Abstract
Background:
The mechanisms underlying the proliferation and apoptosis of glioma cells remain unelucidated. A recent study has revealed that microRNA-92b (miR-92b) inhibits apoptosis of glioma cells via downregulating DKK3. Notably, long noncoding RNA nuclear-enriched abundant transcript 1 (NEAT1) is predicted to have a possible interaction with miR-92b.
Objective:
This study aimed to identify whether NEAT1 affects glioma cell proliferation and apoptosis via regulating miR-92b.
Methods:
The expression of NEAT1 was compared between glioma tissues and adjacent tissues as well as between glioma cells and normal astrocytes using quantitative real-time polymerase chain reaction. Glioma cell proliferation was determined by using the 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide assay, and glioma cell apoptosis was determined by using the flow cytometry.
Results:
The expression of NEAT1 was low in glioma tissues and cells compared to the normal ones. Overexpression of NEAT1 inhibited proliferation and promoted apoptosis of glioma cell lines (U-87 MG and U251). The interaction between NEAT1 and miR-92b was confirmed using RNA immunoprecipitation, RNA pull-down assay, and luciferase reporter assay. Importantly, the tumor suppressor function of overexpressing NEAT1 was achieved by downregulating miR-92b and subsequently upregulating DKK3.
Conclusion:
Our findings indicated that NEAT1 acts as a tumor suppressor in glioma cells, which provides a novel target in overcoming glioma growth.
Keywords: glioma, NEAT1, tumor suppressor, miR-92b, DKK3
Introduction
Glioma is the most common primary brain malignancy in adults, with an annual incidence of 4 to 5 individuals per 100 000 population.1 Surgery, chemotherapy, and radiotherapy are the 3 main therapeutic methods for glioma so far, while the poor outcome has made glioma become one of the leading causes of cancer-related death.2 Thus, there have been a growing number of publications focusing on gene regulatory mechanisms that control glioma growth, from inhibiting proliferation to promoting apoptosis.3,4 However, the mechanisms underlying glioma proliferation and apoptosis remain largely unknown, especially in the regulatory patterns of noncoding RNAs (ncRNAs).
MicroRNAs (miRNAs) are a subset of ncRNAs, with a length of approximately 20 nucleotides, which are known to regulate gene expression in many types of cancers, including leukemia, lung cancer, and breast cancer, and so on.5 After the translocation of the mature miRNAs into the cytoplasm, they bind to DICER and RNA-induced silencing complex (RISC), which includes argonaute (AGO) proteins. With the conjunction to RISC, a guide strand helps to navigate the mature miRNAs to the target messenger RNA (mRNA), consequently resulting in downregulation of target genes.6 In glioma, miR-92b has been reported to inhibit apoptosis of glioma cells via downregulating its target gene——DKK3,7 suggesting miR-92b as an important oncogene in glioma. However, the upstream regulator of miR-92b has not been elucidated.
Long noncoding RNAs (lncRNAs), longer than 200 base pairs, are series of transcripts with no protein-coding function, which belong to the ncRNAs.8 Recently, several lncRNAs have been reported to participate in regulating proliferation and apoptosis of glioma, such as LINC00319,9 HCG11,10 and SNHG20.11 Long noncoding RNA nuclear-enriched abundant transcript 1 (NEAT1) is a critical tumor growth regulator that plays a vital role in many cancers, including breast cancer,12 gastric cancer,13 and hepatic cancer.14 However, its role in glioma has not been completely elucidated yet. As we know, the inhibition of cell proliferation and the promotion of cell apoptosis of glioma are associated with the activation of p53 signaling.15 As NEAT1 is a transcriptional target of p53,16 we assumed that NEAT1 may be involved in the regulation of proliferation and apoptosis of glioma. Notably, NEAT1 is predicted to have a possible interaction with miR-92b by an online software TargetScan.
Therefore, in the current study, the expression of NEAT1 was compared between glioma tissues and adjacent tissues, as well as between glioma cells and normal astrocytes. The results indicated that NEAT1 was significantly downregulated in glioma tissues and cells. Meanwhile, the interaction between NEAT1 and miR-92b was confirmed by using RNA immunoprecipitation, RNA pull-down assay, and luciferase reporter assay. The overexpression of NEAT1 was demonstrated to inhibit proliferation and promote apoptosis of glioma cells via downregulating miR-92b and subsequently upregulating DKK3.
Materials and Methods
Clinical Samples
A total of 20 cases of patients with glioma were enrolled in the study. The glioma tissues and the corresponding adjacent tissues were collected during surgical resection at hospital. All the patients were admitted in hospital from January 2013 to January 2018, including 8 grade I-II tumors, 10 grade III tumors, and 2 grade IV tumors. After the surgery, 20 pairs of fresh frozen tissues were maintained in the −80°C container.
Cell Line, Culture, and Transfection
The normal human astrocytes (NHA; BeNa Culture Collection, Beijing, China) and human glioma cell lines (U-87 MG and U251; Procell Life Science & Technology Co, Ltd., Wuhan, China) were cultured in the Roswell Park Memorial Institute 1640 medium (Sigma-Aldrich, St Louis, Missouri) supplemented with 10% fetal bovine serum (BeNa Culture Collection) at 37°C in an atmosphere of 5% CO2.
The Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, Massachusetts) was used in cell transfection, and the transfection was performed in accordance with the manufacturer’s instructions. The NEAT1-overexpressing vector (pcDNA-NEAT1) and its control (pcDNA), the miR-92b mimic and its control (prenegative control), the miR-92b inhibitor and its control (negative control), and the short hairpin RNA of NEAT1 (shNEAT1) and its control (short hairpin RNA) were synthesized by Genechem (Shanghai, China).
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted from tissues or cells using the TRIzol reagent (Invitrogen, Waltham, Massachusetts). The miRNA First Strand cDNA Synthesis Kit (Gene Copeia, Guangzhou, China) or the All-in-One Fist-Strand cDNA Synthesis Kit (Gene Copeia) was used to synthesize complementary DNAs. Real-time polymerase chain reaction (RT-PCR) was performed by using the miRNAs qPCR Kit (GeneCopeia) or the All-in-One qPCR Mix Kit (GeneCopeia) with CFX96TM Real-Time PCR System (Bio-Rad, Hercules, California); U6 and GAPDH were used as the internal control. The relative RNA expression was calculated by 2−ΔΔCt method.
Western Blot Analysis
Glioma cells were collected and lysed in radioimmunoprecipitation assay (Beytime, Shanghai, China) containing protease inhibitor at 4°C for 30 minutes. Then they were isolated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and transferred onto polyvinylidene difluoride membranes (Thermo Fisher Scientific). The membranes were blocked in Tris-buffered saline buffer with Tween 20 with 5% nonfat milk for 2 hours at 37°C followed by being probed with the primary antibodies against DKK3 (1:1000, ab186409, Abcam, Cambridge, United Kingdom) and β-actin (1:500, ab8226, Abcam) overnight at 4°C. After incubating with secondary antibody (1:2000, ab205719, Abcam), the membranes were exposed to light film (BioMax MR; Kodak, Rochester, New York).
Cell Proliferation Assay
Cell proliferation was evaluated using the 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma-Aldrich) assay. The glioma cells transfected with the indicated vectors were seeded at a density of 2 × 103 cells/well in the 96-well culture plates. The quantification of mitochondrial dehydrogenase activity was measured using the enzymatic conversion of MTT to a colored formazan product. The MTT (10 μL, 10 mg/mL) solution was added to the cells followed by incubation for 4 hours, and the reaction was terminated by removal of the supernatant and addition of 100 μL dimethyl sulfoxide to dissolve the formazan product. The optical density of each well was measured at 570 nm after 30 minutes using a plate reader (ELx808 Bio-Tek Instruments, Winooski, Vermont).
Cell Apoptosis Assay
Cell apoptosis was determined using flow cytometry. In Brief, glioma cells transfected with the indicated vectors were seeded in 6-well plates at a density of 1 × 103 cells/well. After incubation of 24 hours, cells were harvested and washed with phosphate-buffered saline twice. Then the cells were resuspended in Annexin V-fluorescein isothiocyante (Annexin V-FITC; 5 μL) and propidium iodide (5 μL) by Annexin V-FITC Apoptosis Detection Kit (Invitrogen, Carlsbad, California). The apoptotic rate was measured by FACS Calibur (BD, San Jose, CA, USA).
RNA Immunoprecipitation
The RNA immunoprecipitation (RIP) assay was performed using EZ-Magna RIP RNA-binding Protein Immunoprecipitation Kit (Millipore, St Louis, Missouri) according to the manufacturer’s instructions. The U-87 MG cells were lysed using RIP lysis buffer, and cell lysates were incubated with RIP buffer, including magnetic beads conjugated to human anti-Ago2 antibody (ab32381, Abcam), mouse immunoglobulin G (ab190475, Abcam) served as the control. Coprecipitated RNAs were isolated using TRIzol reagent (Takara, Dalian, China) and measured using quantitative RT-PCR (qRT-PCR).
RNA Pull-Down
Full-length of NEAT1 RNA was transcribed into the U-87 MG cells using T7 RNA polymerase. A total of 50 pmol NEAT1 RNA was labeled using biotin and T4 RNA ligase via a PierceTM RNA 3′End Desthiobiotinylation Kit (Thermo Fisher Scientific). The RNA pull-down assay was performed according to the PierceTM Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific). Briefly, the cells were lysed with Pierce IP Lysis Buffer and incubated on ice for 5 minutes. The lysates were centrifuged at 13000×g for 10 minutes, and the supernatant was transferred to a new tube. The labeled RNA was added to 50 μL of beads followed by incubation for 30 minutes at room temperature with agitation. The RNA-bound beads were incubated with the lysates for 60 minutes. The RNA-binding miRNAs were washed and eluted, and the binding miRNAs were detected using qRT-PCR.
Luciferase Reporter Assay
The fragment from lncRNA NEAT1 containing the predicted miR-92b-binding site was amplified by PCR and was cloned into a pmirGLO Dual-luciferase miRNA Target Expression Vector (Promega, Madison, Wisconsin) to construct the reporter vector NEAT1-wild type. To mutate the putative binding site of miR-92b in NEAT1, the sequence of the putative binding site was replaced by using TaKaRa MutanBEST Kit (Takara, Dalian, China). The vectors and miR-92b mimics were cotransfected into 293T cells. The luciferase activity was determined using the dual-luciferase reporter assay system (Promega).
Statistical Analysis
The statistical significance between 2 comparator groups was calculated using the 2-tailed Student t test. The differences of lncRNA expressions between paired tissue sample were evaluated with Wilcox matched pairs signed-ranks test. All the data were analyzed by using SPSS Statistical Package version 18. A P value < .05 was considered as statistically significant.
Results
Expression of LncRNA NEAT1 is Low in Glioma Tissues and Cells
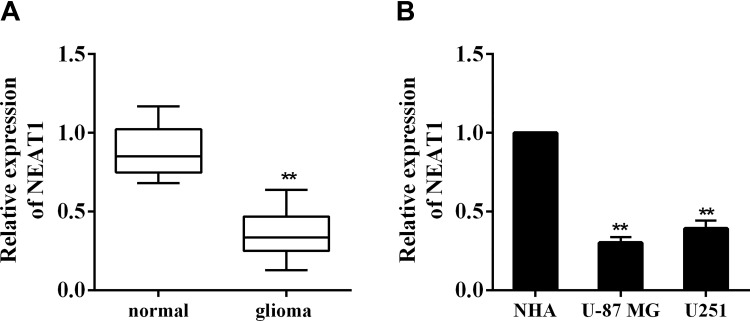
The inhibition of cell proliferation and the promotion of cell apoptosis of glioma are associated with the activation of p53 signaling.15 As a transcriptional target of p53, lncRNA NEAT1 is reported to act like a tumor suppressor,16 although its function in glioma has not been elucidated. To identify the expression of NEAT1 in glioma, we collected glioma tissues and adjacent tissues from 20 patients. The qRT-PCR result indicated that NEAT1 was significantly downregulated in glioma tissues in comparison with normal adjacent tissues (Figure 1A, 0.36 ± 0.14 vs 0.88 ± 0.16, P < .01). Then the expression of NEAT1 was compared in normal astrocytes (NHA) and glioma cell lines (U-87 MG and U251). It showed that NEAT1 was markedly reduced in glioma cell lines than that in astrocytes (Figure 1B, 0.30 ± 0.04 and 0.39 ± 0.05 vs 1, both P < .01). These data indicated that the expression of NEAT1 was low in glioma tissues and cells.
Figure 1.
Expression of lncRNA NEAT1 in glioma tissues and cells. A, Glioma tissues (n = 20) and normal adjacent tissues (n = 20) were collected from glioma patients during surgical resections. The expression of NEAT1 was determined using qRT-PCR. B, The expression of NEAT1 in normal astrocytes (NHA) and glioma cell lines (U-87 MG and U251) was determined using qRT-PCR. **P < .01 vs normal or NHA. lncRNA indicates long noncoding RNA; NEAT1, nuclear-enriched abundant transcript 1; NHA, normal human astrocytes; qRT-PCR, quantitative real-time polymerase chain reaction.
Overexpression of NEAT1 Inhibits Proliferation and Promotes Apoptosis of Glioma Cells
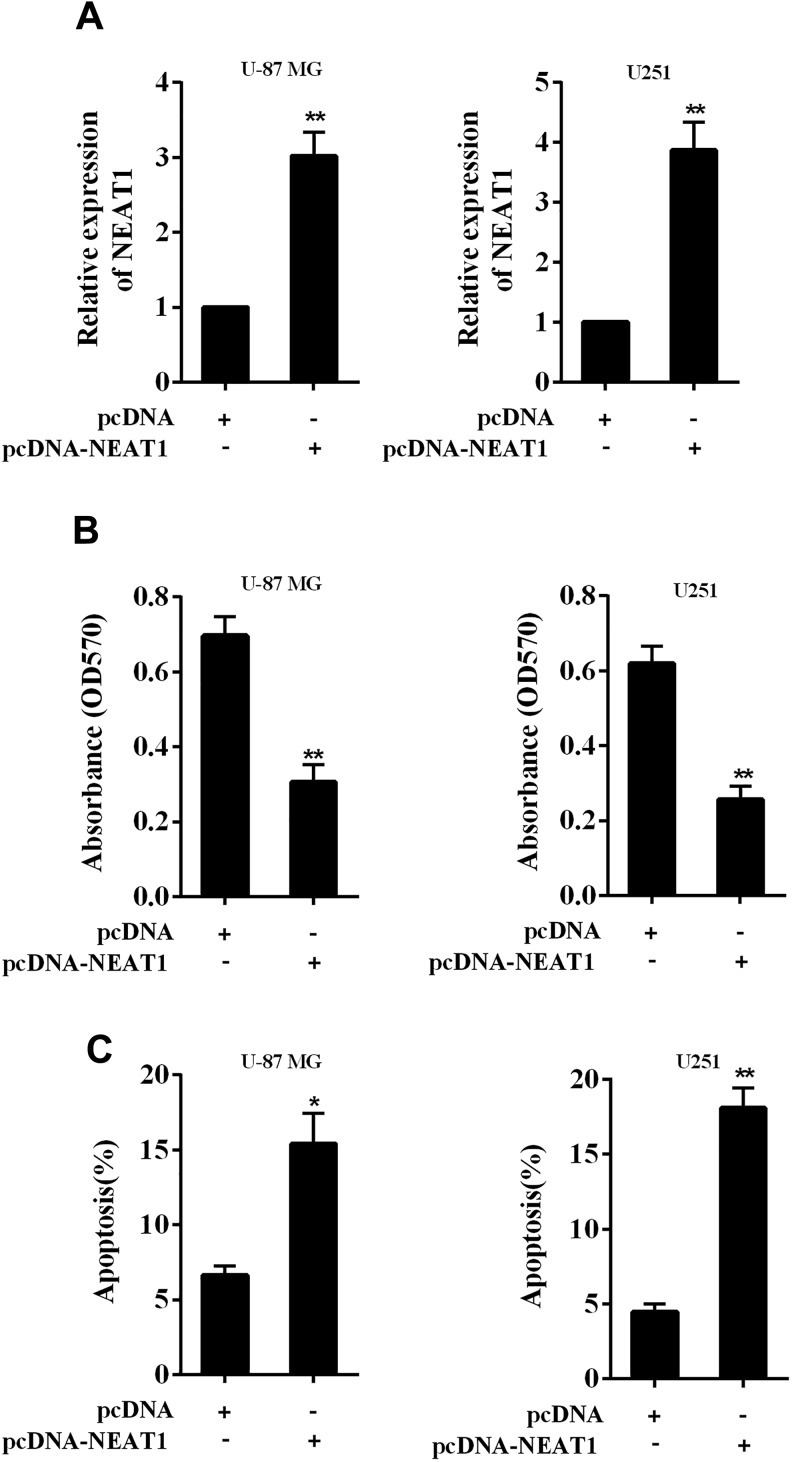
Given that NEAT1 was downregulated in glioma tissues and cells, we further investigated the effect of NEAT1 on glioma cells after overexpression. We overexpressed NEAT1 in glioma cell lines (U-87 MG and U251) using the overexpressing vector of NEAT1 (pcDNA-NEAT1; Figure 2A). The cell proliferation was markedly inhibited after NEAT1 overexpression in both cell lines (Figure 2B, both P < .01), while the cell apoptosis was markedly increased after NEAT1 overexpression (Figure 2C, P < .05 in U-87 MG cells and P < .01 in U251 cells). These data revealed that NEAT1 acts as a tumor suppressor in glioma cells.
Figure 2.
Effect of overexpressing lncRNA NEAT1 in proliferation and apoptosis of glioma cells. A, Glioma cell lines (U-87 MG and U251) were transiently transfected with the NEAT1-overexpressing vector (pcDNA-NEAT1), and the upregulation of NEAT1 after the transfection was confirmed using qRT-PCR. B, The cell proliferation after overexpressing NEAT1 was determined using MTT assay. C, The cell apoptosis after overexpressing NEAT1 was determined using flow cytometry. *P < .05, **P < .01 vs pcDNA. lncRNA indicates long noncoding RNA; MTT, 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide; NEAT1, nuclear-enriched abundant transcript 1; pcDNA, the negative control of pcDNA-NEAT1; qRT-PCR, quantitative real-time polymerase chain reaction.
The Interaction Between NEAT1 and miR-92b
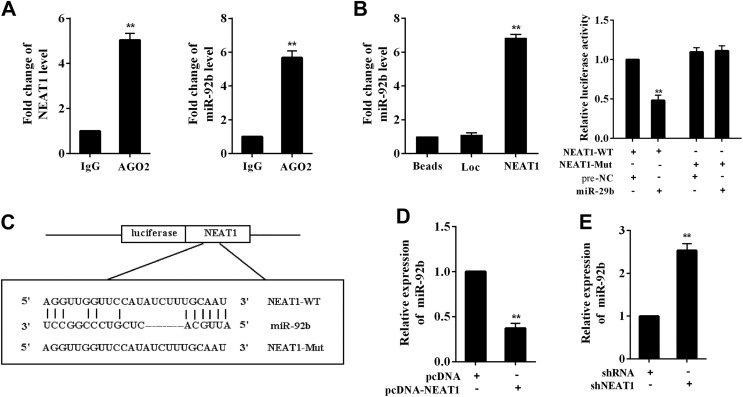
A previous study has reported that miR-92b inhibits glioma cell apoptosis via targeting DKK3.7 Interestingly, the interaction between NEAT1 and DKK3 was predicted by the online software TargetScan. To elucidate such interaction, RIP assay and RNA pull-down assay were performed in U-87 MG cells. In the RIP experiment, the increased expression of NEAT1 and miR-92b in AGO2 immunoprecipitates was detected, suggesting the combination of NEAT1 and miR-92b (Figure 3A). In the RNA pull-down experiment, the expression of miR-92b was significantly increased in the pull-down compounds of NEAT1 (Figure 3B). We also mutated the predicted binding sequences of NEAT1 and miR-92b in the luciferase reporter vector, and the luciferase activity was dramatically reduced in the unmutated vector group after the cotransfection of miR-29b mimic (P < .01), while the luciferase activity in the mutated vector group was not significantly changed (Figure 3C). Meanwhile, NEAT1 expression was negatively correlated with miR-29b using the transfection of NEAT1-overexpressing vector (pcDNA-NEAT1) and the NEAT1-knocking down vector (shNEAT1; Figure 3D and E). These findings indicated the direct binding between NEAT1 and miR-92b in glioma cells.
Figure 3.
Interaction of NEAT1 and miR-92b. The direct binding between NEAT1 and miR-92b was determined using (A) RIP assay and (B) RNA pull-down assay in U-87 MG cells. Immunoglobulin G was used as the control of AGO2. Loc was used as the control of NEAT1. C, The wild type (WT) and the mutant (Mut) luciferase reporter vectors were constructed. The luciferase activity was determined after the cotransfection of WT/Mut vector and miR-92b mimic. D, The expression of miR-92b after overexpressing NEAT1. E, The expression of miR-92b after downregulating NEAT1. Prenegative control (Pre-NC) was used as the control of miR-92b mimic. **P < .01 vs IgG, Loc, NEAT1-WT + pre-NC, pcDNA, or shRNA. IgG indicates immunoglobulin G; NEAT1, nuclear-enriched abundant transcript 1; pcDNA-NEAT1, the overexpression vector of NEAT1; RIP, RNA immunoprecipitation; shNEAT1, short hairpin RNA of NEAT1.
NEAT1 Modulates DKK3 Expression Via Regulating miR-92b
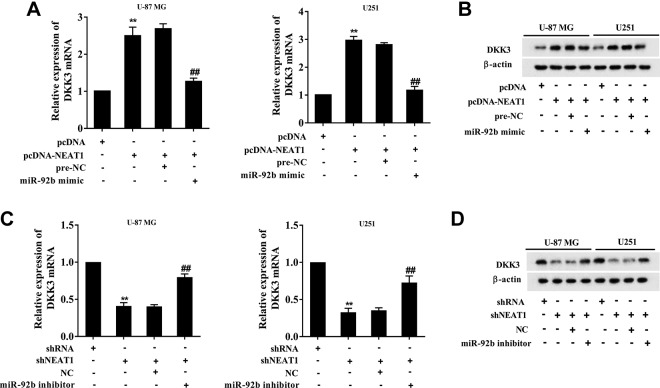
To clarify that whether DKK3 expression was modulated by NEAT1/miR-92b, the glioma cell lines (U-87 MG and U251) were transfected with pcDNA-NEAT1 or cotransfected with miR-92b mimic. The transfection of pcDNA-NEAT1 enhanced the mRNA and protein expressions of DKK3, while the cotransfection negated such response (Figure 4A and B). The glioma cell lines (U-87 MG and U251) were also transfected with shNEAT1 or cotransfected with miR-92b inhibitor. The transfection of shNEAT1 reduced the mRNA and protein expressions of DKK3, while the cotransfection negated such response (Figure 4C and D). These data indicated that NEAT1 promotes DKK3 expression via downregulating miR-92b.
Figure 4.
Nuclear-enriched abundant transcript 1 modulates DKK3 expression via regulating miR-92b. The glioma cell lines (U-87 MG and U251) were transfected with pcDNA-NEAT1 or cotransfected with miR-92b mimic. The (A) mRNA and (B) protein expressions of DKK3 after transfection were determined using qRT-PCR and Western blot analysis, respectively. Prenegative control (Pre-NC) was used as the control of miR-92b mimic. The glioma cell lines (U-87 MG and U251) were also transfected with shNEAT1 or cotransfected with miR-92b inhibitor. The (C) mRNA and (D) protein expressions of DKK3 after transfection were determined using qRT-PCR and Western blot analysis, respectively. Negative control (NC) was used as the control of miR-92b inhibitor. **P < .01 vs pcDNA or shRNA. ## P < .01 vs pcDNA-NEAT1 + pre-NC or shNEAT1 + NC. mRNA indicates messenger RNA; NEAT1, nuclear-enriched abundant transcript 1; shRNA, short hairpin RNA; qRT-PCR, quantitative real-time polymerase chain reaction.
NEAT1 Inhibits Proliferation and Promotes Apoptosis of Glioma Cells Via Downregulating miR-92b and Subsequently Upregulating DKK3
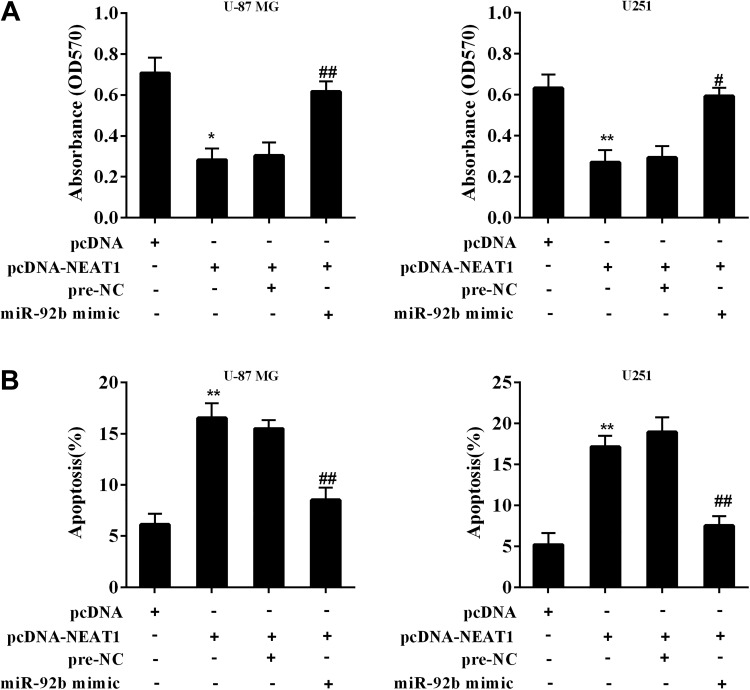
The glioma cell lines (U-87 MG and U251) were transfected with pcDNA-NEAT1 or cotransfected with miR-92b mimic followed by determining cell proliferation and apoptosis using MTT assay and flow cytometry. The cotransfection restored the cell viability which was reduced by NEAT1 overexpression (Figure 5A). The cotransfection reduced the enhancement of the number of apoptotic cells which was raised by NEAT1 overexpression (Figure 5B). Our findings demonstrated that NEAT1 inhibits proliferation and promotes apoptosis of glioma cells via downregulating miR-92b and subsequently upregulating DKK3.
Figure 5.
Effect of NEAT1/miR-92b/DKK3 on glioma cell proliferation and apoptosis. The glioma cell lines (U-87 MG and U251) were transfected with pcDNA-NEAT1 or cotransfected with miR-92b mimic. A, The cell proliferation after transfection was determined using MTT assay. B, The cell apoptosis after transfection was determined using flow cytometry. Prenegative control (Pre-NC) was used as the control of miR-92b mimic. **P < .01 vs pcDNA. ## P < .01 vs pcDNA-NEAT1 + pre-NC. MTT indicates 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide; NEAT1, nuclear-enriched abundant transcript 1.
Discussion
Glioma is the most prevalent primary tumors of the brain and spinal cord, accounting for 50% to 60% of all brain malignancies.17 It can be divided into 4 grades, including grade I tumors such as pilocytic astrocytoma, pleomorphic xanthoastrocytoma, and subependymal giant cell astrocytoma; grade II tumors such as oligodendroglioma and astrocytoma; grade III tumors such as anaplastic oligodendroglioma, anaplastic astrocytoma, anaplastic oligoastrocytoma, and anaplastic ependymoma; and grade IV glioblastoma.18 The median survival time of patients with low-grade glioma (grades II and III) is 3 to 10 years, in comparison with 1.5 years for patients with grade IV.19,20 Overall, the prognosis of glioma is still poor due to the limitation of the current therapeutic methods, including surgery and chemotherapy. Thus, understanding the growth pattern of glioma modulated by gene can possibly provide a new insight in treating glioma. In the current study, we focused on regulation of lncRNA and found a novel tumor suppressor, lncRNA NEAT1, in glioma which has a potential of inhibiting the tumor growth.
Long noncoding RNAs are nonprotein-coding transcripts which are longer than 200 nucleotides. The mechanisms of lncRNAs that regulate gene expression consist of a variety of ways, including the genomic imprinting, the control of transcription or posttranscriptional processing, chromatin modification, and regulation of protein function.21 Therefore, lncRNAs play important roles in a large number of biological processes including development, cell growth, and tumorigenesis. In glioma, several lncRNAs have been reported to affect tumorigenesis in recent years. Li et al9 found that LINC00319 promotes the tumorigenesis of glioma by regulating HMGA2 (high-mobility group AT-hook 2), and the high level of LINC00319 is closely associated with the poor prognosis of patients with glioma. Chen and his colleagues10 demonstrated that lncRNA HCG11 inhibits glioma progression by regulating miR-496/CPEB3 axis. The study conducted by Liu et al11 showed that lncRNA SNHG20 modulated the malignancy of glioma cells via targeting the miR-4486/MDM2/p53 pathway. In our works, we found that lncRNA NEAT1 was dramatically downregulated in glioma tissues and cells in comparison with normal adjacent tissues and astrocytes. Meanwhile, the overexpression of NEAT1 significantly suppressed proliferation and enhanced apoptosis of glioma cells, suggesting the function of NEAT1 in inhibiting glioma progression.
Competing endogenous RNA (ceRNA) mechanism is the most well-identified function of lncRNAs in regulating gene expression. When acting as a ceRNA, lncRNAs can sponge miRNAs thus preventing them to exert the inhibition effect on downstream target mRNAs.22 For example, lncRNA SNHG16 promotes glioma tumorigenicity through sponging miR-373-3p, therefore increasing estimated glomerular filtration rate expressing and subsequently activating phosphatidylinositol 3-kinase/AKT (PI3K/AKT) signaling pathway.23 Similarly, our work found the direct binding between NEAT1 and miR-92b using RIP and RNA pull-down assay. Meanwhile, the luciferase reporter assay indicated that miR-92b expression was negatively correlated with NEAT1. The rescue experiment in glioma cells also indicated the modulation of NEAT1/miR-92b/DKK3 axis. Except for the ceRNA mechanism, lncRNAs are reported to affect protein expression via direct binding. For instance, lncRNA PLAC2 in nucleus can bind with the transcription factor, STAT1, and interact with the RPL36 promoter, thus blocking the cell cycle progression in glioma.24 Inspired by their study, whether NEAT1 modulates protein expressions in glioma deserves further investigations in the future.
NEAT1 is a critical tumor growth regulator that plays a vital role in many cancers. When acting as an oncogene, NEAT1 promotes breast cancer progression via regulating miR-124/STAT3,12 and the high level of NEAT1 expression can predict the metastasis of lymph nodes in various malignant tumors.25 When acting as a tumor suppressor, NEAT1 is repressed in primary chronic myeloid leukemia cells and modulates imatinib-induced apoptosis.26 Consistent with their study, we found the tumor-suppressing effect of NEAT1 in glioma using the transfection of NEAT1 overexpressing vectors. In addition, the result of Zhou et al27 indicated that NEAT1 was downregulated in glioma cells after interfering SRSF1, an oncodriver in glioma via splicing control. Previous studies also indicated that DKK3 knockdown promoted the migration of glioma cells,28 and that DKK3 overexpression enhanced chemosensitivity to temozolomide in glioma.29 Whether NEAT1 affects the migration and chemosensitivity via miR-92b/DKK3 pathway deserves further investigations.
In conclusion, our findings indicated that NEAT1 is downregulated in glioma tissues and cells, and overexpressing NEAT1 suppressed proliferation and promoted apoptosis of glioma cells. Meanwhile, the tumor-suppressing function of NEAT1 was modulated via the miR-92b/DKK3 pathway, suggesting NEAT1 as a novel target in treating glioma.
Footnotes
Authors’ Note: The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request. This study was approved by the Institute Research Medical Ethics Committee of General Hospital of Northern Theater Command (No.2017-59). Written informed consents were obtained from all the patients involved.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the National Natural Science Foundation of China (Grant No. 81671174).
ORCID iD: Guobiao Liang, PhD  https://orcid.org/0000-0002-4350-3877
https://orcid.org/0000-0002-4350-3877
References
- 1. Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 2. Zhang L, Zhang Y. Tunneling nanotubes between rat primary astrocytes and C6 glioma cells alter proliferation potential of glioma cells. Neurosci Bull. 2015;31(3):371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yu M, Yu S, Gong W, Chen D, Guan J, Liu Y. Knockdown of linc01023 restrains glioma proliferation, migration and invasion by regulating IGF-1R/AKT pathway. J Can. 2019;10(13):2961–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang S, Gao M, Yu L. GATAD1 gene amplification promotes glioma malignancy by directly regulating CCND1 transcription. Cancer Med. 2019;8(11):5242–5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. MiRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6(3):235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 7. Li Q, Shen K, Zhao Y, Ma C, Liu J, Ma J. MiR-92b inhibitor promoted glioma cell apoptosis via targeting DKK3 and blocking the Wnt/beta-catenin signaling pathway. J Transl Med. 2013;11(1):302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun X, Luo LH, Zhong KZ, Li JR. LncRNA PCAT29 Suppresses cell proliferation, invasion, and migration in renal carcinoma by regulating FLOT1. Clin Surg Res Comm. 2018;2:35–42. [Google Scholar]
- 9. Li Q, Wu Q, Li Z, et al. Tian, LncRNA LINC00319 is associated with tumorigenesis and poor prognosis in glioma. Eur J Pharmacol. 2019;861:172556. [DOI] [PubMed] [Google Scholar]
- 10. Chen Y, Bao C, Zhang X, Lin X, Huang H, Wang Z. Long non-coding RNA HCG11 modulates glioma progression through cooperating with miR-496/CPEB3 axis. Cell Prolif. 2019;52(5):e12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu J, Cheng LG, Li HG. LncRNA SNHG20 promoted the proliferation of glioma cells via sponging miR-4486 to regulate the MDM2-p53 pathway. Eur Rev Med Pharmacol Sci. 2019;23(12):5323–5331. [DOI] [PubMed] [Google Scholar]
- 12. Pang Y, Wu J, Li X, et al. NEAT1/miR124/STAT3 feedback loop promotes breast cancer progression. Int J Oncol. 2019;55(3):745–754. [DOI] [PubMed] [Google Scholar]
- 13. Zhang J, Guo S, Piao HY, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75(3):379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shaker OG, Abdelwahed MY, Ahmed NA, et al. Evaluation of serum long noncoding RNA NEAT and MiR-129-5p in hepatocellular carcinoma. IUBMB Life. 2019;71(10):1571–1578. [DOI] [PubMed] [Google Scholar]
- 15. Xu LX, Li ZH, Tao YF, et al. Pan, Histone acetyltransferase inhibitor II induces apoptosis in glioma cell lines via the p53 signaling pathway. J Exp Clin Cancer Res. 2014;33:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Idogawa M, Ohashi T, Sasaki Y, Nakase H, Tokino T. Long non-coding RNA NEAT1 is a transcriptional target of p53 and modulates p53-induced transactivation and tumor-suppressor function. Int J Cancer. 2017;140(12):2785–2791. [DOI] [PubMed] [Google Scholar]
- 17. Morgan LL. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2015;17(4):623–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ostrom QT, Gittleman H, Fulop J, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro Oncol. 2015;17(suppl 4):iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30(17):1913–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anderson DM, Anderson KM, Chang CL, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deveson IW, Hardwick SA, Mercer TR, Mattick JS. The dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet. 2017;33(7):464–478. [DOI] [PubMed] [Google Scholar]
- 23. Zhou XY, Liu H, Ding ZB, Xi HP, Wang GW. lncRNA SNHG16 promotes glioma tumorigenicity through miR-373/EGFR axis by activating PI3K/AKT pathway. Genomics. 2019. S0888–7543(18): 30545–30547. [DOI] [PubMed] [Google Scholar]
- 24. Hu YW, Kang CM, Zhao JJ, et al. LncRNA PLAC2 down-regulates RPL36 expression and blocks cell cycle progression in glioma through a mechanism involving STAT1. J Cell Mol Med. 2018;22(1):497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang L, Zhang Z, Yu Z. Long non-coding RNA NEAT1 can predict various malignant tumour lympha node metastasis: a meta-analysis. Artif Cells Nanomed Biotechnol. 2019;47(1):2516–2520. [DOI] [PubMed] [Google Scholar]
- 26. Zeng C, Liu S, Lu S, et al. The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Mol Can. 2018;17(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou X, Li X, Yu L, et al. The RNA-binding protein SRSF1 is a key cell cycle regulator via stabilizing NEAT1 in glioma. Int J Biochem Cell Biol. 2019;113:75–86. [DOI] [PubMed] [Google Scholar]
- 28. Peng G, Yang C, Liu Y, Shen C. MiR-25-3p promotes glioma cell proliferation and migration by targeting FBXW7 and DKK3. Exp Ther Med. 2019;18:769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fujihara T, Mizobuchi Y, Nakajima K, et al. Down-regulation of MDR1 by Ad-DKK3 via Akt/NFkappaB pathways augments the anti-tumor effect of temozolomide in glioblastoma cells and a murine xenograft model. J Neurooncol. 2018;139(2):323–332. [DOI] [PubMed] [Google Scholar]