Abstract
We develop an L-platform/L-scaffold framework we hypothesize may serve as a blueprint to facilitate site-specific RNA-cleaving nucleic acid enzyme design. Building on the L-platform motif originally described by Suslov and coworkers, we identify new critical scaffolding elements required to anchor a conserved general base guanine (“L-anchor”) and bind functionally important metal ions at the active site (“L-pocket”). Molecular simulations, together with a broad range of experimental structural and functional data, connect the L-platform/L-scaffold elements to necessary and sufficient conditions for catalytic activity. We demonstrate that the L-platform/L-scaffold framework is common to five of the nine currently known naturally occurring ribozyme classes (Twr, HPr, VSr, HHr, Psr), and intriguingly from a design perspective, the framework also appears in an artificially engineered DNAzyme (8–17dz). The flexibility of the L-platform/L-scaffold framework is illustrated on these systems, highlighting modularity and trends in the variety of known general acid moieties that are supported. These trends give rise to two distinct catalytic paradigms, building on the classifications proposed by Wilson and coworkers and named for the implicated general base and acid. The “G + A” paradigm (Twr, HPr, VSr) exclusively utilizes nucleobase residues for chemistry, and the “G + M + ” paradigm (HHr, 8–17dz, Psr) involves structuring of the “L-pocket” metal ion binding site for recruitment of a divalent metal ion that plays an active role in the chemical steps of the reaction. Finally, the modularity of the L-platform/L-scaffold framework is illustrated in the VS ribozyme where the “L-pocket” assumes the functional role of the “L-anchor” element, highlighting a distinct mechanism, but one that is functionally linked with the hammerhead ribozyme.
Keywords: RNA catalysis, RNA motif, design, ontology, ribozyme engineering
INTRODUCTION
Nucleolytic ribozymes are small catalytic RNAs that site-specifically cleave the sugar-phosphate backbone of their RNA substrates (Doherty and Doudna 2001; Cochrane and Strobel 2008; Ferré-D'Amaré and Scott 2010; Ward et al. 2014; Lilley 2017; Ren et al. 2017). This reaction is ubiquitous in biology (Lassila et al. 2011) and has importance for biochemical tools and biomedical technology (Vaish et al. 2002; Emilsson et al. 2003; Ausländer et al. 2010; Felletti et al. 2016; Ausländer and Fussenegger 2017; Kobori and Yokobayashi 2018). Currently, there are nine known naturally occurring nucleolytic ribozyme classes: hammerhead (HHr) (Prody et al. 1986; Pley et al. 1994; Scott et al. 1996), hairpin (HPr) (Buzayan et al. 1986; Rupert et al. 2002), hepatitis delta virus (HDVr) (Sharmeen et al. 1988; Ferré-D'Amaré et al. 1998), Varkud satellite (VSr) (Saville and Collins 1990; Suslov et al. 2015), glmS (Winkler et al. 2004; Klein and Ferré-D'Amaré 2006), twister (Twr) (Liu et al. 2014; Roth et al. 2014), twister sister or simply “TS” (TSr) (Weinberg et al. 2015; Liu et al. 2017b), pistol (Weinberg et al. 2015; Ren et al. 2016) (Psr), and hatchet (Htr) (Weinberg et al. 2015) ribozymes. However, molecules of DNA can also be artificially engineered to catalyze RNA cleavage (e.g., 8–17 DNAzyme or 8–17dz) (Santoro and Joyce 1997; Liu et al. 2017a), and these DNAzymes have some potential advantages owing to their greater stability and ease of synthesis (Breaker and Joyce 1994). A broad understanding of the detailed mechanisms of these nucleic acid enzymes provides a foundation from which general design principles may emerge that are transferable to nonbiological contexts (Cech 1992; Ward et al. 2014; Jimenez et al. 2015; Lilley 2017; Ren et al. 2017). A novel comparative analysis of nucleolytic ribozyme active sites has led to new insight into the diverse array of catalytic strategies employed by these systems (Seith et al. 2018) and has motivated closer examination of the platforms that form their catalytic cores.
Recently, Suslov et al. (2015) identified a common active site motif, designated the “L-platform,” in the VSr, HPr, and HHr ribozymes based on structural data available at the time. This four-residue motif involved base stacking interactions that sandwich a strictly conserved guanine and position it adjacent to the scissile phosphate where it is poised to act as a general base in catalyzing the 2′-O-transphosphorylation reaction. As originally described in the context of the VSr, this stem within the L-platform is stabilized by a purine–purine base pair at the base of the “L.”
However, the “L” alone is insufficient to establish a catalytic core for these ribozymes. Further details about how this motif could be expanded to describe the full active site and potentially serve as a generalizable platform to facilitate design are difficult to derive solely from crystallographic data, as these data are usually not sufficient to unambiguously identify the catalytically active states of the enzymes (Gaines and York 2017; Ekesan and York 2019; Gaines et al. 2019; Ganguly et al. 2019a; Kostenbader and York 2019). Rather, rigorous computational simulations together with structural and functional data are required to define a meaningful dynamical ensemble for the active state in solution, as well as other states along the reaction coordinate. It is from these experimentally motivated computational studies combined with a comparative view across known ribozyme classes that predictive insight can be gleaned.
Recently, such studies have been carried out by our group for a number of ribozyme classes, including HHr (Lee et al. 2013; Chen et al. 2017), HPr (Heldenbrand et al. 2014), HDVr (Lee et al. 2016), VSr (Ganguly et al. 2019a), Twr (Gaines et al. 2019), Psr (Kostenbader and York 2019), and TSr (Gaines and York 2017), as well as 8–17dz (Ekesan and York 2019). These studies have provided detailed atomic-level models of the functionally active state, not apparent from the crystal structures, that explain a wide range of experimental data and have enabled identification of common features and new design principles to emerge. Herein, we identify new functionally critical scaffolding elements of the active site architecture that build on the L-platform motif introduced by Suslov et al. (2015). We refer collectively to these elements as the L-scaffold and provide an in-depth characterization of this new composite L-platform/L-scaffold motif (Fig. 1). Additionally, we demonstrate that this motif is common to over half of the currently known naturally occurring ribozyme classes (Twr, HPr, VSr, HHr, Psr), as well as a recently structurally characterized artificially engineered DNAzyme (8–17dz). We also provide details about the structural features of the L-platform/L-scaffold motif and connect them to the necessary and sufficient conditions for catalytic activity.
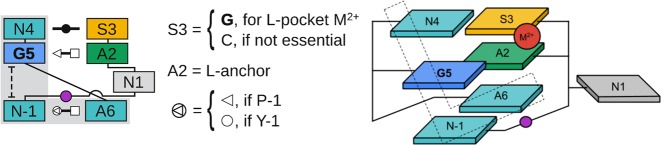
FIGURE 1.
Generalized L-platform/L-scaffold composite motif. Symbolic secondary structure (left) and cartoon-block schematic (right) representations of the generalized L-platform/L-scaffold. Residue numbering and base-pair symbols (Leontis and Westhof 2001; Leontis et al. 2002) are described in the Notation section. Strict conservation of nucleobase identity (e.g., for the general base guanine G5) is shown with bold font. Other nucleotides are shown with ambiguity characters representing apparent trends in preferred nucleobase identity across the ribozyme classes (e.g., N1, A2, and S3). The constituent elements of the L-platform (left: gray background; right: dotted “L”) motif are colored as follows: general base (G5) in blue, stacking/pairing “L” nucleotides (N-1, N4, and A6) in teal. The L-scaffold consists of the L-anchor (A2) in green, L-pocket nucleotide (S3) in yellow, and the scissile phosphate in magenta along with the interactions between those residues and the L-platform. The bulged N1 nucleotide is not colored as it does not generally play a specific functional role (with the exception of twister ribozymes).
The two main structural elements of the L-scaffold are referred to as the “L-anchor” and “L-pocket” (Fig. 1). The L-anchor nucleobase functions to position a conserved G (implicated as the general base) to activate the 2′-OH nucleophile through the interaction of the Hoogsteen edge of the L-anchor with the sugar edge of the G. The L-pocket, on the other hand, enables the formation of a divalent metal ion binding site. We describe distinguishing features of the L-platform/L-scaffold framework that give rise to two distinct catalytic paradigms (expanding the classification proposed by Wilson et al. (2016b) named for the identities of the general base and acid implicated in catalysis. The “G + A” paradigm (Wilson et al. 2016b) (Twr, HPr, VSr) exclusively utilizes nucleobase functional groups for chemistry, and by analogy what we will refer to as the “G + M+” paradigm (HHr, 8–17dz, Psr) involves structuring of the L-pocket for recruitment of a divalent metal ion that plays an active role in the chemical steps of the reaction. We also illustrate how the L-platform/L-scaffold framework enables cross-over of themes associated with each paradigm, such as in the case of VSr that uses the L-pocket within the G + A paradigm to recruit a divalent metal ion that substitutes for the missing nucleobase L-anchor and assists in organizing the active site (without directly being involved in chemistry as in the G + M+ paradigm) (Ganguly et al. 2019a).
We begin by establishing notation to define the prototypical L-platform/L-scaffold framework, and the specific requirements for nucleobase stacking and hydrogen bonding interactions. We then describe distinguishing features of the G + A and G + M+ paradigms, including nucleobase requirements for the L-pocket, and present specific examples for each paradigm. Finally, we provide a comprehensive summary of the mechanisms whereby the L-platform/L-scaffold enables different catalytic strategies, following a recently introduced ontology for discussion of RNA cleavage reactions (Bevilacqua et al. 2019).
NOTATION AND CONVENTIONS
In describing the L-platform/L-scaffold composite motif, we will utilize standard IUPAC nucleic acid nomenclature and ambiguity symbols, as well as abbreviations for edge-to-edge base-pairing families taken from the work of Leontis and Westhof (2001) and Leontis et al. (2002). In the prototypical L-platform/L-scaffold (Fig. 1) the motif is shown with two strands, the substrate strand, and the general base strand. The nucleotide residue positions are then numbered from −1 to 3 for the substrate strand (with the scissile phosphate as position zero) and continue with 4–6 for the general base strand moving in the 5′–3′ direction. This general motif numbering scheme is intended to remove the L-platform/L-scaffold motif from the context of any individual ribozyme class and then number it in a way that allows one to refer to specific nucleotides by their relative positions in the motif and corresponding functional roles. In actuality, the consecutiveness of the bases and number of strands recruited to form this active site motif is variable, although those variations often follow discernible trends. Throughout the text, nucleotide residues will generally be referred to by their motif positions with additional ribozyme specific numbering in parentheses when necessary, and atomic numbering will be indicated by subscripts for distinction. For example, “N1” will indicate a nitrogen atom at atomic position 1 of a nucleotide, whereas “N1” refers to a nucleotide residue in position 1 of the general motif and “N1 (C1.1 in HHr)” indicates N1 in the general motif that corresponds specifically to C1.1 in HHr.
In describing the connection between the L-platform/L-scaffold motif and the specific catalytic strategies enabled by it, we will use a simplified framework originally proposed by Emilsson et al. (2003) and recently expanded to an ontology for facilitating discussion of mechanisms of RNA-cleaving enzymes in precise atomic-level detail (Bevilacqua et al. 2019). For the underlying chemical reaction catalyzed by these ribozymes, a 2′-OH nucleophile becomes activated and makes an in-line attack on the adjacent scissile phosphate, which then proceeds through a pentavalent phosphorane transition state to form 2′,3′-cyclic phosphate and 5′-OH cleavage products. These general catalytic strategies to facilitate RNA cleavage are designated alpha (α) for the in-line fitness of the 2′-OH nucleophile, beta (β) for the electrostatic stabilization/protonation of the nonbridging phosphoryl oxygens (NPOs) of the dianionic phosphorane transition state, gamma (γ) for the deprotonation (activation) of the 2′-OH nucleophile (e.g., by a general base), and delta (δ) for the electrostatic stabilization (neutralization/protonation) of the 5′-O leaving group (e.g., by a general acid). The β, γ, and δ strategies can be further decomposed into primary (1°), secondary (2°), and tertiary (3°) contributions (Bevilacqua et al. 2019).
GENERAL DEFINITION OF THE L-PLATFORM/L-SCAFFOLD MOTIF
Here, we define the generalized prototype L-platform/L-scaffold motif as a series of three base pairs, with one bulged residue, organizing two or more strands into a compact catalytic core shown in Figure 1. A conserved guanine, G5, acts as the general base in catalysis and is sandwiched in the middle of a three-base stack by the N-1 and N4 nucleotides. The N-1 nucleobase (5′ of the scissile phosphate) then sits at the heel of the eponymous “L” and pairs with the Hoogsteen edge of, typically, an adenine denoted as A6. The N1 nucleotide, immediately downstream from the scissile phosphate, is bulged from the motif in the sense that it is not explicitly involved in base-pairing within this three-base-pair stack, independent of consecutiveness. The general base, G5, is then hydrogen bonding along its sugar edge, most commonly in a tSH base pair with A2. Since this interaction is critical for anchoring the central residue of the “L,” residue 2 in the prototypical motif will be referred to as the L-anchor.
Finally, the motif is capped by a cWW base pair (or wobble pair) between S3 and N4. For the ribozymes where a divalent metal ion is critical for function, S3 is conserved as a guanine. The Hoogsteen edge of this guanine then completes a cation binding pocket in conjunction with one of the NPOs of the scissile phosphate as well as one 5′ of the L-anchor (residue 2). This set of ligands defines the L-pocket and consistently promotes the binding of a divalent metal ion within the active site. In contrast, the ribozymes (Twr and HPr) for which divalent metals are not required for catalysis, cytosine at position 3 is the preferred nucleobase. It is interesting to note that this is the only nucleobase that cannot bind a metal along its Hoogsteen edge. These trends lead us to characterize nucleotide 3 as S in the general motif, as a means to highlight how the base identity is correlated with the requirement for divalent metal ions in each of the ribozyme classes. In the next section we discuss how the L-platform/L-scaffold motif can enable RNA cleavage within two catalytic paradigms designated G + A and G + M+ (Wilson et al. 2016b), named for the identities of the general base (guanine) and acid (adenine or a metal ion and/or its ligands).
G + A CATALYTIC PARADIGM
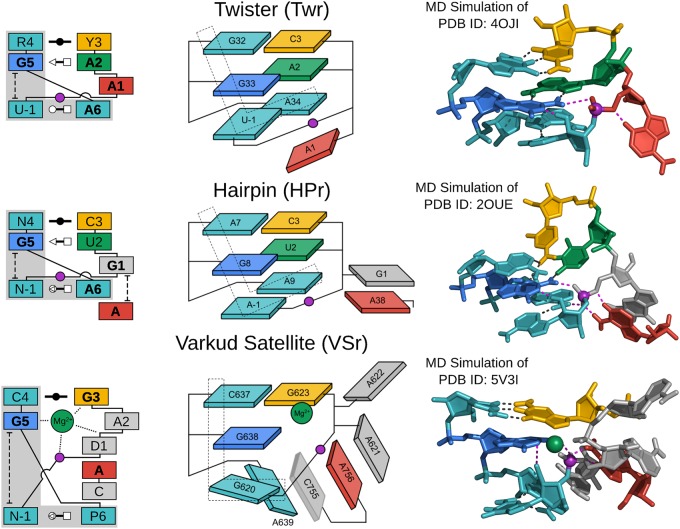
The Twr, HPr, and VSr fit the G + A paradigm and have active sites that exhibit remarkable structural similarities (Fig. 2). Ribozymes in the G + A paradigm utilize the L-platform/L-scaffold framework as an active site architecture to enable the chemical steps of catalysis to be carried out exclusively by nucleobase functional groups. In addition to the strictly conserved G5 that acts as a general base (γ catalysis), a conserved adenine residue acts as a general acid (δ catalysis), either through the N3 (Twr) or the N1 (HPr and VSr) positions. In order for the adenine to act as an effective general acid through either the N1 or N3 heteroatoms (1° δ catalysis), it must have a pKa that is appropriately tuned (2° δ catalysis), and it must be properly positioned for proton donation to the O5′ leaving group (3° δ catalysis).
FIGURE 2.
G + A paradigm. Symbolic secondary structure (left), cartoon-block schematic (middle), and 3D atomic (right) representations of the specific L-platform/L-scaffold composite motif for ribozymes of the “G + A” paradigm (Twr, HPr, and VSr), categorized as such with a guanine (G in blue) and adenine (A in red) implicated as the general base and acid, respectively. Generalized nucleotide residue numbering is used for the secondary structure (left) as in Figure 1, whereas the cartoon-block schematic (middle) uses the canonical residue numbering of the specific ribozyme being illustrated. Base-pair symbols (Leontis and Westhof 2001; Leontis et al. 2002) are described in the Notation section, and the color scheme is the same as in Figure 1: general base (G5) in blue, stacking/pairing “L” residues in teal, L-anchor in green, L-pocket nucleobase in yellow, and the scissile phosphate in magenta. Structural h-bonds are in black and h-bonds implicated in the catalytic mechanism are shown in magenta. The active site metal ion in VSr that plays an organizational role as the L-anchor is shown in green. Bold font is used for residues conserved with a frequency greater than 97% for ribozymes where consensus sequence information is available, as well as residues that are otherwise critical for activity. 3D atomic representations derived from MD simulations of each ribozyme: Twr (Gaines et al. 2019), HPr (Heldenbrand et al. 2014), and VSr (Ganguly et al. 2019a).
G + M+ CATALYTIC PARADIGM
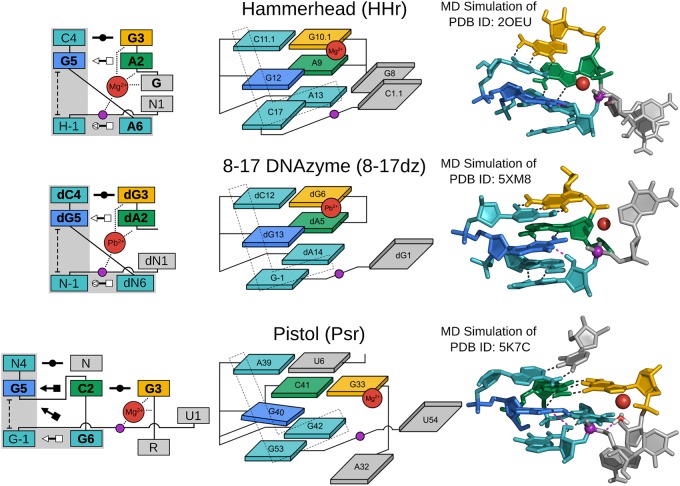
The HHr, 8–17dz, and Psr enable catalytic strategies within the G + M+ paradigm and have similar active site architectures and divalent metal ion binding modes in their active states (Fig. 3). Ribozymes grouped in the G + M+ paradigm all require an L-pocket metal ion binding site that recruits a divalent metal ion to assist in catalysis. The divalent metal ion can aid in stabilizing the O5′ leaving group by providing direct electrostatic stabilization as a Lewis acid (1° δ catalysis), or as a general acid acting through pKa tuning (activation) of a coordinated water molecule or functional group such as a 2′-OH (2° δ catalysis). Finally, the L-pocket metal ion can play a structural role in bringing the required elements for δ catalysis together, orienting the scissile phosphate and positioning the O5′ leaving group (3° δ catalysis).
FIGURE 3.
G + M+ paradigm. Symbolic secondary structure (left), cartoon-block schematic (middle), and 3D atomic (right) representations of the specific L-platform/L-scaffold composite motif for ribozymes and DNAzyme of the “G + M+” paradigm (HHr, 8–17dz, and Psr), categorized as such with a guanine (G in blue) and a metal ion (M+ in red) implicated as the general base and acid, respectively. Generalized nucleotide residue numbering is used for the secondary structure (left) as in Figure 1, whereas the cartoon-block schematic (middle) uses the canonical residue numbering of the specific ribozyme/DNAzyme being illustrated. Base-pair symbols (Leontis and Westhof 2001; Leontis et al. 2002) are described in the Notation section, and the color scheme is the same as in Figure 1: general base (G5/dG5) in blue, stacking/pairing “L” residues in teal, L-anchor in green, L-pocket nucleobase in yellow, and the scissile phosphate in magenta. Structural h-bonds are in black and h-bonds implicated in the catalytic mechanism are shown in magenta. The active site metal ion that plays a role in the chemical steps of the reaction is shown in red. Bold font is used for residues conserved with a frequency greater than 97% for ribozymes where consensus sequence information is available, as well as residues that are otherwise critical for activity. 3D atomic representations derived from MD simulations of each ribozyme: HHr (Lee et al. 2013; Chen et al. 2017), 8–17dz (Ekesan and York 2019), and Psr (Kostenbader and York 2019).
CANONICAL SEQUENCES
The canonical sequences for each ribozyme class, as depicted in the symbolic secondary structure diagrams (Figs. 2, 3, left) were derived from bioinformatics, when available, or through evaluating mutation and in vitro selection experiment results. The canonical sequences are taken from sequence alignments for Twr (Roth et al. 2014), HHr (Perreault et al. 2011), and Psr (Weinberg et al. 2015), whereas the 8–17 DNAzyme sequence, presented in the original crystallographic work (Liu et al. 2017a), was adapted from commonly observed variations and mutagenesis experiments. In a similar fashion, an analysis of mutational and in vitro selection experiments was conducted for HPr and VSr in order to arrive at a canonical sequence for the active site residues.
For VSr, mutations that resulted in a decrease in activity greater than 100-fold (Guo et al. 1993; Wilson et al. 2007) were rejected from the canonical sequence at that position. From the in vitro selection experiments, all nucleobases for which there was more than one sequence that exhibited high activity, that is, >50% activity in the cleavage assay (Andersen and Collins 2000), were included in the canonical sequence. Similar criteria were applied in developing the canonical sequence for HPr. However, there are two nucleotides in the HPr active site for which there is not a strict requirement for a specific nucleobase, the L-anchor (N2) and L-pocket (N3) nucleotides. The mutational data nonetheless suggest that there are clear, but subtle trends for the preferred nucleobases: U2 and C3 depicted in Figure 2. For the L-anchor nucleotide, uracil (the “wild-type” residue) is fastest, followed by adenine (24-fold lower in activity); guanine and cytosine were both observed as 73-fold down in activity (Shippy et al. 1998). However, Pérez-Ruiz et al. (1999) suggest that sequences with cytosine at the N2 position were noncleavable in their assay, which would be consistent with the proposed tHS base-pairing between the L-anchor and the general base guanine (Leontis et al. 2002). For the L-pocket nucleobase in HPr, all nucleobases are tolerated (decrease in activity <10-fold in all cases). However, the observed rate was negatively correlated with the ability for this nucleobase to bind a Mg2+ ion at its Hoogsteen edge (G:N7 > A:N7 > U:O4 > C:“C-H” edge) (Sigel and Sigel 2010), motivating our decision to suggest cytosine in the canonical sequence.
SURVEY OF CATALYTIC STRATEGIES ENABLED BY THE L-PLATFORM/L-SCAFFOLD IN RNA AND DNA ENZYMES
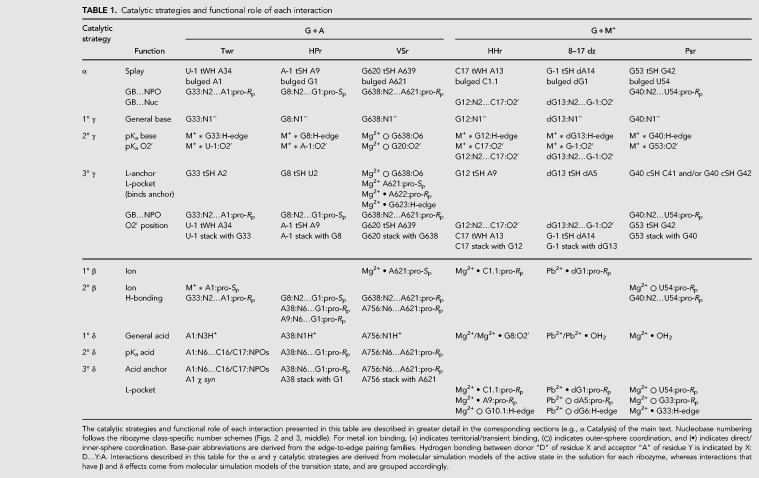
In the previous sections, we introduced the prototypical L-platform/L-scaffold composite motif and the two catalytic paradigms that it supports. Now we will illustrate the tremendous versatility of this motif as a framework for design by providing a comprehensive survey of how these catalytic strategies are exploited to facilitate site-specific RNA cleavage via 2′-O-transphosphorylation in five known ribozyme classes as well as an engineered DNAzyme. Table 1 lists the specific positions and residue interactions that are implicated in each of the catalytic strategies. These are now summarized and discussed within the context of design principles for each of the ribozymes and DNAzyme in Figures 2 and 3.
TABLE 1.
Catalytic strategies and functional role of each interaction
α CATALYSIS
In-line alignment of the nucleophile, scissile phosphate and leaving group is highly correlated with the splaying apart of the two nucleobases flanking the scissile phosphate, N-1, and N1. In the L-platform motif, the N-1 nucleotide is positioned at the heel of the “L” by stacking with the general base, G5, as well as pairing with the nucleobase at position 6 at the toe of the “L.” In the G + A paradigm where exactly two strands form the active site, N1 of the L-scaffold is bulged from the motif by interactions between the L-anchor (residue 2 or the L-pocket Mg2+ in VSr) and the general base. The N1 nucleotide is similarly bulged from the motif in the G + M+ paradigm where additional strands are recruited to position residues 2 and 3. In both paradigms, residue 1 is then involved in ribozyme specific tertiary interactions in order to achieve the requisite splay in conjunction with N-1.
While it has been suggested that ideal in-line alignment alone is unlikely to provide more than a 100-fold increase in the catalytic rate (Emilsson et al. 2003) relative to typical noncatalytic RNAs, constraining the positions of the flanking bases serves not only to improve in-line fitness, but also to localize the reactive atoms within the active site. This is further facilitated by the interaction of the general base exocyclic amine, G5:N2, with an NPO of the scissile phosphate in the G + A paradigm and Psr. Whereas, for HHr and 8–17dz, this same exocyclic amine may instead interact with the nucleophile prolonging the lifetime of its deprotonated form, while helping to align it with the scissile phosphate. As will prove to be a common trend with many of the components of the L-platform/L-scaffold motif, this functional group (G5:N2) likely contributes to multiple catalytic strategies and will be discussed in greater detail throughout the following sections. Finally, it is interesting to note that while the identities of neither N-1 nor N1 are conserved across ribozyme classes, the base-pairing arrangement involving N-1 and the Hoogsteen edge of residue 6 at the base of the “L” is observed in all cases.
β CATALYSIS
In the context of ribozyme catalysis at physiological pH, stabilization of the negative charge accumulation on scissile phosphate NPOs (β catalysis) can be achieved either through direct coordination of a divalent metal ion (1° β) or through hydrogen bond donation by a nucleobase or indirect (outer-sphere) interaction with metal ion (2° β)—and commonly both. While mechanistic pathways that involve direct protonation of an NPO (a form of 1° β catalysis) cannot be definitively ruled out, for the ribozymes considered here, the current body of evidence does not support this as a likely mechanism.
Phosphorothioate substitutions have proven particularly useful in elucidating the interactions with the NPOs, although measurements can be difficult to interpret without complementary site-specific chemical modifications and computational simulations, as was notably the case with the twister ribozyme (Breaker 2017). For Twr, the functional data suggest that direct ion coordination to the scissile phosphate NPO is not required for catalysis (Wilson et al. 2016a). That being said, inner-sphere coordination at the pro-SP position has been observed in some crystal structures (Ren et al. 2014; Košutić et al. 2015) and simulations suggest Twr has an electronegative active site that can attract metal ions nonspecifically to enhance electrostatic stabilization (Gaines et al. 2019). Furthermore, removing the exocyclic amine of G5 (G33 in Twr) by substitution with inosine eliminated the stereospecific thio effect where the wild-type construct was two orders of magnitude slower with sulfur substitution at the pro-RP NPO than at the pro-SP (Wilson et al. 2016a).
However, it was also noted that the addition of thiophilic metals (e.g., Mn2+ or Cd2+) had a negligible impact on the activity of both phosphorothioate substrates, indicating that divalent metal ion interactions with the NPOs are likely to involve indirect coordination, as suggested by the computational modeling (Gaines et al. 2019). In summary, the current model for the twister ribozyme (Table 1) proposes that both the exocyclic amine of G5 (G33 in Twr) donating a hydrogen bond to the pro-RP NPO and nonspecific interactions between monovalent ions and the pro-SP NPO contribute to β catalysis.
In the case of VSr, a functionally important divalent metal ion binds to the pro-SP oxygen, but experiments and simulations suggest the role of this ion is primarily to organize the active site (Ganguly et al. 2019a). It is only in the case of HPr that both NPOs of the scissile phosphate are saturated with hydrogen bonding from nucleobases. Thus, for the ribozymes observed to fit the G + A paradigm, it appears that the pro-RP NPO is involved exclusively in hydrogen bonding, whereas the pro-SP NPO is either available to interact with a metal ion (Twr and VSr) or other hydrogen bonding interactions (HPr). The most notable hydrogen bond donor, in this context, is the exocyclic amine of the general base guanine (G5:N2). As seen in Table 1 this interaction plays multiple roles, being involved not just in β catalysis by stabilizing charge through hydrogen bonding at the transition state, but also in α catalysis (GB-NPO contact) by localizing the reactive atoms in the active site and γ catalysis by both positioning G5 (anchor) and localizing the reactive atoms (GB-NPO contact).
In the G + M+ paradigm, the role of G5:N2 cannot be as cleanly delineated. Crystallographic evidence for Psr suggests that G5:N2 donates a hydrogen bond to the pro-RP NPO, whereas for 8–17dz it is the neutral/protonated G5:N1 position, and for HHr neither hydrogen bond donor along the Watson–Crick edge of G5 is close enough to the pro-RP NPO (though, a water bridged contact could be present). For all three ribozymes, divalent metal ions have not been observed crystallographically directly bound to the pro-RP NPO. However, these crystal structures do not provide a complete, representative picture of the active state in solution. In particular for 8–17dz and HHr, both functional data (Slim and Gait 1991; Wang et al. 1999; Osborne et al. 2005) and simulation (Lee et al. 2013; Ekesan and York 2019) results suggest that a divalent metal ion directly coordinates the pro-RP NPO in the catalytically active state. Furthermore, there is evidence that the pro-RP NPO may bind this metal in the ground state for HHr (Ward and DeRose 2012). Hence, for 8–17dz and HHr, the most likely model of the active state involves a divalent metal ion that directly coordinates the pro-RP NPO. Interestingly, for Psr, there is a large normal thio effect at the pro-RP position of the scissile phosphate that is neither fully rescuable by thiophilic metals, nor by the G5 inosine mutation (G40 in Psr) (Wilson et al. 2019). These data support a model for Psr whereby the L-pocket metal is indirectly coordinating the pro-RP NPO, while G5:N2 donates an additional hydrogen bond to that oxygen. In contrast to the ribozymes in the G + A paradigm, the coordination of the divalent metal ion (either directly or indirectly) to the pro-RP NPO of the scissile phosphate observed in the G + M+ paradigm relieves the functional requirement of G5:N2 to hydrogen bond with the same NPO. It is important to note that, in general, the G5:N2 exocyclic amine may contribute not only to β catalysis, but also to α and γ catalysis (as discussed in their respective sections) making interpretation of G5 inosine mutations challenging without additional functional data and rigorous atomic-level modeling of the active state.
γ CATALYSIS
The L-platform/L-scaffold motif enables activation of the nucleophile (γ catalysis) using primary, secondary, and tertiary contributions that satisfy the following requirements:
1° γ: activation of the nucleophile via abstraction of the proton from the 2′ hydroxyl.
2° γ: dynamic tuning of the pKa values of the general base and nucleophile.
3° γ: spatial localization of the base, nucleophile, and scissile phosphate, and orientation of hydrogen bonds such that nucleophile activation is productive.
Each of these is described in more detail below.
1° γ catalysis
Across all ribozyme classes considered here that employ the L-platform/L-scaffold motif, the prevailing model is one where activation of the nucleophile occurs via abstraction of the proton from the 2′ hydroxyl by the N1 heteroatom of the strictly conserved general base guanine, G5 (Cochrane and Strobel 2008; Wilson and Lilley 2011; Lilley 2017; Liu et al. 2017a; Wilson et al. 2019). Within the admittedly limited data set of all known nucleolytic ribozymes, a guanine proposed to act in this functional role is the most common mechanism for 1° γ catalysis; further including the glmS ribozyme (Soukup 2014) that does not utilize the L-platform/L-scaffold framework.
2° γ catalysis
In order for the G5 general base to activate the nucleophile, it must be deprotonated at the N1 atomic position. In solution, the unperturbed pKa of guanine N1 is 9.2 (Izatt et al. 1971). However, in the ribozyme environment, the guanine is held in an electronegative active site with its WC edge near to the scissile phosphate. This, in the absence of other factors, would likely lead to a considerable pKa up-shift and reduced activity at near-neutral conditions. In addition to the tuning of the G5:N1 pKa, an environment that also increases the acidity of the nucleophile (pKa down-shift) would facilitate proton transfer. The L-platform/L-scaffold enables this to be accomplished by the recruitment of metal ions in both the G + A and G + M+ paradigms.
Metal ion interactions with the G5:O6 position stabilize delocalized charge in the ionized G5− nucleotide causing a down-shift in the pKa. In all systems with the exception of VSr, the electronegative Hoogsteen edge of G5 is left exposed to solvent and attracts metal ions (monovalent and/or divalent) from solution (Mir et al. 2015; Mir and Golden 2016; Chen et al. 2017; Ekesan and York 2019; Gaines et al. 2019; Kostenbader and York 2019). The HHr is a particularly illuminating example. Recent crystallographic work by Golden and coworkers has identified the appearance of a divalent metal ion binding site (“G-site”) at the Hoogsteen edge of the general base guanine in HHr (G12:H-edge in the ribozyme specific numbering scheme and Table 1) at pH 8.5, which was not evident at lower pH (Mir et al. 2015; Mir and Golden 2016), suggesting metal ion binding is correlated with deprotonation. The apparent pKa assigned to the general base in HHr is 8.0, which is among the lowest for the known ribozyme classes. Quantum mechanical calculations and free energy simulations in the absence of G-site binding predicted an up-shift of the microscopic pKa by 3.7 units, whereas with a Mg2+ ion (weakly) bound, the microscopic pKa shifted down by 1.2 units so as to closely align with the apparent pKa value of 8.0 derived from activity-pH profiles (Chen et al. 2017). Similar G-site metal ion binding modes have been observed crystallographically for the Psr (Ren et al. 2016; Nguyen et al. 2017). In VSr, on the other hand, the Hoogsteen edge of G5 is not solvent exposed, but rather makes strong outer-sphere contact with a functionally critical divalent metal ion bound to the pro-SP NPO of the scissile phosphate in addition to the pro-RP NPO of A2 and the Hoogsteen edge of G3 (L-pocket, binds anchor in Table 1; Sood et al. 1998; Kovacheva et al. 2004; Ganguly et al. 2019a). This contact would be expected to tune the G5 pKa, in addition to serving a critical role as the L-anchor to organize the active site, as will be discussed in the 3° γ section below.
Metal ion interactions can also serve to increase the acidity of the O2′ nucleophile facilitating activation (deprotonation). Here again, the electrostatically strained active sites attract metal ions from solution to assist in catalysis. In all of the ribozymes except VSr, monovalent ions from solution are predicted from MD simulations to be territorially bound (Panteva et al. 2015) to the nucleophile and scissile phosphate, and in some cases, form bridging interactions that additionally help to align the nucleophile (Lee et al. 2009; Ekesan and York 2019). In this position, these ions tune (down-shift) the pKa of the nucleophile to increase its acidity and facilitate proton transfer.
It should be emphasized that there are compensating effects related to pKa tuning as a consequence of metal ion interactions in the context of 2° γ catalysis that need to be discussed in terms of both thermodynamics and kinetics. Specifically, in order to enhance catalysis, the thermodynamic gain of pKa down-shifting of the general base upon metal ion binding at its Hoogsteen edge (i.e., increasing the probability of being deprotonated at physiologically relevant pH) cannot be overcompensated by the kinetic penalty of decreased basicity at G5:N1. Similar considerations would apply to balancing the thermodynamic gain of pKa down-shifting of the nucleophile and the kinetic penalty of reduced nucleophilicity. Alternately stated, ribozymes can employ more reactive functional groups that have pKa values shifted from their ideal catalytic pH such that the disadvantage of low abundance (probability) of the active state is partially compensated by higher reactivity. These effects have been considered in recent QM/MM and free energy simulations and found to lead to overall rate enhancement for HHr (Chen et al. 2017), Twr (Gaines et al. 2019), and VSr (Ganguly et al. 2019a). The Hoogsteen edge of guanine is an inherently weak metal ion binding site (Sigel and Sigel 2010; Leonarski et al. 2017), and it is expected that Mg2+ ions are fractionally occupied and have fast exchange rates. Hence, it is possible that metal ion binding kinetics play a role in dynamically tuning the pKa of the general base and 2′-OH to facilitate nucleophile activation.
3° γ catalysis
In addition to base stacking within the “L” of the L-platform motif, the L-anchor and L-pocket components of the L-scaffold have a profound effect on γ catalysis in each of the ribozyme systems. The most common method of anchoring the general base guanine (G5) is through a trans sugar edge/Hoogsteen (tSH) base pair with residue 2 (Fig. 1). Twr, HHr, and 8–17dz each strongly conserve an adenine at position 2, likely because it forms the most stable tSH base pair (three hydrogen bonds) with guanine in this configuration (Leontis et al. 2002). The HPr also anchors the general base via tSH base-pairing and it has been noted that all nucleobases except cytosine are tolerated at position 2 (Pérez-Ruiz et al. 1999), as expected for this base-pair family.
The pistol ribozyme deviates most significantly from the general L-platform/L-scaffold motif, where a cytosine residue (C41 in Psr) is inserted 3′ of the general base and acts as the L-anchor at position 2 in the motif. This shifts G3 (G33) from the prototypical position forming a cWW with N4 (A39) to instead form a cWW pair with C2 (C41). The functional roles of the nucleobases at each of these positions remain the same, despite the unique connectivity and tertiary fold of Psr. With the inserted C41, Psr anchors the general base guanine G5 (G40) along its sugar edge, but through cSH hydrogen bonds with C2:N4 (C41:N4) and/or G6:O6 (G42:O6).
In the case of VSr, residue 2 is a highly conserved adenine (A622) which would be expected to form the previously discussed tHS base pair with the general base G5 (G638). However, recent crystal structures (Suslov et al. 2015; DasGupta et al. 2017) and computational modeling Ganguly et al. 2019a) suggest that this nucleotide (A622) is bulged from the active site along with residue 1 (A621) and does not play the role of the L-anchor. Instead, the general base guanine (G638) in VSr is anchored along its Hoogsteen edge through outer-sphere coordination of a Mg2+ ion bound in the L-pocket, while the bulged N1 and A2 (A621 and A622, respectively) residues support docking of the VSr dimer. The L-pocket binding site is formed by a guanine residue at the 3 position (G623:H-edge) together with the pro-SP and pro-RP NPOs of the scissile and A2 phosphates, respectively (similar to HHr, 8–17dz, and Psr in the G + M+ paradigm).
While the general base is anchored with its WC edge available, the nucleophile must be positioned such that the guanine can deprotonate it. Within the L-platform/L-scaffold framework, there is a single strategy for positioning the nucleophile: a trans base pair involving N-1 and the Hoogsteen edge of nucleobase 6 to form the foot of the “L” of the L-platform (designated “O2′ position” in Table 1). This base pair provides stability for the N-1 residue to stack with the catalytic guanine, G5. It is then the stacking of these two nucleobases, in opposing orientations, that positions the N-1 sugar such that it is well situated for the general base G5 to accept a proton from the O2′ nucleophile. As for the identities of the residues in this base pair, like residue 2, there seems to be a preference for adenine at the 6 position. This is likely due to the flexibility of having both a hydrogen bond donor and acceptor along the Hoogsteen edge of adenine. However, there are numerous combinations of nucleobases that can form either tWH or tSH base pairs. While this full set of base pairs is not strictly isosteric (Leontis et al. 2002), the alignment of the N-1 sugar relative to G5 is similar and thus there is a wide range of variation in the identity of these nucleobases across the ribozyme classes or even within individual classes.
The exocyclic amine of G5 plays an important role in the hydrogen bond network of the L-platform/L-scaffold. Not only does this amine donate an important hydrogen bond to the L-anchor nucleotide, it can also hydrogen bond to one of the NPOs of the scissile phosphate (Twr, HPr, VSr, and Psr) or the nucleophile (HHr and 8–17dz) as discussed previously. In either case, these interactions help to position the nucleophile at a nexus between the general base and the scissile phosphate. Furthermore, this hydrogen bonding network can serve to increase the acidity of the 2′OH nucleophile (similar to the proposed role of Lys41 in RNase A [Raines 1998]) and/or enhance productive hydrogen bonding by elimination of nonproductive, competing hydrogen bond interactions that would hinder its activation by G5:N1− (Seith et al. 2018). Additionally, for the ribozymes that require a divalent metal ion for catalysis (VSr, HHr, Psr, and 8–17dz), the metal ion's interaction with the scissile phosphate may also impact the hydrogen bond network involving the nucleophile and thus contribute to γ catalysis in a similar fashion (Ganguly et al. 2019b). While it is difficult to create experiments that are able to fully decouple these contributions to the various catalytic strategies, theoretical methods, in many instances, are able to integrate constraints that enable their quantitative deconstruction.
δ CATALYSIS
A remarkable feature of the L-platform/L-scaffold motif is its flexibility in supporting different acids, particularly in contrast to the stringent requirement for an invariant general base. Identification of common trends in how the different acids are positioned and utilized provides a foundation for future design focused on tailoring the identity of the general acid. As discussed above, the L-platform/L-scaffold supports both G + A and G + M+ paradigms, differentiated primarily by their distinct mechanisms for δ catalysis—utilizing either a protonated adenine or a divalent metal ion in some way. These distinctions are discussed in terms of 1°, 2°, and 3° contributions to δ catalysis below.
1° δ catalysis
The G + A paradigm, originally coined by Wilson et al. (2016b), groups VSr and HPr, which both use the N1 adenine heteroatom (Jones and Strobel 2003; Kuzmin et al. 2005; Smith and Collins 2007; Suydam et al. 2010) with Twr that uses the N3 adenine heteroatom (Wilson et al. 2016a) for general acid catalysis (Fig. 2). The G + M+ paradigm (Fig. 3) includes HHr, 8–17dz, and Psr, each of which has a metal ion implicated as playing a critical role in general acid catalysis. This divalent ion is recruited to the active site by electrostatic engineering of the previously defined L-pocket (3° δ in Table 1) formed by the Hoogsteen edge of a guanine in position 3 of the L-platform/L-scaffold, along with one of the NPOs of both residue 2 and the scissile phosphate. In HHr, a Mg2+ ion binds in the L-pocket (Wang et al. 1999; Vogt et al. 2006; Lee et al. 2009; Ward and DeRose 2012) and increases the acidity of the 2′-OH of G8 that then can act as the general acid (Blount and Uhlenbeck 2005; Thomas and Perrin 2009), although alternative mechanisms have been suggested where a metal-bound water molecule acts as the acid (Mir et al. 2015; Mir and Golden 2016) and it is possible under different conditions that both pathways are available (Frankel et al. 2017). In 8–17dz and Psr, a water molecule coordinating the divalent metal ion bound in the L-pocket (Pb2+ and Mg2+, respectively) likely acts as the general acid (Liu et al. 2017a; Neuner et al. 2017; Wilson et al. 2019), although simulations suggest that the Pb2+ ion in 8–17dz could also function, at least in part, as a Lewis acid (Ekesan and York 2019).
2° δ catalysis
Similar to the need to tune the pKa of the general base guanine, in the G + A paradigm where the general acid is an adenine, tuning of the pKa can facilitate δ catalysis. However, unlike the guanine general base that required down-shifting of the pKa, the general acid in the case of adenine in the G + A paradigm requires an up-shifted pKa. This is somewhat less challenging in the sense that the negative electrostatic environment of the active site, due to phosphate moieties, facilitates up-shifting the pKa of the general acid by hydrogen bonding with the N6 exocyclic amine of adenine. In Twr the N6 amine of A1 donates dual hydrogen bonds to the NPOs of nucleotide residues 16 and 17 (in the Twr specific numbering scheme), whereas in HPr and VSr, the N6 amine donates a hydrogen bond to the pro-RP oxygen of the scissile phosphate (Ganguly et al. 2019a). In the G + M+ paradigm, there is no obvious special contribution of the L-platform/L-scaffold residues to 2° δ catalysis (although as discussed above, the metal ion itself can promote 2° δ catalysis.
3° δ catalysis
The general acid must be held in a position where it is poised to donate a proton to the O5′ leaving group. In the G + A paradigm, the hydrogen bond interaction between the general acid adenine N6 exocyclic amine and the NPOs discussed above for 2° δ catalysis are also important for holding the general acid in position. In Twr, anchoring of the A1 N6 exocyclic amine is achieved through interactions with the NPOs of two nucleotides involved in a pseudoknot near the active site, together with an uncommon syn orientation about the glycosidic bond of A1. This enables positioning of the general acid adenine, where A1 protonated at N3 is oriented such that it can hydrogen bond with the O5′ leaving group and is thus poised to donate that proton to complete the transphosphorylation reaction. For HPr and VSr, the positioning of the general acid is facilitated by base stacking with the bulged N1 nucleotide of the L-platform/L-scaffold, in addition to hydrogen bonding between the general acid N6 and the pro-RP oxygen of the scissile phosphate.
Role of the L-pocket in the G + M+ paradigm
In the G + M+ paradigm, the positioning of the general acid is enabled by binding of the divalent metal ion in the L-pocket. However, there is some variability in both the positioning of the L-pocket ligands (pro-RP NPO of the scissile phosphate, the Hoogsteen edge of G3, and an additional pro-RP NPO) within the L-platform/L-scaffold as well as the binding modes to those ligands. In HHr, a Mn2+ ion (PDB ID: 2OEU [Martick et al. 2008]), and a Mg2+ ion (PDB ID: 5EAO [Mir and Golden 2016]), each have been observed crystallographically directly coordinated to the N7 of the L-pocket G3 (G10.1 in HHr) as well as the pro-RP NPO of A2 (A9 in HHr). Phosphorothioate-thiophilic metal ion rescue experiments (Wang et al. 1999; Osborne et al. 2005), supported by molecular simulations (Lee et al. 2007, 2008), suggest that in the active state the scissile phosphate acquires a functionally important inner-sphere coordination with a divalent metal ion that has yet to be observed crystallographically.
Original computational studies carried out by Lee et al. (2007, 2008) developed the first rigorous atomic-level model for the active state of HHr whereby the catalytic metal ion occupies a bridging position between the scissile and A9 phosphates (Lee et al. 2009). Crystallographic evidence at the time suggested that a divalent metal ion coordinates G10.1:N7 (L-pocket G3 nucleotide) and the pro-RP of A9 in the ground state, and simulations predicted that this ion can migrate into the bridging position prior to forming the transition state (Lee et al. 2013). In this position, it was discovered the Mg2+ ion forms interactions with the 2′OH of G8, increasing its acidity and enabling it to act as a general acid. Further computational mutagenesis (Lee and York 2010) and quantum mechanical simulations (Wong et al. 2011) lent further support for a metal-activated G8:O2′ to act as the general acid, and experimental studies by Thomas and Perrin (2009) provided convincing evidence the theoretical predictions were correct.
Cd2+ rescue experiments on an extended HHr construct from Schistosoma mansoni indicate that a catalytic metal ion may occupy this binding mode even in the ground state (Ward and DeRose 2012). Due to structural constraints in the active site, this would preclude the metal ion maintaining direct coordination with N7/O6 at the Hoogsteen edge of the L-pocket G3. Rather, outer-sphere coordination with the Hoogsteen edge of G3 is more plausible given that direct coordination of a Mg2+ ion to the N7 positions of nucleobases is quite rare (Leonarski et al. 2017), and substitution of 7-deazaguanine at the G3 position in HHr results in only a modest ∼30-fold decrease in the observed rate (Nakamatsu et al. 2000), leading the authors to conclude that while this site is important (Peracchi et al. 1996; Wang et al. 1999), it is not catalytically indispensable.
In 8–17dz, a Pb2+ ion (PDB ID: 5XM8) was observed with partial occupancy bound at the O6 position of the L-pocket G3 (G6 in 8–17dz) (Liu et al. 2017a). While less biochemical data are available for this system, it has been reported that stereospecific thio substitution at the pro-RP NPO of the scissile phosphate can be used to selectively remove the SP isomer, with the remaining RP isomer being active in the presence of Cd2+ (Huang and Liu 2015). Further, similar to HHr, a 7-deazaguanine substitution at the L-pocket G3 position leads to only a modest ∼25-fold decrease in the observed rate (Peracchi et al. 2005). Together, this suggests the active state of 8–17dz has a catalytic metal ion binding mode similar to that of HHr, involving direct coordination to the pro-RP NPO of the scissile phosphate, and indirect coordination with the Hoogsteen edge of G3.
In Psr, a Mg2+ ion (PDB ID: 5K7C [Ren et al. 2016], 5KTJ [Nguyen et al. 2017], and 6R47 [Wilson et al. 2019]) was modeled as directly coordinated to the N7 of the L-pocket G3 (G33 in Psr), and indirectly coordinates to the pro-RP NPO of the scissile phosphate. Thio substitution experiments indicate that there is a significant, normal effect at the scissile phosphate NPO (Harris et al. 2015), subsequently identified as the pro-RP position (Wilson et al. 2019), that unlike with HHr, is not rescuable by Mn2+ ions. Furthermore, in contrast to both HHr and 8–17dz, loss of inner-sphere coordination with the N7 that occurs upon 7-deazaguanine substitution leads to a minimum ∼300-fold decrease in the observed rate (Neuner et al. 2017) and may be even substantially more detrimental (>104-fold decrease) to activity (Wilson et al. 2019). Taken together, this suggests that the active state of Psr has the catalytic metal ion maintaining direct coordination to the N7 of the L-pocket G3, while making an outer-sphere contact to the pro-RP NPO of the scissile phosphate. This pattern of direct/indirect metal ion coordination for Psr with the L-pocket binding ligands is opposite to that for HHr and 8–17dz.
The functional requirement for direct coordination of a divalent metal ion with the Hoogsteen edge of the L-pocket G3 is, however, not unique to Psr or the G + M+ paradigm. The VSr requires a functionally critical divalent metal to bind in the L-pocket (not to serve as the general acid, but rather the role of the missing L-anchor) and is predicted to directly coordinate the Hoogsteen edge of G3 (G623 in VSr). Similar to Psr, 7-deazaguanine mutation at this position in VSr effectively abolishes activity (Ganguly et al. 2019a). Hence, in the case of Psr and VSr, there is strong evidence that the active state requires direct Mg2+ ion coordination to the N7 position of the L-pocket G3, despite this binding mode being rarely observed crystallographically (Leonarski et al. 2017). As for the remaining ligand defining the L-pocket (prototypically an NPO 5′ of the L-anchor), in the G + M+ paradigm, it is consistently the pro-RP nonbridging oxygen. However, with the L-anchor nucleotide being part of the general base strand in Psr, it is the phosphate 5′ of G3 (rather than A2 in HHr and 8–17dz) that serves as that additional contact for the catalytic metal ion.
CONCLUSION
We present a generalized L-platform/L-scaffold active site architecture that serves as a blueprint to facilitate the rational design of nucleic acid enzymes that catalyze site-specific RNA cleavage through 2′-O-transphosphorylation. We illustrate how the generalized L-platform/L-scaffold is common to five of the nine currently known naturally occurring ribozymes classes (Twr, HPr, VSr, HHr, Psr) as well as a recently structurally characterized artificially engineered DNAzyme (8–17dz). We identify key base-pairing and stacking requirements that enable conserved features to emerge, as well as elements that can tolerate variation both across and within the ribozyme classes to be explored. The L-platform/L-scaffold motif poises an invariant guanine to act as the general base, while leaving the Hoogsteen edge exposed to solvent, enabling recruitment of cations (except for VSr where this is achieved by outer-sphere interactions with a divalent ion bound in the L-pocket). Within this motif, the preference for guanine (over the other naturally occurring nucleobases) to act as the general base is clear. Having both a hydrogen bond donor and an acceptor along the sugar edge provides both stability and flexibility in base-pairing with the L-anchor residue. Along the Watson–Crick edge, the exocyclic amine can also hydrogen bond with the nonbridging oxygens of the scissile phosphate helping to both localize the reactive atoms and stabilize negative charge in the transition state. Finally, the pKa of the N1 site can be dynamically tuned through metal ion interactions at the Hoogsteen edge, facilitating activation of the nucleophile via proton transfer.
In contrast, the identity of the general acid displays almost as much variety as the global folds across the ribozyme classes. Despite the differences, there are still clear trends in how the L-platform/L-scaffold “docks” the various general acids, enabling classification of the ribozymes into one of two paradigms: G + A and G + M+. Within the G + M+ paradigm, a divalent metal implicated in general acid catalysis is bound by a well-defined set of ligands that form the L-pocket. On the other hand, the structural requirements and even the apparent preference for utilizing adenine as the acid remains less clear. Ultimately, the full extent to which the L-platform/L-scaffold can accommodate different general acids in naturally occurring or synthetic contexts remains to be seen.
Furthermore, the L-platform/L-scaffold motif is common to a large fraction of the currently identified small nucleolytic ribozyme classes, but it is by no means a universal platform. The HDV and TSr ribozymes have Mg2+ implicated as the general base and cytosine has been proposed as their general acid. While the glmS ribozyme utilizes a guanine in the general base role, its active site does not conform to the L-platform/L-scaffold architecture, likely due to additional structural requirements for binding the GlcN6P cofactor. A cofactor independent glmS variant has been in vitro evolved (Lau and Ferré-D'Amaré 2013), but retains the wild-type fold and thus does not adopt an L-platform/L-scaffold active site. There are numerous ribozymes both naturally occurring (e.g., hatchet ribozyme) or artificially engineered (e.g., GR5, 10–23, and NaA43 DNAzymes) yet to be structurally characterized and many more yet to be discovered that are certain to provide insight into the general principles of RNA catalysis that could further facilitate the rational design. It is also particularly intriguing that through directed evolution 8–17dz converged on the same L-platform/L-scaffold motif as the naturally occurring ribozymes examined here. This begs the question as to the extent to which design principles such as those defined by the L-platform/L-scaffold motif presented here might also be translated into nonbiological contexts such as with the recently reported Hachimoji RNA/DNA (Hoshika et al. 2019).
ACKNOWLEDGMENTS
The authors are grateful for the financial support provided by the National Institutes of Health (nos. GM62248 to D.M.Y. and GM131568 to J.A.P.). Computational resources were provided by the Office of Advanced Research Computing (OARC) at Rutgers, The State University of New Jersey, the National Institutes of Health under grant no. S10OD012346 and by the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation (nos. ACI-1548562 and OCI-1053575). Additionally, this research is part of the Blue Waters sustained-petascale computing project, which is supported by the National Science Foundation (awards OCI-0725070 and ACI-1238993) and the state of Illinois. Blue Waters is a joint effort of the University of Illinois at Urbana-Champaign and its National Center for Supercomputing Applications.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.071894.119.
REFERENCES
- Andersen AA, Collins RA. 2000. Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol Cell 5: 469–478. 10.1016/S1097-2765(00)80441-4 [DOI] [PubMed] [Google Scholar]
- Ausländer S, Fussenegger M. 2017. Synthetic RNA-based switches for mammalian gene expression control. Curr Opin Biotechnol 48: 54–60. 10.1016/j.copbio.2017.03.011 [DOI] [PubMed] [Google Scholar]
- Ausländer S, Ketzer P, Hartig JS. 2010. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol Biosyst 6: 807–814. 10.1039/b923076a [DOI] [PubMed] [Google Scholar]
- Bevilacqua PC, Harris ME, Piccirilli JA, Gaines C, Ganguly A, Kostenbader K, Ekesan Ş, York DM. 2019. An ontology for facilitating discussion of catalytic strategies of RNA-cleaving enzymes. ACS Chem Biol 14: 1068–1076. 10.1021/acschembio.9b00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Uhlenbeck OC. 2005. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct 34: 415–440. 10.1146/annurev.biophys.34.122004.184428 [DOI] [PubMed] [Google Scholar]
- Breaker RR. 2002. Engineered allosteric ribozymes as biosensor components. Curr Opin Biotechnol 13: 31–39. 10.1016/S0958-1669(02)00281-1 [DOI] [PubMed] [Google Scholar]
- Breaker RR. 2017. Mechanistic debris generated by twister ribozymes. ACS Chem Biol 12: 886–891. 10.1021/acschembio.7b00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR, Joyce GF. 1994. A DNA enzyme that cleaves RNA. Chem Biol 1: 223–229. 10.1016/1074-5521(94)90014-0 [DOI] [PubMed] [Google Scholar]
- Buzayan JM, Gerlach WL, Bruening G. 1986. Nonenzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature 323: 349–353. 10.1038/323349a0 [DOI] [Google Scholar]
- Cech TR. 1992. Ribozyme engineering. Curr Opin Struct Biol 2: 605–609. 10.1016/0959-440X(92)90093-M [DOI] [Google Scholar]
- Chen H, Giese TJ, Golden BL, York DM. 2017. Divalent metal ion activation of a guanine general base in the hammerhead ribozyme: insights from molecular simulations. Biochemistry 56: 2985–2994. 10.1021/acs.biochem.6b01192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane JC, Strobel SA. 2008. Catalytic strategies of self-cleaving ribozymes. Acc Chem Res 41: 1027–1035. 10.1021/ar800050c [DOI] [PubMed] [Google Scholar]
- DasGupta S, Suslov NB, Piccirilli JA. 2017. Structural basis for substrate helix remodeling and cleavage loop activation in the Varkud satellite ribozyme. J Am Chem Soc 139: 9591–9597. 10.1021/jacs.7b03655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty EA, Doudna JA. 2001. Ribozyme structures and mechanisms. Annu Rev Biophys Biomol Struct 30: 457–475. 10.1146/annurev.biophys.30.1.457 [DOI] [PubMed] [Google Scholar]
- Ekesan Ş, York DM. 2019. Dynamical ensemble of the active state and transition state mimic for the RNA-cleaving 8–17 DNAzyme in solution. Nucleic Acids Res 47: 10282–10295. 10.1093/nar/gkz773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson GM, Nakamura S, Roth A, Breaker RR. 2003. Ribozyme speed limits. RNA 9: 907–918. 10.1261/rna.5680603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felletti M, Stifel J, Wurmthaler LA, Geiger S, Hartig JS. 2016. Twister ribozymes as highly versatile expression platforms for artificial riboswitches. Nat Commun 7: 1–8. 10.1038/ncomms12834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Scott WG. 2010. Small self-cleaving ribozymes. Cold Spring Harb Perspect Biol 2: a003574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Zhou K, Doudna JA. 1998. Crystal structure of a hepatitis delta virus ribozyme. Nature 395: 567–574. 10.1038/26912 [DOI] [PubMed] [Google Scholar]
- Frankel EA, Strulson CA, Keating CD, Bevilacqua PC. 2017. Cooperative interactions in the hammerhead ribozyme drive pKa shifting of G12 and its stacked base C17. Biochemistry 56: 2537–2548. 10.1021/acs.biochem.7b00174 [DOI] [PubMed] [Google Scholar]
- Gaines CS, York DM. 2017. Model for the functional active state of the TS ribozyme from molecular simulation. Angew Chem Int Ed 129: 13577–13580. 10.1002/ange.201705608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines CS, Giese TJ, York DM. 2019. Cleaning up mechanistic debris generated by twister ribozymes using computational RNA enzymology. ACS Catal 9: 5803–5815. 10.1021/acscatal.9b01155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Weissman BP, Giese TJ, Li NS, Hoshika S, Rao S, Benner SA, Piccirilli JA, York DM. 2019a. Theory and experiment converge to define the active site configuration and catalytic mechanism of the largest known nucleolytic ribozyme. Nat Chem. 10.1038/s41557-019-0391-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Weissman BP, Piccirilli JA, York DM. 2019b. Evidence for a catalytic strategy to promote nucleophile activation in metal-dependent RNA-cleaving ribozymes and 8–17 DNAzyme. ACS Catal 9: 10612–10617. 10.1021/acscatal.9b02035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HC, De Abreu DM, Tiller ER, Saville BJ, Olive JE, Collins RA. 1993. Nucleotide sequence requirements for self-cleavage of Neurospora VS RNA. J Mol Biol 232: 351–361. 10.1006/jmbi.1993.1395 [DOI] [PubMed] [Google Scholar]
- Harris KA, Lünse CE, Li S, Brewer KI, Breaker RR. 2015. Biochemical analysis of pistol self-cleaving ribozymes. RNA 21: 1852–1858. 10.1261/rna.052514.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldenbrand H, Janowski PA, Giambaşu G, Giese TJ, Wedekind JE, York DM. 2014. Evidence for the role of active site residues in the hairpin ribozyme from molecular simulations along the reaction path. J Am Chem Soc 136: 7789–7792. 10.1021/ja500180q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshika S, Leal NA, Kim MJ, Kim MS, Karalkar NB, Kim HJ, Bates AM, Watkins NE, SantaLucia HA, Meyer AJ, et al. 2019. Hachimoji DNA and RNA: a genetic system with eight building blocks. Science 363: 884–887. 10.1126/science.aat0971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PJJ, Liu J. 2015. Rational evolution of Cd2+-specific DNAzymes with phosphorothioate modified cleavage junction and Cd2+ sensing. Nucleic Acids Res 43: 6125–6133. 10.1093/nar/gkv519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izatt RM, Christensen JJ, Rytting JH. 1971. Sites and thermodynamic quantities associated with proton and metal ion interaction with ribonucleic acid, deoxyribonucleic acid, and their constituent bases, nucleosides, and nucleotides. Chem Rev 71: 439–481. 10.1021/cr60273a002 [DOI] [PubMed] [Google Scholar]
- Jimenez RM, Polanco JA, Lupták A. 2015. Chemistry and biology of self-cleaving ribozymes. Trends Biochem Sci 40: 648–661. 10.1016/j.tibs.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FD, Strobel SA. 2003. Ionization of a critical adenosine residue in the Neurospora Varkud satellite ribozyme active site. Biochemistry 42: 4265–4276. 10.1021/bi020707t [DOI] [PubMed] [Google Scholar]
- Klein DJ, Ferré-D'Amaré AR. 2006. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313: 1752–1756. 10.1126/science.1129666 [DOI] [PubMed] [Google Scholar]
- Kobori S, Yokobayashi Y. 2018. Analyzing and tuning ribozyme activity by deep sequencing to modulate gene expression level in mammalian cells. ACS Synth Biol 7: 371–376. 10.1021/acssynbio.7b00367 [DOI] [PubMed] [Google Scholar]
- Kostenbader K, York DM. 2019. Molecular simulations of the pistol ribozyme: unifying the interpretation of experimental data and establishing functional links with the hammerhead ribozyme. RNA 25: 1439–1456. 10.1261/rna.071944.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Košutić M, Neuner S, Ren A, Flür S, Wunderlich C, Mairhofer E, Vušurović N, Seikowski J, Breuker K, Höbartner C, et al. 2015. A mini-twister variant and impact of residues/cations on the phosphodiester cleavage of this ribozyme class. Angew Chem Int Ed 54: 15128–15133. 10.1002/anie.201506601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacheva YS, Tzokov SB, Murray IA, Grasby JA. 2004. The role of phosphate groups in the VS ribozyme-substrate interaction. Nucleic Acids Res 32: 6240–6250. 10.1093/nar/gkh957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin YI, Costa CPD, Cottrell JW, Fedor MJ. 2005. Role of an active site adenine in hairpin ribozyme catalysis. J Mol Biol 349: 989–1010. 10.1016/j.jmb.2005.04.005 [DOI] [PubMed] [Google Scholar]
- Lassila JK, Zalatan JG, Herschlag D. 2011. Biological phosphoryl-transfer reactions: understanding mechanism and catalysis. Annu Rev Biochem 80: 669–702. 10.1146/annurev-biochem-060409-092741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau MWL, Ferré-D'Amaré AR. 2013. An in vitro evolved glmS ribozyme has the wild-type fold but loses coenzyme dependence. Nat Chem Biol 9: 805–810. 10.1038/nchembio.1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TS, York DM. 2010. Computational mutagenesis studies of hammerhead ribozyme catalysis. J Am Chem Soc 132: 13505–13518. 10.1021/ja105956u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TS, Silva-López C, Martick M, Scott WG, York DM. 2007. Insight into the role of Mg2+ in hammerhead ribozyme catalysis from x-ray crystallography and molecular dynamics simulation. J Chem Theory Comput 3: 325–327. 10.1021/ct6003142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TS, Silva López C, Giambas¸u GM, Martick M, Scott WG, York DM. 2008. Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation. J Am Chem Soc 130: 3053–3064. 10.1021/ja076529e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TS, Giambas¸u GM, Sosa CP, Martick M, Scott WG, York DM. 2009. Threshold occupancy and specific cation binding modes in the hammerhead ribozyme active site are required for active conformation. J Mol Biol 388: 195–206. 10.1016/j.jmb.2009.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TS, Wong KY, Giambas¸u GM, York DM. 2013. Bridging the gap between theory and experiment to derive a detailed understanding of hammerhead ribozyme catalysis. Prog Mol Biol Transl Sci 120: 25–91. 10.1016/B978-0-12-381286-5.00002-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TS, Radak BK, Harris ME, York DM. 2016. A two-metal-ion-mediated conformational switching pathway for HDV ribozyme activation. ACS Catal 6: 1853–1869. 10.1021/acscatal.5b02158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonarski F, D'Ascenzo L, Auffinger P. 2017. Mg2+ ions: do they bind to nucleobase nitrogens? Nucleic Acids Res 45: 987–1004. 10.1093/nar/gkw1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512. 10.1017/S1355838201002515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E. 2002. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res 30: 3497–3531. 10.1093/nar/gkf481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley DMJ. 2017. How RNA acts as a nuclease: some mechanistic comparisons in the nucleolytic ribozymes. Biochem Soc Trans 45: 683–691. 10.1042/BST20160158 [DOI] [PubMed] [Google Scholar]
- Liu Y, Wilson TJ, McPhee SA, Lilley DMJ. 2014. Crystal structure and mechanistic investigation of the twister ribozyme. Nat Chem Biol 10: 739–744. 10.1038/nchembio.1587 [DOI] [PubMed] [Google Scholar]
- Liu H, Yu X, Chen Y, Zhang J, Wu B, Zheng L, Haruehanroengra P, Wang R, Li S, Lin J, et al. 2017a. Crystal structure of an RNA-cleaving DNAzyme. Nat Commun 8: 2006–2015. 10.1038/s41467-017-02203-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wilson TJ, Lilley DMJ. 2017b. The structure of a nucleolytic ribozyme that employs a catalytic metal ion. Nat Chem Biol 13: 508–513. 10.1038/nchembio.2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M, Lee TS, York DM, Scott WG. 2008. Solvent structure and hammerhead ribozyme catalysis. Chem Biol 15: 332–342. 10.1016/j.chembiol.2008.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir A, Golden BL. 2016. Two active site divalent ions in the crystal structure of the hammerhead ribozyme bound to a transition state analogue. Biochemistry 55: 633–636. 10.1021/acs.biochem.5b01139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir A, Chen J, Robinson K, Lendy E, Goodman J, Neau D, Golden BL. 2015. Two divalent metal ions and conformational changes play roles in the hammerhead ribozyme cleavage reaction. Biochemistry 54: 6369–6381. 10.1021/acs.biochem.5b00824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamatsu Y, Warashina M, Kuwabara T, Tanaka Y, Yoshinari K, Taira K. 2000. Significant activity of a modified ribozyme with N7-deazaguanine at g10.1: the double-metal-ion mechanism of catalysis in reactions catalysed by hammerhead ribozymes. Genes Cells 5: 603–612. 10.1046/j.1365-2443.2000.00352.x [DOI] [PubMed] [Google Scholar]
- Neuner S, Falschlunger C, Fuchs E, Himmelstoss M, Ren A, Patel DJ, Micura R. 2017. Atom-specific mutagenesis reveals structural and catalytic roles for an active-site adenosine and hydrated Mg2+ in pistol ribozymes. Angew Chem Int Ed 56: 15954–15958. 10.1002/anie.201708679 [DOI] [PubMed] [Google Scholar]
- Nguyen LA, Wang J, Steitz TA. 2017. Crystal structure of Pistol, a class of self-cleaving ribozyme. Proc Natl Acad Sci USA 114: 1021–1026. 10.1073/pnas.1611191114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne EM, Schaak JE, DeRose VJ. 2005. Characterization of a native hammerhead ribozyme derived from schistosomes. RNA 11: 187–196. 10.1261/rna.7950605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panteva MT, Giambas¸u GM, York DM. 2015. Force field for Mg2+, Mn2+, Zn2+, and Cd2+ ions that have balanced interactions with nucleic acids. J Phys Chem B 119: 15460–15470. 10.1021/acs.jpcb.5b10423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracchi A, Beigelman L, Usman N, Herschlag D. 1996. Rescue of abasic hammerhead ribozymes by exogenous addition of specific bases. Proc Natl Acad Sci USA 93: 11522–11527. 10.1073/pnas.93.21.11522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracchi A, Bonaccio M, Clerici M. 2005. A mutational analysis of the 8–17 deoxyribozyme core. J Mol Biol 352: 783–794. 10.1016/j.jmb.2005.07.059 [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz M, Barroso-delJesus A, Berzal-Herranz A. 1999. Specificity of the hairpin ribozyme: sequence requirements surrounding the cleavage site. J Biol Chem 274: 29376–29380. 10.1074/jbc.274.41.29376 [DOI] [PubMed] [Google Scholar]
- Perreault J, Weinberg Z, Roth A, Popescu O, Chartrand P, Ferbeyre G, Breaker RR. 2011. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput Biol 7: e1002031 10.1371/journal.pcbi.1002031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley HW, Flaherty KM, McKay DB. 1994. Three-dimensional structure of a hammerhead ribozyme. Nature 372: 68–74. 10.1038/372068a0 [DOI] [PubMed] [Google Scholar]
- Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. 1986. Autolytic processing of dimeric plant virus satellite RNA. Science 231: 1577–1580. 10.1126/science.231.4745.1577 [DOI] [PubMed] [Google Scholar]
- Raines RT. 1998. Ribonuclease A. Chem Rev 98: 1045–1066. 10.1021/cr960427h [DOI] [PubMed] [Google Scholar]
- Ren A, Košutić M, Rajashankar KR, Frener M, Santner T, Westhof E, Micura R, Patel DJ. 2014. In-line alignment and Mg2+ coordination at the cleavage site of the env22 twister ribozyme. Nat Commun 5: 5534–5544. 10.1038/ncomms6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren A, Vusurovic N, Gebetsberger J, Gao P, Juen M, Kreutz C, Micura R, Patel D. 2016. Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage. Nat Chem Biol 12: 702–708. 10.1038/nchembio.2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren A, Micura R, Patel DJ. 2017. Structure-based mechanistic insights into catalysis by small self-cleaving ribozymes. Curr Opin Chem Biol 41: 71–83. 10.1016/j.cbpa.2017.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Weinberg Z, Chen AG, Kim PB, Ames TD, Breaker RR. 2014. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat Chem Biol 10: 56–60. 10.1038/nchembio.1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert PB, Massey AP, Sigurdsson ST, Ferré-D'Amaré AR. 2002. Transition state stabilization by a catalytic RNA. Science 298: 1421–1424. 10.1126/science.1076093 [DOI] [PubMed] [Google Scholar]
- Santoro SW, Joyce GF. 1997. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci USA 94: 4262–4266. 10.1073/pnas.94.9.4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville BJ, Collins RA. 1990. A site-specific self-cleavage reaction performed by a novel RNA in neurospora mitochondria. Cell 61: 685–696. 10.1016/0092-8674(90)90480-3 [DOI] [PubMed] [Google Scholar]
- Scott WG, Murray JB, Arnold JRP, Stoddard BL, Klug A. 1996. Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science 274: 2065–2069. 10.1126/science.274.5295.2065 [DOI] [PubMed] [Google Scholar]
- Seith DD, Bingaman JL, Veenis AJ, Button AC, Bevilacqua PC. 2018. Elucidation of catalytic strategies of small nucleolytic ribozymes from comparative analysis of active sites. ACS Catal 8: 314–327. 10.1021/acscatal.7b02976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmeen L, Kuo MY, Dinter-Gottlieb G, Taylor J. 1988. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J Virol 62: 2674–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy R, Siwkowski A, Hampel A. 1998. Mutational analysis of loops 1 and 5 of the hairpin ribozyme. Biochemistry 37: 564–570. 10.1021/bi9721288 [DOI] [PubMed] [Google Scholar]
- Sigel RKO, Sigel H. 2010. A stability concept for metal ion coordination to single-stranded nucleic acids and affinities of individual sites. Acc Chem Res 43: 974–984. 10.1021/ar900197y [DOI] [PubMed] [Google Scholar]
- Slim G, Gait MJ. 1991. Configurationally defined phosphorothioate-containing oligoribonucleotides in the study of the mechanism of cleavage of hammerhead ribozymes. Nucleic Acids Res 19: 1183–1188. 10.1093/nar/19.6.1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Collins RA. 2007. Evidence for proton transfer in the rate-limiting step of a fast-cleaving Varkud satellite ribozyme. Proc Natl Acad Sci USA 104: 5818–5823. 10.1073/pnas.0608864104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood VD, Beattie TL, Collins RA. 1998. Identification of phosphate groups involved in metal binding and tertiary interactions in the core of the Neurospora VS ribozyme. J Mol Biol 282: 741–750. 10.1006/jmbi.1998.2049 [DOI] [PubMed] [Google Scholar]
- Soukup JK. 2014. The structural and functional uniqueness of the glmS ribozyme. Prog Mol Biol Transl Sci 120: 173–193. [DOI] [PubMed] [Google Scholar]
- Suslov NB, DasGupta S, Huang H, Fuller JR, Lilley DMJ, Rice PA, Piccirilli JA. 2015. Crystal structure of the Varkud satellite ribozyme. Nat Chem Biol 11: 840–846. 10.1038/nchembio.1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suydam IT, Levandoski SD, Strobel SA. 2010. Catalytic importance of a protonated adenosine in the hairpin ribozyme active site. Biochemistry 49: 3723–3732. 10.1021/bi100234v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JM, Perrin DM. 2009. Probing general acid catalysis in the hammerhead ribozyme. J Am Chem Soc 131: 1135–1143. 10.1021/ja807790e [DOI] [PubMed] [Google Scholar]
- Vaish NK, Dong F, Andrews L, Schweppe RE, Ahn NG, Blatt L, Seiwert SD. 2002. Monitoring post-translation modification of proteins with allosteric ribozymes. Nat Biotech 20: 810–815. 10.1038/nbt719 [DOI] [PubMed] [Google Scholar]
- Vogt M, Lahiri S, Hoogstraten CG, Britt DR, DeRose VJ. 2006. Coordination environment of a site-bound metal ion in the hammerhead ribozyme determined by 15N and 2H ESEEM spectroscopy. J Am Chem Soc 128: 16764–16770. 10.1021/ja057035p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Karbstein K, Peracchi A, Beigelman L, Herschlag D. 1999. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry 38: 14363–14378. 10.1021/bi9913202 [DOI] [PubMed] [Google Scholar]
- Ward WL, DeRose VJ. 2012. Ground-state coordination of a catalytic metal to the scissile phosphate of a tertiary-stabilized Hammerhead ribozyme. RNA 18: 16–23. 10.1261/rna.030239.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward WL, Plakos K, DeRose VJ. 2014. Nucleic acid catalysis: metals, nucleobases, and other cofactors. Chem Rev 114: 4318–4342. 10.1021/cr400476k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Kim PB, Chen TH, Li S, Harris KA, Lünse CE, Breaker RR. 2015. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol 11: 606–610. 10.1038/nchembio.1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TJ, Lilley DM. 2011. Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid-base catalysis? A critical assessment of available experimental data. RNA 17: 213–221. 10.1261/rna.2473711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TJ, McLeod AC, Lilley DMJ. 2007. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J 26: 2489–2500. 10.1038/sj.emboj.7601698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TJ, Liu Y, Domnick C, Kath-Schorr S, Lilley DMJ. 2016a. The novel chemical mechanism of the twister ribozyme. J Am Chem Soc 138: 6151–6162. 10.1021/jacs.5b11791 [DOI] [PubMed] [Google Scholar]
- Wilson TJ, Liu Y, Lilley DMJ. 2016b. Ribozymes and the mechanisms that underlie RNA catalysis. Front Chem Sci Eng 10: 178–185. 10.1007/s11705-016-1558-2 [DOI] [Google Scholar]
- Wilson TJ, Liu Y, Li NS, Dai Q, Piccirilli JA, Lilley DM. 2019. Comparison of the structures and mechanisms of the pistol and hammerhead ribozymes. J Am Chem Soc 141: 7865–7875. 10.1021/jacs.9b02141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. 2004. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428: 281–286. 10.1038/nature02362 [DOI] [PubMed] [Google Scholar]
- Wong KY, Lee TS, York DM. 2011. Active participation of the Mg2+ ion in the reaction coordinate of RNA self-cleavage catalyzed by the hammerhead ribozyme. J Chem Theory Comput 7: 1–3. 10.1021/ct100467t [DOI] [PMC free article] [PubMed] [Google Scholar]