Abstract
Identification of pheromone receptors plays a central role for uncovering signaling pathways that underlie chemical communication in animals. Here, we describe the synthesis and bioactivity of photoaffinity probes for the ascaroside ascr#8, a sex-pheromone of the model nematode, Caenorhabditis elegans. Structure–activity studies guided incorporation of alkyne- and diazirine-moieties and revealed that addition of functionality in the sidechain of ascr#8 was well tolerated, whereas modifications to the ascarylose moiety resulted in loss of biological activity. Our study will guide future probe design and provides a basis for pheromone receptor identification via photoaffinity labeling in C. elegans.
Introduction
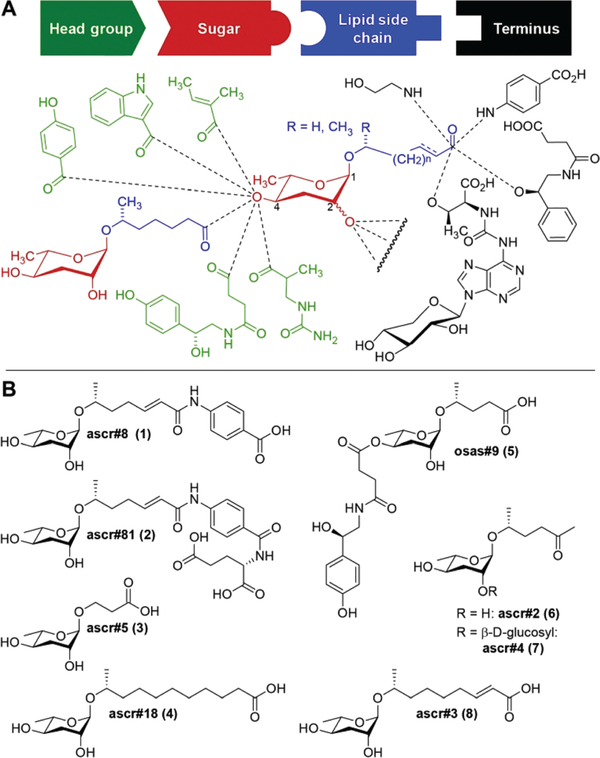
The nematode Caenorhabditis elegans has become an important model for inter-organismal signaling via pheromones. C. elegans and other nematode species communicate with conspecifics via ascarosides, a family of small-molecule pheromones based on the 3,6-dideoxysugar, L-ascarylose, linked to fatty acid-like side chains (Fig. 1). Ascarosides can be further decorated with building blocks from diverse primary metabolic pathways, e.g. the likely folate-derived p-aminobenzoic acid in ascr#8 1 (1), or the neurotransmitter octopamine in osas#9 2 (5). Ascarosides are involved in almost every aspect of the life history of C. elegans, including developmental changes,3–5 aging,6,7 and diverse behaviors e.g. dispersal,8 aggregation,9,10 and mating.11,12 Ascarosides are typically sensed as cocktails of compounds,13 often functioning in synergy.1,8,12 Notably, even small changes in ascaroside structures can result in dramatic differences in biological responses.7
Fig. 1.
Structures of ascaroside pheromones from C. elegans and other nematodes. (A) Overview of structural diversity of ascaroside pheromones. Building blocks from diverse metabolic pathways are combined to furnish a modular library of signaling molecules, with specific examples shown in (B).
Three ascarosides (ascr#2 (6), ascr#3 (8), and ascr#4 (7)) have been shown to synergistically attract males,12 whereas the structurally distinct ascr#8 (1) acts as a potent male attractant even in the absence of other ascarosides.1,11 ascr#8 (1) and the related ascr#81 (2) are unique among identified ascaroside pheromones in that they incorporate a p-aminobenzoate moiety.14,15
Identification of ascaroside receptors has not kept pace with the discovery of new ascarosides and activities, and it appears that multiple G protein-coupled receptors (GPCRs) are involved in sensing individual compounds.16,17 Receptor identification has been hampered in part by the large number of GPCRs in C. elegans, which likely exceeds 1000.18 In order to identify ascaroside receptors, previous studies have employed reverse genetics screens,16,19 as well as quantitative trace locus analyses.20–22 The use of ascaroside-based photoaffinity probes could enable targeted receptor identification and, importantly, has the advantage to demonstrate direct ascaroside-receptor binding. In a previous study, an ascr#2-based photoaffinity probe was used to confirm binding to the GPCR DAF-37, which was originally identified via immunoprecipitation;17 however, the ascr#2 probe used in this study featured extensive structural modifications and correspondingly was much less biologically active than unmodified ascr#2 and thus unlikely to facilitate de novo receptor identification.
Toward identification of receptor(s) of the sex pheromone ascr#8, we aimed to design a photoaffinity probe that would enable covalent linking and receptor pull-down, yet retain most bioactivity. We synthesized four ascr#8 probe designs, two of which retained potent activity. The inactive probes provide insight into structure-activity relationships and may help guide design for probes of other ascaroside pheromones.
Results
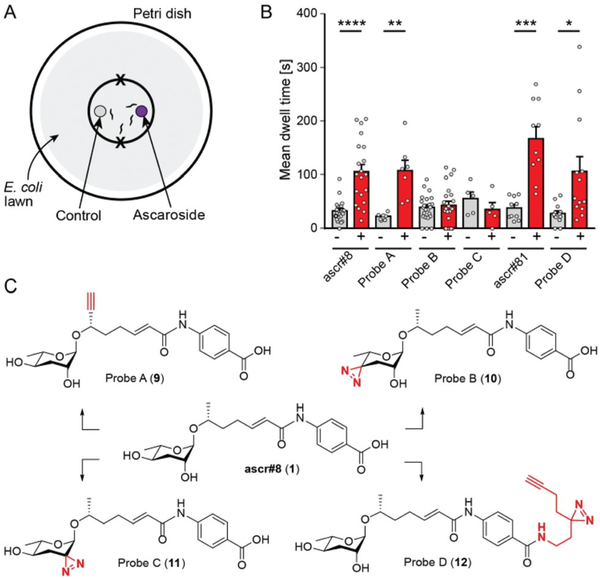
Previous studies showed that ascr#8 elicits a robust attractive response in young adult male C. elegans. We confirmed attraction to ascr#8 (1) using a spot retention assay, in which the dwell time of worms in areas treated either with vehicle control or the chemical of interest is measured (Fig. 2A).1,11,12
Fig. 2.
Response of C. elegans males to ascr#8 and derivatives. (A) Schematic of the spot retention assay. Males are placed on each “X” and scored for time spent in each of the smallest circles. (B) Dwell times of C. elegans males in vehicle controls (−) and ascaroside-treated (+) circles. Data presented as mean ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. (C) Structures of ascr#8 (1) and four probe designs (9–12).
For photoaffinity labeling, the installation of two types of groups into ascr#8 is required, a photoreactive group and a bioorthogonal reactive group for affinity purification. Since large photoreactive groups may interfere with binding between the ligand and its receptor,23 we focused on diazirine derivatives, which have additional advantages including short lifetime following UV irradiation and subsequent high reactivity.24,25 As a click-chemistry handle for affinity purification, we chose an alkyne, since azides may become subject to metabolic reduction.26 To assess whether additional functional groups affect the biological activity of ascr#8, chemical modifications were introduced one at a time, followed by testing the bioactivity of the resulting ascr#8 derivatives.
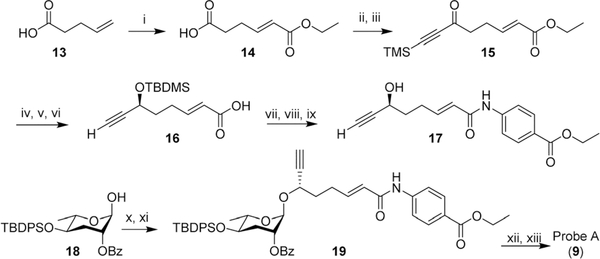
In the initial probe designs, we planned to integrate the click-chemistry moiety as part of the fatty acid side-chain (9). Replacing the ω-methyl group in the side chain with a terminal alkyne seemed most straightforward and least likely to perturb receptor binding. For introduction of the diazirine moiety, we envisioned replacing one of the two hydroxyl groups of the ascarylose, as we felt such modification may cause only minimal changes to the overall size and shape of ascr#8 (1) (Fig. 2). For the synthesis of the alkyne-integrating side chain (Scheme 1), metathesis of 4-pentenoic acid (13) with ethyl acrylate produced intermediate 14, which was converted into the TMS-protected alkyne 15. Following reduction of the ketone, TBDMS protection of the resulting alcohol and hydrolysis of the ester, the acid 16 was coupled to ethyl p-aminobenzoate, followed by TBDMS deprotection. The alkyne-containing side chain 17 was then coupled to protected ascarylose using established procedures, furnishing probe A (9). Given that the alkyne moiety in Probe A is sterically somewhat encumbered, we confirmed the ability of such alkynes to undergo click chemistry using the analogous alkyne derivative of ascr#18 (4) (see ESI†). Probe A was found to elicit levels of attraction comparable to unmodified ascr#8 (Fig. 2B and S1†), indicating the integration of an alkyne moiety in place of the ω-methyl in the side chain does not significantly perturb activity of ascr#8 and may represent a useful entry for the design of other ascaroside receptor probes.
Scheme 1.
Synthesis of probe A (9). (i) ethyl acrylate, Grubb’s second generation catalyst, DCM, 25 °C, 10 h, 75%; (ii) oxalyl chloride, cat. DMF, DCM, 0 °C, 20 min; (iii) bis(trimethylsilyl)acetylene, AlCl3, DCM, 0 °C, 3 h, 26% ~2 steps; (iv) freshly prepared Terashima reagent (see Experimental section), −78 °C, 98%, ee 80%; (v) TBDMSCl, imidazole, DMF, 25 °C, 2.5 h, 94%; (vi) LiOH·H2O, dioxane, H2O, 60 °C, 12 h, 50%; (vii) oxalyl chloride, cat DMF, DCM, 0 °C, 20 min; (viii) benzocaine, DIEA, DCM, 0 °C, 1 h, 87% ~2 steps; (ix) HF in H2O, MeCN, 25 °C, 1 h, 85%; (x) CCl3CN, DBU, DCM, 25 °C, 2 h; (xi) 17, TMSOTf, DCM, 0 °C, 1 h, 50% ~2 steps; (xii) TBAF, THF, 25 °C, 12 h, 93%; (xiii) LiOH·H2O, dioxane, H2O, 60 °C, 12 h, 62%.
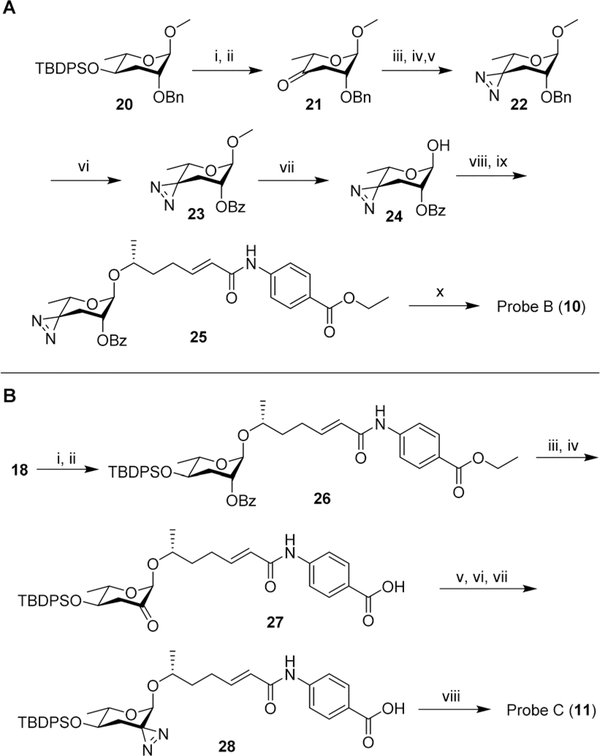
Next, we developed syntheses for introduction of the diazirine at the 2′ and 4′ carbons of ascarylose. Starting from key intermediate (20),27 a diazirine moiety was installed in position 4 in 10 steps, furnishing probe B (10, Scheme 2A). Our approach to probe B necessitated late-stage introduction of benzoyl protection in position 2, since the 2-O-benzoyl-protected derivative of ketone 21 had a strong tendency to eliminate and would not survive under the conditions required to install the diazirine. The 2-O-benzoyl moiety is required for stereochemical control of the subsequent glycosylation step via neighboring group participation (NGP). Conversion of the benzyl ether (Bn) into the benzoyl ester (Bz) after installment of the diazirine group in 22 was achieved through ruthenium(III) chloride-catalyzed oxidation (Scheme 2A). Following a different synthetic strategy, we inserted the diazirine at the 2′ carbon of the ascarylose, furnishing probe C (11, Scheme 2B). In the probe C synthesis, we opted to first establish the glycosidic linkage, followed by benzoyl deprotection, oxidation, and introduction of the diazirine moiety. Desilylation of 28 furnished probe C.
Scheme 2.
(A) Synthesis of probe B (10). (i) TBAF, THF, 25 °C, 8 h, 95%; (ii) PCC, 4 Å molecular sieves, DCM, 25 °C, 4 h, 74%; (iii) 7N NH3 in MeOH, pTsOH, MeOH, 0 °C, 3 h; (iv) NH2OSO3H, 0 °C → rt, 16 h; (v) NEt3, I2 in MeOH titration, 25 °C, 39% ~3 steps; (vi) RuCl3·H2O, NaIO4, DCM : MeCN : H2O = 1: 1: 1, 25 °C, 5 h, 72%; (vii) BBr3, DCM, −78 °C, 30 min, 71% BRSM; (viii) CCl3CN, DBU, DCM, 25 °C, 2 h; (ix) N-(6′R-hydroxy-2′E-heptenoyl)-4-aminobenzoic acid ethyl ester (prepared following previous reported method1), TMSOTf, DCM, 0 °C, 2 h, 57%~2 steps; (x) LiOH·H2O, dioxane, H2O, 60 °C, 3 h, 79%. (B) Synthesis of probe C (11).: (i) CCl3CN, DBU, DCM, 25 °C, 2 h; (ii) N-(6′R-hydroxy-2′E-heptenoyl)-4-aminobenzoic acid ethyl ester (prepared following previous reported method1), TMSOTf, DCM, 0 °C, 2 h, 15% ~2 steps; (iii) LiOH·H2O, dioxane, H2O, 60 °C, 12 h, 42%; (iv) Dess-Martin periodinane, DCM, 25 °C, 12 h, 51%; (v) 7 N NH3 in MeOH, pTsOH, MeOH, 0 °C, 3 h; (vi) NH2OSO3H, 0 °C → rt, 16 h; (vii) NEt3, I2 in MeOH titration, 25 °C; (viii) TBAF, THF, 25 °C, 8 h, 13% ~4 steps.
Testing probes B and C in the spot retention assay, we found that both probes had no activity (Fig. 2B and S1†). In fact, males spent less time in the probe C-treated spot compared to control, although the difference did not reach significance. Loss of biological activity with probe B (10) and probe C (11) suggests that both 2′ and 4′ hydroxyl groups of ascarylose participate in essential interactions during receptor binding. These data support previous studies that demonstrate the importance of these hydroxyl groups in determining the identity of individual ascarosides.8,12
These results led us to reevaluate our design strategy, based on a biological evaluation of naturally occurring ascr#8 derivatives. Recent expansion of the known family of ascarosides has uncovered two derivatives of ascr#8, named ascr#81 and ascr#82.14 In these ascr#8 derivatives, L-glutamic acid or L-glutamyl-L-glutamic acid is attached to the p-aminobenzoate via amide linkages. We asked whether these alterations of the carboxy terminus affect biological activity and tested a synthetic sample of ascr#81 (2) in the spot retention assay. We found that ascr#81 is at least as active as ascr#8 (1) in this assay (Fig. 2B and S1†). In fact, dwell times elicited by ascr#81 were higher than those measured for ascr#8, though the difference did not reach statistical significance.
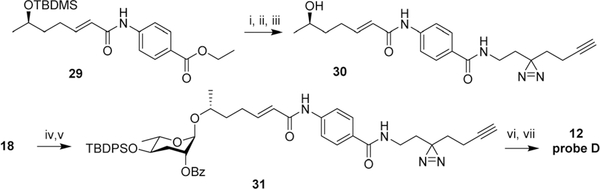
This result suggested that modification of ascr#8 at the carboxy terminus may be well tolerated. Therefore, we revised our probe design to incorporate both the diazirine and alkyne handle into a single moiety linked via an amide to the p-amino-benzoic acid in ascr#8 (probe D (12), Scheme 3). Commercially available 2-(3-(but-3-yn-1-yl)-3H-diazirin-3-yl)ethan-1-amine was incorporated at the terminal carboxylic acid, analogous to the previously described synthesis of ascr#81 (2).14 Gratuitously, bioassays demonstrated that probe D retained biological activity matching that of unmodified ascr#8 (Fig. 2B and S1†).
Scheme 3.
Synthesis of probe D (12). Reagents and conditions: (i) LiOH·H2O, dioxane, H2O, 60 °C, 12 h, 72%; (ii) 2-(3-(but-3-yn-1-yl)-3H-diazirin-3-yl)ethan-1-amine, EDC HCl, DMAP, DCM/DMF, 25 °C, 8 h, 53%; (iii) HF in H2O, MeCN, 25 °C, 1 h, 92%; (iv) CCl3CN, DBU, DCM, 25 °C, 2 h; (v) 30, TMSOTf, DCM, 0 °C, 1 h, 8% ~2 steps; (vi) TBAF, THF, 25 °C, 12 h, 77%; (vii) LiOH·H2O, dioxane, H2O, 60 °C, 12 h, 60%.
Discussion
Together, these results suggest multiple avenues for testing and development of ascaroside receptor probes. The presence of the hydroxyl groups on the ascarylose sugar appears to be essential for maintaining ascaroside activity (Fig. 2, 10, 11).5,14 Introduction of an alkyne moiety at the ω-position of the side chain is well tolerated in the case of ascr#8. Except for ascr#5 (3), all ascarosides for which biological activity has been demonstrated in C. elegans feature an ω-methyl group in the side chain, suggesting that alkyne introduction at this position could be used for probing receptor interactions of biosynthesis of diverse ascarosides in C. elegans. However, in other nematode species, modification of the ω-methyl may be less well tolerated; for example, in Caenorhabditis nigoni, hydroxylation of the m-side chain carbon was found to abolish the attraction of C. nigoni males to ascr#3 (8).28
In the case of ascr#8, use of commercially available 2-(3-(but-3-yn-1-yl)-3H-diazirin-3-yl)ethan-1-amine, as in probe D (12), allowed for inclusion of both the photo-reactive group and bioorthogonal reactive group with relative ease. However, activity of many other ascarosides requires the presence of an unmodified carboxy terminus, and therefore installation of the alkyne at the ω-position of the side chain may be preferable, though a suitable location for the diazirine moiety remains to be found. For the identification of the receptor of ascr#8, our efforts are directed at crosslinking probe D with putative receptor candidates in heterologous expression systems.
Supplementary Material
Acknowledgements
The strain of C. elegans used in this study, CB4088 (him-5 (e1490)), was provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported in part by the NIH (R01GM113692 and R35GM131877 to F.C.S., and R01DC016058 to JS) and startup funds from WPI to JS. We would also like to thank Elizabeth DiLoreto in the Srinivasan Lab for providing comments and edits to early versions of this manuscript.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Electronic supplementary information (ESI) available: Experimental procedures and spectral data for all prepared compounds with copies of 1H NMR and 13C NMR. See DOI: 10.1039/c9ob02099c
Notes and references
- 1.Pungaliya C, Srinivasan J, Fox BW, Malik RU, Ludewig AH, Sternberg PW and Schroeder FC, Proc. Natl. Acad. Sci. U. S. A, 2009, 106, 7708–7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artyukhin AB, Yim JJ, Srinivasan J, Izrayelit Y, Bose N, von Reuss SH, Jo Y, Jordan JM, Baugh LR, Cheong M, Sternberg PW, Avery L and Schroeder FC, J. Biol. Chem, 2013, 288, 18778–18783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeong P-Y, Jung M, Yim Y-H, Kim H, Park M, Hong E, Lee W, Kim YH, Kim K and Paik Y-K, Nature, 2005, 433, 541–545. [DOI] [PubMed] [Google Scholar]
- 4.von Reuss SH and Schroeder FC, Nat. Prod. Rep, 2015, 32, 994–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butcher RA, Nat. Prod. Rep, 2017, 34, 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludewig AH, Izrayelit Y, Park D, Malik RU, Zimmermann A, Mahanti P, Fox BW, Bethke A, Doering F, Riddle DL and Schroeder FC, Proc. Natl. Acad. Sci. U. S. A, 2013, 110, 5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludewig AH, Artyukhin AB, Aprison EZ, Rodrigues PR, Pulido DC, Burkhardt RN, Panda O, Zhang YK, Gudibanda P, Ruvinsky I and Schroeder FC, Nat. Chem. Biol, 2019, 15, 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srinivasan J, von Reuss SH, Bose N, Zaslaver A, Mahanti P, Ho MC, O’Doherty OG, Edison AS, Sternberg PW and Schroeder FC, PLos Biol, 2012, 10, e1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choe A, von Reuss SH, Kogan D, Gasser RB, Platzer EG, Schroeder FC and Sternberg PW, Curr. Biol, 2012, 22, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chute CD and Srinivasan J, Semin. Cell Dev. Biol, 2014, 33, 18–24. [DOI] [PubMed] [Google Scholar]
- 11.Narayan A, Venkatachalam V, Durak O, Reilly DK, Bose N, Schroeder FC, Samuel AD, Srinivasan J and Sternberg PW, Proc. Natl. Acad. Sci. U. S. A, 2016, 113, E1392–E1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PE, Malik RU, Edison AS, Sternberg PW and Schroeder FC, Nature, 2008, 454, 1115–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludewig AH and Schroeder FC, WormBook, 2013, 1–22, DOI: 10.1895/wormbook.1.155.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Artyukhin AB, Zhang YK, Akagi AE, Panda O, Sternberg PW and Schroeder FC, J. Am. Chem. Soc, 2018, 140, 2841–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Reuss SH and Schroeder FC, Nat. Prod. Rep, 2015, 32, 994–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim K, Sato K, Shibuya M, Zeiger DM, Butcher RA, Ragains JR, Clardy J, Touhara K and Sengupta P, Science, 2009, 326, 994–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park D, O’Doherty I, Somvanshi RK, Bethke A, Schroeder FC, Kumar U and Riddle DL, Proc. Natl. Acad. Sci. U. S. A, 2012, 109, 9917–9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bargmann CI, Science, 1998, 282, 2028–2033. [DOI] [PubMed] [Google Scholar]
- 19.Chute CD, DiLoreto EM, Zhang YK, Rayes D, Coyle VL, Choi HJ, Alkema MJ, Schroeder FC and Srinivasan J, bioRxiv, 2018, 275693, DOI: 10.1101/275693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greene JS, Brown M, Dobosiewicz M, Ishida IG, Macosko EZ, Zhang X, Butcher RA, Cline DJ, McGrath PT and Bargmann CI, Nature, 2016, 539, 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene JS, Dobosiewicz M, Butcher RA, McGrath PT and Bargmann CI, eLife, 2016a, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGrath PT, Xu Y, Ailion M, Garrison JL, Butcher RA and Bargmann CI, Nature, 2011, 477, 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang T, Liu Z and Li XD, Chem. Sci, 2015, 6, 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubinsky L, Krom BP and Meijler MM, Bioorg. Med. Chem, 2012, 20, 554–570. [DOI] [PubMed] [Google Scholar]
- 25.Smith E and Collins I, Future Med. Chem, 2015, 7, 159–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasmal PK, Carregal-Romero S, Han AA, Streu CN, Lin Z, Namikawa K, Elliott SL, Köster RW, Parak WJ and Meggers E, ChemBioChem, 2012, 13, 1116–1120. [DOI] [PubMed] [Google Scholar]
- 27.Zhang YK, Sanchez-Ayala MA, Sternberg PW, Srinivasan J and Schroeder FC, Org. Lett, 2017, 19, 2837–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong C, Reilly DK, Bergame C, Dolke F, Srinivasan J and von Reuss SH, J. Org. Chem, 2018, 83, 7109–7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.