condition, as shown by plant images (F) and quantification of anthocyanin accumulation levels (G).
We subsequently investigated whether the enhanced immune responses contribute to DA‐triggered hyper‐sensitivity to Pi deficiency. Exogenous application of SA mimics DA in triggering hyper‐PSR in Pi‐deficient plants, as shown by SA‐induced over‐accumulation of anthocyanin and hyper‐induction of PSR genes (Fig 4C–E; Appendix Fig S4A and B). Also, similar to the observations with DA, SA‐induced over‐accumulation of anthocyanin and hyper‐induction of PSR genes were both substantially impaired in the phr1phl1 mutant compared with wild‐type plants (Fig 4C–E; Appendix Fig S4C). These results indicate that the activation of SA signaling is sufficient to trigger hyper‐PSR in Arabidopsis. In addition, SA accumulation is crucial for DA‐triggered plant hyper‐sensitivity to Pi deficiency, because DA‐induced over‐accumulation of anthocyanin was drastically reduced in transgenic Arabidopsis expressing NahG (Fig 4F and G), which is defective in SA accumulation (van Wees & Glazebrook, 2003). Interestingly, in response to Pi deficiency, PSR gene expression levels in NahG plants were higher than the wild‐type plants and were not further increased by DA (Appendix Fig S4D), which seems to indicate that any strong disruption of SA homeostasis would enhance PSR gene induction.
Because Pi deficiency activates JA‐responsive genes (Khan et al, 2016), we investigated potential involvement of JA in DA‐induced hyper‐PSR. DA not only strongly augmented Pi deficiency‐induced JA biosynthesis and JA‐responsive genes (Fig EV3A), but also caused a 52% increase in JA levels in the Pi‐deficient plants (Fig EV3B). Similar to the effects of SA, exogenous application of methyl jasmonate (MeJA) to Pi‐deficient plants mimicked DA induction of plant hyper‐PSR, and the MeJA‐induced hyper‐PSR also required PHR1/PHL1 (Fig EV3C–E). Thus, in addition to activation of the SA pathway, activation of the JA pathway also contributes to DA‐induced hyper‐PSR in Arabidopsis. However, by contrast to NahG that showed an 80% reduction in DA‐induced anthocyanin levels, the Arabidopsis mutant coi1, which is defective in the JA signaling pathway (Xu et al, 2002), showed a 48% reduction in the anthocyanin level compared with wild‐type plants (Fig EV3F and G). Therefore, it appears that SA plays a more prominent role than JA does in mediating DA‐induced hyper‐PSR in Pi‐deficient plants.
Figure EV3. Deleterious effects caused by DA are due to an activation of immunity in Pi‐deficient plants (related to Fig 4).
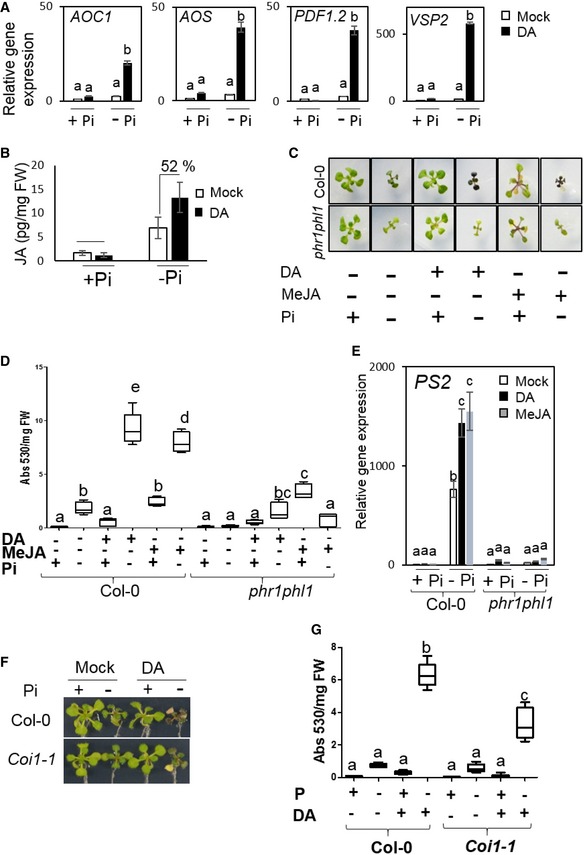
-
ADA induces genes involved in JA biosynthesis and signaling in Arabidopsis grown with Pi deficiency.
-
BDA elevates JA accumulation levels in Pi‐deficient Arabidopsis. Means ± SE of three biological replicates.
-
CImages of plants grown with different treatments using DA (9.7 μg/ml free space) or MeJA (25 μM in plant growth medium).
-
D, EExogenous application of MeJA mimics DA‐induced anthocyanin accumulation (D) and PS2 gene induction (E) patterns in Pi‐deficient plants.
-
F, GCompared with the wild‐type plants, the JA‐insensitive coi1 mutant plants showed altered responses to DA under Pi‐deficiency condition, as shown by plant images (F) and quantification of anthocyanin accumulation levels (G).
DA enhances symbiotic colonization to Pi‐sufficient plants via modulation of immunity
We wondered whether DA, in addition to being deleterious to Pi‐deficient plants, plays additional role in the mutualism between GB03 and the plants with sufficient Pi. Unlike the natural GMVs, DA alone does not trigger growth promotion in plants irrespective to Pi availability (Fig EV4A). However, transcriptome comparison suggested that, in plants grown with sufficient Pi, DA may cause a suppression of plant immunity, because a group of 51 immune‐related genes were identified as DA‐dependent DEGs and 86% of these DEGs were repressed by DA within 48 h (Figs 5A and EV4B; Table EV6). The DA‐repressed immunity genes include NDR1, which is pathogen‐inducible and is required for disease resistance in Arabidopsis (Century et al, 1997). In line with the decreased immune gene expression, DA significantly suppresses Arabidopsis ROS burst induced by microbial PAMPs (Figs 5B and EV4C), such as the immune elicitors flg22 or elf18 (Felix et al, 1999; Kunze et al, 2004). In contrast, PAMP‐induced ROS production was not suppressed by acetoin or 2,3‐butanediol (Fig EV4D), which are two other GMV components that have similar chemical structures to DA. These results suggest that DA helps establishing GB03 association with plants through the modulation of plant immunity.
Figure EV4. DA suppresses plant ROS burst but not PTI gene expression in response to flg22 (related to Fig 5).
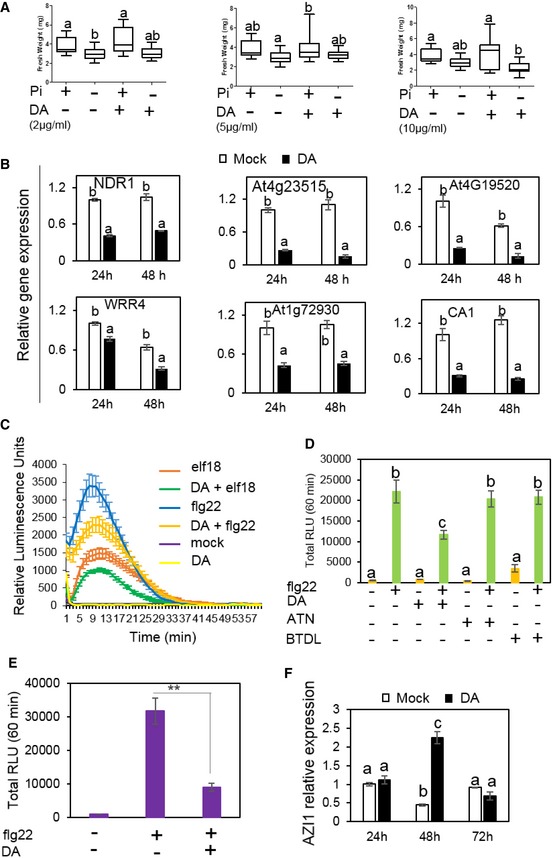
-
AFresh weights measured in plants grown with or without DA treatments. Different letters denote significantly different means at P < 0.05, Tukey's multiple comparison test. The boxplots show representative data from three independent experiments (n = 15). Whiskers represent the min to max data range, and the median is represented by the central horizontal line. The upper and lower limits of the box outline represent the first and third quartiles.
-
BTranscriptional suppression of some immune response‐related genes by DA. qPCR results show values of means ± SE (n = 3), and two biological replicates were analyzed with similar results. Different letters denote significantly different means at P < 0.05, Tukey's multiple comparison test.
-
CDA suppresses plant ROS burst induced by flg22 or elf18. ROS accumulation was represented in relative luminescence units (RLU). Data point indicates mean ± SE (n = 24). Three independent experiments were performed, and similar results were observed. **P < 0.01, Student's t‐test.
-
DDA, but not 2,3‐butanediol (BTDL) or acetoin (ATN), suppresses plant ROS burst induced by flg22 or elf18. Three independent experiments were performed with similar results. Data bar indicates mean ± SE (n = 24) for 60 min. Different letters denote significantly different means at P < 0.05, Tukey's multiple comparison test.
-
EDA suppresses plant ROS burst induced by flg22 in P‐deficient grown plants. Data bar indicates mean ± SE (n = 24) for 60 min. Three independent experiments were performed with similar results. **P < 0.01, Student's t‐test.
-
FDA induces gene expression of AZI1 in P‐deficient grown Arabidopsis plant. Different letters denote significantly different means at P < 0.05, Tukey's multiple comparison test. qPCR data indicate mean ± SE (n = 3), and two biological replicates were analyzed with similar results.
Figure 5. DA enhances symbiotic colonization to Pi‐sufficient plants via modulation of immunity.
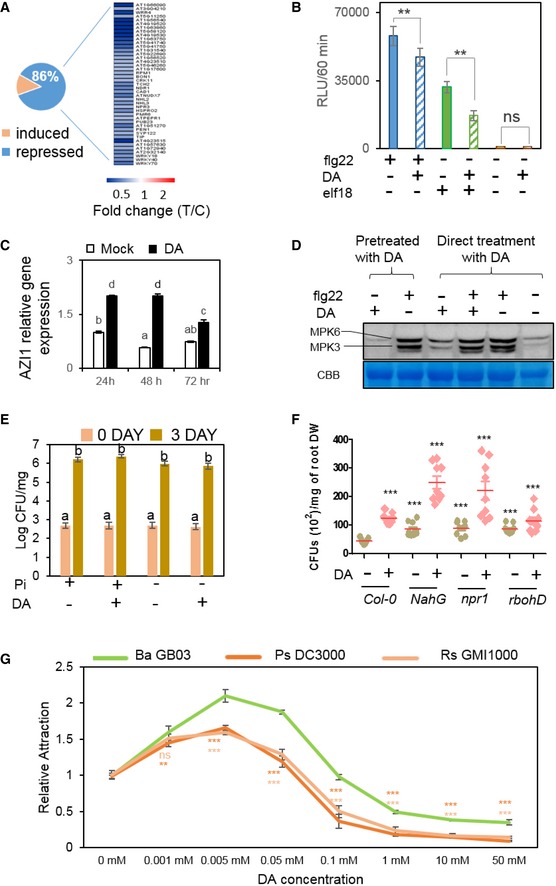
-
AClustering of DA‐regulated immunity‐related genes in Arabidopsis with sufficient Pi, as revealed by RNAseq analysis.
-
BDA suppresses plant ROS burst induced by flg22 or elf18. ROS accumulation was represented in relative luminescence units (RLU). Data bar indicates mean ± SE (n = 24) readings for 60 min. Three independent experiments were performed, and similar results were observed. **P < 0.01, Student's t‐test.
-
CDA induces gene expression of AZI1 in Arabidopsis. Different letters denote significantly different means at P < 0.05, Tukey's multiple comparison test within each group. Bar indicates mean ± SE (n = 3), and two biological replicates were analyzed with similar results.
-
Dflg22‐induced MPK3/MPK6 phosphorylation in Arabidopsis is not decreased by DA. Kinase assays were performed at 5 min after flg22 treatment. Three independent experiments were performed, and similar results were observed.
-
EPlant resistance to the pathogen Pst DC3000 is not reduced by DA. The severity of pathogen infection was measured as colony‐forming units (CFUs) per mg FW plants at 0 and 3 days after inoculation. Data bar indicates mean ± SE (n = 8), and two independent experiments were performed. Different letters denote significantly different means at P < 0.05, Tukey's multiple comparison test within each group.
-
FRoot colonization of GB03 in wild‐type Arabidopsis (Col‐0) and the mutants/transgenic lines npr1, NahG, and rbohd. ***P < 0.001 compared with Col‐0 without DA treatment, n = 9 biological replicates, Student's t‐test.
-
GBacteria chemotaxis with different concentrations of DA. Data point indicates mean± SE (n = 6). Two independent experiments were performed, and similar results were observed. ***P < 0.001, Student's t‐test.
Source data are available online for this figure.
In parallel to the overall down‐regulation of many immune‐associated genes, Arabidopsis AZI1 gene expression was induced by DA (Figs 5C and EV4F; Table EV7). AZI1 is important for systemic acquired resistance through multiple mechanisms, including the priming of plant SA accumulation upon pathogen infection (Jung et al, 2009). More importantly, DA neither decreases flg22‐induced activation of the MAP kinases MPK3 and MPK6 (Fig 5D; Appendix Fig S5A), nor impairs flg22‐triggered induction of immune‐related genes, such as FRK1 and PR1 (Appendix Fig S5B). Consistently, plant resistance to the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 was not reduced by DA (Fig 5E), although it suppresses microbial induction of plant ROS burst. Interestingly, although the levels of SA and JA in Pi‐deficient plants are increased by DA, the resistance to Pst DC3000 is not increased in the plants that are already suffering from hyper‐PSR (Fig 5E), possibly because DA also suppresses ROS production in the Pi‐deficient plants (Fig EV4E), consistent with the observations that ROS play an important role in SA‐mediated defense against P. syringae (Mammarella et al, 2015). Therefore, DA suppresses microbe‐induced ROS production and certain other defense responses in P‐sufficient plants, without sacrificing defense to the examined pathogen Pst DC3000.
To investigate the potential impacts of DA‐mediated suppression of plant immunity, we examined GB03 colonization to Arabidopsis roots. In wild‐type plants that were pre‐treated with DA for 48 h, GB03 colonization to roots was increased by approximately twofold compared with plants without DA pre‐treatment (Fig 5F), demonstrating a positive role of DA in the establishment of a mutualistic association. This conclusion is further supported by the observations that the same treatment of DA does not alter the growth rate of GB03 (Appendix Fig S5C and D). Without DA pre‐treatment, NahG plants and the Arabidopsis mutant npr1, which is defective in SA‐mediated signaling (Cao et al, 1994), both showed higher GB03 colonization rates compared with the wild‐type plants (Fig 5F). Thus, SA‐mediated plant immunity negatively affects the establishment of mutualism between Arabidopsis and GB03. In addition, DA increased GB03 colonization in NahG and npr1 plants (Fig 5F), indicating that DA can further facilitate the association between GB03 and plants in an SA‐independent manner. Meanwhile, the Arabidopsis mutant rbohd, which is defective in the NADPH oxidase RBOHD that is responsible for pathogen‐induced ROS production (Torres et al, 2005), also displayed increased GB03 colonization compared with wild‐type plants when the plants were not pre‐treated with DA (Fig 5F), demonstrating that suppression of plant ROS burst can lead to enhanced GB03 colonization. The association between GB03 and rbohd was not significantly (t‐test, P = 0.0597, n = 9 biological replicates) further increased by DA treatment (Fig 5F). Thus, DA enhances GB03 colonization, and it appears to be largely mediated through suppression of ROS production in plants.
To further investigate the impact of DA on plant–microbe association, we compared GB03 with two pathogens, Pst DC3000 and the root pathogen Ralstonia solanacearum GMI1000, by chemotaxis assays using DA at a wide range of concentrations from 1 to 50 mM (Fig 5G). At low concentrations (0.001, 0.005, and 0.05 mM), DA attracts GB03 more (up to 31%) than the two pathogens. Meanwhile, when applied at high concentrations (1, 10, and 50 mM), DA became deterrent to all three tested bacteria; however, GB03 was much less (up to 64%) deterred compared with the pathogens. Collectively, the chemotaxis assays indicate that DA increases the competitiveness of GB03 over pathogens in terms of bacteria motility. Therefore, DA not only directly enhances GB03 colonization to roots, but also increases competitiveness of GB03 against pathogens, thereby facilitating the establishment of a mutualistic association between GB03 and Pi‐sufficient plants.
These results demonstrate an important function of DA in mediating bacteria interaction with Pi‐sufficient plants, that is, DA can facilitate mutualistic association between plants and beneficial bacteria without sacrificing plant defense against the examined pathogen Pst DC3000. In addition, together with the observations of Pi‐deficient plants, these results demonstrate that DA affects the types of relation between plants and certain rhizobacteria in a way that depends on plant PSR system and phytohormone‐mediated immunity (Fig 6A).
Figure 6. Diacetyl‐mediated plant–microbe interactions highlight the different strategies underlying plant responses to bacteria and fungi in terms of immunity or mutualism.
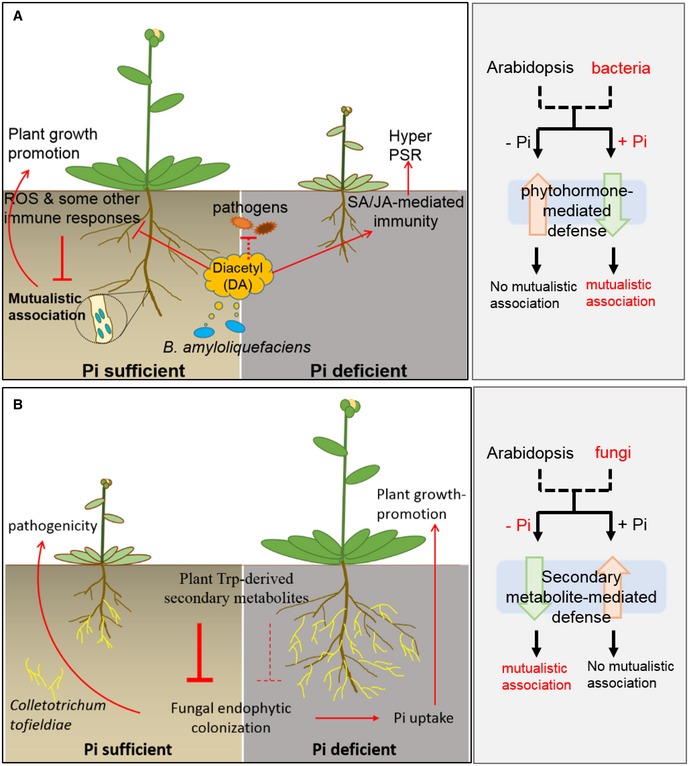
-
A(This study) In phosphate (Pi)‐sufficient plants, diacetyl (DA) partially suppresses the immune system including the production of reactive oxygen species (ROS), primes salicylic acid (SA) accumulation, and increases the competitiveness of GB03 against pathogens, resulting in enhanced symbiont colonization without compromised resistance to the pathogen Pst DC3000. As a result of the mutualistic association, plant growth is promoted. In P‐deficient plants, DA simultaneously elevates the levels of salicylic acid (SA) and jasmonic acid (JA), resulting in strong activation of phytohormone‐mediated immunity and consequently plant hyper‐sensitivity to P deficiency. As a result, plants display stress symptoms.
-
B(Adapted from Hiruma et al, 2016) Pi‐deficient plants allow symbiosis with certain fungi because endophytic fungi can transfer phosphate to plants, whereas Pi‐sufficient plants need no fungi‐assisted Pi uptake and so they deploy Trp‐derived secondary metabolites to defend against fungi invasion.
Discussion
In contrast to the Pi‐dependent mutualistic association between Arabidopsis and B. amyloliquefaciens GB03, symbiosis between Arabidopsis and the fungus C. tofieldiae is established only when the plants are under Pi deficiency, so that the fungus transfers Pi to its host for plant growth promotion, whereas in the presence of sufficient Pi, endophytic colonization of this fungus is restricted by plant Trp‐derived secondary metabolites but not by phytohormone‐mediated defense signaling (Hiruma et al, 2016). Therefore, our findings reveal that plants use different strategies for bacteria, such as B. amyloliquefaciens GB03, and fungi when determining mutualism or immunity (Fig 6). It is possible that Pi‐deficient plants allow symbiosis with fungi because endophytic fungi can transfer phosphate to plants, whereas Pi‐sufficient plants need no fungi‐assisted Pi uptake and so they deploy Trp‐derived secondary metabolites to defend against fungi invasion (Hiruma et al, 2016). In contrast, because rhizobacteria do not transfer phosphate to plants, plants prefer Pi‐sufficient conditions for mutualistic association with these rhizobacteria, whereas Pi‐deficient plants deploy phytohormone‐mediated immunity to ward off the bacteria competitors for Pi uptake (Fig EV4; Appendix Fig S5).
The immune system detects PAMPs and protects plants from being invaded by unwanted microbes. In order to establish mutualistic symbiosis with plants, beneficial microbes need either to avoid producing PAMP or to suppress plant immunity, or both. Our results demonstrate that plant immunity including ROS production plays a negative role in the establishment of mutualism between GB03 and Arabidopsis. DA suppresses ROS production and a set of immune genes in plants, leading to facilitated colonization of GB03. The ROS suppression is important for DA‐mediated establishment of mutualism and is supported by a recent study in Medicago truncatula, in which ROS production was shown to impede rhizobia colonization (Yu et al, 2018). Although it is not clear whether GB03 produces any PAMP, its colonization to roots is facilitated by DA, suggesting that perception of DA can overcome the induction of plant immunity by GB03‐produced PAMPs, if any. Importantly, DA‐mediated modulation of the immune system does not decrease plant defense responses to flg22 or the pathogen Pst DC3000. This phenomenon is probably contributed by a simultaneous combination of DA‐dependent effects, including the specific and partial suppression of plant immune system, priming of SA‐mediated signaling pathway, and direct repelling of pathogens. While chemical treatments clearly demonstrated the biological activity of DA, a B. amyloliquefaciens mutant deficient in DA production might further improve this study. However, that DA is produced from spontaneous decarboxylation of acetolactate makes it unpractical to precisely block the production of DA (Swindell et al, 1996).
It is unclear currently how DA differentially modulates plant immunity. In Caenorhabditis elegans and human cells, DA is sensed by ORD‐10, which encodes a seven‐transmembrane olfactory receptor that responds to DA by activation of a G protein signaling pathway (Sengupta et al, 1996; Zhang et al, 1997). Unlike G protein complexes in animals, plant heterotrimeric G proteins are self‐activating. Arabidopsis RGS1 is a seven‐transmembrane protein, which maintains G protein complex in an inactive state. Activation of FLS2 by flg22 relieves G proteins from the RGS1‐mediated repression and enables ROS production (Liang et al, 2018). Because DA suppresses flg22‐induced ROS production, we hypothesized that DA might function through RGS1‐mediated repression of ROS production. However, this hypothesis is not supported by our unpublished studies on RGS1. Nonetheless, the identity of DA sensor in plants is likely to be revealed by forward genetics in the future. DA is found in a variety of beverages and dairy foods (e.g., Rose, 2017). Since DA influences plant immunity, it would be interesting to test whether DA can also influence immunity of human cells and thereby be involved in either bacteria‐caused diseases or the assembly of internal microbiomes.
Materials and Methods
Plant growth conditions
Wild‐type A. thaliana Col‐0 was used in this study. The mutants and transgenic lines including phr1phl1, NahG, rbohD, and npr1 were all in Col‐0 background. Surface‐sterilized seeds were planted on 1/2‐strength Murashige and Skoog medium (0.5 MS) with 0.7% (w/v) agar and 1.5% (w/v) sucrose, and were vernalized at 4°C in dark for 2 days. Seedlings were grown under sterile conditions with 200 μmol photons/m2/s light, 21 ± 2°C, at a 16‐h light/8‐h dark cycle. Five days after germination, Arabidopsis seedlings were transferred to fresh 0.5 MS medium and other growth medium as indicated in the Figures. For nutrient supplementation experiments, the following nutrients were added individually to 0.05 MS: phosphate KH2PO4; nitrogen NO3NH4, KNO3; sulfur MgSO4; calcium CaCl2; and potassium KCl, in order to reach the concentrations that equal to what they should be in the 0.5 MS medium. We monitored Pi contamination in each batch of agar (Sigma‐Aldrich) and used only those without Pi contamination. Petri dishes containing inner plastic partitions were used to separate different growth medium.
Natural GMV and chemical treatments
One day before transferring seedlings, B. amyloliquefaciens GB03 was cultured in liquid LB medium at 28°C overnight. Shortly after seedlings were transferred to new growth medium, 20 μl of GB03 suspension culture or double‐distilled water (DDW) was applied to the non‐plant side of petri dish plates, containing 0.5 MS or 0.05 MS medium, as indicated in the Figures, with 0.7% (w/v) agar and 1.5% (w/v) sucrose. By positioning plants and bacteria on separate sides of the partitioned petri dish, the effects of GMVs on plants would not be interfered by non‐volatile microbial factors.
For MV treatments using commercial compounds, 100 mM stock aqueous solutions were prepared. We used two systems to do the MV treatments as indicated in Figure Legends and in Table EV1. In one system, the compounds were directly mixed into plant growth medium. In the other system, an aliquot of the stock solution was first mixed with 1 ml lukewarm liquid agar (1%) solution, which was then applied to the non‐plant side of a partitioned petri dish. The agar solution soon cooled down and solidified, so that the volatile compounds would be released in a relatively gradual way and diffuse to reach the plants. The dosages of the compounds were determined according to their production rates reported by Farag et al (2006), which identified GMV components with varying individual abundance ranging from 0.6 ng per 24 h to 614 μg per 24 h. Dosages are as indicated in Figure legends and in Table EV1. The chemical treatments were started shortly after 5‐day‐old seedlings were transferred as above mentioned. Unless stated, all samples for qPCR are collected at 5 DAT. For anthocyanin estimation, leaf surface area, and plant phenotype, data were analyzed from 11 DAT samples.
For salicylic acid (SA) and methyl jasmonate (MeJA) treatments, different concentrations of SA or MeJA, as indicated in the Figures, were applied directly to the plant growth medium.
Quantification of anthocyanin levels
Anthocyanin levels were determined according to Nakata et al (2013). Briefly, shoot fresh weight of 10–15 Arabidopsis seedlings at 11 DAT were measured and then ground with liquid nitrogen. The extraction buffer (45% methanol, 5% acetic acid) was added in the ratio normalized to per mg of fresh weight to the tissues and mixed thoroughly, followed by two rounds of centrifugation at 12,000 × g for 5 min at room temperature. The absorbance of the supernatant obtained was measured at 530 and 637 nm, using a Microplate Reader Thermo Varioskan Flash 13. Anthocyanin contents (Abs530/g F.W.) were calculated by [Abs530 − (0.25 × Abs657)] × volume added.
Trypan blue staining
Trypan blue dye was used to visualize cell death. Leaves of 11 DAT Arabidopsis plants were harvested and put into 1‐ml tube containing trypan blue staining solution (0.4 g trypan blue dissolved in 80 ml PBS) for 1 h. Subsequently, the staining solution was removed and samples were immersed into 98–100% ethanol overnight at room temperature. Then, the ethanol solution was replaced by fresh ethanol until green tissues became completely colorless and leaves were covered with 60% glycerol. Stained leaves were photographed using a Nikon camera coupled to a stereomicroscope.
Chlorophyll fluorescence measurements
The ratio of variable to the maximum chlorophyll fluorescence (Fv/Fm), which indicates the potential quantum yield of PSII photochemistry, was determined with a FluorCam 800MF system (Photon Systems Instruments) following the protocol provided by Photon Systems Instruments.
Detection of acid phosphatase activity
Root acid phosphatase activity was determined according to Ito et al (2015). Briefly, 5‐DAT plants were transferred to 0.5% (w/v) agar containing 0.01% (w/v) 5‐bromo‐4‐chloro‐3‐indolyl phosphate p‐toluidine salt (BCIP). Photographs were taken with a Nikon camera after the appearance of blue color, which was indicative of acid phosphatase activity.
Quantitative RT–PCR
Arabidopsis plants, except for those with flg22 treatment, were grown as described in “Plant growth condition” and collected at 5 DAT (for single time points) or indicated time points. For flg22‐triggered gene expression, 5‐day‐old seedlings were transferred from 0.5 MS containing 1.0% sucrose and 0.7% agar into 0.5 MS liquid medium containing 1.0% sucrose. Five days after transferring, plants were treated with 200 μM DA. At 15 h after DA priming, plants were treated with 0.1 μM flg22 and the samples were subsequently collected at the indicated time points.
Total RNA was isolated using the RNAzol RT RNA Isolation Reagent and was quantified using a NanoDrop ND‐1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). cDNAs were synthesized from 1 μg of total RNA in a 20 μl reaction volume using the (TransScript One‐Step gDNA Removal and cDNA Synthesis SuperMix. Trans). qRT–PCR was carried out using SYBR Green and an iCycler apparatus (Bio‐Rad). Each 20 μl PCR contained 1 μl of diluted cDNA (1:5), 10 μl of SYBR Green SuperMix (Bio‐Rad), and 200 nM of each primer. Experiments were carried out on three biological replicates. Arabidopsis housekeeping gene Actin2 was used for normalization. Primers are listed in Table EV7.
Quantification of miRNAs
Small RNAs of 5 DAT Arabidopsis seedlings were extracted by using RNAzol RT (Molecular Research Center). The abundance of miR399 was quantitatively detected by using TaqMan Small RNA Assays (Applied Bioscience) according to Zhang et al (2014a). Briefly, reverse transcription procedure follows the protocol of TaqMan assays with the following modifications: (i) 400 ng small RNAs were used instead of 100 ng total RNAs; (ii) instead of using 3 μl siRNA RT primer, 2 μl Arabidopsis snoR101 (internal control) RT primer was used in combination with 2 μl miRNA RT primer during the reverse transcription.
Quantification of phosphate content
Phosphate contents were quantified according to Ames (1996). Briefly, shoots and roots of 10–40 Arabidopsis seedlings were weighted amounting to at least 9 mg depending on treatment condition and seedling condition, homogenized in liquid N2, and extracted with 250 μl of 1% acetic acid, followed by tissue lysing by dipping the samples in liquid N2 for 30 s. Debris was pelleted out by centrifugation at 15,871 g for 1 min, and 50 μl of the supernatant was mixed with 700 μl of 10% ascorbic acid + 0.42% ammonium molybdate 4H2O (1 N H2SO4) in the ratio of 1:6 and 250 μl of H2O. The mixture was incubated at 45°C for 20 min followed by spectrophotometric absorbance at 820 nM. Pi content was calculated as 0.01 μM Pi at A820 = 0.26 Ab.
Reactive oxygen species measurements
The measurement of the flg22/elf18‐triggered ROS burst was performed according to Sang and Macho (2017). Briefly, 4‐mm leaf disks of 4‐week‐old seedlings were collected and each disk was placed into a single well of 96‐well plates containing 100 μl of 200 μM DA, BTDL, or ATN in liquid for 15 h. Then, the liquid was removed and replaced by 100 μl of elicitor master mix containing 100 nM flg22 or elf18, 100 μm luminol, and 20 μg/ml HRP (horseradish peroxidase). The ROS burst was detected using a Microplate Reader Thermo Varioskan Flash.
MAP kinase assay
Plants were grown on solid 1/2 MS for 5 days and then transferred into 12‐well plates (three seedlings per well) with liquid 1/2 MS for another 5 days. The 10‐day‐old seedlings were treated with 1 ml water (mock), 200 μM DA, and 100 nM flg22 as indicated in the figures. Seedlings were frozen in liquid N2 at the indicated time points. Samples were ground with liquid N2 and homogenized in MPK extraction buffer [20 mM Tris–HCl, pH 7.5, 5 mM EDTA, 150 mM NaCl, 2 mM DTT, protease inhibitor cocktail (Roche Applied Science) plus phosphatase inhibitor cocktail (Roche Applied Science)]. After centrifugation at 13,523 g for 30 min at 4°C, the supernatant was collected and mixed with 2× SDS loading buffer, and boiled 5 min at 95°C. An aliquot of 20 μl protein of each sample was separated in a 12% SDS–PAGE. Immunoblot analysis was performed using anti‐phospho‐p44/42 MAPK (α‐pTEpY, 1:5,000; Cell Signaling Technology) as the primary antibody and peroxidase‐conjugated goat anti‐rabbit IgG (1:10,000; Sigma‐Aldrich) as the secondary antibody.
Bacteria root colonization
Root colonization test was carried out based on Mafia et al (2009), with slight modifications. Briefly, 16‐day‐old Arabidopsis seedling was exposed to DA or mock condition for 48 h. The seedlings were then pooled as groups of 20 and were transferred to 1.5‐ml tubes containing 1 ml 0.45% NaCl solution with B. amyloliquefaciens GB03 at 1 × 107 cfu/ml. The seedlings with root immersed in the carrier solution were shaken at 150 rpm for 24 h at room temperature. Subsequently, roots were separated from aerial parts and were surface‐sterilized by immersion in ethanol (75%, v/v) for 1 min, washed four times in sterile ddH2O, and ground in the tubes. Four serial dilutions with 0.45% NaCl were performed and were plated on LB solid medium. The fourth dilution was used to count colonies after 24 h of growth at 37°C. The first dilution was centrifuged 30 min at maximum speed and, after the supernatant was discarded, was fully dried in an oven for 48 h to calculate the dry weights. Colony‐forming unit (CFU) numbers were determined per mg of root dry weight. Three independent experiments were performed. Three replicates were performed per condition/genotype.
Chemotaxis test
Diacetyl was tested as chemotaxis conditioner agent of B. amyloliquefaciens GB03, P. syringae pv. tomato (Pst) DC3000, and R. solanacearum GMI1000. The test was performed based on a Boyden chamber test, according to manufacturer's protocols of 8 μm Chemotaxis Assay Kit (CBA‐106; Cell Biolabs, Inc., San Diego, CA, USA). A series of 50, 10, 1, 0.1, 0.05, 0.005, and 0.001 mM of diacetyl solutions were prepared in serum‐free medium DMEM (0.5% BSA; 2 mM CaCl2 and 2 mM MgCl2). Mock conditions were prepared without DA. Bacteria cultures were grown in LB at their optimal temperature and 220 r.p.m. for 12 h, and then centrifuged, washed, and resuspended into 0.45% NaCl solution as the carrier liquid up to 106 cfu/ml. Relative attraction ratio was expressed as relative fluorescence of membrane‐detached cells, which were lysed and quantified using CyQUANT GR fluorescent dye. Fluorescence was measured by a 96‐well plate reader Varioskan Flash (Thermo Fisher Scientific, Inc., NH, USA) at 480/520 nm. Six replicates were performed per condition.
Pathogen inoculation and quantification
Arabidopsis thaliana 5‐day‐old seedlings were transferred to 0.5 MS and P‐deficient medium plates with partition followed by treatment with DA (9.7 μg/ml free space) and kept in growth chamber for 3 days. After 3 days of treatment, seedlings were inoculated with Pst DC3000 by spraying. Pst DC3000 was grown in liquid LB medium, precipitated by centrifugation, and resuspended in 10 mM MgCl2 at 5 × 107 CFU/ml. Silwet L‐77 was added to reach the final concentration of 0.02% before spraying the plates containing the Arabidopsis seedlings. The plates were wrapped with parafilm and kept in the growth chamber for 3 days. The plants were then collected, weighed, ground, and spread on solid LB medium. The bacterial growth was determined based on plant fresh weight. For time 0, samples were collected at 2 h after spray and initial bacterial inoculum was quantified.
RNAseq analysis
Arabidopsis thaliana was grown and treated as described in “Plant growth conditions” section. For experiments with GMVs, plants grown on 0.5 MS and 0.05 MS were harvested at 5 DAT before the stress phenotype was observable. For experiments with DA, plants grown in 0.5 MS medium supplemented with or without 200 μM DA were harvested at 48 h after the seedlings were transferred. RNA was extracted by using Plant RNeasy Kit (Qiagen), and the integrity was monitored using the RNA Nano 6000 Assay of the Agilent Bioanalyzer 2100 system (Agilent). RNA purity and concentration were checked using a NanoDrop ND‐1000 Spectrophotometer.
RNAseq was performed at Core Facility for Genomics, Shanghai Center for Plant Stress Biology, CAS. Two biological replicates of each genotype were generated. Total RNA (1 μg) from each sample was used for library preparation with NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, E7420L) following the manufacturer's instructions. Prepared libraries were assessed for quality using NGS High‐Sensitivity Kit on a Fragment Analyzer (AATI) and for quantity using Qubit 2.0 Fluorometer (Thermo Fisher Scientific).
The RNAseq reads were aligned to the TAIR10 reference genome using TopHat2 with parameters “‐r 50 ‐I 1000 ‐G tair10_ensembl.gtf –b2‐very‐fast” (Kim et al, 2012). DEG lists were identified by Cuffdiff program of Cufflinks with parameters “‐b tair10_ensembl.fa ‐u tair10_ensembl.gtf” and cutoff “log2 fold change > 1 or < −1 and q‐value < 0.05” (Trapnell et al, 2010, 2012; Roberts et al, 2011). Quantitative RT–PCR confirmation of RNAseq DEGs was performed with at least three biological replicates.
Circle diagram of Gene Ontology (GO) enrich analysis was done based on the BioMaps Function Analysis of Virtual Plants 1.3 platform (virtualplant.bio.nyu.edu), using hyper‐geometric distribution as P‐value of over‐representation method and 0.01 as cutoff. Size of circles represents number of genes integrated on each GO category. Color indicates the P‐value cutoff of over‐representation equal or less than the cutoff for each GO category. Darker color indicates higher possibility of each GO category, according to the color scale given by the BioMaps Function Analysis.
Measurements of phytohormone contents
SA and JA levels were determined according to Flors et al (2008) with slight modifications. Briefly, 25 mg of 5 DAT Arabidopsis seedlings was harvested and put into 1.5‐ml tube containing 0.5 ml of 70% MeOH and 2 ng SA‐d4 that was an internal standard. The tubes were vortexed at 1,000 rpm at 10°C for 1 h and centrifuged at 20,000 rcf at 20°C for 10 min. 0.3 ml supernatant was taken and diluted two times with H2O in an HPLC vial, and 50 μl solution was injected into the LC‐MS (Waters Liquid Chromatography ACQUITY UPLC I‐Class coupled with AB SCIEX TripleTOF® 5600+), ACQUITY UPLC BECH C18 1.7 μm VanGuard™ Pre‐Column (2.1 × 5 mm column) followed by Analytical column (ACQUITY UPLC BECH C18 1.7 μm 2.1 × 150 mm). The results were analyzed by PeakView 1.2.
Statistical analysis
Statistical analyses were performed with IBM SPSS software (https://www.ibm.com/analytics/spss-statistics-software). Significant difference between treatments was based on P‐values ≤ 0.05.
Author contributions
HZ designed the project; RJLM and GA set up the experiments with GMV treatment and identified DA; SKS set up SA/JA‐related experiments and perform or coordinate other DA‐related experiments; DH performed experiments for ROS, MAP kinase, and pathogenicity assays; JIV performed experiments for bacteria colonization and chemotaxis; KT analyzed RNAseq raw data for the DEG lists; FY, YS, CS, SZ, YY, XL, YD, WW, JG, WH, and ML participated in the experiments and/or data analyses; and HZ wrote the manuscript with input from C‐PS, J‐KZ, APM, and PWP.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Table EV6
Table EV7
Source Data for Appendix
Review Process File
Source Data for Figure 5
Acknowledgements
We thank Prof. Choong‐Min Ryu (Korea Research Institute of Bioscience & Biotechnology, Korea) for B. amyloliquefaciens GB03, Prof. Vicente Rubio Muñoz (National Center for Biotechnology, CSIC, Spain) for phr1phl1, and Prof. Chanhong Kim (Shanghai Center for Plant Stress Biology, CAS, China) for NahG and npr1. Research in H.Z. Lab has been supported by the Chinese Academy of Sciences (CAS) and by the Thousand Talents Program for Young Scientists, China. R.J.L.M. was supported by the CAS PIFI fellowship.
The EMBO Journal (2020) 39: e102602
See also: https://doi.org/10.15252/embj.2019104144 (January 2020)
Data availability
The RNAseq data from this publication have been deposited to the NCBI's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE138478 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138478).
References
- Ames BN (1996) Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol 8: 115–118 [Google Scholar]
- Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R (2013) Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci USA 110: E1621–E1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Halane MK, Gassmann W, Stacey G (2017) The role of plant innate immunity in the Legume‐Rhizobium symbiosis. Annu Rev Plant Biol 28: 535–561 [DOI] [PubMed] [Google Scholar]
- Castrillo G, Teixeira PJ, Paredes SH, Law TF, de Lorenzo L, Feltcher ME, Finkel OM, Breakfield NW, Mieczkowski P, Jones CD et al (2017) Root microbiota drive direct integration of phosphate stress and immunity. Nature 543: 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ (1997) NDR1, a pathogen‐induced component required for Arabidopsis disease resistance. Science 12: 1963–1965 [DOI] [PubMed] [Google Scholar]
- Choi SK, Jeong H, Kloepper JW, Ryu CM (2014) Genome sequence of Bacillus amyloliquefaciens GB03, an active ingredient of the first commercial biological control product. Genome Announc 30: 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- Farag MA, Ryu CM, Sumner LW, Paré PW (2006) GC‐MS SPME profiling of rhizobacterial volatiles reveals prospective inducers of growth promotion and induced systemic resistance in plants. Phytochemistry 67: 2262–2268 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Flors V, Ton J, van Doorn R, Jakab G, García‐Agustín P, Mauch‐Mani B (2008) Interplay between JA, SA and ABA signaling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola . Plant J 54: 81–92 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate‐starvation response in Arabidopsis . Curr Biol 22: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Hacquard S, Kracher B, Hiruma K, Münch PC, Garrido‐Oter R, Thon MR, Weimann A, Damm U, Dallery JF, Hainaut M et al (2016) Survival trade‐offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat Commun 7: 11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma K, Gerlach N, Sacristán S, Nakano RT, Hacquard S, Kracher B, Neumann U, Ramírez D, Bucher M, O'Connell RJ et al (2016) Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165: 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Nozoye T, Sasaki E, Imai M, Shiwa Y, Shibata‐Hattam M, Ishige T, Fukui K, Ito K, Nakanishi H et al (2015) Strigolactone regulates anthocyanin accumulation, acid phosphatases production and plant growth under low phosphate condition in Arabidopsis . PLoS ONE 10: e0119724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT (2009) Priming in systemic plant immunity. Science 3: 89–91 [DOI] [PubMed] [Google Scholar]
- Khan GA, Vogiatzaki E, Glauser G, Poirier Y (2016) Phosphate deficiency induces the jasmonate pathway and enhances resistance to insect herbivory. Plant Physiol 171: 632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee S, Ryu CM (2012) Interspecific bacterial sensing through airborne signals modulates locomotion and drug resistance. Nat Commun 4: 1809 [DOI] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wu QY, Sun YL, Wang LY, Yang XH, Meng QW (2010) Overexpression of chloroplastic monodehydroascorbate reductase enhanced tolerance to temperature and methyl viologen‐mediated oxidative stresses. Physiol Plant 139: 421–434 [DOI] [PubMed] [Google Scholar]
- Liang X, Ma M, Zhou Z, Wang J, Yang X, Rao S, Bi G, Li L, Zhang X, Chai J et al (2018) Ligand‐triggered de‐repression of Arabidopsis heterotrimeric G proteins coupled to immune receptor kinases. Cell Res 28: 529–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XM, Zhang H (2015) The effects of bacterial volatile emissions on plant abiotic stress tolerance. Front Plant Sci 6: 774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B, Kamilova F (2009) Plant‐growth‐promoting rhizobacteria. Annu Rev Microbiol 63: 541–556 [DOI] [PubMed] [Google Scholar]
- Mafia RG, Alfenas AC, Ferreira EM, Binoti DHB, Mafia GMV, Mounteer AH (2009) Root colonization and interaction among growth promoting rhizobacteria isolates and eucalypts species. Revista Árvore 33: 1–9 [Google Scholar]
- Mammarella ND, Cheng Z, Fu ZQ, Daudi A, Bolwell GP, Dong X, Ausubel FM (2015) Apoplastic peroxidases are required for salicylic acid‐mediated defense against Pseudomonas syringae . Phytochemistry 112: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FM, Uroz S, Barker DG (2017) Ancestral alliances: plant mutualistic symbioses with fungi and bacteria. Science 356: 6340 [DOI] [PubMed] [Google Scholar]
- Mehterova N, Balazadehc S, Hillee J, Tonevaa V, Mueller‐Roeberc B, Gechev T (2012) Oxidative stress provokes distinct transcriptional responses in the stress‐tolerant atr7 and stress‐sensitive loh2 Arabidopsis thaliana mutants as revealed by multi‐parallel quantitative real‐time. Plant Physiol Biochem 59: 20–29 [DOI] [PubMed] [Google Scholar]
- Nakata M, Mitsuda N, Herde M, Koo AJ, Moreno JE, Suzuki K, Howe GA, Ohme‐Takagi MA (2013) bHLH‐type transcription factor, ABA‐INDUCIBLE BHLH‐TYPE TRANSCRIPTION FACTOR/JA‐ASSOCIATED MYC2‐LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis . Plant Cell 25: 1641–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paré PW, Zhang H, Aziz M, Xie X, Kim MS, Shen X, Zhang J (2011) Beneficial rhizobacteria induce plant growth: mapping signaling networks in Arabidopsis In Biocommunication in soil microorganisms, soil biology, Witzany G. (ed.), Vol. 23, pp 403–412. Berlin Heidelberg: Springer‐Verlag; [Google Scholar]
- Pieterse CM, Zamioudis C, Berendsen RL, Weller DM, Van Wees SC, Bakker PA (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52: 347–375 [DOI] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L (2011) Identification of novel transcripts in annotated genomes using RNA‐Seq. Bioinformatics 27: 2325–2329 [DOI] [PubMed] [Google Scholar]
- Rose CS (2017) Early detection, clinical diagnosis and management of lung disease from exposure to diacetyl. Toxicology 10.1016/j.tox.2017.03.019 [DOI] [PubMed] [Google Scholar]
- Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Paré PW, Kloepper JW (2003) Bacterial volatiles promote growth in Arabidopsis . Proc Natl Acad Sci USA 100: 4927–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang YY, Macho AP (2017) Analysis of PAMP‐triggered ROS burst in plant immunity. Methods Mol Biol 1578: 143–153 [DOI] [PubMed] [Google Scholar]
- Sasse J, Martinoia E, Northen T (2008) Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci 23: 25–41 [DOI] [PubMed] [Google Scholar]
- Sengupta P, Chou JH, Bargmann CI (1996) odr‐10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell 84: 899–909 [DOI] [PubMed] [Google Scholar]
- Somssich IE, Wernert P, Kiedrowski S, Hahlbrock K (1996) Arabidopsis thaliana defense‐related protein ELI3 is an aromatic alcohol:NADP+ oxidoreductase. Proc Natl Acad Sci USA 93: 14199–14203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell SR, Benson KH, Griffin HG, Renault P, Ehrlich SD, Gasson MJ (1996) Genetic manipulation of the pathway for diacetyl metabolism in Lactococcus lactis . Appl Environ Microbiol 62: 2641–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose‐specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL (2005) Pathogen‐induced, NADPH oxidase‐derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana . Nat Genet 37: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees SC, Glazebrook J (2003) Loss of non‐host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J 33: 733–742 [DOI] [PubMed] [Google Scholar]
- Wu CH, Derevnina L, Kamoun S (2018) Receptor networks underpin plant immunity. Science 22: 1300–1301 [DOI] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D (2002) The SCF(COI1) ubiquitin‐ligase complexes are required for jasmonate response in Arabidopsis . Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Dong X (2014) Perception of the plant immune signal salicylic acid. Curr Opin Plant Biol 20: 64–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Xiao A, Dong R, Fan Y, Zhang X, Liu C, Wang C, Zhu H, Duanmu D, Cao Y et al (2018) Suppression of innate immunity mediated by the CDPK‐Rboh complex is required for rhizobial colonization in Medicago truncatula nodules. New Phytol 220: 425–434 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chou JH, Bradley J, Bargmann CI, Zinn K (1997) The Caenorhabditis elegans seven‐transmembrane protein ODR‐10 functions as an odorant receptor in mammalian cells. Proc Natl Acad Sci USA 94: 12162–12167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kim MS, Krishnamachari V, Payton P, Sun Y, Grimson M, Farag MA, Ryu CM, Allen R, Melo IS et al (2007) Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis . Planta 226: 839–851 [DOI] [PubMed] [Google Scholar]
- Zhang H, Xie X, Kim MS, Kornyeyev DA, Holaday S, Pare PW (2008) Soil bacteria augment Arabidopsis photosynthesis by decreasing glucose sensing and abscisic acid levels in planta. Plant J 56: 264–273 [DOI] [PubMed] [Google Scholar]
- Zhang H, Sun Y, Xie X, Kim MS, Dowd SE, Paré PW (2009) A soil bacteria regulates plant acquisition of iron via deficiency‐inducible mechanisms. Plant J 58: 568–577 [DOI] [PubMed] [Google Scholar]
- Zhang H, Tang K, Qian W, Duan CG, Wang B, Zhang H, Wang P, Zhu X, Lang Z, Yang Y et al (2014a) An Rrp6‐like protein positively regulates noncoding RNA levels and DNA methylation in Arabidopsis . Mol Cell 54: 418–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Lu S, Liu D (2014b) A major root‐associated acid phosphatase in Arabidopsis, AtPAP10, is regulated by both local and systemic signals under phosphate starvation. J Exp Bot 65: 6577–6588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Oldroyd GE (2017) Plant signalling in symbiosis and immunity. Nature 15: 328–336 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Table EV6
Table EV7
Source Data for Appendix
Review Process File
Source Data for Figure 5
Data Availability Statement
The RNAseq data from this publication have been deposited to the NCBI's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE138478 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138478).


