A scalable recombineering procedure for whole-gene editing in plants via homologous recombination in bacteria was used to tag 62 auxin-related genes in 123 transgenic Arabidopsis lines.
Abstract
Gene functional studies often rely on the expression of a gene of interest as transcriptional and translational fusions with specialized tags. Ideally, this is done in the native chromosomal contexts to avoid potential misexpression artifacts. Although recent improvements in genome editing have made it possible to directly modify the target genes in their native chromosomal locations, classical transgenesis is still the preferred experimental approach chosen in most gene tagging studies because of its time efficiency and accessibility. We have developed a recombineering-based tagging system that brings together the convenience of the classical transgenic approaches and the high degree of confidence in the results obtained by direct chromosomal tagging using genome-editing strategies. These simple, scalable, customizable recombineering toolsets and protocols allow a variety of genetic modifications to be generated. In addition, we developed a highly efficient recombinase-mediated cassette exchange system to facilitate the transfer of the desired sequences from a bacterial artificial chromosome clone to a transformation-compatible binary vector, expanding the use of the recombineering approaches beyond Arabidopsis (Arabidopsis thaliana). We demonstrated the utility of this system by generating more than 250 whole-gene translational fusions and 123 Arabidopsis transgenic lines corresponding to 62 auxin-related genes and characterizing the translational reporter expression patterns for 14 auxin biosynthesis genes.
INTRODUCTION
The last few years have witnessed dramatic advances in high-throughput experimental and computational approaches to investigate the molecular mechanisms behind biological processes. Nevertheless, certain types of information-rich functional data are still exceedingly tedious and time-consuming to obtain. Thus, any experimental approaches that require in vivo expression of the gene of interest (GOI) to, for example, gather high-resolution spatiotemporal expression patterns, determine protein subcellular localization, or identify protein–protein and protein–DNA/RNA complexes still heavily rely on classical restriction enzyme or recombination-based cloning strategies. Although these classical approaches are simple and accessible and therefore widely used, they have several limitations regarding scalability and may suffer from uncertainty when trying to capture the native expression patterns and levels of the genes under investigation. This uncertainty comes from technical considerations and limited functional knowledge when choosing the DNA sequences to be included in the construct, with the risk that some unknown, but important, regulatory sequences may be left out. This is not a trivial problem when the native expression pattern of a GOI needs to be imposed on the tagged gene. In the absence of a strict criterion, more or less arbitrary lengths of DNA sequences (typically from 1 to 4 kb of sequences upstream and 1 kb or less of sequences downstream of the start and stop codon, respectively) or all of the intergenic sequences flanking the GOI are usually chosen. These strategies, however, do not guarantee that all regulatory sequences are captured. Genetic complementation of a mutant line is relied upon to confirm that the expression patterns of the generated transgene accurately reflect that of the corresponding native gene. This time-consuming and not fully foolproof approach is, however, not possible when either a mutant line is not available or, more commonly, when the mutant does not display any detectable phenotype. The obvious solution to this problem is to increase the size of the sequences flanking the GOI that would be included in the transgene or, even better, to insert the tag or the desired modification in the GOI directly in its native chromosomal location. Although the latter genome-editing approach is the highly desirable gold standard and the number of reports of precise gene editing in plants is constantly increasing (Cermak et al., 2015; Begemann et al., 2017; Yu et al., 2017; Dahan-Meir et al., 2018; Li et al., 2018), as reviewed by Soyars et al. (2018), the transgenic approach is still the most widely used methodology to generate plants expressing genes carrying a tag or other modifications that facilitate their visualization or biochemical characterization. Classical transgenic approaches are not ideal either, as they become tedious and inefficient as the size of the DNA fragments used increases.
To overcome some of the limitations of traditional transgenic approaches, a system involving highly efficient homologous recombination in bacterial strains engineered to express the Exo, Beta, and Gam proteins from the lambda phage known as the lambda red recombineering system (Yu et al., 2000; Copeland et al., 2001) has been developed. The high efficiency of this recombineering system has made it an essential tool in bacterial genome engineering (Isaacs et al., 2011), allowing for the rapid, efficient, and simultaneous editing of hundreds of loci in the bacterial genomes. Although the lambda red system has not been shown to work in eukaryotic cells, DNA from multicellular organisms can be efficiently modified using this system when introduced into recombineering-ready Escherichia coli strains. Thus, recombineering has been successfully used to generate genome-wide collections of fluorescently tagged proteins in several model organisms, such as Drosophila and Caenorhabditis elegans (Sarov et al., 2012, 2016). In addition to E. coli recombineering strains (Warming et al., 2005), several other system-specific elements are required in order to make this technology accessible to the research community. First, a collection of sequence-indexed genomic clones covering the whole genome of the organism of interest needs to be available. This is essential to easily identify a clone containing a GOI and the flanking sequences containing all of the putative regulatory sequences for that gene. In the case of plants, the reintroduction of these large genomic DNA fragments into the plant genome typically requires the use of Agrobacterium-mediated transformation. This imposes the additional requirement that the vector carrying the large genomic DNA fragments should be compatible with Agrobacterium-mediated transformation. Alternatively, the large DNA fragments from a bacterial artificial chromosome (BAC) need to be transferred to a suitable binary vector (Bitrián et al., 2011). In addition, the unrestricted availability of a set of reusable recombineering cassettes suitable for the insertion of tags commonly used in plant research at any position in any GOI, as well as tools that allow for the generation of custom-designed tagging cassettes or the introduction of any other sequence modifications in the GOIs, is essential for the widespread use of this technology among plant biologists. Finally, robust and simple protocols to facilitate the use of recombineering in any plant biology research laboratory with a standard molecular biology setup, as well as scalable pipelines that allow for the implementation of this technology to entire gene families, pathways, or the even the whole genome, are essential for the plant community to take full advantage of the benefits offered by recombineering technology.
We previously demonstrated that recombineering could be used to generate whole-gene translational fusions and point mutations in genes harbored in transformation-ready bacterial artificial chromosomes (TACs) and that these large TAC clones could be used for Agrobacterium-mediated transformation (Zhou et al., 2011). However, this original system has several limitations. First, it requires a sequence-indexed collection of TAC clones, in practice restricting its use to Arabidopsis (Arabidopsis thaliana). This system also uses classical recombineering cassettes based on the selectable galactokinase (galK) system (Warming et al., 2005), which relies on specialized media and expensive reagents (Warming et al., 2005). In addition, the relatively low efficiency of the contra-selection steps used to replace the galK gene by the tag of interest precludes this approach from being scaled up and requires significant troubleshooting when first adopted in a laboratory.
Here, we present a set of tools and protocols that overcome all of these limitations. The plant recombineering kit we describe allows for the use of standard media and antibiotic selection, and it provides a set of ready-to-use tags and a vector that can be utilized to convert any tag of interest into a recombineering-ready cassette. Importantly, a set of plasmids and cassettes has been generated to facilitate the transfer of tens of thousands of base pairs from a BAC to a high-capacity binary vector, opening this technology to many plant species for which sequence-indexed genomic clones covering the genome are available. Finally, we compiled sequence information from two Arabidopsis TAC libraries into a public genome browser, allowing for the easy identification of TAC clones containing the Arabidopsis GOI. All of the vectors and cassettes required to carry out recombineering experiments in plants are available via the Arabidopsis Biological Resource Center (ABRC), while the JAtY and Kazusa TAC libraries (Hirose et al., 2015) are available from the ABRC and RIKEN BioResource Research Center public stock centers. To demonstrate the utility of this system, we tagged more than 250 genes with different tags. We have made publicly available 123 transgenic lines corresponding to 62 genes through the ABRC and Nottingham Arabidopsis Stock Centre. Among these lines are those corresponding to β-glucuronidase (GUS) translational fusions of all members of the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1)/TAR and YUCCA (YUC) auxin biosynthetic enzyme families implicated in the production of the auxin indole-3-acetic acid (IAA) from amino acid Trp via indole-3-pyruvic acid (IPyA). The characterization of these lines in the roots and hypocotyls of seedlings grown under different pharmacological treatments, as well as untreated inflorescences and flowers, provides a general map of the auxin biosynthetic machinery in a limited set of tissues and conditions.
RESULTS
Generation of Excisable Antibiotic-Based Recombineering Cassettes
Classical recombineering strategies (Figure 1A; Warming et al., 2005; Zhou et al., 2011) rely on two consecutive recombineering steps (Figures 1B to 1F). In the first step, a positive/negative selectable marker such as galK is inserted into the genomic location to be modified (Figure 1D), followed by a second recombineering step where galK is substituted by the desired tag or replacement sequence (Figure 1F). One drawback of this time-consuming approach is that the negative selection step is prone to false positives (Warming et al., 2005; Zhou et al., 2011). In addition, several colonies per construct are often produced, which need to be tested to identify a true recombination product with the desired changes. An alternative approach to reduce the number of recombineering reactions needed has been the use of bifunctional recombineering cassettes that contain both the tag to be inserted in the GOI and a positive/negative selectable marker (Figures 1G to 1J). This selectable marker is flanked by Flippase (FLP) recognition target sites (FRTs; Figure 1G; Tursun et al., 2009), thus enabling marker removal by activating the expression of a FLP recombinase with very high efficiency (Figure 1J; Warming et al., 2005). In this alternative recombineering system, the positive selectable marker is first used to identify insertion events of the recombineering cassette in the GOI (Figure 1H). An inducible FLP recombinase already engineered in some E. coli recombineering strains, such as SW105, is then used to trigger the excision of the selectable marker, leaving behind just the reporter gene and a 36-nucleotide FRT scar (Figure 1J). The identification of these excision events can be facilitated by the loss of the galK activity, which inhibits bacterial growth in the presence of 2-deoxygalactose (Warming et al., 2005).
Figure 1.
Recombineering Process Comparing Classical and New Accelerated Strategies.
(A) Schematic representation of the basic concept of recombineering where the lambda red proteins Exo, Beta, and Gam mediate the integration of a linear fragment of DNA electroporated into a recombineering E. coli strain carrying a GOI in a BAC. During this process, the 5′-3′ exonuclease Exo produces 3′-protruding ends in the linear DNA that, upon binding to Beta, find homology regions of as little as 40 nucleotides in the GOI and mediate the integration of the linear DNA molecule. Gam inhibits several E. coli nucleases, preventing the degradation of the linear DNA. After cells with the modified DNA are selected, the whole BAC is transferred to Agrobacterium and used for plant transformation.
(B) The first step in any recombineering experiment is the identification of a genomic clone (typically a TAC or a BAC) containing the gene or sequences of interest.
(C) In the classical galK-based system, the galK positive/negative selectable marker is amplified using a pair of primers that contain at least 40 nucleotides of sequence corresponding to the sequence flanking the desired insertion site in the target genomic DNA clone. In this example, the amplification of the galK cassette with the GS1 and GS2 primers will result in the production of an amplicon (GS1-galK-GS2). The GS1 and GS2 sequences in the amplicon will target the galK selectable marker to the desired location in the gene, in this particular example, the 3′ of the gene just before the stop codon.
(D) Electroporation of this amplicon in a recombineering competent E. coli strain such as SW105 and the selection of the galK-positive colonies in minimal medium supplemented with Gal as the only carbon source will result in a clone containing the galK marker at the desired location in the GOI, in this particular example, immediately upstream of the stop codon. Because of the slow growth of bacteria in minimal medium, this process takes up to 7 d.
(E) Using the same set of primers used to amplify the galK cassette, a TAG/AnyDNA cassette (such as GFP) is amplified to generate the amplicon GS1-TAG/AnyDNA-GS2.
(F) As before, the GS1 and GS2 sequences will target the amplicon to the desired location, replacing the galK by the TAG/AnyDNA sequence. This sequence replacement can be accomplished by electroporating the GS1-TAG/AnyDNA-GS2 amplicon into the recombineering cells carrying the GOI tagged with galK and selecting for clones that lost galK in minimum medium supplemented with 2-deoxygalactose. Again, this may take up to 7 d due to the slow growth of bacteria in minimal medium. Only galK-negative colonies will survive in the presence of this chemical.
(G) The faster and user-friendly bifunctional cassette system combines the selectable marker (such as an antibiotic resistance gene) and the tag of interest in a single cassette. By flanking the sequences of the selectable marker with the FRTs, the selectable marker sequence can be readily removed post-insertion by a highly efficient in vivo FLP reaction. UA, universal adaptor.
(H) Similar to the classical approach, the bifunctional large cassette, GS1-5′UA-TAG/AnyDNA-FRT-galK/AmpR-FRT-5′UA-GS2 is first amplified with a pair of primers, GS1 and GS2, to add the gene-specific sequences that will target the recombineering cassette to the desired location in the gene. UA, universal adaptor.
(I) By electroporating this cassette into the recombineering E. coli strain SW105 containing the GOI and selecting for, in this example, ampicillin-resistant clones, the bacteria with the desired construct can be efficiently and rapidly identified. The use of regular LB and antibiotic selection allows for the identification of the bacteria with the desired construct in as little as 2 d. UA, universal adaptor.
(J) Finally, the induction of FLP recombinase already engineered in the SW105 strain would result in the removal of the sequences corresponding to the selectable marker (bottom), leading to the tag containing the reporter or epitope of interest followed by a 36-nucleotide (nt)-long FRT-containing scar that encodes 12 extra amino acids. The approximate time period required for each step is indicated. The GS1 primer should have the following structure: 5′-40 nt just upstream of the nucleotide, after which you want to insert your tag, followed by the 5′UA sequence -GGAGGTGGAGGTGGAGCT-3′. Similarly, the GS2 primer should have the structure: 5′-40 nt corresponding to the reverse complement of the sequence just downstream of the nucleotide, in front of which you want to insert your tag, followed by the 3′UA sequence GGCCCCAGCGGCCGCAGCAGCACC-3′. UA, universal adaptor.
With the ultimate goal of facilitating the use of recombineering and to allow for increased throughput, we have adopted and improved the bifunctional cassettes containing both a selectable marker and a tag of interest. Although initially we chose the classical galK selectable marker to generate these bifunctional cassettes due to its counter-selectable capabilities (Warming et al., 2005; Zhou et al., 2011), we later generated a simplified and easier-to-use antibiotic-based excisable bifunctional recombineering cassette (Alonso and Stepanova, 2014). This cassette not only eliminates the need for using minimal medium used in the galK selection and contra-selection steps but also better exploits the high efficiency of the FLP-based excision system already engineered into the SW105 recombineering strain genome. Here, we have expanded the collection of bifunctional recombineering cassettes to a total of 11. These new antibiotic-based cassettes consist of several of the most commonly used tags in plants and an antibiotic resistance gene flanked by the FRT sites (Figure 1G; Supplemental Data Set 1). In addition to simplifying and accelerating the selection of recombination events, these ampicillin- or tetracycline-based recombineering cassettes are not only compatible with the selectable markers of two end-sequenced TAC libraries in Arabidopsis (Zhou et al., 2011; Hirose et al., 2015) but also with the most popular plant BAC libraries that use kanamycin or chloramphenicol as the antibiotic for selection in bacteria (Budiman et al., 2000; Yuan et al., 2000; Zhang et al., 2016). In addition, several fluorescent protein genes and GUS tags incorporated in these new cassettes have been codon optimized for high expression in plants (Supplemental Data Set 1).
Importantly, all our recombineering cassettes share the same 5′ and 3′ universal adaptor sequences (Supplemental Data Set 1). These sequences, which are common to all of our constructs, serve three purposes. First, these adaptor sequences allow for the use of the same set of gene-specific 60-mer primers to tag a GOI with any of the different tags in the collection. Second, the in-frame adaptor sequences encode a poly-Gly and a poly-Ala linker, providing a flexible connection and thus minimizing conformational interferences between the protein of interest and the corresponding tag (Tian et al., 2004). And third, these adaptors have been designed to allow the same cassettes to be used in N-terminal, C-terminal, or internal translational fusion experiments.
Although the new antibiotic-based recombineering cassettes make the generation of the translational fusions much simpler and more efficient, they do not allow for the same level of flexibility as provided by the classical galK system. Thus, for example, the counter-selectable properties of galK can be used, once inserted in the GOI, to generate replacement recombination events between the native sequence and any linear DNA fragment flanked by short (>40-nucleotide) homology arms (Figure 1F). By contrast, this sort of sequence modification cannot be generated with our native excisable antibiotic-based system where one recombineering cassette needs to be constructed for each new tag. In order to bypass this limitation and, at the same time, to further facilitate the generation of new recombineering cassettes, we have developed two new recombineering cassettes: a Universal tag-generator cassette, where the counter-selectable marker RPSL based on the gene encoding Ribosomal Protein S12 (Russell and Dahlquist, 1989) allows for the selection of DNA replacement events in the presence of streptomycin; and a galK-FRT-Amp-FRT cassette, where galK can be used as a contra-selectable marker (Figure 2B; Supplemental Data Set 1). These two cassettes can be used to facilitate the addition of new tags to our collection of bifunctional recombineering cassettes by simply replacing the RPSL or galK sequences with the sequence of a new tag (Figures 2A to 2C; Supplemental Protocol) or to generate nearly any type of gene-editing event, from single nucleotide modifications to large deletions, by replacing the whole cassette with the sequence of interest via recombineering (Figures 2D to 2H; Supplemental Protocol; Stepanova et al., 2011; Brumos et al., 2018). As a proof of concept, we used the Universal tag-generator cassette to create a new RFP recombineering cassette and the galK-FTR-Amp-FRT cassette to generate the GFP, mCherry, and 3xMYC recombineering cassettes (Supplemental Data Set 1).
Figure 2.
Schematic Representation of Two Applications for the tag-generator Cassette.
(A) to (H) A tag-generator cassette consisting of the negative selectable marker gene RPSL and the positive selectable marker AmpR conferring ampicillin resistance can be used for the easy generation of new bifunctional recombineering cassettes containing any desired tag (see [A] to [C]), or to perform precise gene editing (such as introducing point mutations, deletions, or insertions) in the GOI (see [D] to [H]). To facilitate the use of this tag-generator cassette, in addition to the negative (RPSL) and positive (AmpR) selectable markers, the construct contains the 5′ and 3′ universal adaptors (UAs) that allow for the amplification of any recombineering cassette in our collection and the TGR sequence that allows for the in-frame insertion of any tag, making it possible to use the resulting cassettes in tagging experiments at any position in the GOI (N-terminal, C-terminal, or internal). Finally, this cassette also includes FRT sites flanking the sequences conferring ampicillin resistance (AmpR), allowing for the precise and efficient elimination of the selectable marker gene post-insertion (A). The tag-generator cassette can be used to construct new recombineering cassettes (see [B] and [C]). A ready-to-use SW105 E. coli strain containing a TAC clone that harbors the tag-generator cassette has been constructed (A). Using the primers TGF (5′-TAAAAAGGGTTCTCGTTGCTAAGGAGGTGGAGGTGGAGCT-3′ in-frame with 20 nucleotides of the 5′ end of the new tag) and TGR (5′-GAAAGTATAGGAACTTCCCACCTGCAGCTCCACCTGCAGC-3′ in-frame with 20 nucleotides that anneal to the 3′ end of the tag of interest), the tag of interest (TAG/AnyDNA) can be amplified, generating the 5′UA-TAG/AnyDNA-TGR amplicon (B). By electroporating this amplicon in the SW105 recombineering strain carrying the tag-generator cassette and selecting for the absence of RPSL (streptomycin-resistant colonies), a new bifunctional recombineering cassette for the tag of interest will be obtained (C). The tag-generator cassette can also be used in a two-step recombination procedure similar to the classical galK approach to generate any type of sequence modification, such as seamless insertion of a tag, introduction of point mutations, and so on. In this case, the process starts with the identification of the genomic clone containing the GOI (D). Using GS1 and GS2 primers (Figure 1) to PCR-amplify the tag-generator cassette, an amplicon containing the sequences flanking the point where the gene editing will take place is obtained (E). By electroporating this amplicon in SW105 recombineering cells carrying the BAC or TAC clone with the desired gene and selecting for ampicillin-resistant colonies, the GOI is tagged with the tag-generator cassette (F). Next, a replacement DNA construct containing the edited sequence (point mutations, deletions, insertions, and so on; depicted as a red box in [H]) flanked by long regions of homology to the GOI (100 to 200 bp on each side of the region to be edited are recommended) is produced, typically by commercial DNA synthesis (G). When designing these constructs, it is important to consider that recombination can take place at any point within the regions of homology between the replacement sequence and the GOI tagged with the tag-generator cassette (bottom). By electroporating the replacement DNA and selecting for colonies resistant to streptomycin, the desired final product is obtained (H).
Recombineering-Based Trimming and Transfer of Large Genomic Constructs from BACs to Binary Vectors
The ability to precisely edit the sequence of a GOI in the context of a large BAC has the great advantage of capturing distant regulatory sequences (even tens of thousands of base pairs away), thus preserving the native expression patterns in the transgene reporter fusions. The use of BACs containing the GOI as the source of the genomic sequences to be edited has, however, several critical drawbacks. First, the researcher does not have the flexibility to choose the exact DNA regions flanking the GOI that would be included in the final construct, as this would be determined by the sequences already present in the selected BAC clone. Second, the choice of sequence-indexed BACs containing the GOI is limited to clones available in the BAC collection. And third, in most plant species (with likely the sole exception of Arabidopsis), the BAC clone collections that have been mapped back to the genome cannot be directly used for Agrobacterium-mediated transformation, as the vectors used in various genome sequencing efforts lack the features for propagation in Agrobacterium and for the subsequent transfer of DNA from this bacteria to the plant genome. To circumvent these limitations, we have developed a set of antibiotic-selection–based recombineering trimming cassettes (Figure 3A; Supplemental Data Set 1) that allow for the efficient elimination of undesired sequences flanking the GOI (Figures 3C and 3D; Supplemental Protocol). This simple trimming procedure allows the researcher to precisely define the DNA regions flanking the GOI to be included in the final construct (assuming a BAC clone containing the desired regions has been identified), thus eliminating extra genes that may cause phenotypic alterations when present in a copy-number excess. An added advantage of this strategy is that by reducing the size of the final construct, the transferring efficiency of the desired edited sequences to the plant nuclear genome is also increased (Zhou et al., 2011; Brumos et al., 2018).
Figure 3.
Schematic Representation of Two Applications for the Trimming Cassettes.
(A) to (F) Two trimming cassettes, one conferring tetracycline resistance (FRT2-Tet-FRT2 trimming cassette) and another conferring ampicillin resistance (FRT5-Amp-FRT5 trimming cassette), have been generated to facilitate the elimination of undesired sequences in TAC or BAC clones ([A] to [D]), as well as for the efficient transfer of large fragments of DNA from BAC clones to binary vectors ( [E] and [F]). To make these actions possible, each antibiotic selectable marker in the trimming cassettes is flanked by a different pair of orthogonal FRT sequences, FRT2 or FRT5, which not only allow for the elimination of the antibiotic resistance sequences after the trimming process ([C] and [D]) but also for the efficient in vivo transfer of large fragments of DNA from a BAC or TAC clone to a modified binary vector ( [E] and [F]). The first step in the process of trimming a genomic sequence is to identify a BAC or TAC clone carrying the GOI (A). Using DNA for the ready-to-use FRT2-Tet-FRT2 and FRT5-Amp-FRT5 trimming cassettes as PCR templates and two pairs of primers, FRT2F/FRT2R, and FRT5F/FRT5R, two amplicons containing the sequences of the trimming cassettes flanked by 40 nucleotides (nt) homologous to the sequences flanking the region to be deleted in the target genomic DNA are produced by PCR (B). Electroporating these amplicons into electrocompetent SW105 cells carrying the TAC clone harboring the GOI and selecting for colonies resistant to both ampicillin and tetracycline results in the replacement of the undesired genomic DNA sequences by the trimming cassette sequences (C). Inducing the expression of the FLP recombinase present in the genome of the SW105 cells results in the elimination of the ampicillin and tetracycline selectable sequences, leaving behind a single FRT2 and FRT5 site at each flank, respectively (D). The trimming product obtained in (D) contains the desired genomic DNA fragment flanked by two orthogonal FRT sites, opening the possibility of using cassette-exchange strategies to move this potentially large (at least up to 78 kb) DNA from the original BAC/TAC to a binary vector. To generate binary vectors suitable for this cassette-exchange reaction, a derivative of the Gateway pDONR221 vector containing the negative selectable marker SacB flanked by the head-to-toe FRT2 and FRT5 sites was generated (E). Using this new vector, the FRT2-SacB-FRT5 cassette can be easily transferred to any attR1-attR2–containing destination vector such as pGWB1 (E). To transfer the genomic DNA fragment flanked by the FRT2 and FRT5 sites to the pGWB1-FRT2-SacB-FRT5 vector, SW105 cells carrying the trimmed BAC or TAC clone (D) can be electroporated with the pGWB1-FRT2-SacB-FRT5 vector. In the presence of Suc (negative selection for the SacB gene) and hygromycin (positive selection for the pGWB1 backbone), the product of a successful cassette-exchange reaction can be efficiently selected (F). Dark green arrows indicate resistant genes that work both in plants and bacteria. The primers used to amplify the trimming cassettes have the following structure: FRT2 F: 5′-40 nt corresponding to the sequence upstream of the nucleotide in front of which one wants to insert the FRT2 site followed by the sequence -TTCAAATATGTATCCGCTCA-3′. FRT2 R: 5′-40 nt corresponding to the reverse complement sequence downstream of the nucleotide after which one wants to insert the FRT2 site followed by the sequence -TTACCAATGCTTAATCAGTG-3′. FRT5 F: 5′-40 nt corresponding to the sequence upstream of the nucleotide in front of which one wants to insert the FRT5 site-AACGAATGCTAGTCTAGCTG-3′. FRT5 R: 5′-40 nt corresponding to the reverse complement sequence downstream of the nucleotide after which one wants to insert the FRT5 site-TTAGTTGACTGTCAGCTGTC-3′.
In addition to the antibiotic resistance markers present in these trimming cassettes, we also included two sets of orthogonal FRT sites (FRT2 in the tetracycline and FRT5 in the ampicillin cassettes, respectively; Schlake and Bode, 1994), thus allowing for the removal of the antibiotic resistance genes once the trimming has been completed (Figure 3D). Importantly, after the FLP-mediated excision of the antibiotic resistance genes, the two remaining FRT2 and FRT5 sites left in the construct display a head-to-tail orientation. As illustrated in Figures 3E and 3F and the Supplemental Figure, this FRT configuration allows for the transfer of the selected DNA flanked by the FRTs to an engineered binary vector through an in vivo cassette-exchange reaction (Figures 3E and 3F; Supplemental Figure; Turan et al., 2013). To carry out the transfer of BAC DNA to any Gateway-compatible binary vector, we constructed a pDONR221-based entry clone with the negative selectable marker SacB flanked by the FRT2 and FRT5 sites in the same head-to-tail configuration as in the trimmed BAC (Figure 3E; Supplemental Data Set 1). This FRT2-SacB-FRT5 cassette can now be transferred to any attR1-attR2–containing destination vector using the standard LR Gateway recombination system, making it capable of accepting an FRT2/FRT5-flanked insert from any BAC clone (Figure 3F).
One possible advantage of this in vivo FLP-based cassette exchange system relative to the in vitro systems such as Gateway is the higher upper size limit of DNA fragments that can be routinely mobilized between vectors (Supplemental Protocol). Using this strategy, we generated pGWB1-FRT2-SacB-FRT5 as a standard destination vector for our FLP-mediated cassette exchange reactions (Figure 3E; Supplemental Data Set 1). To test the efficiency of the FLP-based system to exchange large DNA fragments, we tested the ability to T-DNA fragments of ∼16, ∼37, and ∼78 kb from a BAC containing the YUC9-GUS translation fusion gene to the pGWB1-FRT2-SacB-FRT5 binary vector. Although we were able to transfer all three DNA fragments, we found that the efficiency of the transfer (measured as the number of colonies obtained and the integrity of the DNA fragment in those colonies) dropped considerably as the DNA fragment size increased (Table 1). Perhaps this was due to the compromised stability of very large constructs in a multicopy plasmid such as pGWB1 not designed to hold such large DNA inserts. To overcome such limitation, we engineered pYLTAC17, a low-copy vector designed for the generation of large-insert genomic TAC libraries (Liu et al., 2002), to carry the exchange cassette FRT2-SacB-FRT5, allowing for the transfer, stable propagation, and plant transformation of large fragments of DNA originally carried in a BAC clone (Supplemental Figure; Supplemental Data Set 1). Importantly, we observed both an increase in the number of colonies obtained and in the integrity of the transferred DNA when using this vector compared with the results obtained with pGWB1-FRT2-SacB-FRT5. Furthermore, to expand the spectrum of BAC libraries that can be used as a DNA donor in this system, we introduced aadA, an aminoglycoside 3′-adenylyltransferase gene that confers spectinomycin and streptomycin resistance in both in E. coli and Agrobacterium, in addition to the kanamycin-resistance gene already present in the pYLTAC17-FRT2-SacB-FRT5-Spect vector (see “Methods”; Supplemental Figure; Supplemental Data Set 1). We also generated a second version of this vector, pYLTAC17-FRT2-SacB-FRT5-Spect-Kan, where the Bar gene for phosphinothricin (Basta) resistance has been replaced by the NPTII gene for kanamycin selection in planta (Supplemental Figure; Supplemental Data Set 1). Using the pYLTAC17-FRT2-SacB-FRT5-Spect plasmid side by side with the pGWB1-FRT2-SacB-FRT5 vector, the efficiency of DNA transfer from the BAC to pYLTAC17-FRT2-SacB-FRT5-Spect was higher than that to pGWB1-FRT2-SacB-FRT5-Spect as the acceptor vector, especially for DNA fragments as large as 78 kb.
Table 1. Efficiency of DNA Transfer from the BAC IGF F20D22 to pGWB1-FRT2-SacB-FRT5 and pYLTAC17-FRT2-SacB-FRT5-Spec-Kan Vectors.
| Experiment | pGWB1-FRT2-SacB-FRT5 | pYLTAC17-FRT2-SacB-FRT5-Spec-Kan | |||||
|---|---|---|---|---|---|---|---|
| Colonies Analyzed (Total No.) | Positive Colonies | % | Colonies Analyzed (Total No.) | Positive Colonies | % | ||
| First experiment | JMA2364 (∼16 kb) | 10 (72) | 8 | 80 | 10 (>1000) | 10 | 100 |
| JMA2365 (∼37 kb) | 7 (40) | 5 | 71.4 | 10 (>1000) | 10 | 100 | |
| JMA2366 (∼78 kb) | 3 (4) | 1 | 33.3 | 10 (>1000) | 10 | 100 | |
| Second experiment | JMA2364 (∼16 kb) | 10 | 10 | 100 | 10 (>1000) | 10 | 100 |
| JMA2365 (∼37 kb) | 10 | 9 | 90 | 10 (>1000) | 10 | 100 | |
| JMA2366 (∼78 kb) | 6 | 3 | 50 | 10 (>1000) | 9 | 90 | |
Recombineering in 96-Well Format Using Highly Efficient FLP-Based Marker Excision Cassettes
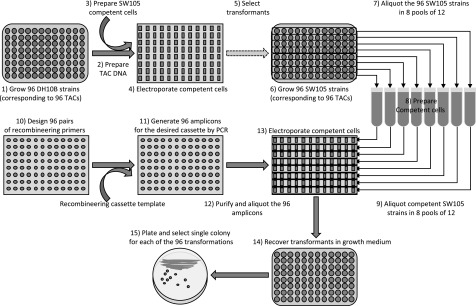
In the post-genome era, with thousands of gene sequences available, scalability represents a key element of any experimental procedure that aims to facilitate gene functional analysis. With the goal of developing a simple pipeline to process 96 recombineering samples in parallel (see “Methods”; Figure 4), we identified various bottlenecks. The foremost challenge was the development of an efficient method to transfer 96 TAC clones from the original E. coli strain DH10B to the recombineering E. coli strain SW105. This problem was addressed by growing the 96 DH10B strains in a 96-deep-well plate in 96-well format (Supplemental Protocol). The next critical step that needed to be scaled up was the insertion of the tag in the desired locations in each of the 96 selected genes. PCRs with a 60-mer primer pair containing 40 nucleotides flanking the insertion site of the GOI and 20 nucleotides corresponding to the universal adaptors flanking the recombineering cassette were used to obtain the 96 gene-indexed recombineering amplicons. Key for the implementation of this procedure was the experimental design of a strategy that would allow for the efficient introduction of 96 different recombineering cassettes into 96 different SW105 strains carrying the individual BACs of interest without the need to individually prepare electrocompetent cells for each of the 96 SW105 strains. This was achieved by preparing electrocompetent cells from pools of 12 strains corresponding to a full row in the 96-well plate in such a way that 96 TAC clones were represented in eight nonredundant pools of competent cells per plate (Figure 4; Supplemental Protocol). Because of the high sequence specificity of the recombination events, only those cells in a pool carrying the gene corresponding to the particular gene-indexed recombineering amplicon can undergo recombination and therefore acquire the selectable marker encoded in the cassette. It is important to point out that caution should be taken so that each gene to be tagged in a pool is present only in one of the TACs in that pool. For each of the 96 parallel recombination experiments, the fidelity of the recombination events was assayed by PCR using gene-specific primers flanking the selected insertion site with an efficiency of ∼100% (Supplemental Protocol), as we have reported previously (Zhou et al., 2011; Supplemental Protocol. The selectable marker was efficiently removed in all 96 strains in parallel by the activation of the FLP gene with l-Ara (for details, see Supplemental Protocol). For the first 96 constructs, we used the recombineering cassette containing the galK marker flanked by the FRT sites with the idea that the positive/negative selection of galK could be used to select for galK- clones after the FLP-mediated excision. Because of the extremely high efficiency of excision, we found the galK contra-selection unnecessary, as the desired excision events for most clones could be identified without the need for contra-selection. In fact, the analysis of three independent clones for each construct was sufficient in most cases to find at least one excision event lacking any undesired mutation.
Figure 4.
Schematic Representation of the 96-Well-Format Recombineering Pipeline.
The process starts by growing 96 DH10B strains carrying the desired TAC clones (the best TAC clones from the two available Arabidopsis libraries for any given gene can be found in our genome browser at https://brcwebportal.cos.ncsu.edu/plant-riboprints/ArabidopsisJBrowser) in a 96-deep-well plate (1). The cells are pelleted by centrifugation and a 96-well-format alkaline-lysis DNA miniprep protocol is used to obtain DNA for the corresponding 96 TACs (2). Electrocompetent SW105 cells are prepared and aliquoted into a 96-well electroporator cuvette (3). DNA for each of the selected 96 TAC clones is added to the electroporation cuvette wells and electroporated into the SW105 competent cells (4). After the electroporation, cells are resuspended in LB and transferred to a 96-deep-well plate where they are allowed to recover before they are plated in selection medium. Individual clones grown in selection medium are tested by PCR and arranged back into a 96-well format (dashed arrow indicates that several steps are not shown; [5]). The SW105 strains carrying 96 TAC clones selected in step 5 are grown overnight in a 96-deep-well plate (6). Cells from the overnight culture are used to inoculate eight cultures corresponding to pools of 12 clones each (7). Electrocompetent cells from each of the eight pools of 12 clones are prepared (8). Aliquots of cells from each pool are placed into the wells of the corresponding rows of the 96-well electroporation cuvette. For example, from pool 1, 12 identical aliquots would be placed in each of the wells of the first row of the 96-well electroporator cuvette and so on (9). In parallel, a pair of 60-mers per gene are designed (primer sequences for generating N- and C-terminal amplicons for any gene and any of our ready-to-use recombineering cassettes can be obtained from our genome browser at https://brcwebportal.cos.ncsu.edu/plant-riboprints/ArabidopsisJBrowser; [10]) and used to generate the corresponding 96 amplicons using the DNA from one of our ready-to-use cassettes as a template (11). The amplicons are purified by simple chloroform extraction and ethanol precipitation in a 96-well plate (12). The corresponding 96 amplicons are added to the electrocompetent cells and electroporated in the 96-well electroporation cuvette (13). As before, the cells are resuspended in LB and transferred to a 96-deep-well plate to allow them to recover (14). The cells from each transformation are then streaked onto LB plates with the proper antibiotic (15). Individual colonies (one or two per construct) are examined by colony PCR using a combination of gene- and tag-specific primers, and the integrity and fidelity of the recombination is checked by PCR-fragment sequencing.
The first 96 genes (Supplemental Data Set 2) corresponding to hormone-related genes were tagged using a modified version of a previously developed (Tursun et al., 2009) Venus-FRT-galK-FRT cassette (Supplemental Data Set 1) where we added the universal adapters (Tian et al., 2004). From these 96 selected genes, we failed to generate the desired constructs in two cases where, after sequencing three independent clones, we were not able to identify a construct with the desired modifications. For 17 additional genes, we had to sequence two clones to find the desired mutation-free construct, and in three cases, a third clone had to be sequenced. As we described previously (Zhou et al., 2011), most of the observed mutations were found in the sequences corresponding to the long oligos used to amplify the recombineering cassettes. After sequence verification, 80 of 94 clones were successfully transferred to recA- Agrobacterium strain UIA143 pMP90 (Hamilton, 1997) using our 96-well-plate pipeline described above (see “Methods”; Figure 4). In the 14 cases in which we did not succeed in transferring the TAC clone to Agrobacterium, we observed Agrobacterium colonies growing in kanamycin-selection medium, but they tested negative for the presence of the tagged gene by PCR.
Importantly, the efficiency of FLP-based excision of the galK cassette was ∼100%, even in the absence of counter-selection conditions, indicating that the positive/negative selectable marker galK could be replaced by a much more convenient antibiotic-based, positive-only selection marker, allowing for the use of standard growth medium (instead of the minimal medium required in the galK system). In addition to lowering the complexity and cost of the recombineering experiments, the use of antibiotic-based cassettes also significantly reduces the time required for E. coli to grow in the selection medium, decreasing from 5 d for galK selection in M63 minimal medium to 2 d (as the recombineering strains need to be grown at 32°C to avoid the induction of the lambda red proteins) for antibiotic-based selection in standard Luria-Bertani (LB) medium (Figure 1). To test the utility of these antibiotic-based recombineering cassettes, we generated the Universal AraYpet-FRT-Amp-FRT cassette and used it to tag another set of 96 hormone-related genes (Supplemental Data Sets 1 and 3). In this second experiment, we included most of the genes in the shikimate- and shikimate-derived metabolic pathways, focusing on those related to auxin biosynthesis. Similar to the galK-based system, we were able to obtain mutation-free constructs for most of the genes (89 of 96) and transfer them to Agrobacterium in 79 of the 89 cases. Although we are not sure why Agrobacterium transformation failed for 10 genes, in a follow-up study, we found that by adding the aadA gene (Sandvang, 1999) as a second antibiotic selectable marker, we could eliminate false positives during the transfer of large TAC clones from E. coli to Agrobacterium, thus improving the efficiency of selection of TAC clones in Agrobacterium to ∼100%.
We used all 159 Venus or Ypet constructs transferred to Agrobacterium to transform Arabidopsis via the highly efficient floral dip method but replaced the Suc with Glc to prevent toxicity in some Agrobacterium strains where the SacB gene was still active (Zhou et al., 2011). To facilitate the plant transformation process of the large number of constructs generated, we grew the 159 Agrobacterium strains on solid medium (two 150-mm Petri dishes per construct) and harvested the Agrobacterium cells in transformation medium just before performing the floral dip method (for details, see “Methods”). Of these 159 constructs, we generated Arabidopsis transgenic lines for 33 genes (two independent lines for 31 of these genes and one single line for the other two genes; Supplemental Data Sets 2 and 3). This subset of lines was selected based on an initial screen of young T1 seedlings with positive fluorescence signal and subsequent PCR confirmation of the desired genotype. We decided to prioritize this relatively small subset of genes due to the resources that would be needed (and the logistical challenges that would be involved) in propagating, making homozygous, and subsequently characterizing several lines per construct for which there was no evidence of detectable fluorescence and therefore future utility was readily available.
The lack of detectable expression of the reporter gene could be due to several factors. The rates of deletions of TAC constructs during the plant transformation process could be as high as 70% for large constructs but are negligible for constructs smaller than 25 kb (Zhou et al., 2011). This is, however, an unlikely explanation for most of the cases for which we failed to detect fluorescence, as we examined the progenies from an average of 28 T1s per construct (Supplemental Data Sets 2 to 4). Based on the estimate of a maximum deletion rate of 70% (Zhou et al., 2011), this number of lines should have been enough to include at least some T1 lines without the truncations. Furthermore, we did not see a correlation between the TAC size and the detection of fluorescence, with an average TAC size of ∼65 kb for both fluorescence-positive and -negative constructs (Supplemental Data Sets 2 to 4). Another and probably more significant factor is the low expression/accumulation levels of many of the tagged proteins, since we did not preselect our list of genes based on their expression levels, but only on their roles in hormone biology. In fact, we observed a direct correlation between published RNA levels in seedling roots and the detection of root fluorescence in our reporter lines. Thus, the average mRNA expression level for the GOI in young seedling roots was ∼2000 for the fluorescence-positive lines and ∼850 for the fluorescence-negative lines (Supplemental Data Sets 2 to 4). Nevertheless, to offset the possible problems due to both TAC size and low expression, we could either identify transgenic lines containing the whole transgene harboring both ends of the T-DNA by PCR, as we have done previously (Zhou et al., 2011), or we could trim distal genomic sequences unlikely to contain regulatory elements affecting the expression of the GOI but present in the original TAC clones.
Much more difficult to circumvent is the problem of lack of detectable fluorescence signal due to low levels of expression of the selected genes. To try to alleviate these two problems derived from using large TAC clones and weak florescence signal from genes expressed at low levels, we selected 87 of previously tagged genes related to auxin biosynthesis, transport, and response but for which we did not detected clear fluorescence signal in our previous experiments, and we generated new recombineering constructs tagged with three copies of the bright fluorescent protein gene Ypet (Supplemental Data Sets 1 and 4). Toward this end, we generated a new codon-optimized, FLP-based, ampicillin-resistant, excisable recombineering cassette (Supplemental Data Set 1). At the same time, we trimmed all of these new constructs to reduce the insert to just the tagged gene and 15 kb of flanking sequences (10 kb upstream of ATG and 5 kb downstream of the stop codon) using the trimming tools described in the section "Recombineering-Based Trimming and Transfer of Large Genomic Constructs from BACs to Binary Vectors." Using this approach, we were able to detect signals for 16 additional genes. Although we decided to trim these clones to an arbitrary size of 10 kb upstream and 5 kb downstream of the GOI, it is important to note that the tools presented here allow the researcher the flexibility to select the upstream- and downstream-sequence sizes of their choice (as long as a TAC or BAC clone containing such sequences is available). A balance between the advantages (such as increasing the probability of including potential regulatory sequences) and disadvantages (such as possible artifacts due to the increased copy number of certain genes or the greater probability of truncations of the T-DNA during plant transformation) of including large upstream and downstream regions flanking the GOI should be evaluated case by case by the researcher.
Characterizing the Expression Patterns of TAA1/TAR and YUC Genes
To further demonstrate the utility of this scalable recombineering system, we used a new codon-optimized GUS recombineering cassette to tag the 14 auxin biosynthetic genes of the IPyA pathway (Supplemental Data Set 4): TAA1, TAR1, TAR2, and YUC1 to YUC11. TAA1 and TARs encode Trp aminotransferases that catalyze the synthesis of IPyA from Trp (Stepanova et al., 2008; Tao et al., 2008), whereas YUC1 to YUC11 are flavin monooxygenases that convert IPyA to IAA (Sugawara et al., 2009; Stepanova et al., 2011). We generated transgenic plants for all 14 genes and examined the expression patterns of translational fusions in seedlings and reproductive tissues (Figures 5 to 7 ).
Figure 5.
GUS Staining Patterns of Translational Recombineering Fusions of Auxin Biosynthesis Genes and DR5:GUS in Roots.
Seedlings were germinated for 3 d in the dark in control AT medium or in AT medium supplemented with 10 μM NPA, 10 μM ACC, 10 μM NPA + 10 μM ACC, or 50 nM NAA. Samples were optically cleared with ClearSee. Bar = 100 μm.
Figure 7.
GUS Staining Patterns of Translational Recombineering Fusions of Auxin Biosynthesis Genes and DR5:GUS in Inflorescences and Flowers.
Images of individual flowers represent the enlarged versions of the boxed areas of inflorescences. Red arrows mark the GUS activity domains of interest highlighted in the text. The samples of DR5:GUS and the TAA1 recombineering fusion with GUS were optically cleared with ClearSee to enable visualization of GUS activity in the ovules and developing seeds. Black bars in the inflorescence images = 2.5 mm. White bars in the flower pictures = 250 μm.
In roots of 3-d-old dark-grown seedlings germinated in control AT medium, we detected the expression of translational fusions with GUS for TAA1 and 4 of 11 YUCs (YUC3, YUC7, YUC8, and YUC9) in the primary root meristem, as well as TAA1 and YUC6 in the pre-vasculature (Figure 5). Treatment with the auxin transport inhibitor naphthylphthalamic acid (10 µM NPA), ethylene precursor 1-aminocyclopropane-1-carboxylic acid (10 µM ACC), NPA and ACC combined, or synthetic auxin naphthaleneacetic acid (50 nM NAA) enabled us to detect the expression of all three TAA1/TAR genes and 9 of 11 YUCs in roots, except for YUC1 and YUC10, which were not expressed in distal regions of the primary root of 3-d-old etiolated plants in any of the conditions tested. ACC treatment upregulated TAA1, TAR1, TAR2, YUC2, YUC3, YUC4, YUC5, YUC6, YUC8, YUC9, and YUC11, which is consistent with the induction of the auxin responsive reporter DR5:GUS (Figure 5) and the known stimulatory effect of ethylene on auxin biosynthesis in roots (Stepanova et al., 2005, 2007; Růžička et al., 2007; Swarup et al., 2007). NPA treatment increased the expression of TAA1, TAR2, YUC3, YUC5, YUC7, YUC8, and YUC11 in germinating seedlings, and accordingly DR5, but the domains of NPA-triggered GUS activity were different for different genes. For example, for TAA1 and YUC5, GUS staining in NPA-treated seedlings was visible in the root elongation zones, suggesting that local auxin production is activated in this part of the root in response to the inhibition of polar auxin transport. Furthermore, the expression of TAA1 in the developing vasculature and of TAR2 in the stele was also enhanced by NPA. The expression domains of YUC3 and YUC8 in NPA became dramatically expanded in the primary root meristems, presumably leading to increased local production and accumulation of auxin in these tissues, as witnessed by the extensive widening of the DR5:GUS domains. The shift of the DR5 maximum is in agreement with the previously reported broadening of the stem cell niche under NPA treatment (Sabatini et al., 1999). The re-patterning of meristematic tissues is triggered by the increased levels of IAA trapped in the auxin-producing cells, with similar outcomes described for root meristems in plants exposed to the exogenous synthetic auxin 2,4-D (Sabatini et al., 1999).
Combined NPA plus ACC treatment had additive or synergistic effects on the expression of TAA1, TAR1, TAR2, YUC3, and YUC9 or in the case of YUC5, YUC6, and YUC8, phenocopied the single NPA treatments (Figure 5). Interestingly, in some cases, combined NPA plus ACC treatment resulted in the loss of some of the subdomains of expression visible with ACC alone (e.g., GUS staining in root hairs for YUC2, YUC3, YUC4, YUC5, and YUC6) or led to a shift in the domain of GUS activity, as observed for TAR1. Finally, NAA treatment upregulated TAA1 in the root elongation zone, TAR2 in the stele and root cap, YUC3 in the entire root tip, YUC6 in the vasculature, YUC2 (mildly) in the root meristematic region, and DR5:GUS in the vasculature and root meristem, suggesting that exogenous auxin can remodel endogenous auxin biosynthesis patterns. Of the 12 genes whose expression was detectable in roots, only YUC7 was not prominently responsive to any of the treatments tested (Figure 5).
In shoots of 3-d-old etiolated seedlings, TAA1, TAR2, and five YUC genes, YUC1, YUC3, YUC4, YUC5, and YUC6, were expressed in control medium, whereas the expression of TAR1, YUC2, YUC7, YUC8, YUC10, and YUC11 became detectable in seedlings exposed to ACC, NPA plus ACC, and/or NAA (Figure 6). The spatial domains of GUS reporter activity varied for different auxin biosynthesis genes. For example, under control conditions, TAA1, YUC1, YUC4, and, to a lesser extent TAR2, had defined expression in the shoot apical meristem. TAA1 and YUC6 were active in the hypocotyl vasculature, whereas TAA1, TAR2, and YUC6 had some activity in the cotyledon vasculature. YUC4 and YUC5 showed complementary expression patterns along the cotyledon perimeter. YUC4 expression was concentrated in the distal end of the cotyledon, and YUC5 was active along the edge of the cotyledon without overlapping with the YUC4 domain (Figure 6). These well-defined expression patterns of GUS fusions suggest that local auxin is produced in specific tissues by the combinatorial action of several Trp aminotransferases and flavin-containing monooxygenases that together contribute to establishing the morphogenic gradients of auxin.
Figure 6.
GUS Staining Patterns of Translational Recombineering Fusions of Auxin Biosynthesis Genes and DR5:GUS in Shoots.
Seedlings were germinated for 3 d in the dark in control AT medium or in AT medium supplemented with 10 μM NPA, 10 μM ACC, 10 μM NPA + 10 μM ACC, or 50 nM NAA. Samples were optically cleared with ClearSee. Bar = 200 μm.
Of the pharmacological treatments tested in shoots of 3-d-old etiolated seedlings, the addition of ACC to the growth medium had the greatest effect on auxin gene activity, inducing 9 of the 14 genes of the IPyA pathway, specifically TAR2, YUC2, YUC3, YUC4, YUC5, YUC6, YUC7, YUC8, and YUC10 (Figure 6). Remarkably, in the presence of NPA plus ACC, all nine genes showed patterns and levels of expression indistinguishable from those in NPA alone, suggesting that NPA blocks the effect of ACC in shoots and that ACC may exert its effect by inducing polar auxin transport, a notion consistent with prior reports (Růžička et al., 2007; Swarup et al., 2007). By contrast, the poorly expressed YUC11 displayed barely detectable activity in both ACC and NPA plus ACC, but not in NPA alone. Of the 13 auxin biosynthesis genes whose expression was detectable in shoots (all but YUC9), only YUC1 was not notably responsive to any of the four pharmacological treatments (Figure 6).
We also tested the expression of the 14 auxin biosynthesis genes in the inflorescences and flowers of soil-grown plants (Figure 7). TAA1 showed predominant expression in young gynoecia, especially in developing ovules, and somewhat milder expression in the transmitting tract and ovules of older gynoecia (red arrows in Figure 7). In young anthers, TAA1 exhibited a broad domain of expression, but as the anthers matured, the domain of TAA1 activity became more restricted, concentrating at the distal tips of these organs (red arrows in Figure 7). TAR1 and TAR2 were also expressed in the anthers of young flowers, and TAR2 was also expressed in the gynoecium and in the petals and sepal abscission zones of mature flowers (red arrows in Figure 7). Complementing the expression of TAR2 in the abscission zones of older floral organs were multiple YUC genes (all but YUC9 and YUC11; red arrows in Figure 7). DR5:GUS and all members of the YUC family showed varying degrees of activity in anthers, with immature male reproductive organs in YUC2 and YUC6 lines displaying the most prominent GUS activity, predominantly in stages 8 to 13 flowers (staging according to Smyth et al., 1990; Alvarez-Buylla et al., 2010). In older flowers, YUC8 showed localized expression in the upper region of the stamen filament at the junction with the anther (red arrows in Figure 7). Remarkably, only YUC4 was clearly active in the gynoecia among all YUC family members (Villarino et al., 2016), specifically in the stigmatic tissue (red arrows in Figure 7). DR5:GUS, on the other hand, exhibited well-defined domains of expression in the ovules and developing seeds of older gynoecia (red arrows in Figure 7). None of the YUCs or TAA1/TARs were prominently expressed in older anthers, petals, or sepals (Figure 7). While we cannot exclude the possibility that some of the auxin biosynthesis genes are mildly active in those tissues, the expression levels of these enzyme genes fell below our detection limit.
DISCUSSION
Recombineering
High-efficiency homologous recombination mediated by the expression of specific phage proteins in bacteria, the process also known as recombineering, is an invaluable tool for high-throughput genome editing in bacteria (Isaacs et al., 2011). Although recombineering can equal and, in some respects, surpass the popular clustered regularly interspaced short palindromic repeats (CRISPR)-Cas systems as a precise genome-editing tool in bacteria, this system has not been efficient in eukaryotic cells. Nevertheless, the power of recombineering has been widely used in eukaryotic model systems such as C. elegans (Sarov et al., 2006; Tursun et al., 2009) and Drosophila (Venken et al., 2008; Ejsmont et al., 2009; Sarov et al., 2016) to generate genome-wide collections of whole-gene translational fusions, thus opening doors to obtaining high-confidence gene expression landscapes in these organisms. As these whole-gene translational fusions are likely to capture most, if not all, of the regulatory sequences of a gene, it is not surprising that whenever systematic comparisons between classical and whole-gene recombineering-based, translational fusions have been performed, the superiority of the recombineering results has been clearly established (Sarov et al., 2012). Although no such systematic analysis has been performed in plants, our anecdotal experience in Arabidopsis also suggests that recombineering-based, whole-gene translational reporters are better at reflecting the native gene expression patterns in plants. Thus, for example, the expression profiles of classical translational fusions for the auxin biosynthetic gene TAA1 that passed the gold-standard quality control step of complementing the mutant phenotype (Yamada et al., 2009) are quite different from the expression domains observed with recombineering-based constructs (Stepanova et al., 2008). Importantly, we recently showed that the recombineering-based, whole-gene reporter fusions that included the introns of TAA1, but not classical translational fusion constructs that did not include the introns, were able to complement a larger array of phenotypes examined under different conditions and in different tissues and mutant backgrounds (Brumos et al., 2018). Although somewhat anecdotal, the case of TAA1 is not the only one reported, as the expression patterns deduced from an AUX1 recombineering construct better explain the role of this gene in auxin redistribution in the root than the classical promoter-fusion constructs (Band et al., 2014). Furthermore, in this study, we showed that the recombineering-construct–derived expression patterns of TAA1 and several YUC genes (YUC1, YUC2, YUC3, YUC4, YUC5, and YUC7) are different from that previously reported using classical promoter fusions (Cheng et al., 2006; Yamada et al., 2009; Lee et al., 2011; Chen et al., 2014; Kasahara, 2015; Challa et al., 2016; Brumos et al., 2018; Xu et al., 2018) but in close agreement with published mRNA levels (Supplemental Data Sets 5 and 6). Again, although no systematic or comprehensive comparison has yet been performed in plants, the few examples described here in a plant with a relatively small and compact genome such as Arabidopsis, as well as the systematic analysis in C. elegans, strongly argue for the use of caution when inferring native expression patterns from translational fusions that harbor only a few kilobases of genomic DNA flanking the GOI and do not include the potential regulatory sequences present in introns, coding, and untranslated 5′ and 3′ regions. It is logical to think that the need for the use of large genomic regions that include the GOI and the corresponding flanking sequences should be even greater in plants with larger (i.e., less compact) genomes. Ideally, direct tagging of the GOI in its chromosomal context should produce the most reliable expression patterns and should be considered the gold standard, but the current technology is not yet efficient enough to be widely adopted. It is likely that in the same way that the constant advances in the CRISPR-Cas system technologies have made the introduction of mutations in a particular gene almost routine in many plant research laboratories, the precise editing and insertion/replacement of sequences may also become habitual in the near future. At this point, however, recombineering is perhaps one of the best alternatives, as it offers a relatively simple way to generate translational fusions and other types of gene-editing events in the pseudo-chromosomal context of large BACs. Nonetheless, to take full advantage of the power of recombineering, experimental-system-specific resources and tools need to be developed (Venken et al., 2006, 2009; Poser et al., 2008; Ejsmont et al., 2009; Tursun et al., 2009; Sarov et al., 2016).
In the past, we and others have shown that recombineering could be used to create precise gene modifications in the context of large DNA constructs in plants (Stepanova et al., 2008, 2011; Bitrián et al., 2011; Zhou et al., 2011; Péret et al., 2012, 2013; Band et al., 2014; Fábregas et al., 2015; Han et al., 2015; Worden et al., 2015; Bhosale et al., 2018; Brumos et al., 2018; Yanagisawa et al., 2018; Gómez et al., 2019). Despite the obvious advantages of using large fragments of DNA to ensure that most, if not all, regulatory sequences have been captured, and the relative ease by which different types of modifications can be introduced in large DNA clones such as BACs or TACs, recombineering has, at present, not been widely embraced by the plant community. Although there are probably several reasons for this, the extra labor and time required to generate recombineering constructs, the limited access to sequenced TAC libraries, the difficulty of working with large DNA constructs, and so on are among the likely factors.
To eliminate some of these potential obstacles for adopting recombineering and thus to make this technology more accessible, we have developed and made freely available a new set of tools and resources. A collection of recombineering cassettes that contain both a commonly used tag (such as GFP, GUS, and so on) and an antibiotic resistance marker have been generated (Supplemental Data Set 1). In these cassettes, the sequences of the antibiotic resistance gene can be precisely removed with ∼100% efficiency using the FRT sites flanking the sequence by inducing a FLP recombinase integrated in the recombineering SW105 strain of E. coli. Using this set of recombineering cassettes not only makes the procedure much faster and cheaper but also extremely efficient and simple, all while avoiding the use of complicated and expensive bacterial minimal growth media. Limited access to transformation-ready BACs containing the GOI could have also limited the adoption of this technology. To eliminate this potential problem, we have deposited a copy of the JAtY library developed at the John Innes Centre by Dr. Ian Bancroft’s group in the ABRC. This, together with the recent publication of the sequence information for several thousand clones of the Kazusa TAC collection (Hirose et al., 2015), also available via the ABRC and RIKEN, and our Genome Browser tool (https://brcwebportal.cos.ncsu.edu/plant-riboprints/ArabidopsisJBrowser/) and dedicated MATLAB application (https://github.com/Alonso-Stepanova-Lab/Recombineering-App) to identify the best TAC clone and design a set of primers to tag any given gene, should significantly improve the accessibility and use of recombineering in plants.
To extend the use of recombineering beyond Arabidopsis, we also developed another set of recombineering cassettes and binary vectors for the efficient transfer of large fragments of DNA from a BAC to high-capacity, transformation-ready vectors, such as derivatives of pYLTAC17. This opens the possibility of using recombineering in any transformable plant species for which a BAC library covering the whole genome has been at least end sequenced. Previous work from the Csaba Koncz group has implemented the use of gap-repair cloning to T-DNA from a BAC to binary vectors (Bitrián et al., 2011). Although this is a clever and relatively simple approach, it requires the cloning of different genomic DNA fragments in a binary vector for each GOI, limiting its convenience and scalability. Our cassette-exchange approach expands the ability to use recombineering to other plant species. In addition, it allows for scalability and the use of very large DNA fragments (>75 kb) originally present in a BAC clone for plant transformation.
Finally, our antibiotic-based positive/negative selection cassettes (such as the Universal tag-generator cassette) provide a simple way to convert any existing tag into a recombineering-ready cassette. Thus, although our toolset comes with a collection of reporter tags ready to be used in gene expression analysis experiments, other types of specialized tags (such as those for the study of protein–protein interactions, protein–DNA or protein–RNA complexes, and so on) can be easily converted into recombineering cassettes using our tag-generator tool. This same tag-generator tool can also be utilized to generate more sophisticated gene edits in the context of a BAC clone. In these types of experiments, the tag generator cassette is first inserted in the location near the point where the change needs to be introduced using positive selection for ampicillin. The whole cassette can then be replaced by the sequence of choice by selecting against RPSL in the presence of streptomycin. The only limitation of the type of modification that can be made using this approach is the size of the DNA fragment used to replace the Universal tag-generator cassette due the inverse relationship between the size of a linear DNA fragment and its electroporation efficiency into E. coli. However, most applications only require the use of up to a few thousand base pairs as replacement DNA, and fragments of such sizes can be efficiently transformed into the recombineering E. coli strains. Thus, although the tag generator cassette is functionally equivalent to the classic galK cassette, it has the clear advantage of requiring simple LB medium and highly efficient antibiotic resistance instead of complicated and expensive minimum media and 2-deoxygalactose metabolic selection required for use of the galK-based systems. In summary, the toolset and resources described in this work should make it possible for any molecular biology research laboratory, and even teaching laboratories equipped for basic bacterial growth and PCR amplification capabilities, to carry out large arrays of gene-editing experiments by recombineering.
To further demonstrate the utility of the developed tools and resources, we implemented an experimental pipeline for tagging by recombineering 96 genes in parallel. Although very high-throughput protocols have previously been developed for the generation of genome-wide translational fusions in Drosophila and C. elegans, we have opted for an intermediate throughput where individual clones after each transformation or recombination event are tested. We believe that the approach described here is better when a relatively small number of genes are being tagged, as it ensures that final constructs will be obtained for most, if not all, of the genes of interest. In addition, this technology relies on robust procedures commonly used in standard molecular biology laboratories and uses strains and tools that are freely available (such as the toolset described here and the recombineering E. coli SW105 strain accessible through the National Cancer Institute). The testing steps in solid medium and the preparation of competent cells in pools, however, could be eliminated, as has been done previously by others (Sarov et al., 2012, 2016), to further increase the throughput of the procedure. Importantly, many of the tools developed in this work could be directly utilized in a future genome-scale project. However, rather than develop an ultra-high-throughput procedure, we have focused on setting up a robust and scalable protocol (suitable for tagging a single gene to a few hundred genes) that we believe provides a good balance between simplicity, accessibility, and throughput.
Auxin Biosynthesis
Auxin gradients play key roles in plant growth and development. In the past, the morphogenic auxin gradients have mainly been explained by the combined action of auxin transport and signaling/response (reviewed in Vanneste and Friml, 2009). Only in the past few years has the contribution of local auxin production been associated with the generation and maintenance of the morphogenic auxin maxima (Stepanova et al., 2008; Brumos et al., 2018; Zhao, 2018). Our present work characterizing the general expression patterns of all the genes involved in IAA production through the IPyA pathway, the main route of auxin biosynthesis in Arabidopsis (Mashiguchi et al., 2011; Stepanova et al., 2011), elucidates the spatiotemporal patterns of auxin production by defining the domains of activity of every TAA1/TAR and YUC gene in a limited set of tissues and developmental stages.
The establishment and maintenance of the shoot and root apical meristems is governed by auxin gradients generated by the joint action of local auxin biosynthesis and transport (Brumos et al., 2018; Wang and Jiao, 2018). Our observations indicate that in the shoot apical meristem, auxin is locally synthesized by the Trp aminotransferases TAA1 and TAR2 and the flavin monooxygenases YUC1 and YUC4. In roots, TAA1, YUC3, YUC7, YUC8, and YUC9 are responsible for the production of IAA in the stem cell niche of the root apical meristem. These observations are in agreement with recent single-cell RNA-sequencing assays profiling the developmental landscape of Arabidopsis roots (Zhang et al., 2019), where YUC3, YUC8, and YUC9 are included in the stem cell niche clusters.
In roots, ethylene triggers local auxin biosynthesis, leading to an increase in auxin levels and the inhibition of root elongation (Stepanova et al., 2005, 2007, 2008; Růžička et al., 2007; Swarup et al., 2007; Brumos et al., 2018). Higher order mutants of the TAA1/TAR and YUC gene families (Stepanova et al., 2008; Mashiguchi et al., 2011) display root-specific, ethylene-insensitive phenotypes. However, the specific genes involved in the local production responsible for the boost in auxin levels, particularly in the root elongation zone, have not been yet identified. Here, we discovered that multiple genes of the IPyA pathway (TAA1, TAR1, TAR2, YUC3, YUC5, YUC8, and YUC11) were induced in roots treated with the ethylene precursor ACC, with TAA1, YUC3, and, to a lesser degree, YUC5 displaying clear upregulation in the elongation zone. This observation suggests that auxin locally produced by these genes in the elongation zone may contribute to the arrest of root growth in the presence of ethylene. In addition, other ethylene-inducible TARs and YUCs may also contribute to the ethylene-triggered, auxin-mediated inhibition of root growth, as auxin transport also plays an important role in the ethylene responses in roots via the transcriptional induction of the auxin transporter genes AUX1, PIN1, PIN2, and PIN4 by ethylene (Růžička et al., 2007).
Our survey of auxin gene expression patterns in the recombineering fusions has unexpectedly uncovered the ACC-triggered induction of multiple auxin biosynthesis genes in the shoots of etiolated seedlings. As many as 9 of the 14 genes investigated, including TAR2, YUC2, YUC3, YUC4, YUC5, YUC6, YUC7, YUC8, and YUC10, were upregulated to different degrees by the ethylene precursor ACC in the hypocotyls and/or cotyledons, suggesting that a boost in auxin levels contributes to the ethylene-induced shortening of hypocotyls and/or inhibition of cotyledon expansion (Vaseva et al., 2018). To date, the effect of ethylene on auxin biosynthesis has been extensively investigated only in roots (Stepanova et al., 2005, 2007, 2008; Růžička et al., 2007; Swarup et al., 2007; Brumos et al., 2018). Having the recombineering reporter lines available for all major auxin biosynthetic pathway genes opens doors not only to the study of auxin production in seedlings but also to the dissection of spatiotemporal patterns of local auxin biosynthesis in all organs and tissues under a myriad of different conditions, genotypes, and treatments.
In fact, an inquiry into the spatial distribution of the expression of auxin biosynthetic genes in reproductive organs uncovered anthers, gynoecia, and developing ovules and seeds as the major sites of auxin biosynthesis. What is perhaps unexpected is that in the flowers, the strongest YUC gene activity (and consequently the expression of the auxin-responsive reporter DR5:GUS) is concentrated almost exclusively in the male reproductive organs (in the anthers), whereas TAA1 is predominantly active in the female organs (in the gynoecia). These observations suggest that some of the product of the TAA1/TAR-catalyzed biochemical reaction, IPyA, which serves as a substrate for YUCs to produce the auxin IAA, may be transported within the flowers out of the gynoecia, for example, to the anthers. As IPyA is a highly labile compound, at least in vitro (Tam and Normanly, 1998), determining whether and how it moves within the plant may be challenging. Alternatively, YUC expression in the gynoecia may simply be below our detection limit, or the conversion of IPyA to IAA may not be the rate-limiting bottleneck step in every tissue that makes auxin. Nonetheless, some IPyA is likely made directly in young anthers, as TAA1, TAR1, and TAR2 all show some activity in those organs. The local anther-made IPyA, together with the pool of IPyA potentially transported from the gynoecia, can then be utilized by multiple anther-expressed YUCs to produce auxin to contribute to pollen maturation, pre-anthesis filament elongation, and anther dehiscence (Cecchetti et al., 2008). Our prior work (Brumos et al., 2018) indicated that the spatiotemporal misregulation of TAA1 expression in developing flowers, which is expected to shift the domains of local IPyA and hence auxin production, results in flower infertility, highlighting the importance of the specific patterns of auxin gene activity for proper flower development. With the new recombineering resources at hand, we can now start dissecting the roles of individual TAA1/TAR and YUC family members in the development of flowers and other organs and tissues in Arabidopsis.
Several of the translational reporters generated in this study by recombineering, specifically those for TAA1, YUC1, YUC2, YUC3, YUC4, YUC5, and YUC7, behave differently from the previously published translational and/or transcriptional reporters for the same genes (Supplemental Data Set 6; Cheng et al., 2006; Yamada et al., 2009; Lee et al., 2011; Chen et al., 2014; Kasahara, 2015; Challa et al., 2016; Brumos et al., 2018; Xu et al., 2018). For example, in primary roots, translational TAA1 fusions that do not include introns are mainly expressed in the stele (Yamada et al., 2009; Brumos et al., 2018), whereas the recombineering construct is active in the quiescent center (QC) and pro-vasculature (this work; Stepanova et al., 2008; Brumos et al., 2018). The YUC3 promoter fusion is the strongest in the elongation zone of the primary root (Chen et al., 2014), but the recombineering construct is predominantly detected in the QC, as well as in the columella initials and the root cap (this work). For YUC5, prominent QC expression is observed for transcriptional fusions (Chen et al., 2014), but not for translational fusions generated by recombineering (this work), yet both constructs are active along the edges of the cotyledons (Challa et al., 2016). Although lacking sufficient spatial resolution for a proper comparison, the recombineering results for YUC3 and YUC5 are consistent with the mRNA levels reported by Transcription Variation Analysis (TraVA; Supplemental Data Set 5; Klepikova et al., 2016). For YUC7, a transcriptional fusion is mildly active in the proximal regions of the root, but is not detectable in the root meristem (Lee et al., 2011), whereas the recombineering construct for this gene is highly active in the QC and the root cap (this work). Analogous discrepancies are seen in the reproductive organs. For example, in mature flowers, YUC1 is detectable in the flower abscission zones only with a recombineering translational fusion (this work), but not with a transcriptional reporter (Cheng et al., 2006). YUC2 promoter fusion shows expression in young flowers, specifically in the valves of gynoecia, the pedicels, flower organ abscission zones, and petals, and no detectable expression in mature flowers (Cheng et al., 2006), whereas a recombineering translational fusion construct for this gene is active in the anthers of young flowers and in the abscission zones of petals and sepals in mature flowers (this work). Again, the expression pattern of YUC2 in anthers of young flowers observed in the recombineering lines is consistent with the relatively high mRNA levels observed in these tissues (Supplemental Data Set 5). For YUC4, both transcriptional and translational fusions are expressed in the female reproductive structures, specifically in the stigmas, but in the male reproductive structures, the transcriptional reporters are active only in the distal tips of the anthers (Cheng et al., 2006), whereas the recombineering-generated translational fusions have more ubiquitous, uniform activity throughout the entire anthers (this work). In addition, transcriptional reporters for YUC4 are detected in young flower buds at the base of floral organs (Cheng et al., 2006), but this domain of activity is not readily observed in the recombineering-generated translational reporter fusions (this work).
The differences in the expression patterns and levels of the previously published promoter-only transcriptional fusions or intronless translational reporters versus the new recombineering lines are likely due to the lack of some key regulatory elements in the former that are now captured in the latter. Again, the recombineering constructs include much larger upstream (10-kb) and downstream (5-kb) regions of the genes and possess the full coding regions with all of the introns. The presence of introns may not only provide important transcriptional regulatory sequences, as we have shown in the case of TAA1 (Brumos et al., 2018), but also produce a diverse population of mRNAs due to alternative splicing, as may be the case for YUC4 (Kriechbaumer et al., 2012). Differences in the length, content, and structure of the transcripts can lead to differences in RNA stability and localization, resulting in variable expression levels and patterns (Kriechbaumer et al., 2012). It is also not uncommon to see discrepancies in the expression patterns of transcriptional versus translational fusions, even for constructs that harbor identical promoter fragments. For example, the activities of YUC1 and YUC4 reporters produced by classical cloning approaches differ for the transcriptional versus translational constructs, with translational reporters showing less activity specifically in young flowers than their respective transcriptional fusions (Xu et al., 2018).
In summary, the differences in expression between classical and recombineering constructs can be explained by the presence of transcriptional regulatory elements missing in the classical constructs and thus affecting overall reporter expression patterns. Consistent with the argument, we see a good, although not perfect, correlation between the mRNA levels available via the TraVA database (Supplemental Data Set 5; Klepikova et al., 2016) and the GUS activity patterns observed with the recombineering lines. On the other hand, in the cases when the mRNA levels and GUS expression disagree, translational regulation, protein movement, and/or degradation could be responsible for the discrepancies. For example, protein turnover plays a major role in the expression of auxin coreceptor proteins Aux/IAAs, with translational fusions for these genes, unlike transcriptional reporters or Aux/IAA mRNA, being hard to detect due to rapid Aux/IAA protein degradation in the presence of auxin (Tiwari et al., 2001; Zhou et al., 2011). Finally, we cannot exclude the possibility that the discrepancies in gene expression patterns between studies may also arise from differences in plant growth conditions or sample processing between laboratories, as even for the same constructs and transgenic lines, different laboratories report different spaciotemporal patterns of activity, as is the case for YUC1 and YUC4 promoter fusions (Cheng et al., 2006; Banasiak et al., 2019). Regardless of the molecular or environmental underpinnings of the expression profile differences between previously published classical and newly generated recombineering reporters, the expression patterns obtained with the latter constructs should better reflect those of the native gene and thus represent a more reliable source of gene functional information.
METHODS
General Recombineering Procedures
Recombineering experiments were performed as described previously by Alonso and Stepanova, (2014). In brief, Escherichia coli recombineering strain SW105 cells carrying the TAC or BAC of interest were grown overnight at 32°C in LB supplemented with the antibiotic needed to select for the corresponding BAC or TAC. Overnight culture (1 mL) was used to inoculate 50 mL of LB plus antibiotic in a 250-mL flask, followed by incubation at 32°C for 2 to 3 h with constant shaking. The lambda red recombineering system was activated by incubating the cells in a water bath at 42°C with constant shaking for 15 min. The cells were immediately cooled down in a water-ice bath, and electrocompetent cells were then prepared (Alonso and Stepanova, 2015). The cells were electroporated with the PCR-amplified recombineering cassette, allowed to recover in LB for 1 h at 32°C, and plated on an LB plate with the corresponding selection. After a 2-d incubation at 32°C, the presence of the recombination event in the primary transformants was confirmed by colony PCR using a gene-specific primer and a primer specific for the inserted cassette. Primer sequences are provided in Supplemental Data Sets 2 to 4. The FLP reaction was performed by growing the SW105 cells harboring the TAC or BAC clone with the desired recombineering cassette already inserted in the location of interest overnight at 32°C under constant shaking in LB medium supplemented with the necessary antibiotics to select for the BAC or TAC. Fresh LB medium (1 mL) with the antibiotic necessary to select for the BAC or TAC backbone was inoculated with 50 μL of overnight culture and 10 μL of 10% (w/v) l-Ara. The cells were grown for 3 h at 32°C with constant shaking. A sterile toothpick was dipped into the culture and used to streak a few cells onto a fresh LB plate supplemented with the antibiotic necessary to select for the BAC or TAC backbone with the goal of obtaining isolated colonies. Colonies were then tested by colony PCR to confirm the elimination of the FRT-flanked DNA sequences. To ensure that the modification in the GOI was correct, the test PCR product was sequenced using the corresponding test oligos.
Commercial DNA synthesis services (IDT) were used to obtain the following sequences: Universal GUS-FRT-Amp-RFP, Universal RPSL-Amp, and Universal tag-generator cassettes as well as the Universal AraYpet and the Universal 3xAraYpet fluorescent protein genes. The sequences of these cassettes are provided in Supplemental Data Set 1.
Recombineering and Trimming of the 3xYPET and GUS Cassettes in a 96-Well Format
The basic recombineering procedures (Alonso and Stepanova, 2015) were followed during the parallel processing of 96 constructs with the following modifications. The 96 DH10B strains carrying the GOIs were inoculated in 96 1-mL LB kanamycin cultures in a 96-deep-well plate and grown overnight. TAC DNA was extracted by regular alkaline lysis (Alonso and Stepanova, 2014) using 12 strips of eight 1-mL tubes. In parallel, freshly prepared SW105 electrocompetent cells (Alonso and Stepanova, 2015) were aliquoted into the 96 electroporation wells of a 96-well electroporation plate (BTX Electroporation Systems). Forty microliters of competent cells and 3 μL of DNA were mixed in the cuvette and electroporated as described previously by Alonso and Stepanova, (2015). The cells were transferred to a 96-deep-well plate and incubated at 32°C with shaking for 1 to 2 h to recover. The cells were collected by centrifugation and plated onto LB kanamycin plates. After confirming the presence of the TAC clones using the testing primers by PCR (Supplemental Data Sets 2 to 4), glycerol stocks for the 96 SW105 strains were generated. Using these stocks, 96 cultures were grown overnight in a 96-deep-well plate. Eight sets of 12 strains were pooled to inoculate eight 1-liter flasks prefilled with 250 mL of LB kanamycin. The pooled cultures were grown for 3 h, heat shocked at 42°C, and used to prepare electrocompetent cells as described previously by Alonso and Stepanova, (2015). In parallel, 96 amplicons corresponding to the desired recombineering cassette (Universal Venus-FRT-galK-FRT, Universal AraYpet-FRT-Amp-FRT, or Universal 3xAraYpet-FRT-Amp-FRT) were obtained using the DNA template for the cassette and the corresponding recombineering primers (Supplemental Data Sets 2 to 4). PCR fragments were purified by chloroform extraction and ethanol precipitation. PCR DNA was resuspended in 20 μL of water, and 3 μL aliquots were used for electroporation in the 96-well electroporation cuvette. Recovery, plating, and testing were also done as described in the section "General Recombineering Procedures" except that LB kanamycin and ampicillin plates were used for selection. Test PCR products were sequenced to confirm the integrity and fidelity of the recombination events. For trimming, the TetR gene was amplified from the FRT2-Tet-FRT2 trimming cassette to generate 96 trimming amplicons using the primers replaLB-tet Universal and one of the 96 Gene-DelRight primers (Supplemental Data Sets 2 to 4 and 7). The 96-well format recombination procedure was done as described in the section "General Recombineering Procedures", except that the recombination events were selected in LB plates supplemented with kanamycin and tetracycline. The insertions were confirmed using the LBtest and the corresponding TestDelRight primer (Supplemental Data Sets 2 to 4 and 7). A second round of trimming was performed using 96 amplicons obtained by amplifying the AmpR gene from the FRT5-Amp-FRT5 trimming cassette with primers replaRB-amp Universal and one of the 96 Gene-DelLeft recombineering primers. After confirming the trimming by PCR using the primers testRB and the corresponding TestDelLeft oligo, the second antibiotic resistance gene, aadA, an aminoglycoside 3′-adenylyltransferase gene that confers spectinomycin and streptomycin resistance in both in E. coli and Agrobacterium, was introduced into the trimmed constructs by recombineering using an amplicon obtained by amplifying the Kan-Spec cassette using the primers Spect-Kan-testF and Spect-Kan-testR (Supplemental Data Set 7). Plasmid DNAs for the 96 strains obtained were prepared by alkaline lysis using 12 strips of eight 1-mL tubes and electroporated into electrocompetent Agrobacterium using the same 96-well procedure as described at the beginning of this section. Agrobacterium selection was done using LB plates supplemented with kanamycin and spectinomycin.
Generation of the Universal Venus-FRT-galK-FRT Cassette
The Universal AraYpet was utilized as a template with primers PEO1F and PEO1R. This amplicon was inserted in JAtY clone JAtY68N23 using the classical galK system as described by Zhou et al., (2011) to generate the Universal AraYpet cassette. The Venus-FRT-galK-FRT sequences from the pBalu6 were amplified using the primers VenusPeo1F and VenusPeo1R (Supplemental Data Set 7), and this PCR product was used to replace the Ypet sequences in the Universal AraYpet cassette by recombineering, as described by Zhou et al., (2011).
Generation of the Universal mCherry-FRT-galK-FRT Cassette
A strategy similar to that used to generate the Universal Venus-FRT-Galk-FRT was utilized to produce the Universal mCherry-FRT-galK-FRT cassette, but in this case the primers CherryPeo1F and VenusPeo1R (Supplemental Data Set 7) were used to amplify the mCherry FRT-galK-FRT sequences from pBalu8. This PCR product then served to replace the Ypet sequences in the Universal AraYpet cassette by recombineering as described by Zhou et al., (2011).
Generation of the Universal AraYpet-FRT-Amp-FRT Cassette
The ampicillin resistance gene AmpR and the corresponding promoter were amplified from pBluescript SK- with primers PEO1FRTAmpF and PEO1FRTAmpR (Supplemental Data Set 7). This PCR product was then used in a recombineering reaction to insert the FRT-Amp-FRT sequences into the Universal AraYpet cassette to generate the Universal AraYpet-FRT-Amp-FRT cassette.
Generation of the Universal AraYpet-FRT-TetA-FRT Cassette
The tetracycline resistance gene TetA and the corresponding promoter sequences were amplified from genomic DNA of the recombineering strain of E. coli SW102 with primers PEO1FRTtetAF and PEO1FRTtetRAR (Supplemental Data Set 7). This PCR product was then used in a recombineering reaction to insert the FRT-Tet-FRT sequences into the Universal AraYpet cassette to generate the Universal AraYpet-FRT-TetA-FRT cassette.
Generation of the Universal 3xAraYpet-FRT-Amp-FRT Cassette
The Universal 3xAraYpet sequence was commercially synthesized by IDT and utilized as a template for PCR with primers IAA5F and IAA5R (Supplemental Data Set 7). The obtained amplicon was inserted into the JAtY61G08 clone using the classical galK recombineering approach as described by Zhou et al., (2011) to generate the Universal 3xAraYpet cassette. The FRT-Amp-FRT sequences from the Universal AraYpet-FRT-Amp-FRT cassette were amplified with primers 3YpetFAFF1 and 3YpetFAFR1 (Supplemental Data Set 7) and inserted into the Universal 3xAraYpet cassette to create the Universal 3xAraYpet-FRT-Amp-FRT cassette.
Generation of the Universal-RFP-FRT-Amp-FRT Cassette
The Universal tag-generator cassette was commercially synthesized by IDT and amplified with primers PCL5_STOP_5UA and 3UA_PCL3 (Supplemental Data Set 7). The resulting amplicon was inserted into the tomato BAC clone HBa0079M15 by recombineering (Zhou et al., 2011). The RFP DNA was amplified from the pUBC-RFP-DEST vector (Grefen et al., 2010) with primers PCL5_5UA_RFP-f and UR_RFP-r (Supplemental Data Set 7). The resulting product was used in a recombineering reaction to replace the RPSL sequence in the BAC clone containing the Universal tag-generator cassette.
Generation of the pDONR221-FRT2-SacB-FRT5 and pGWB1-FRT2-SacB-FRT5 Vectors
The SacB gene was amplified from the BiBAC2 vector (Hamilton, 1997) with primers FRT2SLongNew and FRT5Long (Supplemental Data Set 7). The PCR product was cloned into pDONR221 to create the pDONR221-FRT2-SacB-FRT5 vector. Spacer sequences for the orthogonal FRTs were obtained from a published source (Schlake and Bode, 1994). The pGWB1-FRT2-SacB-FRT5 vector was generated by transferring the FRT2-SacB-FRT5 sequences from the pDONR221-FRT2-SacB-FRT5 to the pGWB1 binary vector using a Gateway LR reaction.
Generation of the Kan-Spec Cassette
The aadA sequences were amplified from the pTF101 vector with primers SpectFKan and SpectRKan (Supplemental Data Set 7) and inserted into the JAtY63D14 clone to generate the Kan-Spec cassette template. Primers Spect-Kan-testF and Spect-Kan-testR (Supplemental Data Set 7) can be used to amplify the Kan-Spec cassette from the Kan-Spec cassette template.
Generation of the pYLTAC17-FRT2-SacB-FRT5-Spec Vector
The RPSL-Amp sequences were amplified from the Universal RPSL-Amp cassette with primers replaRBAmpRPSL and replaLBTetRPSL (Supplemental Data Set 7). The resulting PCR product was used to replace all of the Arabidopsis (Arabidopsis thaliana) genomic and SacB sequences in the JAtY56F21 clone by recombineering (Zhou et al., 2011), producing the pYLTAC17-RPSL-Amp vector. Next, the FRT2-SacB-FRT2 cassette was amplified from the pDONR221-FRT2-SacB-FRT5 vector with primers replaRBSacBM13F and replaLBSacBM13R (Supplemental Data Set 7) and utilized in a recombineering reaction to replace the RPSL-Amp sequence in pYLTAC17-RPSL-Amp to generate the pYLTAC17-FRT2-SacB-FRT5 vector. Next, the bacterial aadA gene to confer spectinomycin and streptomycin resistance was amplified from the Spec-Kan cassette by PCR with primers Spec-Kan-testF and Spec-Kan-testR (Supplemental Data Set 7) and integrated into the pYLTAC17-FRT2-SacB-FRT5 vector by recombineering to generate the final pYLTAC17-FRT2-SacB-FRT5-Spec vector.
Generation of the pYLTAC17-FRT2-SacB-FRT5-Spec-Kan Vector
The RPSL-Amp sequences were amplified from the Universal RPSL-Amp cassette with primers pYLTAC-RPSL-Amp-f2 and pYLTAC-RPSL-Amp-r2 (Supplemental Data Set 7). The resulting PCR product was used to replace the Act1 5′-Bar-Nos 3′ cassette in pYLTAC17-FRT2-SacB-FRT5 to generate the pYLTAC17-FRT2-SacB-FRT5-RPSL-Amp. Next, the Kan resistance cassette for plant selection was amplified from pGWB1 by PCR with primers pYLTAC-Kan-f2 and pYLTAC-Kan-r2 (Supplemental Data Set 7), and the resulting PCR product was utilized to replace the RPSL-Amp sequence in pYLTAC17-FRT2-SacB-FRT5-RPSL-Amp to generate pYLTAC17-FRT2-SacB-FRT5-Kan. Finally, the bacterial aadA gene to confer spectinomycin and streptomycin resistance was amplified from the Spec-Kan cassette by PCR with primers Spec-Kan-testF and Spec-Kan-testR (Supplemental Data Set 7) and integrated into the pYLTAC17-FRT2-SacB-FRT5-Kan vector by recombineering to generate the final pYLTAC17-FRT2-SacB-FRT5-Spec-Kan vector.
Generation of the FRT2-Tet-FRT2 Trimming Cassette
The TetA resistance gene was amplified from the Universal AraYpet-FRT-TetA-FRT cassette with primers FRT2-Tet-F (JAtY Universal) and FRT2-Tet-R (EIN3del; Supplemental Data Set 7) and inserted into the TAC clone JAtY63D14 by recombineering.
Generation of the FRT5-Amp-FRT5 Trimming Cassette
The ampicillin resistance gene AmpR was amplified with primers FRT5-Amp-R (BeloBAC11Right universal) and FRT5-Amp-PIN5delR (Supplemental Data Set 7). The resulting PCR product was used as a template in a second PCR with primers FRT5-Amp-EIN3delR and FRT5-Amp-R (Supplemental Data Set 7), and the resulting PCR product was used in a recombineering reaction resulting in the insertion of the FRT5-Amp-FRT5 cassette into TAC clone JAtY63D14.
Generation of the Universal galK-FRT-Amp-FRT Cassette
The galK sequence was amplified from the Universal Venus-FRT-galK-FRT cassette with primers UnigalK F and UniGalk R (Supplemental Data Set 7). The resulting PCR product was utilized in a recombineering reaction to replace the AraYpet sequences from the Universal AraYpet-FRT-Amp-FRT cassette, resulting in the Universal galK-FRT-Amp-FRT cassette.
Generation of the Universal GFP-FRT-Amp-FRT, Universal mCherry-FRT-AmpFRT, and Universal 3xMYC-FRT-Amp-FRT Cassettes
The GFP, mCherry, and 3xMYC sequences were amplified with primer pairs UniGFP F/UniGFP R, mCherryAmp F/mCherryAmp R, and Uni3XMYC F/Uni3XMYC R, respectively (Supplemental Data Set 7). Each of the PCR products was used in an independent recombineering experiment to replace the galK sequence of the Universal Galk-FRT-Amp-FRT cassette with the sequences of each of these new tags.
Generation of the Universal AraYpet-3xMYC-FRT-Amp-FRT Cassette
The 3xMYC-FRT-Amp-FRT sequence was amplified from the Universal 3xMYC-FRT-Amp-FRT cassette with primers Ypet-3xMYC and Uni3xMYC R (Supplemental Data Set 7). The corresponding PCR product was inserted immediately after the Ypet sequence to generate the Universal AraYpet-3xMYC-FRT-Amp-FRT cassette.
Examining the in Vivo Efficiency of an Exchange Cassette Reaction
To evaluate the efficiency of transferring large DNA fragments from a BAC clone to two different binary vectors, pGWB1-FRT5-SaB-FRT5 and pYLTAC17-FRT2-SacB-FRT5-Spec, the YUC9 gene (At1g04180) was tagged with GUS at the C terminus by a recombineering reaction in which the Universal AraGus-FRT-Amp-FRT cassette was amplified with primers InsertGUS-Amp-f and InsertGUS-Amp-r (Supplemental Data Set 7). To generate DNA sequences (containing YUC9-GUS) of different sizes flanked by the FRT2 and FRT5 sites, the FRT2-Tet-FRT2 trimming and FRT5-Amp-FRT5 trimming cassettes were inserted 10, 25, or 57 kb upstream and 5, 11, and 20 kb downstream of the YUC9 gene, respectively, by recombineering, where the FRT2-Tet-FRT2 trimming cassette was amplified with primer pairs Up-10kb_FRT2_f/Up-10kb_FRT2_r, Up-25kb_FRT2_f/Up-25kb_FRT2_r, and Up-57kb_FRT2_f/Up-57kb_FRT2_r, and the FRT5-Amp-FRT5 trimming cassette was amplified with primer pairs Down-5kb_FRT5_f/Down-5kb_FRT5_r, Down-11kb_FRT5_f/Down-11kb_FRT5_r, and Down-20kb_FRT5_f/Down-20kb_FRT5_r (Supplemental Data Set 7). After inserting the corresponding PCR fragments in the BAC clone and removing the antibiotic resistance genes in a FLP reaction, three clones were generated that harbor the FRT2-FRT5–flanked genomic DNA fragments containing the YUC9 gene tagged with GUS at the C terminus and 10 kb upstream plus 5 kb downstream, 25 kb upstream plus 11 kb downstream, or 57 kb upstream plus 20 kb downstream of YUC9. The in vivo cassette exchange reaction was performed by electroporating E. coli SW105 competent cells carrying one of the three YUC9-GUS constructs and grown in the presence of 0.1% (w/v) l-Ara for 3 h prior to starting the process of preparing the cultures for electroporation. After electroporation with either the pGWB1-FRT2-SacB-FRT5 or pYLTAC17-FRT2-SacB-FRT5-Spec vector, the cells were allowed to recover for additional 3 h at 32°C in LB medium supplemented with 0.1% (w/v) l-Ara. Clones containing the binary vectors carrying the YUC9-GUS genomic sequences delimited by the FRT2 and FRT5 sites were selected on LB plates supplemented with 10% (w/v) Suc (to select against the unmodified binary vectors) and either kanamycin (50 mg/mL) and hygromycin (200 mg/mL) to select for the pGWB1-based plasmids or kanamycin (50 mg/mL) and spectinomycin (50 mg/mL) to select for the pYLTAC17-derived vectors, respectively.
Plant Transformation and Fluorescence Analysis
Transformation of Agrobacterium tumefaciens strain UIA143 pMP90 (Hamilton, 1997) with the tagged constructs of interest was performed by electroporation as described by Alonso and Stepanova, (2015). The resulting colonies were re-streaked on LB plates supplemented with the appropriate antibiotic, and single colonies were tested by PCR with gene-specific primers to confirm presence of the construct. Fresh colonies were inoculated into 5 mL of liquid LB kanamycin, grown with shaking overnight at 28°C, and the resulting saturated cultures were split into two and plated on two 150-mm LB kanamycin plates. After two nights at 30°C, the cells were scraped off with a spatula and resuspended in 100 mL of liquid transformation solution [1× Murashige and Skoog, pH 6.0, 1% (w/v) Glc spiked with 200 µL/L Silvett-77]. Wild-type Arabidopsis plants grown in soil under a 16-h fluorescent light/8-h dark cycle until the inflorescences were ∼15 cm long were transformed with the resulting cultures using the floral dip method (Clough and Bent, 1998). Plants were allowed to recover under a plastic dome for 24 to 48 h and then uncovered and grown to maturity. T1 plants were selected in 20 µg/mL phosphinothricin in AT plates [1× MS, 1% (w/v) Suc, pH 6.0, with KOH, 7% (w/v) Bacto-agar] and propagated in soil (50:50 mix of Sun Gro gemination and propagation mixes) under a 16-h light/8-h dark cycle. T3 plants homozygous for the constructs were confirmed by genotyping with a combination of tag-specific (Ypet or GUS) and gene-specific primers.
Fluorescence analysis of Ypet- and 3xYpet-tagged lines was performed under a Zeiss Axioplan microscope in the T1, T2, and/or T3 generations, focusing on the expression patterns in 3-d-old etiolated seedlings. T3 lines homozygous for the tagged constructs listed in Supplemental Data Sets 2 to 4 were donated to the ABRC.
GUS Staining and Optical Clearing of Plant Tissues
Seeds of homozygous T3 and/or T4 GUS-tagged lines were sterilized with 50% (v/v) commercial bleach spiked with Triton to break seed clumps, washed five or more times with sterile water to remove bleach, resuspended in melted and precooled sterile 0.7% (w/v) low-melting point agarose, and plated on plain AT plates or plates supplemented with 10 µM ACC, 10 µM NPA, 10 µM ACC plus 10 µM NPA, or 50 nM NAA. After 3 d at 4°C to equalize germination, the seedlings were exposed to light for 1 to 2 h at room temperature to restart the clock and germinated at 22°C for 3 d in the dark. The seedlings were fixed in cold 90% acetone and immediately stained for GUS overnight as described by Stepanova et al., (2005). For flower and inflorescence analysis, transgenic T3 and T4 lines homozygous for the transgenes were grown in soil under a 16-h light/8-h dark cycle. Tips of inflorescences (∼3 cm) were excised with small scissors, fixed in cold 90% (v/v) acetone, stored overnight at −20°C to help remove chlorophyll, stained for GUS as described by Stepanova et al., (2005), and stored in 70% ethanol for several additional days to remove residual chlorophyll prior to imaging. Etiolated 3-d-old seedlings were fixed in 90% (v/v) acetone and optically cleared using freshly prepared ClearSee solution (Kurihara et al., 2015) for at least 7 d. Images of inflorescences mounted on AT plates were taken with Q Capture software on a 5.0 RTV digital camera (Q Imaging) under a Leica MZ12.5 stereomicroscope. To examine roots, hypocotyls, and flowers, samples were mounted on glass slides and imaged with the same camera and software on an AxioSkop2 Plus microscope (Zeiss) with Nomarski optics.
Quick Practical Guide for Standard Recombineering Applications
To make recombineering more accessible to plant researchers, we provide abbreviated instructions on how to implement this method using the resources and tools reported in this study. We also provide a more detailed step-by-step protocol in the Supplemental Protocol.
Perform a Standard Gene Tagging Experiment with One of the Universal Recombineering Cassettes of the Collection
(1) In order to insert a tag from the collection using our Universal recombineering cassettes, a TAC (using our Genome Browser at https://brcwebportal.cos.ncsu.edu/plant-riboprints/ArabidopsisJBrowser/ or MatLab application https://github.com/Alonso-Stepanova-Lab/Recombineering-App for Arabidopsis) or BAC clone containing a GOI needs to be identified. (2) Next, the TAC/BAC DNA is isolated and transferred into the SW105 recombineering strain via electroporation. (3) Recombineering and testing primers for the GOI are designed using our Genome Browser or by generating the following forward and reverse primers: Recombineering-F: 5′-40 nucleotides identical to the sequence immediately upstream of the desired insertion point for the tag followed by the sequence -GGAGGTGGAGGTGGAGCT-3′; Recombineering-R: 5′-reverse complement of the 40 nucleotides immediately downstream of the desired insertion point followed by the sequence -GGCCCCAGCGGCCGCAGCAG-3′. (4) The next step is to generate a recombineering amplicon using these primers, any of the recombineering cassettes with the tag of interest as a template, and a proofreading polymerase. (5) The amplicon is then inserted into the desired location by recombineering as described in the section “General Recombineering Procedures”). (6) To test the resulting colonies, test primers flanking the insertion site and regular Taq polymerase are used for colony PCRs on the recombineering products. (7) Once a desired clone is identified, the antibiotic selection sequences in the tag are removed using an in vivo FLP reaction as described in the section “General Recombineering Procedures”). (8) Finally, the construct is verified by re-sequencing of the integrated DNA tag and the genomic DNA-tag junction sites using the test primers.
Perform a Trimming Experiment of a TAC or BAC Clone Using the FRT2-tet-FRT2 and FRT5-Amp-FRT5 Cassettes
(1, 2) The first two steps of the procedure are the same as described for standard gene tagging. (3) The FRT2-Tet-FRT2 cassette is then amplified from the template using primers FRT2-Tet-FRT2-F and FRT2-Tet-FRT2-R. The forward FRT2-Tet-FRT2-F primer is comprised of 5′-40 nucleotides identical to the 40 nucleotides upstream of the sequence to be deleted at the 5′ end of the clone, followed by the sequence AACGAATGCTAGTCTAGCTG-3′. For example, when using the JAtY or KAZUSA TAC, we use the primer NewreplaRBFRT2-Tet 5′-TATATTGCTCTAATAAATTTTTGGCGCGCCGGCCAATTAGGCCCGGGCGG-TTCAAATATGTATCCGCTCATG-3′. Similarly, the reverse FRT2-Tet-FRT2-R primer consists of 5′-40 nucleotides with the reverse complement sequence just downstream of the sequence to be deleted at the 3′ end of the TAC or BAC clone, followed by the sequence TTACCAATGCTTAATCAGTG-3′. (4) In parallel, the FRT5-Amp-FRT5 cassette is amplified using primers FRT5-Amp-FRT5F and FRT5-Amp-FRT5R. These consist of 5′-40 nucleotides identical to the 40 nucleotides upstream of the sequence to be deleted at the 3′ end of the TAC/BAC clone, followed by the sequence AACGAATGCTAGTCTAGCTG-3′ for the forward primer; and 5′- 40 nucleotides with reverse complement to the 40 nucleotides downstream of the sequence to be deleted at the 3′ end of the TAC/BAC clone, followed by the sequence TTAGTTGACTGTCAGCTGTC-3′ for the reverse primer. When using the JAtY or KAZUSA libraries, the primer NewreplaLBFRT5-Amp 5′-TTAGTTGACTGTCAGCTGTCCTTGCTCCAGGATGCTGTTTTTGACAACGG-TTAGTTGACTGTCAGCTGTC-3′ is used as a reverse primer. After generating these amplicons, steps 5 to 8 from the standard gene tagging section are followed to trim and confirm the desired deletions.
Generate a Recombineering Cassette for a New Tag Using the Universal Tag-Generator Cassette
To generate a new tag, the Universal tag-generator cassette is provided as a ready-to-use SW105 strain carrying the tomato BAC clone HBa0079M15 harboring the Universal tag-generator cassette (Figure 2A). The following steps are required to generate any new tag. (1) The tag of interest (e.g., LUCIFERASE) is amplified from a DNA template with a proofreading polymerase and the primers tag-generatorF and tag-generatorR. These consist of the sequence 5′-TAAAAAGGGTTCTCGTTGCTAAGGAGGTGGAGGTGGAGCT-, followed by the first 20 nucleotides of the tag of interest-3′ (it is important that the sequence of the tag starts with the first nucleotide of the first codon of the tag) for the forward primer; and 5′-GAAAGTATAGGAACTTCCCACCTGCAGCTCCACCTGCAGC-, followed by the reverse complement of the last 20 nucleotides before the stop codon of the tag of interest-3′ for the reverse primer. (2) To replace the RPSL marker in the Universal tag-generator cassette with the Tag-generator amplicon from the previous step, a standard recombineering protocol is as described in the section “General Recombineering Procedures”), except that the positive recombinant colonies are selected in LB medium supplemented with streptomycin to select against the RPSL gene. (3) The sequence integrity of the new tag, as well as that of the recombination sites, is confirmed by sequencing a PCR product using primers flanking the new tag.
Perform a Sequence Replacement/Deletion Using the Universal Tag-Generator or Universal RPSL-Amp Cassettes
(1, 2) The first two steps of the procedure are the same as above in the standard gene tagging section. (3) The Universal tag-generator or Universal RPSL-Amp amplicons are generated using a replacementF primer with the sequence 5′-40 nucleotides upstream of the sequence to be modified, followed by GGAGGTGGAGGTGGAGCT-3′ and a replacementR primer with the sequence 5′-40 nucleotides reverse complementary to the 40 nucleotides immediately downstream of the sequence to be modified GGCCCCAGCGGCCGCAGCAG-3′. (4) The sequence to be modified in the TAC/BAC clone is replaced with the amplicon from step (3) using the standard recombineering protocol (see General Recombineering Procedures) selecting for ampicillin-resistant colonies. (5) A DNA fragment is commercially synthesized that contains the sequence with the desired modifications/deletions flanked by at least 40 nucleotides (preferably, 100 to 200 nucleotides) of the sequences homologous to the sequences flanking the inserted amplicon. (6) The replacement sequences from the previous step are used to replace the amplicon inserted in step (4) using the same recombineering procedures (see the section "Generate a Recombineering Cassette for a New Tag Using the Universal Tag-Generator Cassette") to replace RPSL with the tag of interest.
Accession Numbers
Sequence data from this article can be found in Supplemental Data Sets 1 to 7. Genome Browser and MatLab application to assist with the TAC clone selection and primer design can be found at https://brcwebportal.cos.ncsu.edu/plant-riboprints/ArabidopsisJBrowser/ and https://github.com/Alonso-Stepanova-Lab/Recombineering-App, respectively. The ABRC stock numbers for the recombineering cassettes and seeds for the transgenic lines are given in Supplemental Data Sets 1 to 4.
Supplemental Data
Supplemental Figure. Schematic representation of high-capacity cassette-exchange-ready binary vectors and their applications.
Supplemental Data Set 1. Sequences of the different recombineering cassettes.
Supplemental Data Set 2. General information for the clones generated with the 5′UA-Venus-FRT-GalK-FRT-3′UA cassette.
Supplemental Data Set 3. General information for the clones generated with the 5′UA-Ypet-FRT-Amp-FRT-3′UA cassette.
Supplemental Data Set 4. General information for the clones generated with the 5′UA-3xYpet/GUS-FRT-Amp-FRT-3′UA cassette.
Supplemental Data Set 5. Comparison of mRNA (TraVA data) and GUS (this work) expression levels.
Supplemental Data Set 6. Summary of expression pattern discrepancies between GUS expression in this work and previously published expression patterns.
Supplemental Data Set 7. List of primers used to generate the different cassettes and constructs not included in Supplemental Data Sets 1 to 3.
Supplemental Protocol. Recombineering step-by-step.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jeonga Yun, Clara Alonso-Stepanova, Cierra Clark, Katherine Hobbet, and Emily Nelson for technical assistance; Robert Franks for making equipment available for this research; Oliver Hobert for providing the Venus-FRT-galK-FRT cassette; Ian Bancroft for providing the JAtY TAC collection; and Donald Court and the National Cancer Institute for providing the SW105 strain. This work was supported by the National Science Foundation (grants DBI 0820755 and MCB 0519869 to J.M.A.; MCB 0923727 and IOS 1444561 to J.M.A. and A.N.S.; IOS 1650139 to A.N.S. and J.M.A.; and IOS 1750006 to A.N.S.); and the Spanish Ministry of Science, Culture and Sports Programa de Movilidad fellowship (grant PRX14/00419 to M.A.P.-A.).
AUTHORS CONTRIBUTIONS
J.M.A., A.N.S., J.B., C.Z., and M.A.P.-A. designed the experiments and performed research. J.M.A., A.N.S., J.B., and C.Z. wrote the article. Y.G., D.S., and A.P.P. assisted in research.
References
- Alonso J.M., Stepanova A.N. (2014). Arabidopsis transformation with large bacterial artificial chromosomes. Methods Mol. Biol. 1062: 271–283. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N. (2015). A recombineering-based gene tagging system for Arabidopsis. Methods Mol. Biol. 1227: 233–243. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla E.R., Benitez M., Corvera-Poire A., Chaos Cador A., de Folter S., Gamboa de Buen A., Garay-Arroyo A., Garcia-Ponce B., Jaimes-Miranda F., Perez-Ruiz R.V., Pineyro-Nelson A., Sanchez-Corrales Y.E. (2010). Flower development. The Arabidopsis Book 8: e0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banasiak A., Biedron M., Dolzblasz A., Berezowski M.A. (2019). Ontogenetic changes in auxin biosynthesis and distribution determine the organogenic activity of the shoot apical meristem in pin1 mutants. Int. J. Mol. Sci. 20: E180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band L.R., et al. (2014). Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell 26: 862–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann M.B., Gray B.N., January E., Gordon G.C., He Y., Liu H., Wu X., Brutnell T.P., Mockler T.C., Oufattole M. (2017). Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci. Rep. 7: 11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhosale R., et al. (2018). A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nat. Commun. 9: 1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitrián M., Roodbarkelari F., Horvath M., Koncz C. (2011). BAC-recombineering for studying plant gene regulation: Developmental control and cellular localization of SnRK1 kinase subunits. Plant J. 65: 829–842. [DOI] [PubMed] [Google Scholar]
- Brumos J., Robles L.M., Yun J., Vu T.C., Jackson S., Alonso J.M., Stepanova A.N. (2018). Local auxin biosynthesis is a key regulator of plant development. Dev. Cell 47: 306–318. [DOI] [PubMed] [Google Scholar]
- Budiman M.A., Mao L., Wood T.C., Wing R.A. (2000). A deep-coverage tomato BAC library and prospects toward development of an STC framework for genome sequencing. Genome Res. 10: 129–136. [PMC free article] [PubMed] [Google Scholar]
- Cecchetti V., Altamura M.M., Falasca G., Costantino P., Cardarelli M. (2008). Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20: 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Baltes N.J., Cegan R., Zhang Y., Voytas D.F. (2015). High-frequency, precise modification of the tomato genome. Genome Biol. 16: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa K.R., Aggarwal P., Nath U. (2016). Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28: 2117–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Dai X., De-Paoli H., Cheng Y., Takebayashi Y., Kasahara H., Kamiya Y., Zhao Y. (2014). Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55: 1072–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20: 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Copeland N.G., Jenkins N.A., Court D.L. (2001). Recombineering: A powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2: 769–779. [DOI] [PubMed] [Google Scholar]
- Dahan-Meir T., Filler-Hayut S., Melamed-Bessudo C., Bocobza S., Czosnek H., Aharoni A., Levy A.A. (2018). Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 95: 5–16. [DOI] [PubMed] [Google Scholar]
- Ejsmont R.K., Sarov M., Winkler S., Lipinski K.A., Tomancak P. (2009). A toolkit for high-throughput, cross-species gene engineering in Drosophila. Nat. Methods 6: 435–437. [DOI] [PubMed] [Google Scholar]
- Fábregas N., Formosa-Jordan P., Confraria A., Siligato R., Alonso J.M., Swarup R., Bennett M.J., Mahonen A.P., Cano-Delgado A.I., Ibanes M. (2015). Auxin influx carriers control vascular patterning and xylem differentiation in Arabidopsis thaliana. PLoS Genet. 11: e1005183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez M.D., Fuster-Almunia C., Ocana-Cuesta J., Alonso J.M., Perez-Amador M.A. (2019). RGL2 controls flower development, ovule number and fertility in Arabidopsis. Plant Sci. 281: 82–92. [DOI] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Hamilton C.M. (1997). A binary-BAC system for plant transformation with high-molecular-weight DNA. Gene 200: 107–116. [DOI] [PubMed] [Google Scholar]
- Han S.W., Alonso J.M., Rojas-Pierce M. (2015). REGULATOR OF BULB BIOGENESIS1 (RBB1) is involved in vacuole bulb formation in Arabidopsis. PLoS One 10: e0125621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y., et al. (2015). The Arabidopsis TAC Position Viewer: A high-resolution map of transformation-competent artificial chromosome (TAC) clones aligned with the Arabidopsis thaliana Columbia-0 genome. Plant J. 83: 1114–1122. [DOI] [PubMed] [Google Scholar]
- Isaacs F.J., et al. (2011). Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science 333: 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H. (2015). Current aspects of auxin biosynthesis in plants. Biosci. Biotechnol. Biochem. 80: 34–42. [DOI] [PubMed] [Google Scholar]
- Klepikova A.V., Kasianov A.S., Gerasimov E.S., Logacheva M.D., Penin A.A. (2016). A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 88: 1058–1070. [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V., Wang P., Hawes C., Abell B.M. (2012). Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J. 70: 292–302. [DOI] [PubMed] [Google Scholar]
- Kurihara D., Mizuta Y., Sato Y., Higashiyama T. (2015). ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142: 4168–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Jung J.H., Han D.Y., Seo P.J., Park W.J., Park C.M. (2011). Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta 235: 923–938. [DOI] [PubMed] [Google Scholar]
- Li J., Zhang X., Sun Y., Zhang J., Du W., Guo X., Li S., Zhao Y., Xia L. (2018). Efficient allelic replacement in rice by gene editing: A case study of the NRT1.1B gene. J. Integr. Plant Biol. 60: 536–540. [DOI] [PubMed] [Google Scholar]
- Liu Y.G., Liu H., Chen L., Qiu W., Zhang Q., Wu H., Yang C., Su J., Wang Z., Tian D., Mei M. (2002). Development of new transformation-competent artificial chromosome vectors and rice genomic libraries for efficient gene cloning. Gene 282: 247–255. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 18512–18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B., et al. (2012). AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietra S., Gustavsson A., Kiefer C., Kalmbach L., Horstedt P., Ikeda Y., Stepanova A.N., Alonso J.M., Grebe M. (2013). Arabidopsis SABRE and CLASP interact to stabilize cell division plane orientation and planar polarity. Nat. Commun. 4: 2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser I., et al. (2008). BAC TransgeneOmics: A high-throughput method for exploration of protein function in mammals. Nat. Methods 5: 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C.B., Dahlquist F.W. (1989). Exchange of chromosomal and plasmid alleles in Escherichia coli by selection for loss of a dominant antibiotic sensitivity marker. J. Bacteriol. 171: 2614–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K., Ljung K., Vanneste S., Podhorská R., Beeckman T., Friml J., Benková E. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. [DOI] [PubMed] [Google Scholar]
- Sandvang D. (1999). Novel streptomycin and spectinomycin resistance gene as a gene cassette within a class 1 integron isolated from Escherichia coli. Antimicrob. Agents Chemother. 43: 3036–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M., Barz C., Jambor H., Hein M.Y., Schmied C., Suchold D., Stender B., Janosch S. (2016). A genome-wide resource for the analysis of protein localisation in Drosophila. eLife 5: e12068 Available at: 10.7554/eLife.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M., et al. (2012). A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150: 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M., Schneider S., Pozniakovski A., Roguev A., Ernst S., Zhang Y., Hyman A.A., Stewart A.F. (2006). A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat. Methods 3: 839–844. Available at: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
- Schlake T., Bode J. (1994). Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry 33: 12746–12751. [DOI] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyars C.L., Peterson B.A., Burr C.A., Nimchuk Z.L. (2018). Cutting edge genetics: CRISPR/Cas9 editing of plant genomes. Plant Cell Physiol. 59: 1608–1620. [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Hoyt J.M., Hamilton A.A., Alonso J.M. (2005). A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17: 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Dolezal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Likhacheva A.V., Alonso J.M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Robles L.M., Novak O., He W., Guo H., Ljung K., Alonso J.M. (2011). The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973. Available at: 10.1105/tpc.111.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S., Hishiyama S., Jikumaru Y., Hanada A., Nishimura T., Koshiba T., Zhao Y., Kamiya Y., Kasahara H. (2009). Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 5430–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Perry P., Hagenbeek D., Van Der Straeten D., Beemster G.T., Sandberg G., Bhalerao R., Ljung K., Bennett M.J. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19: 2186–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam Y.Y., Normanly J. (1998). Determination of indole-3-pyruvic acid levels in Arabidopsis thaliana by gas chromatography-selected ion monitoring-mass spectrometry. J. Chromatogr. A 800: 101–108. [DOI] [PubMed] [Google Scholar]
- Tao Y., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G.W., et al. (2004). High-throughput fluorescent tagging of full-length. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.B., Wang X.J., Hagen G., Guilfoyle T.J. (2001). AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turan S., Zehe C., Kuehle J., Qiao J., Bode J. (2013). Recombinase-mediated cassette exchange (RMCE) - a rapidly-expanding toolbox for targeted genomic modifications. Gene 515: 1–27. [DOI] [PubMed] [Google Scholar]
- Tursun B., Cochella L., Carrera I., Hobert O. (2009). A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS One 4: e4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., Friml J. (2009). Auxin: A trigger for change in plant development. Cell 136: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Vaseva I.I., Qudeimat E., Potuschak T., Du Y., Genschik P., Vandenbussche F., Van Der Straeten D. (2018). The plant hormone ethylene restricts Arabidopsis growth via the epidermis. Proc. Natl. Acad. Sci. USA 115: E4130–E4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K.J., Carlson J.W., Schulze K.L., Pan H., He Y., Spokony R., Wan K.H., Koriabine M., de Jong P.J., White K.P., Bellen H.J., Hoskins R.A. (2009). Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6: 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K.J., He Y., Hoskins R.A., Bellen H.J. (2006). P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751. [DOI] [PubMed] [Google Scholar]
- Venken K.J., Kasprowicz J., Kuenen S., Yan J., Hassan B.A., Verstreken P. (2008). Recombineering-mediated tagging of Drosophila genomic constructs for in vivo localization and acute protein inactivation. Nucleic Acids Res. 36: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino G.H., Hu Q., Manrique S., Flores-Vergara M., Sehra B., Robles L., Brumos J., Stepanova A.N., Colombo L., Sundberg E., Heber S., Franks R.G. (2016). Transcriptomic signature of the SHATTERPROOF2 expression domain reveals the meristematic nature of Arabidopsis gynoecial medial domain. Plant Physiol. 171: 42–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jiao Y. (2018). Auxin and above-ground meristems. J. Exp. Bot. 69: 147–154. Available at: 10.1093/jxb/erx299. [DOI] [PubMed] [Google Scholar]
- Warming S., Costantino N., Court D.L., Jenkins N.A., Copeland N.G. (2005). Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden N., et al. (2015). CESA TRAFFICKING INHIBITOR inhibits cellulose deposition and interferes with the trafficking of cellulose synthase complexes and their associated proteins KORRIGAN1 and POM2/CELLULOSE SYNTHASE INTERACTIVE PROTEIN1. Plant Physiol. 167: 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., et al. (2018). SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J. 37: e97499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Greenham K., Prigge M.J., Jensen P.J., Estelle M. (2009). The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 151: 168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa M., Alonso J.M., Szymanski D.B. (2018). Microtubule-dependent confinement of a cell signaling and actin polymerization control module regulates polarized cell growth. Curr. Biol. 28: 2459–2466. [DOI] [PubMed] [Google Scholar]
- Yu D., Ellis H.M., Lee E.C., Jenkins N.A., Copeland N.G., Court D.L. (2000). An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97: 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q.H., Wang B., Li N., Tang Y., Yang S., Yang T., Xu J., Guo C., Yan P., Wang Q., Asmutola P. (2017). CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci. Rep. 7: 11874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q., Liang F., Hsiao J., Zismann V., Benito M.I., Quackenbush J., Wing R., Buell R. (2000). Anchoring of rice BAC clones to the rice genetic map in silico. Nucleic Acids Res. 28: 3636–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2016). Building two indica rice reference genomes with PacBiol. long-read and Illumina paired-end sequencing data. Sci. Data 3: 160076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.Q., Xu Z.G., Shang G.D., Wang J.W. (2019). A Single-Cell RNA Sequencing Profiles the Developmental Landscape of Arabidopsis Root. Molecular Plant 12: 648–660. [DOI] [PubMed] [Google Scholar]
- Zhao Y. (2018). Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Biol. 69: 417–435. [DOI] [PubMed] [Google Scholar]
- Zhou R., Benavente L.M., Stepanova A.N., Alonso J.M. (2011). A recombineering-based gene tagging system for Arabidopsis. Plant J. 66: 712–723. [DOI] [PubMed] [Google Scholar]