Graphical abstract
Abbreviations: PPIs, protein-protein interactions; RF, Random Forest; Y2H, yeast two-hybrid; MS, mass spectroscopy; ML, machine learning; CT, Conjoint Triad; AC, Auto Covariance; LD, Local Descriptor; SGD, stochastic gradient descent; SVM, Support Vector Machine; Adaboost, Adaptive Boosting; MLP, Multiple Layer Perceptron; RBF, radial basis function; ACC, Accuracy; MCC, Matthews correlation coefficient; ROC, Receiver Operating Characteristic; PR, Precision-Recall; AUC, area under the ROC curve; AUPRC, area under the PR curve
Keywords: Human-virus interaction, Protein-protein interaction, Prediction, Embedding, Doc2vec, Machine learning
Highlights
-
•
We proposed a doc2vec + RF classifier for predicting human-virus PPIs.
-
•
Doc2vec can effectively capture more context information from protein sequences.
-
•
The proposed method revealed better performance than several existing predictors.
Abstract
The identification of human-virus protein-protein interactions (PPIs) is an essential and challenging research topic, potentially providing a mechanistic understanding of viral infection. Given that the experimental determination of human-virus PPIs is time-consuming and labor-intensive, computational methods are playing an important role in providing testable hypotheses, complementing the determination of large-scale interactome between species. In this work, we applied an unsupervised sequence embedding technique (doc2vec) to represent protein sequences as rich feature vectors of low dimensionality. Training a Random Forest (RF) classifier through a training dataset that covers known PPIs between human and all viruses, we obtained excellent predictive accuracy outperforming various combinations of machine learning algorithms and commonly-used sequence encoding schemes. Rigorous comparison with three existing human-virus PPI prediction methods, our proposed computational framework further provided very competitive and promising performance, suggesting that the doc2vec encoding scheme effectively captures context information of protein sequences, pertaining to corresponding protein-protein interactions. Our approach is freely accessible through our web server as part of our host-pathogen PPI prediction platform (http://zzdlab.com/InterSPPI/). Taken together, we hope the current work not only contributes a useful predictor to accelerate the exploration of human-virus PPIs, but also provides some meaningful insights into human-virus relationships.
1. Introduction
Virus infections still pose a major threat to human health. As of the World Health Organization (WHO), HIV/AIDS causes the deaths of one million people in 2016. World-wide dengue fever cases have continuously increased in recent decades [1], pointing to 50 million annual cases that cause 25,000 deaths [2], [3]. The investigation of the human-virus interactome is therefore increasingly important, leading to extensive efforts to determine the ways viruses infect, hijack and utilize host functions to carry out their own life activities. Within the complex human-virus interaction system, protein-protein interactions (PPIs) serve as a foundation of cell communication between human and viruses and play a vital role for viral infections and host immune responses [4], [5]. As a consequence, in-depth exploration of human-virus PPIs is critical for a thorough understanding of a virus’ pathogenesis, providing an essential foundation for the development of effective therapeutic and prevention strategies to combat diseases.
Experimental techniques for PPI identification have been developed in the past decades. While PPIs can be determined individually by using various genetic, biochemical and biophysical methods, high-throughput experimental techniques such as yeast two-hybrid (Y2H) and mass spectroscopy (MS) allowed the determination of PPIs on a large scale [6], [7], [8] that have been widely utilized to infer protein functions and understand corresponding biological processes. However, such high-throughput experimental screens are mainly applied to identify intraspecies PPIs [9], [10], [11], while interspecies interactomes remained relatively understudied. Moreover, the experimental determination of PPIs is typically time-consuming, laborious and hard to obtain complete protein interactomes. Therefore, efficient computational methods for PPI prediction can complement experimental techniques by providing experimentally testable hypothesis and exclude protein pairs with low interacting probability to limit the range of PPI candidates.
A plethora of computational methods for PPI prediction have been developed, traditionally utilizing interolog mapping [12], [13] and domain-domain/motif interaction-based inference [14], [15], [16]. Apart from sequence information, protein 3D structures [17], [18] and gene co-expression relationships [19] have also been used to predict PPIs, although protein structures and expression data of query protein pairs are generally hard to obtain. With the technical advance of machine learning (ML) and the availability of known PPIs, ML-based methods have been intensively employed to predict PPIs. Briefly, ML-based methods train a binary classifier using known PPIs to distinguish interacting and non-interacting protein pairs from query samples [20]. Although various heterogeneous information or evidences as features can be integrated to provide a predictive framework, most ML-based methods utilize protein sequence information.
Although mainly focusing on the prediction of intraspecies PPIs [21], [22], [23], ML-driven PPI prediction approaches are increasingly applied to determine interspecies PPIs [24], [25], [26], such as interactions between human and viral proteins [20], [27], [28], [29]. Encoding protein sequence information, most schemes account for residue physicochemical properties of protein sequences, yet ignore the relationships between amino acid segments as a function of the context of whole protein sequences. Moreover, nearly all of the constructed models are designed for certain individual virus species, limiting their generalizability to other human host-virus systems. Currently, tens of thousands of human-virus PPIs have been experimentally determined, providing an unprecedented abundance of data to develop generalizable ML-based methods to predict interactions of proteins of human and any virus.
To create a ML model for human-virus PPI prediction, the key step is to conduct feature encoding which converts human and viral protein sequences to fixed-dimensional vectors. For PPI prediction, some common sequence encoding schemes such as Conjoint Triad (CT) [30], Auto Covariance (AC) [31] and Local Descriptor (LD) [32], [33], [34], [35] are widely used, in which residue-specific physicochemical properties or interaction effects have been taken into account to some extent. However, there are two shortcomings for these manually constructed feature vectors. One is that such methods usually fail to sufficiently consider semantic information (such as the order of residues) in entire sequences. The other one is that they ignore potential information from the large quantity of unlabeled protein sequences while these information can represent very important properties of proteins.
To capture semantic information of residues in entire sequences as much as possible, word/document embedding techniques were recently developed. The word embedding uses vectors to represent words which are learned from the contexts of words in a given document. One of the widely used word embedding models is word2vec which uses a shallow two-layer neural network to learn word vectors [36]. As an extension of word2vec, doc2vec was developed to learn document-based embeddings for entire sentences, paragraphs, or documents [37]. Recently, such word/document embedding representation approaches have been used to process biological sequences [38], [39], [40]. Here, each protein sequence can be reviewed as a sentence and broken to multiple overlapping/non-overlapping residue segments regarded as words (i.e. k-mers) that were used to train word2vec/doc2vec models. To learn the semantic information as much as possible, a large protein dataset (e.g., the UniProt database) was often used. Such learned protein embeddings can be further used to train various ML classifiers for biological prediction tasks. In the real applications of protein classification, note that the advantage of doc2vec over word2vec has been reported [39]. Therefore, we attempted to introduce doc2vec into the prediction of human-virus PPIs. To our best knowledge, the doc2vec embedding technique has not been reported in the interspecies protein interaction predictions.
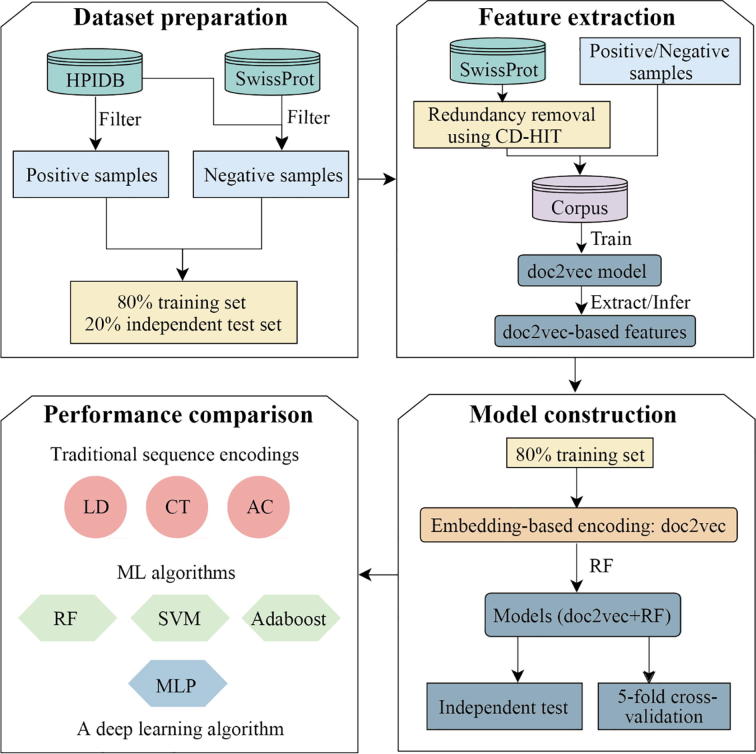
Here, we introduce a computational pipeline (Fig. 1) that is based on a protein sequence embedding-based ML method, allowing us to predict human-virus PPIs. In particular, we consider human-virus PPIs as positive samples and compile negative PPI samples to construct a training dataset and an independent test set. We train a doc2vec model with such training data as well as a large number of unlabeled protein sequences to learn protein features that allow a reliable prediction of human-virus protein interactions, utilizing a Random Forest (RF) approach. Through 5-fold cross-validation and independent tests, we extensively compare the results of our prediction framework with other popular sequence encoding schemes and ML algorithms, suggesting that our pipeline significantly outperforms other approaches. Moreover, we also rigorously benchmark our prediction framework against existing human-virus PPI prediction methods. Finally, our sequence embedding-based ML method is freely accessible to the community through an online webserver (http://zzdlab.com/InterSPPI/).
Fig. 1.
Workflow of our computational pipeline to predict human-virus PPIs. In the dataset preparation step, we constructed positive and negative data samples, utilizing human-virus protein interaction data from HPIDB as well as SwissProt database. Furthermore, we randomly sampled 80% as training data, while remining data was used as an independent test set. In the feature extraction step, we formed a corpus of sequence information from such protein data to train a doc2vec model, allowing us to extract/infer protein sequence specific features. Representing 80% of interactions between proteins through such feature embeddings as training data we used Random Forests (RF) to predict protein interactions using 5-fold cross-validation and independent test sets (remaining 20% of interaction data). In the final step, we compared our doc2vec + RF model with combinations of different encoding schemes such as the Conjoint Triad (CT), Local Descriptor (LD) and Auto Covariance (AC) and widely used ML methods such as Support Vector Machine (SVM), Multiple Layer Perceptron (MLP) and Adaptive Boosting (Adaboost).
2. Materials and methods
2.1. Data set construction
We downloaded host-pathogen PPI data from the Host-Pathogen Interaction Database (HPIDB; version 3.0) [41] that contains manually curated host-pathogen interactions and also integrates corresponding molecular interactions from other public protein interaction databases. To obtain high-quality PPI samples, we excluded interactions from large-scale MS experiments that have been experimentally observed only once because the MS experiments generally identify protein complexes rather than binary interactions [42]. Further excluding non-physical interactions, redundant PPIs, and interactions between proteins with less than 30 amino acids, more than 5000 amino acids or non-standard amino acids, we obtained 22,653 experimentally verified human-virus PPIs as a positive sample set. Regarding the construction of negative samples, previous studies have shown that completely random pairing may introduce sizeable amounts of noise, limiting the usability of such PPIs as negative samples sets. As an alternative, the ‘Dissimilarity-Based Negative Sampling’ method [43], accounts for sequence similarity of viral proteins. For example, if viral proteins A and B are similar (sequence identity > 0.3) [44] and A interacts with host protein C, protein pair B-C should not be considered a potential negative sample. Following these guidelines, we randomly selected viral proteins from the positive sample set and human proteins as of the SwissProt database [45] and sampled human and viral protein pairs as a non-interacting, negative PPI set that do not occur in potential positive sample sets. Specifically, the ratio of positive to negative samples was 1:10. Further, we divided our samples into a training set (80%) and an independent test set (20%) for model training and performance assessment, respectively. To reduce sampling bias caused by sample partition, we randomly constructed 3 different training and independent test sets.
2.2. Doc2vec model
In the unsupervised doc2vec embedding learning framework, feature representation of continuous protein sequences is based on the assumption that a set of protein sequences comprises a ‘document’. In particular, each sequence is considered a sentence written in a biological language, suggesting that the corresponding biological function can be semantically interpreted [46]. As for training data (termed as corpus), we utilized non-redundant protein sequences with lengths between 30 and 5000 amino acids from the SwissProt database [45] where CD-HIT was employed to removing redundancy (sequence identity ≤ 0.5) [44] and sequences in our positive/negative PPI samples. Considering the doc2vec model training requests a large size of corpus and previous studies have suggested that a larger corpus often results in a better and more robust performance, the sequence identity threshold of 0.5 deems reasonable. After the above filtering steps, we obtained 291,726 proteins as a corpus for the doc2vec model training. Following previous works [31], [40], we broke such amino acid sequences into non-overlapping residue segments (k-mers) as biological words. Then we used these k-mer residue segments (words) and the complete sequences (sentences) to train the doc2vec model (Fig. S1). The distributed-memory (DM) model architecture was adopted to train the doc2vec model, allowing us to represent each word through context words and the sentence vector. All the word and sentence vectors were trained by using stochastic gradient descent (SGD) and backpropagation to update weight parameters iteratively [36]. After training, the output sentence vectors were used as our protein sequence features.
The doc2vec model training and inference were implemented using the Python library Gensim [47]. We optimized hyperparameters (e.g., k-mers and the dimensionality of output vectors) using 5-fold cross-validation. In particular, we trained a Random Forest (RF) classifier on the PPI training data using different lengths of k-mers, where k was ranging from 2 to 7 and considered different dimensions of output vectors (number of hidden layer neurons {16, 32, 64, 128, 256}).
2.3. Parameter optimization for ML algorithms
We mainly used RF to train PPI prediction models, an ensemble learning method where classification trees are constructed using different bootstrap samples of the data (‘bagging’). In addition, random forests change how classification trees are constructed by splitting each node, using the best among a predictor subset randomly chosen at that node (‘boosting’). While we kept default parameters, we set the number of trees in the forest (n_estimators) to 1500 while the criterion of selecting predictor features was set as ‘entropy’. We also compared corresponding results with three other popular ML algorithms, including Support Vector Machine (SVM), Adaptive Boosting (Adaboost) and one of deep learning architectures named Multiple Layer Perceptron (MLP). These algorithms were implemented by utilizing the Python-based ML library scikit-learn [48] and deep learning library keras (https://keras.io/), respectively. For all the ML-based algorithms, parameters were optimized through the GridSeachCV function, using cross-validation sets and considering the ‘neg_log_loss’ scoring function as assessment criterion.
SVM performs classification by mapping low-dimensional inputs into a high-dimensional feature space through a kernel function. Here, we chose the radial basis function (RBF) and optimized parameters C, γ, ranging between [2−5, 215] and [2−15, 23], respectively. Due to the computational costs of SVM, we only utilized one fifteenth of the training samples to optimize parameters. AdaBoost is a meta-algorithm for establishing a strong classifier by combining the outputs of multiple weak classifiers (decision trees) into a weighted sum, benefitting cases that were misclassified by weak classifiers. We optimized the maximum number of trees to 50, while the optimized learning rate was set to 0.01. The deep learning method MLP is a feedforward neutral network consisting of an input layer, hidden layers and an output layer. MLP trains the classifier by supervised backpropagation and utilizes nonlinear activation functions to distinguish linearly indivisible data. Here, we used two hidden layers with 128 and 64 neurons, and adopted ‘ReLU’ as the activation function. Moreover, the mini-batch size and the learning rate was set to 64 and 0.0001, respectively. To avoid over-fitting, we used dropout layers as regularizers. For the output layer, the activation function ‘sigmoid’ was utilized to retrieve normalized probabilities between 0 and 1.
2.4. Other popular sequence-based encoding schemes
2.4.1. Conjoint Triad (CT)
Based on the physicochemical properties of their side chains, 20 amino acids are clustered into seven groups (AGV, DE, FILP, HNQW, KR, MSTY and C). Replacing each amino acid in a protein sequence with the corresponding group number, the frequency of each conjoint triad in the protein sequence is determined through a sliding window. As a consequence, a protein pair is finally represented by a 686-dimensional (7 × 7 × 7 × 2) vector [30].
2.4.2. Local Descriptor (LD)
Similar to CT encoding, the seven groups of amino acids are also used in LD. Briefly, LD divides a protein sequence into ten local regions to further extract features of each subregion, mainly reflecting local characteristics of the underlying protein [34]. Each region is represented by three features that reflect the characteristics of seven amino acid groups. The three features are Composition (C), Transition (T), and Distribution (D), where C represents the composition of each amino acid group, T reflects the composition of any two amino acid groups, and D represents the distribution of the first, 25%, 50%, 75%, and 100% of the total number of amino acids. In each region, the corresponding dimensionality for C, T and D is 7, 21 and 35, respectively. Therefore, the final dimension of the LD encoding for a protein pair is 1260 [(7 + 21 + 35) × 10 × 2].
2.4.3. Auto Covariance (AC)
AC encoding [31] accounts for correlations and interactions between variables at different positions, widely applied to coding protein sequences [49], [50]. In this study, we employed seven residue physicochemical properties (Table S1) to represent the protein feature. AC features of protein sequences can be inferred by
, where n is the length of the protein sequence X, lag represents the sequence distance between residues and is the normalized jth physicochemical property value of the ith amino acid. In this way, protein sequences with variable sequence lengths can be encoded into vectors with a fixed dimension, . As for protein interactions, a protein pair was represented by concatenating the AC vectors of two proteins. Here, we set lag to 30, transforming a protein pair into a 420-dimensional (30 × 7 × 2) vector.
In addition to the singular sequence encodings, we also simultaneously considered a combination of above three sequence encodings by concatenating these schemes to form a 2366-dimensional (1260 + 686 + 420) vector (LD_CT_AC).
2.5. Performance evaluation
We used both 5-fold cross-validation and an independent test to compare the performance of different computational frameworks. To ensure significance of our results, we randomly selected samples for three times, the final result is the average performance of the three replicates. Furthermore, the following commonly used measurements such as Recall (Sensitivity), Specificity, Accuracy (ACC), Precision, F1-score, Matthews correlation coefficient (MCC), were utilized to evaluate the performances of the proposed prediction model. The corresponding formulae are as follows:
and
where TP, FP, TN and FN represent the number of true positives, false positives, true negatives and false negatives, respectively. To achieve a more intuitive and effective evaluation of the models, we plotted the Receiver Operating Characteristic (ROC) curve and considered the area under the curve (AUC). In addition, we considered the Precision-Recall (PR) curve and the corresponding area under the PR curve (AUPRC), that is commonly employed to assess classification performance when the positive and negative samples are imbalanced [51]. In general, the closer the value of AUC/AUPRC is to 1, the better the performance of the prediction model is. All ROC/PR curves were determined with the R package ROCR [52].
3. Results and discussion
3.1. The performance of doc2vec + RF
Here, we introduced a sequence embedding technique called doc2vec to convert protein sequences into feature vectors, allowing us to construct a RF classifier to predict human-virus PPIs. To achieve best performance, we optimized the length of k-mers in doc2vec ranging from 2 to 7 through performance comparison of the corresponding RF models for PPI prediction. In terms of AUPRC and AUC, 4-mers and 5-mers provided better performance using 5-fold cross-validation, and 5-mers yielded the highest AUPRC value (Table S2). Thus, we employed 5-mers for our final doc2vec model construction. Moreover, the vector size of the doc2vec features was also optimized. Specifically, we observed that the dimensionality of 32 can roughly achieve best performance for the prediction of human-virus PPIs, implying the low dimensionality and high efficiency of the doc2vec encoding.
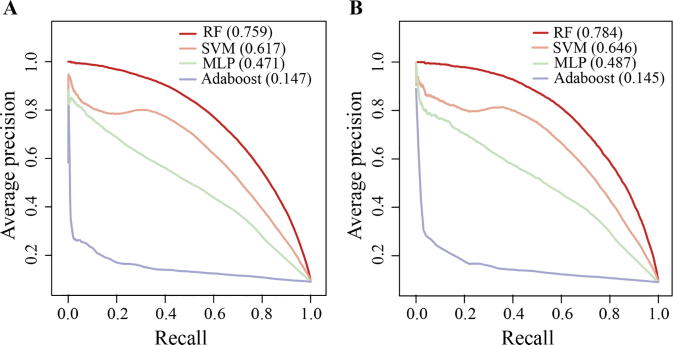
In general, the combination of doc2vec with 5-mers and vector size 32 and RF (doc2vec + RF) provided excellent performance as the corresponding AUPRC values were 0.759 and 0.784 when we applied 5-fold cross-validation and used independent tests, respectively (Fig. 2). At a recall control of 80%, the corresponding precision value in the 5-fold cross-validation and independent test was 54.77% and 58.82%, respectively. The performance results were corroborated by the corresponding ROC curves in Fig. S2 where doc2vec + RF achieved an AUC = 0.947 for the 5-fold cross-validation and AUC = 0.954 for the independent test, suggesting that the embedding technique effectively transferred information encoded in protein sequences to the task of human-virus PPI prediction.
Fig. 2.
Performance of various classifiers in predicting human-virus PPIs based on doc2vec encoding. Areas under the Precision-Recall curves (AUPRC) indicate that Random Forests (RF) outperformed Support Vector Machine (SVM), Multiple Layer Perceptron (MLP) and Adaptive Boosting (Adaboost) (A) applying 5-fold cross-validation and (B) using an independent test set.
3.2. Comparison with the computational frameworks of doc2vec + other ML algorithms
To benchmark the performance of doc2vec in the other ML algorithms, we compared RF with widely used ML algorithms (SVM and Adaboost) and a deep learning method (MLP). For a fair comparison, all the ML classifiers were trained on the same dataset and evaluated on both of the 5-fold cross-validation and independent tests. In this work, we assessed the performance mainly depending on the AUPRC values as the ratio of positive to negative training sets is highly unbalanced (1:10). Here, we tested the performance of different ML models on the 5-fold cross-validation (Fig. 2A), we found that RF clearly outperformed SVM (AUPRC = 0.617; one tailed t-test, p-value = 6.47 × 10−7), MLP (AUPRC = 0.471; one tailed t-test, p-value = 5.12 × 10−8) and Adaboost (AUPRC = 0.147; one tailed t-test, p-value = 4.77 × 10−7). Similar performance ranks can be observed using the independent test sets (Fig. 2B; one tailed t-test, p-value = 1.30 × 10−3, 1.74 × 10−5 and 7.47 × 10−9, respectively). Additionally, ROC curves of each ML classifier using 5-fold cross-validation and independent tests in Fig. S2 confirm our initial observations. Collectively, the RF classifier outperformed the other popular ML algorithms based on the doc2vec encoding.
3.3. Comparison with other popular sequence encoding schemes
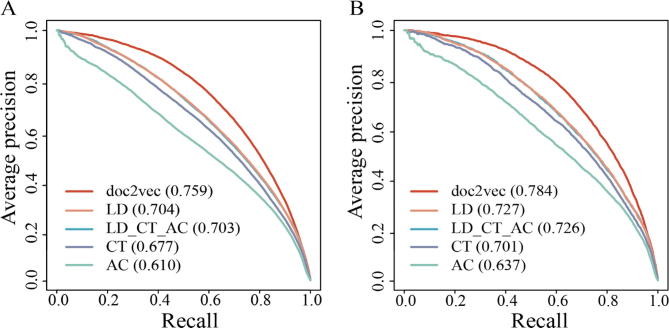
To benchmark the performance of the doc2vec encoding, we trained the RF models based on the other three commonly used sequence encoding schemes (AC, CT and LD). In general, the doc2vec-based RF framework outperformed other encoding schemes using 5-fold cross-validation as well as independent tests (Fig. 3 and Fig. S3; one tailed t-test, all the p-values < 0.01). Notably, the concatenation of the three encoding schemes failed to provide better performance, as results were only comparable to the individual LD encoding, implying that the incorporation of feature vectors did not increase the ratio of signal to noise effectively. Altogether, the doc2vec encoding outperformed the other popular sequence-based encodings based on the RF classifier.
Fig. 3.
Performance of RF classifier in predicting human-virus PPIs based on different sequence-based encoding schemes. Areas under the Precision-Recall curves (AUPRC) indicate that doc2vec encoding provided best prediction performance compared to a combination of Local Descriptor (LD), Conjoint Triad (CT) and Auto Covariance (AC) as well as these encoding techniques separately (A) applying 5-fold cross-validation and (B) using an independent test set.
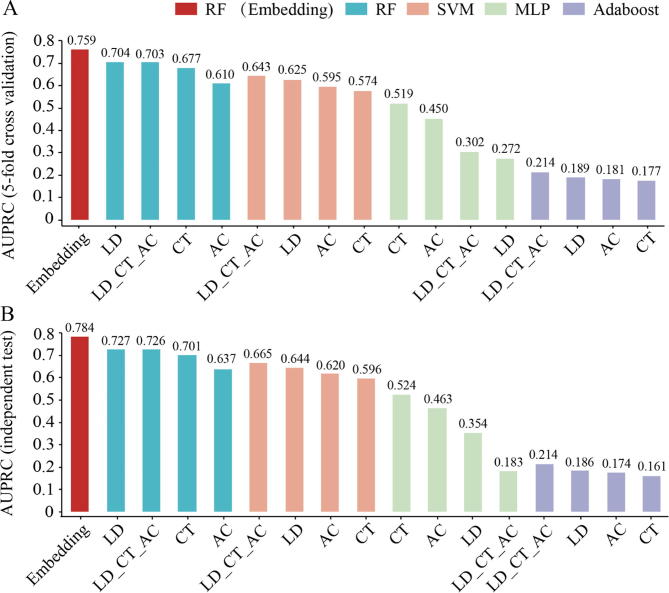
To explore whether doc2vec + RF is an optimal computational framework, we examined combinations of the other algorithms (SVM, Adaboost and MLP) with those popular sequence-based encoding schemes (AC, CT, LD and LD_CT_AC). In Fig. 4, we observed that the AUPRC of doc2vec + RF was 5.5 and 5.7 percentage points higher than that of the second best performing combination (LD + RF; one tailed t-test, p-value = 3.64 × 10−5 and 1.33 × 10−4), when we considered results obtained with 5-fold cross-validation and independent sets (corresponding curves are shown in Figs. S4 and S5). Generally, we observed that combinations of sequence embeddings with RF outperformed other ML methods, with SVM leading MLP and Adaboost.
Fig. 4.
Performance of various combinations of ML algorithms and sequence-based encoding schemes in predicting human-virus PPIs. Areas under the Precision-Recall curves (AUPRC) show that our pipeline that combined doc2vec embedding and Random Forests (RF) outperforms other combinations, (A) applying 5-fold cross-validation and (B) using an independent test. Considering the computational costs of SVM, note that only half of the whole samples were used to train and assess the SVM classifiers.
3.4. Comparison with existing human-virus PPI prediction methods
To further assess our method, we compared our method with three existing prediction methods for human-virus PPIs, including Barman et al.’s method [53], Alguwaizani et al.’s method [54] and DeNovo [43]. Barman et al.’s method uses three common ML methods including SVM, RF, and Naïve Bayes to predict human-virus PPIs based on integrative features such as domain-domain association, network topology and sequence information. After data preprocessing, 1035 positive samples from VirusMINT and 1035 negative samples by negative sampling were used to train and test models through 5-fold cross-validation. As for Alguwaizani et al.’s work, the authors utilized simple features such as the repeat patterns and composition of amino acids to characterize protein sequences for human-virus PPI prediction. Then they also used the SVM algorithm to train their model and compared their model with Barman et al.’s method on the same data set through 5-fold cross-validation. To allow a fair comparison, we first used the identical data set to train our new doc2vec model to infer doc2vec-based features, and retrained our RF-based model using their samples based on 5-fold cross-validation. Notably, Table 1 indicates that our doc2vec-based RF model outperformed Alguwaizani et al.’s SVM model and Barman et al.’s method in terms of most of the performance measures.
Table 1.
Performance comparison of our doc2vec + RF model with Alguwzizani et al.’s and Barman et al.’s methods using Barman et al.’s dataset.
| Method | SN (%) | SP (%) | ACC (%) | PPV (%) | NPV (%) | MCC | AUC | F1 (%) |
|---|---|---|---|---|---|---|---|---|
| Our model | 81.85 | 76.45 | 79.17 | 77.83 | 80.67 | 0.584 | 0.871 | 79.79 |
| Alguwzizani et al.’s SVMa,b | 73.72 | 83.48 | 78.60 | 81.69 | 76.06 | 0.575 | 0.847 | 77.50 |
| Barman et al.’s SVMa,c,d | 67.00 | 74.00 | 71.00 | 72.00 | NA | 0.440 | 0.730 | 69.41 |
| Barman et al.’s RFa,c,d | 55.66 | 89.08 | 72.41 | 82.26 | NA | 0.480 | 0.760 | 66.39 |
The performance was assessed through 5-fold cross-validation.
The corresponding values were retrieved from [54].
The corresponding values were retrieved from [53].
NA means the corresponding parameter is not available. SN: Sensitivity; SP: Specificity; ACC: Accuracy; PPV: Positive Predictive Value (PPV = Precision); NPV: Negative Predictive Value (NPV = TN/(TN + FN)); MCC: Matthews Correlation Coefficient; AUC: the area under the ROC curve; F1 = 2 × (Precision × Recall)/(Precision + Recall).
Regarding the DeNovo method, the authors proposed a domain/motif-based SVM method to predict human-virus PPIs. To compare with DeNovo, we rebuilt our doc2vec and RF model based on the dataset used in DeNovo. Then, we assessed the performance of our reconstructed model on the test set from DeNovo containing 425 positive samples and 425 negative samples. Note that Alguwaizani et al also compared their model against the DeNovo’s model based on the datasets of DeNovo, which has also allowed us to further compare our model with Alguwaizani et al.’s method and DeNovo simultaneously through the DeNovo test set. As shown in Table 2, our model outperformed DeNovo and Alguwaizani et al.’s method considering all performance metrics on the DeNovo’s test set.
Table 2.
Performance comparison of our doc2vec + RF model with DeNovo and Alguwzizani et al.’s method using the test set of DeNovo.
| Method | SN (%) | SP (%) | ACC (%) | PPV (%) | NPV (%) | MCC | AUC | F1 (%) |
|---|---|---|---|---|---|---|---|---|
| Our model | 90.33 | 96.17 | 93.23 | 95.99 | 90.74 | 0.866 | 0.981 | 93.07 |
| Alguwzizani et al.’s SVMa,b | 86.35 | 86.59 | 86.47 | 86.56 | 86.39 | 0.729 | 0.926 | NA |
| DeNovob,c | 80.71 | 83.06 | 81.90 | NA | NA | NA | NA | NA |
The corresponding values were retrieved from [54].
NA means the corresponding parameter is not available.
The corresponding values were retrieved from [43]. SN: Sensitivity, SP: Specificity, ACC: Accuracy, PPV: Positive Predictive Value (PPV = Precision); NPV: Negative Predictive Value (NPV = TN/(TN + FN)); MCC: Matthews Correlation Coefficient; AUC: the area under the ROC curve; F1 = 2 × (Precision × Recall)/(Precision + Recall).
3.5. Cross-species prediction comparison
To further demonstrate the generalization capabilities of our models, we also conducted cross-species prediction experiments between human and viral proteins of three viral species (i.e., H1N1, HIV-1 and EBV). Taking H1N1 as an example, cross-species testing means that we test the prediction performance of human-H1N1 PPIs using the model in which the known human-H1N1 PPIs are totally precluded from the training. Among the 22,653 human-virus PPIs, the number of PPIs between human and H1N1, HIV-1, EBV is 1877, 2215 and 3454, respectively. In brief, we first trained three predictive models based on the datasets excluding the interactions involving the above three viruses respectively. Then, the human-virus PPIs involved in the three viruses were utilized as the test sets to assess the predictive power of each model. To have a robust assessment, we also performed three repeats by sampling. Although cross-species PPI predictions showed a considerable decrease in performance, our model still outperformed other sequence encoding schemes-based ML methods (Table S3). To explore the reasons for the performance decline, we examined the BLAST sequence alignments between viral proteins in training sets and test sets. Boxplots of BLAST E-values in Fig. S6 indicated that H1N1 proteins shared higher sequence similarity with viral proteins in the training set, achieving better performance in predicting human-H1N1 PPIs. Collectively, our results confirmed a reasonably good generalization ability of the proposed method. However, prediction accuracy will be inevitably decreased when the query viral protein is not in the training set or has a low similarity with viral proteins in the training set.
3.6. Webserver implementation
To facilitate the research community, we also built a webserver that provides access to the proposed doc2vec-based RF method, which is freely available at our host-pathogen PPI prediction platform (http://zzdlab.com/InterSPPI/). The prediction model was built based on an unbalanced human host-virus PPI dataset with positive-to-negative ratio 1:10 and trained with the whole training set. The webserver was implemented with CentOS 7.4 and Apache 2.4.6. Users can submit human-virus protein sequence pairs in FASTA format. The webserver will automatically calculate the interaction probability of the query protein pair. Three thresholds to determine whether two proteins interact are provided, which correspond to specificity controls at 99%, 95% and 90%, respectively. Note that the proposed method was optimally designed to process proteins with sequence lengths more than 30 amino acids and less than 5000 amino acids. As we know, human small proteins also perform important functional roles in many biological processes [55], and thus the prediciton issue of small proteins interacting viral proteins should be taken into account in our future work.
4. Conclusions
In this work, we developed a doc2vec embedding-based RF classifier in predicting human-virus PPIs. We observed that our computational framework significantly outperformed computational framework combinations of other widely used ML algorithms and commonly-used sequence encoding schemes. Stringent benchmarking experiments further showed that the proposed method was fully comparable to and often outperformed those existing state-of-the-art human-virus PPI prediction methods. Our results demonstrate that the representation of proteins through feature embedding can allow us to capture more context information from protein sequences, significantly improving prediction performance. We anticipate that our work can provide a useful tool to identify potential interactions between human and viral proteins, further guiding hypothesis-driven experimental efforts to determine proteins involved in human-virus interactions and interrogating the associated functional roles.
As for future developments, the application of deep learning methods has been booming in the past several years, prompting researchers to design deep learning architectures to predict intraspecies PPIs [30], [56], [57]. Furthermore, other features such as protein structural information and host PPI network topology also play an increasingly important role for the prediction of host-pathogen PPIs [25], [58]. By fully accounting for these technical advances, more powerful computational frameworks will be developed to propel human-virus PPI prediction to the next level.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the National Key Research and Development Program of China (2017YFC1200205).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.12.005.
Contributor Information
Stefan Wuchty, Email: wuchtys@cs.miami.edu.
Ziding Zhang, Email: zidingzhang@cau.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Amarasinghe A., Kuritsky J.N., Letson G.W., Margolis H.S. Dengue virus infection in Africa. Emerg Infect Dis. 2011;17:1349–1354. doi: 10.3201/eid1708.101515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guzman M.G., Halstead S.B., Artsob H., Buchy P., Farrar J., Gubler D.J. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8:S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gubler D.J. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang S, Fu C, Lian X, Dong X, Zhang Z. Understanding human-virus protein-protein interactions using a human protein complex-based analysis framework. mSystems 2019;4:e00303–18. [DOI] [PMC free article] [PubMed]
- 5.Dyer M.D., Murali T.M., Sobral B.W. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 7.Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoemaker B.A., Panchenko A.R. Deciphering protein-protein interactions. Part I. experimental techniques and databases. PLoS Comput Biol. 2007;3 doi: 10.1371/journal.pcbi.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 10.Ito T., Tashiro K., Muta S., Ozawa R., Chiba T., Nishizawa M. Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci U S A. 2000;97:1143–1147. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L. Systematic identification of protein complexes in saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 12.Yu H., Luscombe N.M., Lu H.X., Zhu X., Xia Y., Han J.D. Annotation transfer between genomes: protein–protein interologs and protein–DNA regulogs. Genome Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He F., Zhang Y., Chen H., Zhang Z., Peng Y.L. The prediction of protein-protein interaction networks in rice blast fungus. BMC Genomics. 2008;9:519. doi: 10.1186/1471-2164-9-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyer M.D., Murali T.M., Sobral B.W. Computational prediction of host-pathogen protein-protein interactions. Bioinformatics. 2007;23:i159–166. doi: 10.1093/bioinformatics/btm208. [DOI] [PubMed] [Google Scholar]
- 15.Singhal M., Resat H. A domain-based approach to predict protein-protein interactions. BMC Bioinf. 2007;8:199. doi: 10.1186/1471-2105-8-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang A., He L., Wang Y. Prediction of GCRV virus-host protein interactome based on structural motif-domain interactions. BMC Bioinf. 2017;18:145. doi: 10.1186/s12859-017-1500-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q.C., Petrey D., Deng L., Qiang L., Shi Y., Thu C.A. Structure-based prediction of protein–protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinha R., Kundrotas P.J., Vakser I.A. Docking by structural similarity at protein-protein interfaces. Proteins. 2010;78:3235–3241. doi: 10.1002/prot.22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge H., Liu Z., Church G.M., Vidal M. Correlation between transcriptome and interactome mapping data from saccharomyces cerevisiae. Nat Genet. 2001;29:482–486. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- 20.Qi Y., Tastan O., Carbonell J.G., Klein-Seetharaman J., Weston J. Semi-supervised multi-task learning for predicting interactions between HIV-1 and human proteins. Bioinformatics. 2010;26:i645–i652. doi: 10.1093/bioinformatics/btq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen R., Yu H., Greenbaum D., Kluger Y., Krogan N.J., Chung S. A bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 22.Ben-Hur A., Noble W.S. Kernel methods for predicting protein-protein interactions. Bioinformatics. 2005;21:i38–i46. doi: 10.1093/bioinformatics/bti1016. [DOI] [PubMed] [Google Scholar]
- 23.Scott M.S., Barton G.J. Probabilistic prediction and ranking of human protein-protein interactions. BMC Bioinf. 2007;8:239. doi: 10.1186/1471-2105-8-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang S., Li H., He H., Zhou Y., Zhang Z. Critical assessment and performance improvement of plant-pathogen protein-protein interaction prediction methods. Brief Bioinform. 2019;20:274–287. doi: 10.1093/bib/bbx123. [DOI] [PubMed] [Google Scholar]
- 25.Lian X., Yang S., Li H., Fu C., Zhang Z. Machine-learning-based predictor of human-bacteria protein-protein interactions by incorporating comprehensive host-network properties. J Proteome Res. 2019;18:2195–2205. doi: 10.1021/acs.jproteome.9b00074. [DOI] [PubMed] [Google Scholar]
- 26.Ahmed I., Witbooi P., Christoffels A. Prediction of human-Bacillus anthracis protein-protein interactions using multi-layer neural network. Bioinformatics. 2018;34:4159–4164. doi: 10.1093/bioinformatics/bty504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dyer M.D., Murali T.M., Sobral B.W. Supervised learning and prediction of physical interactions between human and HIV proteins. Infect Genet Evol. 2011;11:917–923. doi: 10.1016/j.meegid.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emamjomeh A., Goliaei B., Zahiri J., Ebrahimpour R. Predicting protein-protein interactions between human and hepatitis C virus via an ensemble learning method. Mol BioSyst. 2014;10:3147–3154. doi: 10.1039/c4mb00410h. [DOI] [PubMed] [Google Scholar]
- 29.Cui G., Fang C., Han K. Prediction of protein-protein interactions between viruses and human by an SVM model. BMC Bioinf. 2012;13:S5. doi: 10.1186/1471-2105-13-S7-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun T., Zhou B., Lai L., Pei J. Sequence-based prediction of protein protein interaction using a deep-learning algorithm. BMC Bioinf. 2017;18:277. doi: 10.1186/s12859-017-1700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang K.K., Wu Z., Bedbrook C.N., Arnold F.H. Learned protein embeddings for machine learning. Bioinformatics. 2018;34:2642–2648. doi: 10.1093/bioinformatics/bty178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L., Xia J.F., Gui J. Prediction of protein-protein interactions from protein sequence using local descriptors. Protein Pept Lett. 2010;17:1085–1090. doi: 10.2174/092986610791760306. [DOI] [PubMed] [Google Scholar]
- 33.Cui J., Han L.Y., Li H., Ung C.Y., Tang Z.Q., Zheng C.J. Computer prediction of allergen proteins from sequence-derived protein structural and physicochemical properties. Mol Immunol. 2007;44:514–520. doi: 10.1016/j.molimm.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 34.Davies M.N., Secker A., Freitas A.A., Clark E., Timmis J., Flower D.R. Optimizing amino acid groupings for GPCR classification. Bioinformatics. 2008;24:1980–1986. doi: 10.1093/bioinformatics/btn382. [DOI] [PubMed] [Google Scholar]
- 35.Tong J.C., Tammi M.T. Prediction of protein allergenicity using local description of amino acid sequence. Front Biosci. 2008;13:6072–6078. doi: 10.2741/3138. [DOI] [PubMed] [Google Scholar]
- 36.Le Q., Mikolov T. Distributed representations of sentences and documents. Proc Int Conf Mach Learn. 2014;14:1188–1196. [Google Scholar]
- 37.Lau J.H., Baldwin T. An empirical evaluation of doc2vec with practical insights into document embedding generation. Proc Workshop Represent Learn NLP. 2016;1:78–86. [Google Scholar]
- 38.Ng P. dna2vec: consistent vector representations of variable-length k-mers. arXiv preprint arXiv 2017;1701.06279.
- 39.Kimonthi D, Soni A, Biyani P, Hogan JM. Distributed representations for biological sequence analysis. arXiv preprint arXiv 2016;1608.05949.
- 40.Asgari E., Mofrad M.R. Continuous distributed representation of biological sequences for deep proteomics and genomics. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0141287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ammari M.G., Gresham C.R., McCarthy F.M., Nanduri B. HPIDB 2.0: a curated database for host-pathogen interactions. Database (Oxford) 2016;2016:baw103. doi: 10.1093/database/baw103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das J., Yu H. HINT: high-quality protein interactomes and their applications in understanding human disease. BMC Syst Biol. 2012;6:92–103. doi: 10.1186/1752-0509-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eid F.E., Elhefnawi M., Heath L.S. DeNovo: virus-host sequence-based protein-protein interaction prediction. Bioinformatics. 2016;32:1144–1150. doi: 10.1093/bioinformatics/btv737. [DOI] [PubMed] [Google Scholar]
- 44.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res 2017;45:D158–D169. [DOI] [PMC free article] [PubMed]
- 46.Heinzinger M, Elnaggar A, Wang Y, Dallago C, Nechaev D, Matthes F, et al. Modeling the language of life – deep learning protein sequences. bioRxiv 2019:614313. [DOI] [PMC free article] [PubMed]
- 47.Řehůřek R, Sojka P. Software framework for topic modelling with large corpora. In Proceedings of LREC 2010 Workshop on New Challenges for NLP Frameworks 2010;45–50.
- 48.Pedregosa F., Varoquaux G., Gramfort A., Mueller V., Thirion B., Grisel O. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 49.You Z.H., Lei Y.K., Zhu L., Xia J., Wang B. Prediction of protein-protein interactions from amino acid sequences with ensemble extreme learning machines and principal component analysis. BMC Bioinf. 2013;14:S10. doi: 10.1186/1471-2105-14-S8-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Y., Yu L., Wen Z., Li M. Using support vector machine combined with auto covariance to predict protein-protein interactions from protein sequences. Nucleic Acids Res. 2008;36:3025–3030. doi: 10.1093/nar/gkn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis J, Goadrich M. The relationship between precision-recall and ROC curves. In Proceedings of the 23rd international conference on machine learning 2006;233–240.
- 52.Sing T., Sander O., Beerenwinkel N., Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 53.Barman R.K., Saha S., Das S. Prediction of interactions between viral and host proteins using supervised machine learning methods. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0112034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alguwaizani S., Park B., Zhou X., Huang D.S., Han K. Predicting interactions between virus and host proteins using repeat patterns and composition of amino acids. J Healthc Eng. 2018;2018:1391265. doi: 10.1155/2018/1391265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plaza S., Menschaert G., Payre F. In search of lost small peptides. Annu Rev Cell Dev Biol. 2017;33:391–416. doi: 10.1146/annurev-cellbio-100616-060516. [DOI] [PubMed] [Google Scholar]
- 56.Hashemifar S., Neyshabur B., Khan A.A., Xu J. Predicting protein-protein interactions through sequence-based deep learning. Bioinformatics. 2018;34:i802–i810. doi: 10.1093/bioinformatics/bty573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan X.Y., Zhang Y.N., Shen H.B. Large-scale prediction of human protein−protein interactions from amino acid sequence based on latent topic features. J Proteome Res. 2010;9:4992–5001. doi: 10.1021/pr100618t. [DOI] [PubMed] [Google Scholar]
- 58.Lasso G., Mayer S.V., Winkelmann E.R., Chu T., Elliot O., Patino-Galindo J.A. A structure-informed atlas of human-virus interactions. Cell. 2019;178:1526–1541. doi: 10.1016/j.cell.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.