Comprehensive analysis of Lys-63 polyubiquitination sheds light on its importance to plant growth and development and identifies the machinery driving this posttranslational modification.
Abstract
The lack of resolution when studying the many different ubiquitin chain types found in eukaryotic cells has been a major hurdle to our understanding of their specific roles. We currently have very little insight into the cellular and physiological functions of Lys-63 (K63)–linked ubiquitin chains, although they are the second most abundant forms of ubiquitin in plant cells. To overcome this problem, we developed several large-scale approaches to characterize (1) the E2-E3 ubiquitination machinery driving K63-linked ubiquitin chain formation and (2) K63 polyubiquitination targets to provide a comprehensive picture of K63 polyubiquitin networks in Arabidopsis (Arabidopsis thaliana). Our work identified the ubiquitin-conjugating enzymes (E2s) UBC35/36 as the major drivers of K63 polyubiquitin chain formation and highlights the major role of these proteins in plant growth and development. Interactome approaches allowed us to identify many proteins that interact with the K63 polyubiquitination-dedicated E2s UBC35/36 and their cognate E2 variants, including more than a dozen E3 ligases and their putative targets. In parallel, we improved the in vivo detection of proteins decorated with K63-linked ubiquitin chains by sensor-based proteomics, yielding important insights into the roles of K63 polyubiquitination in plant cells. This work strongly increases our understanding of K63 polyubiquitination networks and functions in plants.
INTRODUCTION
Posttranslational modification of proteins by the 76–amino acid polypeptide ubiquitin (Ub) is crucial for regulating protein stability, activity, localization, or interactions with partners (Mukhopadhyay and Riezman, 2007). Ubiquitination involves the sequential action of three classes of enzymes: Ub-activating enzymes, Ub-conjugating enzymes (E2s), and Ub ligases (E3s). The activities of these enzymes ultimately result in the covalent attachment of Ub to a Lys (K) residue in the target protein. Polyubiquitin (polyUb) chains are formed by the further attachment of Ub moieties linked together by one of the seven Lys residues present in a Ub molecule (K6, K11, K27, K29, K33, K48, and K63) or by the N-terminal Met in the form of head–tail linear repeats (Peng et al., 2003; Pickart and Fushman, 2004; Kirisako et al., 2006). PolyUb chains exhibit different topologies and are associated with diverse biological functions (Woelk et al., 2007). K48-linkage of Ub moieties triggers the degradation of target proteins by the 26S proteasome (Pickart and Fushman, 2004). Much less is known about the other polyUb chain linkages (Pickart and Fushman, 2004; Kirisako et al., 2006; Woelk et al., 2007). K63-linked Ub chains do not induce proteasome-dependent degradation. The roles of polyubiquitination involving residue K63 from Ub (hereafter referred to as K63 polyubiquitination) have been widely studied in yeast and mammals, including roles in the endocytosis of plasma membrane proteins, DNA damage responses, and more marginally autophagy and signaling (Woelk et al., 2007; Adhikari and Chen, 2009; Komander and Rape, 2012).
Extensive work in nonplant model organisms has demonstrated that K63 polyUb chains form via two distinct mechanisms. First, HECT-type E3s have the ability to directly transfer Ub moieties to target proteins and thus catalyze the formation of specific linkage types regardless of the E2 (Kim and Huibregtse, 2009; Sheng et al., 2012). For non-HECT E3s, K63 polyUb is dictated by the E2 UBC13 (UBIQUITIN CONJUGATING ENZYME13) and involves a heterodimeric complex composed of UBC13 and the UEV1/MMS2 E2 variants (E2v), which lack the active-site Cys residue. Mechanistically, K63 polyUb chain formation requires the active-site Cys of UBC13 to be covalently bound to a first donor Ub, while UEV1/MMS2 binds noncovalently to a second acceptor Ub to make residue K63 of Ub available for chain elongation (Hodge et al., 2016). E2v also provide specificity to the UBC13-mediated K63 polyUb. In humans for example, the UBC13/MMS2 heterodimer drives K63 polyUb in the nucleus and DNA damage responses, while UBC13/UEV1A is involved in cytoplasmic K63-linked chain formation and drives nuclear factor-κB signaling (NF-κB), a crucial process controlling cytokine production, inflammation, and immunity (Hofmann and Pickart, 1999, 2001; Andersen et al., 2005).
Plant genomes have a relatively low number of HECT E3s, with only 7 HECT E3s encoded in Arabidopsis (Arabidopsis thaliana) among a total of 1500 E3s (Romero-Barrios and Vert, 2018). In comparison, humans possess 600 E3s with ∼30 HECT E3 members. By contrast, Arabidopsis has more UBC13-type E2s and UEV1 E2v than humans, with two UBC13 E2s (UBC35/36) among the 37 Arabidopsis E2s and four UEV1 E2v (UEV1A to UEV1D). Similar to what has been described in humans, plant UEV1s confer specificity to K63 polyubiquitination events, with UEV1B and UEV1C primarily accumulating in the nucleus and UEV1A mostly accumulating in the cytosol in Brachypodium distachyon (Guo et al., 2016).
The characterization of Arabidopsis T-DNA mutant alleles of the K63 polyubiquitination machinery has shed light on its participation to several biological processes. The ubc35-1 ubc36-1 and uev1d mutants both show hypersensitivity to DNA-damaging agents (Wen et al., 2008; Pan and Schmidt, 2014), similar to their yeast and mammalian counterparts. The ubc35-1 mutant is unable to form branched root hairs upon iron starvation and mis-expresses genes regulated by iron, pointing to a role of UBC35 in iron metabolism (Li and Schmidt, 2010). ubc35-1 ubc36-1 also shows shorter primary roots, fewer lateral roots and root hairs, and lower sensitivity to exogenously applied auxin than the wild type (Wen et al., 2014). Finally, UBC13-type E2s from Arabidopsis and tomato (Solanum lycopersicum), or tomato UEV1s, have been connected to pathogen responses (Mural et al., 2013; Turek et al., 2018; Wang et al., 2019). Even though many pathways appear to be targeted by the loss of UBC35/36 or UEV1A to UEV1D, the rather mild developmental defects observed in the corresponding loss-of-function mutants argue either for a nonessential role of K63 polyubiquitination in plants or for the prominent role of HECT-type E3s in the global landscape of K63 polyubiquitination despite their low numbers.
Until recently, only a few plant proteins were reported to be K63 polyubiquitinated, although K63 polyubiquitination represents the second most abundant ubiquitination type after K48-linked chains (Kim et al., 2013). In most cases, these are membrane proteins for which K63 polyubiquitination drives their endocytosis and degradation in the vacuole (Romero-Barrios and Vert, 2018). Advanced mass spectrometry approaches using tandem ubiquitin binding entity (TUBE)-based affinity purification of ubiquitinated proteins or using combined fractional diagonal chromatography (COFRADIC) identified thousands of ubiquitinated proteins and Ub attachment sites within (Kim et al., 2013). However, both strategies fail to provide information on Ub linkages, limiting our ability to tackle the roles of K63 polyUb chains with precision. Using a K63 polyUb sensor based on yeast VPS27 that recognizes K63-linked Ub chains (Sims et al., 2012; van Wijk et al., 2012), we recently identified ∼100 Arabidopsis proteins as part of the first plant K63 polyubiquitome (Johnson and Vert, 2016). This analysis revealed a tight link between K63 polyubiquitination and membrane proteins, likely underlying their degradation through endocytosis. Another striking connection was established with ribosomal proteins, which is consistent with the reported role of K63 polyubiquitination in sustaining translation efficiency under stress conditions in yeast (Silva et al., 2015; Back et al., 2019). Therefore, such a powerful sensor-based proteomic approach provided the first glimpse of the K63 polyubiquitination networks in plants and has the great potential to uncover unsuspected roles of this still poorly characterized posttranslational modification. Here, to increase our resolution of K63 polyubiquitination networks, we conducted several complementary large-scale approaches to probe the composition and functional importance of the K63 polyubiquitination machinery as well as its targets in Arabidopsis. This analysis allowed us not only to provide a better picture of K63 polyubiquitination-dependent processes in plants but also to identify roles of K63 polyubiquitination in various processes such as nuclear import, splicing, and DNA structure/topology.
RESULTS
K63 Polyubiquitination Catalyzed by UBC35/36 Is Essential for Plant Growth and Development
To functionally characterize the importance of K63 polyubiquitination and evaluate the contribution of UBC13 homologs to the formation of K63 polyUb chains in plants, we isolated T-DNA knockout alleles for the two Arabidopsis UBC13 genes UBC35 and UBC36 (Supplemental Figure 1A). The ubc35-1 allele (WiscDsLox323H12) harbored no mRNA for UBC35 and is therefore a null mutant (Supplemental Figure 1B), which is consistent with previous reports (Li and Schmidt, 2010; Wen et al., 2014). By contrast, the ubc36-1 allele (SALK_047381) still showed UBC36 mRNA accumulation albeit to a lesser extent than its wild-type counterpart (Supplemental Figure 1B). Although the same ubc36-1 allele was previously reported to be a knockout (Wen et al., 2014), we obtained evidence that ubc36-1 is a knockdown allele. Regardless, both ubc35-1 and ubc36-1 showed no macroscopic phenotype (Supplemental Figure 1C). We also confirmed that the ubc35-1 ubc36-1 double mutant still accumulated UBC36 mRNA (Supplemental Figure 1D).
To examine the consequences of loss-of-function of UBC13 in plants, we isolated a new ubc36 knockout allele (GABI_836B11) showing no UBC36 mRNA accumulation (Figure 1A; Supplemental Figure 1A), which we named ubc36-2. Similar to ubc35-1, ubc36-2 showed no macroscopic phenotype despite being impaired in the accumulation of K63 polyubiquitinated proteins (Figures 1B and 1C). The drop in K63 polyubiquitinated protein accumulation observed in both mutants supports the idea that Arabidopsis E2 enzymes UBC35 and UBC36 are the functional homologs of UBC13 from yeast and act redundantly to catalyze the formation of K63 polyUb chains. To evaluate the functional consequences of a total loss of UBC13-dependent K63 polyubiquitination in plants, we crossed ubc35-1 and ubc36-2 plants. In contrast to ubc35-1 ubc36-1, the ubc35-1 ubc36-2 double mutant completely lacked UBC35 and UBC36 transcripts (Figure 1A). Very strong growth defects were observed for homozygous ubc35-1 ubc36-2 mutants, with most plants arresting after germination. The penetrance of the mutations was not complete, since ∼5% of homozygous ubc35-1 ubc36-2 plants developed to the point of producing a few seeds but were devoid of detectable K63 polyubiquitinated proteins (Figures 1B, 1D, and 1E). F3 seeds germinated poorly and did not develop beyond the cotyledon stage, further pointing to the major role of UBC13-type E2s (Figure 1F). These phenotypes are overall much more dramatic than the previously reported phenotypes of ubc35-1 ubc36-1 plants (Li and Schmidt, 2010; Wen et al., 2014), which is consistent with the finding that ubc36-1 used in these published studies is not a null allele. The severity of the growth defects of ubc35-1 ubc36-2 is also in accordance with the finding that plants expressing ubiquitination-defective forms of proteins known to be modified with K63 polyUb chains, such as the metal transporter IRT1, are strongly impaired (Barberon et al., 2011; Dubeaux et al., 2018). Altogether, our work sheds light on the crucial roles of UBC13-type E2s and K63 polyUb chain formation in plant growth and development.
Figure 1.
Characterization of Arabidopsis Loss-of-Function Mutants for UBC35 and UBC36.
(A) RT-PCR analysis monitoring the accumulation of UBC35 and UBC36 transcripts in the wild-type (WT), ubc35-1, ubc36-2, and ubc35-1 ubc36-2 plants. Amplification of ACTIN2 (ACT2) was used as a loading control.
(B) Accumulation of K63 polyubiquitinated proteins in total extracts from the wild type (WT), ubc35-1, ubc36-2, and ubc35-1 ubc36-2 visualized by immunoblot analysis using Apu3 K63 polyUb-specific antibodies. Detection of tubulin served as a loading control. The sizes of marker proteins in kilodaltons are shown.
(C) Phenotypes of 3-week-old wild-type (WT), ubc35-1, and ubc36-2 plants grown in soil.
(D) Phenotypes of the 5-week-old wild-type (WT) and ubc35-1 ubc36-2 F2 plants grown in soil.
(E) Fertility defects of ubc35-1 ubc36-2. Arrows point to aborted seeds in siliques.
(F) Phenotypes of the 15-d-old wild-type (WT) and ubc35-1 ubc36-2 F3 seedlings grown in vitro.
Genomic Responses Dependent on K63 PolyUb Chain Formation
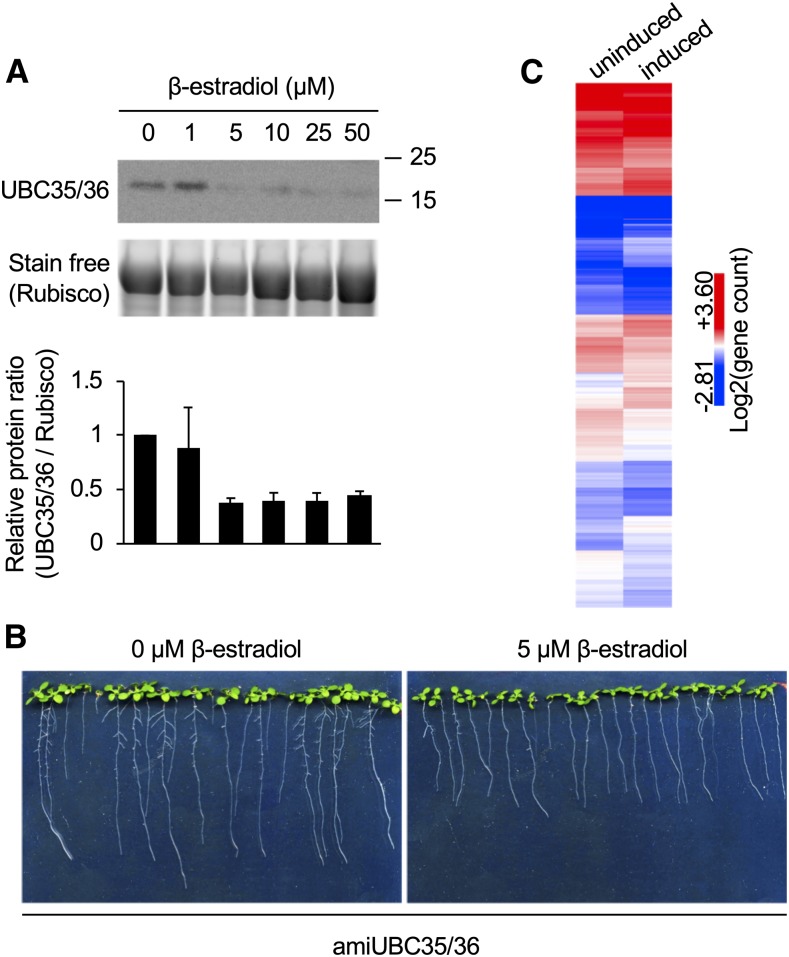
To pinpoint the cellular or physiological functions that are dependent on the formation of K63 polyUb chains, we sought to characterize the genomic responses to the loss of UBC13-type of E2s. Considering the extreme severity of the ubc35-1 ubc36-2 double mutant phenotype, we generated transgenic lines expressing an artificial microRNA targeting both UBC35 and UBC36 (amiUBC35/36) under the control of an estradiol-inducible promoter. amiUBC35/36-expressing lines grown on β-estradiol–containing plates showed a robust decrease in UBC13 protein accumulation (Figure 2A). Downregulation of UBC35/36 expression resulted in reduced root growth (Figure 2B), similar to what has been observed in ubc35-1 ubc36-1 plants and consistent with the finding that amiUBC35/36 plants still possess ∼25% of the wild-type levels of total UBC13-type E2s. To evaluate the global changes associated with the downregulation of UBC35/36, we performed RNA sequencing (RNA-seq) analysis of mock-treated and estradiol-induced amiUBC35/36 lines. A profound genomic reprogramming occurred upon downregulation of UBC35 and UBC36 gene expression, with ∼700 of genes upregulated and ∼1000 genes downregulated (P < 0.01, fold change >1.5; Figure 2C; Supplemental Data Sets 1 and 2). Gene ontology (GO) enrichment analysis indicated that upregulated genes in the amiUBC35/36 lines were significantly enriched in the GO terms response to abiotic (light, oxidative stress, nutrition, and heat) and biotic (oomycetes, insects, and fungi) stresses, while terms corresponding to RNA processing were underrepresented (Supplemental Data Set 3). GO terms corresponding to glucosinolate biosynthesis, cell-cycle microtubule-based processes, and RNA processing were either over- or under-represented among downregulated genes (Supplemental Data Set 4). Therefore, the biological processes associated with these terms represent putative cellular functions requiring K63 polyubiquitination involving target proteins yet to be identified. Taken together, the genomic perturbations associated with impairment in the formation of K63 polyUb chains target multiple environmental stress responses as well as endogenous cellular functions, further pointing to the essential role of K63 polyUb in plant growth, development, and responses to the environment.
Figure 2.
Characterization of Inducible amiUBC35/36 Lines.
(A) Influence of amiUBC35/36 expression on UBC35/36 protein accumulation. Total proteins were extracted from 12-d-old seedlings grown on mock or β-estradiol medium. Ub-unloaded UBC35/36 protein accumulation was monitored using anti-UBC13 antibodies. Stain free visualization of total proteins was used as a loading control. The sizes of marker proteins in kilodaltons are shown. Quantification shows UBC35/36 protein accumulation relative to loading control, normalized to the mock-treated condition. Results are presented as mean of two independent experiments. Error bars represent sd. Rubisco, ribulose-1,5-bis-phosphate carboxylase/oxygenase.
(B) Phenotypes of amiUBC35/36 lines grown for 12 d on mock (left) or plates containing 5 µM β-estradiol (right).
(C) Heatmap of differentially regulated genes upon downregulation of UBC35/36 expression. Differentially expressed genes were clustered using Euclidean distance. Related to Figure 1.
Interactome-Based Approach to Study K63 PolyUb Networks
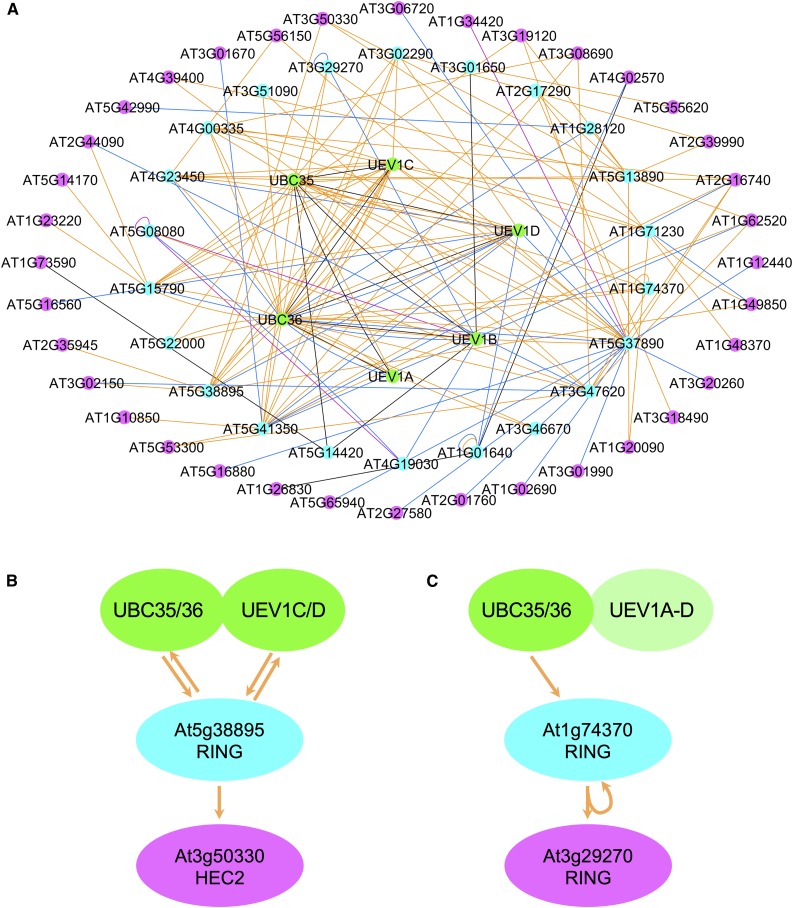
Having established that K63 polyUb chain formation requires both UBC35/36 E2s and participates in a wide variety of processes, we sought to identify E3 Ub ligases that interact with UBC35 and UBC36. This information is instrumental to better understand the biological roles associated with K63 polyubiquitination. We previously investigated the yeast two-hybrid (Y2H) first generation Arabidopsis interactome map AI-1, generated using a library of ∼8000 Arabidopsis open reading frames (ORFs; 8k_space), for proteins that interact with UBC36 and UEV1A, UEV1C, or UEV1D (UBC35 and UEV1B were not present in the original 8k_space; Arabidopsis Interactome Mapping Consortium, 2011). This study identified six E3 ligases among a total of 13 primary interacting proteins (Johnson and Vert, 2016). To extend the analysis of K63 polyUb networks, we determined protein–protein interactions (PPIs) between UBC35/36 and UEV1A to UEV1D (used as bait) and 12,000 Arabidopsis proteins encoded by sequence-verified ORFs (12k_space; InterAtome) in the Y2H mapping liquid pipeline (Monachello et al., 2019). Such screening of bait proteins against individual ORFs indeed allows the detection of weakly interacting proteins or proteins with low abundance that would be masked in library-based Y2H approaches. The complete ORF library encodes close to 50% of the Arabidopsis proteome and contains hundreds of factors belonging to the ubiquitination machinery, including ∼450 E3 ligases (Supplemental Table 1). This data set allowed us to identify 24 PPIs (Figure 3A; Supplemental Data Set 5). The complete list of interactions and annotations can be used to build networks with Cytoscape and to extract relevant information about K63 polyUb networks and subnetworks reported in the present study (Supplemental Data Set 6). Both UBC35 and UBC36 interacted with the four E2v UEV1A to UEV1D (Figure 3B; Supplemental Data Set 6). These E2v were previously shown to interact with UBC36 (Wen et al., 2008), thereby validating our interactome. Most of the PPIs identified in the original interactome for UBC/UEV1 were also picked up in our new interactome (11 of 13; Figure 3C; Supplemental Data Set 5; Johnson and Vert, 2016), but we have now uncovered 13 new PPIs for UBC35/36 and UEV1A to UEV1D. Overall, 13 E3 Ub ligases, mostly from the RING/U-box family, have been identified. These proteins are therefore likely involved in the selection of substrates to be modified with K63 polyUb chains. Two other proteins related to Ub and protein degradation, but not belonging to the E3 ligase category, were isolated: the COP9 signalosome subunit CSN5A and the otubain-like deubiquitinase. Overall, analysis of the combination of the 12,000 and the 8000 previously published ORF data sets, together with literature-curated interactions, yielded 28 PPIs for the UBC13-based K63 polyubiquitination machinery, with 13 E3 ligases potentially involved in substrate decoration with K63-linked Ub chains (Supplemental Data Set 7).
Figure 3.
Identification of UBC35/36-UEV1A to UEV1D Primary Interacting Proteins by Interactome Analysis.
(A) Primary interacting proteins of UBC35/36-UEV1A to UEV1D identified by Y2H interactome analysis using the 12k_space ORFs from Arabidopsis. Interactions between UBC35/36-UEV1A/B/C/D–interacting proteins are also shown.
(B) Respective interactions between E2 and E2 variants driving K63 polyubiquitination.
(C) Venn diagram showing the overlap between UBC35/36-UEV1A to UEV1D primary interacting proteins coming from the 8k_space (Arabidopsis Interactome Mapping Consortium, 2011) and the 12k_space. Edge color: blue, Arabidopsis Interactome Map (Arabidopsis Interactome Mapping Consortium, 2011); purple, Plant Immune System Network (Mukhtar et al., 2011); black, literature-curated interactions; orange, this study.
Further screening of the ORF library using primary interactants of UBC35/36 and UEV1A to UEV1D identified many secondary interacting proteins (Supplemental Figure 2, Supplemental Data Set 6). Considering that a few proteins act as major hubs and gather hundreds of interactions each, we removed such hubs to better highlight the network of interactions (Figure 4A). Of particular interest is the identification of several partners of UBC35/36-UEV1A/B/C/D–interacting E3s, which presumably represent E3 targets undergoing K63 polyubiquitination. As an example of E2-E3-target reconstitution, the RING E3 ligase encoded by the At5g38895 gene interacts with UBC35 and UBC36 as well as with UEV1C and UEV1D, and also partners with the basic-helix-loop-helix transcription factor HEC2 (Figure 4B). HEC proteins are known to control the female reproductive tract, shoot apical meristem development, and photomorphogenesis in Arabidopsis (Gremski et al., 2007; Schuster et al., 2015). Our interactome analyses suggest that HEC2-dependent functions in plant development may be regulated by K63 polyubiquitination. Overall, the interactome-based characterization presented here yields important perspective into the biological functions of K63 polyubiquitination by identifying molecular actors at stake in pathways and responses shown to require this posttranslational modification.
Figure 4.
Identification of UBC35/36-UEV1A to UEV1D Secondary Interacting Proteins by Interactome Analysis.
(A) Proteins interacting with primary interactants of UBC35/36-UEV1A-D from Figure 2 are shown in pink.
(B) and (C) Examples of E2-E3-target reconstitution for At5g38895 (B) and At1g74370 (C). Edge color: blue, Arabidopsis Interactome Map (Arabidopsis Interactome Mapping Consortium, 2011); purple, Plant Immune System Network (Mukhtar et al., 2011); black, literature-curated interactions; orange, this study.
Identification of K63 Polyubiquitinated Proteins by Sensor-Based Proteomics
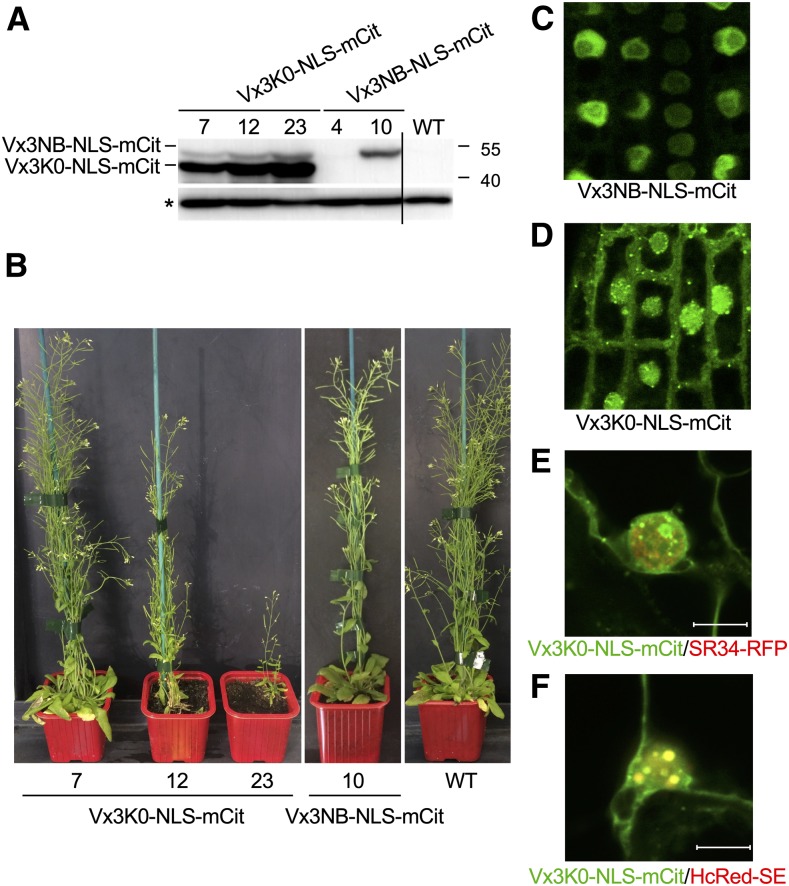
To obtain more direct insights into K63 polyubiquitination, we continued our effort to catalog proteins decorated with K63 polyUb chains in vivo. Using a K63 polyUb-specific sensor to immunopurify proteins modified with K63-linked polyUb chains, we previously isolated ∼100 proteins as part of the K63 polyubiquitome in plants (Johnson and Vert, 2016). To provide a deeper coverage of K63 polyubiquitinated proteins in vivo, we took two complementary approaches in this study. The first strategy was to use a longer column and longer gradient to improve peptide separation during chromatography, thus increasing resolution. The second approach took advantage of transgenic lines expressing a modified version of the K63 polyUb sensor. The original Vx3K0-GFP sensor line shows a mostly cytosolic localization, with enrichment in the plasma membrane and endosomes, but this sensor is globally excluded from the nucleus in Arabidopsis plants (Johnson and Vert, 2016). However, we now know that UBC35 and UBC36, the major determinants of K63 polyUb chain formation, localize to both the cytosol and nucleus (Supplemental Figures 3A to 3C). We therefore created a sensor that is also able to reach the nucleus thanks to the fusion with a nuclear localization signal (Vx3K0-NLS-mCit), together with its nonbinding control (Vx3NB-NLS-mCit). We screened the monoinsertional homozygous transgenic lines for Vx3K0-NLS-mCit and Vx3NB-NLS-mCit based on their expression levels (Figure 5A). Consistent with a previous report, the point mutations introduced in Vx3NB created a mobility shift compared with Vx3K0 (Johnson and Vert, 2016). Care was taken to select lines showing low expression levels for Vx3K0-NLS-mCit to avoid detrimental effects of competing with endogenous K63 polyUb binding proteins (Figure 5B).
Figure 5.
Characterization of Vx3K0-NLS and Vx3NB-NLS K63 PolyUb Sensor Lines.
(A) Immunoblot analysis monitoring the accumulation of the Vx3K0-NLS-mCitand Vx3NB-NLS-mCit proteins in different mono-insertional homozygous transgenic lines using anti-GFP antibodies. The wild-type (WT) plants served as a negative control. The asterisk indicates a nonspecific band used as a loading control. The sizes of marker proteins in kilodaltons are shown.
(B) Phenotypes of the 5-week-old soil-grown wild-type (WT), Vx3K0-NLS-mCit, and Vx3NB-NLS-mCit transgenic lines. Lines Vx3K0-NLS-mCit 7 and Vx3NB-NLS-mCit 10 were selected for further analysis.
(C) to (D) Subcellular localization of Vx3K0-NLS-mCit (C) and Vx3NB-NLS-mCit (D) sensors in the primary roots of Arabidopsis stable transgenic lines. Bar = 10 µm.
(E) and (F) Colocalization analysis between Vx3K0-NLS-mCit and SR34-RFP (E) or HcRed-SERRATE (F) using transient expression in N. benthamiana leaves. Representative images are shown (n = 10). The overlay between the green (mCit) and red (RFP or Hc-Red) channels is shown. Colocalization is revealed by the yellow color observed for Vx3K0-NLS-mCit and HcRed-SE nuclear bodies. Bar = 10 µm.
Observation of the selected transgenic lines by confocal microscopy indicated that the Vx3NB-NLS nonbinding control sensor localized exclusively to the nucleus (Figure 5C). By contrast, the Vx3K0-NLS binding sensor accumulated in the nucleus and cytosol, likely due to partial retention to cytosolic K63 polyubiquitinated proteins immediately after translation (Figure 5D). Vx3K0-NLS was also enriched in dotted structures, which were similar to the FM4-64–positive endosomes observed in Vx3K0-mCit lines (Johnson and Vert, 2016), as well as in nuclear foci. These locations are consistent with the tight connection we and others previously uncovered between K63 polyubiquitination and membrane protein dynamics (Romero-Barrios and Vert, 2018). To decipher the biological relevance of such Vx3K0-NLS–positive nuclear foci, we evaluated the possible colocalization of known proteins marking nuclear foci (Lorković et al., 2008; Raczynska et al., 2014). Vx3K0-NLS-mCit did not colocalize significantly with the general splicing factor SERINE/ARGININE-RICH PROTEIN SPLICING FACTOR34 (SR34-RFP) in nuclei (Pearson’s correlation coefficient = 0.34; Figure 5E; Supplemental Figure 4A). However, extensive overlap with the HcRed-SERRATE protein was observed in nuclear foci (Pearson’s correlation coefficient = 0.88; Figure 5F; Supplemental Figure 4B); this protein plays pivotal roles in various aspects of RNA metabolism, including microRNA biogenesis, constitutive and alternative splicing, biogenesis of noncoding RNAs, and RNA transport and stability. These findings again point to the likely requirement for K63 polyUb-decorated proteins in nuclear processes involving RNA metabolism.
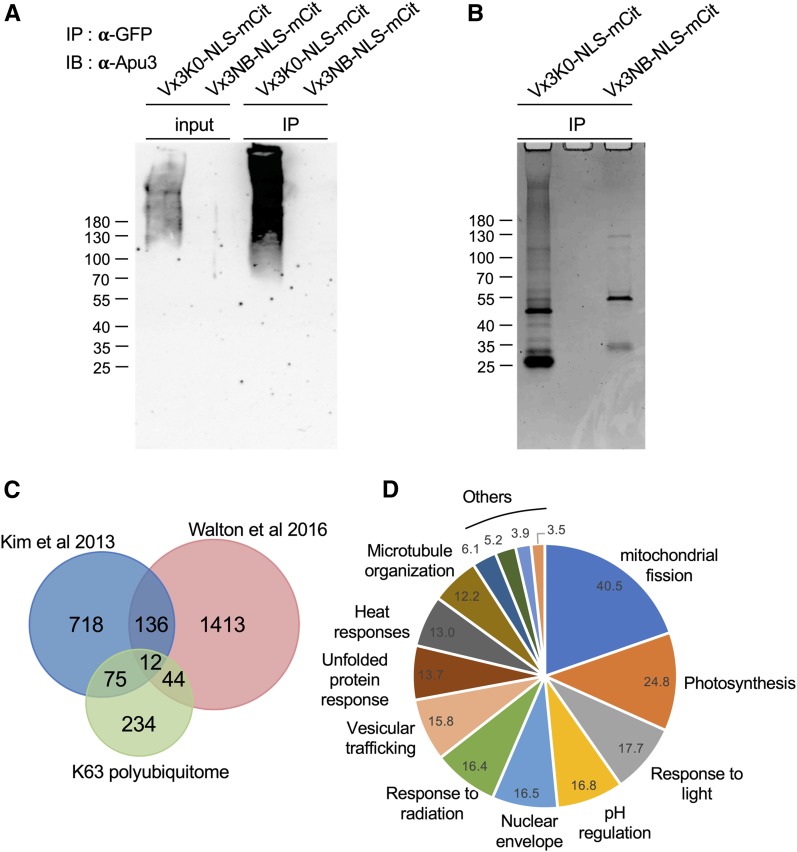
To specifically isolate proteins carrying K63 polyUb chains in vivo and to eliminate possible copurified proteins, we prepared total extracts from Vx3K0-NLS-mCit and Vx3NB-NLS-mCit seedlings using the stringent radioimmunoprecipitation assay (RIPA) buffer, as described previously by Johnson and Vert, (2016). Vx3K0-NLS and Vx3NB-NLS were successfully immunoprecipitated using GFP-coupled microbeads (Supplemental Figure 5). K63 polyUb-decorated proteins were specifically recognized by Vx3K0-NLS, as visualized by probing immunoprecipitates with Apu3 antibodies (Figure 6A). The Ub sensor protected ubiquitinated proteins from deubiquitination in the cell and during the extraction process, as observed previously by Hjerpe et al. (2009) and Johnson and Vert (2016), allowing proteins carrying K63 polyubiquitin chains to accumulate in the input fraction (Figure 6A). To identify the proteins bound by Vx3K0-NLS-mCit, we subjected the immunoprecipitates to electrophoretic separation and Coomassie blue staining (Figure 6B; Supplemental Figure 6), followed by in-gel trypsin digestion and mass spectrometry analyses. A protein was considered for the K63 polyUb proteome catalog if two or more different matching peptides with protein and peptide thresholds of 95% or more were identified in Vx3K0-NLS data sets and absent from the negative control. Using these criteria and after eliminating contaminants, we identified close to 400 Arabidopsis proteins as potentially K63 polyubiquitinated (Supplemental Data Set 8), strongly increasing our resolution in identifying K63 polyubiquitination.
Figure 6.
Isolation and Analysis of the Arabidopsis K63 Ubiquitome.
(A) Sensor-based immunoprecipitation of proteins carrying K63 polyUb chains. Immunoprecipitation was performed using anti-GFP antibodies on RIPA buffer-solubilized protein extracts from mono-insertional homozygous plants expressing Vx3K0-NLS-mCit and the negative control Vx3NB-NLS-mCit. Extracts were subjected to immunoblotting with anti-GFP antibodies (see Supplemental Figure 6) to assess immunoprecipitation efficiency, and with anti-K63 polyUb Apu3 antibodies. The sizes of marker proteins in kilodaltons are shown. IB, immunoblotting; IP, immunoprecipitation.
(B) Coomassie blue staining of Vx3K0-NLS-mCit and Vx3NB-NLS-mCit immunoprecipitates shown in (A).
(C) Venn diagram showing the overlap between proteins carrying K63 polyUb chains purified by sensor-based proteomics and the nonlinkage-resolutive TUBE and COFRADIC ubiquitomes (Kim et al., 2013; Walton et al., 2016).
(D) GO term enrichment of K63 polyubiquitinated proteins identified by mass spectrometry. The enrichment fold is shown.
The recently established TUBE and COFRADIC ubiquitomes serve as the reference list of ubiquitinated proteins (Kim et al., 2013; Walton et al., 2016), although these studies provide no hint about the possible Ub linkages. Among the K63 polyubiquitome, 87 and 56 proteins were already identified as ubiquitinated in the TUBE and COFRADIC proteomes, respectively (Figure 6C). Approximately 50 proteins are also common to the Vx3K0-NLS and Vx3K0 data sets, including many plasma membrane proteins (ATPases, transporters, channels). GO enrichment analysis of the Vx3K0-NLS ubiquitome revealed an over-representation of GO terms that overlapped with our previous Vx3K0 data set, such as GO terms related to metabolism, vesicular trafficking, and transport across membranes (Figure 6D; Supplemental Data Set 9). More interesting is the finding that, using the Vx3K0-NLS sensor, we identified many nuclear proteins that were not identified in our first K63 polyubiquitome (Table 1; Supplemental Data Set 10). GO annotations of the nuclear K63 polyubiquitome revealed enrichment of terms corresponding to nuclear structure, nuclear import/export, chromosome organization, or splicing (Supplemental Data Set 11). In particular, we identified several proteins linked to RNA binding, unwinding, or splicing, which is consistent with the enrichment of Vx3K0-NLS in SERRATE-positive nuclear foci. The nuclear import machinery also appears to be under the control of K63 polyubiquitination, with several nuclear pore complex and factors involved in nuclear cargo recognition and transport (importins, RAN GTPase Activated Protein). Proteins involved in nuclear positioning and the maintenance of nuclear shape, such as SAD1/UNC-84 DOMAIN PROTEIN1 (SUN1), SUN2, LITTLE NUCLEI1 (LINC1), and LINC4, were also found, raising the question about the role of K63 polyubiquitination in these processes. Finally, several histones were found to be K63 polyubiquitinated, including HISTONE VARIANT H2AX, for which K63 polyubiquitination has been tightly linked to DNA damage responses in yeast and mammals (Mattiroli et al., 2012). Overall, these findings demonstrate our ability to better detect K63 polyubiquitinated proteins using Vx3K0-NLS–based proteomics and our improved peptide separation setup, providing excellent resolution of K63 polyUb-dependent processes in plants.
Table 1. Examples of Proteins and Biological Processes Involving Nuclear Proteins Targeted by K63 Polyubiquitination Identified in this study.
| Gene | Name | Description |
|---|---|---|
| RNA | ||
| AT1G06220 | MEE5 | Homolog of splicing factor Snu114 |
| AT1G20960 | BRR2 | DEAD/DExH box ATP-dependent RNA helicase |
| AT1G72730 | DEA(D/H)-box RNA helicase family | |
| AT1G80070 | PRP8 | Pre-mRNA splicing |
| AT3G58510 | DEA(D/H)-box RNA helicase | |
| AT5G07350 | TSN1 | RNA binding protein with nuclease activity |
| AT5G26742 | ATRH3 | DEAD box RNA helicase |
| Nuclear transport | ||
| AT1G14850 | NUP155 | Nucleoporin |
| AT1G68910 | WIT2 | Nuclear envelope docking of RANGAP |
| AT1G79280 | NUA | Nuclear pore anchor |
| AT2G05120 | NUP133 | Nucleoporin |
| AT3G10650 | Nucleoporin involved in mRNA export | |
| AT3G63130 | RANGAP1 | GTPase activating protein involved in nuclear export |
| AT4G16143 | IMPA-2 | Importin alpha isoform 2 |
| AT5G20200 | Nucleoporin-like protein | |
| Chromatin | ||
| AT1G07660 | H4 | Histone H4 |
| AT1G52740 | H2A.9 | Histone H2A |
| AT2G28720 | H2B.3 | Histone H2B |
| AT1G08880 | H2AX | Histone H2AX |
| AT3G06400 | CHR11 | Chromatin remodeling factor |
| AT3G45980 | H2B.9 | Histone H2B |
| AT5G59910 | H2B.11 | Histone H2B |
| Nucleus structure | ||
| AT1G67230 | LINC1 | Peripheral nuclear coiled-coil protein |
| AT3G10730 | SUN2 | Cytoskeletal-nucleoskeletal bridging complex |
| AT5G04990 | SUN1 | Cytoskeletal-nucleoskeletal bridging complex |
| AT5G65770 | LINC4 | Peripheral nuclear coiled-coil protein |
Blank cells indicate no data.
Validation of Nuclear Targets of K63 Polyubiquitination
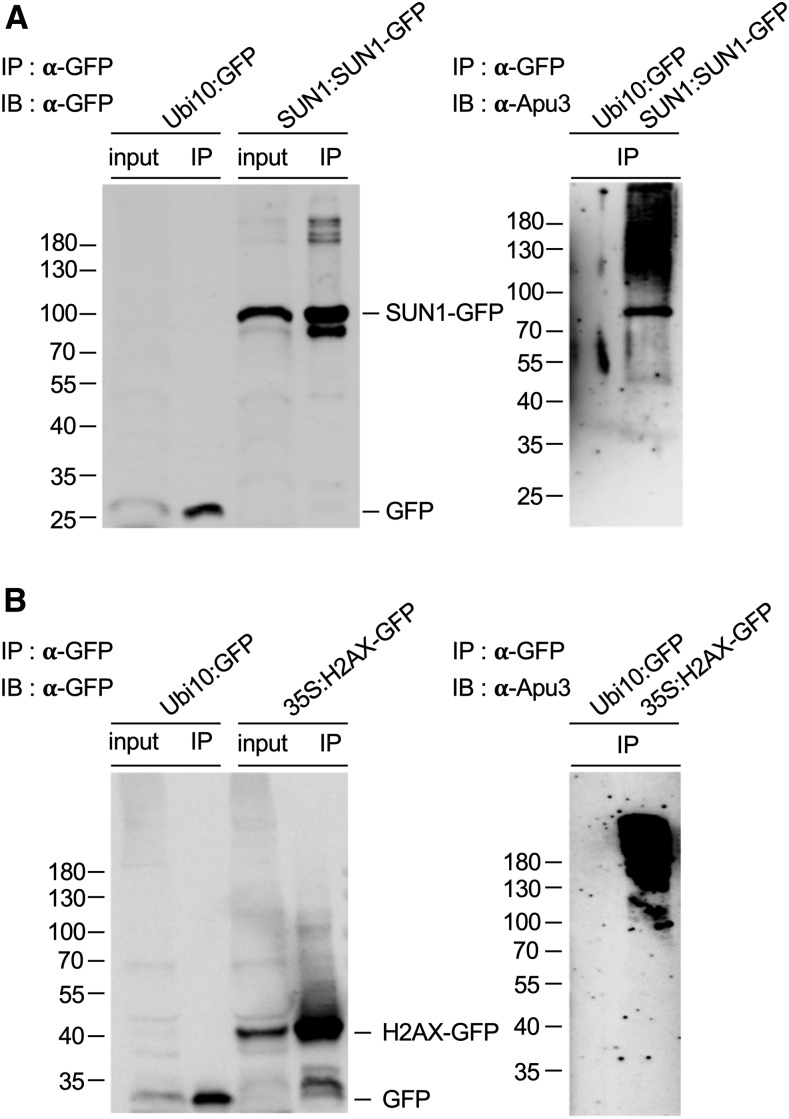
Since the proteins included in the K63 polyubiquitome were identified based on their ability to be recognized by the Vx3K0-NLS K63 polyUb chain sensor, we decided to confirm the presence of K63 polyUb chains on these proteins using another approach. We first used SUN1 and H2AX as a test case. We expressed SUN1 and H2AX in stable Arabidopsis transgenic lines as GFP fusions, extracted the fusion proteins using RIPA buffer amended with deubiquitinase inhibitors, and subjected them to immunoprecipitation using GFP antibodies coupled to magnetic microbeads. We probed the immunoprecipitates with GFP antibodies to confirm immunoprecipitation of the protein of interest and with Apu3 antibodies that specifically recognize K63 polyUb chains. The free GFP negative control showed no signal using Apu3, greatly contrasting with the typical high molecular weight smear observed for SUN1 and H2AX (Figures 7A and 7B). This result confirms that both proteins are indeed decorated with K63-linked Ub chains in planta and validates our K63 polyubiquitome. As an alternative method for rapidly validating our mass spectrometry data, we also tested the transient expression of SUN2, MATERNAL EFFECT EMBRYO ARREST5 (MEE5), and the histone H2B in wild tobacco (Nicotiana benthamiana) leaves. All three proteins showed nuclear localization, with SUN2 localized to the nuclear envelope, MEE5 to the nucleoplasm, and H2B to the nucleolus as well as in nuclear foci (Figures 8A, 8C, and 8E). While the free GFP control yielded no signal upon detection with Apu3 antibodies, all three immunoprecipitated proteins showed a high molecular weight smear (Figures 8B, 8D, and 8F), confirming that these proteins are K63 polyubiquitinated in vivo.
Figure 7.
Validation of in Planta K63 Polyubiquitination for Nuclear Hits Identified by Sensor-Based Proteomics.
(A) and (B) In vivo K63 polyubiquitination of SUN1 and H2AX. Immunoprecipitation was performed using anti-GFP antibodies on RIPA buffer-solubilized protein extracts from mono-insertional homozygous SUN1:SUN1-GFP (A) and 35S:H2AX-GFP (B) transgenic lines. Plants expressing free GFP were used as a negative control. Input and immunoprecipitation samples were subjected to immunoblotting with anti-GFP and anti-K63 polyUb antibodies (Apu3). The sizes of marker proteins in kilodaltons are shown. IB, immunoblotting, IP, immunoprecipitation.
Figure 8.
Validation of K63 Polyubiquitination for Nuclear Hits Identified by Sensor-Based Proteomics Using Transient Expression in Wild Tobacco.
(A), (C), and (E) Subcellular localization of SUN2-GFP (A), MEE5-GFP (C), and H2B.11-GFP (E) transiently expressed in N. benthamiana leaves. Bar = 10 µm.
(B), (D), and (F) In vivo K63 polyubiquitination of transiently expressed SUN2 (B), MEE5-GFP (D), and H2B.11-GFP (F). Immunoprecipitation was performed using anti-GFP antibodies on RIPA buffer-solubilized protein extracts. N. benthamiana plants expressing free GFP were used as a negative control (0). Eluates were subjected to immunoblotting with anti-GFP and anti-K63 polyUb antibodies (Apu3). The sizes of marker proteins in kilodaltons are shown. IB, immunoblotting, IP, immunoprecipitation.
DISCUSSION
The existence of many possible polyubiquitination linkages remains a hurdle in the analysis of the biological roles of Ub (Komander and Rape, 2012). Only recently have scientists started to tackle the functions of subtypes of Ub modifications using antibodies or sensors able to recognize specific linkages, or deubiquitinase able to trim certain chains. In this work, we developed several large-scale approaches to provide a better understanding of K63 polyUb-dependent processes and the corresponding machinery.
Detailed genetic, biochemical, and structural analyses in yeast and humans demonstrated that K63 polyUb chain formation is under the control of the UBC13-type E2s and their UEV1 E2v partners (Hodge et al., 2016). By interacting with E3s that select specific substrates to be modified, UBC13-type E2s and E2v directly transfer Ub moieties to target proteins as K63 polyUb chains. Besides UBC13/UEV1, HECT E3s also have the ability to catalyze K63 polyUb chain formation independently of E2s (Kim and Huibregtse, 2009; Sheng et al., 2012). However, the contribution of UBC13-type E2s to K63 polyUb chain formation was previously thought to be rather modest considering the mild developmental defects (reduced root hair number and bifurcation in response to low iron levels) and altered responses to low iron levels, cold, pathogens, and auxin shown by UBC13-type E2 mutants (Li and Schmidt, 2010; Wen et al., 2014; Wang et al., 2019). Here, we demonstrated that ubc36-1 is only a knockdown allele, preventing the assessment of the true contribution of UBC13-type E2s. Using a new ubc36-2 null allele, we now show that UBC35 and UBC36 are absolutely essential for producing K63-linked polyUb chains in plants and are crucial for plant growth and development. This translates into the altered expression of hundreds of genes involved in the cell cycle, metabolism, and RNA- or microtubule-based processes, consequently affecting biotic and abiotic stress responses upon the loss of K63 polyUb chain formation. Such a prominent role of UBC35/36 also suggests that HECT-type E3s, only seven of which have been found in Arabidopsis, have much more limited and likely specialized functions in the K63 polyubiquitination of a few targets.
UBC13-type E2s act in concert with E3s that are responsible for the recruitment of substrate(s) to be K63 polyubiquitinated. Considering the vast number of E3s encoded by plants genomes, deciphering the identity of E3s that select substrates destined to be K63 polyUb-decorated has been a challenge. To address this issue, we searched for proteins able to interact with UBC35/36 as well as their E2v UEV1A to UEV1D and identified more than a dozen Arabidopsis E3s likely involved in K63 polyubiquitination. This compares to the limited number of E3s that have been shown to target proteins for K63-linked ubiquitination in humans, with the best examples being RNF8, RNF168, Mdm2, TRAF6, cIAP1/2, CHIP, Parkin, UCHL1, TRAF2, and ITCH, together with the HECT E3s HectH9 and NEDD4-2 (Adhikary et al., 2005; Bertrand et al., 2008; Lim and Lim, 2011; Mattiroli et al., 2012). Most of the Arabidopsis E3s uncovered as UBC35/36 or UEV1A to UEV1D interactors are RING/U-Box E3 ligases, with the exception of a BTB-POZ E3 ligase. These findings support the conclusions of previous studies on E2-E3 pairing, revealing interactions between UBC35 and UBC36 with RING and plant U-box E3s (Kraft et al., 2005; Turek et al., 2018).
Surprisingly, no F-box proteins were found to interact with the K63 polyUb E2 machinery, although hundreds are present in the ORF library. This may be explained by the limited capacity of Y2H analysis to detect interactions between UBC35/36 or UEV1A to UEV1D and factors from multi-subunit E3 ligases. However, since no interaction was observed between UBC35/36 or UEV1A to UEV1D and the E2-interacting subunit RBX1 from F-box–containing Cullin-RING E3 ligase CRL1 complexes either, we believe that instead, F-box proteins rarely engage in substrate selection for K63 polyubiquitination. Another important finding is that UBC35 and UBC36 essentially interact with the same E3s, indicating that there is no subfunctionalization of these E2s. Global analysis of the ability of Arabidopsis E2s to interact with a subset of RING E3s already suggested that both UBC35 and UBC36 bind in vitro to the same RING members (Kraft et al., 2005), which is consistent with our current findings. Instead, UEV1 proteins have undergone diversification and specialization in both humans and plants. MMS2 drives UBC13-dependent K63 polyUb in the nucleus and DNA damage responses, while UEV1A is involved in cytoplasmic K63-linked chain formation and drives nuclear factor-κB signaling (Hofmann and Pickart, 1999, 2001; Andersen et al., 2005). Similarly, in B. distachyon, UEV1B and UEV1C primarily accumulate in the nucleus, while UEV1A mostly accumulates in the cytosol (Guo et al., 2016).
Besides UBC35/36-UEV1A/B/C/D–E3 pairing, crucial information when studying ubiquitination processes includes the identities of E3 targets. By reiterating the screening of the 12k_space ORFs with UBC35/36- or UEV1A to UEV1D–interacting E3s, we identified many putative substrates of the K63 polyubiquitination machinery. This allowed us to reconstitute several E2/E2v-E3-target cascades. In addition to such cascades, our interactome also shed light on possible regulators of UBC35/36-UEV1A/B/C/D–interacting E3s, which possibly function in a K63 polyUb-independent manner. For example, the RING protein encoded by the At1g74370 gene interacts with UBC35/UBC36, with itself, and with another RING E3 ligase (At2g29270; Figure 4C). The latter E3 may either be regulated by K63 polyubiquitination catalyzed by the UBC35/36-RINGAt1g74370 machinery, or may instead be a direct regulator of RINGAt1g74370 independently of K63 polyUb chain formation. Globally, validation of the presence of K63 polyUb chains on possible E3 targets is necessary to discriminate between these two scenarios. Regardless, the interactome-based characterization of UBC35/36/UEV1A to UEV1D performed in this study provided crucial insights into K63 polyubiquitination networks.
While our work provided a high-resolution snapshot of the K63 polyubiquitination machinery, there are still several limitations that need to be addressed to broaden our view of K63 polyUb networks. (1) Although well suited to pick up interactions between UBC35/36 or UEV1A to UEV1D with E3s and their targets, our Y2H interactome is restricted to the 12,000 ORFs available. Approximately two-thirds of plant E3s and half of the Arabidopsis proteome remain to be tested, which may reveal additional participants in K63 polyUb formation. (2) Y2H analysis is not the most appropriate approach for probing interactions with membrane-bound or organelle-localized (with the exception of the nucleus) proteins, thereby providing a restricted view on K63 polyUb networks. (3) The identification of non-E3 proteins as interacting with UBC35/36 or UEV1A to UEV1D is puzzling, as these proteins are devoid of typical E2-interacting domains such as RING or U-box domains. These hits may therefore represent indirect interactions bridged by endogenous yeast proteins, for example. Alternatively, these non-E3 hits may use uncharacterized domains to interact with UBC35/36 or UEV1A to UEV1D and may therefore be genuine regulators of E2s. A good example is the otubain-like thioesterase, which interacts with UBC36 and whose OTUB1 human counterpart binds to UBC13 and Ub and inhibits UBC13 catalytic activity (Nakada et al., 2010). (4) Some E3s have the ability to interact with several E2s, thereby recruiting targets to be decorated with different chain types. For example, human Mdm2, RNF8, and cIAP1/2 can trigger both K63- and K48-linked polyubiquitination, while TRAF6 and RNF168 are the only two known E3s that selectively target substrate proteins for K63-linked polyubiquitination (Wang et al., 2012). One should therefore keep in mind that targets identified in our interactome may not represent genuine K63 polyubiquitination targets but may instead be modified with other linkage types. The ability to detect K63 polyubiquitination of targets in planta consequently appears to an essential step for validation.
Using an improved Vx3K0-based sensor and mass spectrometry workflow, we have greatly increased the repertoire of Arabidopsis proteins known to be decorated with K63 polyUb chains in vivo. Although K63 polyUb-modified proteins were greatly enriched in our pipeline, a very large majority of peptides resulting from their tryptic digestion do not carry Ub footprints, which renders the detection of modified peptides difficult, as already reported (Johnson and Vert, 2016). Enriching Vx3K0-NLS–purified proteins for ubiquitinated peptides using Gly-Gly antibodies, which recognize Ub remnants after trypsin digestion (Kim et al., 2011; Udeshi et al., 2013), should strongly improve the detection of K63 polyubiquitination sites. Regardless, our current study reveals a strong connection between K63 polyubiquitination and processes such as the dynamics of organelles and endomembrane (fission of mitochondria, vesicular trafficking), the organization of the nucleus (nuclear envelope, DNA topology, microtubules, and transcription), and photosynthesis.
Although global themes clearly emerged from our RNA-seq interactome, and proteomic analysis of the global functions dependent on K63 polyubiquitination, there is very limited overlap between the different data sets. This finding is expected for RNA-seq, which does not monitor K63 polyubiquitination per se but rather identifies genes whose expression is directly or indirectly modulated by K63 polyubiquitination. The comparison of our proteomic and interactome data sets yielded 16 common proteins. While this analysis provided invaluable information about the entire cascade required to K63 polyubiquitinate these proteins, the overlap overall appears quite restricted. However, mass spectrometry and Y2H approaches are known to generate largely nonoverlapping candidate lists (Gavin et al., 2002) due to inherent differences in the processes monitored. For example, interactions involving membrane proteins cannot be probed in our Y2H interactome. Both approaches, combined with the analysis of amiUBC35/36 transcriptomes, should therefore be considered complementary methods for establishing K63 polyubiquitination networks.
Altogether, the mapping of K63-linked Ub networks greatly increases our understanding of Ub in plants and highlights the contributions of K63 polyUb to a wide array of cellular and physiological functions.
METHODS
Plant Materials and Growth Conditions
Single Arabidopsis thaliana (Arabidopsis) ubc35-1 (WiscDsLox323H12) and ubc36-1 (SALK_047381) mutants were described in a previous study (Li and Schmidt, 2010). ubc36-2 (GABI_836B11) was obtained from Nottingham Arabidopsis Stock Centre and genotyped to isolate homozygous mutants. ubc35-1 ubc36-2 was obtained after crossing, selfing, and genotyping F2 plants.
The wild-type, ubc35-1, ubc36-1, ubc36-2, ubc35-1 ubc36-1, and ubc35-1 ubc36-2 plants and the transgenic lines 35S:UBC35-GFP, 35S:UBC36-GFP, Ubi10:Vx3K0-NLS-mCit, Ubi10:Vx3NB-NLS-mCit, Ubi10:GFP, Ubi10:H2AX-GFP, and Ubi10:H2B.11-GFP were grown under sterile conditions on vertical plates containing half-strength Linsmaier and Skoog medium at 21°C. Plants were cultivated under a 16-h light/8 h dark cycle with a light intensity of 90 µmol m−2 s−1 using Philips 17W F17T8/TL741 bulbs. Plants were grown in soil to obtain progeny or to assess macroscopic phenotypes.
Constructs
ORFs corresponding to UBC35 and UEV1B were amplified and cloned into the pDONR207 vector by Gateway cloning using primers listed in Supplemental Data Set 12. LR recombinations were performed using the pDEST-AD-CYH2 and pDEST-DB destination vectors, allowing N-terminal fusions with the activation domain (AD) or the DNA binding (DB) domain of the GAL4 transcription factor (Dreze et al., 2010). Other baits (UBC36, UEV1A, UEV1C, and UEV1D) were retrieved from the destination vectors derived from the Arabidopsis Interactome (Arabidopsis Interactome Mapping Consortium, 2011).
Vx3K0 and Vx3NB were a kind gift of Robert Cohen (Colorado State University, Fort Collins, CO). The Vx3K0 and Vx3NB cassettes, and UBC35, UBC36, H2AX, and H2B.11 cDNAs were amplified by PCR with primers containing attB1 and attB2 Gateway recombination sites (Supplemental Data Set 12) and cloned into pDONR-P1P2. Expression vectors were constructed by multisite Gateway recombination of the pB7m34GW destination vector with pDONR-P4P1r-pUbi10 or pDONR-P4P1r-p35S for promoters; pDONR-P1P2-UBC35, pDONR-P1P2-UBC36, pDONR-P1P2-Vx3K0, pDONR-P1P2-Vx3NB, pDONR-P1P2-GFP, pDONR-P1P2-H2AX, and pDONR-P1P2-H2B.11 for genes of interest; and pDONR-P2rP3-NLS-mCitrine for fluorescent reporters. MEE5 (At1g06220) was amplified by PCR and cloned at the SmaI site under the control of the 35S promoter in the pPZP212-derived pCH3-GFP vector. SUN1:SUN1-GFP or SUN2:SUN2-GFP binary vectors were kindly provided by Katja Graumann (Oxford Brookes University, Oxford, England). Constructs for the generation of artificial microRNAs targeting UBC35 and UBC36 were prepared as described (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) using primers listed in Supplemental Data Set 12.
Stable transgenic lines were generated by the floral dip method (Clough and Bent, 1998). For each construct, several independent mono-insertional homozygous T3 transgenic lines were selected. Transient expression in wild tobacco (Nicotiana benthamiana) followed routine procedures.
Mutant Genotyping and Characterization
The ubc36-2 and ubc35-1 ubc36-2 mutants were genotyped using primers listed in Supplemental Data Set 12. Primers for the ACTIN2 gene were used as a control. Detection of UBC35/36 transcripts was performed by RT-PCR of total RNA using a Veriti Thermal cycler (Applied Biosystem). Briefly, 1 µg of RNA was subjected to RT using a High Capacity Reverse Transcription kit from Applied Biosystems. PCR was performed using primers for UBC35 (22 cycles), UBC36 (22 cycles), and ACTIN2 (20 cycles). RT-PCR analyses were done in duplicate.
Protein Extraction and Protein Gel Blot Analysis
Total proteins were extracted from a pool of 12-d-old plants. Proteins were separated on a 10% Bis-Tris NuPage gel (Thermo Fisher Scientific) and transferred onto a nitrocellulose membrane. For protein detection, the following antibodies were used: monoclonal anti-GFP horseradish peroxidase coupled (130-091-833, 1/5000; Miltenyi Biotech), anti-UBC13 (37-1100, 1/1500; Thermo Fisher Scientific), anti-Ub (P4D1 3936, 1/2500; Cell Signaling), anti-K63 polyUb (Apu3 05-1308, 1/2000; Millipore), and anti-tubulin (AS10 681, 1/5000; Agrisera). To ascertain that no signal was carried over from one immunodetection to another, each antibody was incubated on independent membranes. Immunoblot experiments were done in duplicate.
Confocal Microscopy
Plant samples were mounted in water and imaged under a Leica TCS SP8 confocal laser-scanning microscope. Images were taken of the root tips from 7-d-old Arabidopsis plants grown in the light or wild tobacco leaves. To image mCit, RFP, and HcRed, 514- and 561-nm laser lines were used. Detection settings were kept constant in individual sets of experiments to allow for a comparison of the expression and localization of reporter proteins. Colocalization studies were performed using the Coloc2 plugin of ImageJ. Fluorescence intensity profiles were also obtained using ImageJ.
Whole-Genome RNA Profiling and Bioinformatic Analysis
Roots from noninduced and induced amiUBC35/36 seedlings grown for 12 d were collected and pooled, and total RNA was extracted using an RNeasy kit and DNase treatment according to the supplier’s instructions (Qiagen). RNA-seq experiments were performed in triplicate. Total RNA was checked for integrity on a bioanalyzer using an RNA 6000 Pico kit (Agilent Technologies). PolyA RNA-seq libraries were constructed from 600 ng of total RNA using a TruSeq Stranded mRNA kit (Illumina) according to the manufacturer’s recommendations. Libraries were pooled in equimolar proportions and sequenced (Single Read 75 pb) on an Illumina NextSeq500 instrument, using a NextSeq 500 High Output 75 Cycles kit. Demultiplexing was performed (bcl2fastq2 V2.15.0) and adapters were removed (Cutadapt1.9.1). Reads were mapped onto the Arabidopsis genome (The Arabidopsis Information Resource 10 [TAIR10] version 2017) with TopHat2. Mapped reads were assigned to features using featureCounts 1.5.0-p2, and differential analysis was performed using DESeq2. Hierarchical clustering was performed with Hierarchical Clustering Explorer (www.cs.umd.edu/hcil/hce) on normalized Log2(gene count) using Euclidean distance. GO enrichment analysis was performed using TAIR (PANTHER GO-Slim Biological Process, GO Ontology database released 2019).
Interactome Analyses
The Y2H high-throughput binary interactome mapping pipeline used here was performed in liquid medium as reported previously (Monachello et al., 2019) and is an adaptation of the solid medium protocol used in the AI-1 project (Dreze et al., 2010). Briefly, low copy number yeast expression vectors expressing DB-X and AD-Y hybrid proteins and the two Y2H strains Saccharomyces cerevisiae Y8930 and Y8800 were used. Yeast genome-integrated GAL2-ADE2 and LYS2:GAL1-HIS3 were used as reporter genes. Expression of the GAL1-HIS3 reporter gene was tested using 1 mM 3-amino-1,2,4-triazole, a competitive inhibitor of the HIS3 gene product. Yeast strains Y8800 MATa and Y8930 MATα were transformed with AD-Y and DB-X constructs, respectively. Prior to Y2H screening, the DB-X strains were tested for their ability to auto-activate GAL1-HIS3 reporter gene expression in the absence of AD-fusion protein. In case of autoactivation, DB-X constructs were removed from the baits and assayed in a reverse screen against the DB-AtORFeome library using their AD-X fusion.
Yeast strains expressing DB-X bait were individually grown (30°C for 72 h) in 5 mL of selective medium (Sc-Leu) and pooled (maximum of 50 individual bait yeast strains). The 127 96-well plates of the (AD)-AtORFeome library were replicated into 32 384-well plates filled with selective medium (Sc-Trp) using the colony picker Qpix2 XT and incubated at 30°C for 72 h. DB-bait pools and the AD-library were replicated into mating plates filled with YEPD medium and incubated at 30°C for 24 h. Mating plates were then replicated into screening plates filled with Sc-Leu-Trp-His + 1 mM 3-amino-1,2,4-triazole medium and incubated at 30°C for 5 d. Only diploid yeast cells with interacting pairs could grow in this medium. In order to identify primary positives cultures, the OD600 values of the yeast cultures were measured using a Tecan Infinite M200 PRO microplate reader. Cultures identified as harboring positive interactions were picked from selective medium and protein pairs were identified by depooling of DB-baits in a similar but targeted matricial liquid assay in which all DB-baits were individually tested against all positive AD-proteins. Identified pairs were cherry picked and checked by PCR and DNA sequencing. Interaction networks were designed with Cytoscape.
Immunoprecipitation
Immunoprecipitation experiments were performed using 12-d-old-seedlings from the wild-type, GFP, Vx3K0-NLS-mCit, and Vx3NB-NLS-mCit plants. Care was taken to use transgenic lines harboring similar amounts of transgene-expressed proteins. To extract total proteins, tissues were ground in liquid nitrogen and resuspended in ice-cold RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% [v/v] Igepal, 0.5% [w/v] sodium deoxycholate, and 0.1% [w/v] SDS) amended with protease inhibitors and 20 mM N-ethylmaleimide (NEM) to inhibit deubiquitinases during protein preparation. The samples were solubilized at 4°C for 30 min before centrifugation at 14,000g for 15 min at 4°C to eliminate cell debris. Supernatants were subjected to immunoprecipitation with a µMACS GFP Isolation kit (Miltenyi Biotec). To extract nuclear proteins and validate nuclear hits from the K63 ubiquitome, tissues were ground in liquid nitrogen and resuspended in ice-cold extraction buffer A (20 mM Tris-HCl, pH 8.0, 0.4 M Suc, 10 mM MgCl2, 5 mM β-mercaptoethanol, 20 mM NEM, and protease inhibitors). After filtration, the extracts were centrifuged at 100g for 20 min at 4°C, and the pellet was resuspended in ice-cold extraction buffer B (10 mM Tris-HCl, pH 8.0, 0.25 M Suc, 10 mM MgCl2, 5 mM β-mercaptoethanol, 1% [v/v] Triton X-100, 20 mM NEM, and protease inhibitors). The extracts were centrifuged at 4000g for 10 min at 4°C, and the pellet was resuspended in ice-cold extraction buffer C (10 mM Tris-HCl, pH 8.0, 1.7 M Suc, 2 mM MgCl2, 5 mM β-mercaptoethanol, 0.15% [v/v] Triton X-100, 20 mM NEM, and protease inhibitors). The nuclei were pelleted at 20,000g before being lysed in nuclear lysis buffer (50 mM Tris-HCl, pH 8.0, 0.1% [w/v] SDS, 10 mM EDTA, 20 mM NEM, protease inhibitors, and 10 units of DNase I). After centrifugation for 10 min at 10,000g, the supernatant corresponded to the total nuclear fraction. Total nuclear fractions were subjected to immunoprecipitation with a µMACS GFP Isolation kit (Miltenyi Biotec). Immunoprecipitates were eluted off of the beads using Laemmli buffer. The samples were boiled for 2 min at 95°C and separated on a 10% Bis-Tris NuPage Gel (Thermo Fisher Scientific).
Mass Spectrometry and Data Analyses
Immunoprecipitates were separated for 10 min on a 10% Bis-Tris NuPage Gel (Life Technologies) and subjected to Coomassie Blue staining before the bands were cut off. In-gel trypsin digestion, reduction, and alkylation followed previously published procedures (Shevchenko et al., 2006; Szabó et al., 2018). Peptides were analyzed separately by nano-liquid chromatography–tandem mass spectrometry in a Triple-TOF 4600 mass spectrometer (AB Sciex) coupled to a nanoRSLC ultra performance liquid chromatography system (Thermo Fisher Scientific) equipped with a trap column (Acclaim PepMap100C18, 75 μm i.d.× 2 cm, 3 μm) and an analytical column (Acclaim PepMapRSLCC18, 75 μm i.d.× 50 cm, 2 μm, 100 Å). Peptides were loaded at 5 µL/min with 0.05% trifluoroacetic acid in 5% acetonitrile and separated at a flow rate of 300 nL/min with a 5 to 35% solvent B gradient for 120 min. Solvent A was 0.1% formic acid in water, and solvent B was 0.1% formic acid in 100% acetonitrile. Nano-liquid chromatography–tandem mass spectrometry experiments were conducted using the data-dependent acquisition method by selecting the 20 most intense precursors for collision-induced dissociation fragmentation with Q1 quadrupole set at low resolution for better sensitivity. The identification of K63 ubiquitome by mass spectrometry was done in duplicate.
Raw data were processed with MS Data Converter software (AB Sciex) to generate .mgf data files, and protein identifications were performed using the MASCOT search engine (Matrix Science, London, England) against the TAIR10 database (version 2017) with carbamidomethylation of cysteines set as the fixed modification and oxidation of methionines and ubiquitinylation (Gly-Gly) of Lys as the variable modifications. Peptide and fragment tolerance was, respectively, set at 20 ppm and 0.05 D. Results were analyzed with Scaffold 4.0 software (Proteome Software). Proteins were considered to be part of the K63 ubiquitome when identified using at least two unique peptides at a 95% probability level for both peptides and proteins. GO enrichment analysis of the target proteins from the K63 ubiquitome was performed using TAIR (PANTHER GO-Slim Biological Process, GO Ontology database released 2019). The subcellular localizations of proteins were predicted using SUBA3 (Tanz et al., 2013).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: UBC35 (At1g78870), UBC36 (At1g16890), H2AX (At1g08880), H2B.11 (At5g59910), MEE5 (At1g06220), SUN1 (At5g04990), SUN2 (At3g10730), ACTIN2 (At3g18780). The identifiers of the T-DNA mutants used in this study are ubc35-1 (WiscDsLox323H12), ubc36-1 (SALK_047381), and ubc36-2 (GABI_836B11). RNA-seq data were deposited at Array Express (accession no. E-MTAB-8437; www.ebi.ac.uk/arrayexpress).
Supplemental Data
Supplemental Figure 1. Characterization of Arabidopsis loss-of-function mutants for UBC35 and UBC36.
Supplemental Figure 2. K63 polyubiquitin networks revealed by interactome analysis.
Supplemental Figure 3. Subcellular localization of UBC35 and UBC36.
Supplemental Figure 4. Co-localization analysis of Vx3K0-NLS-mCit with SR34-RFP and HcRed-SERRATE.
Supplemental Figure 5. Immunoprecipitation of the Vx3K0-NLS-mCit and Vx3NB-NLS-mCit sensors.
Supplemental Figure 6. Coomassie blue staining of Vx3K0-NLS-mCit and Vx3NB-NLS-mCit immunoprecipitates.
Supplemental Table 1. Representation of E3 ligases in the Arabidopsis genome, as reported by Mazzucotelli et al. (2006), and in the 12,000 ORF library.
Supplemental Data Set 1. List of genes upregulated in estradiol-induced amiUBC35/36 lines compared with non-induced (padj<0.01, Fold change >1.5).
Supplemental Data Set 2. List of genes downregulated in estradiol-induced amiUBC35/36 lines compared with non-induced (padj<0.01, Fold change >1.5).
Supplemental Data Set 3. Gene ontology (GO) enrichment analysis on upregulated genes in estradiol-induced amiUBC35/36 lines.
Supplemental Data Set 4. Gene ontology (GO) enrichment analysis on downregulated genes in estradiol-induced amiUBC35/36 lines.
Supplemental Data Set 5. List of primary interacting proteins identified with UBC35/36 and UEV1A-D used as baits against the 12,000 ORF library.
Supplemental Data Set 6. List of interacting proteins identified by ORF-based yeast two hybrid interactome within this study (12k_space, InterATOME; 8k_space, AI1; LCI, literature-curated interaction).
Supplemental Data Set 7. List of interacting proteins identified with UBC35/36 and UEV1A-D combing the 12,000 ORF library, the published Arabidopsis 8000 ORF-based interactome and literature-curated interactions.
Supplemental Data Set 8. List of proteins identified by sensor-based proteomics using the Vx3K0-NLS-mCit sensor line.
Supplemental Data Set 9. Gene ontology (GO) enrichment analysis on proteins identified by sensor-based proteomics.
Supplemental Data Set 10. List of nuclear proteins identified by sensor-based proteomics using the Vx3K0-NLS-mCit sensor line.
Supplemental Data Set 11. Gene ontology (GO) enrichment analysis on nuclear proteins identified by sensor-based proteomics.
Supplemental Data Set 12. List of primers used in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work has benefited from the expertise of the high-throughput sequencing and imaging core facilities of the Institute for Integrative Biology of the Cell (Gif-sur-Yvette, France) and the Plant Science Research Laboratory (Castanet-Tolosan, France). We are thankful to David Cornu from the Institute for Integrative Biology of the Cell SICaPS facility for mass spectrometry analysis. We also thank Robert Cohen (Colorado State University, Fort Collins, CO, USA) and Katja Graumann (Oxford Brookes University, Oxford, England) for sharing the K63 sensor constructs and SUN1:SUN1-GFP or SUN2:SUN2-GFP binary vectors, respectively. This work was supported by the Marie Curie Action (grant PCIG-GA-2012-334021 to G.V.), the Agence Nationale de la Recherche (grant ANR-13-JSV2-0004-01 to G.V.), the French Laboratory of Excellence project “TULIP” (ANR-10-LABX-41 and ANR-11-IDEX-0002-02 to G.V.), and “Saclay Plant Sciences-SPS” (ANR-10-LABX-0040-SPS to G.V., D.M., and C.L.).
AUTHOR CONTRIBUTIONS
N.R.-B. characterized the sensor lines and performed the proteomic analysis; D.M. performed the interactome analysis; U.D. carried out the colocalization experiments with nuclear body markers; A.W. generated amiUBC35/36 lines and performed the RNA-seq experiments; H.S.C. analyzed the RNA-seq data; A.C. isolated and characterized the ubc35 and ubc36 mutants; A.J. studied the subcellular localization of UBC35 and UBC36; C.L. supervised and analyzed the interactome data; and G.V. conceived the project, designed the experiments, analyzed the data, and wrote the article.
References
- Adhikari A., Chen Z.J. (2009). Diversity of polyubiquitin chains. Dev. Cell 16: 485–486. [DOI] [PubMed] [Google Scholar]
- Adhikary S., et al. (2005).. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123: 409–421. [DOI] [PubMed] [Google Scholar]
- Andersen P.L., Zhou H., Pastushok L., Moraes T., McKenna S., Ziola B., Ellison M.J., Dixit V.M., Xiao W. (2005). Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J. Cell Biol. 170: 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium. (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back S., Gorman A.W., Vogel C., Silva G.M. (2019). Site-specific K63 ubiquitinomics provides insights into translation regulation under stress. J. Proteome Res. 18: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 108: E450–E458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand M.J., Milutinovic S., Dickson K.M., Ho W.C., Boudreault A., Durkin J., Gillard J.W., Jaquith J.B., Morris S.J., Barker P.A. (2008). cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 30: 689–700. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dreze M., Monachello D., Lurin C., Cusick M.E., Hill D.E., Vidal M., Braun P. (2010). High-quality binary interactome mapping. Methods Enzymol. 470: 281–315. [DOI] [PubMed] [Google Scholar]
- Dubeaux G., Neveu J., Zelazny E., Vert G. (2018). Metal sensing by the irt1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol. Cell 69: 953–964. [DOI] [PubMed] [Google Scholar]
- Gavin A.C., et al. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Gremski K., Ditta G., Yanofsky M.F. (2007). The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134: 3593–3601. [DOI] [PubMed] [Google Scholar]
- Guo H., Wen R., Wang Q., Datla R., Xiao W. (2016). Three Brachypodium distachyon Uev1s promote Ubc13-mediated Lys63-linked polyubiquitination and confer different functions. Front. Plant Sci. 7: 1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R., Aillet F., Lopitz-Otsoa F., Lang V., England P., Rodriguez M.S. (2009). Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10: 1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge C.D., Spyracopoulos L., Glover J.N. (2016). Ubc13: The Lys63 ubiquitin chain building machine. Oncotarget 7: 64471–64504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann R.M., Pickart C.M. (1999). Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96: 645–653. [DOI] [PubMed] [Google Scholar]
- Hofmann R.M., Pickart C.M. (2001). In vitro assembly and recognition of Lys-63 polyubiquitin chains. J. Biol. Chem. 276: 27936–27943. [DOI] [PubMed] [Google Scholar]
- Johnson A., Vert G. (2016). Unraveling K63 polyubiquitination networks by sensor-based proteomics. Plant Physiol. 171: 1808–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.Y., Scalf M., Smith L.M., Vierstra R.D. (2013). Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.C., Huibregtse J.M. (2009). Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29: 3307–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J., Harper J.W., Gygi S.P. (2011). Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44: 325–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Kamei K., Murata S., Kato M., Fukumoto H., Kanie M., Sano S., Tokunaga F., Tanaka K., Iwai K. (2006). A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25: 4877–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Rape M. (2012). The ubiquitin code. Annu. Rev. Biochem. 81: 203–229. [DOI] [PubMed] [Google Scholar]
- Kraft E., Stone S.L., Ma L., Su N., Gao Y., Lau O.S., Deng X.W., Callis J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139: 1597–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Schmidt W. (2010). A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant J. 62: 330–343. [DOI] [PubMed] [Google Scholar]
- Lim K.L., Lim G.G. (2011). K63-linked ubiquitination and neurodegeneration. Neurobiol. Dis. 43: 9–16. [DOI] [PubMed] [Google Scholar]
- Lorković Z.J., Hilscher J., Barta A. (2008). Co-localisation studies of Arabidopsis SR splicing factors reveal different types of speckles in plant cell nuclei. Exp. Cell Res. 314: 3175–3186. [DOI] [PubMed] [Google Scholar]
- Mattiroli F., Vissers J.H., van Dijk W.J., Ikpa P., Citterio E., Vermeulen W., Marteijn J.A., Sixma T.K. (2012). RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150: 1182–1195. [DOI] [PubMed] [Google Scholar]
- Mazzucotelli E., Belloni S., Marone D., De Leonardis A., Guerra D., Di Fonzo N., Cattivelli L., Mastrangelo A. (2006). The e3 ubiquitin ligase gene family in plants: Regulation by degradation. Curr. Genomics 7: 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monachello D., Guillaumot D., Lurin C. (2019). A pipeline for systematic yeast 2-hybrid matricial screening in liquid culture. Protocol Exchange. 10.21203/rs.2.9948/v1 [Google Scholar]
- Mukhopadhyay D., Riezman H. (2007). Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205. [DOI] [PubMed] [Google Scholar]
- Mukhtar, et al. (2011). Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mural R.V., Liu Y., Rosebrock T.R., Brady J.J., Hamera S., Connor R.A., Martin G.B., Zeng L. (2013). The tomato Fni3 lysine-63-specific ubiquitin-conjugating enzyme and suv ubiquitin E2 variant positively regulate plant immunity. Plant Cell 25: 3615–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada S., et al. (2010). Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466: 941–946. [DOI] [PubMed] [Google Scholar]
- Pan I.C., Schmidt W. (2014). Functional implications of K63-linked ubiquitination in the iron deficiency response of Arabidopsis roots. Front. Plant Sci. 4: 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J.E., Thoreen C.C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S.P. (2003). A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21: 921–926. [DOI] [PubMed] [Google Scholar]
- Pickart C.M., Fushman D. (2004). Polyubiquitin chains: Polymeric protein signals. Curr. Opin. Chem. Biol. 8: 610–616. [DOI] [PubMed] [Google Scholar]
- Raczynska K.D., Stepien A., Kierzkowski D., Kalak M., Bajczyk M., McNicol J., Simpson C.G., Szweykowska-Kulinska Z., Brown J.W., Jarmolowski A. (2014). The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 42: 1224–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Barrios N., Vert G. (2018). Proteasome-independent functions of lysine-63 polyubiquitination in plants. New Phytol. 217: 995–1011. [DOI] [PubMed] [Google Scholar]
- Schuster C., Gaillochet C., Lohmann J.U. (2015). Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 142: 3343–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y., et al. (2012). A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Mol. Cell. Proteomics 11: 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. (2006). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1: 2856–2860. [DOI] [PubMed] [Google Scholar]
- Silva G.M., Finley D., Vogel C. (2015). K63 polyubiquitination is a new modulator of the oxidative stress response. Nat. Struct. Mol. Biol. 22: 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims J.J., Scavone F., Cooper E.M., Kane L.A., Youle R.J., Boeke J.D., Cohen R.E. (2012). Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat. Methods 9: 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó Á., Papin C., Cornu D., Chélot E., Lipinszki Z., Udvardy A., Redeker V., Mayor U., Rouyer F. (2018). Ubiquitylation dynamics of the clock cell proteome and TIMELESS during a circadian cycle. Cell Reports 23: 2273–2282. [DOI] [PubMed] [Google Scholar]
- Tanz S.K., Castleden I., Hooper C.M., Vacher M., Small I., Millar H.A. (2013). SUBA3: A database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res. 41: D1185–D1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek I., Tischer N., Lassig R., Trujillo M. (2018). Multi-tiered pairing selectivity between E2 ubiquitin-conjugating enzymes and E3 ligases. J. Biol. Chem. 293: 16324–16336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udeshi N.D., Svinkina T., Mertins P., Kuhn E., Mani D.R., Qiao J.W., Carr S.A. (2013). Refined preparation and use of anti-diglycine remnant (K-ε-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics 12: 825–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk S.J., Fiskin E., Putyrski M., Pampaloni F., Hou J., Wild P., Kensche T., Grecco H.E., Bastiaens P., Dikic I. (2012). Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Mol. Cell 47: 797–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton A., et al. (2016). It’s time for some “site” seeing: novel tools to monitor the ubiquitin landscape in Arabidopsis thaliana. Plant Cell 28: 6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Gao Y., Li L., Jin G., Cai Z., Chao J.I., Lin H.K. (2012). K63-linked ubiquitination in kinase activation and cancer. Front. Oncol. 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., et al. (2019). Arabidopsis UBC13 differentially regulates two programmed cell death pathways in responses to pathogen and low-temperature stress. New Phytol. 221: 919–934. [DOI] [PubMed] [Google Scholar]
- Wen R., Torres-Acosta J.A., Pastushok L., Lai X., Pelzer L., Wang H., Xiao W. (2008). Arabidopsis UEV1D promotes lysine-63-linked polyubiquitination and is involved in DNA damage response. Plant Cell 20: 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen R., Wang S., Xiang D., Venglat P., Shi X., Zang Y., Datla R., Xiao W., Wang H. (2014). UBC13, an E2 enzyme for Lys63-linked ubiquitination, functions in root development by affecting auxin signaling and Aux/IAA protein stability. Plant J. 80: 424–436. [DOI] [PubMed] [Google Scholar]
- Woelk T., Sigismund S., Penengo L., Polo S. (2007). The ubiquitination code: A signalling problem. Cell Div. 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]