Abstract
Bone marrow–derived mesenchymal stem cells (MSCs) differentiate into osteoblasts upon stimulation by signals present in their niche. Because the global signaling cascades involved in the early phases of MSCs osteoblast (OB) differentiation are not well-defined, we used quantitative mass spectrometry to delineate changes in human MSCs proteome and phosphoproteome during the first 24 h of their OB lineage commitment. The temporal profiles of 6252 proteins and 15,059 phosphorylation sites suggested at least two distinct signaling waves: one peaking within 30 to 60 min after stimulation and a second upsurge after 24 h. In addition to providing a comprehensive view of the proteome and phosphoproteome dynamics during early MSCs differentiation, our analyses identified a key role of serine/threonine protein kinase D1 (PRKD1) in OB commitment. At the onset of OB differentiation, PRKD1 initiates activation of the pro-osteogenic transcription factor RUNX2 by triggering phosphorylation and nuclear exclusion of the histone deacetylase HDAC7.
The skeleton is a dynamic tissue constantly undergoing bone matrix resorption and formation, two coupled processes mediated by osteoclastic and osteoblastic cells, respectively (Parfitt et al. 2011). Decreased osteoblastic bone formation relative to osteoclastic bone resorption is the cellular basis of age- and osteoporosis-related bone loss (Feng and McDonald 2011). Osteoblasts differentiate from skeletal stem cells, also known as stromal or mesenchymal stem cells (MSCs), which are present within the bone marrow stroma. MSCs are among the most suitable cell types for cell therapy applications due to their ease of isolation, multilineage differentiation potential, their immunomodulatory and regeneration promoting properties, as well as their excellent safety record (Jafari et al. 2014; Zaher et al. 2014). Therefore, MSCs have been used in an increasing number of clinical trials for enhancing tissue repair in different contexts such as regeneration of skeletal defects (Andrzejewska et al. 2019). Determining the signaling mechanisms that regulate MSCs commitment and differentiation into the bone-forming osteoblastic cells can pave way for the development of novel approaches for enhancing bone formation and treating osteoporotic bone loss (Abdallah et al. 2015).
The current model of osteoblast (OB) differentiation from stem cells is based on a large number of in vitro studies and consists of a progressive differentiation sequence that includes phases of cell proliferation followed by cell maturation, extracellular matrix production and, lastly, matrix mineralization (Lian and Stein 1995). Although this model is useful as an overview, there is a need for delineating the molecular and signaling events underlying these phases. Several previous studies have used a reductionist approach to determine factors and signaling pathways that are important for OB differentiation and identified a number of key signaling cascades including Wnt, Hedgehog, Notch, and Bone morphogenetic proteins (BMPs) (Abdallah et al. 2015; Hojo et al. 2015). However, such approaches fail to provide information regarding how signaling events are initiated and integrated in a coherent, well-orchestrated biological process.
Over the last decade, mass spectrometry (MS)-based proteomics has evolved as a powerful platform for studying complex biological processes at a systemwide level. We and others have previously used MS-based quantitative proteomics using stable isotope labeling by amino acids in cell culture (SILAC) to provide a global overview of human embryonic stem cell proteomes and phosphoproteomes during differentiation (Prokhorova et al. 2009; Van Hoof et al. 2009; Rigbolt et al. 2011a; Rigbolt and Blagoev 2012), to monitor receptor tyrosine kinase signaling in MSCs differentiation to osteoblasts (Kratchmarova et al. 2005), and to determine secreted factors during OB differentiation (Kristensen et al. 2012), as well as proteome and secretome variations during muscle cell differentiation (Henningsen et al. 2010; Le Bihan et al. 2015). Here, we combined SILAC-based quantitative proteomic and phosphoproteomic analyses to elucidate the signaling events occurring during the first 24 h of OB differentiation of human MSCs, identifying more than 6000 proteins and 15,000 phosphorylation sites.
Results
Temporal profiling of the proteome and phosphoproteome during hMSC commitment to osteoblastic lineage
SILAC-labeled hMSCs were either left unstimulated or induced to undergo OB differentiation for 30 min, 1, 6, or 24 h (Fig. 1A). Cell lysates were separated into nuclear and cytoplasmic fractions and each combined into two different SILAC pools allowing comparable quantitation of proteins and phosphorylation events at the five time points. Cell lysates were either subjected to 1D gel electrophoresis followed by in-gel digestion for proteome determination or to enrichment of phosphopeptides before LC-MS/MS measurements for delineation of phosphorylation events at a site-specific level (Fig. 1A).
Figure 1.
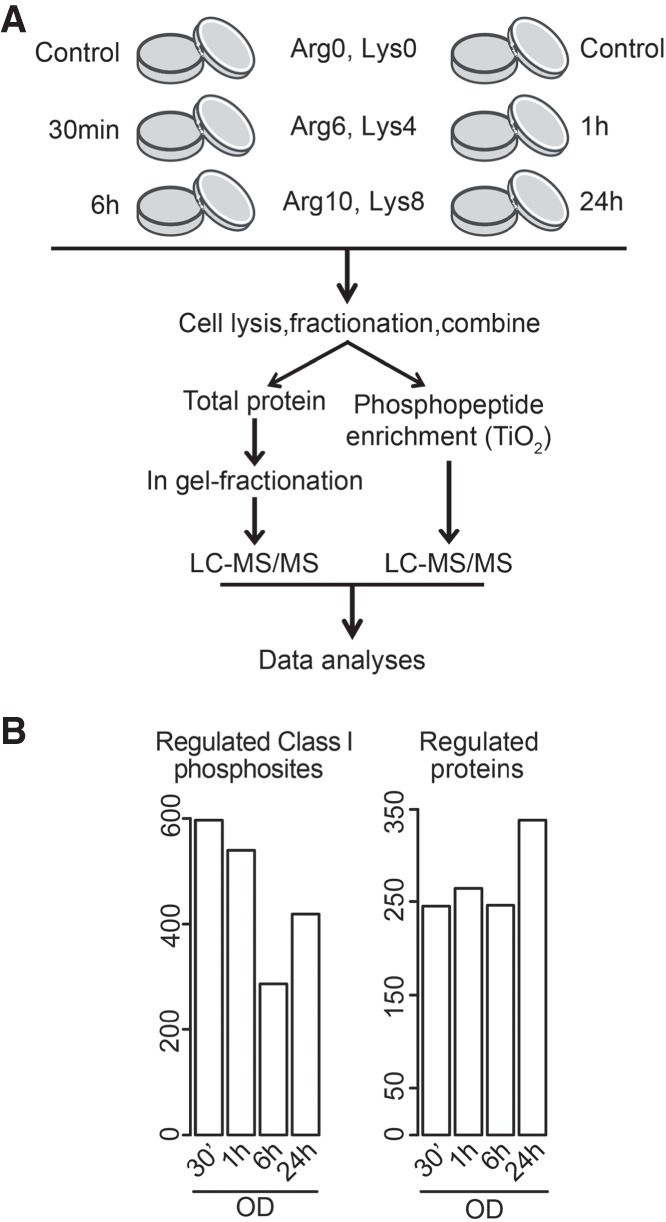
Quantitative mass spectrometry revealed widespread changes during the initial period of osteoblast commitment. (A) Flow diagram of the quantitative proteomic and phosphoproteomic experiment. Human MSCs were labeled with the indicated combinations of arginine (Arg) and lysine (Lys) SILAC amino acids, induced to undergo osteoblast differentiation, and harvested at indicated time points. After cell lysis and fractionation, proteins were pooled, digested, and either analyzed by LC-MS/MS or enriched for phosphorylated species followed by LC-MS/MS analysis. (B) Numbers of significantly changed phosphorylation sites (left) and proteins (right) at indicated time points during osteoblastogenesis relative to undifferentiated cells.
We identified 6252 proteins (Supplemental Table S1), of which 565 showed changes in at least one time point (P < 0.05 with at least 1.5-fold increase or decrease). The number of regulated proteins increased from an average of 250 proteins during the first 6 h to nearly 350 at 24 h (Fig. 1B). The phosphoproteomic analysis identified 15,059 unique phosphorylation sites with an overall distribution of 88% on serine, 10% on threonine, and 2% on tyrosine residues (Supplemental Table S2). Quantitative information was obtained for 10,930 sites, of which 797 displayed significant changes in abundance in at least one time point (P < 0.05 with at least twofold increase or decrease compared to unstimulated cells). In contrast to the observed changes in protein abundances, the highest number of altered phosphorylation sites was observed shortly after induction of differentiation, followed by a drop at 6 h, before a secondary boost at 24 h, hinting to the presence of two signaling waves during early OB differentiation (Fig. 1B).
The proteomic landscape changed within the 24 h after induction of OB differentiation
To examine the dynamics of protein expression at a global level, we used unsupervised fuzzy c-means clustering (Supplemental Fig. S1A). Because the majority of the protein alterations occurred at later time points, we focused on the data from the 6- and 24-h time points. Proteins grouped into six clusters with a general down-regulation (Clusters 1 and 3), up-regulation (Clusters 2 and 6), or transient changes (Clusters 4 and 5) (Supplemental Fig. S1A). Analyses using Gene Ontology (GO) (Ashburner et al. 2000) revealed enrichment for several biological processes and molecular functions in each individual cluster (Supplemental Fig. S1B,C). We found enrichment in different processes related to “extracellular matrix” in Clusters 1 and 2 that represent early responsive proteins. Proteins related to “collagen metabolism” and “extracellular matrix disassembly” were found in Cluster 1 (down-regulated at 6 h and 24 h after induction), whereas proteins associated with “extracellular matrix organization” were found in Cluster 2 (up-regulated 6 h and 24 h after the induction). Proteins involved in “DNA replication and regulation of cell proliferation” were also enriched in Cluster 2, suggesting that the initial phase of MSC osteoblast differentiation is associated with enhanced cell division, which is in accordance with previous findings (Lian and Stein 1995; Marumoto et al. 2017).
Some of the regulated proteins showed significant changes only 24 h after the onset of differentiation (Cluster 6). Several of the proteins in this cluster were associated with the transforming growth factor beta (TGFB) and Hippo pathways indicating increased contribution of these signaling networks in the differentiation process at this later time point. A significant fraction of the proteins underwent only a temporal change in their expression (Clusters 4 and 5). Proteins linked with Toll-like receptor signaling were overrepresented in Cluster 4, characterized by a transient decrease, whereas proteins involved in “transcriptional regulation” were enriched in the group with temporary elevated levels (Cluster 5).
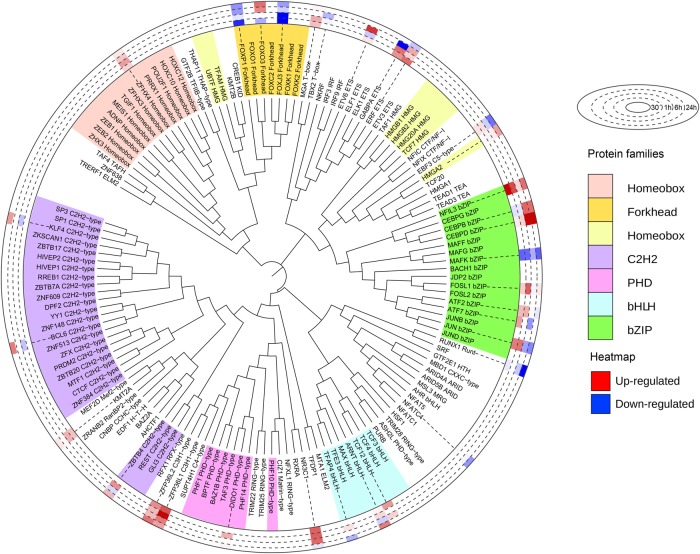
Because transcription factors play pivotal roles in cellular differentiation responses, we extracted and quantified transcription factors present within the identified protein clusters. We found 140 transcription factors with DNA-binding motifs according to their GO terms (Fig. 2), of which 32 showed temporal changes. Among these, 22 were regulated at either protein or phosphosite level and were linked with terms related to cell differentiation, bone mineral density, and ossification (Supplemental Fig. S2). For example, CCAAT enhancer binding protein beta (CEBPB) and delta (CEBPD), which are members of the basic leucine zipper (bZIP) family, were up-regulated at the 6- and 24-h time points. The two transcription factors are known to interact and positively regulate RUNX family transcription factor 2 (RUNX2)-mediated gene expression (Gutierrez et al. 2002). Fork-head transcription factors were in general down-regulated with the exception of FOXO3, which was slightly up-regulated at 6 and 24 h in agreement with a role of this factor in protecting osteoblasts from oxidative stress and apoptosis (Ambrogini et al. 2010). Furthermore, the zinc-finger proteins ZFP36L1 and ZFP36L2 were also found increased at the protein level (Fig. 2). These two factors are known to be up-regulated in response to several extracellular signals such as TGFB, TNFA, EGF, and glucocorticoids. Both have been reported to play important roles in cell differentiation (Stumpo et al. 2009; Chen et al. 2015; Tseng et al. 2017) and, for ZFP36L1, to favor OB differentiation over adipogenesis (Tseng et al. 2017).
Figure 2.
The levels of several transcription factors are altered during early osteoblast commitment. Identified and quantified transcription factors were ordered by the phylogenetic distance of their DNA-binding domains and aligned using iTOL (Letunic and Bork 2019). Histogram in the outer ring represents the regulation at protein level during induction of osteoblastic differentiation. Color codes for the different families are shown on the right.
Changes in the phosphoproteome revealed two intracellular kinase-signaling waves associated with OB lineage commitment of hMSC
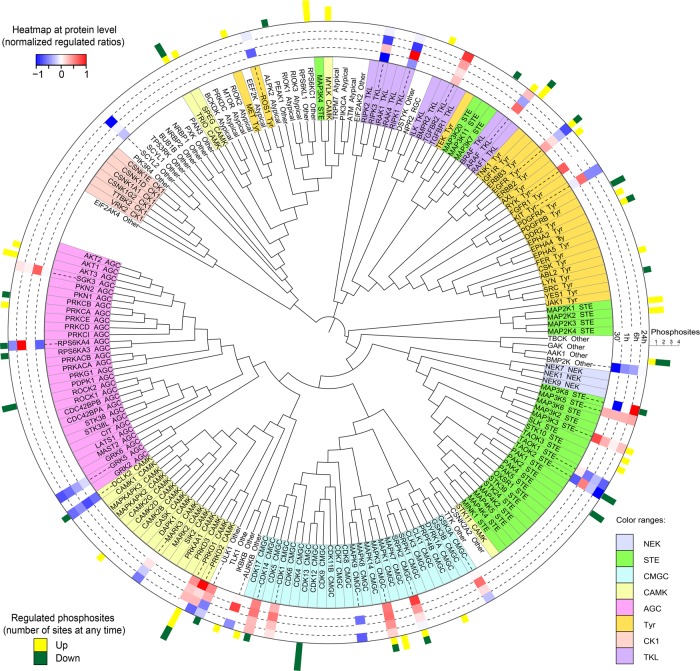
Various members from most of the kinase families were represented in our data set, covering 187 quantified kinases: 30 were altered at the protein level, and 45 showed changes in their phosphorylation status (Fig. 3). To assess the importance of the identified fluctuations, we selected 10 kinases hitherto not associated with osteoblast differentiation and examined the effect of their individual knockdown on the conversion of hMSCs into osteoblasts. Of these kinases, eight were regulated at their level of expression and two at their phosphorylation status. Knockdown of seven of the 10 candidates had a significant impact on alkaline phosphatase activity at day 6 of differentiation (Fig. 4). Moreover, silencing of the kinases identified as possible negative regulators by quantitative proteomics, that is, down-regulated in the MS data set, led to stimulation of osteoblast conversion, whereas knockdown of the potential positive regulators resulted in reduced differentiation.
Figure 3.
Commitment to the osteoblast lineage leads to numerous alterations in kinase expression and phosphorylation status. Identified and quantified kinases were ordered by the phylogenetic distance of their kinase domains and aligned using iTOL. Color codes for distinct kinase families are shown in the right bottom corner. Histogram in the inner ring represents the regulation at protein level at the corresponding time points, whereas bars on outer ring show number and direction of regulated phosphorylation sites.
Figure 4.
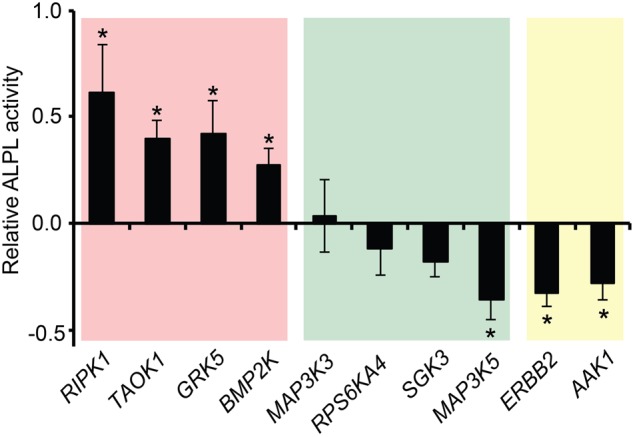
Knockdown of regulated kinases alter osteoblast differentiation. Before induction of differentiation, hMSCs were transfected with siRNAs targeting indicated kinases. Six days postinduction, the degree of differentiation was scored by measuring alkaline phosphatase (ALPL) activity, displayed relative to the scrambled siRNA–transfected control hMSCs: (*) P < 0.05, Student's t-test. Background coloring is based on the observed quantitative proteomics changes during differentiation: (red) decreased protein level; (green) increased protein level; (yellow) altered phosphorylation.
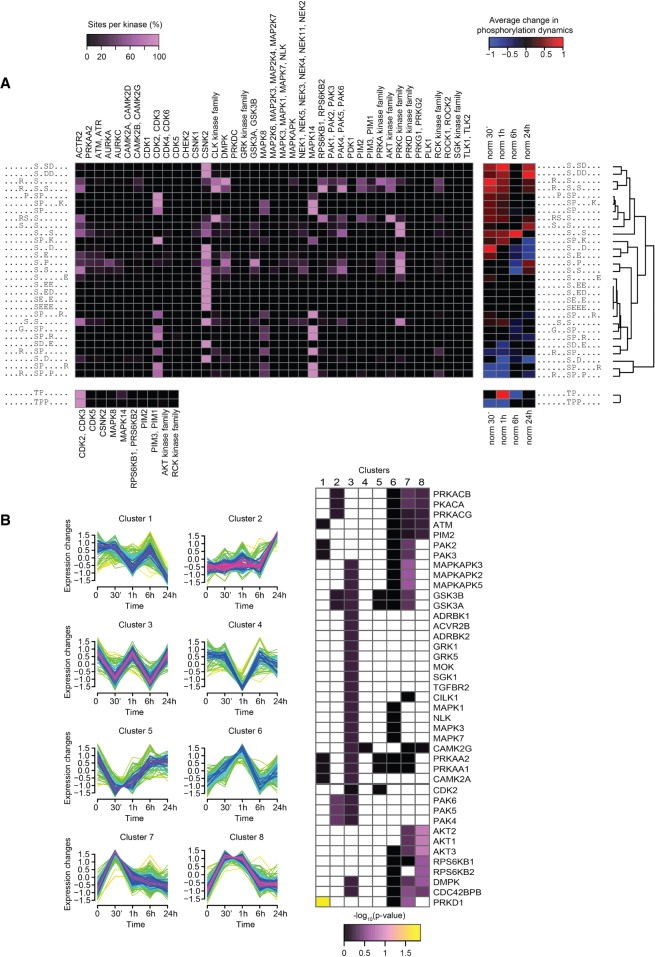
We next extracted overrepresented linear phosphorylation motifs from the identified phosphorylation sites and possible kinases capable of phosphorylating these motifs (Fig. 5A, left). The predictions were then correlated with the quantified dynamics of the corresponding phosphorylation sites (Fig. 5A, right). This depiction allows identification of kinases affiliated with highly regulated motifs and their dynamic changes during OB differentiation. The prediction identified the mitogen-activated protein kinase 14 (MAPK14, also known as p38) as a putative kinase for many of the regulated sites (Fig. 5A) and indeed MAPK14 is an important known regulator of OB differentiation (Rodríguez-Carballo et al. 2016). However, other MAPK members could not be excluded as possible candidates because the phosphorylation motif is shared among them. We identified activating phosphorylation sites on two of these kinases, MAPK3 and MAPK1 (also known as ERK1 and ERK2, respectively), both peaking at 30 min (Supplemental Table S2). Phosphorylation of MAPK14 itself was not detected in the MS analyses, but western blot analysis revealed that, in contrast to ERKs, MAPK14 maintained high levels of activation even at 1 h (Supplemental Fig. S3A).
Figure 5.
Motif analyses revealed kinases with likely involvement in osteoblastogenesis. (A) Significantly overrepresented linear phosphorylation motifs were identified with Motif X and matched to kinases predicted with NetworKIN (Linding et al. 2008), in which an increase in purple intensity equals an increase in the percentage of the given site that is predicted to be phosphorylatable by the given kinase (left). The identified motifs were hierarchically clustered according to the average phosphorylation dynamics of the sites matching each motif. The average change in phosphorylation of sites matching the motif in each time point is color-coded blue for decreased or red for increased compared to undifferentiated cells. Only average phosphorylation changes with a mean significantly different from zero on confidence level 0.01 were included. (B) Class I sites were clustered according to their temporal dynamics (left). Fisher's exact test with Benjamini–Hochberg adjustment was used to identify kinases with significant overrepresentation of substrates within any of the clusters (right).
To obtain further insights into the involvement of MAPKs, we extracted the 1287 predicted phosphorylation sites for the MAPK family present in our data set, clustered them according to their dynamic changes, and assessed for overrepresented biological processes in the corresponding clusters (Supplemental Fig. S3B,C). We observed that sites correlating with the dynamics of MAPK14 activation, that is, with increased phosphorylation at the first two time points after stimulation and peaking at 1 h (Cluster 3), were enriched in categories associated with OB lineage commitment, for example, “osteoblast development” and “positive regulation of osteoblast differentiation.” This was not the case for proteins belonging to clusters that resemble the activation profile of the ERKs with its maximum effect after 30 min. This argues that despite several of the MAPK family members were activated, MAPK14 was crucial in mediating the early steps in OB conversion of hMSCs. In this Cluster 3, we also identified other terms associated with MAPK14 activity such as “transcription from RNA polymerase II promoter” and “positive regulation of transcription from RNA polymerase II promoter,” including sites Ser320 in FOSL2, Ser666 in HCFC1, Ser90 and Ser100 in JUND, Ser250 in NFIX, Ser727 in STAT1, and Ser371 in TP53BP1. The recruitment of RNA polymerase II complex to regulate transcription requires MAPK14 activity (Ferreiro et al. 2010); furthermore, this process has also been related with the chromatin remodeling necessary for OB differentiation (Villagra et al. 2006).
Besides MAPK14, we identified other kinases possibly involved in the regulation of commitment to osteoblastic lineage. Several putative sites for AKT protein kinases and their downstream targets, RPS6KB1 (ribosomal protein S6 kinase B1) and RPS6KB2, were observed to be regulated immediately following OB induction. Conversely, the promiscuous casein kinase 2 alpha catalytic subunits (CSNK2A1–3) were found to be a possible kinase for many of the overrepresented but not regulated sites (Fig. 5A).
As a complementary approach, we grouped regulated phosphorylation sites into eight clusters using fuzzy c-means (Fig. 5B). Although some phosphorylations were only affected at 24 h (Cluster 2), most sites underwent changes during earlier time points and returned close to baseline levels (Clusters 3–8), indicating temporal and stage-specific effects. We then performed separate KEGG pathways and kinase substrate enrichment analyses to explore for discrete signaling pathways and individual kinases, respectively, with significant overrepresentation within any of the clusters. We observed enrichment of several receptor tyrosine kinase (RTK) signaling pathways, for example, EGF, VEGFA, and insulin in Clusters 6–8 (Supplemental Table S3). In agreement with the preceding findings arguing for an involvement of the MAPKs and the placement of these kinases as nodal points in RTK networks, MAPK signaling was also tightly associated with the same clusters (Supplemental Table S3). Besides MAPK, other signaling cascades known to be involved in OB differentiation, like MTOR and FOXO pathways, were enriched in these clusters as well (Teixeira et al. 2010; Pramojanee et al. 2014; Dai et al. 2017). An additional set of sites that showed increased phosphorylation at the onset of differentiation (Cluster 8) were affiliated with another nodal point kinase family, AKT (Fig. 5B), reflecting the role of the AKT signaling in the initial phase of OB differentiation. Collectively, Clusters 6–8 could therefore be assigned to a first wave of phosphorylation events and appear to be a compilation of several signaling cascades.
Regarding the pathways associated with the second activation wave, sites targeted by group II of the PAK (p21 [RAC1] activated kinase) family of kinases (PAK4, PAK5, and PAK6) were overrepresented in Cluster 2 containing sites with up-regulation at 24 h with little or no fluctuations at earlier time points (Fig. 5B). Although this group of kinases has been implicated in several biological processes and tumor formation, its role in OB biology remains unexplored (Wells and Jones 2010). Of note, proteins involved in Hippo and TGFB signaling were up-regulated at their protein levels at this latest time point (Supplemental Fig. S1), arguing for an involvement of these signaling pathways in the second kinase activation wave as well. A pattern therefore emerges, in which several intertwined kinase-signaling pathways commonly initiated by RTKs are activated, and their downstream effectors MAPK and AKT act as the radial points during the first wave. As these routes cling off, a more defined program is initiated, involving PAK, TGFΒ, and Hippo signaling networks.
PRKD1 regulates osteoblast differentiation via HDAC7 phosphorylation
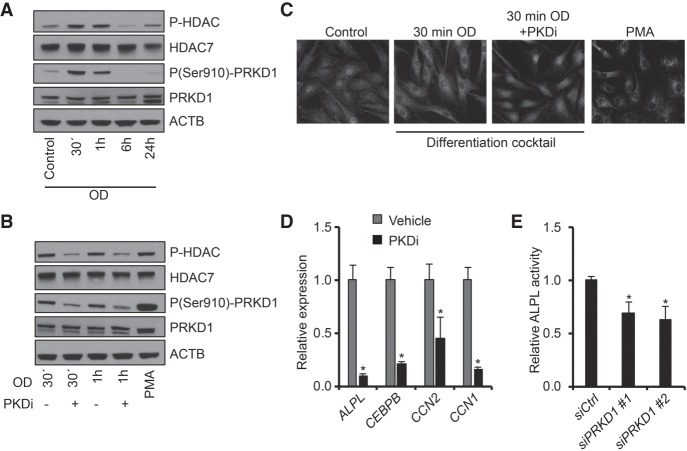
From the kinase prediction analyses, serine/threonine protein kinase D1 (PRKD1) showed the highest enrichment with substrates predominantly found in Clusters 1 and 7 (Fig. 5B). In accordance, western blotting with phospho-specific antibody revealed that the activating Ser910 phosphorylation on PRKD1 was indeed augmented 30 min after induction and declined to baseline levels at the 6- and 24-h time points (Fig. 6A; Supplemental Fig. S4A). We therefore assessed the involvement of PRKD1 in the commitment of hMSCs to osteoblastic cells using a PRKD1-specific chemical inhibitor, CRT0066101 (Harikumar et al. 2010). The inhibition of PRKD1 had only a mild effect on the early phosphorylation status of MAPK14, a canonical target of PRKD1 (Supplemental Fig. S3A). We also examined the effect of stimulating hMSCs with phorbol myristate acetate (PMA), which is a potent activator of several members of the PKC family of kinases, including PRKD1. Despite the very strong activation of PRKD1 (Fig. 6B; Supplemental Fig. S4B), the PMA treatment of the cells did not augment the phosphorylation of MAPK14 (Supplemental Fig. S3A), further indicating that MAPK14 is not the main target of PRKD1 in the initial stages of OB lineage commitment.
Figure 6.
PRKD1 regulates HDAC7 localization in immortalized hMSCs. (A) Western blot images of HDAC and PRKD1 phosphorylation levels during osteoblast differentiation, n = 2. (B) Western blot of HDAC and PRKD1 phosphorylation 30 min and 1 h after induction of osteoblastic differentiation, with and without PRKD1 inhibitor (PKDi), n = 2. (C) Detection of HDAC7 in unstimulated hMSCs (Control), 30 min after osteoblastic induction (30 min OD) with and without PKDi or PMA-treated hMSCs, n = 3. (D) mRNA levels of CTGF, CEBPD, CYR61, and ALPL in relative expression 24 h after osteoblastic differentiation with and without PKDi. (E) Quantitation of alkaline phosphatase (ALPL) activity on day 6 of osteoblast differentiation of hMSC transfected with control nontargeting siRNA (siCtrl) or two independent siRNAs targeting PRKD1 (siPRKD1 #1, siPRKD1 #2). For D and E: (*) P < 0.05, Student's t-test; n = 3.
It has previously been shown that PRKD1 regulates the cytoplasmic-nuclear shuttling of members of the class IIa histone deacetylases and transcriptional repressors (HDAC4, −5 and −7) by direct phosphorylation (Dequiedt et al. 2005; Parra et al. 2005; Sinnett-Smith et al. 2014). We therefore hypothesized that PRKD1 may regulate OB differentiation through phosphorylation and relocalization of one or more of the HDACs. Temporal profile of the phosphorylation status of the residue in the three HDACs (HDAC4-Ser632/HDAC5-Ser661/HDAC7-Ser486) controlling for their nuclear-cytoplasmic shuttling and targeted by PRKD1, was virtually identical to the activation pattern of the kinase (Fig. 6A; Supplemental Fig. S4A). In support of PRKD1 being responsible for this phosphorylation, CRT0066101 treatment resulted in decreased HDAC phosphorylation, whereas activation of PRKD1 by PMA increased the phosphorylation of HDACs (Fig. 6B; Supplemental Fig. S4B). Although the phospho-specific antibody recognizes all three HDACs when phosphorylated at these positions, the phosphorylation dynamics of HDAC4 Ser632 and HDAC5 Ser661 were quantified in the MS analyses and neither showed significant changes (Supplemental Table S2), indicating that the PRKD1 effect is mediated mainly through phosphorylation of HDAC7 Ser486. Because this phosphorylation site is known to regulate HDAC cellular localization (Parra et al. 2005), we assessed the effect of the PRKD1 inhibitor on the distribution of HDAC7 in hMSCs. Upon induction of OB differentiation, HDAC7 was translocated from the nucleus to the cytoplasm in vehicle treated cells. This relocalization was completely prevented by the PRKD1 inhibitor (Fig. 6C; Supplemental Fig. S4C), suggesting that PRKD1 regulates OB commitment of hMSCs through phosphorylation and nuclear exclusion of the RUNX2-repressor HDAC7. In support, the mRNA levels of four RUNX2 target genes and early osteoblast gene markers were all significantly decreased by the PRKD1 inhibitor (Fig. 6D). In addition, siRNA-mediated knockdown of PRKD1 in hMSCs resulted in reduced alkaline phosphatase activity on day 6 of differentiation (Fig. 6E), further indicating the functional importance of PRKD1 for the OB differentiation.
Discussion
Lineage commitment and differentiation of hMSCs into osteoblastic cells is a complex cellular process. Our analyses provide a first global view of the complex proteomic and phosphoproteomic changes during the initial 24 h of hMSCs transition to the OB lineage. We identified 6252 proteins and 15,059 unique phosphorylation sites during the early steps of OB conversion. The data exposed that osteoblast commitment involves complex makeover already at the first 24 h, including changes in phosphoproteins associated with cell proliferation, extracellular matrix, and vascular remodeling, all known to be important for efficient OB differentiation (Lian and Stein 1995).
By modulating gene expression, transcription factors are responsible for a large part of the dynamics in the proteome. We found that the expression of several members of the bZIP family was increased during OB differentiation, arguing for a strong involvement of these transcription factors in regulating OB commitment. Both CEBPB and CEBPD were up-regulated, and both have previously been shown to be involved in osteoblastic lineage commitment through cooperative actions with RUNX2 (Gutierrez et al. 2002; Tominaga et al. 2008). In contrast, another bZIP factor devoid of a transactivating domain, namely MAFK, was down-regulated at all time points (Fig. 2). It is possible that MAFK restricts OB differentiation because it has the ability to bind and repress other bZIPs and especially MAF (Kataoka et al. 1996), which plays an essential role in OB development through interplay with RUNX2 (Nishikawa et al. 2010). Moreover, we observed temporal inhibition of the NF-kB transcription factors and Rho activity (Supplemental Fig. S1). These observations are also in agreement with previous data showing a strong inhibitory effect on OB differentiation by the two signaling pathways (Novack 2011; Strzelecka-Kiliszek et al. 2017).
The dynamics of regulated phosphorylation sites and protein expression changes indicated the existence of two consecutive signaling waves, the first peaking immediately after induction of differentiation followed by a second commencing 24 h after initiation (Supplemental Fig. S5). During the first wave, a myriad of signaling cascades are activated, including a number of RTK pathways. The exact contribution of each signaling component awaits further analyses, but it is likely that their mutual crosstalk is of pivotal importance for a balanced and timed initiation of OB differentiation. This is exemplified by the fact that several of the phosphorylated proteins are embedded in more than one pathway. RAF1 and TSC2, when phosphorylated on Ser29 and Ser1319, respectively, are both key nodes in interaction between MTOR and MAPK pathways (Mendoza et al. 2011). Other sites such as Ser2 in RRAGC or Ser226 in MAP2K2 are known to have the potential to activate each of the pathways independently. Both pathways can converge on the same transcription factors such as FOXO (Mendoza et al. 2011), whose localization is also affected by STAT3 (Oh et al. 2012), which we find phosphorylated at Ser727 as well (Supplemental Table S2). Collectively, numerous pathways with several shared nodal points are activated at this first wave during differentiation.
The second wave of signaling appears to rely on a completely different set of signaling cascades (Supplemental Fig. S5). Of the lesser studied but highly enriched pathways in our data, the up-regulation of the Hippo pathway suggests strong involvement of this cascade 24 h after induction of differentiation (Lee et al. 2015). This concurs with an up-regulation of the expression of transforming growth factor beta receptor 2 (TGFBR2), arguing for crosstalk between these two pathways in the OB commitment as they also cooperate in other biological processes (Luo 2017). KEGG-based enrichment analysis of phosphorylation sites augmented at this later stage suggested activation of the DNA repair machinery as well (Supplemental Table S3). The DNA damage response was previously associated with differentiation processes (Sherman et al. 2011), arguing that maintaining genomic integrity is also essential for the progress of OB conversion. In our data we found XPC, known member of pluripotency-maintaining complex (Cattoglio et al. 2015), to become phosphorylated at Ser883 and Ser884. The LIG1 and LIG3 proteins, reported to be involved in human neural crest stem cell differentiation (Newman et al. 2017), were phosphorylated at Ser51 and Thr244, respectively, as well (Supplemental Table S2). Moreover, substrates for the group II of the PAK kinases were increasingly phosphorylated at the 24 h time point. These kinases are at least partially activated by the small GTPases, CDC42 and RAC1. CDC42 has been shown to induce chondrogenesis, but not OB differentiation, downstream from TGFB stimulation through the group I PAK member PAK1 and MAPK14 (Wang et al. 2016). It is therefore conceivable that the diverging groups of this family of kinases integrates upstream signaling events and balances osteoblast versus chondrocyte lineage commitment.
As a follow up on the global changes during OB differentiation, we identified a mechanistic involvement of PRKD1. At first, two complementary approaches that predict kinases responsible for the dynamic changes in the phosphoproteome at the onset of OB differentiation revealed pivotal roles for MAPK14 and PRKD1 kinases. Members of the MAPK family, and notably MAPK14, are well known for their involvement in OB lineage commitment (Rodríguez-Carballo et al. 2016). PRKD1 has also at several instances been described as a key mediator of the bone morphogenetic protein (BMP)-induced OB differentiation, mainly through activation of the MAPK14 (Lemonnier et al. 2004; Celil and Campbell 2005; Kim et al. 2011). In our study, the addition of a PRKD1 inhibitor did not affect the level of the activating phosphorylation of MAPK14, suggesting independent effects of the two kinases within the first hours of OB differentiation. In agreement with a previous study focused on BMP signaling (Jensen et al. 2009), we observed a PRKD1-dependent nuclear exclusion of the RUNX2-repressive HDAC7. It is therefore likely that part of the stimulatory effect of PRKD1 on OB differentiation is mediated through the relief of RUNX2 from HDAC7-mediated repression.
Our study shows that lineage commitment of hMSCs can be conceptualized as coordinated successive waves of interacting signaling pathways, where the “choir is more important than the solo” (Kassem and Bianco 2015). From a translation perspective, our findings suggest a possibility for a more flexible, multiple protein targeting approach to better control hMSCs lineage commitment to osteoblastic cells and bone regeneration, an important application in regenerative medicine.
Methods
Cell culture, SILAC labeling, and differentiation
As a model for human primary MSCs, we used hMSC-TERT cell line that has been immortalized by ectopic expression of telomerase reverse transcriptase gene (hTERT) (Simonsen et al. 2002). For simplicity, we will refer to the hMSC-TERT as hMSCs throughout the paper. For SILAC experiments, cells were labeled with L-arginine (Arg0) and L-lysine (Lys0) for the unstimulated cells, L-arginine-13C6 (Arg6) and L-lysine-2H4 (Lys4) for the 30-min and 1-h conditions, or L-arginine-13C615N4 (Arg10) and L-lysine-13C615N2 (Lys8) for the 6- and 24-h conditions (Cambridge Isotope Laboratories) at concentrations of 28 and 42 mg/L, respectively. Differentiation of hMSCs cells into the osteoblastic lineage was performed as described previously (Kratchmarova et al. 2005). For PRKD1 inhibition, CRT0066101 (Abcam) was added 30 min before the induction of OB differentiation at a final concentration of 10 µM. For PRKD1 activation, phorbol myristate acetate (PMA) (Sigma-Aldrich) was used at a final concentration of 150 ng/mL.
Sample processing for LC-MS/MS analysis
Two triple-encoding SILAC experiments were performed in parallel for each replicate (total of three biological replicates). Cells were stimulated for 30 min, 1 h, 6 h, or 24 h, rinsed twice with cold PBS and lysed in 50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 0.1% sodium deoxycholate, and 1 mM EDTA supplemented with phosphatase and protease inhibitors (Roche). Nuclei were pelleted by centrifugation for 10 min at 10,000g at 4°C. Cytoplasmic proteins in the supernatant were precipitated using acetone and resuspended in 10 mM Tris, pH 7.5, with 6 M urea and 2 M thiourea. The same buffer, supplemented with Benzonase, was used for resuspension of nuclear proteins. Protein concentrations were determined using Bradford (Bio-Rad) according to the manufacturer's instructions. Samples were combined in a 1:1:1 ratio (L:M:H). Proteins were reduced with 10 mM dithiothreitol and alkylated with 55 mM iodoacetamide. For protein identification, 100 µg protein of each SILAC pool were separated using 1D SDS-polyacrylamide gel electrophoresis on NuPAGE Novex gels (Life Technologies). The gel was stained with Colloidal Blue (Life Technologies), cut into pieces, and digested with trypsin (Promega) overnight at 37°C. Resulting peptide mixture was extracted and desalted using modified Stage tips as described elsewhere (Akimov et al. 2011). The remainder of the protein lysates was used for phosphopeptide enrichment before MS analysis. Proteins were digested in solution for 4 h at room temperature with LysC (Wako Chemicals) followed by trypsin overnight (Akimov et al. 2018), used in a ratio of 1:160 and 1:200, respectively, relative to protein content. Peptide solutions were acidified with trifluoroacetic acid to a final concentration of 0.1% and cleared by centrifugation (20,000g, 5 min). Phosphopeptides were enriched and fractionated using TiO2 as described earlier (Rigbolt et al. 2011a).
LC-MS/MS analysis
Peptides were separated using an in-house packed fused silica column with a length of 15 cm, an inner diameter of 75 µm and reverse phase material (3 µm C18, ReproSil-Pur C18 AQ) (Dr. Maisch HPLC GmbH, Ammerbuch Germany). EASY-nLC (Proxeon), Agilent 1100, or Agilent 1200 systems were used, generating linear gradients of 90 min (solvent A was 0.5% acetic acid in water, and solvent B was 80% Acetonitrile and 0.5% acetic acid) at a constant flow of 250 nL/min. Mass spectrometry was performed in positive ion mode using LTQ-Orbitrap XL for total protein coverage (data-dependent acquisition for isolation and fragmentation of the 10 most intense ions, CID fragmentation) and LTQ-Orbitrap Velos and Q-Exactive for phospho-enriched samples (data-dependent acquisition for isolation and fragmentation of the 10 most intense ions for Velos and the 12 most intense for Q-Exactive, HCD fragmentation) (all from Thermo Fisher Scientific).
Data analysis is described in detail in Supplemental Methods. Briefly, all MS raw files from the three biological replicas were processed together using the MaxQuant with FDR of 1% set for both protein and site identification. Protein with shared peptides were combined in protein groups, and both unique and razor peptides were used for quantitation.
Clustering of protein and sites based on their temporal profiles
Phosphosite SILAC ratios were normalized where possible by dividing the phosphosite ratio with total protein ratio to ensure that changes were caused by the dynamics of the site and not the protein. Resulting values were log2 transformed and subjected to unsupervised clustering with the fuzzy c-means algorithm implemented in the GProX software (Rigbolt et al. 2011b). Only sites with a complete profile, P < 0.05 (Significance A, MaxQuant) and a ratio with at least a twofold increase or decrease in at least one time point were considered. Enrichment analysis for kinase substrates within the clusters were performed using Fisher's exact test with Benjamini–Hochberg adjustment for multiple testing (Benjamini and Hochberg 1995). Protein ratios were subjected to the same method, designating as changing proteins with P < 0.05 (Significance A, MaxQuant) with at least 1.5-fold increase or decrease in at least one point. Details for additional bioinformatic analyses are provided in Supplemental Methods.
Immunocytochemistry and western blotting
Immunocytochemistry was performed as previously described (Sanchez-Quiles et al. 2017). Briefly, cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with horse serum. Cells were incubated with mouse anti-HDAC7 antibody (Santa Cruz Biotechnology sc-74563), rinsed, incubated with anti-mouse/Cy3 secondary antibody (Jackson ImmunoResearch 715-165-150) plus DAPI (Sigma-Aldrich), and rinsed before mounted on cover slides. Images were acquired using an Observer Z1 microscope (Zeiss). See Supplemental Methods for western blotting details.
Quantitative real-time polymerase chain reaction (qRT-PCR) and siRNA transfections
Relative mRNA levels were measured by qRT-PCR as previously described (Hallenborg et al. 2016; Supplemental Methods). A reverse transfection protocol using Lipofectamine 2000 (Invitrogen) was used for the siRNA transfections. hMSC cultures were trypsinized, and 18,000 cells/cm2 were reverse-transfected with two independent siRNAs targeting the kinases (Supplemental Methods) in two biological replicates for each siRNA. For PRKD1, the experiment was performed in triplicate for each of its siRNAs.
Quantitation of alkaline phosphatase (ALPL) activity
To quantify alkaline phosphatase activity, p-nitrophenyl phosphate (Fluka) was used as substrate and to correct for differences in cell number, ALPL activity data were normalized to cell viability measurements as described previously (Jafari et al. 2017). Briefly, CellTiter-Blue reagent (Promega) was added to the normal culture medium, incubated for 1 h at 37°C, and fluorescent intensity (560EX/590EM) was measured using FLUOstar Omega multimode microplate reader (BMG Labtech). Cells were then washed with Tris-buffered saline (TBS), fixed in 3.7% formaldehyde, 90% ethanol at room temperature for 30 sec, incubated with substrate (1 mg/mL of p-nitrophenyl phosphate in 50 mM NaHCO3, pH 9.6, and 1 mM MgCl2) for 20 min at 37°C, and the absorbance measured at 405 nm, using FLUOstar Omega multimode microplate reader.
Data access
The mass spectrometric data generated in this study have been submitted to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD011001.
Supplementary Material
Acknowledgments
This work was in part funded by the Lundbeck Foundation, Danish National Research Foundation (DNRF grant no. 141), Innovation Fund Denmark, Danish Strategic Research Council, Danish Medical Research Council, Novo Nordisk Foundation (NNF18OC0052768), and Danish Council for Technology and Production Sciences (DFF, 8022-00051). The work was also supported in part by the Villum Foundation through the Villum Center for Bioanalytical Sciences. We thank the Danish National Mass Spectrometry Platform for Functional Proteomics (PRO-MS) for instrument support and assistance.
Author contributions: I.B.-H., K.T.G.R., P.H., V.S.-Q., I.S., A.J., and V.A. performed the experiments. I.B.-H., K.T.G.R., I.K., J.D., and B.B. analyzed data. I.K., J.D., M.K., and B.B. supervised the project. I.B.-H., P.H., M.K., and B.B. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.248286.119.
References
- Abdallah BM, Jafari A, Zaher W, Qiu W, Kassem M. 2015. Skeletal (stromal) stem cells: an update on intracellular signaling pathways controlling osteoblast differentiation. Bone 70: 28–36. 10.1016/j.bone.2014.07.028 [DOI] [PubMed] [Google Scholar]
- Akimov V, Rigbolt KT, Nielsen MM, Blagoev B. 2011. Characterization of ubiquitination dependent dynamics in growth factor receptor signaling by quantitative proteomics. Mol Biosyst 7: 3223–3233. 10.1039/c1mb05185g [DOI] [PubMed] [Google Scholar]
- Akimov V, Barrio-Hernandez I, Hansen SVF, Hallenborg P, Pedersen AK, Bekker-Jensen DB, Puglia M, Christensen SDK, Vanselow JT, Nielsen MM, et al. 2018. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat Struct Mol Biol 25: 631–640. 10.1038/s41594-018-0084-y [DOI] [PubMed] [Google Scholar]
- Ambrogini E, Almeida M, Martin-Millan M, Paik JH, Depinho RA, Han L, Goellner J, Weinstein RS, Jilka RL, O'Brien CA, et al. 2010. FoxO-mediated defense against oxidative stress in osteoblasts is indispensable for skeletal homeostasis in mice. Cell Metab 11: 136–146. 10.1016/j.cmet.2009.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrzejewska A, Lukomska B, Janowski M. 2019. Concise review: mesenchymal stem cells: from roots to boost. Stem Cells 37: 855–864. 10.1002/stem.3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Cattoglio C, Zhang ET, Grubisic I, Chiba K, Fong YW, Tjian R. 2015. Functional and mechanistic studies of XPC DNA-repair complex as transcriptional coactivator in embryonic stem cells. Proc Natl Acad Sci 112: E2317–E2326. 10.1073/pnas.1505569112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celil AB, Campbell PG. 2005. BMP-2 and insulin-like growth factor-I mediate Osterix (Osx) expression in human mesenchymal stem cells via the MAPK and protein kinase D signaling pathways. J Biol Chem 280: 31353–31359. 10.1074/jbc.M503845200 [DOI] [PubMed] [Google Scholar]
- Chen MT, Dong L, Zhang XH, Yin XL, Ning HM, Shen C, Su R, Li F, Song L, Ma YN, et al. 2015. ZFP36L1 promotes monocyte/macrophage differentiation by repressing CDK6. Sci Rep 5: 16229 10.1038/srep16229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Xu Z, Ma X, Niu N, Zhou S, Xie F, Jiang L, Wang J, Zou W. 2017. mTOR/Raptor signaling is critical for skeletogenesis in mice through the regulation of Runx2 expression. Cell Death Differ 24: 1886–1899. 10.1038/cdd.2017.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequiedt F, Van Lint J, Lecomte E, Van Duppen V, Seufferlein T, Vandenheede JR, Wattiez R, Kettmann R. 2005. Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor–induced Nur77 expression and apoptosis. J Exp Med 201: 793–804. 10.1084/jem.20042034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, McDonald JM. 2011. Disorders of bone remodeling. Annu Rev Pathol 6: 121–145. 10.1146/annurev-pathol-011110-130203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreiro I, Barragan M, Gubern A, Ballestar E, Joaquin M, Posas F. 2010. The p38 SAPK is recruited to chromatin via its interaction with transcription factors. J Biol Chem 285: 31819–31828. 10.1074/jbc.M110.155846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez S, Javed A, Tennant DK, van Rees M, Montecino M, Stein GS, Stein JL, Lian JB. 2002. CCAAT/enhancer-binding proteins (C/EBP) β and δ activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J Biol Chem 277: 1316–1323. 10.1074/jbc.M106611200 [DOI] [PubMed] [Google Scholar]
- Hallenborg P, Siersbaek M, Barrio-Hernandez I, Nielsen R, Kristiansen K, Mandrup S, Grontved L, Blagoev B. 2016. MDM2 facilitates adipocyte differentiation through CRTC-mediated activation of STAT3. Cell Death Dis 7: e2289 10.1038/cddis.2016.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar KB, Kunnumakkara AB, Ochi N, Tong Z, Deorukhkar A, Sung B, Kelland L, Jamieson S, Sutherland R, Raynham T, et al. 2010. A novel small-molecule inhibitor of protein kinase D blocks pancreatic cancer growth in vitro and in vivo. Mol Cancer Ther 9: 1136–1146. 10.1158/1535-7163.MCT-09-1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henningsen J, Rigbolt KT, Blagoev B, Pedersen BK, Kratchmarova I. 2010. Dynamics of the skeletal muscle secretome during myoblast differentiation. Mol Cell Proteomics 9: 2482–2496. 10.1074/mcp.M110.002113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo H, Ohba S, Chung U-il. 2015. Signaling pathways regulating the specification and differentiation of the osteoblast lineage. Regen Ther 1(Supplement C): 57–62. 10.1016/j.reth.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari A, Harkness L, Zaher W, Kassem M. 2014. Adult stromal (skeletal, mesenchymal) stem cells: advances towards clinical applications. In Adult stem cells (ed. Turksen K), pp. 359–373. Springer, New York. [Google Scholar]
- Jafari A, Qanie D, Andersen TL, Zhang Y, Chen L, Postert B, Parsons S, Ditzel N, Khosla S, Johansen HT, et al. 2017. Legumain regulates differentiation fate of human bone marrow stromal cells and is altered in postmenopausal osteoporosis. Stem Cell Rep 8: 373–386. 10.1016/j.stemcr.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen ED, Gopalakrishnan R, Westendorf JJ. 2009. Bone morphogenic protein 2 activates protein kinase D to regulate histone deacetylase 7 localization and repression of Runx2. J Biol Chem 284: 2225–2234. 10.1074/jbc.M800586200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassem M, Bianco P. 2015. Skeletal stem cells in space and time. Cell 160: 17–19. 10.1016/j.cell.2014.12.034 [DOI] [PubMed] [Google Scholar]
- Kataoka K, Noda M, Nishizawa M. 1996. Transactivation activity of Maf nuclear oncoprotein is modulated by Jun, Fos and small Maf proteins. Oncogene 12: 53–62. [PubMed] [Google Scholar]
- Kim DY, Jung MS, Park YG, Yuan HD, Quan HY, Chung SH. 2011. Ginsenoside Rh2(S) induces the differentiation and mineralization of osteoblastic MC3T3-E1 cells through activation of PKD and p38 MAPK pathways. BMB Rep 44: 659–664. 10.5483/BMBRep.2011.44.10.659 [DOI] [PubMed] [Google Scholar]
- Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M. 2005. Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science 308: 1472–1477. 10.1126/science.1107627 [DOI] [PubMed] [Google Scholar]
- Kristensen LP, Chen L, Nielsen MO, Qanie DW, Kratchmarova I, Kassem M, Andersen JS. 2012. Temporal profiling and pulsed SILAC labeling identify novel secreted proteins during ex vivo osteoblast differentiation of human stromal stem cells. Mol Cell Proteomics 11: 989–1007. 10.1074/mcp.M111.012138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan MC, Barrio-Hernandez I, Mortensen TP, Henningsen J, Jensen SS, Bigot A, Blagoev B, Butler-Browne G, Kratchmarova I. 2015. Cellular proteome dynamics during differentiation of human primary myoblasts. J Proteome Res 14: 3348–3361. 10.1021/acs.jproteome.5b00397 [DOI] [PubMed] [Google Scholar]
- Lee J, Youn BU, Kim K, Kim JH, Lee DH, Seong S, Kim I, Han SH, Che X, Choi JY, et al. 2015. Mst2 controls bone homeostasis by regulating osteoclast and osteoblast differentiation. J Bone Miner Res 30: 1597–1607. 10.1002/jbmr.2503 [DOI] [PubMed] [Google Scholar]
- Lemonnier J, Ghayor C, Guicheux J, Caverzasio J. 2004. Protein kinase C-independent activation of protein kinase D is involved in BMP-2-induced activation of stress mitogen-activated protein kinases JNK and p38 and osteoblastic cell differentiation. J Biol Chem 279: 259–264. 10.1074/jbc.M308665200 [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47: W256–W259. 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JB, Stein GS. 1995. Development of the osteoblast phenotype: molecular mechanisms mediating osteoblast growth and differentiation. Iowa Orthop J 15: 118–140. [PMC free article] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Pasculescu A, Olhovsky M, Colwill K, Bork P, Yaffe MB, Pawson T. 2008. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res 36(Database issue): D695–D699. 10.1093/nar/gkm902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K. 2017. Signaling cross talk between TGF-β/Smad and other signaling pathways. Cold Spring Harb Perspect Biol 9: a022137 10.1101/cshperspect.a022137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto A, Milani R, da Silva RA, da Costa Fernandes CJ, Granjeiro JM, Ferreira CV, Peppelenbosch MP, Zambuzzi WF. 2017. Phosphoproteome analysis reveals a critical role for hedgehog signalling in osteoblast morphological transitions. Bone 103: 55–63. 10.1016/j.bone.2017.06.012 [DOI] [PubMed] [Google Scholar]
- Mendoza MC, Er EE, Blenis J. 2011. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci 36: 320–328. 10.1016/j.tibs.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman EA, Chukkapalli S, Bashllari D, Thomas TT, Van Noord RA, Lawlor ER, Hoenerhoff MJ, Opipari AW, Opipari VP. 2017. Alternative NHEJ pathway proteins as components of MYCN oncogenic activity in human neural crest stem cell differentiation: implications for neuroblastoma initiation. Cell Death Dis 8: 3208 10.1038/s41419-017-0004-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K, Nakashima T, Takeda S, Isogai M, Hamada M, Kimura A, Kodama T, Yamaguchi A, Owen MJ, Takahashi S, et al. 2010. Maf promotes osteoblast differentiation in mice by mediating the age-related switch in mesenchymal cell differentiation. J Clin Invest 120: 3455–3465. 10.1172/JCI42528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novack DV. 2011. Role of NF-κB in the skeleton. Cell Res 21: 169–182. 10.1038/cr.2010.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HM, Yu CR, Dambuza I, Marrero B, Egwuagu CE. 2012. STAT3 protein interacts with class O Forkhead transcription factors in the cytoplasm and regulates nuclear/cytoplasmic localization of FoxO1 and FoxO3a proteins in CD4+ T cells. J Biol Chem 287: 30436–30443. 10.1074/jbc.M112.359661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfitt M, Qiu S, Palnitkar S, Rao DS. 2011. Abnormal bone remodeling in patients with spontaneous painful vertebral fracture. J Bone Miner Res 26: 475–485. 10.1002/jbmr.239 [DOI] [PubMed] [Google Scholar]
- Parra M, Kasler H, McKinsey TA, Olson EN, Verdin E. 2005. Protein kinase D1 phosphorylates HDAC7 and induces its nuclear export after T-cell receptor activation. J Biol Chem 280: 13762–13770. 10.1074/jbc.M413396200 [DOI] [PubMed] [Google Scholar]
- Pramojanee SN, Phimphilai M, Chattipakorn N, Chattipakorn SC. 2014. Possible roles of insulin signaling in osteoblasts. Endocr Res 39: 144–151. 10.3109/07435800.2013.879168 [DOI] [PubMed] [Google Scholar]
- Prokhorova TA, Rigbolt KT, Johansen PT, Henningsen J, Kratchmarova I, Kassem M, Blagoev B. 2009. Stable isotope labeling by amino acids in cell culture (SILAC) and quantitative comparison of the membrane proteomes of self-renewing and differentiating human embryonic stem cells. Mol Cell Proteomics 8: 959–970. 10.1074/mcp.M800287-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt KT, Blagoev B. 2012. Quantitative phosphoproteomics to characterize signaling networks. Semin Cell Dev Biol 23: 863–871. 10.1016/j.semcdb.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B. 2011a. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal 4: rs3 10.1126/scisignal.2001570 [DOI] [PubMed] [Google Scholar]
- Rigbolt KT, Vanselow JT, Blagoev B. 2011b. GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol Cell Proteomics 10: O110.007450 10.1074/mcp.O110.007450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Carballo E, Gámez B, Ventura F. 2016. p38 MAPK signaling in osteoblast differentiation. Front Cell Dev Biol 4: 40 10.3389/fcell.2016.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Quiles V, Akimov V, Osinalde N, Francavilla C, Puglia M, Barrio-Hernandez I, Kratchmarova I, Olsen JV, Blagoev B. 2017. Cylindromatosis tumor suppressor protein (CYLD) deubiquitinase is necessary for proper ubiquitination and degradation of the epidermal growth factor receptor. Mol Cell Proteomics 16: 1433–1446. 10.1074/mcp.M116.066423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MH, Bassing CH, Teitell MA. 2011. Regulation of cell differentiation by the DNA damage response. Trends Cell Biol 21: 312–319. 10.1016/j.tcb.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen JL, Rosada C, Serakinci N, Justesen J, Stenderup K, Rattan SI, Jensen TG, Kassem M. 2002. Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat Biotechnol 20: 592–596. 10.1038/nbt0602-592 [DOI] [PubMed] [Google Scholar]
- Sinnett-Smith J, Ni Y, Wang J, Ming M, Young SH, Rozengurt E. 2014. Protein kinase D1 mediates class IIa histone deacetylase phosphorylation and nuclear extrusion in intestinal epithelial cells: role in mitogenic signaling. Am J Physiol Cell Physiol 306: C961–C971. 10.1152/ajpcell.00048.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka-Kiliszek A, Mebarek S, Roszkowska M, Buchet R, Magne D, Pikula S. 2017. Functions of Rho family of small GTPases and Rho-associated coiled-coil kinases in bone cells during differentiation and mineralization. Biochim Biophys Acta 1861(5 Pt A): 1009–1023. 10.1016/j.bbagen.2017.02.005 [DOI] [PubMed] [Google Scholar]
- Stumpo DJ, Broxmeyer HE, Ward T, Cooper S, Hangoc G, Chung YJ, Shelley WC, Richfield EK, Ray MK, Yoder MC, et al. 2009. Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood 114: 2401–2410. 10.1182/blood-2009-04-214619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira CC, Liu Y, Thant LM, Pang J, Palmer G, Alikhani M. 2010. Foxo1, a novel regulator of osteoblast differentiation and skeletogenesis. J Biol Chem 285: 31055–31065. 10.1074/jbc.M109.079962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga H, Maeda S, Hayashi M, Takeda S, Akira S, Komiya S, Nakamura T, Akiyama H, Imamura T. 2008. CCAAT/enhancer-binding protein β promotes osteoblast differentiation by enhancing Runx2 activity with ATF4. Mol Biol Cell 19: 5373–5386. 10.1091/mbc.e08-03-0329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng KY, Chen YH, Lin S. 2017. Zinc finger protein ZFP36L1 promotes osteoblastic differentiation but represses adipogenic differentiation of mouse multipotent cells. Oncotarget 8: 20588–20601. 10.18632/oncotarget.15246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof D, Muñoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, Mummery CL, Krijgsveld J. 2009. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5: 214–226. 10.1016/j.stem.2009.05.021 [DOI] [PubMed] [Google Scholar]
- Villagra A, Cruzat F, Carvallo L, Paredes R, Olate J, van Wijnen AJ, Stein GS, Lian JB, Stein JL, Imbalzano AN, et al. 2006. Chromatin remodeling and transcriptional activity of the bone-specific osteocalcin gene require CCAAT/enhancer-binding protein β-dependent recruitment of SWI/SNF activity. J Biol Chem 281: 22695–22706. 10.1074/jbc.M511640200 [DOI] [PubMed] [Google Scholar]
- Wang JR, Wang CJ, Xu CY, Wu XK, Hong D, Shi W, Gong Y, Chen HX, Long F, Wu XM. 2016. Signaling cascades governing Cdc42-mediated chondrogenic differentiation and mensenchymal condensation. Genetics 202: 1055–1069. 10.1534/genetics.115.180109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CM, Jones GE. 2010. The emerging importance of group II PAKs. Biochem J 425: 465–473. 10.1042/BJ20091173 [DOI] [PubMed] [Google Scholar]
- Zaher W, Harkness L, Jafari A, Kassem M. 2014. An update of human mesenchymal stem cell biology and their clinical uses. Arch Toxicol 88: 1069–1082. 10.1007/s00204-014-1232-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.