Abstract
Genome-wide association studies indicate that many disease susceptibility regions reside in non-protein-coding regions of the genome. Long noncoding RNAs (lncRNAs) are a major component of the noncoding genome, but their biological impacts are not fully understood. Here, we performed a CRISPR interference (CRISPRi) screen on 2263 epidermis-expressed lncRNAs and identified nine novel candidate lncRNAs regulating keratinocyte proliferation. We further characterized a top hit from the screen, progenitor renewal associated non-coding RNA (PRANCR), using RNA interference–mediated knockdown and phenotypic analysis in organotypic human tissue. PRANCR regulates keratinocyte proliferation, cell cycle progression, and clonogenicity. PRANCR-deficient epidermis displayed impaired stratification with reduced expression of differentiation genes that are altered in human skin diseases, including keratins 1 and 10, filaggrin, and loricrin. Transcriptome analysis showed that PRANCR controls the expression of 1136 genes, with strong enrichment for late cell cycle genes containing a CHR promoter element. In addition, PRANCR depletion led to increased levels of both total and nuclear CDKN1A (also known as p21), which is known to govern both keratinocyte proliferation and differentiation. Collectively, these data show that PRANCR is a novel lncRNA regulating epidermal homeostasis and identify other lncRNA candidates that may have roles in this process as well.
Whole-exome sequencing has accelerated the discovery into genetic causes of disease. However, the majority of whole-exome studies do not identify a causative mutation (Yang et al. 2013), potentially reflecting the fact that protein-coding regions comprise only ∼1%–2% of the human genome and indicating the potential contribution of non-protein-coding mutations. Consistent with this, genome-wide association studies (GWAS) indicate that >93% of disease-linked single-nucleotide polymorphisms (SNPs) map to the noncoding genome (Tak and Farnham 2015). These observations therefore suggest a potential underappreciated disease relevance of noncoding elements such as enhancers and noncoding RNAs (Zhang and Lupski 2015).
A major component of the noncoding genome are long noncoding RNAs (lncRNAs), transcribed RNA elements >200 nucleotides (nt) with no apparent protein-coding potential (Wilusz et al. 2009; Da Sacco et al. 2012). Compared with their protein-coding counterparts, lncRNA transcripts have a lower expression level and show strong tissue-specific expression patterns, implying potential functions particular to specific biological states (Derrien et al. 2012; Liu et al. 2017). To date, nearly 28,000 human lncRNAs have been cataloged (Hon et al. 2017), but only about 200 have been functionally characterized (Quek et al. 2015). This is in part due to the challenges of studying lncRNAs. They are less sequence-conserved than proteins (Kellis et al. 2014), and some functional lncRNAs are primate- or even human-specific (Awan et al. 2017). This limits the application of classical genetic systems, such as murine models, to studying human lncRNAs. In addition, the majority of lncRNAs are functional in only one cell type (Liu et al. 2017). Therefore, the understanding of lncRNAs requires examination in the appropriate cell/tissue context. Lastly, it has been a matter of ongoing debate whether noncoding elements are broadly functional at all or whether many of these elements have minimal biological significance (Doolittle 2013).
A recent evaluation of lncRNA expression across species and development indicates that lncRNAs are dynamically expressed and conserved in organs, suggesting that they have evolved specialized functions in human tissues (Sarropoulos et al. 2019). Here, we use human epidermis as a model system for evaluating the function of lncRNAs in tissue homeostasis. In the epidermis, there is a dynamic, ongoing balance between progenitor proliferation and differentiation. Genetic disruptions of proliferation or differentiation disrupt this homeostasis and underlie common diseases such as eczema, psoriasis, and keratinocyte cancers, which collectively impact >20% of the population (Lopez-Pajares et al. 2013). Understanding the potential role of lncRNAs in epidermal homeostasis therefore has broad relevance to human health. Using the human epidermis as a model also offers several unique advantages. First, the skin is accessible, allowing collection of primary tissue and cells for study. The ex vivo culture conditions for primary human progenitor keratinocytes are well developed and permit complex genetic manipulations. Finally, epidermal progenitors can be reconstituted into organotypic tissue, allowing studies in a three-dimensional context (Oh et al. 2013). These advantages are particularly useful in studying lncRNAs, whose phenotypes in cultured cells do not always extrapolate to primary tissue or in vivo contexts (Bassett et al. 2014). In this report, we perform a CRISPR interference (CRISPRi) screen to systematically identify functional lncRNAs that have roles in human epidermal homeostasis and characterize the tissue and molecular phenotype of a novel lncRNA hit from the screen.
Results
Transcriptome analysis and CRISPRi screen to identify functional lncRNAs
The epidermis consists primarily of organized layers of keratinocytes. Stem/progenitor keratinocytes reside in the innermost layer atop a basement membrane and are capable of self-renewal or differentiation. Differentiating keratinocytes detach from the basement membrane and migrate upward to form the suprabasal layers, which serve as a structural and functional barrier. At the outermost layers, keratinocytes are enucleated to form a cornified layer, which is eventually sloughed off the surface. To sustain homeostasis, the epidermis dynamically balances cell renewal, differentiation, and cell loss.
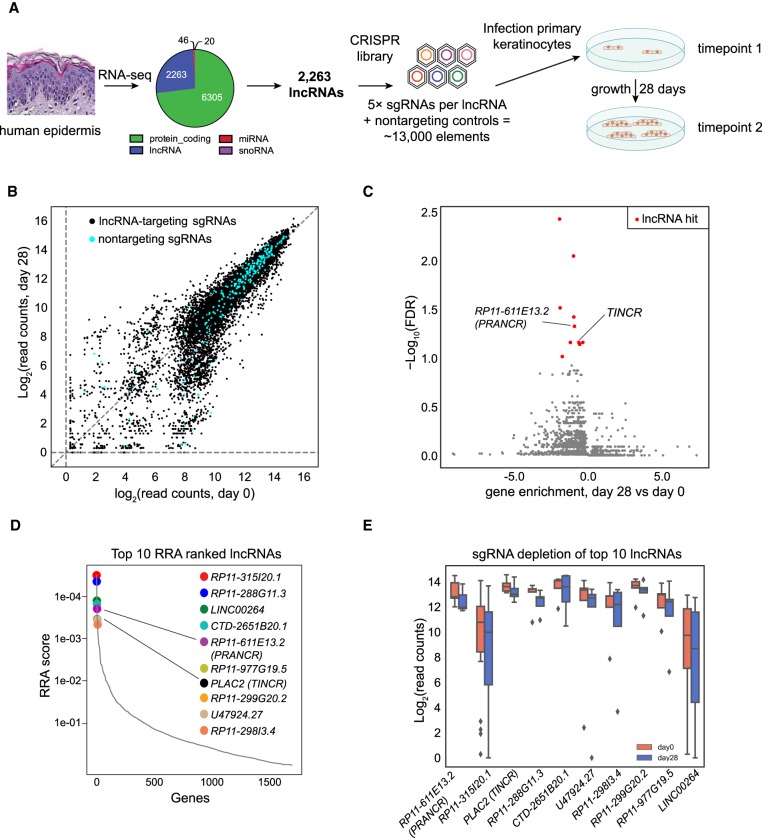
To identify lncRNAs regulating tissue homeostasis, we first focused on identifying lncRNAs governing epidermal progenitor renewal. As expression level is the most significant predictor of functional lncRNAs (Liu et al. 2017), we combined RNA expression profiling of human epidermis with a CRISPRi screen (Fig. 1A). By using RNA-sequencing data from duplicate biological samples of clinically normal human epidermis (Sun et al. 2015), we identified 8634 gene transcripts at an RPKM of >1. Classification of expressed transcripts into protein-coding lncRNAs, microRNAs, and small nucleolar RNAs, led to the assignment of 2263 elements as lncRNAs (Fig. 1A). The average expression level of all lncRNAs was lower than that of protein-coding genes (Supplemental Fig. S1), consistent with previous reports (Derrien et al. 2012; Liu et al. 2017; Tuck et al. 2018).
Figure 1.
Discovery of lncRNAs controlling proliferation of epidermal progenitors. (A) Schematic of CRISPRi library design strategy and the CRISPRi screen. RNA sequencing identified 2263 lncRNAs with RPKM > 1. Five sgRNAs were designed for each target lncRNA transcript, together with 250 nontargeting controls, to form the CRISPRi library. (B) Scatter plots of sgRNA abundance at day 0 and day 28 of the screen. Nontargeting sgRNAs are shown in blue. (C) FDR values of each lncRNA candidate, as calculated by MAGeCK. A discovery threshold for positive hits (red) was defined by lncRNAs with similar or stronger FDR values as a known positive control, TINCR (previously known as PLAC2) (FDR threshold = <0.1). Gene enrichment represents average log-scale enrichment of sgRNAs changing concordantly with the lncRNA selection. (D) The robust ranking aggregation (RRA) scores of top lncRNA screen hits. (E) Normalized read counts of sgRNAs of the top 10 ranked lncRNA hits comparing postscreen (day 28) versus prescreen (day 0) abundance. Center lines represent median values; box limits represent the interquartile range; whiskers each extend 1.5 times the interquartile range; and dots represent outliers.
To systematically interrogate the potential roles of these lncRNAs in epidermal progenitor growth, we designed a CRISPRi screen. In a classic version of CRISPRi, a catalytically dead (d)Cas9 protein is fused to a KRAB transcriptional repressor domain, which is guided to its genomic target by a single guide RNA (sgRNA) (Qi et al. 2013). This system has proven useful for loss-of-function targeting of lncRNAs, which are not reliably inactivated by the short indels generated by CRISPR-Cas9 nuclease approaches (Liu et al. 2017).
We constructed a custom sgRNA library composed of five independent sgRNAs against each of the 2263 lncRNAs using Sequence Scan for CRISPR (SSC) (Xu et al. 2015) and included 250 nontargeting sgRNA controls. dCas9-expressing keratinocytes were generated by lentiviral transduction of a dCas9-KRAB expression cassette into the clone 103 epidermal keratinocyte cell line (Sun et al. 2015) and selection of a high-expressing clone (see Methods). Into these keratinocytes, the CRISPRi sgRNA library was transduced at a multiplicity of infection of 0.3, selected cells were propagated in culture, and genomic DNA collected at the initial time point and after 28 d of continuous proliferation (Fig. 1A). Deep sequencing was used to quantify sgRNA representation at each time point. After normalization and mean-variance modeling, we determined enriched and depleted sgRNAs (Fig. 1B). The experiment was performed in technical and biological duplicates, which both showed high reproducibility (R2 of 0.91 and 0.86, respectively) (Supplemental Fig. S1).
CRISPRi screen identifies novel lncRNAs regulating progenitor replication
We assessed screening results using Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout (MAGeCK), a computational analysis tool developed to robustly identify essential gene elements from CRISPR screens (Li et al. 2014). The MAGeCK algorithm uses a mean-variance model to test if sgRNA abundances differ significantly before versus after the screen with respect to neutral variation of sgRNA abundance, as assessed by the nontargeting sgRNAs. A robust rank aggregation (RRA) algorithm (lncRNA gene ranking) computes P-values, false-discovery rates (FDRs) and RRA scores (Li et al. 2014) based on screen results.
We chose to initially benchmark our screen results against TINCR, one of the few well-characterized lncRNAs known to be involved in epidermal homeostasis (Kretz et al. 2013). To do so, we considered lncRNAs scored with an FDR value similar to or lower than TINCR (FDR = 0.07; threshold at FDR < 0.10) as positive hits. We also stipulated that at least three independent sgRNAs against a lncRNA candidate “hit” must change consistently within the screen. By using these relatively strict parameters, we identified nine novel candidate lncRNA hits, all of which were positive regulators of proliferation (red dots in Fig. 1C). We excluded two pseudogenes (MEMO1P1 and GUSBP4) because their high sequence homology to protein-coding genes complicates the use of RNA interference and downstream analysis. The remaining candidates were ranked, rendering our candidate lncRNA list (Fig. 1D,E). These lncRNA hits had higher expression values than the global lncRNA average (Supplemental Fig. S1), consistent with expression levels being an indicator of functional lncRNAs (Liu et al. 2017).
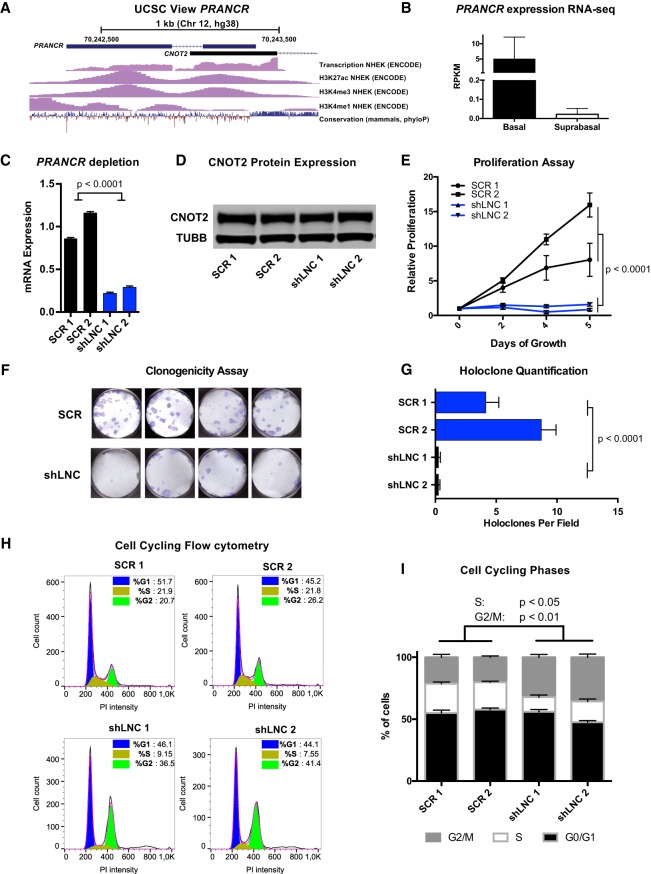
To further validate the screen, we aimed to further characterize the phenotype and function of a top candidate. We chose to focus on RP11-611E13.2 for several reasons. First, ENCODE data showed that its transcriptional start site (TSS) is enriched with histone 3 lysine 27 acetylation (H3K27ac) and histone 3 lysine 4 trimethylation (H3K4me3) in neonatal human epidermal keratinocytes (Fig. 2A), a pattern consistent with active genes (Kellis et al. 2014). In addition, our RNA-seq data on the human epidermis included subcategorization of basal versus suprabasal transcripts by laser capture microdissection, which allowed us to evaluate counts from basal (progenitor) and suprabasal (differentiated) epidermal layers (Sun et al. 2015). RP11-611E13.2 is more highly expressed in the basal layers of the epidermis (average RPKM of 5 in basal layers vs. 0.02 in suprabasal layers), consistent with its expression where epidermal progenitors are actively dividing (Fig. 2B). Based on these observations and the screening results, we termed this lncRNA progenitor renewal associated non-coding RNA (PRANCR) and performed a more detailed evaluation of its phenotype and function.
Figure 2.
PRANCR is a novel epidermal lncRNA and is essential for keratinocyte proliferation and cell cycle progression. (A) Schematic of PRANCR locus on Chromosome 12, with UCSC tracks for transcription, H3K27ac, H3K4me3, and H3K4me1 from the ENCODE Project, and conservation tracks from phyloP. (B) PRANCR expression in basal and suprabasal layers from microdissected human epidermis. Bars, RPKM values with SEM; n = 2. (C) PRANCR mRNA expression in control (scrambled; SCR) and PRANCR-depleted (shLNC) progenitors. Bars, mean with SEM; n = 4; expression compared with one-way ANOVA. (D) Western blot for CNOT2, a protein expressed divergently from the PRANCR genomic locus, in SCR and shLNC progenitor cells. (E) Proliferation assay of control versus PRANCR-depleted progenitors, measured with a fluorescence-based cell quantification assay. Plotted values represent relative increase at each time point relative to day 0. n = 4; dots represent mean value with SEM. Comparisons performed by two-way ANOVA. (F) Holoclone assay of control and PRANCR-depleted keratinocytes. Representative images are shown. (G) Quantification of holoclones from control and PRANCR-depleted keratinocytes; bars, mean with SEM; n = 12 each. Differences tested using one-way ANOVA. (H) Flow cytometric analysis of cell cycle using propidium iodide DNA staining, comparing normal and PRANCR-depleted human epidermal keratinocytes. Graphs represent FlowJo analysis of the flow cytometric results of ≥10,000 cells using the Dean–Jett–Fox model for each cell replicate. (I) Quantification of cell cycling phases based on data from experiments in three independent keratinocyte lines, represented in H and Supplemental Fig. S3. Bars, mean with SEM; n = 3 independent keratinocyte cell lines, with ≥10,000 cells per line. Comparisons performed by one-way ANOVA followed by Dunnett's multiple comparison test. (SCR1/2) Scrambled short hairpin 1 or 2; (shLNC1/2) short hairpin RNA 1 or 2 targeting PRANCR.
PRANCR is essential for proliferation and clonogenicity of primary keratinocytes
CRISPRi can block genome elements by establishing a repressive chromatin state at the genomic locus, by blocking transcription of a functional RNA, or both. To distinguish between these possibilities, we used short hairpin RNA (shRNA)–mediated knockdown to deplete PRANCR RNA without interfering with the genomic locus. Of the six independent, nonoverlapping shRNAs (shLNCs), five resulted in PRANCR RNA depletion ranging from 30%–78% (Supplemental Fig. S2A). We used the two shLNCs that achieved the greatest PRANCR knockdown for most downstream experiments (Fig. 2C).
PRANCR is transcribed divergently from the same promoter region as the protein-coding gene CNOT2 (Fig. 2A). Therefore, we performed western blotting to evaluate if CNOT2 protein levels were altered with PRANCR shRNA-mediated depletion. We observed no change in CNOT2 expression using multiple independent shRNAs against PRANCR (Fig. 2D; Supplemental Fig. S2), arguing against the phenotype arising from changing expression of the divergently transcribed gene. We also evaluated if the phenotype identified by the CRISPRi screen could be recapitulated with PRANCR RNAi-mediated depletion by assessing the proliferation of keratinocyte progenitors. Our results confirmed markedly inhibited keratinocyte proliferation with PRANCR knockdown (Fig. 2E), a result also observed for all five effective shRNAs (Supplemental Fig. S2B). Together, these results support a primary role for PRANCR lncRNA in causing the observed phenotype.
Next, we evaluated whether PRANCR influenced the stem cell potential of epidermal progenitors. In human skin, epidermal keratinocytes can be classified into different populations with varying clonogenic potential (Barrandon and Green 1987). Holoclones, the population with the greatest renewal and proliferative capacity, are critical for long-term epidermal renewal. To assess the impact of PRANCR on clonogenic potential, control versus PRANCR-depleted keratinocytes were seeded on fibroblast feeders. PRANCR depletion resulted in a significantly reduced number of holoclones, showing an intrinsic loss of clonogenic capacity resulting from loss of PRANCR (Fig. 2F,G).
The robust phenotype of reduced proliferative capacity prompted us to investigate how PRANCR depletion impacted cell cycle progression. To quantitate cell cycle state, we performed flow cytometric analysis of propidium iodide (PI)–stained cells. Upon depletion of PRANCR (Fig. 2H), we observed a decrease in the proportion of cells in the S phase (∼8%–9% vs. 21%–22% in control cells) and an increase in the proportion of cells in G2/M phase (∼37%–41% vs. 21%–26%), with minimal changes in the fraction of cells in the G1/G0 phase (∼44%–46% vs. 45%–52%). Independent biological replicates confirmed these findings (summarized in Fig. 2I; Supplemental Fig. S3). An increase of cells in G2/M phase can lead to induction of apoptosis. Therefore, we assessed whether PRANCR depletion promoted apoptosis. We quantitated apoptosis induction by flow cytometric analysis of FITC–Annexin V labeling of phosphatidylserine (PS) exposure (Supplemental Fig. S2). We observed no difference in apoptosis induction upon PRANCR depletion. Together, these experiments established PRANCR as a novel lncRNA essential for epidermal progenitor renewal, proliferation, and clonogenic potential, without effects on cell apoptosis.
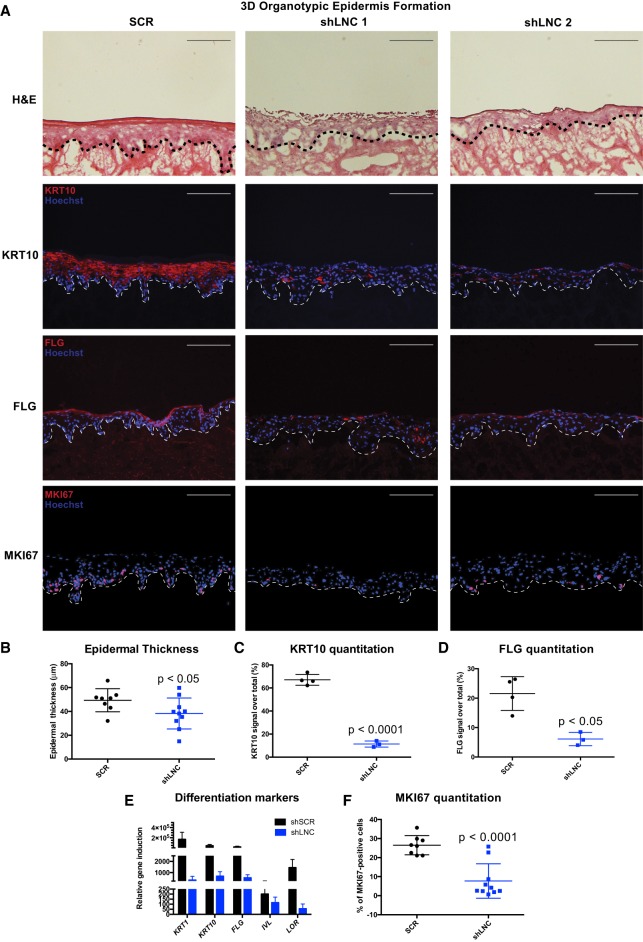
PRANCR is required for formation of organotypic stratified epidermis
Organotypic epidermal tissue displays superior correlation to in vivo biological skin states compared with cultured cells (Ridky et al. 2010). We therefore sought to assess the impact of PRANCR depletion in organotypic human epidermal tissue. Tissues were generated with PRANCR-depleted and control (scrambled) progenitors (Fig. 3, hematoxylin and eosin stains). PRANCR-depleted progenitors displayed disrupted architecture of the outermost epidermal layers (Fig. 3A, hematoxylin and eosin stains). PRANCR-depleted epidermis was notably thinner than controls (Fig. 3B). Proper organization of the outermost epidermal layers is essential for skin barrier function and the ability to prevent evaporative water loss. Corroborating the histological findings, immunofluorescence of epidermal differentiation markers keratin 10 (KRT10) and filaggrin (FLG) showed a deficiency in expression of structural and barrier proteins required for functional epidermis (Fig. 3A,C,D). We assessed mRNA expression levels of the epidermal differentiation markers keratin 1, keratin 10, involucrin, filaggrin, and loricrin cornified envelope precursor protein, critical genes disrupted in human skin diseases (O'Driscoll et al. 2002; Virtanen et al. 2003; Smith et al. 2006). Their expression was consistently reduced in PRANCR-depleted epidermis (Fig. 3E). Finally, to assess progenitor replication at the basal epidermal stratum, we performed staining for the replication marker MKI67 (Guillaud et al. 1991), which revealed a marked decrease in MKI67-positive cells in PRANCR-depleted epidermis (Fig. 3A,F). Collectively, these data indicate that PRANCR impacts epidermal tissue homeostasis by regulating both proliferation and differentiation.
Figure 3.
PRANCR is required for proliferation and differentiation in stratified epidermis. (A) Hematoxylin and eosin staining (top); immunofluorescence of the differentiation proteins KRT10 and FLG (middle), and immunofluorescence of proliferation marker MKI67 (bottom) in control and PRANCR-depleted (shLNC) epidermal tissue. Nuclei are stained in blue (Hoechst 33342). Scale bars, 100 µm. (B) Quantitation of epidermal thickness. Each dot represents the average of three measurements per image at fixed positions. Error bars, mean with SD; n = 8 in control and n = 10 in PRANCR knockdown. Differences evaluated using Student's t-test. (C,D) KRT10 and FLG quantitation as a percentage of the total fluorescence signal. Dots represent the average intensities measured from different images taken for each tissue. Error bars, mean with SD; n = 4 tissues in control and n = 3 tissues in PRANCR knockdown. (E) RNA expression in PRANCR-depleted epidermis versus control. Bars, mean with SEM; n = 4. (F) MKI67 quantitation as a percentage of total cells. Error bars, mean with SD; n = 8 in control and n = 10 in PRANCR knockdown. Differences evaluated using Student t-test. (SCR) Scrambled short hairpin; (shLNC1/2) short hairpin RNA 1 or 2 targeting PRANCR.
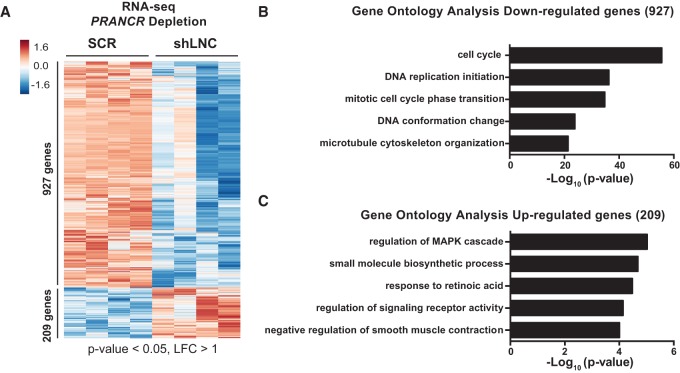
PRANCR regulates expression of cell cycle and MAPK pathway genes in trans
To investigate how PRANCR controls keratinocyte progenitor function, we performed whole-transcriptome sequencing on control and PRANCR-depleted primary keratinocytes. After differential expression analysis using DESeq (Anders and Huber 2010) and based on thresholds of absolute log2 fold change (LFC) of one and a P-value of <0.05, we identified 1136 differentially expressed genes (DEGs) in PRANCR knockdown cells (Fig. 4A). Most of these genes (927) showed decreased expression. Gene Ontology (GO) analysis using Metascape (Tripathi et al. 2015) revealed enrichment in GO terms related to the cell cycle, mitotic cell phase transition, and DNA replication initiation (Fig. 4B). The 209 up-regulated genes showed enrichment for several pathways, most notably genes related to MAPK pathway signaling (Fig. 4C). MAPK signaling is integral to keratinocyte renewal and differentiation (Scholl et al. 2007), raising the possibility that the PRANCR depletion phenotype might function through this pathway.
Figure 4.
PRANCR regulates expression of cell cycle and MAPK cascade genes in trans. (A) RNA-seq analysis of PRANCR-depleted keratinocytes. RNA was harvested 72 h after transduction of shRNAs. Hierarchical clustering of differentially expressed genes (greater than twofold change) in PRANCR-depleted samples (shLNC; n = 4; biological replicates of two independent PRANCR-targeting shRNAs) versus control samples (SCR; n = 4; biological replicates of two independent scrambled shRNAs). (B,C) Gene Ontology (GO) terms of genes that are down-regulated (B) or up-regulated (C) upon depletion of PRANCR.
lncRNAs can regulate gene expression of adjacent (cis) and/or distal (trans) genes (Ulitsky and Bartel 2013). To characterize how PRANCR impacts global gene transcription, we assessed the genomic location of the DEGs. Our results suggest no disproportionate enrichment for DEGs on Chromosome 12, where PRANCR resides (P-value for down-regulated genes P = 0.167 and for up-regulated genes P = 0.051; hypergeometric test) (Supplemental Fig. S4). We also observed no significant change of expression for transcripts immediately upstream of and downstream from PRANCR (CNOT2 and MYRFL). In addition to assessing PRANCR-depleted cells, we evaluated expression of the five cis-adjacent genes in PRANCR-depleted organotypic epidermis. We also found, as observed in the cultured cell context, that the cis-adjacent genes displayed no consistent directional changes in organotypic tissue (Supplemental Fig. S4). Based on these results, we conclude that PRANCR does not display significantly enriched cis-regulation and affects genes in trans.
PRANCR regulates expression of E2F- and FOXM1-targeted genes
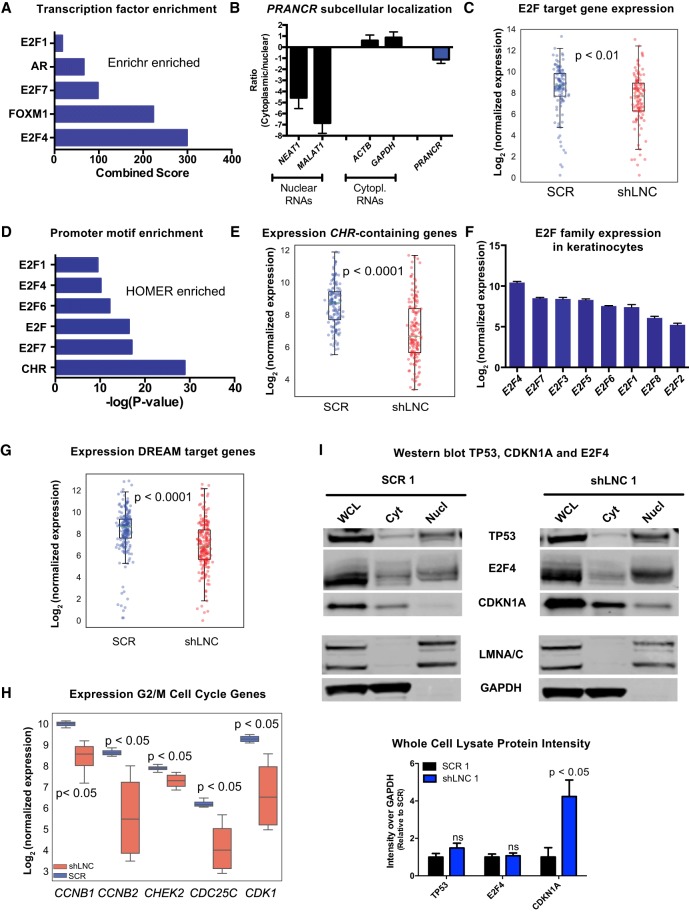
lncRNAs can affect the expression of distal genes by controlling transcriptional complexes that globally impact gene expression (Long et al. 2017). The analysis tool Enrichr (Chen et al. 2013b; Kuleshov et al. 2016) integrates genome-wide ChIP experiments in the ChEA (Lachmann et al. 2010) and ENCODE Project databases (The ENCODE Project Consortium 2012) to identify transcription factors involved in the control of a gene set. For PRANCR down-regulated DEGs, Enrichr identified E2F4 and FOXM1 as the most enriched transcription factors (Fig. 5A). These transcription factors are prominent components of two distinct, but closely related protein complexes that govern cell cycle gene expression (Engeland 2018). E2F transcription factors have a predominantly nuclear localization (Magae et al. 1996) and are essential to epidermal development (Ivanova et al. 2005). To determine whether PRANCR displayed similar localization, subcellular fractionation was performed, which confirmed the enrichment of PRANCR transcripts in the nucleus (Fig. 5B). Next, we specifically examined the expression of E2F target genes (Bracken et al. 2004) among the genes altered with the PRANCR knockdown (Fig. 5C). This analysis confirmed that known E2F transcription factor target genes were markedly down-regulated upon PRANCR depletion (P-value = 0.0074), consistent with a model of PRANCR affecting expression of genes targeted by the E2F family of transcription factors.
Figure 5.
PRANCR regulates expression of cell cycle genes containing the CHR promoter element. (A) Combined score of transcription factor enrichment of genes down-regulated upon PRANCR depletion using Enrichr gene set enrichment analysis. (B) PRANCR mRNA expression in cytoplasmic and nuclear cell fractions, measured by qRT-PCR, compared with known cytoplasmic (ACTB and GAPDH) and nuclear RNAs (NEAT1 and MALAT1). Bars, mean with SEM; n = 4 cell lines. (C) Normalized expression of E2F target genes. For box plots, dots represent mean expression level (n = 4) of each gene, lines represent median values of the gene set, box limits denote interquartile range, and whiskers extend 1.5 times the interquartile range. Differences evaluated with Student's t-test. (D) Motif enrichment of down-regulated DEGs using HOMER. (E) Normalized expression of genes containing a CHR promoter element. (F) Normalized expression of E2F family members in primary keratinocytes (n = 4; mean with SD). (G) Normalized expression of DREAM target genes. (H) Normalized expression of G2/M genes. (I) Western blot of TP53, E2F4, and CDKN1A in whole-cell lysate, cytoplasmic (Cyt), and nuclear (Nucl) fractions in control (SCR) and PRANCR-depleted cells (shLNC1). LMNA/C (nuclear) and GAPDH (cytoplasmic) represent experimental fractionation controls. Lower panel shows quantification of results by densitometry after normalization of total protein levels and represents the average of three biological replicates (Supplemental Fig. S6). Results evaluated with paired Student's t-test.
PRANCR regulates expression of cell cycle genes containing a CHR promoter element
We further explored the role of PRANCR in gene expression regulation by applying HOMER motif analysis, which identifies regulatory motifs enriched in the promoters of a gene set (Heinz et al. 2010). For the genes down-regulated by PRANCR knockdown, this analysis identified enrichment of E2F binding sites (Fig. 5D). Additionally, this analysis suggested an even stronger role for the cell cycle genes homology region (CHR), a DNA element present in promoters of many cell cycle genes (Müller and Engeland 2010) that are bound by E2F4- and/or FOXM1-containing protein complexes (Chen et al. 2013c; Fischer et al. 2014). Gene expression analysis of 148 late cell cycle genes harboring evolutionary conserved CHR elements (Müller et al. 2014) confirmed a highly significant and consistent down-regulation of these genes in PRANCR-depleted keratinocytes (P-value = 3.2 × 10−15) (Fig. 5E).
Analysis of RNA-seq data indicated that E2F4 is the most highly expressed E2F family member in primary keratinocytes (Fig. 5F). Recent studies indicate that the expression of cell cycle genes with CHR sites is repressed indirectly by TP53, via CDKN1A and E2F4, which is collectively called the TP53-CDKN1A-DREAM-CHR pathway (Quaas et al. 2012). Therefore, we tested whether genes of this specific pathway are affected by PRANCR knockdown. The gene expression profile of 210 genes in the TP53-CDKN1A-DREAM-CHR pathway (Fischer et al. 2016) indicated that expression of these target genes is strongly repressed upon PRANCR depletion (P-value = 2.9 × 10−10) (Fig. 5G). Consistent with the observed G2/M arrest upon PRANCR knockdown (Fig. 2I), down-regulation of genes through this pathway appears to be important for G2/M cell cycle control (Fischer et al. 2016). Additionally, prominent examples of TP53-DREAM genes involved in G2/M checkpoint control—CHEK2, CDK1, CCNB1, CCNB2, and CDC25C (Engeland 2018)—were impaired with PRANCR knockdown (P-value < 0.05) (Fig. 5H). As the regulation of expression of CHR-containing genes is a critical shared mechanism across different cell types (Müller and Engeland 2010), we also examined whether PRANCR functions similarly in another cell type. We depleted PRANCR in primary human fibroblasts and observed impaired proliferation as well as reduced expression of G2/M checkpoint control genes CCNB1, CCNB2, CDC25C, and CDK1 (Supplemental Fig. S5). Together, our results support a model in which PRANCR regulates expression of late cell cycle genes containing CHR sites. This may represent a general mechanism that is functional in multiple cell types and tissues.
PRANCR alters CDKN1A expression and localization
Finally, to gain insight to how PRANCR may interact with the TP53-CDKN1A-DREAM/E2F4-CHR pathway, we evaluated if PRANCR depletion altered protein levels or subcellular localization of TP53, CDKN1A, or E2F4 (Fig. 5I). TP53 and E2F4 expression were relatively unchanged, but CDKN1A expression increased approximately fourfold with PRANCR depletion (Fig. 5I; Supplemental Fig. S6). CDKN1A was also more highly represented in nuclear fractions upon PRANCR depletion (Fig. 5I; Supplemental Fig. S6). CDKN1A has a dual role in the epidermis to negatively regulate both proliferation and differentiation (Devgan et al. 2006), a phenotype that mirrors what we observed with PRANCR depletion. Together, the transcriptomic evidence and phenotypic similarities suggest an initial model by which PRANCR regulates epidermal homeostasis by modulating CDKN1A expression and nuclear localization.
Discussion
The noncoding genome has important undiscovered roles in human development and disease (Yang et al. 2013; Tak and Farnham 2015; Zhang and Lupski 2015). However, the biological significance of most noncoding genetic elements is still unknown. Genome-wide screens are a valuable approach to systematically evaluate their potential functions. In this report, we performed a CRISPRi screen in human keratinocytes to identify lncRNAs controlling epidermal progenitor replication, a fundamental process underlying skin homeostasis. Applying a relatively stringent threshold, we identified nine lncRNAs that regulate progenitor renewal and represent a foundation for follow-up studies to understand involvement of lncRNAs in this process.
A more detailed characterization of a top candidate, PRANCR, reveals a novel lncRNA that is required for both proliferation and clonogenicity of epidermal progenitors, as well as tissue stratification/differentiation. We found that experimental PRANCR depletion leads to up-regulation of total and nuclear CDKN1A, and we hypothesize that this may reflect a principal mechanism by which PRANCR controls progenitor replication. CDKN1A promotes formation of the E2F4-containing DREAM complex, which binds E2F and CHR promoter motifs to repress target gene transcription of cell cycle genes (Quaas et al. 2012; Fischer et al. 2014). In contrast, when CDKN1A levels are low, this protein complex switches to a FOXM1-containing MMB complex that activates late cell cycle genes with a CHR motif (Sadasivam et al. 2012; Chen et al. 2013c; Engeland 2018). PRANCR may therefore govern keratinocyte progenitor cell cycle through CDKN1A-mediated regulation of both E2F- and FOXM1-targeted genes.
Further supporting this overarching hypothesis is the observation that PRANCR and CDKN1A both share negative regulatory impacts on both epidermal proliferation and differentiation. Generally, proliferation and differentiation are envisioned as opposing fates in a cell. For instance, the epidermal transcription factor ZNF750 forms unique protein complexes that promote differentiation and block proliferation (Boxer et al. 2014). High levels of CDKN1A in the epidermis, however, inhibits both self-renewal and differentiation of keratinocytes (Missero et al. 1996; Topley et al. 1999; Devgan et al. 2006), similar to what is observed with PRANCR depletion. Although early CDKN1A induction leads to cell cycle arrest, persistent elevated expression suppresses differentiation through activation of the MAPK cascade (Devgan et al. 2006). Consistent with this, RNA-seq of PRANCR-depleted cells revealed up-regulation of genes regulating MAPK signaling (Fig. 4C).
These findings naturally raise additional questions: PRANCR depletion leads to increased CDKN1A expression, but does expression of PRANCR down-regulate CDKN1A? This would oppose the function of TP53, which is known to activate CDKN1A, and raises the possibility that PRANCR is pro-oncogenic. Both E2F and CHR motifs have been reported to be central elements in key genes associated with cancer signaling (Paci et al. 2017), which are orchestrated by TP53 (Engeland 2018). In addition, our initial characterization examined expression of PRANCR in bulk culture. However, it is possible that PRANCR expression changes dynamically during progression through the cell cycle and/or in response to contextual cues. Future single cell and in vivo–level experiments will aim to answer these questions and further elucidate the role of PRANCR in the epidermis.
Our study shows the value of CRISPRi screens to provide insight into the functional noncoding genome. Studying lncRNAs presents several challenges. Accurate identification of lncRNA transcriptional coordinates is critical for CRISPRi screens, which function optimally by targeting regions close to the TSS (Gilbert et al. 2014). Different annotation systems show variability in lncRNA annotations, and to best address these differences, we combined RefSeq, UCSC, and ENCODE/GENCODE annotations to identify and map lncRNAs. Since the initiation of our work, the FANTOM5 consortium reported a comprehensive atlas of human lncRNA genes with high-confidence 5′ ends using cap analysis gene expression (CAGE) (Hon et al. 2017). These efforts have improved the mapping of lncRNA TSS, which will further improve efficiency in the design and function of future lncRNA CRISPR screens.
Disruptions of epidermal homeostasis underlie many skin diseases. PRANCR and other epidermal lncRNAs may contribute to the pathogenesis of these conditions by controlling cell proliferation and differentiation, as well as other fundamental biological processes that form the basis of human tissue development and health. The CRISPRi lncRNA screening strategy presented here can also be adapted to interrogate the potential roles of PRANCR and other epidermal lncRNAs in processes such as carcinogenesis, cell–cell communication, and response to microbes. These genome-scale screening approaches will help elucidate the functions of the vast noncoding genome in human tissue development and disease.
Methods
Primary keratinocyte culture
Primary epidermal keratinocytes were isolated from discarded neonatal foreskin from circumcisions, collected upon written informed consent under an institutional review board protocol approved by the University of California, San Diego. Cells were isolated based on the protocol described previously (Aasen and Izpisua Belmonte 2010) and propagated in 50:50 mixture (“50:50 media”) of K-SFM and 154 media (Life Technologies) with recommended supplements and 1× antibiotic-antimycotic (Thermo Fisher Scientific), at 37°C and 5% CO2.
RNA sequencing of human skin biopsies
We analyzed RNA sequencing performed previously on clinically normal sun-protected human skin (Sun et al. 2015). Reads were aligned to the hg19 genome assembly using TopHat (Trapnell et al. 2009). At the time of study design, hg19 was chosen as a reference because of its inclusion of more complete ENCODE and Epigenomics Mapping Consortium data sets. The choice of reference sequence does not significantly alter principal conclusions: PRANCR is present in both hg19 and hg38. These RNA-seq data consisted of laser microdissected epidermis from two unrelated individuals. To obtain a high-confidence transcriptome, we integrated the transcript annotation with priority of RefSeq > UCSC > ENCODE/GENCODE databases and provided only one transcript annotation if it was defined in multiple databases. Transcripts belonging to the same gene were merged, and reads per kilobase per million (RPKM) were assigned to each gene. Genes with an average RPKM < 1 were excluded. The categorization of each gene as protein-coding, lncRNA, miRNA, or snoRNA is based on their annotation in RefSeq, UCSC, and ENCODE/GENCODE databases.
sgRNA library design
The CRISPR library was generated with sgRNAs designed against each of the 2263 epidermally expressed lncRNAs using the SSC algorithm optimized for CRISPRi (Xu et al. 2015). The genomic interval from −50 to +450 relative to the TSS was used for each target lncRNA transcript. All potential sgRNAs were sorted by efficiency score, and negative scores were discarded. For each lncRNA transcript, the top five scoring sgRNAs were selected, and if any of the top five scoring sgRNAs overlapped by >5 nt, the lower scoring sgRNA was replaced with the next highest-scoring sgRNA targeting this lncRNA. Once sgRNA selection was complete, candidate sgRNAs were evaluated for the presence of a “G” nucleotide in the −20 position to facilitate efficient transcription by the Pol III promoter. If the −20 position was not a “G,” then a “G” was substituted at this position. For negative control sgRNAs, 250 sequences of randomly generated 19-mers were generated and verified for their inability to match human genome sequences. A “G” nucleotide was then prepended to each 19-mer.
Construction of the CRISPR-Cas9 sgRNA library
All designed library sgRNA sequences were prepended and appended with linker sequences (5′ linker: CTTGTGGAAAGGACGAAACACC; 3′ linker: GTTTAAGAGCTATGCTGGAAACAGC) to facilitate polymerase chain reaction amplification and to serve as invariant sequence overhangs for InFusion cloning (Clontech). The oligonucleotides were synthesized (CustomArray) and delivered as a single oligonucleotide pool. The pool was amplified for 15 cycles (forward primer: ATCTTGTGGAAAGGACGAAACA; reverse primer: CTGTTTCCAGCATAGCTCTTAAAC) with CloneAmp HiFi PCR premix (Clontech). The entire PCR reaction was resolved on a 2% agarose gel, and the product was retrieved by gel slice isolation. A lentiviral vector pSICO-(F + E) was derived from the vector pSLQ1651 (Addgene plasmid 51024) (Chen et al. 2013a) by insertion of a 1.9-kb BsmBI stuffer fragment between the U6 promoter and downstream tracrRNA. InFusion cloning was performed to assemble the sgRNA library into BsmBI-digested pSICO-(F + E). Lentivirus was generated by the transfection of lentiviral helper plasmids and the CRISPRi plasmid library into 293T cells, and lentiviral supernatant was collected 48 h afterward. Supernatant was concentrated using Lenti-X concentrator (Clontech) and frozen at −80°C in replicate aliquots. One aliquot was thawed, and infection titration was performed on keratinocytes to determine appropriate dosing to achieve a multiplicity of infection of 0.3.
CRISPRi screen
pLEX-KRAB-dCas9-Blast was created by cloning the KRAB-dCas9 open reading frame (Addgene plasmid 60954) (Gilbert et al. 2014) into the pLEX-MCS vector (Thermo Fisher Scientific) using the BamHI/XhoI restriction sites. The puromycin-resistance cassette was swapped for a blasticidin-resistance cassette, yielding pLEX-KRAB-dCas9-Blast. Clone 103 keratinocytes (Sun et al. 2015) were infected with pLEX-KRAB-dCas9-Blast in the presence of 3 µg/mL of polybrene, selected in 2 µg/mL of blasticidin for 72 h, and then expanded in limited dilution plating to isolate individual clones. Expanded clones were evaluated by western blot to select clones with the highest-expressing KRAB-dCas9; 107 cells from a KRAB-dCas9–expressing keratinocyte line were infected with the titrated CRISPRi library at MOI 0.3 and selected for 48 hours with puromycin. Starting cell numbers were chosen to achieve >300× sgRNA overrepresentation. After selection, 4 × 106 cells were reserved and snap frozen at the preselection time point. The remaining cells were distributed onto 150-mm tissue culture plates at 106 cells per plate and grown in 50:50 media. Cells were monitored visually every day and were split upon reaching ∼70% confluence and replated at a minimum of 4 × 106 cells to maintain >300× library overrepresentation. The screen was taken to 28 d, which allows enough keratinocytes cell doublings to detect significant changes in sgRNA abundance, and cells were harvested at the endpoint. The screen was performed in technical duplicates on two different keratinocyte clones. Genomic DNA was isolated from cell pellets using a Genomic DNA isolation kit (Qiagen). sgRNAs were quantitated by amplification from genomic DNA using PrimeStar (Clontech) using primers that flanked the sgRNA sequence (forward primer: ACACGACGCTCTTCCGATCTTGTGGAAAGGACGAAACACC; reverse primer: GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCGCTGTTTCCAGCATAGCTCTTAA). To increase the complexity of the amplicons and facilitate improved clustering on the Illumina sequencer, we used a mixture of forward primers with staggered-length scramble sequences (ACACGACGCTCTTCCGATCTNNTGTGGAAAGGACGAAACACC, ACACGACGCTCTTCCGATCTNNNNTGTGGAAAGGACGAAACACC, ACACGACGCTCTTCCGATCTNNNNNNTGTGGAAAGGACGAAACACC). The first round of PCR was performed for 20 cycles from a minimum of 16 µg genomic DNA to assure genomic DNA oversampling and to reduce sgRNA amplification bias. The resulting product was column-purified with a PCR purification kit (Macherey-Nagel), and the entire product was introduced into a second round of PCR for seven cycles to introduce Illumina sequencing primers and unique barcodes for each experiment (forward: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT; reverse: CAAGCAGAAGACGGCATACGAGAT < Illumina index 6-bp barcode > GTGACTGGAGTTCAGACGTG). Libraries were quantified using the NEBNext library quantitation kit (New England Biolabs) and mixed in equimolar ratios for sequencing on an Illumina HiSeq 4000 using a read length of 75 bp.
Analysis of CRISPRi screen results
MAGeCK (0.5.5) was used to analyze the screening sequencing data (Li et al. 2014). The quality of raw data in FASTQ format was evaluated using FastQC, and the sequencing reads were adapter trimmed using Cutadapt before alignment (Martin 2011). Reads were then mapped to the screening sgRNA library without tolerating any mismatches, and the raw read counts of sgRNAs of all samples were merged into a count matrix, which was automatically performed in the MAGeCK software with the “count” command (sgRNA ranking). The MAGeCK “test” command was then used to identify the negatively and positively selected lncRNAs. Instead of computing the size factor from all sgRNAs (the default normalization method for MAGeCK), the size factor was estimated from the negative control sgRNAs to gain a more realistic estimation. All the other analyses and outputs of MAGeCK were according to the default parameters. During the final step of MAGeCK algorithm, a RRA computes P-values, FDRs, and RRA scores (Li et al. 2014) to rank the interrogated lncRNA genes. To select the most promising lncRNA candidates, lncRNAs with statistical parameters comparable or better than the positive control TINCR (FDR = 0.07) were filtered (FDR <0.1 and >2 sgRNAs changing concordantly with the lncRNA phenotype).
RNA interference–mediated gene knockdown
For short hairpin-targeted gene knockdown of PRANCR, shRNAs were cloned into the pLKO.1 vector (the RNAi Consortium). Lentivirus was generated by transfection of both packaging and transfer plasmids into 293T cells using Lipofectamine 3000 (Life Technologies). Supernatants containing lentivirus were collected 48 h after transfection and concentrated with Lenti-X Concentrator (Clontech) and stored at −80°C. For knockdown, 5.0 × 105 keratinocytes were infected with scrambled control or PRANCR-targeting shRNAs in medium containing 3 µg/mL polybrene and incubated overnight. Infected cells were selected in medium supplemented with 1 µg/mL puromycin. The shRNA sequences targeting PRANCR are as follows: shLNC1, 5′-CACTTTGAATGACAACGATTT-3′; shLNC2, 5′-TACTTCACTCCTTTAAGTTTC-3′. Scrambled shRNA sequences are as follows: SCR1, 5′-CCTAAGGTTAAGTCGCCCTCG-3′; SCR2, 5′-GCAAGCTGACCCTGAAGTTCA-3′.
Cell proliferation assay
To assess cell proliferation rates, 5000 cells were plated on a 24-well plate in duplicate for each condition and each time point. Media was changed every 48 h. At each time point, cell abundance was assessed using alamarBlue (Thermo Fisher Scientific). At each time point, alamarBlue reagent was added following the manufacturer's instructions, and fluorescence was measured following 2-h incubation at 37°C using the SpectraMax iD3 microplate reader (Molecular Devices). To compare between conditions, fluorescent signals at the start of the experiment (day 0) were set to one. Subsequently, relative proliferation was measured relative to the day 0 fluorescence signal, as changes in fluorescence are directly proportional to the cell number.
Holoclone assay
3T3 fibroblasts were treated with mitomycin C (15 µg/mL) for 2 h at 37°C. Next, 500,000 mitomycin C–treated fibroblasts were seeded onto six-well plates and incubated overnight. The next day, 300 keratinocytes were seeded onto each well. Clones were propagated for up to 15 d with media changed every 3–4 d. At the endpoint, fibroblasts were dislodged by vigorously washing with PBS. Holoclones were fixed in ice-cold methanol: acetone (1:1) for 3 min, stained with 0.02% crystal violet for 2 min, and destained with more than three washes of PBS. Cells were air-dried and imaged by scanning. Holoclones detection and counting were then performed on these scans using ImageJ software.
Cell cycle analysis
Cells were cultured in low-serum medium for 24 h after puromycin selection. Cell cycle analysis was performed using the cell cycle phase determination kit (Cayman Chemical) according to the manufacturer's instructions. In short, 1 × 105 cells were washed and fixed overnight at −20°C. Thereafter, cells were stained with PI and incubated for 30 min in the dark, and at least 10,000 cells per condition were measured by flow cytometry using the Guava easyCyte 8HT (Millipore). Analysis of the resulting FCS2.0 files was performed with FlowJo software, and flow cytometric data were fit using the built-in Dean-Jett-Fox univariate model to assess the relative distribution of cells over the different cell cycle phases.
Organotypic culture
Air-dried devitalized human dermis was mounted onto 1.7-cm × 1.7-cm supports, and 500,000 keratinocytes were seeded onto the basement membrane. Tissue was grown in media (Gangatirkar et al. 2007) at an air-liquid interface over a course of 7 d, with medium changed daily. Half of the final tissue was collected in TRIzol for RNA isolation (Supplemental Methods), and the other half was embedded in O.C.T. media (Sakura) and sectioned on a cryostat at 7-µm thickness. Sections were visualized with hematoxylin/eosin or immunofluorescence (Supplemental Methods). Epidermal thickness was measured at three fixed sites across the tissue using ImageJ, measured from the basement membrane and the most superficial aspect of the stratum corneum. The percentage of MKI67-positive cells was counted using ImageJ using the Analyze particles feature on both DAPI-positive and MKI67-positive cells. For quantitation of KRT10 and FLG, the total (Hoechst and KRT10/FLG) and KRT10/FLG fluorescent signals were quantified using ImageJ (“Threshold color” and “Measure” features), and the %KRT10/FLG was measured as the ratio of the KRT10/FLG signal (“area”) over the total fluorescent signal.
PRANCR knockdown RNA-seq analysis
Primary keratinocytes from two independent donors were infected with two scrambled and two PRANCR-targeting shRNAs. After 72 h, following complete negative (noninfected) puromycin selection, total RNA was isolated using the Direct-zol RNA kit (Zymo research) and treated with DNase I, and mRNA was enriched by oligo(dT) magnetic beads (Invitrogen). Sequencing was performed on the Illumina HiSeq 4000 using 50-bp single-end reads. Raw data qualities were evaluated by FastQC. Sequence reads are mapped to the human reference genome (hg19) using STAR (Dobin et al. 2013). Read counts of each gene were collected into a matrix, and the differential expression analysis was performed using DESeq (Anders and Huber 2010). After identifying DEGs, GO analysis was performed using Metascape (Tripathi et al. 2015).
Subcellular RNA fractionation
Measurement of the abundance of nuclear and cytoplasmic mRNA was performed as described previously (Wang et al. 2006). In short, cells were lysed, nuclei were pelleted by centrifugation, and the supernatant was collected as cytoplasmic fraction. Next, RT-PCR was performed on both fractions, followed by qPCR quantification of the relative abundance of specific mRNA transcripts in both fractions. NEAT1 and MALAT1 mRNAs were used as a positive control for nuclear localization; ACTB and GAPDH mRNAs, as positive controls for cytoplasmic localization. Primer sequences can be found in the Supplemental Methods.
Protein isolation and western blot
Whole-cell protein lysates were prepared in RIPA buffer and quantitated with the BCA assay (Pierce). To separate cultured cells into cytoplasmic and nuclear/cytoskeletal fractions, we used the cell fractionation kit (Cell Signaling Technologies) according to the manufacturer's instructions. Proteins were analyzed using western blotting (Supplemental Methods). Quantification was performed using Image Studio software (LI-COR Biosciences).
Data access
The raw and processed sequencing data generated in this study are available at the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE125400.
Supplementary Material
Acknowledgments
This work was supported by the National Key R&D Program of China (2017YFA0102900 to K.Q.), the National Natural Science Foundation of China grants (81788101, 91640113, 91940306, and 31771428 to K.Q.), the Fundamental Research Funds for the Central Universities (to K.Q.), the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (K08AR067853 to B.K.S.), and the Doris Duke Charitable Foundation (Clinical Scientist Development award 2019088 to B.K.S.). We thank the USTC Supercomputing Center and the School of Life Science Bioinformatics Center for providing supercomputing resources for this project. The funding sources had no input on the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author contributions: B.K.S. and K.Q. conceived of the project; P.C., A.B.C.O., K.Q., and B.K.S. designed the experiments; P.C., A.B.C.O., B.C., M.A.I., and B.K.S. performed the experiments and data acquisition. P.C., A.B.C.O., W.Z., B.H., K.Q., and B.K.S. analyzed and interpreted the data. A.B.C.O. and B.K.S. wrote the first draft of the manuscript, and P.C. and K.Q. participated in manuscript revision. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.251561.119.
References
- Aasen T, Izpisua Belmonte JC. 2010. Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem cells. Nat Protoc 5: 371–382. 10.1038/nprot.2009.241 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awan HM, Shah A, Rashid F, Shan G. 2017. Primate-specific long non-coding RNAs and microRNAs. Genomics Proteomics Bioinformatics 15: 187–195. 10.1016/j.gpb.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon Y, Green H. 1987. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci 84: 2302–2306. 10.1073/pnas.84.8.2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras TR, Haerty W, et al. 2014. Considerations when investigating lncRNA function in vivo. eLife 3: e03058 10.7554/eLife.03058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer LD, Barajas B, Tao S, Zhang J, Khavari PA. 2014. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev 28: 2013–2026. 10.1101/gad.246579.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, Helin K. 2004. E2F target genes: unraveling the biology. Trends Biochem Sci 29: 409–417. 10.1016/j.tibs.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. 2013a. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155: 1479–1491. 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. 2013b. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Müller GA, Quaas M, Fischer M, Han N, Stutchbury B, Sharrocks AD, Engeland K. 2013c. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol Cell Biol 33: 227–236. 10.1128/MCB.00881-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Sacco L, Baldassarre A, Masotti A. 2012. Bioinformatics tools and novel challenges in long non-coding RNAs (lncRNAs) functional analysis. Int J Mol Sci 13: 97–114. 10.3390/ijms13010097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devgan V, Nguyen BC, Oh H, Dotto GP. 2006. p21WAF1/Cip1 suppresses keratinocyte differentiation independently of the cell cycle through transcriptional up-regulation of the IGF-I gene. J Biol Chem 281: 30463–30470. 10.1074/jbc.M604684200 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. 2013. Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci 110: 5294–5300. 10.1073/pnas.1221376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeland K. 2018. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ 25: 114–132. 10.1038/cdd.2017.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Steiner L, Engeland K. 2014. The transcription factor p53: not a repressor, solely an activator. Cell Cycle 13: 3037–3058. 10.4161/15384101.2014.949083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Quaas M, Steiner L, Engeland K. 2016. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res 44: 164–174. 10.1093/nar/gkv927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangatirkar P, Paquet-Fifield S, Li A, Rossi R, Kaur P. 2007. Establishment of 3D organotypic cultures using human neonatal epidermal cells. Nat Protoc 2: 178–186. 10.1038/nprot.2006.448 [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159: 647–661. 10.1016/j.cell.2014.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaud P, Vermont J, Seigneurin D. 1991. Automatic classification of cells in cell cycle phases based on Ki-67 antigen quantification by fluorescence microscopy. Cell Prolif 24: 481–491. 10.1111/j.1365-2184.1991.tb01176.x [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, et al. 2017. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543: 199–204. 10.1038/nature21374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova IA, D'Souza SJ, Dagnino L. 2005. Signalling in the epidermis: the E2F cell cycle regulatory pathway in epidermal morphogenesis, regeneration and transformation. Int J Biol Sci 1: 87–95. 10.7150/ijbs.1.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, et al. 2014. Defining functional DNA elements in the human genome. Proc Natl Acad Sci 111: 6131–6138. 10.1073/pnas.1318948111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. 2013. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493: 231–235. 10.1038/nature11661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma'ayan A. 2010. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26: 2438–2444. 10.1093/bioinformatics/btq466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15: 554 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, et al. 2017. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355: aah7111 10.1126/science.aah7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Wang X, Youmans DT, Cech TR. 2017. How do lncRNAs regulate transcription? Sci Adv 3: eaao2110 10.1126/sciadv.aao2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Pajares V, Yan K, Zarnegar BJ, Jameson KL, Khavari PA. 2013. Genetic pathways in disorders of epidermal differentiation. Trends Genet 29: 31–40. 10.1016/j.tig.2012.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magae J, Wu CL, Illenye S, Harlow E, Heintz NH. 1996. Nuclear localization of DP and E2F transcription factors by heterodimeric partners and retinoblastoma protein family members. J Cell Sci 109: 1717–1726. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Missero C, Di Cunto F, Kiyokawa H, Koff A, Dotto GP. 1996. The absence of p21Cip1/WAF1 alters keratinocyte growth and differentiation and promotes ras-tumor progression. Genes Dev 10: 3065–3075. 10.1101/gad.10.23.3065 [DOI] [PubMed] [Google Scholar]
- Müller GA, Engeland K. 2010. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J 277: 877–893. 10.1111/j.1742-4658.2009.07508.x [DOI] [PubMed] [Google Scholar]
- Müller GA, Wintsche A, Stangner K, Prohaska SJ, Stadler PF, Engeland K. 2014. The CHR site: definition and genome-wide identification of a cell cycle transcriptional element. Nucleic Acids Res 42: 10331–10350. 10.1093/nar/gku696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll J, Muston GC, McGrath JA, Lam HM, Ashworth J, Christiano AM. 2002. A recurrent mutation in the loricrin gene underlies the ichthyotic variant of Vohwinkel syndrome. Clin Exp Dermatol 27: 243–246. 10.1046/j.1365-2230.2002.01031.x [DOI] [PubMed] [Google Scholar]
- Oh JW, Hsi TC, Guerrero-Juarez CF, Ramos R, Plikus MV. 2013. Organotypic skin culture. J Invest Dermatol 133: 1–4. 10.1038/jid.2013.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paci P, Colombo T, Fiscon G, Gurtner A, Pavesi G, Farina L. 2017. SWIM: a computational tool to unveiling crucial nodes in complex biological networks. Sci Rep 7: 44797 10.1038/srep44797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaas M, Müller GA, Engeland K. 2012. p53 can repress transcription of cell cycle genes through a p21WAF1/CIP1-dependent switch from MMB to DREAM protein complex binding at CHR promoter elements. Cell Cycle 11: 4661–4672. 10.4161/cc.22917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek XC, Thomson DW, Maag JL, Bartonicek N, Signal B, Clark MB, Gloss BS, Dinger ME. 2015. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res 43: D168–D173. 10.1093/nar/gku988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridky TW, Chow JM, Wong DJ, Khavari PA. 2010. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat Med 16: 1450–1455. 10.1038/nm.2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivam S, Duan S, DeCaprio JA. 2012. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev 26: 474–489. 10.1101/gad.181933.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarropoulos I, Marin R, Cardoso-Moreira M, Kaessmann H. 2019. Developmental dynamics of lncRNAs across mammalian organs and species. Nature 571: 510–514. 10.1038/s41586-019-1341-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl FA, Dumesic PA, Barragan DI, Harada K, Bissonauth V, Charron J, Khavari PA. 2007. Mek1/2 MAPK kinases are essential for mammalian development, homeostasis, and Raf-induced hyperplasia. Dev Cell 12: 615–629. 10.1016/j.devcel.2007.03.009 [DOI] [PubMed] [Google Scholar]
- Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, Liao H, Evans AT, Goudie DR, Lewis-Jones S, et al. 2006. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet 38: 337–342. 10.1038/ng1743 [DOI] [PubMed] [Google Scholar]
- Sun BK, Boxer LD, Ransohoff JD, Siprashvili Z, Qu K, Lopez-Pajares V, Hollmig ST, Khavari PA. 2015. CALML5 is a ZNF750- and TINCR-induced protein that binds stratifin to regulate epidermal differentiation. Genes Dev 29: 2225–2230. 10.1101/gad.267708.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak YG, Farnham PJ. 2015. Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin 8: 57 10.1186/s13072-015-0050-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topley GI, Okuyama R, Gonzales JG, Conti C, Dotto GP. 1999. p21WAF1/Cip1 functions as a suppressor of malignant skin tumor formation and a determinant of keratinocyte stem-cell potential. Proc Natl Acad Sci 96: 9089–9094. 10.1073/pnas.96.16.9089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, et al. 2015. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 18: 723–735. 10.1016/j.chom.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck AC, Natarajan KN, Rice GM, Borawski J, Mohn F, Rankova A, Flemr M, Wenger A, Nutiu R, Teichmann S, et al. 2018. Distinctive features of lincRNA gene expression suggest widespread RNA-independent functions. Life Sci Alliance 1: e201800124 10.26508/lsa.201800124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. 2013. lincRNAs: genomics, evolution, and mechanisms. Cell 154: 26–46. 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen M, Smith SK, Gedde-Dahl T Jr., Vahlquist A, Bowden PE. 2003. Splice site and deletion mutations in keratin (KRT1 and KRT10) genes: unusual phenotypic alterations in Scandinavian patients with epidermolytic hyperkeratosis. J Invest Dermatol 121: 1013–1020. 10.1046/j.1523-1747.2003.12534.x [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhu W, Levy DE. 2006. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods 39: 356–362. 10.1016/j.ymeth.2006.06.010 [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23: 1494–1504. 10.1101/gad.1800909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Xiao T, Chen CH, Li W, Meyer CA, Wu Q, Wu D, Cong L, Zhang F, Liu JS, et al. 2015. Sequence determinants of improved CRISPR sgRNA design. Genome Res 25: 1147–1157. 10.1101/gr.191452.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, et al. 2013. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 369: 1502–1511. 10.1056/NEJMoa1306555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Lupski JR. 2015. Non-coding genetic variants in human disease. Hum Mol Genet 24: R102–R110. 10.1093/hmg/ddv259 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.