Abstract
Cell fate commitment involves the progressive restriction of developmental potential. Recent studies have shown that this process requires not only shifts in gene expression but also an extensive remodeling of the epigenomic landscape. To examine how chromatin states are reorganized during cellular specification in an in vivo system, we examined the function of pioneer factor TFAP2A at discrete stages of neural crest development. Our results show that TFAP2A activates distinct sets of genomic regions during induction of the neural plate border and specification of neural crest cells. Genomic occupancy analysis revealed that the repertoire of TFAP2A targets depends upon its dimerization with paralogous proteins TFAP2C and TFAP2B. During gastrula stages, TFAP2A/C heterodimers activate components of the neural plate border induction program. As neurulation begins, TFAP2A trades partners, and TFAP2A/B heterodimers reorganize the epigenomic landscape of progenitor cells to promote neural crest specification. We propose that this molecular switch acts to drive progressive cell commitment, remodeling the epigenomic landscape to define the presumptive neural crest. Our findings show how pioneer factors regulate distinct genomic targets in a stage-specific manner and highlight how paralogy can serve as an evolutionary strategy to diversify the function of the regulators that control embryonic development.
The reiterative use of regulatory proteins is a common feature of embryonic development (Raible 2006; Taylor and LaBonne 2007; Davidson 2009). A finite number of signaling systems and transcription factors are continuously deployed in the genetic programs that generate cellular diversity. Although post-translational modifications and molecular interactions have been shown to modulate protein function (Slattery et al. 2011; Lee et al. 2012), we still have a limited understanding of how transcriptional regulators are able to perform specific tasks within different developmental contexts. This is pertinent for studies on pioneer factors, which are hypothesized to continuously reorganize chromatin states during the progressive stages of cell fate commitment (Heinz et al. 2010; Zaret and Carroll 2011).
The neural crest is a useful system to study context-specific functions of transcriptional regulators (Simões-Costa and Bronner 2015). This stem cell population gives rise to many cellular derivatives in the vertebrate embryo, including melanocytes, peripheral nerves, bone, and cartilage (Le Douarin and Kalcheim 1999). Neural crest formation is orchestrated by a modular gene regulatory network (Meulemans and Bronner-Fraser 2004; Sauka-Spengler and Bronner-Fraser 2008; Simões-Costa and Bronner 2015). This genetic program is initiated during gastrulation, with the induction of the neural plate border, a region of the ectoderm that contains neural crest, neural, and placodal progenitors (Groves and LaBonne 2014). In the early neurula, a subset of cells at the neural plate border become specified as the bona fide neural crest. These sequential steps in the gene regulatory network are defined by the coexpression of genes within the temporally defined modules (Simões-Costa and Bronner 2015), allowing for the progressive commitment to a neural crest fate. This feature of the gene regulatory network affords us with a tractable platform to explore the context-specific functions of developmental genes.
A number of factors participate in multiple network modules and may play separate roles at distinct steps of neural crest formation. One example is the pioneer transcription factor TFAP2A. TFAP2A is associated with permissive chromatin states in neural crest cells, characterized by high levels of EP300 and H3K27ac (Rada-Iglesias et al. 2012). TFAP2A is able to bind to nucleosome DNA (Grossman et al. 2018; Fernandez Garcia et al. 2019), and it exerts a strong effect on chromatin accessibility according to DNase I hypersensitivity analysis (Sherwood et al. 2014). Furthermore, Tfap2a knockout mice display craniofacial malformations and embryonic lethality (Schorle et al. 1996). In humans, missense mutations in the TFAP2A gene result in branchio-oculo-facial syndrome, characterized by cleft palate and other craniofacial abnormalities (Milunsky et al. 2008). Lastly, functional studies in Xenopus embryos indicate that TFAP2A acts reiteratively as a regulator of both neural plate border induction and neural crest specification (de Crozé et al. 2011). Although TFAP2A has been described as a critical regulator of neural crest formation, the mechanisms driving its target specificity during induction and specification remain elusive.
TFAP2A belongs to the TFAP2 family of transcription factors, consisting of five paralogous proteins that bind to DNA as dimers (Eckert et al. 2005). Several lines of evidence suggest that TFAP2A's related paralogs TFAP2B and TFAP2C also play roles during neural crest formation. Genetic studies in zebrafish have shown that Tfap2a and Tfap2c work together to promote neural crest formation (Li and Cornell 2007). Also, in mice, double conditional knockout mutants of Tfap2a/Tfap2b are depleted of melanocytes and possess craniofacial defects, suggesting a synergistic effect of these factors in promoting neural crest differentiation (Seberg et al. 2017; Van Otterloo et al. 2018). Furthermore, mutations in the TFAP2B gene cause Char syndrome, an autosomal dominant disorder associated with patent ductus arteriosus and facial dysmorphism (Satoda et al. 2000).
Here, we explore the hypothesis that TFAP2 paralogs work in concert to progressively define the molecular identity of the presumptive neural crest. We use stage-specific functional analysis and genomic profiling to characterize distinct functions of TFAP2A, TFAP2B, and TFAP2C during neural crest formation. Next, we define the timing of TFAP2 heterodimerization during neural crest induction and specification. Finally, we manipulate the timing of TFAP2-paralog expression to test the idea that TFAP2A target specificity is mediated by the assembly of distinct heterodimers, allowing for the diversification of protein function in these two developmental contexts.
Results
TFAP2A is independently required for induction of the neural plate border and specification of neural crest cells
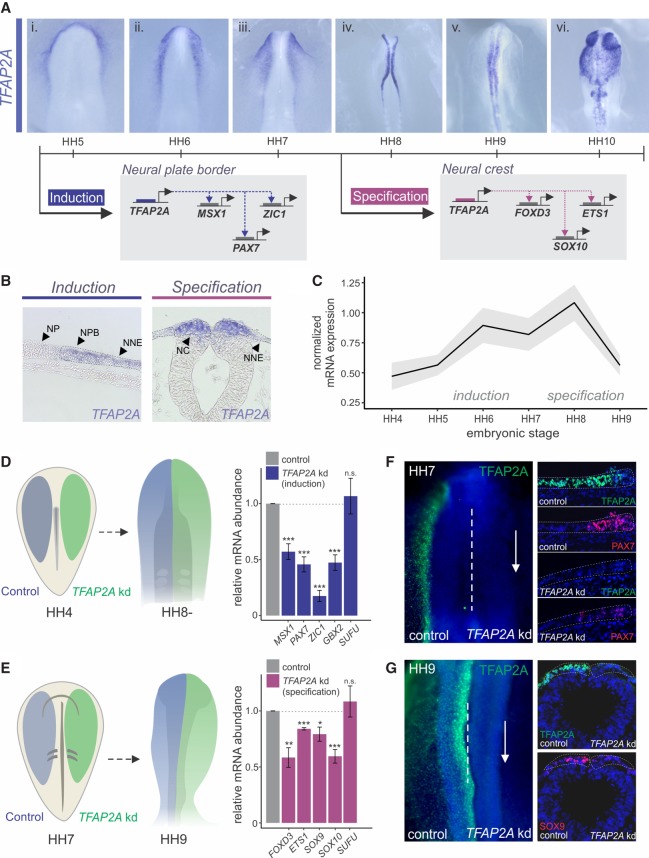
To examine the expression pattern of TFAP2A in the early chick embryo, we performed whole-mount in situ hybridization at distinct developmental time points (Fig. 1A). During gastrulation (Hamburger and Hamilton stage [HH] 5), TFAP2A expression was detected in the neural plate border region and the nonneural ectoderm (Fig. 1Ai). Upon neurulation, TFAP2A expression was enhanced in both premigratory and migratory neural crest (Fig. 1Aii–vi,B), as described in previous reports in other model systems (Zhang et al. 2006). Consistent with this, TFAP2A displayed strong colocalization with both induction and specification markers (Supplemental Fig. S1A). To examine temporal shifts in TFAP2A expression, we used quantitative reverse transcription PCR (RT-PCR). Analysis of avian embryos at six developmental stages (HH4–HH9) revealed a bimodal pattern of gene expression, with peaks in mRNA expression at the stages of induction and specification (Fig. 1C).
Figure 1.
TFAP2A is required for both induction and specification of neural crest cells. (A) Whole-mount in situ hybridization for TFAP2A in chick embryos at six developmental stages (HH5–HH10). TFAP2A is enriched in both premigratory and migratory neural crest. Previous studies have shown regulation by TFAP2A of neural plate border genes MSX1, PAX7, and ZIC1 and for neural crest genes FOXD3, ETS1, and SOX10. (B) Transverse sections displaying TFAP2A expression in the neural plate border and the neural crest. (C) Whole-embryo quantitative RT-PCR for TFAP2A at six developmental stages (HH4–HH9) depicted as normalized mRNA expression relative to the housekeeping gene, HPRT1. (D,E) Electroporation scheme for stage-specific knockdown of TFAP2A. (D) Embryos were bilaterally electroporated with control and TFAP2A DsiRNA reagents at HH4 and screened for an induction phenotype at HH8−. Quantitative RT-PCR for induction genes MSX1, PAX7, ZIC1, and GBX2 in control versus TFAP2A DsiRNA-treated sides of bilaterally electroporated embryos, represented as fold change compared with control. Embryos were surveyed at HH8− (n = 11). (E) Embryos electroporated at HH7 were screened for a specification phenotype at HH9. Quantitative RT-PCR for specification genes, FOXD3, ETS1, SOX9, and SOX10 in control versus TFAP2A DsiRNA-treated sides of bilaterally electroporated embryos, represented as fold change compared with control. Embryos were surveyed at HH9 (n = 10). The neural marker SUFU was included as a control to ensure defects were neural crest specific. (F) Immunohistochemistry (whole-mount and transverse sections) for TFAP2A (green), as well as the neural plate border PAX7 (red), upon bilateral TFAP2A DsiRNA treatment. Orientation of knockdown sections has been flipped for comparison. (G) Immunohistochemistry (whole-mount and transverse sections) for TFAP2A (green) and the neural crest marker SOX9 (red) upon bilateral TFAP2A DsiRNA treatment. (HH) Hamburger Hamilton; (NP) neural plate; (NPB) neural plate border; (NNE) nonneural ectoderm; (NC) neural crest; (kd) knockdown; (n.s.) not significant. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001 (for number of embryos analyzed, see also Supplemental Table S1).
Previous studies have placed TFAP2A at distinct modules of the gene regulatory network, suggesting that it acts reiteratively in both induction and specification (Fig. 1A; Nikitina et al. 2008; de Crozé et al. 2011). To confirm that TFAP2A is independently required for these steps of neural crest formation in avian embryos, we performed stage-specific knockdown experiments. We transfected the ectoderm of avian embryos with a Dicer-substrate short interfering RNA (DsiRNA) targeted to TFAP2A at distinct developmental time points (Fig. 1D,E). These treatments were administered unilaterally, and the resulting phenotypes were evaluated by comparing the control versus the knockdown side of the embryo (Simões-Costa et al. 2015). Quantitative RT-PCR revealed a significant decrease in the expression of neural plate border genes (MSX1, PAX7, ZIC1, and GBX2) (Fig. 1D) and neural crest specification genes (FOXD3, ETS1, SOX9, and SOX10) (Fig. 1E) following DsiRNA treatment at the respective developmental stages. These results were further confirmed by immunohistochemistry, which showed disruption in expression of both PAX7 (Fig. 1F) and SOX9 (Fig. 1G). Although the DsiRNA treatment resulted in a robust knockdown of TFAP2A protein and mRNA (Fig. 1F,G; Supplemental Fig. S1B), it did not grossly effect cell proliferation or death (Supplemental Fig. S1C). Taken together, these results indicate that TFAP2A has independent functions in neural crest formation and is required for the establishment of both induction and the specification regulatory states.
TFAP2A regulates distinct sets of genomic targets during cell fate commitment
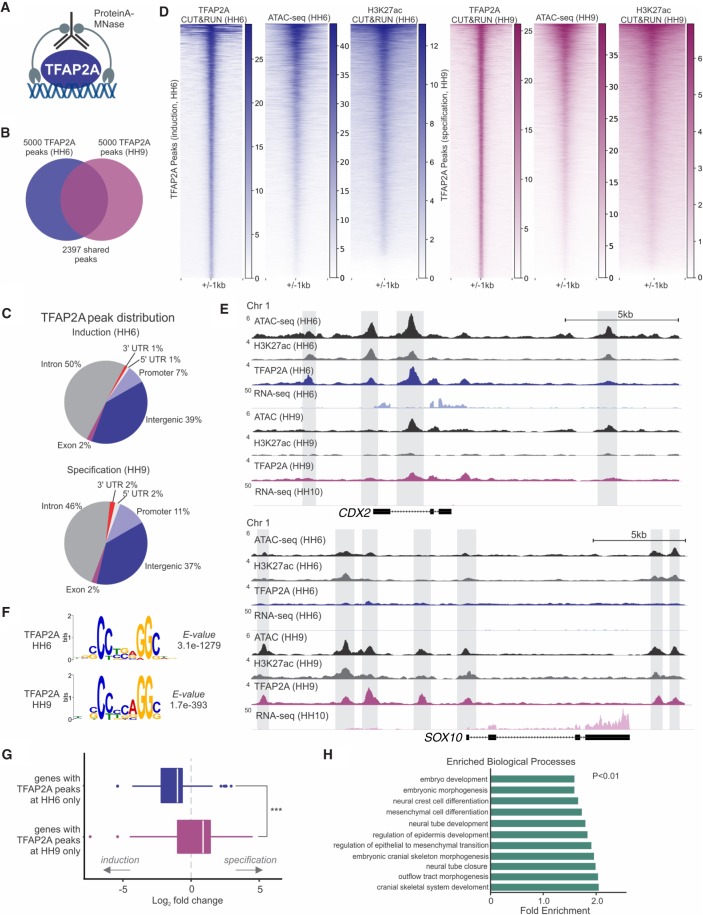
To define the stage-specific functions of TFAP2A in neural crest formation, we conducted genome-wide mapping of transcription factor occupancy with cleavage under targets and release using nuclease (CUT&RUN) (Fig. 2A; Skene and Henikoff 2017). We performed this assay at two developmental time points corresponding to neural plate border induction (HH6) and neural crest specification (HH9), using antibodies targeted to TFAP2A and the active histone mark H3K27ac. We also performed ATAC-seq at these two respective stages to assess changes in chromatin accessibility. Our CUT&RUN data sets for TFAP2A identified 8442 and 18,046 high-confidence (P ≤ 0.001) peaks at HH6 and HH9, respectively (Supplemental Fig. S2A; Supplemental Table S2). This large number of binding events was consistent with previous reports of TFAP2A acting as a critical regulator of the genome of neural crest cells (Rada-Iglesias et al. 2012). Pairwise Pearson's correlation analysis of TFAP2A and H3K27ac CUT&RUN data sets found a strong correlation between stage-matched biological replicates (Supplemental Fig. S2B,C). CUT&RUN also allowed for profiling of TFAP2A binding events with superior resolution (library fragments averaged ∼100–120 bp) (Supplemental Fig. S2D) and signal-to-noise ratio (Supplemental Fig. S2E) than that obtained with ChIP-seq (Rada-Iglesias et al. 2012; Seberg et al. 2017). De novo motif enrichment analysis of both induction and specification peaks yielded the TFAP2A consensus binding sequence (G/C)CCNNNGG(G/C) (Williams and Tjian 1991; Mohibullah et al. 1999) as the highest-ranking motif, which was centrally enriched in the CUT&RUN peaks (Fig. 2F; Supplemental Fig. S2F,H).
Figure 2.
TFAP2A is associated with distinct sets of cis-regulatory elements at sequential steps of neural crest formation. (A) CUT&RUN was used to assess global binding by TFAP2A during induction (HH6) and specification (HH9) in developing chick embryos. (B) Venn diagram of the top 5000 significantly enriched TFAP2A peaks present during induction and specification, 2397 of which are maintained between both developmental time points. (C) Pie charts depicting genomic distribution of TFAP2A peaks at induction (HH6) and specification (HH9). (D) Heatmaps displaying TFAP2A CUT&RUN, ATAC-seq, and H3K27ac CUT&RUN signal at TFAP2A peaks during induction (HH6) and specification (HH9). (E) ATAC, CUT&RUN, and RNA-seq profiles at the loci of the induction gene CDX2 and specification gene SOX10. (F) De novo motif enrichment analysis via MEME-ChIP shows a strong enrichment for the TFAP2A binding motif from both HH6 and HH9 CUT&RUN data sets. Values indicate significance of motif occurrence as reported by MEME. (G) Boxplot displaying the log2 fold change distribution of differentially expressed genes (Padj < 0.1) between HH6 and HH10, which are associated with TFAP2a binding. Genes with HH6-only peaks are enriched during induction, whereas genes with HH9-only peaks are enriched during specification. Significance was assessed via an unpaired two-tailed t-test. (H) Bar plot showing significantly enriched Gene Ontology (GO) terms from GO analysis of genes with annotated TFAP2A peaks (see also Supplemental Table S3). (HH) Hamburger Hamilton; (kb) kilobase. (***) P ≤ 0.001.
Analysis of the top 5000 peaks from each data set, which showed the greatest enrichment for TFAP2A, revealed that ∼50% of the peaks were shared between HH6 and HH9, whereas the other 50% represent stage-specific binding events (Fig. 2B). TFAP2A peak distributions relative to genomic features were similar at both developmental stages, with 46%–50% intronic peaks, 37%–39% intergenic peaks, and 7%–11% of peaks localized to promoter regions (Fig. 2C). Consistent with the pioneer function of TFAP2A, regions occupied by the transcription factor showed enriched chromatin accessibility and H3K27ac signal (Fig. 2D; Creyghton et al. 2010). These results indicate that there is a global shift in TFAP2A occupancy, as well as in chromatin accessibility and activation, during the transition from induction to specification.
Closer inspection of TFAP2A peaks revealed association with the loci of many neural plate border and neural crest genes. Nearest gene assignment of TFAP2A peaks followed by Gene Ontology (GO) analysis showed an enrichment of factors involved in neural crest, embryonic development, and cell fate commitment (Fig. 2H; Supplemental Table S3). TFAP2A occupancy also correlated with the expression dynamics of neural crest genes. For instance, CDX2, a neural plate border marker that is down-regulated upon neural crest specification (Sanchez-Ferras et al. 2012), is highly enriched for TFAP2A binding during neural plate border induction but is devoid of any binding at later stages. Similarly, SOX10, which begins to be expressed at HH8 and has a neural crest–specific expression pattern (Betancur et al. 2010), is associated with TFAP2A during neural crest specification but not during induction (Fig. 2E). Consistent with this, analysis of components of the neural crest gene regulatory network indicated that early and late TFAP2A dynamic peaks, that is, those that are present at induction or specification only, are preferentially associated with induction and specification genes, respectively (Supplemental Table S4). In addition, genes expressed during both induction and specification, such as MSX1, are bound by TFAP2A at both time points (Supplemental Fig. S2G).
The association of TFAP2A with changes in chromatin accessibility and H3K27ac signal in the loci of neural crest genes (Fig. 2E) suggests that this dynamic binding underlies changes in the epigenomic landscape of neural crest cells. To further explore this possibility, we performed differential binding analysis of TFAP2A peaks during induction and specification. This analysis identified 1672 and 2319 differentially bound peaks (fold change > <2 and FDR < 0.05) that represent putative enhancer elements subject to TFAP2A regulation in a stage-specific manner (Supplemental Fig. S3A; Supplemental Table S5). We found that several of these loci correspond to previously characterized neural crest enhancers, including Sox10e1 and Snai2 enh-13 (Supplemental Fig. S3A; Betancur et al. 2010; Williams et al. 2019). Furthermore, we also tested a number of novel regulatory regions to determine whether they may be acting as bona fide enhancers, several of which were present in the loci of genes associated with neural crest and embryonic development (Supplemental Fig. S3B; Supplemental Table S6). We cloned regulatory regions that displayed dynamic TFAP2A binding in pTK:eGFP (Uchikawa et al. 2003) and transfected these constructs in gastrula-stage embryos. We observed that these enhancers were active in the neural plate border (HH6) and neural crest (HH9), consistent with the timing of TFAP2A binding (Supplemental Fig. S3C).
Finally, to test if the timing of TFAP2A binding is associated with temporal patterns of gene expression, we performed RNA-seq on neural crest cells at two stages corresponding to induction (HH6) and specification (HH10). By comparing gene expression levels between these two stages, we found that target gene expression correlates with TFAP2A occupancy. Genes that are associated with TFAP2A at early stages are more likely to be enriched at the induction, whereas late TFAP2A binding is associated with higher mRNA levels during specification (Fig. 2E,G). This dynamic regulation suggests differential binding in a stage-dependent manner to drive the expression of distinct target genes. In conjunction with our functional analysis (Fig. 1), these results indicate a dual role for TFAP2A during induction and specification in that it activates distinct gene regulatory modules as neural crest cells become progressively committed.
Sequential requirement for TFAP2 paralogs in the presumptive neural crest
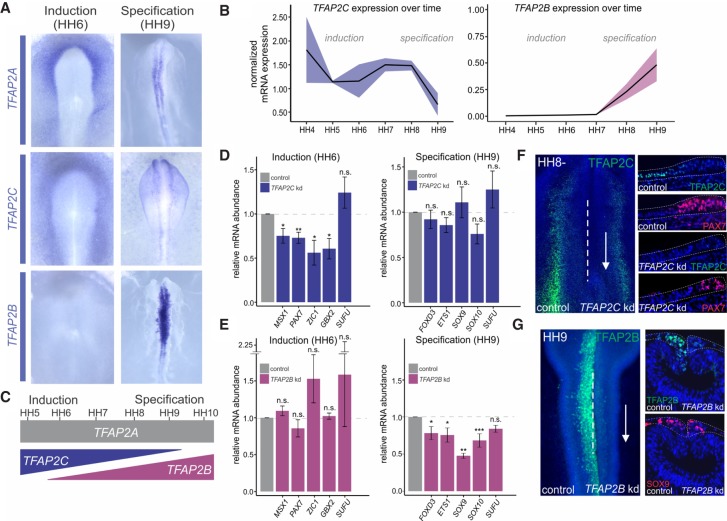
Our genomic analysis shows that TFAP2A controls distinct cis-regulatory elements in a stage-specific manner. We next investigated how this factor is able to target different sets of enhancers during induction and specification. Because members of the TFAP2 family of transcription factors bind to DNA as dimers, we hypothesized that other TFAP2 paralogs may also contribute to regulate neural plate border induction and neural crest specification. To test this, we assessed the spatiotemporal expression patterns of TFAP2C and TFAP2B in the early chick embryo. We found that although TFAP2C was enriched at the neural plate border, it was lost in the neural crest lineage at later stages and its expression became restricted to the presumptive epidermis. Conversely, TFAP2B transcripts were not detected until just before the onset of specification, when the gene became robustly expressed in the neural crest lineage (Fig. 3A; Supplemental Fig. S4A,B). Quantitative RT-PCR analysis revealed opposing trends in mRNA expression dynamics, with TFAP2C levels decreasing upon the onset of TFAP2B expression (Fig. 3B). The temporal patterns in TFAP2C and TFAP2B expression suggest that these paralogs may play discrete roles in neural crest development, regulating induction and specification, respectively. Also, the expression patterns of TFAP2C and TFAP2B are contained within the TFAP2A expression domain, suggesting subfunctionalization of these paralogs over the course of evolution (Fig. 3C; Ohno 1970).
Figure 3.
TFAP2B and TFAP2C show complementary expression patterns and are required for induction and specification. (A) In situ hybridization for TFAP2A, TFAP2C, and TFAP2B in whole-mount chick embryos during induction (HH6) and specification (HH9). (B) Whole-embryo quantitative RT-PCR for TFAP2C and TFAP2B at six developmental stages (HH4–HH9) displays normalized mRNA expression relative to the housekeeping gene HPRT1. (C) TFAP2C and TFAP2B only partially recapitulate the expression pattern of TFAP2A. (D) Quantitative RT-PCR for induction genes MSX1, PAX7, ZIC1, and GBX2 (n = 6) and specification genes FOXD3, ETS1, SOX9, and SOX10 (n = 6), in control versus TFAP2C morpholino-treated sides of bilaterally electroporated embryos, represented as fold change compared with control. Phenotypes were surveyed at HH8− and HH9, respectively. (E) Quantitative RT-PCR for induction genes MSX1, PAX7, ZIC1, and GBX2 (n = 8) and specification genes FOXD3, ETS1, SOX9, and SOX10 (n = 8), in control versus TFAP2B DsiRNA-treated sides of bilaterally electroporated embryos, represented as fold change compared with control. Phenotypes were surveyed at HH8− and HH9, respectively. The neural marker SUFU was included as a control to ensure defects were neural crest specific. (F) Immunohistochemistry (whole-mount and transverse sections) for TFAP2C (green), as well as the induction marker PAX7 (red), in bilaterally transfected TFAP2C morpholino-treated embryos. Orientation of knockdown sections has been flipped for comparison. (G) Immunohistochemistry (whole-mount and transverse sections) for TFAP2B (green), as well as the specification marker SOX9, upon bilateral control versus TFAP2B DsiRNA treatment. (B,D,E) Error bars, SE. (HH) Hamburger Hamilton; (kd) knockdown; (n.s.) not significant. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001 (for number of embryos analyzed, see also Supplemental Table S1).
To assess the function of TFAP2C and TFAP2B in neural crest development, we performed time-controlled knockdowns of each paralog and surveyed their effects on neural crest development. We used the same stage-specific transfection scheme used in our analysis of TFAP2A function (Fig. 1D,E), and quantified gene expression of target genes with quantitative RT-PCR. TFAP2C knockdown during induction resulted in a down-regulation of the neural plate border markers MSX1, PAX7, ZIC1, and GBX2 but had no significant effects on the expression of specification genes FOXD3, ETS1, SOX9, and SOX10 (Fig. 3D). Conversely, TFAP2B knockdown produced no effects on the neural plate border but resulted in the disruption of the specification program (Fig. 3E). These phenotypes were confirmed by immunohistochemistry for the neural plate border induction marker PAX7 (Basch et al. 2006) and the neural crest specification marker SOX9 (Fig. 3F,G; Supplemental Fig. S4C; Mori-Akiyama et al. 2003). Double knockdown of both TFAP2A and TFAP2C in induction, as well as TFAP2A and TFAP2B in specification, resulted in a similar down-regulation of neural plate border and neural crest markers, respectively (Supplemental Fig. S4E,F). Although the knockdown methods used resulted in a robust down-regulation of the targeted proteins (Fig. 3F,G), we did not observe changes in cell death following embryo transfection (Supplemental Fig. S4D). These results indicate that TFAP2C and TFAP2B regulate induction and specification, respectively, and raise the possibility that TFAP2 paralogs regulate different steps of neural crest formation.
TFAP2 paralogs cooperate to regulate induction and specification
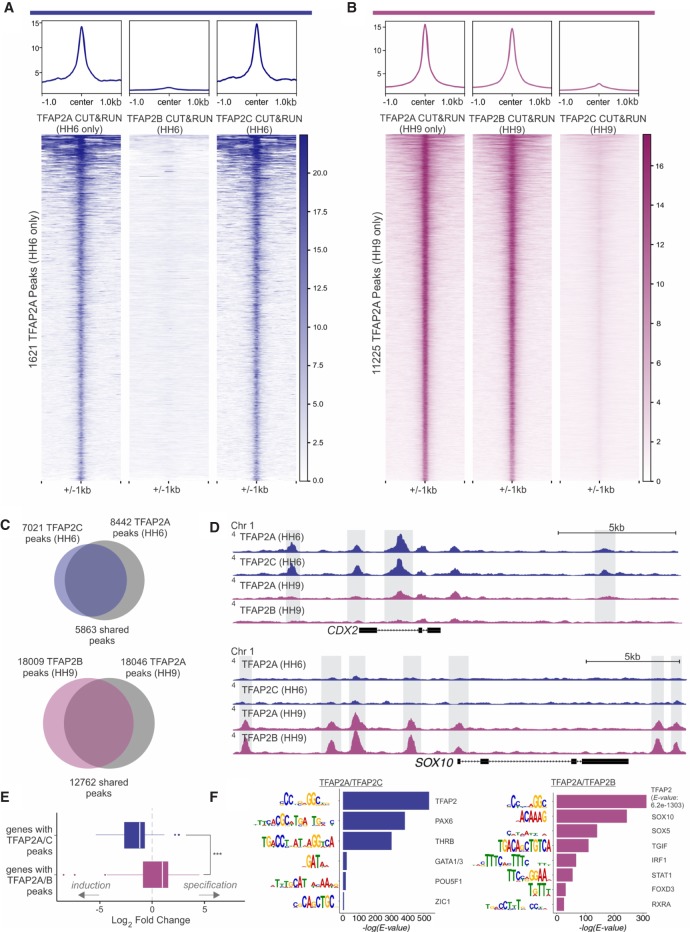
Our previous data showed that TFAP2C and TFAP2B overlap with TFAP2A both in expression and function at different stages of neural crest development. These findings led us to postulate that TFAP2C and TFAP2B cooperate with TFAP2A to regulate discrete sets of neural crest enhancers during induction and specification. To test this, we performed CUT&RUN for both TFAP2C and TFAP2B at time points corresponding to induction (HH6) and specification (HH9), using paralog-specific antibodies (Supplemental Fig. S5A–C). By surveying the genomic occupancy of each gene at both stages, we found that TFAP2 paralogs are consistently associated with the same genomic regions as TFAP2A (Fig. 4A,B). We identified 7021 peaks for TFAP2C at HH6 (induction), 5863 of which are co-occupied by TFAP2A (Fig. 4C; Supplemental Table S3). Analysis of TFAP2B binding during neural crest specification identified 18,009 peaks, of which 12,762 showed co-occupancy with TFAP2A (Fig. 4C; Supplemental Table S3). When we examined dynamic TFAP2A peaks, we observed that they were occupied by distinct paralog pairs. Early (HH6) TFAP2A peaks were enriched for TFAP2C occupancy but had no TFAP2B binding (Fig. 4A). Conversely, late (HH9) TFAP2A peaks were associated with TFAP2B but only displayed background TFAP2C signal (Fig. 4B). Because our CUT&RUN peaks display a single, centrally enriched TFAP2 motif (Supplemental Fig. S2F), the co-occupancy of putative enhancers by distinct paralogs suggests that they are acting as heterodimers.
Figure 4.
TFAP2A/C have shared regulatory targets in induction, whereas TFAP2A/B have shared targets in specification. (A) Profiles and heatmaps displaying signal for TFAP2A, TFAP2B, and TFAP2C at induction-specific TFAP2A peaks (1621 peaks). (B) Profiles and heatmaps displaying signal for TFAP2A, TFAP2B, and TFAP2C at specification-specific TFAP2A peaks (11,225 peaks). (C) At induction (HH6), CUT&RUN analysis identified 8442 TFAP2A peaks and 7021 TFAP2C peaks, 5863 of which co-occupied by both TFAP2A and TFAP2C. During specification, 18,046 TFAP2A peaks and 18,009 TFAP2B peaks were detected, with an overlap of 12,762 peaks occupied by both TFAP2A and TFAP2B. (D) CUT&RUN profiles for TFAP2A/C at induction (HH6) and TFAP2A/B and at specification (HH9) in the loci of CDX2 and SOX10. (E) Boxplot displaying distribution of genes significantly enriched (Padj < 0.1) at HH6 versus HH10 that have TFAP2A/C peaks TFAP2A/B in their loci. Genes with associated TFAP2A/C peaks are enriched during induction, whereas genes annotated with TFAP2A/B peaks are enriched during specification. (F) Rank-ordered results of motif enrichment analysis of TFAP2A/C peaks and TFAP2A/B peaks via MEME-ChIP. Values indicate significance of motif occurrence as reported by MEME (redundant motifs have been omitted). (HH) Hamburger Hamilton; (kb) kilobase. (***) P ≤ 0.001.
The results described above indicate that TFAP2 paralogs coregulate active regions in the neural crest genome in stage-specific manner and suggest that TFAP2A changes binding partners as cells transition from induction to specification. This was confirmed by the analysis of individual loci of neural plate border and neural crest genes. For example, CDX2, which is enriched for TFAP2A binding during induction, is also bound by TFAP2C at this same developmental time point. However, binding of both factors is depleted at later stages (HH9) (Fig. 4D; Supplemental Fig. S6C). In contrast, SOX10, which displays TFAP2A binding only during neural crest specification, is also bound by TFAP2B at this time point (Fig. 4D; Supplemental Fig. S6C). The repertoire of genomic targets of TFAP2C and TFAP2B is dynamic and changes extensively during induction and specification (Supplemental Fig. S6A,B). Furthermore, genes that are bound by TFAP2A during both induction and specification, such as MSX1, also show binding first by TFAP2C and then by TFAP2B (Supplemental Fig. S6C). Cumulatively, thousands of loci in the neural plate border and neural crest show dynamic regulation by TFAP2A, which shares regulatory targets with TFAP2C and TFAP2B (Supplemental Table S2).
To test if co-occupancy of TFAP2 paralogs is associated with gene expression, we again used RNA-seq data sets from HH6 and HH10. Consistent with our previous analysis of TFAP2A dynamic peaks, we found that genes that are associated with TFAP2A/C are more likely to be enriched at the induction stages. Conversely, TFAP2A/B binding correlates with higher mRNA levels during specification (Fig. 4E). According to these results, we postulate that TFAP2 heterodimers act as a molecular switch to regulate the transition from induction to specification during neural crest formation. Motif enrichment analysis of TFAP2C and TFAP2B suggests that the target specificity of heterodimers depends upon the recruitment of different cofactors. Analysis of TFAP2A/C peak loci (1-kb region flanking the peak summit) revealed a significant enrichment for binding sites of other factors involved in the formation of the neural plate border, such as PAX, GATA1/3, and ZIC1 (Fig. 4F). TFAP2A/B peaks, on the other hand, are populated with sites for SOX10, SOXD, and FOXD3 (Honoré et al. 2003; Badis et al. 2009; Jolma et al. 2013), which are important components of the specification program (Fig. 4F; Dottori et al. 2001; Honoré et al. 2003). Consistent with previously published results showing that TFAP2 heterodimers recognize the same DNA sequence (Eckert et al. 2005), similar TFAP2 consensus binding motifs (Fig. 4F; Supplemental Fig. S6D) were retrieved from both data sets.
To further explore the hypothesis that TFAP2 factors participate with different cofactors during discrete stages of neural crest formation, we scanned individual enhancers from our differential binding analysis (Supplemental Fig. S3) for binding motifs of predicted cofactors such as PAX, ZIC1, SOX10, and FOXD3. When the sequences for these cofactor binding sites were mutated in our expression constructs, the enhancers were no longer capable of driving reporter gene expression (Supplemental Fig. S7). To quantify wild-type and mutant enhancer activity in neural crest cells, we cotransfected our enhancer reporter constructs with the neural crest reporter (Tfap2aE1:mCherry) and used flow cytometry measure GFP reporter fluorescence within the mCherry-positive neural crest cell population. Every enhancer tested displayed decreased reporter fluorescence in neural crest cells upon mutation of cofactor binding sites (Supplemental Fig. S7). Thus, TFAP2 heterodimers may cooperate with other factors that are part of the gene regulatory network to activate distinct sets of cis-regulatory enhancers and drive the transition from induction to specification.
A molecular switch based on sequential heterodimerization of TFAP2 factors
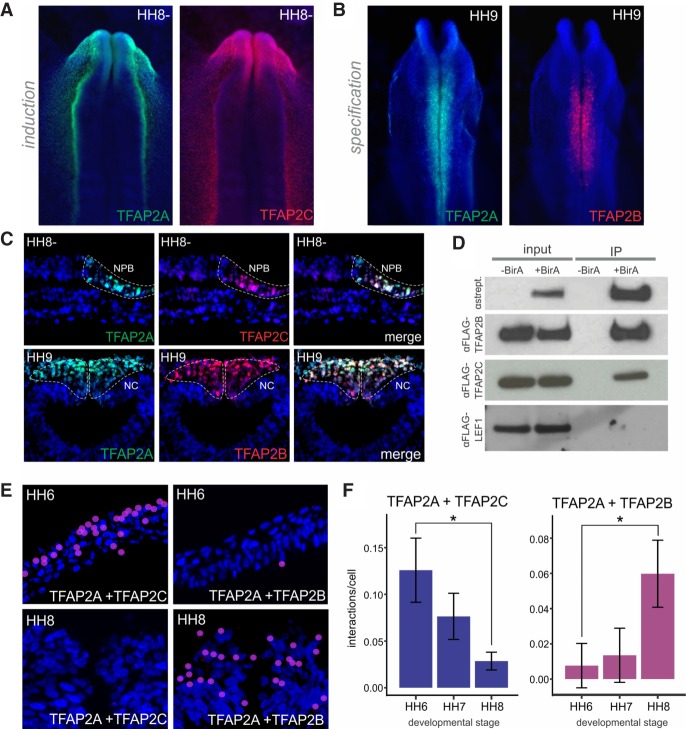
Thus far, our findings suggest that TFAP2A/C activate the neural plate border induction program and that TFAP2A/B activate the specification module of the neural crest gene regulatory network. Thus, we next tested the hypothesis that TFAP2 paralogs act as a molecular switch, trading heterodimerization partners to control distinct steps in neural crest formation. According to this model, TFAP2A/C heterodimers mediate neural plate border induction, whereas TFAP2A/B heterodimers promote neural crest specification. Immunohistochemistry of HH6 and HH9 chick embryos confirmed colocalization of TFAP2A and TFAP2C in neural plate border cells (Fig. 5A,C). At later stages, TFAP2A expression was detected in both the neural crest and the nonneural ectoderm (Fig. 5B,C), whereas TFAP2C was restricted to the ectoderm alone (Supplemental Fig. S8A,B). In contrast, although TFAP2B was not expressed at early stages, it was highly enriched in the neural crest during specification, overlapping with the TFAP2A expression domain (Fig. 5B,C; Supplemental S8A,B). Notably, coexpression of TFAP2B and TFAP2C was only detected transiently, at a discrete stage just before the onset of neural crest specification (Supplemental Fig. S8C).
Figure 5.
TFAP2A switches heterodimeric partners during neural crest development. (A) Immunohistochemistry for TFAP2A (green) and TFAP2C (red) shows strong colocalization at the neural plate border. (B) Immunohistochemistry for TFAP2A (green) and TFAP2B (red) shows colocalization in the neural crest. (C) Transverse sections showing overlap in expression of TFAP2A and TFAP2C at the neural plate border and nonneural ectoderm, and TFAP2A and TFAP2B at the dorsal neural tube. (D) Coimmunoprecipitation of Avi-tagged TFAP2A and FLAG-tagged TFAP2B, TFAP2C, and LEF1. Constructs for biotagged proteins were transfected into chick embryos, and pulldown was conducted with or without the presence of the BirA enzyme. In the presence of BirA, biotinylated TFAP2A coimmunoprecipitates both FLAG-TFAP2B and FLAG-TFAP2C but is unable to pull down LEF1. (E) Proximity ligation assays (PLAs) visualized in transverse sections of the neural plate border and the neural crest. Positive interactions, indicated by red puncta, are enriched for TFAP2A/C at the neural plate border and TFAP2A/B at the neural crest. (F) Quantification of PLA puncta in embryos at three developmental stages (HH6–HH8) reveals a graded decrease in the number of interactions per cell between TFAP2A and TFAP2C over time, as well as an increase in TFAP2A/TFAP2B interactions over time. The number of interactions was normalized to those detected in the neural plate/ventral neural tube to account for differences in background fluorescence. Error bars, SE. (HH) Hamburger Hamilton; (NPB) neural plate border; (NC) neural crest. (*) P ≤ 0.05 (for number of embryos analyzed, see also Supplemental Table S1).
To provide a direct demonstration that TFAP2 paralogs form heterodimers, we performed coimmunoprecipitation (co-IP) with in vivo biotinylation. We cotransfected avian embryos with an Avi-tagged TFAP2A expression construct and a vector driving expression of the BirA enzyme (Beckett et al. 1999). Pulldown of TFAP2A with streptavidin beads resulted in co-IP of both FLAG-tagged TFAP2B and TFAP2C, confirming that these factors form heterodimers in avian embryos (Fig. 5D). However, TFAP2A does not coimmunoprecipitate FLAG-tagged LEF1, a WNT signaling effector active in the presumptive neural crest (Bhattacharya et al. 2018). To survey the spatiotemporal interactions among the endogenous TFAP2 proteins, we performed proximity ligation assays (PLAs) for TFAP2A/C and TFAP2A/B in both the neural plate border and the neural crest of developing chick embryos. This technique uses two primary antibodies raised in different species in combination with species-specific PLA secondary antibodies, each with an attached unique short DNA strand. When the PLA probes are in close proximity (<14 nm), the DNA strands are able to ligate and amplify, resulting in the production of red puncta visible in tissue sections (Supplemental Fig. S8F; Söderberg et al. 2006; Simões-Costa et al. 2015). An enrichment of PLA puncta was observed for TFAP2A/C in the neural plate border and TFAP2A/B in the neural crest (Fig. 5E; Supplemental Fig. S8F). Quantification of the number of interactions across three developmental stages (HH6–HH8) revealed a gradual decrease in TFAP2A/C dimers in the presumptive neural crest. Conversely, we observed a graded increase in TFAP2A/B interactions (Fig. 5F) as the cells become specified. As a negative control, we performed PLAs for TFAP2A and MSX1, which is expressed throughout neural crest development but was not predicted to be a TFAP2A cofactor via motif enrichment analysis (Fig. 4F). The antibody used for this analysis has also been previously shown to function in PLA (Simões-Costa et al. 2015). As a positive control, we performed PLAs for the coactivator protein EP300, which has previously been shown to interact with TFAP2A (Bragança et al. 2002). Although interactions between TFAP2A and EP300 were detected at all time points analyzed, little to no interactions were detected between TFAP2A and MSX1 (Supplemental Fig. S8D,E). These experiments confirm the presence of distinct TFAP2 heterodimers in the neural plate border and the neural crest. In conjunction with our genomic analysis, this indicates that TFAP2A switches partners to drive the transition from induction to specification. Accordingly, we propose that TFAP2 paralogs operate as a molecular switch that operates to reorganize the epigenomic landscape of neural crest cells, driving the sequential activation of gene regulatory network modules to progressively define the presumptive neural crest.
Manipulating the TFAP2 molecular switch to control timing of neural crest specification
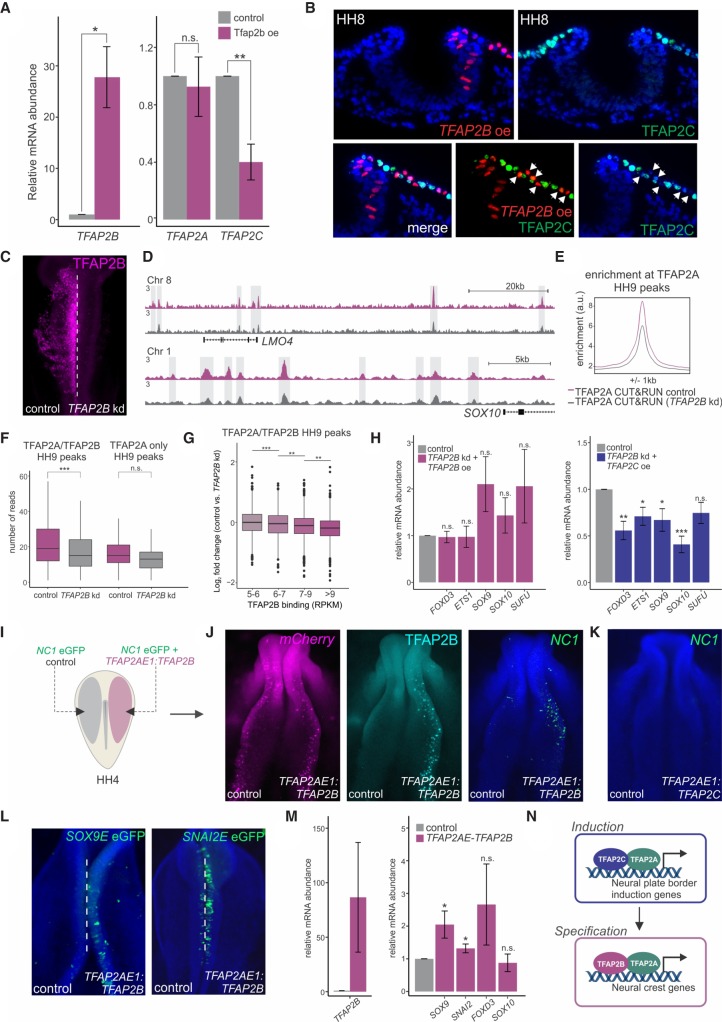
To understand the control of the TFAP2 molecular switch, we investigated how TFAP2B displaces TFAP2C to drive the assembly of TFAP2A/B heterodimers. Our CUT&RUN analysis suggests that TFAP2B is activated not only by genes within the induction program (Simoes-Costa and Bronner 2016) but also by TFAP2A/C heterodimers. This is shown by the presence of multiple TFAP2A/C peaks within the TFAP2B locus (Supplemental Table S2). Because TFAP2C mRNA levels rapidly decrease upon the onset of TFAP2B expression (Supplemental Fig. 3B), we postulated the existence of a repressive mechanism. To test this, we used unilateral electroporations to transfect embryos with a TFAP2B expression vector (Fig. 6A). TFAP2B overexpression resulted in the strong down-regulation of TFAP2C mRNA levels in the neural folds, whereas TFAP2A expression remained unchanged (Fig. 6A). To confirm that this effect was cell autonomous, we drove ectopic expression of TFAP2B in the nonneural ectoderm of HH8 embryos, which retain high levels of TFAP2C at HH8 (Fig. 6B). Histological analysis revealed specific loss of TFAP2C in the cells that received the TFAP2B expression vector. Thus, TFAP2B-mediated repression of TFAP2C likely drives the assembly of TFAP2A/B heterodimers. Because we observe association of TFAP2B with the TFAP2C locus during specification in our CUT&RUN data sets (Supplemental Table S2), it is likely that this inhibitory interaction is direct.
Figure 6.
Manipulation of the TFAP2 molecular switch causes premature specification. (A) Quantitative RT-PCR for TFAP2B, TFAP2A, and TFAP2C in control versus TFAP2B overexpression sides of bilaterally electroporated embryos. (B) Transverse sections displaying immunohistochemistry for TFAP2C (green) under TFAP2B overexpression (red). White arrowheads indicate a cell-autonomous decrease in TFAP2C protein levels. (C) Immunostaining for TFAP2B upon TFAP2B DsiRNA treatment. CUT&RUN for TFAP2A was performed in TFAP2B DsiRNA-treated embryos at HH9 to assess TFAP2A occupancy upon strong down-regulation of the TFAP2B protein. (D) CUT&RUN profiles for TFAP2A at the LMO4 and SOX10 loci under control versus TFAP2B knockdown conditions. (E) Profiles displaying enrichment of TFAP2A at TFAP2A peaks during specification in control versus DsiRNA-treated sides of bilaterally electroporated embryos. (F) Boxplots displaying read counts of control versus TFAP2B DsiRNA-treated TFAP2A CUT&RUN data sets at genomic regions corresponding to TFAP2B-dependent and -independent specification peaks (16,945 and 494 peaks, respectively). Statistical significance was determined via an unpaired two-tailed t-test. (G) Boxplots displaying log2 fold change in TFAP2A binding in control versus TFAP2B knockdown conditions across four levels of TFAP2B enrichment (5–6: 1185 peaks; 6–7: 1059 peaks; 7–9: 1752 peaks; more than 9: 1720 peaks). Statistical significance was determined via one-way ANOVA with Tukey's test. (H) Quantitative RT-PCR for the specification genes FOXD3, ETS1, SOX9, and SOX10 in embryos electroporated with a TFAP2B DsiRNA and rescued with a TFAP2AE1:TFAP2B or TFAP2AE1:TFAP2C expression construct. Phenotypes were surveyed at HH9. The neural marker SUFU was included as a control to ensure defects were neural crest specific. (I) Electroporation scheme for reprogramming of neural crest enhancer, NC1. HH4 embryos were bilaterally electroporated with the FOXD3 enhancer reporter NC1-eGFP, in addition to a construct driving neural crest–specific overexpression of TFAP2B (TFAP2AE1:TFAP2B). (J) mCherry reporter, Immunostaining for TFAP2B and the NC1 enhancer reporter in embryos bilaterally electroporated with TFAP2AE1:TFAP2B. Ectopic expression of TFAP2B in early presumptive neural crest cells jumpstarts the specification program, as displayed by an increase in NC1 enhancer activity. (K) NC1 enhancer reporter in embryos electroporated with TFAP2AE1:TFAP2C. Ectopic expression of TFAP2C is unable to activate NC1 enhancer activity. (L) Additional neural crest enhancers, Sox9E and Snai2E, display increased reporter activity upon premature TFAP2B expression. (M) Quantitative RT-PCR for TFAP2B, as well as the specification genes SOX9, SNAI2, FOXD3, and SOX10 in FACS-sorted mCherry+ cells from control versus TFAP2AE1:TFAP2B-electroporated sides of bilaterally electroporated embryos. Premature expression of TFAP2B results in a significant increase in expression of SOX9 and SNAI2. (N) Model for regulation of neural plate border induction and neural crest specification by TFAP2 heterodimer pairs. (A,G,L) Error bars, SE. (HH) Hamburger Hamilton; (kd) knockdown; (oe) overexpression; (kb) kilobase. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001 (for number of embryos analyzed, see also Supplemental Table S1).
Next, we surmised that upon strong down-regulation of the TFAP2B protein, heterodimerization will be hindered and TFAP2A will not be efficiently targeted to specification loci. To test this idea, we performed CUT&RUN for TFAP2A in the presence of a TFAP2B DsiRNA. To ensure the data sets were comparable, we performed this experiment on bilaterally electroporated embryos and compared TFAP2A binding in the control versus the TFAP2B knockdown side of the embryo (Fig. 6C). This analysis revealed a decrease in TFAP2A occupancy at specification peaks (Fig. 6E). Notably, this decrease in occupancy was also specific to those TFAP2A peaks occupied by heterodimers (Fig. 6F), as TFAP2B-independent binding was not affected. Furthermore, peaks that displayed greater enrichment for TFAP2B binding showed a stronger reduction in the knockdown condition (Fig. 6G). A closer look at the loci of the neural crest specification genes LMO4 and SOX10 showed decreased binding by TFAP2A within numerous cis-regulatory elements (Fig. 6D). Together, these data support the idea that TFAP2B is required for the recruitment of TFAP2A to specification loci.
Given this specific requirement of TFAP2B for regulation of specification peaks, we next conducted rescue experiments to test if TFAP2C was able to rescue TFAP2B loss of function. We performed stage-specific knockdown of TFAP2B during neural crest specification (as shown in Fig. 3E) in combination with neural crest–specific overexpression of either TFAP2B or TFAP2C. To achieve neural crest–specific overexpression, we used the TFAP2A enhancer TFAP2AE1, which is active in the prospective neural crest HH6 onward (Supplemental Fig. S9), to drive early TFAP2B or TFAP2C expression in the neural crest lineage. As expected, TFAP2B overexpression was able to rescue the effects of TFAP2B knockdown, as shown by quantitative RT-PCR for the specification genes FOXD3, ETS1, SOX9, and SOX10 (Fig. 6H). In contrast, overexpression of TFAP2C does not adequately rescue the expression of specification genes (Fig. 6H). These results indicate that TFAP2B is specialized in its ability to promote neural crest specification. This is likely through the recruitment of TFAP2A to specification-specific genomic loci.
Finally, we tested the molecular switch by manipulating the timing of TFAP2B expression. Because TFAP2B is able to repress TFAP2C expression, premature introduction of the former should be sufficient to drive the formation of TFAP2A/B dimers and aberrantly activate the specification program. We unilaterally transfected the TFAP2AE1:TFAP2B vector into avian embryos with the NC1 enhancer (Fig. 6I), a reporter of neural crest specification isolated from the FOXD3 gene (Simões-Costa et al. 2012). As predicted by our model, this manipulation resulted in a premature onset of specification, with early activity of NC1 in the reprogrammed side of the embryo (Fig. 6J). Embryos transfected with TFAP2AE1:TFAP2C displayed no such reprogramming (Fig. 6K). Furthermore, we found that premature TFAP2B expression is also capable of activating other neural crest specification enhancers, including Sox9 enh-345 and Snai2 enh-13 (Fig. 6L; Williams et al. 2019). To determine whether this precocious enhancer activation is accompanied by increased expression of neural crest specification genes, we FACS-sorted neural crest cells from bilaterally electroporated embryos transfected with TFAP2AE1:TFAP2B or a TFAP2AE1:mCherry control and performed quantitative RT-PCR for the specification genes SOX9, SNAI2, FOXD3, and SOX10. We observed that genes within the specification program, SOX9 and SNAI2, showed a significant increase in gene expression upon premature TFAP2B expression (Fig. 6M). This shows that we are able to manipulate the TFAP2 molecular switch to alter the timing of neural crest specification.
Discussion
Context-specific functions of transcriptional regulators are modulated by a number of mechanisms, including interactions with various cofactors, post-translational modifications, and, in the case of some proteins, distinctive dimerization capabilities (Hai and Curran 1991; Lee et al. 2012; Lukoseviciute et al. 2018). Here we describe a molecular switch based on the heterodimerization of pioneer transcription factors that drives the progression between sequential steps of cell fate commitment. Our functional and genomic analyses of TFAP2A, TFAP2C, and TFAP2B show that these paralogs work together to promote overarching changes in the epigenomic landscape of progenitor cells (Fig. 6N).
TFAP2A is considered an essential regulator of neural crest development (Rada-Iglesias et al. 2012), acting to establish active chromatin states. By demonstrating that TFAP2A activates different genomic regions by exchanging its dimerization partners, we describe how a complex gene regulatory program is restructured during cell state transitions. We propose that this TFAP2-mediated molecular switch operates based on the intracellular availability of paralogous proteins. First, TFAP2A cooperates with TFAP2C to activate the neural plate border induction program. TFAP2A/C heterodimers, along with induction genes (Simoes-Costa and Bronner 2016), subsequently activate the transcription of TFAP2B specifically in neural crest cells. Once expressed, TFAP2B represses transcription of TFAP2C, resulting the dissolution of TFAP2A/C heterodimers and driving TFAP2A to exchange partners (Supplemental Fig. S10). We tested this regulatory subcircuit by manipulating the timing of TFAP2B expression. Consistent with our model, we found that introducing TFAP2B in the prospective neural crest cells results in premature specification (Fig. 6J,M).
This model for TFAP2 regulation differs from what has been proposed by previous studies examining the function of these paralogs in the zebrafish and mouse neural crest (Hoffman et al. 2007; Li and Cornell 2007; Seberg et al. 2017). Additive genetic ablation of TFAP2 paralogs results in stronger effects in neural crest derivatives than targeting an individual gene, suggesting these factors have redundant function (Van Otterloo et al. 2018). Our results, however, show that TFAP2 heterodimers have distinct genomic targets, indicating the existence of paralog-specific functions during neural crest development. Although we show TFAP2B is uniquely suited to work with TFAP2A during specification, further experiments will be required to define the full spectrum of paralog-specific functions. Our results do not exclude the possibility that some level of functional redundancy exists, which would explain the additive effects observed in genetic ablation studies.
Despite their functional divergence, much of the preference for specific DNA sequences has been maintained among TFAP2 paralogs. Motif enrichment analysis identified only subtle differences between the motifs occupied by each pair of paralogs (Fig. 4F). However, co-occurring transcription factor motif analysis revealed that TFAP2 heterodimers may be cooperating with distinct pioneer factors during induction (e.g., GATA1/2/3), and specification (FOXD3) (Kadauke et al. 2012; Iwafuchi-Doi and Zaret 2014; Lukoseviciute et al. 2018). Indeed, our results suggest that differences in target specificity maybe be determined by recruitment of regulatory partners that cooperate during the binding and activation of cis-regulatory elements (Supplemental Fig. S7). This would mean that TFAP2C and TFAP2B interact with different cofactors, forming larger protein complexes that are recruited to either induction or specification enhancer regions. The data sets assembled in our study provide a useful platform for future studies on the structure and function of these regulatory regions.
Finally, our model for the genomic control of neural crest specification also has implications for vertebrate. The TFAP2 family of transcription factors has been hypothesized to play an important role in the evolutionary origins of neural crest cells (Meulemans and Bronner-Fraser 2002; Yu et al. 2008). This family consists of five paralogs, which are thought to have arisen via progressive genome duplications (Eckert et al. 2005). TFAP2A, which is considered the ancestral paralog, is expressed in both vertebrate and nonvertebrate chordates in the nonneural ectoderm and neural plate border, suggesting a conserved role of this factor in early stages of development (Meulemans and Bronner-Fraser 2002; Yu et al. 2008). However, all vertebrate species (agnathans and gnathostomes) display TFAP2A expression in the neural crest. The co-option of TFAP2A by neural crest cells was concomitant with progressive gene duplications, which lead to the appearance of other TFAP2 genes. Although only one TFAP2 gene has been identified in the cephalochordate, amphioxus (Yu et al. 2008), and the urochordate, Ciona intestinalis (Imai et al. 2017), at least three TFAP2 paralogs have been discovered in lampreys, a lower vertebrate species (Van Otterloo et al. 2012; Smith et al. 2018). Therefore, it is likely that the appearance of the TFAP2-mediated molecular switch coincided with the emergence of the neural crest in ancient vertebrates. Thus, the TFAP2 family provides an excellent example of how subfunctionalization among paralogs may be used as an evolutionary strategy to diversify the functional repertoire of regulatory factors that control embryonic development.
Methods
Embryo transfection and perturbation experiments
Chick embryos at HH4-5 were transfected with morpholinos, DsiRNAs and DNA constructs by ex ovo electroporation, as previously described (Simões-Costa et al. 2015). Detailed protocols may be found in the Supplemental Methods.
Immunohistochemistry
For whole-mount immunohistochemistry, embryos were dissected from the filter paper after fixation and washed in TBS containing 0.1% Triton X-100 and 1% DMSO (TBTD). Embryos were blocked for 2 h in TBTD supplemented with 10% donkey serum and incubated in primary antibody diluted in blocking solution, overnight at 4°C.
Expression vectors
The TFAP2A, TFAP2B, and TFAP2C expression constructs were assembled by insertion of the full-length cDNA sequence of avian TFAP2A/B/C in a pCI-H2B-RFP backbone. All coding sequences were PCR amplified from an HH8 cDNA library. Enhancer reporter constructs were assembled as described (Simões-Costa et al. 2012), in which enhancer regions were cloned into ptk eGFP (Uchikawa et al. 2003), such that enhancer activity drives GFP expression (for primers used to amplify enhancer sequences, see Supplemental Table S4). All enhancer sequences were PCR amplified from a chicken gDNA library. The Tfap2aE1:Tfap2b and Tfap2aE1:Tfap2c constructs were cloned from the PCI-H2B-RFP-Tfap2b/c overexpression vectors by exchanging the CAG promoter with a Tfap2aE1 insert.
Quantitative RT-PCR
To quantify TFAP2A, TFAP2B, and TFAP2C expression in embryos over time, whole embryos were dissected and lysed directly in 100 µL of lysis buffer from the RNAqueous-micro total RNA isolation kit (Thermo Fisher Scientific AM1931). RNA was purified according to the kit's protocol. cDNA was synthesized from total RNA using the SuperScript III first-strand synthesis system (Thermo Fisher Scientific 18080051). Gene perturbation experiments were performed as previously described (Bhattacharya et al. 2018), and detailed methods are described in the Supplemental Methods.
ATAC-seq
For isolation of neural crest cells, embryos were transfected with 1 µg/µL of the enhancer reporter construct PTK-AP2aE1-eGPF (Uchikawa et al. 2003). To obtain neural crest cells from different stages, embryos were cultured until HH6 (8 h) and HH9 (13–14 h) and screened for robust GFP expression in neural crest cells. Embryo heads were dissected and incubated in Accumax (Accutase SCR006) cell dissociation solution for 40 min at room temperature (RT) under mild agitation. At least 20,000 GFP+ cells from each stage were sorted into 200 µL of HANKS buffer supplemented with 0.5% BSA. ATAC-seq library preparation was performed as previously described (Buenrostro et al. 2015). Detailed methods and data analysis are described in the Supplemental Methods.
CUT&RUN
Neural plate border/neural crest dissections were performed on HH6/HH9 embryos (10 embryos/20 neural folds per experiment). Cells were dissociated in Accumax (Accutase SCR006) cell dissociation solution for 20 min at RT under mild agitation. CUT&RUN experiments were performed as previously described (Skene and Henikoff 2017). Protein A–MNase was kindly provided by Dr. Steve Henikoff. CUT&RUN for TFAP2A upon TFAP2B knockdown was performed in the same manner, only using embryos electroporated unilaterally with a DsiRNA for TFAP2B (IDT). Neural crest dissections were performed on HH9 electroporated embryos, and CUT&RUN was conducted on pooled tissues from the control versus the knockdown sides of the embryos.
CUT&RUN library preparation
CUT&RUN libraries were prepared using the NEBNext ultra II DNA library prep kit (New England Biolabs E7645). Quality control of prepared libraries was conducted using an ABI 3730xl DNA analyzer for fragment analysis. Libraries were pooled to equimolar concentrations using the KAPA library quant kit ROX low qPCR mix (Roche 07960336001) and sequenced with paired-end 37-bp reads on an Illumina NextSeq 500 instrument. The data analysis pipeline is described in the Supplemental Methods.
RNA-seq
For isolation of neural crest cells, embryos were transfected with 1 µg/µL of an enhancer of the TFAP2A gene (Tfap2aE1) (Attanasio et al. 2013) cloned into PTK-eGFP (Uchikawa et al. 2003). To obtain neural crest cells from different stages, embryos were cultured until HH6 (8 h) and HH10 (13–14 h) and screened for robust GFP expression in neural crest cells. Embryo heads were dissected and incubated in Accumax (Accutase SCR006) cell dissociation solution for 40 min at RT under mild agitation. At least 5000 GFP+ and GFP− cells from each stage were sorted directly into 100 µL of lysis buffer from the RNAqueous-micro total RNA isolation kit (Thermo Fisher Scientific AM1931). Total RNA was isolated according to the kit's protocol. RNA was poly(A) selected using the NEBNext Poly(A) mRNA magnetic isolation module (New England Biolabs E7490). TruSeq-barcoded RNA-seq libraries were generated with the NEBNext ultra II directional RNA library prep kit (New England Biolabs E7760) and sequenced on an Illumina NextSeq 500 instrument with single-end 75-bp reads. The data analysis pipeline is described in the Supplemental Methods.
Proximity ligation assays
To identify interactions between TFAP2A and TFAP2B/TFAP2C/EP300/MSX1, we used the Duolink fluorescence approach (Sigma-Aldrich DUO92101). Embryos of different developmental stages were fixed with phosphate buffer (PB) containing 4% PFA for 20 min and cryosectioned in OCT compound (VWR). Primary antibody pairs used are listed in the Supplemental Methods. The assay was performed according to the manufacturer's instructions. PLA-positive puncta were quantified with a fluorescent microscope. To quantify number of puncta per cell, we specified a region of interest (ROI) at the neural plate border/dorsal neural tube in addition to the neural plate/ventral neural tube to be used as a control. Number of cells (indicated via DAPI staining) and number of puncta were counted within each ROI, and the number of puncta were divided by the number of cells. The number of puncta/cell in the neural plate/ventral neural tube was subtracted from that of the neural plate border/dorsal neural tube to account for variation in background fluorescence between tissue sections/experiments.
Coimmunoprecipitation
For co-IP experiments, the coding sequences of TFAP2A, TFAP2B, TFAP2C, and LEF1 were fused to AVI and FLAG tags and cloned into pCI-H2B-RFP. PCI-AVI-Tfap2a was coelectroporated into stage HH4 embryos with PCI-FLAG-Tfap2b, PCI-FLAG-Tfap2c, PCI-FLAG-Lef1, as well as with or without a construct containing the BirA enzyme coding sequence. Embryos were incubated until stage HH8 and collected for IP. Electroporation efficiency was confirmed by RFP expression before dissection. Nuclear extraction, co-IP, and western blot was conducted as previously described (Simões-Costa et al. 2015), using MyOne streptavidin T1 Dynabeads (Thermo Fisher Scientific).
Data access
All genomic data sets generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE126880.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH), National Institute of Dental and Craniofacial Research, grants R00DE024232 and 1R01DE028576 and the Meinig Family Foundation to M.S.-C., and NIH, Eunice Kennedy Shriver National Institute of Child Health and Human Development, grant 1F31HD097927-01 and the Cornell Center for Vertebrate Genomics Scholars Program to M.R.
Author contributions: M.S.-C. and M.R. conceived and designed the experimental approach. M.R. performed the experiments and analyzed the data. M.S.-C. and M.R. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.249680.119.
References
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, et al. 2013. Fine tuning of craniofacial morphology by distant-acting enhancers. Science 342: 1241006 10.1126/science.1241006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. 2009. Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723. 10.1126/science.1162327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI. 2006. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature 441: 218–222. 10.1038/nature04684 [DOI] [PubMed] [Google Scholar]
- Beckett D, Kovaleva E, Schatz PJ. 1999. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci 8: 921–929. 10.1110/ps.8.4.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. 2010. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc Natl Acad Sci 107: 3570–3575. 10.1073/pnas.0906596107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Rothstein M, Azambuja AP, Simoes-Costa M. 2018. Control of neural crest multipotency by Wnt signaling and the Lin28/let-7 axis. eLife 7: e40556 10.7554/eLife.40556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragança J, Swingler T, Marques FI, Jones T, Eloranta JJ, Hurst HC, Shioda T, Bhattacharya S. 2002. Human CREB-binding protein/p300-interacting transactivator with ED-rich tail (CITED) 4, a new member of the CITED family, functions as a co-activator for transcription factor AP-2. J Biol Chem 277: 8559–8565. 10.1074/jbc.M110850200 [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109: 21.29.1–21.29.9. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. 2009. The regulatory genome: gene regulatory networks in development and evolution Elsevier, Burlington, MA. [Google Scholar]
- de Crozé N, Maczkowiak F, Monsoro-Burq AH. 2011. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proc Natl Acad Sci 108: 155–160. 10.1073/pnas.1010740107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dottori M, Gross MK, Labosky P, Goulding M. 2001. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development 128: 4127–4138. [DOI] [PubMed] [Google Scholar]
- Eckert D, Buhl S, Weber S, Jäger R, Schorle H. 2005. The AP-2 family of transcription factors. Genome Biol 6: 246 10.1186/gb-2005-6-13-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Garcia M, Moore CD, Schulz KN, Alberto O, Donague G, Harrison MM, Zhu H, Zaret KS. 2019. Structural features of transcription factors associating with nucleosome binding. Mol Cell 75: 921–932.e6. 10.1016/j.molcel.2019.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Engreitz J, Ray JP, Nguyen TH, Hacohen N, Lander ES. 2018. Positional specificity of different transcription factor classes within enhancers. Proc Natl Acad Sci 115: E7222–E7230. 10.1073/pnas.1804663115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, LaBonne C. 2014. Setting appropriate boundaries: fate, patterning and competence at the neural plate border. Dev Biol 389: 2–12. 10.1016/j.ydbio.2013.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai T, Curran T. 1991. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci 88: 3720–3724. 10.1073/pnas.88.9.3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman TL, Javier AL, Campeau SA, Knight RD, Schilling TF. 2007. Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. J Exp Zool B Mol Dev Evol 308B: 679–691. 10.1002/jez.b.21189 [DOI] [PubMed] [Google Scholar]
- Honoré SM, Aybar MJ, Mayor R. 2003. Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Dev Biol 260: 79–96. 10.1016/S0012-1606(03)00247-1 [DOI] [PubMed] [Google Scholar]
- Imai KS, Hikawa H, Kobayashi K, Satou Y. 2017. Tfap2 and Sox1/2/3 cooperatively specify ectodermal fates in ascidian embryos. Development 144: 33–37. 10.1242/dev.142109 [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS. 2014. Pioneer transcription factors in cell reprogramming. Genes Dev 28: 2679–2692. 10.1101/gad.253443.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. 2013. DNA-binding specificities of human transcription factors. Cell 152: 327–339. 10.1016/j.cell.2012.12.009 [DOI] [PubMed] [Google Scholar]
- Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. 2012. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150: 725–737. 10.1016/j.cell.2012.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C. 1999. The neural crest, 2nd ed. Cambridge University Press, Cambridge. [Google Scholar]
- Lee PC, Taylor-Jaffe KM, Nordin KM, Prasad MS, Lander RM, LaBonne C. 2012. SUMOylated SoxE factors recruit Grg4 and function as transcriptional repressors in the neural crest. J Cell Biol 198: 799–813. 10.1083/jcb.201204161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cornell RA. 2007. Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Dev Biol 304: 338–354. 10.1016/j.ydbio.2006.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukoseviciute M, Gavriouchkina D, Williams RM, Hochgreb-Hagele T, Senanayake U, Chong-Morrison V, Thongjuea S, Repapi E, Mead A, Sauka-Spengler T. 2018. From pioneer to repressor: Bimodal foxd3 activity dynamically remodels neural crest regulatory landscape in vivo. Dev Cell 47: 608–628.e6. 10.1016/j.devcel.2018.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. 2002. Amphioxus and lamprey AP-2 genes: implications for neural crest evolution and migration patterns. Development 129: 4953–4962. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. 2004. Gene-regulatory interactions in neural crest evolution and development. Dev Cell 7: 291–299. 10.1016/j.devcel.2004.08.007 [DOI] [PubMed] [Google Scholar]
- Milunsky JM, Maher TA, Zhao G, Roberts AE, Stalker HJ, Zori RT, Burch MN, Clemens M, Mulliken JB, Smith R, et al. 2008. TFAP2A mutations result in branchio-oculo-facial syndrome. Am J Hum Genet 82: 1171–1177. 10.1016/j.ajhg.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohibullah N, Donner A, Ippolito JA, Williams T. 1999. SELEX and missing phosphate contact analyses reveal flexibility within the AP-2α protein: DNA binding complex. Nucleic Acids Res 27: 2760–2769. 10.1093/nar/27.13.2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori-Akiyama Y, Akiyama H, Rowitch DH, de Crombrugghe B. 2003. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc Natl Acad Sci 100: 9360–9365. 10.1073/pnas.1631288100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitina N, Sauka-Spengler T, Bronner-Fraser M. 2008. Dissecting early regulatory relationships in the lamprey neural crest gene network. Proc Natl Acad Sci 105: 20083–20088. 10.1073/pnas.0806009105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Springer-Verlag, Berlin, NY. [Google Scholar]
- Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J. 2012. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell 11: 633–648. 10.1016/j.stem.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible DW. 2006. Development of the neural crest: achieving specificity in regulatory pathways. Curr Opin Cell Biol 18: 698–703. 10.1016/j.ceb.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Sanchez-Ferras O, Coutaud B, Djavanbakht Samani T, Tremblay I, Souchkova O, Pilon N. 2012. Caudal-related homeobox (Cdx) protein-dependent integration of canonical Wnt signaling on paired-box 3 (Pax3) neural crest enhancer. J Biol Chem 287: 16623–16635. 10.1074/jbc.M112.356394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoda M, Zhao F, Diaz GA, Burn J, Goodship J, Davidson HR, Pierpont ME, Gelb BD. 2000. Mutations in TFAP2B cause Char syndrome, a familial form of patent ductus arteriosus. Nat Genet 25: 42–46. 10.1038/75578 [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. 2008. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol 9: 557–568. 10.1038/nrm2428 [DOI] [PubMed] [Google Scholar]
- Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. 1996. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 381: 235–238. 10.1038/381235a0 [DOI] [PubMed] [Google Scholar]
- Seberg HE, Van Otterloo E, Loftus SK, Liu H, Bonde G, Sompallae R, Gildea DE, Santana JF, Manak JR, Pavan WJ, et al. 2017. TFAP2 paralogs regulate melanocyte differentiation in parallel with MITF. PLoS Genet 13: e1006636 10.1371/journal.pgen.1006636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Hashimoto T, O'Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, Gifford DK. 2014. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol 32: 171–178. 10.1038/nbt.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões-Costa M, Bronner ME. 2015. Establishing neural crest identity: a gene regulatory recipe. Development 142: 242–257. 10.1242/dev.105445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M, Bronner ME. 2016. Reprogramming of avian neural crest axial identity and cell fate. Science 352: 1570–1573. 10.1126/science.aaf2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões-Costa MS, McKeown SJ, Tan-Cabugao J, Sauka-Spengler T, Bronner ME. 2012. Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is encrypted in the genome. PLoS Genet 8: e1003142 10.1371/journal.pgen.1003142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões-Costa M, Stone M, Bronner ME. 2015. Axud1 integrates Wnt signaling and transcriptional inputs to drive neural crest formation. Dev Cell 34: 544–554. 10.1016/j.devcel.2015.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6: e21856 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, et al. 2011. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147: 1270–1282. 10.1016/j.cell.2011.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Timoshevskaya N, Ye C, Holt C, Keinath MC, Parker HJ, Cook ME, Hess JE, Narum SR, Lamanna F, et al. 2018. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat Genet 50: 270–277. 10.1038/s41588-017-0036-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3: 995–1000. 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Taylor KM, LaBonne C. 2007. Modulating the activity of neural crest regulatory factors. Curr Opin Genet Dev 17: 326–331. 10.1016/j.gde.2007.05.012 [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. 2003. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell 4: 509–519. 10.1016/S1534-5807(03)00088-1 [DOI] [PubMed] [Google Scholar]
- Van Otterloo E, Li W, Garnett A, Cattell M, Medeiros DM, Cornell RA. 2012. Novel Tfap2-mediated control of soxE expression facilitated the evolutionary emergence of the neural crest. Development 139: 720–730. 10.1242/dev.071308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Otterloo E, Li H, Jones KL, Williams T. 2018. AP-2α and AP-2β cooperatively orchestrate homeobox gene expression during branchial arch patterning. Development 145: dev157438 10.1242/dev.157438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T, Tjian R. 1991. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev 5: 670–682. 10.1101/gad.5.4.670 [DOI] [PubMed] [Google Scholar]
- Williams RM, Candido-Ferreira I, Repapi E, Gavriouchkina D, Senanayake U, Ling ITC, Telenius J, Taylor S, Hughes J, Sauka-Spengler T. 2019. Reconstruction of the global neural crest gene regulatory network in vivo. Dev Cell 51: 255–276.e7. 10.1016/j.devcel.2019.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JK, Meulemans D, McKeown SJ, Bronner-Fraser M. 2008. Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Res 18: 1127–1132. 10.1101/gr.076208.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241. 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Luo T, Sargent TD. 2006. Expression of TFAP2β and TFAP2γ genes in Xenopus laevis. Gene Expr Patterns 6: 589–595. 10.1016/j.modgep.2005.11.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.