A C2H2-type zinc finger protein gene containing an EAR motif inhibits root hair initiation and elongation by suppressing the expression of genes known to play important roles in root hair initiation and elongation.
Abstract
Cys2His2-like fold group (C2H2)–type zinc finger proteins promote root hair growth and development by regulating their target genes. However, little is known about their potential negative roles in root hair initiation and elongation. Here, we show that the C2H2-type zinc finger protein named ZINC FINGER PROTEIN1 (AtZP1), which contains an ERF-associated amphiphilic repression (EAR) motif, negatively regulates Arabidopsis (Arabidopsis thaliana) root hair initiation and elongation. Our results demonstrate that AtZP1 is highly expressed in root hairs and that AtZP1 inhibits transcriptional activity during root hair development. Plants overexpressing AtZP1 lacked root hairs, while loss-of-function mutants had longer and more numerous root hairs than the wild type. Transcriptome analysis indicated that AtZP1 downregulates genes encoding basic helix-loop-helix (bHLH) transcription factors associated with root hair cell differentiation and elongation. Mutation or deletion of the EAR motif substantially reduced the inhibitory activity of AtZP1. Chromatin immunoprecipitation assays, AtZP1:glucocorticoid receptor (GR) induction experiments, electrophoretic mobility shift assays, and yeast one-hybrid assays showed that AtZP1 directly targets the promoters of bHLH transcription factor genes, including the key root hair initiation gene ROOT HAIR DEFECTIVE6 (RHD6) and root hair elongation genes ROOT HAIR DEFECTIVE 6-LIKE 2 (RSL2) and RSL4, and suppresses root hair development. Our findings suggest that AtZP1 functions downstream of GL2 and negatively regulates root hair initiation and elongation, by suppressing RHD6, RSL4, and RSL2 transcription via the GL2/ZP1/RSL pathway.
INTRODUCTION
Plant cell differentiation is a highly complex process that is precisely regulated in time and space, providing a critical control mechanism for the plant life cycle (Kang et al., 2013). Root hairs are specialized structures produced from root epidermal cells (Peterson and Farquhar, 1996). Root hairs expand the root surface area in the soil, facilitate plant growth and the absorption of nutrients and water, help anchor roots to the soil, and mediate interactions with soil-borne microbes (Böhme et al., 2004; Tanaka et al., 2014). In-depth studies of root hairs have theoretical and practical significance for improving crop nutrient use, water absorption and utilization, and crop yields and quality and are also important for further elucidating the mechanisms underlying cell fate, cell development, and programmed cell death (Bernhardt et al., 2003; Cao et al., 2013; Li et al., 2014).
Root hair development is broadly divided into four stages: root hair cell fate determination, initiation, elongation (tip growth), and maturation (Gilroy and Jones, 2000; Lee and Cho, 2013). Depending on the species of plant, root hairs form from root epidermal cells in different ways; these ways are divided into three categories based on cell location. The first category is random, that is, any epidermal cell may develop into a root hair; most dicotyledonous plants and ferns produce root hairs this way (Clowes, 2000; Pemberton et al., 2001; Tominaga-Wada et al., 2013; Tominaga-Wada and Wada, 2014). The second category is asymmetric cell differentiation; that is, during the later period of epidermal stem cell division, the meristem produces two sizes of epidermal cells, and only the short epidermal cells can divide into root hairs. Root hairs of monocotyledonous plants, the lower ferns, and primitive angiosperms belong to this category (Kim et al., 2006; Kim and Dolan, 2011). For plants in the third category, including cruciferous plants such as Arabidopsis (Arabidopsis thaliana, only epidermal cells located above two cortical cells can develop into root hairs; these epidermal cells are called hair cells (H cells). By contrast, epidermal cells located above only one cortical cell develop into normal epidermal cells, called nonhair cells (N cells; Dolan et al., 1994; Grierson and Schiefelbein, 2002; Lucas et al., 2013). The Arabidopsis root cortex generally comprises eight cells on which approximately 8 H cells and 14 N cells exist. In some cases, N cells may also be transformed into H cells known as ectopic root hair cells (Dolan et al., 1994; Schiefelbein et al., 2009; Lucas et al., 2013).
During the root hair cell fate decision stage, a core combinatorial regulatory complex including the WD40 repeat protein TRANSPARENT TESTA GLABRA1 (TTG1), MYB transcription factor WEREWOLF (WER) or MYB23, and basic helix-loop-helix (bHLH) transcription factor GLABRA3 (GL3) or ENHANCER OF GLABRA3 (EGL3) forms the WER-GL3/EGL3-TTG transcriptional complex. The accumulation of this complex and its expression in N cells positively regulate the homeodomain-Leu-zipper transcription factor GL2, promoting N cell fate specification (Rerie et al., 1994; Lee and Schiefelbein, 1999; Walker et al., 1999; Bernhardt et al., 2003; Schiefelbein, 2003; Pesch and Hülskamp, 2004; Ueda et al., 2005; Ishida et al., 2008; Lin et al., 2015). By contrast, the R3-type MYB transcription factor CAPRICE (CPC) in N cells can move to an area near H cells. CPC competes with WER and binds to the GL3/EGL3-TTG1 complex, thereby inhibiting the expression of GL2 and thus maintaining H cell fate in root epidermal cells (Ishida et al., 2008; Bruex et al., 2012; Kang et al., 2013; Lin et al., 2015). Because GL2 is predominantly expressed in N cells, it is usually considered to be a negative regulator of root hair development, and the activation or inhibition of GL2 determines the fate of root hair development. Therefore, in Arabidopsis, GL2 is thought to represent an important genetic switch in cell fate determination and differentiation during root hair pattern formation (Galway et al., 1994; Masucci et al., 1996; Lin et al., 2015).
Once root hair cell fate has been determined, bHLH-type genes downstream of GL2 play important roles in root hair initiation and elongation. RHD6 and RSL1 (the closest putative paralog of RHD6) encode transcription factors belonging to the class I RSL family that are specifically expressed in H cells. RHD6 and RSL1 are functionally overlapping genes that play a major role in root hair initiation (Masucci and Schiefelbein, 1994; Heim et al., 2003; Menand et al., 2007; Proust et al., 2016). RSL2 and RSL4 are transcription factors belonging to the class II RSL family that play overlapping roles in root hair elongation, with RSL4 having a major role in this process (Yi et al., 2010; Proust et al., 2016). LJRHL1-LIKE1 (LRL1), LRL2, and LRL3, in the LRL I subfamily, play overlapping roles in root hair elongation (Grierson et al., 2014; Schiefelbein et al., 2014; Salazar-Henao et al., 2016), whereas LRL4 and LRL5, in the LRL II subfamily, are negative regulators of root hair elongation (Honkanen and Dolan, 2016).
In addition to MYB-, bZIP-, and bHLH-type transcription factors, Cys2His2-like fold group (C2H2)–type transcription factors also play important roles in this process. Most C2H2-type transcription factors have a unique zinc finger protein structure, with most containing one to four conserved zinc finger protein motifs (QALGGH; Laity et al., 2001; Luo et al., 2012a, 2012b). Moreover, a few zinc finger proteins contain ERF-associated amphiphilic repression (EAR) motifs. The EAR motif in plants is present in the class II AP2/ERF proteins and the C terminus of the C2H2-type zinc finger proteins (transcription factor IIIA [TFIIIA] class), and it reduces both the underlying transcription level of the reporter gene and the transcriptional activation activity of other transcription factors (Kazan, 2006). For example, the C-terminal inhibitory residues of AtERF4 are DLDLNL, the C-terminal inhibitory residues of SUPMAN are DLDLEL, and the C-terminal inhibitory residues of AtZFP10 and AtZFP11 are DLELRL; if a mutation occurs in this domain, the protein’s inhibitory function decreases or disappears (Dinkins et al., 2002, 2003). These proteins may function as transcriptional repressors in gene regulatory networks (Ohta et al., 2001; Ciftci-Yilmaz and Mittler, 2008). The first C2H2-type zinc finger protein discovered, GLABROUS INFLORESCENCE STEMS (GIS), plays an important role in trichome development in Arabidopsis (Gan et al., 2006). GIS homologs such as GIS2, GIS3, ZINC FINGER PROTEIN8 (ZFP8), ZFP5, and ZFP6 also participate in Arabidopsis trichome growth and development (Gan et al., 2007a, 2007b; An et al., 2011; Zhou et al., 2012, 2013; Yan et al., 2014; Sun et al., 2015). More importantly, ZFP5 is a key transcription factor that positively regulates root hair development during the root hair cell fate decision stage (An et al., 2011, 2012).
It is clear that C2H2-type zinc finger proteins play important roles in regulating epidermal cell differentiation. To date, only C2H2-type zinc finger proteins that positively regulate root hair growth and development during the fate decision stage have been identified, whereas negative regulators of this process have not. Moreover, no C2H2 zinc finger proteins have been shown to regulate root hair initiation and elongation. In a study aimed at identifying genes associated with salt gland initiation in Limonium bicolor, a gene encoding a C2H2 zinc finger protein similar to Arabidopsis AtZP1 was found to be differentially expressed, but its function in epidermal cell development is unclear (Yuan et al., 2015).
In this study, we demonstrate that the C2H2-type zinc finger protein AtZP1, which contains an EAR motif, negatively regulates root hair development. Molecular, genetic, and biochemical analyses establish that AtZP1 regulates root hair initiation and elongation by directly targeting bHLH transcription factor genes RHD6, RSL2, and RSL4 and is positively regulated by GL2. These results provide insights into the pathway of root hair cell developmental.
RESULTS
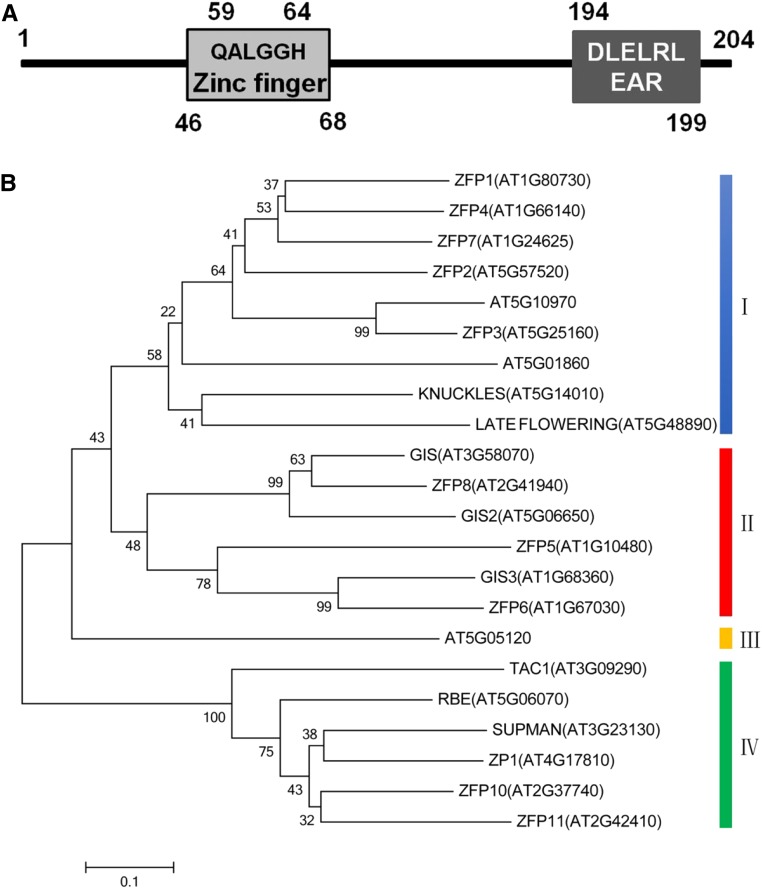
AtZP1 Is a Typical C2H2-Type Zinc Finger Protein Containing an EAR Motif
The AtZP1 gene contains a 615-bp coding sequence (including the stop codon) encoding a 22-kD protein with 204 amino acids. The AtZP1 protein contains one typical C2H2-type zinc finger domain between amino acids 46 and 68 and the conserved sequence QALGGH inside the zinc finger between amino acids 59 and 64. An EAR motif (DLELRL sequence) was present between amino acids 194 and 199 in AtZP1 (Figure 1A). We performed amino acid sequence alignment with MAFFT and found high sequence similarity between AtZP1 and various other C2H2 zinc finger proteins in Arabidopsis. Phylogenetic analysis revealed that different C2H2-type zinc finger protein genes were divided into four branches, while AtZP1 and GIS family genes were divided into IV and II subgroups, respectively, suggesting that they are significantly different in function (Supplemental Files 1 and 2). These results indicate that AtZP1 is a typical C2H2-type zinc finger protein that is unlike GIS family proteins, which promote epidermal cell differentiation.
Figure 1.
Bioinformatics Analysis of AtZP1.
(A) Conserved domain and sequence analysis of AtZP1. Different numbers (1 to 204) and lines represent all the contiguous amino acids of AtZP1 from N terminus to C terminus. Boxes represent conserved domains of the AtZP1 protein. The C2H2 zinc finger domain is the amino acids from 46 to 68, the QALGGH sequence is from 59 to 64, and the EAR motif is from 194 to 199.
(B) Phylogenetic relationships among different C2H2 zinc finger protein family members in Arabidopsis. Phylogenetic analysis of 22 C2H2 zinc finger proteins using the neighbor-joining method. The proteins were divided into four groups (I to IV) corresponding to different branches in the neighbor-joining tree. Bar = 0.1.
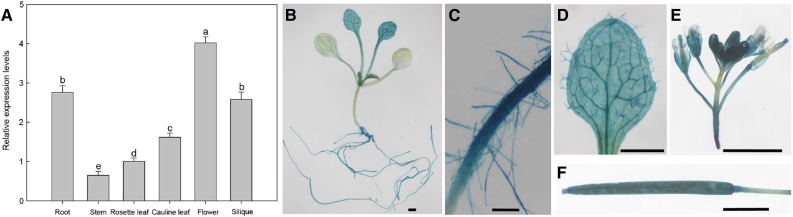
AtZP1 Is Expressed in Root and Epidermal Cells
We analyzed the relative expression levels of AtZP1 in different tissues by RT-qPCR. AtZP1 was expressed mainly in flowers, roots, and siliques (Figure 2A). Subcellular localization in Nicotiana benthamiana showed that AtZP1 is expressed in the nucleus (Supplemental Figure 1). We generated AtZP1 promoter–GUS transgenic lines to further examine the expression pattern of AtZP1. β-Glucuronidase (GUS) staining revealed that AtZP1 was expressed mainly in young leaves and roots (Figure 2B), particularly in maturing trichomes and leaf veins (Figure 2D), the root meristem zone, and the root hairs (Figure 2C), flowers (Figure 2E), and siliques (Figure 2F). These results suggest that AtZP1 has a range of roles throughout plant development, including root hair growth and development.
Figure 2.
Expression Analysis of AtZP1.
(A) Relative expression levels of AtZP1 in different Arabidopsis tissues. Data represent the means of three biological replicates ± sd. Statistical significance was determined by one-way ANOVA, Duncan’s multiple range test. Significant differences at P < 0.05 are represented by different letters (a to e) above the bars.
(B) to (F) GUS analysis of AtZP1 promoter activity in young seedlings (B), mature roots (C), epidermal cells of young leaves (D), flowers (E), and siliques (F). Bar in (B), (D), (E), and (F) = 0.5 cm; bar in (C) = 200 μm.
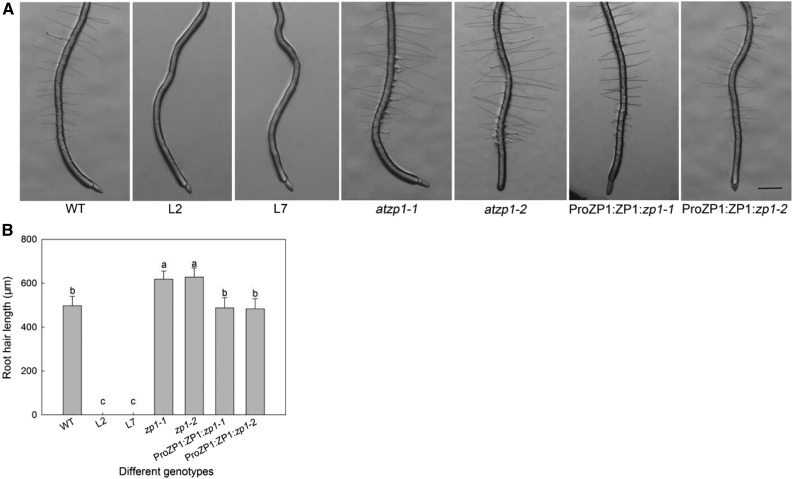
AtZP1 Is a Negative Regulator of Root Hair Development in Arabidopsis
To explore the role of AtZP1 in mediating root hair growth and development, we generated AtZP1 overexpression lines in which the gene was controlled by the Cauliflower mosaic virus 35S promoter, as well as loss-of-function atzp1 mutants (via clustered regularly interspaced short palindromic repeats [CRISPR]-Cas9) and AtZP1 complementation lines in the atzp1 mutant background under the control of the AtZP1 promoter (Supplemental Figure 2). Compared with the wild type (Figure 3A), the overexpression lines lacked root hairs and the atzp1 mutants had considerably longer root hairs. Complementation of atzp1 restored the root hair phenotype to almost that of the wild type (Figures 3A and 3B). The percentage of H cells throughout the epidermis did not significantly differ in the overexpression, loss-of-function, or complementation lines compared with the wild type (Supplemental Table 1). However, the percentage of root hairs was significantly reduced in the overexpression plants (no root hairs) and significantly increased in the mutants, with complementation restoring the number of root hairs in the mutant to the wild-type levels. These results indicate that AtZP1 does not affect H cell differentiation during the root hair cell fate decision stage but negatively regulates root hair initiation and elongation.
Figure 3.
Root Hair Phenotypes of Wild Type, Overexpression Lines (L2 and L7), Loss-of-Function atzp1 Mutants (zp1-1 and zp1-2), and AtZP1 Complementation Lines in the atzp1 Mutant Background (ProZP1:ZP1:zp1-1 and ProZP1:ZP1:zp1-2).
(A) Root hair phenotypes of the wild type (WT), overexpression lines (L2 and L7), atzp1 mutants (zp1-1 and zp1-2), and AtZP1 complementation lines in the atzp1 mutant background (ProZP1:ZP1:zp1-1 and ProZP1:ZP1:zp1-2). Bar = 400 μm.
(B) Average root hair length in the wild type (WT), overexpression lines (L2 and L7), atzp1 mutants (zp1-1 and zp1-2), and complementation lines in the atzp1 mutant background (ProZP1:ZP1:zp1-1 and ProZP1:ZP1:zp1-2). Error bars indicate sd (n = 20). Statistical significance was determined by one-way ANOVA, Duncan’s multiple range test. Significant differences at P < 0.01 are represented by different letters (a to c) in different columns.
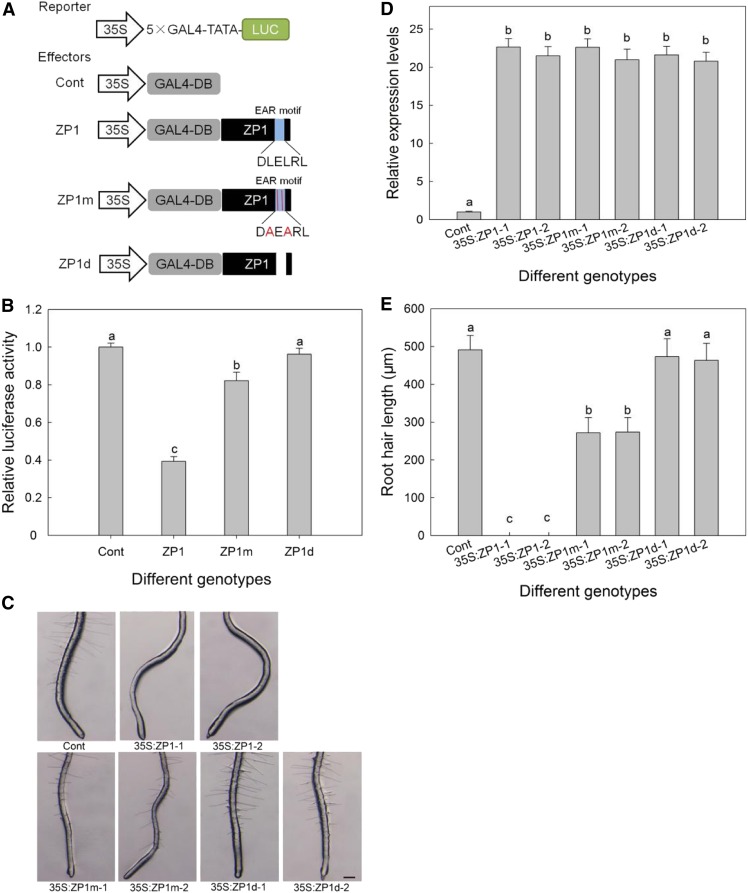
AtZP1 Mainly Acts as a Transcriptional Repressor in the Regulation of Root Hair Development
To investigate whether AtZP1 plays a role in transcriptional activation or transcriptional inhibition of root hair development, we used the luciferase transient expression method to analyze the transcriptional activity of AtZP1. The AtZP1 coding sequence was fused to the GAL4 DNA binding domain to construct the effector, and the luciferase (LUC) gene was fused to the GAL4 binding site to construct the reporter plasmid, both driven by the 35S promoter (Figure 4A). Bioluminescence analysis revealed that in the presence of AtZP1, luciferase activity decreased more than twofold (Figure 4B), indicating that AtZP1 has strong inhibitory activity. Either mutation of the EAR motif of AtZP1 (ZP1m, a Leu-to-Ala substitution) or the deletion of this motif (ZP1d) significantly reduced the inhibitory effect of AtZP1 (Figure 4B), indicating that AtZP1 regulates the expression of downstream genes as a transcriptional repressor and that the EAR motif plays a key role in its inhibitory activity.
Figure 4.
AtZP1 Is a Transcriptional Repressor of Root Hair Growth and Development.
(A) Schematic representation of the reporter and various effector constructs used in the luciferase activity assay. Red font represents mutated amino acids.
(B) Bioluminescence (showing relative luciferase activity) in Arabidopsis protoplasts. Error bars indicate sd (n = 3). Statistical significance was determined by one-way ANOVA, Duncan’s multiple range test. Significant differences at P < 0.01 are represented by different letters (a to c) above the bars. Cont, control group.
(C) Root hair phenotypes of the controls (the wild type; Cont), 35S:ZP1 lines, 35S:ZP1m lines, and 35S:ZP1d lines. Bar = 200 μm.
(D) Relative expression levels of AtZP1 in different types of transgenic Arabidopsis lines determined by RT-qPCR; UBQ10 was used for the internal reference. Cont indicates the control group (the wild-type Arabidopsis). Data are means of three biological replicates ± sd. Statistical significance was determined by one-way ANOVA, Duncan’s multiple range test. Significant differences at P = 0.05 are represented by different letters (a and b) above the bars.
(E) Average root hair length for the controls (the wild type; Cont), 35S:ZP1 lines, 35S:ZP1m lines, and 35S:ZP1d lines. Statistical significance was determined by one-way ANOVA, Duncan’s multiple range test. For each column, significant differences at P < 0.01 are represented by different letters (a to c) above the bars.
To further demonstrate the repressive effect of AtZP1 on root hair development, we generated 35S:ZP1 (carrying the complete EAR motif), 35S:ZP1m (carrying two site mutations in the EAR motif), and 35S:ZP1d (with the EAR motif deleted) transgenic lines (Figure 4D). The 35S:ZP1 lines lacked root hairs, the 35S:ZP1m lines generated a reduced number of root hairs, and 35S:ZP1d lines had a similar phenotype to the control (the wild type; Figure 4C). Moreover, compared with the 35S:ZP1 lines, root hair elongation was only partially inhibited in the 35S:ZP1m lines (Figure 4E).
Furthermore, the percentage of H cells in the root epidermis of the 35S:ZP1, 35S:ZP1m, and 35S:ZP1d lines was similar to that of the control (Supplemental Table 2). Further investigation of root hair percentage at the H cell and N cell position indicated that mutating the EAR motif in AtZP1 significantly relieved the inhibition of root hair growth from the H cell position (Supplemental Table 2). However, the relationship between AtZP1 and root hair patterning genes is unclear.
AtZP1 Possibly Targets bHLH Genes RHD6, RSL2, and RSL4
We investigated the transcriptional changes in two different 3-d-old overexpression lines. Many differentially expressed genes were identified (Supplemental Figure 3A). Analysis using the Biological Networks Gene Ontology tool and Gene Ontology (GO) enrichment analysis showed that the downregulated genes are involved in pathways such as trichoblast differentiation, root hair cell differentiation, and root hair elongation (Supplemental Figure 3B). Three downregulated genes are involved in root hair initiation—the bHLH transcription factor gene RHD6, ROOT HAIR-SPECIFIC GENE18 (RHS18), and MORPHOGENESIS OF ROOT HAIR6 (MRH6)—along with 38 root-hair-elongation–related genes such as the bHLH transcription factor genes RSL2, RSL4, LRL1, and LRL3, the genes EXPANSIN A7 (EXPA7) and CAN OF WORMS1 (COW1), and root-hair-specific genes, but we did not detect changes in the expression of genes involved in the root hair cell fate decision complex, such as CPC/WER-GL3/EGL3-TTG and GL2 (Supplemental Figure 3C).
We selected 11 differentially expressed genes to verify the accuracy of the RNA sequencing (RNA-seq) results and found a high correlation coefficient between the two data sets, confirming the reproducibility and accuracy of our RNA-seq data (Supplemental Figure 4A). We also examined the relative expression levels of RHD6, RSL2, RSL4, LRL1, and LRL3 in the wild-type plants, the overexpression lines, and the mutants by RT-qPCR. The results were consistent with the RNA-seq data (Supplemental Figure 4B). Therefore, we further explored the interactions with bHLH transcription factors such as RHD6, RSL2, RSL4, LRL1, and LRL3 with AtZP1.
AtZP1 Directly Suppresses the Expression of RHD6, RSL2, and RSL4
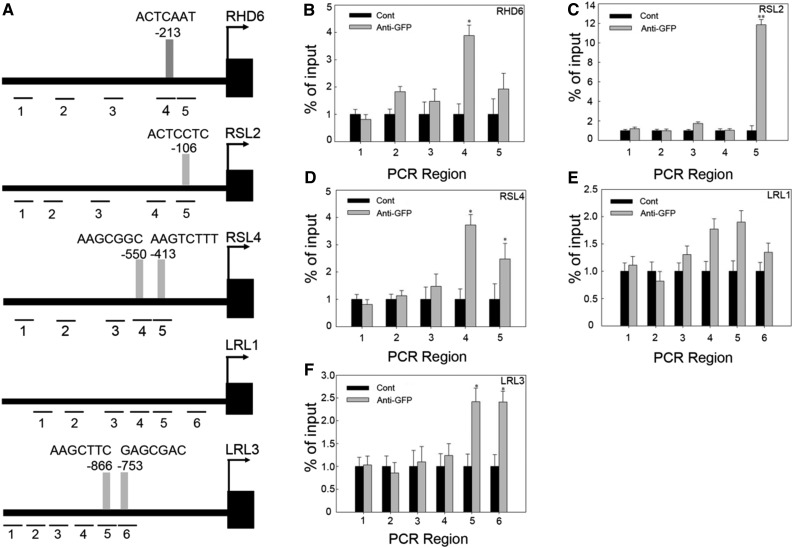
To investigate whether AtZP1 directly regulates these bHLH transcription factors in vivo, we performed a chromatin immunoprecipitation (ChIP) experiment using 3-d-old ProAtZP1:AtZP1:atzp1-GFP seedlings. We selected several sequences similar to A[AG/CT]CNAC, the possible binding site of AtZP1 in the promoter regions of bHLH transcription factors ∼3 kb upstream of the start codon: these possible binding sequences were designated regions 1 to 6, respectively (Figure 5A). The ChIP assay showed that promoter fragments 4 (ACTCAAC) of RHD6, 5 (ACTCCTC) of RSL2, 4 (AAGCGGC) and 5 (AAGTCTTT) of RSL4, 5 (AAGCTTC) and 6 (GAGCGAC) of LRL3, and 4 (AAGCCAC) and 5 (AAGCTAG) of LRL1 were enriched with GFP antibody, whereas the data support an interaction with the other promoters, but not with LRL1 (Figures 5B to 5F). These results demonstrate that AtZP1 directly or indirectly binds to the promoters of these five genes to control root hair initiation and elongation.
Figure 5.
ChIP Analysis Showing That AtZP1 Binds to the A[AG/CT]CNAC Regions of bHLH Gene Promoters.
(A) Conserved sequence A[AG/CT]CNAC (gray bars) and various promoter positions in the candidate bHLH genes examined by ChIP-PCR (lines 1 to 6). The positions in the promoters are relative to the start codons of the bHLH genes. Numbers (1 to 6) and lines represent the possible binding sequences of AtZP1 in the different promoter region. Different bars represent the binding sequence of AtZP1 at the corresponding position of the promoters; numbers and bases show the position and sequence information of AtZP1 binding to the promoter.
(B) to (F) ChIP-PCR enrichment (fold) of the RHD6 gene promoter regions (B), ChIP-PCR enrichment (fold) of the RSL2 gene promoter regions (C), ChIP-PCR enrichment (fold) of the RSL4 gene promoter regions (D), ChIP-PCR enrichment (fold) of the LRL1 gene promoter regions (E), ChIP-PCR enrichment (fold) of the LRL3 gene promoter regions (F). The ChIP experiment was performed with wild-type (Cont) and ProAtZP1:AtZP1:GFP transgenic Arabidopsis. Error bars indicate ± sd from three biological repeats. Values are relative to each Cont value. *P < 0.05 and **P < 0.01 (Student’s t test) represent significant differences from the control value.
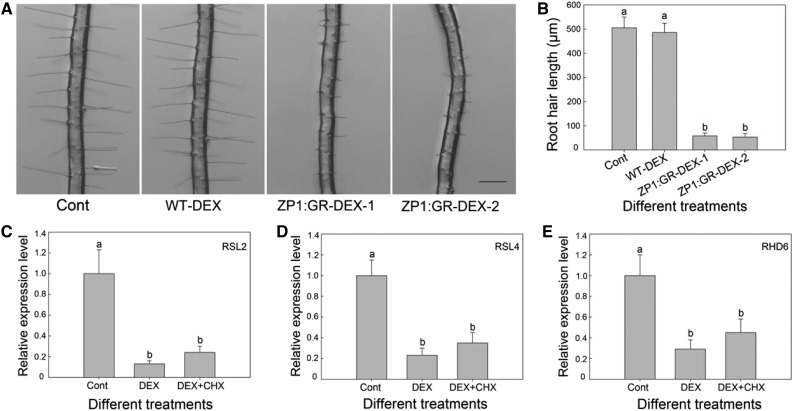
To determine whether AtZP1 regulates root hair development by directly repressing the expression of these bHLH transcription factor genes, we generated AtZP1:GR transgenic plants in the atzp1 background driven by the 35S promoter. AtZP1 expression was immediately induced in the transgenic plants by exogenous application of dexamethasone (DEX; Supplemental Figure 5). When 2-d-old seedlings were transferred to plates containing DEX for 2 d, root hair growth in wild-type seedlings did not significantly differ from that of the control (the wild-type seedlings under normal growth conditions), whereas this treatment significantly suppressed root hair growth in the p35S:AtZP1:GR plants compared with the control (Figure 6A). In addition, root hairs were significantly shorter in p35S:AtZP1:GR seedlings treated with DEX than in control seedlings (Figure 6B). On measuring the relative expression levels of the bHLH transcription factor genes, we found that RHD6, RSL2, and RSL4 were significantly downregulated in the roots of 3-d-old DEX-treated AtZP1:GR seedlings (Figures 6C to 6E).
Figure 6.
AtZP1 Represses the Expression of RSL2, RSL4, and RHD6.
(A) Root hair phenotypes of the wild-type and AtZP1:GR plants after transfer to DEX-containing medium for 2 d. The control shows the wild-type Arabidopsis on standard medium. Bar = 300 μm.
(B) Root hair length of the wild-type and AtZP1:GR plants after transfer to DEX-containing medium for 2 d. Six biological replicates were performed. Error bars indicate sd (n = 6). Statistical significance was determined by one-way ANOVA, Duncan’s multiple range test. Significant differences at P < 0.01 are represented by different letters (a and b) above the bars.
(C) to (E) RT-qPCR analysis of RSL2 (C), RSL4 (D), and RHD6 (E) expression after AtZP1 activation in the presence of DEX or CHX+DEX. Three biological replicates were performed. Error bars indicate sd (n = 3). Statistical significance was determined by one-way ANOVA, Duncan’s multiple range test. Significant differences at P < 0.05 are represented by different letters (a and b) above the bars. Cont, control group.
To investigate whether RHD6, RSL2, and RSL4 are the direct targets of AtZP1, we added the same concentration of the translation inhibitor cycloheximide (CHX) to the DEX induction system. RHD6, RSL2, and RSL4 transcript levels were also significantly suppressed by DEX+CHX treatment compared with those in the control group. These results suggest that AtZP1 directly targets RHD6, RSL2, and RSL4 to regulate root hair initiation and elongation (Figures 6C to 6E; Supplemental Figures 6 and 7).
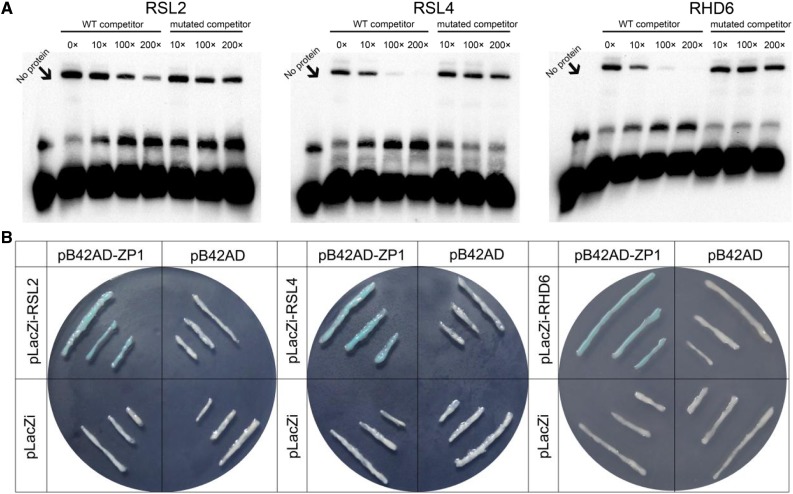
To verify that AtZP1 binds directly to the A[AG/CT]CNAC sequence of the RHD6 (ACTCAAC), RSL2 (ACTCCTC), and RSL4 (AAGTCTTT) promoters in vitro, we performed electrophoretic mobility shift assays (EMSAs) and yeast one-hybrid analysis. In the EMSA, recombinant AtZP1 tagged with glutathione S-transferase with a molecular mass of ∼50 kD was successfully expressed and purified. When the AtZP1 recombinant protein was incubated with biotin-labeled A[AG/CT]CNAC probes, clear signals were observed for RHD6, RSL2, and RSL4. To verify the specificity of the AtZP1 binding, we performed competition experiments. Unlabeled competitors (10- to 200-fold the amount of biotinylated probe) were added to the binding buffer. The interaction signal was significantly reduced by the addition of 100- and 200-fold unlabeled probe. However, when A[AG/CT]CNAC-mutated probes (10- to 200-fold) were used as competitors, no obvious changes in binding signals were detected. These results indicate that AtZP1 specifically binds the promoter sequences of RHD6, RSL2, and RSL4 (Figure 7A). For the yeast (Saccharomyces cerevisiae) one-hybrid assay, LacZ activity was detected in yeast cells cotransfected with different combinations of vectors. These yeast cells grew on SD-Trp-Ura medium, but only cells harboring the pB42AD-ZP1 and pLacZi-A[AG/CT]CNAC vector combination turned blue when grown in SD-Trp-Ura medium containing Z-buffer/X-Gal. By contrast, yeast cells harboring the control combinations of pB42AD and pLacZi, pB42AD-ZP1 and pLacZi, and pB42AD and pLacZi-A[AG/CT]CNAC did not turn blue even after a long incubation. These results help confirm that AtZP1 specifically binds to the A[AG/CT]CNAC sequence in the promoters of RHD6, RSL2, and RSL4 (Figure 7B).
Figure 7.
EMSAs and Yeast One-Hybrid Analysis.
(A) EMSAs showing the binding of AtZP1 to the A[AG/CT]CNAC regions of bHLH gene promoters. glutathione S-transferase–tagged AtZP1 and biotinylated A[AG/CT]CNAC probe were used. The competitor was nonbiotinylated A[AG/CT]CNAC at 10-, 100-, and 200-fold the amount of biotinylated probe. WT, wild type.
(B) Interaction of AtZP1 with the bHLH gene promoters verified by yeast one-hybrid analysis. Detection of LacZ activity in cotransformed yeast with different expression vectors pB42AD-ZP1 and pLacZi-A[AG/CT]CNAC. Three cotransformation combinations, pB42AD and pLacZi, pB42AD and pLacZi-A[AG/CT]CNAC, and pB42AD-ZP1 and pLacZi, were used as the controls.
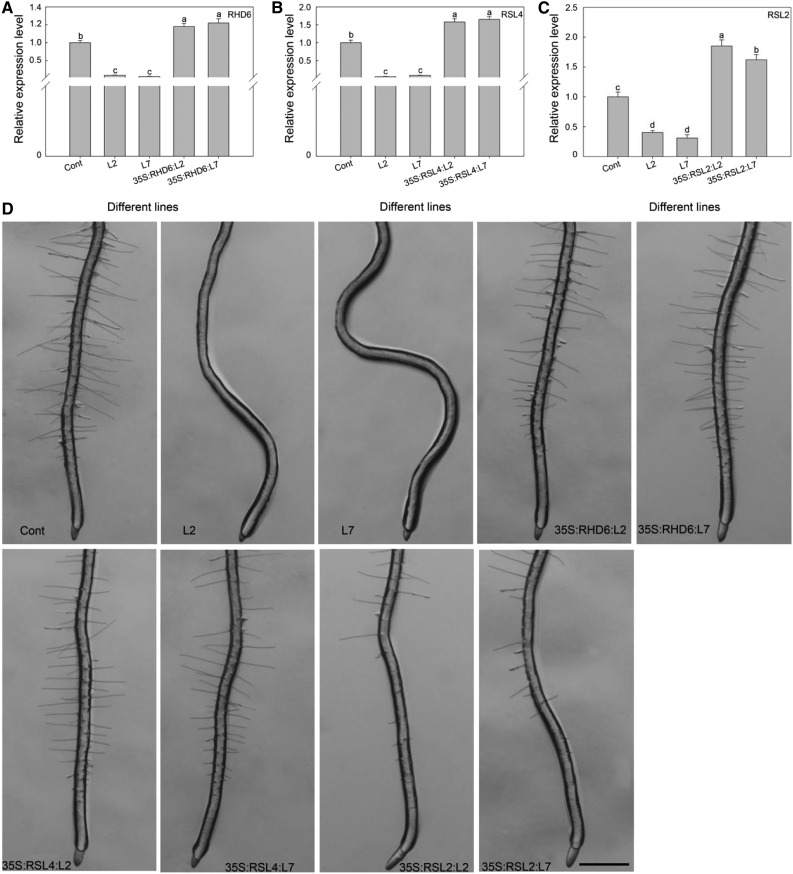
To further determine the genetic relationship between AtZP1- and bHLH-type transcription factors, we overexpressed RHD6, RSL4, and RSL2 in AtZP1-overexpressing plants lacking root hairs. The expression levels of RHD6, RSL4, and RSL2 achieved or exceeded the expression level of the controls in different transgenic lines (Figure 8A). Moreover, the transgenic lines grew differing amounts of root hairs, in a pattern indicating that RHD6, RSL4, and RSL2 restored the root hair loss phenotype at least partially toward wild-type levels (Figures 8A to 8D).
Figure 8.
AtZP1 Functions in the Same Genetic Pathway as RHD6, RSL4, and RSL2.
(A) to (C) Relative expression levels of RHD6 (A), RSL4 (B), and RSL2 (C) in different lines. Three biological replicates were performed. Error bars indicate sd (n = 3). Statistical significance was determined by one-way ANOVA, Duncan’s multiple range test. Significant differences at P < 0.05 are represented by different letters (a to d) above the bars.
(D) Root hair phenotype of the 5-d-old wild-type (Cont) plants; AtZP1 overexpression lines (L2 and L7); and RHD6, RSL4, and RSL2 overexpression lines in the L2 and L7 background. Bar = 500 μm.
AtZP1 Is Regulated by GL2
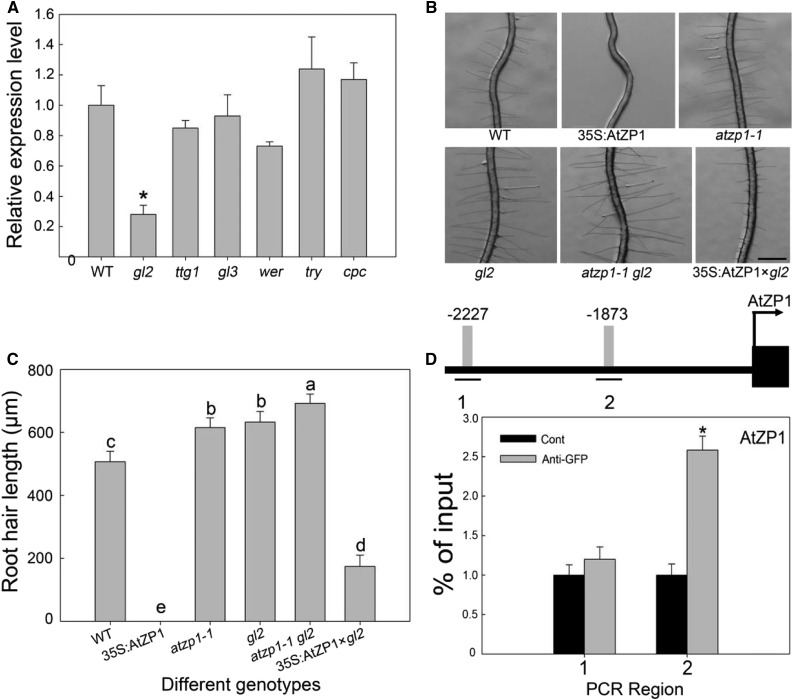
The above-mentioned experiments demonstrated that AtZP1 regulates root hair initiation and elongation by directly binding to the promoter regions of key transcription factor genes RHD6, RSL2, and RSL4. However, the upstream gene regulating AtZP1 expression was unknown. TTG1, GL3, GL2, WER, TRY, and CPC are key genes in root hair cell fate decision. To explore the relationship between AtZP1 and root hair cell-fate–determining genes, particularly GL2, we examined the relative expression levels of AtZP1 in the wild type, gl2, ttg, gl3, wer, try, and cpc plants. AtZP1 was significantly downregulated (more than twofold) in the gl2 mutants, indicating that AtZP1 is positively regulated by GL2, a negative regulator of root hair development (Figure 9A).
Figure 9.
AtZP1 Is Regulated by GL2.
(A) Expression analysis of AtZP1 in various mutants and the wild type (WT). Three biological replicates were performed. Error bars indicate sd (n = 3). Statistical significance was determined by Student’s t tests. *P < 0.05 represents a significant difference.
(B) Root hair phenotypes of the wild-type (WT), 35S:AtZP1, atzp1-1, gl2, atzp1-1 gl2, and 35S:AtZP1×gl2 plants. Bar = 500 μm.
(C) Mean root hair lengths of the wild-type (WT), 35S:AtZP1, atzp1-1, gl2, atzp1-1 gl2, and 35S:AtZP1×gl2 genotypes. Six biological replicates were performed. Error bars indicate sd (n = 6). Statistical significance was determined by one-way ANOVA, Duncan’s multiple range test. Significant differences at P < 0.01 are represented by different letters (a to e) above the bars.
(D) Schematic diagram of the AtZP1 promoter region (3 kb from the start codon, L1 box sequence TAAATGT [gray bars] and its location [lines marked 1 and 2]) and ChIP-PCR enrichment (fold) of the regions (the ChIP experiment was performed with the wild type [Cont] and ProGL2:GL2:GFP transgenic Arabidopsis). The error bars indicate the ± sds from three biological repeats. The values are relative to each Cont value. *P < 0.05 (Student’s t test) represents significant differences from the control value. Numbers (1 and 2) and lines represent the possible binding sequences of GL2 in the different promoter region of AtZP1. Different bars represent the binding sequence of GL2 at the corresponding position of the AtZP1 promoter, numbers (−2227 and −1873) show the position information of AtZP1 promoter.
Finally, to further investigate the relationship between AtZP1 and GL2, we crossed the AtZP1 mutant atzp1-1 with the gl2 mutant. The F2 offspring double mutants of this cross had longer root hairs than their parents (Figure 9C). We also crossed the AtZP1-overexpressing line without root hairs as the female parent with the gl2 mutant as the male parent. The F2 offspring double mutants of this cross had root hairs (Figure 9B). The 3-kb promoter region of AtZP1 contains two GL2-gene-specific recognition sites known as L1 boxes, and the ChIP assay showed that the promoter fragment of AtZP1 was enriched with anti-GFP antibody. These results demonstrate that GL2 directly binds to the AtZP1 promoter to control root hair initiation and elongation (Figure 9D).
DISCUSSION
AtZP1 Is an Unusual C2H2-Type Transcriptional Repressor Zinc Finger Protein That Regulates Root Hair Development
AtZP1 is a C2H2-type zinc finger protein in the TFIIIA superfamily (Benjamin, 2000). AtZP1 contains a plant-specific QALGGH motif (Figure 1) and, surprisingly, it also contains an EAR motif (DLELRL sequence) at its C terminus (Figure 1); this motif generally acts as a transcriptional repressor domain during plant development (Hiratsu et al., 2003; Matsui et al., 2008; Niu et al., 2015). Our luciferase activity assay demonstrated that AtZP1 indeed functions as a transcriptional repressor during root hair development (Figure 4). Plants overexpressing AtZP1 lacked root hairs in the primary root, whereas atzp1 mutants exhibited a significantly increased number of roots hairs, which were elongated (Figures 3A and 3B). In transgenic plants harboring AtZP1 with the EAR motif removed, there was no difference in root hair development from the wild type (Supplemental Table 2). These results suggest that AtZP1 acts as a transcriptional repressor during root hair development and that the EAR motif is the main inhibitory domain in AtZP1.
The C2H2-type zinc finger family is one of the largest gene families in plants. These genes, encoding transcriptional regulators, comprise ∼176 members in Arabidopsis (Laity et al., 2001; Ciftci-Yilmaz and Mittler, 2008; An et al., 2012a). The basic features of AtZP1 differ from those of other family members. Phylogenetic analysis revealed that AtZP1 and GIS family genes are mainly distributed in two separate branches (Figure 1B). GIS and its homologs, such as ZFP8, GIS2, ZFP5, ZFP6, and GIS3, act as transcriptional activators during trichome and root hair development in Arabidopsis (Gan et al., 2007b, 2007a; An et al., 2012b; Zhou et al., 2012, 2013; Yan et al., 2014; Sun et al., 2015). Unlike AtZP1, GIS family proteins do not contain inhibitory domains. These results suggest that these two C2H2 zinc finger protein subfamilies play opposite roles in regulating plant epidermal cell development.
AtZP1 Negatively Regulates Root Hair Development during Root Hair Initiation and Elongation
Our analyses of root hair length and density (Figures 3A and 3B) and the proportion of root epidermal cells that are H cells (Supplemental Table 1) in plants of different genotypes indicated that AtZP1 acts primarily in root hair initiation and elongation rather than in root hair cell fate decision. Transcriptome analysis further demonstrated that AtZP1 significantly repressed the expression of genes involved in trichoblast differentiation, root hair cell differentiation, and root hair elongation (Supplemental Figure 3B). During root hair cell fate specification, no changes were found in the expression of positive regulatory genes CPC and TRY, the negative regulatory genes WER, GL3, EGL3, and TTG1, or their downstream gene GL2 (Supplemental Figure 3C; Rerie et al., 1994; Masucci et al., 1996; Lee and Schiefelbein, 1999; Walker et al., 1999; Schellmann et al., 2002; Bernhardt et al., 2003; Schiefelbein, 2003; Kirik et al., 2004; Pesch and Hülskamp, 2004; Ueda et al., 2005; Simon et al., 2007; Tominaga et al., 2008; Lin et al., 2015). By contrast, key root hair initiation genes such as RHD6, RHS18, and MRH6 and key root hair elongation genes such as RSL2, RSL4, LRL1, and LRL3 were significantly downregulated during this stage of development (Supplemental Figure 3C; Menand et al., 2007; Yi et al., 2010; Pires et al., 2013; Marzol et al., 2017). These findings suggest that AtZP1 functions in root hair initiation and elongation by regulating downstream genes such as the RSL genes, implying that the role of AtZP1 is quite different from that of other zinc finger proteins in terms of its location in the root hair development signaling pathway (Yi et al., 2010; Marzol et al., 2017).
AtZP1 Directly Binds to the Promoters of RHD6, RSL2, and RSL4 and Thereby Modulates the Downstream Genes for Root Hair Initiation and Elongation
bHLH transcription factors play a central role in regulating root hair growth and development (Rymen et al., 2017). The RSL and LRL family genes function in root hair initiation and elongation and regulate the expression of many of their downstream genes during this process (Yi et al., 2010; Salazar-Henao et al., 2016; Hwang et al., 2017). Transcription factors can bind to the promoter regions of downstream genes to regulate their expression. The differentially expressed bHLH genes contain several A[AG/CT]CNAC binding sequences for C2H2-type zinc finger proteins in their promoter regions (Sakai et al., 1995; Kubo et al., 1998; An et al., 2012). In our in vivo ChIP experiments, AtZP1 bound to the promoter regions of bHLH transcription factors genes RHD6, RSL2, RSL4, LRL1, and LRL3. Interestingly, most of the binding-site sequences were near the transcription start sites of the downstream bHLH genes, and the binding-site sequences differed among bHLH transcription factors (Figure 5). These results suggest that AtZP1 directly or indirectly binds to the promoters of these bHLH transcription factor genes in vivo (Hwang et al., 2017).
We then used the AtZP1:GR system to screen for direct target genes of AtZP1 (Zuo and Chua, 2000; An et al., 2012). Among the bHLH transcription factor genes, only RHD6, RSL4, and RSL2 were significantly downregulated under both CHX and DEX treatment, indicating that they are direct target genes of AtZP1 (Figure 6). EMSAs and yeast one-hybrid assays indicated that AtZP1 interacts with the A[AG/CT]CNAC elements in the promoter regions of RHD6, RSL4, and RSL2 (Figure 7). These results provide direct evidence that AtZP1 regulates root hair initiation and elongation by directly regulating the expression of bHLH transcription factor genes such as RHD6, RSL4, and RSL2.
Once the fate of root hair cells has been decided, the class I RSL bHLH transcription factors RHD6 and RSL1 control root hair initiation during the earliest stage of development (Masucci and Schiefelbein, 1994; Menand et al., 2007; Kim and Dolan, 2016; Proust et al., 2016; Kim et al., 2017). Overexpressing RHD6 in the AtZP1 overexpression line background restored the root hair phenotype to the wild type in some extent (Figure 8), illustrating the important role of RHD6 in root hair initiation and function downstream of the AtZP1 signaling pathway.
Root hair length is determined by the continuous growth of the root hair tip (Knox et al., 2003). RSL4 is a direct target gene of RHD6, a bHLH transcription factor belonging to the class II RSL subfamily that promotes the earliest stage of root hair elongation along with its orthologous gene RSL2. An RSL4 loss-of-function mutant has significantly reduced root hair length and density, whereas plants overexpressing RSL4 have the opposite phenotype (Yi et al., 2010). Furthermore, rsl2 mutants exhibit root hair branching (Menand et al., 2007), indicating that RSL4 and RSL2 are required for the proper localization of root hairs in root epidermal cells and for maintaining root hair elongation (Franciosini et al., 2017). RSL4 is expressed in the trichoblast elongation region, and RSL4 protein levels determine the final size of the root hair (Yi et al., 2010; Datta et al., 2015). Plants expressing RSL4 driven by the GL2 promoter in the wild type or rhd6 mutant backgrounds exhibited root hair development from trichoblasts. This finding suggests that the function of RSL4 in inducing root hair growth and development may be independent of the RHD6/RSL1 signaling pathway (Hwang et al., 2017).
Overexpressing RSL4 in the AtZP1-overexpression background also restored the root hair phenotype to that of the wild type to some extent; the effect of this gene was weaker than that of RHD6 but stronger than that of RSL2 (Figure 8), demonstrating that RSL4 is under the control of the AtZP1 signaling pathway. Many genes controlling root hair morphogenesis are regulated by RSL4 (Won et al., 2009; Yi et al., 2010; Vijayakumar et al., 2016). Yi et al. (2010) identified 88 downstream genes and Vijayakumar et al. (2016) identified 34 direct target genes of RSL4, respectively, including genes encoding cell wall modification proteins, phosphatidylinositol transfer proteins, and vesicle transport–related proteins, which are required for root hair elongation (Yi et al., 2010; Vijayakumar et al., 2016). RSL4 also promotes the production of reactive oxygen species by regulating the expression of its downstream NADPH oxidases, C and J (also known as RESPIRATORY BURST OXIDASE HOMOLOG [RBOH] proteins), and several class III apoplastic peroxidases (Hwang et al., 2017; Marzol et al., 2017). These RSL4-regulated genes function in reactive oxygen species homeostasis, metabolism, cell wall synthesis and modification, and signaling pathways, and they comprise the minimal subset of genes required for root hair growth and development. Most genes that were downregulated during root hair elongation are also downstream genes of RSL4 (Supplemental Figure 3C); these genes might be indirectly regulated by AtZP1 (Hwang et al., 2017). These results indicate that AtZP1 mainly plays a negative regulatory role in the RHD6/RSL4/RSL2 pathway.
AtZP1 Is Regulated by the Root Hair Fate Determination Protein GL2
RHD6, RSL1 RSL2, LRL1, and LRL2 are directly negatively regulated by GL2 during root hair development (Lin et al., 2015). GL2 is generally considered to be a negative regulator of root hair growth and development and is mainly expressed in N cells, and gl2 mutants produce ectopic, long root hairs (Lin et al., 2015). In trichoblasts, the loss of function of GL2 leads to RHD6 expression (Hwang et al., 2017). The functions of RSL1, RSL2, LRL1, and LRL2 are also inhibited by GL2 in N cells (Lin et al., 2015). In other words, a pathway has been identified that is negatively regulated by GL2 comprising the downstream bHLH initiation and elongation genes involved in root hair development.
Our study revealed that AtZP1 has a similar function as GL2. What is the relationship between AtZP1 and GL2? The expression level of AtZP1 was significantly downregulated in the gl2 mutant, indicating that AtZP1 functions downstream of GL2 and is positively regulated by GL2. In addition, in N cells, PHOSPHOLIPASE Dz1 (PLDz1) is directly inhibited by GL2, and the ectopic expression of this gene in N cells results in the initiation of root hairs at N cells, indicating that PLDz1 is involved in root hair growth and development (Ohashi et al., 2003). CELLULOSE SYNTHASE5 and XYLOGLUCAN ENDOTRANSGLYCOSYLASE17 are two other target genes of GL2 that function in polysaccharide synthesis, but their specific roles in root hair development require further study (Tominaga-Wada et al., 2009). Analysis of the progeny of atzp1 and gl2 (Figure 9B) demonstrated that AtZP1 and GL2 are both negative regulators of this process. By contrast, analysis of the progeny of the AtZP1 overexpression line and the gl2 mutant (Figure 9B) indicated that AtZP1 and GL2 function in the same signaling pathway and that GL2 functions upstream of AtZP1. Moreover, the presence of the L1 box sequence in the promoter region of AtZP1 and the results of ChIP analysis suggest that AtZP1 is a direct target gene of GL2 (Figure 9D). Therefore, AtZP1 is a previously unidentified target gene of GL2 and is an essential component of the root hair growth and development signaling pathway.
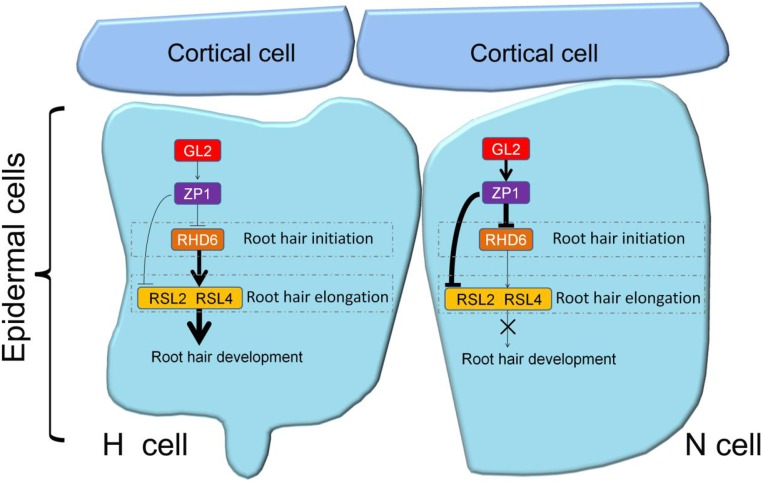
Based on these findings, we propose a model for the role of AtZP1 in regulating root hair development. According to this model, root hair development is regulated by the GL2/ZP1/RSL module in N cell files. GL2 is mainly expressed in this type of cell. AtZP1 is positively regulated by GL2, and the downstream genes of AtZP1 whose expression is repressed by AtZP1 are divided into two categories. The class I RSL gene RHD6 forms the first category and is predominantly responsible for root hair initiation. Class II RSL genes RSL4 and RSL2 form the second category, which are mainly responsible for root elongation. In this signaling pathway, AtZP1 inhibits the initiation and elongation of root hairs in N cells. In H cells, GL2 is rarely expressed; consequently, its downstream gene AtZP1 is rarely expressed. Therefore, AtZP1 together with GL2 cannot inhibit the expression of root hair initiation and elongation genes, leading to root hair growth (Figure 10).
Figure 10.
A Model Showing How AtZP1 Regulates the Expression of bHLH Transcription Factor Genes to Promote Root Hair Development.
GL2 positively regulates AtZP1, and AtZP1 directly negatively regulates RHD6, RSL4, and RSL2. RHD6 directly regulates RSL4. The thickness of the arrow represents the relative level of gene expression.
METHODS
Plant Materials and Growth Conditions
The wild-type Arabidopsis (Arabidopsis thaliana) ecotype Colombia-0 was used as the control in this study. The Wer (CS6349), gl2 (CS67762), ttg1 (CS8065), cpc (CS6399), try (CS6518), and gl3 (CS67763) mutants were obtained from the Arabidopsis Biological Resources Center. The plants were grown at 18 to 22°C under a light intensity of 120 μmol m−2 s−1 with Philips F25T/TL841 25-W bulbs, a 16-h/8-h light/dark photoperiod, and 70% relative humidity. The Arabidopsis seeds were disinfected with 75% (v/v) ethanol and 95% (v/v) ethanol three times for 4 min and twice for 1 min, respectively, and washed several times with sterile distilled water. The seeds were sown in one-half strength Murashige and Skoog Medium (MS) medium, incubated at 4°C in darkness for 3 to 4 d, and cultured vertically under normal light/dark conditions. For transformant screening, the appropriate concentration of antibiotic was added to one-half strength MS medium (50 mg L−1 kanamycin, 50 mg L−1 hygromycin). Seedlings showing antibiotic resistance were transplanted into small pots filled with soil.
Cloning and Bioinformatics Analysis
The coding sequences of AtZP1 were acquired from The Arabidopsis Information Resource website and cloned. Online tools from National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/), PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), SMART (http://smart.embl-heidelberg.de/), and ExPASy (https://www.expasy.org/) were used for nucleic acid sequence and protein sequence analysis and functional domain prediction (Luo et al., 2012a). MEGA5.1 software was used for phylogenetic tree construction by the neighbor-joining method, with statistical support for the nodes from at least 1000 trials.
Expression Pattern of AtZP1 in Different Tissues
Arabidopsis tissues including roots, stems, rosette leaves, flowers, and pods were collected and used to measure the relative expression levels of AtZP1. The samples were immediately frozen in liquid nitrogen and stored at −80°C prior to analysis. For RT-qPCR, three biological replicates were performed. AtZP1 and UBQ10 were used as internal controls and were amplified using primers listed in Supplemental Table 3 (Li et al., 2015).
Promoter–GUS Reporter Analysis
The AtZP1pro:GUS lines were constructed in the wild-type background. The 2-kb target sequence upstream of the start codon was amplified using appropriate forward and reverse primers (Supplemental Table 4), inserted into the pENTR/D-TOPO vector (Invitrogen; http://www.invitrogen.com), and recombined into the destination vector pBGWFS7 using the Gateway linear recombination (LR) reaction (Omar and Ismail, 2016). T0 transgenic plants were selected using the herbicide BASTA and confirmed by PCR amplification. T3 generation homozygous plants were used for analysis. Histochemical GUS analysis was performed as described previously by Lin et al., (2015). The tissues were immersed in 90% (v/v) acetone for 0.5 h at room temperature and washed three times with 0.1 M Na2HPO4, pH 7.0. The samples were placed under a vacuum for 0.5 h, incubated at 37°C overnight in GUS staining solution, dehydrated in 70% (v/v) ethanol three to five times, and viewed under a dissecting microscope (Troll and Lindsley, 1955; Jefferson et al., 1987; Lin et al., 2015).
Subcellular Localization Assay of AtZP1
The coding sequence of AtZP1 was obtained using appropriate forward and reverse primers (Supplemental Table 5). AtZP1 was cloned into the pENTR/D-TOPO Gateway vector and transferred into the pB7FWG2 destination expression vector by LR reaction under the control of the Cauliflower mosaic virus 35S promoter. The vector was transformed into Agrobacterium tumefaciens GV3101 and used to infect Nicotiana benthamiana epidermal cells. A two-photon laser scanning confocal microscope (TCS S8MP, Leica) was used to determine the fluorescent signals of the zinc finger protein AtZP1 in subcellular organelles (Wunder et al., 2013).
Generation of AtZP1 Overexpression Lines
To produce the AtZP1 overexpression plants, the coding sequence of AtZP1 was amplified using appropriate forward and reverse primers (Supplemental Table 6) and cloned into pENTR/D-TOPO Gateway vector. The coding sequence was inserted into the destination vector pB7WG2D by LR reaction (Karimi et al., 2002; Lim et al., 2016) and transferred into Agrobacterium GV3101. The plants were transformed by Agrobacterium-mediated transformation (Wang et al., 2011).
Isolation of AtZP1 Loss-of-Function Mutants by CRISPR-Cas9
To produce the AtZP1 loss-of-function mutants, the gene-editing tool CRISPR-Cas9 was used to knockout AtZP1. The targets were screened using the online tool CRISPR-PLANT (http://www.genome.arizona.edu/crispr/CRISPRsearch.html). The coding sequence (626 bp) of AtZP1 targets carrying the appropriate restriction enzyme sites (KpnI) were amplified using the pCBC-DT1T2 vector as a template using two pairs of primers (Supplemental Table 7). The target fragments of AtZP1 were ligated into final expression vector pHEC401 driven by the U6-26 and U6-29 promoters. The pHEC401-2gR vector was transformed into the wild-type Arabidopsis by Agrobacterium-mediated transformation (Gao et al., 2013; Li et al., 2013; Lei et al., 2014).
Generation of the AtZP1 Complementation Line in the atzp1 Mutant Background
The entire AtZP1 genomic fragment, including the promoter region and the complete coding sequence, was amplified with appropriate forward and reverse primers (Supplemental Table 8), using the wild-type Arabidopsis DNA as a template. The fragment was directly ligated into the expression vector pBGWFS7 with no promoter and transformed into the atzp1 mutant to obtain the AtZP1 complementation line.
Observation of Root Hair Phenotypes and Measurement of Root Hair Length
Root hair phenotypes were visualized using a dissecting microscope (SZX16, Olympus; Zhao et al., 2016) using ∼5-d-old seedlings; ∼4-mm root tip sections were photographed with a digital camera.
Root hair length was measured using ImageJ software. For each genotype, the 20 longest root hairs were measured, and at least 15 seedlings were used (An et al., 2012b). To determine the number of H and N cells in the root epidermis in various genotypes, two regions along the root of a 5-d-old seedling were randomly selected using a fluorescence microscope (ECLIPSE 80i). For each region, five cells closely connected to the H position and five cells closely connected to the N position were selected to count H cells and N cells. For each phenotype, 10 seedlings were selected; thus, the results represent 100 cells (5 cells × 2 locations × 10 plants) in the H position and 100 cells in the N position (An et al., 2012b).
Transient Luciferase Expression Assay
The reporter plasmid of GAL4-LUC was constructed as reported previously by Ikeda et al. (2009). For the effector plasmids, AtZP1 and its various mutant sequences were amplified, and NdeI and SalI cleavage sites were added to the forward and reverse primers, respectively (Supplemental Table 9). The coding sequences of AtZP1, ZP1m (EAR-motif amino acid mutation), and ZP1d (EAR-motif amino acid deletion) were ligated to the pGBKT7 vector (Clontech). The various coding region sequences and the binding domain fusion sequences were then amplified using specific primers (Supplemental Table 9) and ligated to the final expression vector p2GW7 using the LR reaction. A transient expression assay was performed with Arabidopsis protoplasts as described previously by Asai et al. (2002). For each experiment, 5 μg of reporter plasmid and 4 μg of effector plasmid were used to transform Arabidopsis protoplasts, and 0.5 μg of pRLC plasmid was used as an internal control to normalize the activity of the reporter gene (Wang et al., 2010). To test the effects of different types of AtZP1 mutant genes on root hair phenotype, the coding regions of ZP1, ZP1m, and ZP1d were amplified and cloned into the p2GW7 expression vector.
RNA Extraction and Transcriptomic Profiling
Total RNA was isolated from the primary roots (5 mm to the root tip) of the 3-d-old wild type and two independent AtZP1 overexpression lines without root hairs using a Total Plant RNA Extraction Kit (Karroten). The RNA concentration and integrity were assessed using a NanoDrop ND-2000 spectrophotometer and agarose gel electrophoresis, respectively (Sui et al., 2015; Vijayakumar et al., 2016). The libraries were constructed according to the high-throughput Illumina Strand-Specific RNA Sequencing Library scheme (Zhong et al., 2011). Briefly, after sufficient RNA was prepared, the mRNA was enriched using magnetic beads with oligo(dT). The purified mRNA was cleaved into short fragments using fragmentation buffer. Using the mRNA as a template, first-strand cDNA was synthesized using random hexamers (hexameric random primers), and deoxynucleotide triphosphates, buffer, DNA polymerase I, and RNase H were added to synthesize the second-strand cDNA. Sequencing adapters were connected to the sequencing fragments after purification using a QiaQuick PCR Kit. The fragment size was determined by agarose gel electrophoresis, followed by PCR amplification. The library was sequenced using the Illumina HiSeq 2500 platform at BioMarker Technologies . RNA-seq data for the control (wild type) and experimental samples (overexpression lines) were obtained from two biological replicates. Reads with adaptors, low-quality reads, and reads with more than 5% unknown bases were filtered out to generate nonredundant clean reads for subsequent sequence assembly. TopHat version 2.0.10 was used to map the clean reads to the Arabidopsis reference genome (Trapnell et al., 2009). To estimate the read count of each gene, the mapping results were filtered to maintain only unique mapped reads before being piped into Cuffdiff (http://cole-trapnell-lab.github.io/cufflinks/). The reads per kilobase per million mapped reads value was calculated using the internal script of the count table based on Cuffdiff’s output. To detect the presence of a particular transcript, the reads per kilobase per million mapped reads threshold value was set to 0.1. DESeq was used to detect differentially expressed genes (Anders and Huber, 2010), and false discovery rate <0.01 and fold change ≥2 were used as the criteria to identify differentially expressed genes.
The assembled sequences were compared with several databases including the National Center for Biotechnology Information nonredundant (nr; Pruitt et al., 2007), GO (Ashburner et al., 2000), Clusters of Orthologous Group (COG; Tatusov et al., 2000), Swiss-Prot (Apweiler et al., 2004), and Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al., 2004) databases using BLAST with E-value ≤ 1e−10 as the standard (Altschul et al., 1997).
GO annotation of unigenes was performed using the Blast2 GO program (Conesa et al., 2005), and the GO functions were classified and mapped using WEGO software (Ye et al., 2006). GO terms were classified into three categories, molecular function, cell fraction, and biological process, providing a broad overview for understanding gene function from a macroscopic angle (Ashburner et al., 2000). The Biological Networks Gene Ontology plug-in for Cytoscape was used to determine GO categories significantly over-represented among misregulated genes (Maere et al., 2005). The COG online comparison tool was used to annotate the COG functions of unigenes. The KEGG Web server (http://www.genome.jp/kegg/) was used to assign KEGG pathways to the assembled sequences. Both the KEGG orthology assignments and KEGG pathways were populated with KEGG orthology assignments. To compare the gene lists, the HYPERGEO.DIST function in Excel software (Microsoft) was used for statistical analysis. The metadata file comprises the following SRA accessions: SRR8750820, SRR8750821, SRR8750824, SRR8750825, SRR8750822, and SRR8750823 (https://submit.ncbi.nlm.nih.gov/subs/sra/).
The relative expression levels of 11 randomly selected differentially expressed genes identified by RNA-seq were tested by RT-qPCR. Genes and primers are listed in Supplemental Table 10.
Obtaining p35S:AtZP1:GR Transgenic Lines and DEX and CHX Treatment
The AtZP1 coding sequence (no stop codon) was amplified using a forward primer containing a MLuI site at the 5′ end and a reverse primer containing an ApaI site at the 5′ end (Supplemental Table 11). The amplified sequence and the pGreen 0244 empty expression vector were simultaneously digested with MLuI and ApaI and ligated together to construct the AtZP1-pGreen 0244-GR fusion expression vector. The GR expression vector was transformed into the atzp1 mutant to obtain GR:AtZP1:atzp1 transgenic lines. For the treatment, a solution of 20 mM DEX and 20 mM CHX dissolved in ethanol was produced and stored until use. GR:AtZP1:atzp1 transgenic plants were cultured on standard MS medium for 3 d. For DEX treatment, 3-d-old transgenic seedlings were treated with a final concentration of 20 μM DEX in MS liquid medium. For DEX and CHX treatment, 3-d-old transgenic seedlings were treated with a final concentration of 20 μM DEX and 20 μM CHX in MS liquid medium. For the control group, 3-d-old transgenic seedlings were treated with a final concentration of 20 μM ethanol in MS liquid medium. At 12 h after treatment, the root tips were sampled and subjected to RT-qPCR analysis (Vijayakumar et al., 2016).
Yeast One-Hybrid Assay
The A[AG/CT]CNAC sequence is a possible binding domain of C2H2 transcription factors (Sakai et al., 1995; Kubo et al., 1998). The binding sequences in the promoter region of bHLH transcription factor genes (ACTCCTC for RSL2, AAGCGGC for RSL4, ACTCAAT for RHD6) were repeated three times in series, artificially synthesized into single-stranded DNA, and re-natured into double-stranded DNA. EcoRI and XhoI restriction endonuclease sites were added to the 5′ and 3′ ends of the synthetic chain, respectively. The binding sequence and the pLacZi vector were digested with the same restriction endonuclease and ligated to construct the pLacZi-RSL2/RSL4/RHD6 recombinant vector. The AtZP1 coding sequence was amplified using a forward primer containing an EcoRI site and a reverse primer containing an XhoI site (Supplemental Table 12). The amplified sequence and pB42AD vector were digested with EcoRI and XhoI and ligated to form the pB42AD:ZP1 expression vector. Four different vector combinations—pB42AD and pLacZi, pB42AD:ZP1 and pLacZi, pB42AD and pLacZi:RSL2/RSL4/RHD6, and pLacZi:RSL2/RSL4/RHD6 and pB42AD:ZP1—were cotransformed into AH109 yeast cells. The yeast was cultured for 2 to 3 d in SD-Trp-Ura double deficiency medium. Single clones harboring different vector combinations were transferred to SD-Trp-Ura medium containing Z-buffer/X-Gal and incubated at 30° for 0.5 to 8 h, and then color changes in the yeast cells were observed (Han et al., 2016).
ChIP and RT-qPCR Analyses
ChIP analysis was performed using an EZ-ChIP Kit (Upstate) according to the manufacturer’s protocol (Meng et al., 2017). The ProAtZP1:AtZP1 sequence was amplified by forward and reverse primers (Supplemental Table 13) and ligated into the GFP expression vector pBGWFS7 without a promoter by LR reaction. The 2.1-kb GL2 promoter fragment was cloned to construct the GL2 promoter-driven GL2-GFP vector (Ohashi et al., 2003; Lin et al., 2015), and the PCR products encoding the GL2 promoter, GFP, and GL2 were connected together into the three-fragment recombination vector pK7m34GW (Invitrogen; Lin et al., 2015). Three-day-old ProZP1:ZP1:atzp1-1-GFP and ProGL2-GFP:GL2:gl2 transgenic seedlings were used for ChIP analysis. For each experimental group, ∼0.3 g of primary root tissue was harvested and cross-linked in GB buffer (0.4 M Suc, 10 mM Tris, pH 8.0, 1 mM EDTA, pH 8.0, and 1 mM phenylmethylsulfonyl fluoride) containing 1% (v/v) formaldehyde under a vacuum at 4°C for 10 min. The cross-linking reaction was stopped by adding 1 M Gly under a vacuum at 4°C for 5 min. Following the addition of protease inhibitor cocktail 2, the sample was rinsed twice with PBS buffer. The material was thoroughly ground in liquid nitrogen, and the chromatin was resuspended and sheared into 200- to 1000-bp fragments by sonication. The DNA fragments were immunoprecipitated with anti-GFP antibody (catalog no. 11814460001, Roche Diagnostics) and analyzed by RT-qPCR as described previously (Zhou et al., 2009; Cheng et al., 2014). The primers used for qPCR were designed based on the A[AG/CT]CNAC sequence listed in Supplemental Tables 14 to 18 (Sakai et al., 1995; Kubo et al., 1998). The “% of input” values of the ChIP-qPCR fragments were calculated in the transgenic plants and the control as described previously (Hwang et al., 2017).
Preparation of AtZP1 Protein and EMSA
To produce AtZP1 protein for EMSA, the coding sequence of AtZP1 was amplified using a forward primer containing a BamHI site and a reverse primer containing a SalI site (Supplemental Table 19). The sequence was digested with BamHI and SalI and inserted into the pGEX-4T-1 vector. Escherichia coli strain BL21 (DE3) cells were treated with 0.25 M isopropyl β-d-1-thiogalactopyranoside at 20°C for 5 h to induce AtZP1 protein expression. The E. coli cells were enriched by centrifugation at 5000 rpm (Centrifuge 5810 R, Eppendorf) for 15 min at 4°C and dissolved in B-PER buffer (Thermo Fisher Scientific). The mixed proteins in the cleavage products were bound to agarose containing GSH at 4°C for 2 h and rinsed slowly 8 to 10 times with Tris-buffered saline buffer. The bound proteins were dissolved in elution buffer at 4°C and purified to only AtZP1 protein, as confirmed by SDS-PAGE electrophoresis. EMSA was performed using a Light Shift Chemiluminescent EMSA Kit (Thermo Fisher Scientific) following the manufacturer’s protocol. The pGEX-4T-1-ZP1 recombinant protein and biotin probe were used for analysis. Biotinylated probe containing the A[AG/CT]CNAC binding sequence was designed based on the corresponding promoter regions of RSL2, RSL4, and RHD6 (Supplemental Table 19). Synthetic and annealed double-stranded probes were used at a concentration of 20 pmol mL−1, and 10-, 100-, and 200-fold levels of nonbiotinylated probe were used as the competitor. The mutated competitors were generated by replacing three bases in the binding elements (RSL2, ACTCCTC to CCTACTA; RSL4, AAGCGGC to CAGAGGA; RHD6, ACTCAAT to CCTAAAA). AtZP1 was reacted with biotin-labeled probe or competitor probe in binding buffer at room temperature for 0.5 h. The protein–probe complexes were separated by 8% polyacrylamide gel electrophoresis at 100 V for 2.5 h, transferred to a nylon membrane by electrophoresis at 84 mA for 1.5 h, and cross-linked twice using 254 nm UV light. The results were detected using a Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher Scientific; Hwang et al., 2017).
Expression of RHD6, RSL4, and RSL2 in AtZP1 Overexpression Plants
To produce the RHD6, RSL4, and RSL2 gene restored plants under the background of AtZP1 overexpression lines, the coding sequences of RHD6, RSL4, and RSL2 were amplified using the forward primer containing a KpnI site at the 5′ end and the reverse primer containing a BamHI site at the 5′ end (Supplemental Table 20), cloned into the 35S promoter vector pCAMBIA-1300, and transferred into Agrobacterium GV3101. To express RHD6, RSL4, and RSL2 in AtZP1 overexpressing plants, the AtZP1 overexpression plants were transformed by Agrobacterium-mediated transformation, and the transgenic lines were selected based on hygromycin resistance (Wang et al., 2011).
Statistical Analysis
The statistical results are described as means ± sd, where n is the number of biological replicates. The data were analyzed using the statistical software SPSS 17.0 (SPSS). One-way ANOVA and Student’s t test were used as specified packages. Different letters and asterisks in the tables and figures indicate significant difference among the means (at 0.05 or 0.01) by Duncan’s test or Student’s t test.
Accession Numbers
The gene sequences used this study can be found in The Arabidopsis Information Resource under the following accession numbers: ACT7 (AT5G09810), CPC (AT2G46410), EGL3 (AT1G63650), GL2 (AT1G79840), GL3 (AT5G41315), GIS (AT3G58070), GIS2 (AT5G06650), GIS3 (AT1G68360), KNUCKLES (AT5G14010), LATE (AT5G48890), LRL1 (AT2G24260), LRL2 (AT4G30980), LRL3 (AT5G58010), RBE (AT5G06070), RHD6 (AT1G66470), RSL2 (AT4G33880), RSL4 (AT1G27740), SUPERMAN (AT3G23130), TAC1 (AT3G09290), TTG1 (AT5G24520), WER (AT5G14750), ZFP1 (AT1G80730), ZFP2 (AT5G57520), ZFP3 (AT1G80730), ZFP4 (AT1G66140), ZFP5 (AT1G10480), ZFP6 (AT1G67030), ZFP7 (AT5G57520), ZFP8 (AT2G41940), ZFP10 (AT2G37740), ZFP11 (AT2G42410), ZP1 (AT4G17810), and UBQ10 (AT4G05320).
Supplemental Data
Supplemental Figure 1. Identification of transgenic lines. (Supports Figure 3.)
Supplemental Figure 2. The subcellular localization of AtZP1 in Nicotiana benthamiana epidermal cells. (Supports Figure 2.)
Supplemental Figure 3. AtZP1 represses the expression of genes associated with root hair development in different lines (3 d). (Supports Figures 5, 6, and 7.)
Supplemental Figure 4. Validation of RNA-Seq results by RT-qPCR. (Supports Figures 5–7.)
Supplemental Figure 5. Producing the p35S:AtZP1:GR transgenic lines. (Supports Figure 6.)
Supplemental Figure 6. RT-qPCR analysis of CPC, WER, GL3, EGL3, TTG, and GL2 expression of WT and AtZP1 overexpression lines. (Supports Figures 5–7.)
Supplemental Figure 7.. RT-qPCR analysis of RSL2, RSL4, and RHD6 expression of ZP1-GR lines after CHX treatment. (Supports Figure 6.)
Supplemental Table 1. H cell specification in the root epidermis and root hair and non-root hair cell production in various genotypes
Supplemental Table 2. H cell specification in the root epidermis and root hair and non-root hair cell production in various lines overexpressing ZP1 with a mutated EAR motif.
Supplemental Table 3. Primers used for expression pattern analysis of AtZP1.
Supplemental Table 4. Primers used for promoter GUS reporter analysis.
Supplemental Table 5. Primers used for the subcellular localization assay of AtZP1
Supplemental Table 6. Primers used to generate AtZP1 overexpression lines.
Supplemental Table 7. Primers used for AtZP1 loss-of-function mutants by CRISPR-Cas9.
Supplemental Table 8. Primers used to generate the AtZP1 complementation line.
Supplemental Table 9. Primers used for transient luciferase expression assay.
Supplemental Table 10. Comparison of genes identified by RNA-Seq and their primer sequences.
Supplemental Table 11. Primers used to obtain p35S:AtZP1-GR transgenic lines and RT-qPCR genes.
Supplemental Table 12. Primers used for yeast one-hybrid assay.
Supplemental Table 13. Primers used to construct the GFP transgenic line.
Supplemental Table 14. Possible AtZP1 binding sites in the RHD6 gene promoter region and primers used for ChIP analysis.
Supplemental Table 15. Possible AtZP1 binding sites in the RSL2 gene promoter region and primers used for ChIP analysis.
Supplemental Table 16. Possible AtZP1 binding sites in the RSL4 gene promoter region and primers used for ChIP analysis.
Supplemental Table 17. Possible AtZP1 binding sites in the LRL1 gene promoter region and primers used for ChIP analysis.
Supplemental Table 18. Possible AtZP1 binding sites in the LRL3 gene promoter region and primers used for ChIP analysis.
Supplemental Table 19. Binding sequence primers of AtZP1 to the promoter regions of target genes used in EMSA.
Supplemental Table 20. Primers used to amplify the RHD6, RSL4, and RSL2 genes.
Supplemental File 1. Sequence alignments used to generate the phylogeny presented in Figure 1B obtaining by MAFFT program
Supplemental File 2. Tree file used to generate the phylogeny in Figure 1B obtaining by MEGA5.1 software
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the National Natural Science Research Foundation of China (grants 31570251, 31600200,and 31770288), the Shandong Province Key Research and Development Plan (grants 2017CXGC0313 and 2016GNC113012), the Natural Science Research Foundation of Shandong Province (grants ZR2014CZ002 and ZR2017MC003), and the Higher Educational Science and Technology Program of Shandong Province (grants J15LE08 and J17KA136).
AUTHOR CONTRIBUTIONS
G.H., D.Z., Z.G., and B.W. designed the research; G.H., X.W., X.D., C.W., N.S., J.G., F.Y., X.L., Y.Z., Z.C., and Z.M. performed the experiments; all authors analyzed the data; and G.H. and B.W. wrote the article, with contributions from the other authors.
Footnotes
Articles can be viewed without a subscription.
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An L., Zhou Z., Su S., Yan A., Gan Y. (2012a). GLABROUS INFLORESCENCE STEMS (GIS) is required for trichome branching through gibberellic acid signaling in Arabidopsis. Plant Cell Physiol. 53: 457–469. [DOI] [PubMed] [Google Scholar]
- An L., Zhou Z., Sun L., Yan A., Xi W., Yu N., Cai W., Chen X., Yu H., Schiefelbein J., Gan Y. (2012b). A zinc finger protein gene ZFP5 integrates phytohormone signaling to control root hair development in Arabidopsis. Plant J. 72: 474–490. [DOI] [PubMed] [Google Scholar]
- An L., Zhou Z., Yan A., Gan Y. (2011). Progress on trichome development regulated by phytohormone signaling. Plant Signal. Behav. 6: 1959–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R., et al. (2004). UniProt: The universal protein knowledgebase. Nucleic Acids Res. 32: D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.-L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Ashburner M., et al. The Gene Ontology Consortium. (2000). Gene ontology: Tool for the unification of biology. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhme K., Li Y., Charlot F., Grierson C., Marrocco K., Okada K., Laloue M., Nogué F. (2004). The Arabidopsis COW1 gene encodes a phosphatidylinositol transfer protein essential for root hair tip growth. Plant J. 40: 686–698. [DOI] [PubMed] [Google Scholar]
- Benjamin L. (2000). Genes VII. (New York: Oxford University Press; ). [Google Scholar]
- Bernhardt C., Lee M.M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439. [DOI] [PubMed] [Google Scholar]
- Bruex A., Kainkaryam R.M., Wieckowski Y., Kang Y.H., Bernhardt C., Xia Y., Zheng X., Wang J.Y., Lee M.M., Benfey P., Woolf P.J., Schiefelbein J. (2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Chen C., Zhang D., Shu B., Xiao J., Xia R. (2013). Influence of nutrient deficiency on root architecture and root hair morphology of trifoliate orange (Poncirus trifoliata L. Raf.) seedlings under sand culture. Sci. Hortic. (Amst.) 162: 100–105. [Google Scholar]
- Cheng Z.J., Zhao X.Y., Shao X.X., Wang F., Zhou C., Liu Y.G., Zhang Y., Zhang X.S. (2014). Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 26: 1053–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S., Mittler R. (2008). The zinc finger network of plants. Cell. Mol. Life Sci. 65: 1150–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes F. (2000). Pattern in root meristem development in angiosperms. New Phytol. 146: 83–94. [Google Scholar]
- Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. (2005). Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Datta S., Prescott H., Dolan L. (2015). Intensity of a pulse of RSL4 transcription factor synthesis determines Arabidopsis root hair cell size. Nat. Plants 1: 15138. [DOI] [PubMed] [Google Scholar]
- Dinkins R.D., Pflipsen C., Collins G.B. (2003). Expression and deletion analysis of an Arabidopsis SUPERMAN-like zinc finger gene. Plant Sci. 165: 33–41. [Google Scholar]
- Dinkins R., Pflipsen C., Thompson A., Collins G.B. (2002). Ectopic expression of an Arabidopsis single zinc finger gene in tobacco results in dwarf plants. Plant Cell Physiol. 43: 743–750. [DOI] [PubMed] [Google Scholar]
- Dolan L., Duckett C.M., Grierson C., Linstead P., Schneider K., Lawson E., Dean C., Poethig S., Roberts K. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474. [Google Scholar]
- Franciosini A., Rymen B., Shibata M., Favero D.S., Sugimoto K. (2017). Molecular networks orchestrating plant cell growth. Curr. Opin. Plant Biol. 35: 98–104. [DOI] [PubMed] [Google Scholar]
- Galway M.E., Masucci J.D., Lloyd A.M., Walbot V., Davis R.W., Schiefelbein J.W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166: 740–754. [DOI] [PubMed] [Google Scholar]
- Gan Y., Liu C., Yu H., Broun P. (2007b). Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134: 2073–2081. [DOI] [PubMed] [Google Scholar]
- Gan Y., Kumimoto R., Liu C., Ratcliffe O., Yu H., Broun P. (2006). GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell 18: 1383–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y., Yu H., Peng J., Broun P. (2007a). Genetic and molecular regulation by DELLA proteins of trichome development in Arabidopsis. Plant Physiol. 145: 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Yan P., Shen W., Li X., Zhou P., Li Y. (2013). Modular construction of plasmids by parallel assembly of linear vector components. Anal. Biochem. 437: 172–177. [DOI] [PubMed] [Google Scholar]
- Gilroy S., Jones D.L. (2000). Through form to function: Root hair development and nutrient uptake. Trends Plant Sci. 5: 56–60. [DOI] [PubMed] [Google Scholar]
- Grierson C., Nielsen E., Ketelaarc T., Schiefelbein J. (2014). Root hairs. Arabidopsis Book 12: e0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C., Schiefelbein J. (2002). Root hairs. The Arabidopsis Book 1: e0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Wu M., Cao L., Yuan W., Dong M., Wang X., Chen W., Shang F. (2016). Characterization of OfWRKY3, a transcription factor that positively regulates the carotenoid cleavage dioxygenase gene OfCCD4 in Osmanthus fragrans. Plant Mol. Biol. 91: 485–496. [DOI] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747. [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739. [DOI] [PubMed] [Google Scholar]
- Honkanen S., Dolan L. (2016). Growth regulation in tip-growing cells that develop on the epidermis. Curr. Opin. Plant Biol. 34: 77–83. [DOI] [PubMed] [Google Scholar]
- Hwang Y., Choi H.-S., Cho H.-M., Cho H.-T. (2017). Tracheophytes contain conserved orthologs of a basic helix-loop-helix transcription factor that modulate ROOT HAIR SPECIFIC genes. Plant Cell 29: 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Mitsuda N., Ohme-Takagi M. (2009). Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21: 3493–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59: 365–386. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32: D277–D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.H., Song S.-K., Schiefelbein J., Lee M.M. (2013). Nuclear trapping controls the position-dependent localization of CAPRICE in the root epidermis of Arabidopsis. Plant Physiol. 163: 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kazan K. (2006). Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 11: 109–112. [DOI] [PubMed] [Google Scholar]
- Kim C.M., Dolan L. (2011). Root hair development involves asymmetric cell division in Brachypodium distachyon and symmetric division in Oryza sativa. New Phytol. 192: 601–610. [DOI] [PubMed] [Google Scholar]
- Kim C.M., Dolan L. (2016). ROOT HAIR DEFECTIVE SIX-LIKE class I genes promote root hair development in the grass Brachypodium distachyon. PLoS Genet. 12: e1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.M., Han C.-D., Dolan L. (2017). RSL class I genes positively regulate root hair development in Oryza sativa. New Phytol. 213: 314–323. [DOI] [PubMed] [Google Scholar]
- Kim D.W., Lee S.H., Choi S.-B., Won S.-K., Heo Y.-K., Cho M., Park Y.-I., Cho H.-T. (2006). Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18: 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Simon M., Huelskamp M., Schiefelbein J. (2004). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268: 506–513. [DOI] [PubMed] [Google Scholar]
- Knox K., Grierson C.S., Leyser O. (2003). AXR3 and SHY2 interact to regulate root hair development. Development 130: 5769–5777. [DOI] [PubMed] [Google Scholar]
- Kubo K.-i., Sakamoto A., Kobayashi A., Rybka Z., Kanno Y., Nakagawa H., Takatsuji H., Takatsuji H. (1998). Cys2/His2 zinc-finger protein family of petunia: Evolution and general mechanism of target-sequence recognition. Nucleic Acids Res. 26: 608–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity J.H., Lee B.M., Wright P.E. (2001). Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11: 39–46. [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483. [DOI] [PubMed] [Google Scholar]
- Lee R.D.-W., Cho H.-T. (2013). Auxin, the organizer of the hormonal/environmental signals for root hair growth. Front. Plant Sci. 4: 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y., Lu L., Liu H.-Y., Li S., Xing F., Chen L.-L. (2014). CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7: 1494–1496. [DOI] [PubMed] [Google Scholar]
- Li J.-F., Norville J.E., Aach J., McCormack M., Zhang D., Bush J., Church G.M., Sheen J. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31: 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Song A., Gao C., Wang L., Wang Y., Sun J., Jiang J., Chen F., Chen S. (2015). Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep. 34: 1365–1378. [DOI] [PubMed] [Google Scholar]
- Li T., Lin G., Zhang X., Chen Y., Zhang S., Chen B. (2014). Relative importance of an arbuscular mycorrhizal fungus (Rhizophagus intraradices) and root hairs in plant drought tolerance. Mycorrhiza 24: 595–602. [DOI] [PubMed] [Google Scholar]
- Lim S.-H., Song J.-H., Kim D.-H., Kim J.K., Lee J.-Y., Kim Y.-M., Ha S.-H. (2016). Activation of anthocyanin biosynthesis by expression of the radish R2R3-MYB transcription factor gene RsMYB1. Plant Cell Rep. 35: 641–653. [DOI] [PubMed] [Google Scholar]
- Lin Q., Ohashi Y., Kato M., Tsuge T., Gu H., Qu L.-J., Aoyama T. (2015). GLABRA2 directly suppresses basic helix-loop-helix transcription factor genes with diverse functions in root hair development. Plant Cell 27: 2894–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas W.J., et al. (2013). The plant vascular system: Evolution, development and functions 55: 294–388. [DOI] [PubMed] [Google Scholar]
- Luo X., Bai X., Zhu D., Li Y., Ji W., Cai H., Wu J., Liu B., Zhu Y. (2012a). GsZFP1, a new Cys2/His2-type zinc-finger protein, is a positive regulator of plant tolerance to cold and drought stress. Planta 235: 1141–1155. [DOI] [PubMed] [Google Scholar]
- Luo X., Cui N., Zhu Y., Cao L., Zhai H., Cai H., Ji W., Wang X., Zhu D., Li Y., Bai X. (2012b). Over-expression of GsZFP1, an ABA-responsive C2H2-type zinc finger protein lacking a QALGGH motif, reduces ABA sensitivity and decreases stomata size. J. Plant Physiol. 169: 1192–1202. [DOI] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449. [DOI] [PubMed] [Google Scholar]
- Marzol E., Borassi C., Juárez S.P.D., Mangano S., Estevez J.M. (2017). RSL4 takes control: multiple signals, one transcription factor. Trends Plant Sci 22: 553–555. [DOI] [PubMed] [Google Scholar]
- Masucci J.D., Rerie W.G., Foreman D.R., Zhang M., Galway M.E., Marks M.D., Schiefelbein J.W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260. [DOI] [PubMed] [Google Scholar]
- Masucci J.D., Schiefelbein J.W. (1994). The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin-and ethylene-associated process. Plant Physiol. 106: 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K., Umemura Y., Ohme-Takagi M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55: 954–967. [DOI] [PubMed] [Google Scholar]
- Menand B., Yi K., Jouannic S., Hoffmann L., Ryan E., Linstead P., Schaefer D.G., Dolan L. (2007). An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480. [DOI] [PubMed] [Google Scholar]
- Meng W.J., Cheng Z.J., Sang Y.L., Zhang M.M., Rong X.F., Wang Z.W., Tang Y.Y., Zhang X.S. (2017). Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29: 1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L., Lin H., Zhang F., Watira T.W., Li G., Tang Y., Wen J., Ratet P., Mysore K.S., Tadege M. (2015). LOOSE FLOWER, a WUSCHEL-like Homeobox gene, is required for lateral fusion of floral organs in Medicago truncatula. Plant J. 81: 480–492. [DOI] [PubMed] [Google Scholar]
- Ohashi Y., Oka A., Rodrigues-Pousada R., Possenti M., Ruberti I., Morelli G., Aoyama T. (2003). Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300: 1427–1430. [DOI] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar A.F., Ismail I. (2016). Isolation of Persicaria minor sesquiterpene synthase promoter and its deletions for transgenic Arabidopsis thaliana In AIP Conference Proceedings. (American Institute of Physics: AIP Publishing; ), p. 20020. [Google Scholar]
- Pemberton L.M., Tsai S.-L., Lovell P.H., Harris P.J. (2001). Epidermal patterning in seedling roots of eudicotyledons. Ann. Bot. (Lond.) 87: 649–654. [Google Scholar]
- Pesch M., Hülskamp M. (2004). Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr. Opin. Genet. Dev. 14: 422–427. [DOI] [PubMed] [Google Scholar]
- Peterson R.L., Farquhar M.L. (1996). Root hairs: Specialized tubular cells extending root surfaces. Bot. Rev. 62: 1–40. [Google Scholar]
- Pires N.D., Yi K., Breuninger H., Catarino B., Menand B., Dolan L. (2013). Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc. Natl. Acad. Sci. USA 110: 9571–9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proust H., Honkanen S., Jones V.A., Morieri G., Prescott H., Kelly S., Ishizaki K., Kohchi T., Dolan L. (2016). RSL class I genes controlled the development of epidermal structures in the common ancestor of land plants. Curr. Biol. 26: 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R. (2007). NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35: D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie W.G., Feldmann K.A., Marks M.D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8: 1388–1399. [DOI] [PubMed] [Google Scholar]
- Rymen B., Kawamura A., Schäfer S., Breuer C., Iwase A., Shibata M., Ikeda M., Mitsuda N., Koncz C., Ohme-Takagi M. (2017). ABA suppresses root hair growth via OBP4 transcriptional-regulator repression of the RSL2 promoter. Plant Physiol. 173: 1750–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Medrano L.J., Meyerowitz E.M. (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378: 199–203. [DOI] [PubMed] [Google Scholar]
- Salazar-Henao J.E., Vélez-Bermúdez I.C., Schmidt W. (2016). The regulation and plasticity of root hair patterning and morphogenesis. Development 143: 1848–1858. [DOI] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., Thumfahrt J., Jürgens G., Hülskamp M. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21: 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J. (2003). Cell-fate specification in the epidermis: A common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 6: 74–78. [DOI] [PubMed] [Google Scholar]
- Schiefelbein J., Huang L., Zheng X. (2014). Regulation of epidermal cell fate in Arabidopsis roots: The importance of multiple feedback loops. Front. Plant Sci. 5: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J., Kwak S.-H., Wieckowski Y., Barron C., Bruex A. (2009). The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. J. Exp. Bot. 60: 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Lee M.M., Lin Y., Gish L., Schiefelbein J. (2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 311: 566–578. [DOI] [PubMed] [Google Scholar]
- Sui N., Yang Z., Liu M., Wang B. (2015). Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genomics 16: 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Zhang A., Zhou Z., Zhao Y., Yan A., Bao S., Yu H., Gan Y. (2015). GLABROUS INFLORESCENCE STEMS3 (GIS3) regulates trichome initiation and development in Arabidopsis. New Phytol. 206: 220–230. [DOI] [PubMed] [Google Scholar]
- Tanaka N., Kato M., Tomioka R., Kurata R., Fukao Y., Aoyama T., Maeshima M. (2014). Characteristics of a root hair-less line of Arabidopsis thaliana under physiological stresses. J. Exp. Bot. 65: 1497–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V. (2000). The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28: 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga R., Iwata M., Sano R., Inoue K., Okada K., Wada T. (2008). Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135: 1335–1345. [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R., Iwata M., Sugiyama J., Kotake T., Ishida T., Yokoyama R., Nishitani K., Okada K., Wada T. (2009). The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. Plant J. 60: 564–574. [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R., Nukumizu Y., Sato S., Wada T. (2013). Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS One 8: e54019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R., Wada T. (2014). Regulation of root hair cell differentiation by R3 MYB transcription factors in tomato and Arabidopsis. Front. Plant Sci. 5: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troll W., Lindsley J. (1955). A photometric method for the determination of proline. J. Biol. Chem. 215: 655–660. [PubMed] [Google Scholar]
- Ueda M., Koshino-Kimura Y., Okada K. (2005). Stepwise understanding of root development. Curr. Opin. Plant Biol. 8: 71–76. [DOI] [PubMed] [Google Scholar]
- Vijayakumar P., Datta S., Dolan L. (2016). ROOT HAIR DEFECTIVE SIX-LIKE4 (RSL4) promotes root hair elongation by transcriptionally regulating the expression of genes required for cell growth. New Phytol. 212: 944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A.R., Davison P.A., Bolognesi-Winfield A.C., James C.M., Srinivasan N., Blundell T.L., Esch J.J., Marks M.D., Gray J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Avci U., Nakashima J., Hahn M.G., Chen F., Dixon R.A. (2010). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. USA 107: 22338–22343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhao Q., Chen F., Wang M., Dixon R.A. (2011). NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J. 68: 1104–1114. [DOI] [PubMed] [Google Scholar]
- Won S.-K., Lee Y.-J., Lee H.-Y., Heo Y.-K., Cho M., Cho H.-T. (2009). Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol. 150: 1459–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunder T., Liu Q., Aseeva E., Bonardi V., Leister D., Pribil M. (2013). Control of STN7 transcript abundance and transient STN7 dimerisation are involved in the regulation of STN7 activity. Planta 237: 541–558. [DOI] [PubMed] [Google Scholar]
- Yan A., Wu M., Zhao Y., Zhang A., Liu B., Schiefelbein J., Gan Y. (2014). Involvement of C2H2 zinc finger proteins in the regulation of epidermal cell fate determination in Arabidopsis. J. Integr. Plant Biol. 56: 1112–1117. [DOI] [PubMed] [Google Scholar]
- Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L., Wang J. (2006). WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 34: W293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K., Menand B., Bell E., Dolan L. (2010). A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 42: 264–267. [DOI] [PubMed] [Google Scholar]
- Yuan F., Lyu M.J., Leng B.Y., Zheng G.Y., Feng Z.T., Li P.H., Zhu X.G., Wang B.S. (2015). Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 38: 1637–1657. [DOI] [PubMed] [Google Scholar]
- Zhao P., Sokolov L.N., Ye J., Tang C.-Y., Shi J., Zhen Y., Lan W., Hong Z., Qi J., Lu G.-H., Pandey G.K., Yang Y.H. (2016). The LIKE SEX FOUR2 regulates root development by modulating reactive oxygen species homeostasis in Arabidopsis. Sci. Rep. 6: 28683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Joung J.-G., Zheng Y., Chen Y.-r., Liu B., Shao Y., Xiang J.Z., Fei Z., Giovannoni J.J. (2011). High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb. Protoc. 2011: 940–949. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhang X., Kang X., Zhao X., Zhang X., Ni M. (2009). SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell 21: 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., An L., Sun L., Gan Y.(2012). ZFP5 encodes a functionally equivalent GIS protein to control trichome initiation. Plant Signal. Behav. 7: 28–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Sun L., Zhao Y., An L., Yan A., Meng X., Gan Y. (2013). Zinc Finger Protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. New Phytol. 198: 699–708. [DOI] [PubMed] [Google Scholar]
- Zuo J., Chua N.-H. (2000). Chemical-inducible systems for regulated expression of plant genes. Curr. Opin. Biotechnol. 11: 146–151. [DOI] [PubMed] [Google Scholar]