ABSTRACT
Autophagy, characterized by the elevator of autophagy-related gene 14 (ATG14) and the dysregulation of autophagy-related proteins, contributes to the cisplatin (DDP) resistance in ovarian cancer. Forkhead box protein P1 (FOXP1), which is a well-defined transcription factor, is reported to have the oncogenic effect on ovarian cancer. This study aims to identify the effect of miR-29c-3p/FOXP1/ATG14 pathway in regulating autophagy and DDP resistance in ovarian cancer. The expressions of miR-29c-3p, FOXP1, ATG14 and autophagy-related proteins were detected in DDP-sensitive ovarian cancer cell lines (SKOV3 and A2780) and DDP-resistant cell lines (SKOV3/DDP and A2780/DDP). Cell viability was detected using the MTT assay. The therapeutic effect of miR-29c-3p overexpression was observed in the xenograft model of nude mice.Compared with DDP-sensitive cells, miR-29c-3p was decreased in DDP-resistant cells, and an enhancement of FOXP1, ATG14, autophagy, and drug resistance was shown in DDP-resistant cells. The anti-resistant effect of miR-29c-3p was observed as overexpressing miR-29c-3p inhibited cell viability of DDP-resistant cells. Moreover, FOXP1 was a target of miR-29c-3p, which was confirmed by the luciferase reporter assay, and ATG14 was transactivated by FOXP1, which was confirmed by the ChIP assay. Overexpression of miR-29c-3p increased DDP sensitivity by downregulating FOXP1/ATG14 in vitro. The tumor volume was reduced after the injection of miR-29c-3p-overexpressing SKOV3/DDP cells in vivo. Overexpression of miR-29c-3p inhibited autophagy and DDP resistance partly via downregulating FOXP1/ATG14 pathway, suggesting miR-29c-3p as a novel target in overcoming DDP resistance in ovarian cancer.
KEYWORDS: Ovarian cancer, miR-29c-3p, FOXP1, ATG14, autophagy
1. Introduction
Ovarian cancer is one of the most common gynecologic malignancies, and the associated mortality rate is the fifth highest among gynecologic malignancies worldwide [1]. Chemotherapy is the first choice in ovarian cancer treatment, but tumor cells are vulnerable to drug resistance [2]. Nearly 70% of ovarian cancer patients are sensitive to platinum chemotherapy, which is used as first-line treatment [3]. Due to platinum resistance, the 5-year survival rate for ovarian cancer patients is less than 30% [3]. Platinum resistance has become a key obstacle to the successful treatment of ovarian cancer, and the underlying mechanism remains unclear. Autophagy is the main way in which organisms remove damaged, aging, degenerative, and nonfunctional proteins and organelles, and it is a common mechanism under various physiological and pathological conditions [4]. Studies showed that autophagy was closely related to the occurrence and development of tumors [5], and that autophagy affected the sensitivity of tumor cells to chemotherapeutic drugs [6]. In recent years, many studies demonstrated that autophagy was associated with chemoresistance in ovarian cancer. Zhang et al. [7] reported that TXNDC17 increased resistance of ovarian cancer to paclitaxel by inducing autophagy. Wang et al. [8] revealed that knockdown of ERK reduced cisplatin (DDP) resistance by inhibiting autophagy in ovarian cancer. Thus, inhibition of autophagy might increase the sensitivity of ovarian cancer cells to chemotherapeutic drugs.
Forkhead box gene P1 (FOXP1), a member of the FOXP subfamily of transcription factors, plays a role in normal embryonic development, myocardial cell development, and speech formation in humans [9]. According to recent evidence, FOXP1 appears to be associated with malignancy [10]. For example, Wang et al. [11] found that downregulation of FOXP1 suppressed hepatocarcinoma cell proliferation by regulating the G1/S phase of the cell cycle. Takayama et al. [12] showed that FOXP1 impeded the development of prostate cancer by inhibiting tumor proliferation and migration. A number of studies reported that FOXP1 played a role in ovarian cancer [13,14]. However, the molecular mechanism underlying the regulation of FOXP1 in chemoresistance of ovarian cancer is unclear.
MicroRNAs (miRNAs) are a group of small noncoding 21–25 nucleotide RNAs that regulate gene expression by targeting mRNAs at the post-transcriptional level [15]. Studies revealed that miRNAs were involved in the occurrence, development, and chemotherapy resistance of cancer [16]. Iwagami et al. [17] reported that miR-320c played a role in gemcitabine resistance of pancreatic cancer cells through SMARCC1. Zhou et al. [18] found that miR-203 promoted oxaliplatin resistance by downregulating ATM in colorectal cancer cells. Yin et al. [19] revealed that miR-204-5p elevated colorectal cancer sensitivity to chemotherapy by repressing RAB22A. Studies also found that various miRNAs, such as miR-497 [20], miR-134 [21], miR-130a [22], and miR-374a [22], regulated drug resistance in ovarian cancer. The miR-29 family, including miR-29a, miR-29b, and miR-29c, is reported to be closely related to drug resistance in osteosarcoma [23] and breast cancer [24]. In ovarian cancer, the downregulation of miR-29 increases DDP resistance in cancer cells [25]. In the pre-experiment, the downregulation of miR-29c in DDP-resistant ovarian cancer cell line was more significant than the downregulations of miR-29a and miR-29b. Adding that miR-29c-3p was downregulated in ovarian cancer tissues [26], miR-29c-3p was chosen for further investigations.
The present study investigated the association between miR-29c-3p and FOXP1 in ovarian cancer cells to explore the function of miR-29c-3p/FOXP1 in autophagy and DDP resistance of ovarian cancer.
2. Material and methods
2.1. Ethics statement
All animal experiments were approved by the Animal Care and Use Committee of The First Affiliated Hospital of Zhengzhou University.
2.2. Cell culture
A DDP-sensitive ovarian cancer cell lines (SKOV3 and A2780) were purchased from the American Type Culture Collection (ATCC, USA). A DDP-resistant ovarian cancer cell lines (SKOV3/DDP and A2780/DDP) were purchased from Shanghai Xinyu Biological Technology Co. Ltd. All the cell lines were cultured in RPMI-1640 medium (Gibco, U.S.) containing 10% FBS (Gibco), 100 U/mL of penicillin, and 100 U/mL of streptomycin (Invitrogen, U.S.). DDP (0.2 μg/mL) was added to the medium of DDP-resistant cell lines to maintain resistance.
2.3. Plasmid construction
The coding sequence of FOXP1 or ATG14 was amplified using PCR and inserted into pcDNA3.0 (Invitrogen; Thermo Fisher Scientific, Inc.). PCR amplification was performed as follows: 94 °C for 3 min, followed by 30 cycles of 94 °C for 40 sec, 56 °C for 45 sec and 72 °C for 60 sec, followed by terminal elongation. A DNA Engine Opticon 2 Real-Time Cycler (MJ Research, Inc.) and Taq DNA Polymerase (Invitrogen; Thermo Fisher Scientific, Inc.) were used. Insertion accuracy was confirmed via direct Sanger sequencing.
2.4. Cell transfection
Cell transfection was performed using Lipofectamine 2000 (Thermo Fisher Scientific, U.S.) reagent, according to the manufacturer’s instructions. Briefly, cells were cultured in 96-well plates and transfected with appropriate expression vectors (miR-29c-3p inhibitor, miR-29c-3p mimic, pcDNA-FOXP1, si-FOXP1, pcDNA-autophagy-related gene 14 (ATG14), si-ATG14, and their control vectors) using Lipofectamine 2000 (Thermo Fisher Scientific). The expression vectors used in this study were supplied by Applied Biological Materials (ABM, Canada). Cells in each well were transfected with 100 nM miR-29c-3p mimic or 200 nM miR-29c-3p inhibitor. The transfection concentration of siRNA-ATG14 and siRNA-FOXP1 was 50 nM. The transfection quantity of pcDNA-ATG14 and pcDNA-FOXP1 was 2 μg. The oligonucleotide sequences of miR-29c-3p inhibitor and negative control (NC) are 5ʹ-UAACCGAUUUCAAAUGGUGCUA-3ʹ and 5ʹ- UCUACUCUUUCUAGGAGGUUGUGA-3ʹ, respectively. The oligonucleotide sequences of miR-29c-3p mimic and pre-NC are shown as follows: miR-29c-3p mimic, sense: 5ʹ-UAGCACCAUUUGAAAUCGGUUA-3ʹ, anti-sense: 5ʹ-UAACCGAUUUCAAAUGGUGCU A-3ʹ; pre-NC, sense: 5ʹ-UCACAACCUCCUAGAAAGAGUAGA-3ʹ, anti-sense: 5ʹ-UCUACUC UUUCUAGGAGGUUGUGA-3ʹ. The sequences of siRNA-FOXP1, siRNA-ATG14, and si-control are 5ʹ-GCGAAGATTTCCAATCATT-3ʹ, 5ʹ-GGCAAAUCUUCGACGAUCC CAUAUA-3ʹ, and 5ʹ-TTCTCCGAACGTGTCACGT-3ʹ, respectively.
2.5. Cell treatment groups
The SKOV3 or A2780 cells were divided into a number of groups, as follows: (1) An NC group and miR-29c-3p inhibitor group. The cells were treated with various concentrations of DDP (0, 1.25, 2.5, 5.0, 7.5, and 15 μg/mL) after transfection. (2) A pcDNA group and pcDNA-FOXP1 group. (3) An NC group, miR-29c-3p inhibitor group, miR-29c-3p inhibitor + si-control group, and miR-29c-3p inhibitor + si- FOXP1 group. (4) An NC group, miR-29c-3p inhibitor group, miR-29c-3p inhibitor + si-control group, miR-29c-3p inhibitor + si-FOXP1 group, and miR-29c-3p inhibitor + si-FOXP1 + pcDNA-ATG14 group. After transfection, the cells were treated with 5 nM rapamycin (Sigma, U.S.), an autophagy agonist. (5) An NC group, miR-29c-3p inhibitor group, miR-29c-3p inhibitor + si-control group, miR-29c-3p inhibitor + si-FOXP1 group, and miR-29c-3p inhibitor + si-FOXP1 + pcDNA-ATG14 group. The cells were treated with 5 μg/mL of DDP after transfection and incubated for 24 h.
The DDP-resistant cells were divided into the following groups: (1) A pre-NC group and miR-29c-3p mimic group. The cells were treated with various concentrations of DDP (0, 1.25, 2.5, 5.0, 7.5, and 15 μg/mL) after transfection. (2) An si-control group and si-FOXP1 group. (3) A pre-NC group, miR-29c-3p mimic group, miR-29c-3p mimic + pcDNA group, and miR-29c-3p mimic + pcDNA-FOXP1 group. (4) A pre-NC group, miR-29c-3p mimic group, miR-29c-3p mimic + pcDNA group, miR-29c-3p mimic + pcDNA-FOXP1 group, and miR-29c-3p mimic + pcDNA-FOXP1 + si-ATG14 group. After transfection, the cells were treated with 10 mM 3-methyladenine (3-MA) (Sigma), an autophagy inhibitor. (5) A pre-NC group, miR-29c-3p mimic group, miR-29c-3p mimic + pcDNA group, miR-29c-3p mimic + pcDNA-FOXP1 group, and miR-29c-3p mimic + pcDNA-FOXP1 + si-ATG14 group. The cells were treated with 5 μg/mL DDP after transfection and incubated for 24 h.
2.6. Quantitative real-time polymerase chain reaction (qRT-PCR)
According to the manufacturer’s instructions, Trizol (Invitrogen) was used to extract total RNA from tissues or cells. The total RNA was then reverse transcribed into cDNA using a cDNA Synthesis Kit (Thermo Fisher Scientific). A qRT-PCR assay was performed to determine mRNA levels of miR-29c-3p, FOXP1, and ATG14 using SYBR Green Master Mix (Takara, Shiga, Japan). The samples were incubated at 95 °C for 10 min for initial denaturation, and then subjected to 40 PCR cycles, each consisting of 95 °C for 15 sec and 60 °C for 60 sec. U6 was used as the internal control of miR-29c-3p, and β-actin was used as the internal control of mRNAs. The 2−ΔΔCt method was used to analyze mRNA levels. The primers used in the study were shown in Table 1.
Table 1.
The primers used in qRT-PCR.
| Gene name | Primer sequences |
|---|---|
| miR-29c-3p | Forward: 5ʹ-CTGACCTTAGCACCATTTGAAATC-3’ |
| Reverse: 5ʹ-TATCGTTGTACTCCACTCCTTGAC-3’ | |
| FOXP1 | Forward: 5′-CTTGCTCAAGGCATGATTCC-3′ |
| Reverse: 5′-CCTTGGTTCGTCAGCCAGTA-3′ | |
| ATG14 | Forward: 5′-ATGAGCGTCTGGCAAATCTT-3′ |
| Reverse: 5′-CCCATCGTCCTGAGAGGTAA-3′ | |
| U6 | Forward: 5′-CTCGCTTCGGCAGCACATATACT-3′ |
| Reverse: 5′-ACGCTTCACGAATTTGCGTGTC-3′ | |
| β-actin | Forward: 5ʹ-AGTGTGACGTGGACATCCGCAAAG-3ʹ |
| Reverse: 5′-ATCCACATCTGCTGGAAGGTGGAC-3′ |
2.7. Western blot assay
Total proteins were extracted from tissues or cells using RIPA buffer (Bolingkewei, Beijing, China). These proteins were separated by 12% SDS-PAGE and transferred onto PVDF membranes (Millipore, U.S.). After being blocked with 5% skim milk at room temperature for 1 h, the membranes were incubated overnight at 4° C with the following primary antibodies: anti-FOXP1 antibody (1:100, Cell Signaling Technology), anti-ATG14 antibody (1:1000, Proteintech, U.S.), anti- microtubule-associated protein 1 light chain 3 (LC3-I) antibody (1:2000, Cell Signaling Technology), anti-LC3-II antibody (1:2000, Cell Signaling Technology), anti-sequestosome 1 (P62) antibody (1:100, Cell Signaling Technology), anti-Beclin 1 antibody (1:2000, Boshide Biotech, Wuhan, China), anti-multidrug resistance gene 1 (MDR-1) antibody (1:50, Sigma), anti-H3 antibody (1:1000, Abcam) and anti-β-actin antibody (1:400, Santa Cruz). The next day, the membranes were incubated with horseradish peroxidase-conjugate antihuman or antirabbit secondary antibody (1:2000, Santa Cruz) for 1 h at room temperature. An EZ-ECL kit (Beyotime, Shanghai, China) was used to detect the band.
2.8. MTT assay
An MTT (Sigma) assay was used to measure cell viability. The cells were seeded in 96-well plates (2 × 104 cells/well). After transfection, the cells were treated with different concentrations of DDP and cultured for 24 h. Then, 20 μl of MTT (Sigma) were added to each well and cultured with the cells for an additional 4 h. Finally, dimethylsulfoxide (150 μl) was added to each well, and the absorbance of the cells was measured using a microplate reader (Molecular Devices, U.S.). Cell viability was calculated as the absorbance of the treated cells relative to that of untreated cells.
2.9. Dual luciferase reporter (DLR) assay
Bioinformatics software (TargetScan and microRNA. org) was used to analyze the binding sites of miR-29c-3p and the 3ʹUTR of FOXP1. A luciferase reporter plasmid (LRP) containing wild-type (WT) and mutant (Mut) 3′UTRs of FOXP1 was constructed according to the binding sites predicted by the software. The SKOV3 and SKOV3/DDP cells were co-transfected with a FOXP1 3ʹUTR-WT reporter vector or FOXP1 3ʹUTR-Mut reporter vector using Lipofectamine 2000 (Invitrogen). After transfection for 24 h, the cells were lysed. A DLR assay system was then used to detect luciferase activity, according to the manufacturer’s instructions.
2.10. Chromatin immunoprecipitation (ChIP) assay
A ChIP kit (Millipore) was used to detect protein-DNA interactions following the manufacturer’s protocols. Briefly, the cells were seeded in culture dishes at a density of 1 × 105 cells/mL and fixed with 1% formaldehyde for 10 min at room temperature. After washing twice with PBS, the cells were lysed with 1% SDS containing a protease inhibitor cocktail. RNA fragments were then disrupted and centrifuged, followed by incubation with FOXP1 and IgG antibody overnight at 4° C. Protein A/G was added to the complexes and incubated for 2 h at 4° C. After washing with high and low salt buffers, the complexes were eluted from the beads using elution buffer. The protein–DNA complexes were then retro-crosslinked at 65° C for 4 h. RNA was extracted using Trizol (Invitrogen), and the PCR products were electrophoresed on a regular agarose gel.
2.11. Animal experiment
Nude mice (aged 4–5 weeks) were purchased from Bioray Laboratories Inc., Shanghai, China. The mice were randomly divided into a pre-NC group (control group, n = 5) and miR-29c-3p mimic group (n = 5). In the miR-29c-3p mimic group, SKOV3/DDP cells (5 × 107 cells/mL) transfected with an miR-29c-3p mimic were subcutaneously injected into nude mice (200 µl/mouse). The pre-NC group was injected with an SKOV3/DDP cell suspension. DDP (4 mg/kg) was injected into the nude mice every 3 days after the tumoral diameters had increased to 5 mm. The sizes of the tumors were measured every 3 days. After 21 d, the mice were killed, and the protein expression of FOXP1, ATG14, LC3-I, LC3-II, P62, Beclin 1, and MDR-1 in tumor tissues was measured using a Western blot assay.
2.12. Statistical analysis
SPSS 17.0 software was used for data analysis. The student’s t test or an analysis of variance was used to analyze the differences between groups. All experiments were performed at least three times, and all data are expressed as mean ± standard deviation (SD). A value of P < 0.05 was considered statistically significant.
3. Results
3.1. Expressions of miR-92c-3p, FOXP1, and autophagy-related proteins in DDP-sensitive cells and DDP-resistant cells
In this experiment, we measured the expressions of autophagy-related molecules in the DDP-sensitive cell lines (SKOV3 and A2780) and in the DDP-resistant cell lines (SKOV3/DDP and A2780/DDP). Compared with the DDP-sensitive cells, miR-29c-3p expression was significantly decreased in DDP-resistant cells (Figure 1(a and d)). The protein expression levels of FOXP1, ATG14, and MDR-1, were higher in DDP-resistant cells than in DDP-sensitive cells (Figure 1(b and e). Meanwhile, the FOXP1 expression in nucleus was increased in DDP-resistant cells, suggesting more FOXP1 was transported from cytoplasm to nucleus to act as a transcription factor (Figure 1(b and e)). The LC3-Ⅱ/LC3-Ⅰ ratio and Beclin 1 protein levels were increased in DDP-resistant cells, whereas P62 protein expression was decreased in these cells (Figure 1(c and f)). These data indicated that miR-92c-3p was downregulated, while FOXP1 was upregulated in DDP-resistant cells, and that the autophagy was induced in DDP-resistant cells.
Figure 1.
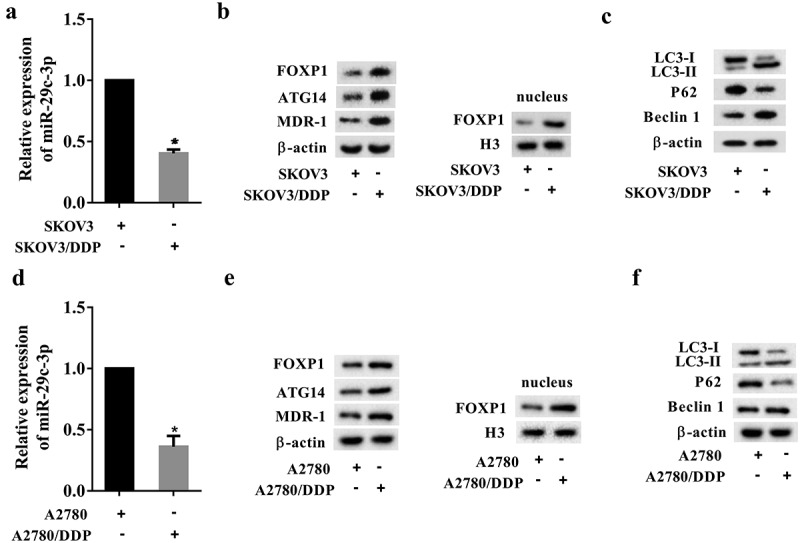
Expressions of miR-92c-3p, FOXP1, autophagy-related proteins, and drug-resistant proteins in ovarian cancer cells. (a) The expression of miR-29c-3p in the DDP-sensitive cell line (SKOV3) and in the DDP-resistant cell line (SKOV3/DDP) was detected using qRT-PCR. (b) The expression of FOXP1, ATG14, and MDR-1, and the expression of FOXP1 in nucleus were detected using Western blot assays. (c) The expression of autophagy-related proteins (LC3-I, LC3-II, P62, and Beclin 1) was detected using Western blot assays. (d) The expression of miR-29c-3p in the DDP-sensitive cell line (A2780) and in the DDP-resistant cell line (A2780/DDP) was detected using qRT-PCR. (e) The expression of FOXP1, ATG14, and MDR-1, and the expression of FOXP1 in nucleus were detected using Western blot assays. (f) The expression of autophagy-related proteins (LC3-I, LC3-II, P62, and Beclin 1) was detected using Western blot assays. Three independent experiments with biological repeats. *P < 0.05, vs. SKOV3 or A2780.
3.2. Effect of miR-29c-3p on cell viability of DDP-treated ovarian cancer cells
To examine the effect of miR-29c-3p on cell viability of DDP-treated ovarian cancer cells, the DDP-sensitive cell lines (SKOV3 and A2780) and the DDP-resistant cell lines (SKOV3/DDP and A2780/DDP) were treated with DDP at various concentrations for 24 h according to the inhibitory concentration 50 (IC50) of each cells (Supplemental Figure 1), and the MTT assay was performed to evaluate cell viability. Compared with the control group, miR-29c-3p inhibitor enhanced cell viability of DDP-sensitive cells (Figure 2(a and c)), indicating that the miR-29c-3p inhibitor increased the resistance of DDP-sensitive cells to DDP. The cell viability of DDP-resistant cells was significantly reduced by miR-29c-3p mimic compared with the control group (Figure 2(b and d)), indicating that miR-29c-3p mimic increased the sensitivity of DDP-resistant cells to DPP. Together, these data demonstrated the anti-resistant effect of miR-29c-3p in ovarian cells.
Figure 2.
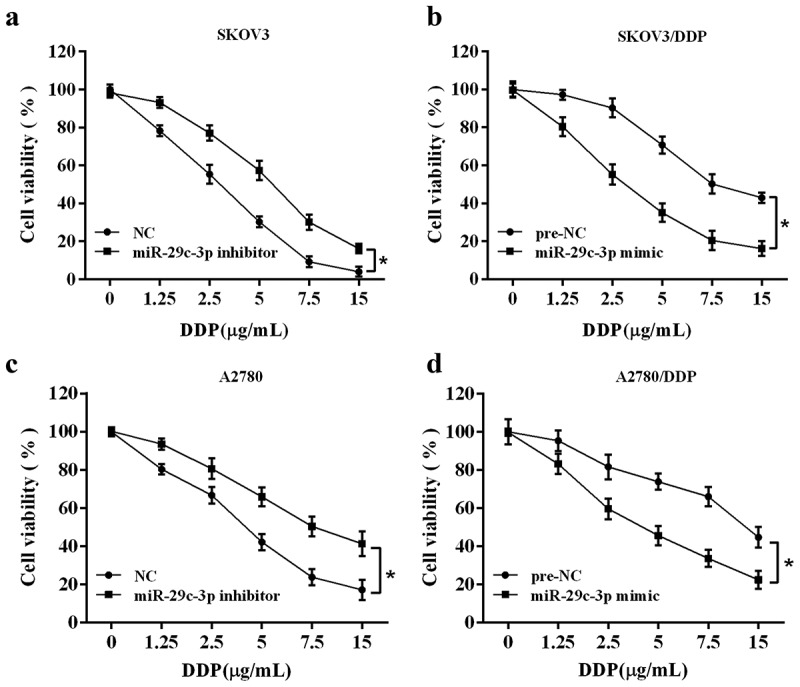
Effect of miR-29c-3p on cell viability of DDP-treated ovarian cancer cells. (a) The DDP-sensitive cell line (SKOV3) was transfected with miR-29c-3p inhibitor followed by the DDP treatment with increasing concentrations (0, 1.25, 2.5, 5, 7.5, and 15 μg/mL) for 24 h. Cell viability was detected using the MTT assay. (b) The DDP-resistant cell line (SKOV3/DDP) was transfected with miR-29c-3p mimic followed by the DDP treatment with increasing concentrations (0, 1.25, 2.5, 5.0, 7.5, 15 μg/mL) for 24 h. Cell viability was detected using the MTT assay. (c) The DDP-sensitive cell line (A2780) was transfected with miR-29c-3p inhibitor followed by the DDP treatment with increasing concentrations (0, 1.25, 2.5, 5, 7.5, and 15 μg/mL) for 24 h. Cell viability was detected using the MTT assay. (b) The DDP-resistant cell line (A2780/DDP) was transfected with miR-29c-3p mimic followed by the DDP treatment with increasing concentrations (0, 1.25, 2.5, 5.0, 7.5, 15 μg/mL) for 24 h. Cell viability was detected using the MTT assay. Three independent experiments with biological repeats. *P < 0.05, vs. negative control (NC) or pre-NC.
3.3. miR-29c-3p directly regulates FOXP1 expression
The results of the bioinformatics analysis (TargetScan and microRNA.org) showed that the 3ʹUTR of FOXP1 contained a sequence binding to miR-29c-3p (Figure 3(a)). An LRP containing WT and Mut 3ʹUTRs of FOXP1 was constructed according to the binding sites predicted by the software. SKOV3 cells were co-transfected with an miR-29c-3p inhibitor and LRP, and SKOV3/DDP cells were co-transfected with an miR-29c-3p mimic and LRP. The results of a DLR assay revealed markedly increased luciferase activity of the 3ʹUTR of WT FOXP1 in SKOV3 cells co-transfected with the miR-29c-3p inhibitor compared with the NC group. In contrast, there was no significant change in the luciferase activity of the 3ʹUTR of Mut FOXP1 (Figure 3(b)). As expected, the miR-29c-3p inhibitor upregulated the mRNA and protein expression of FOXP1 in SKOV3 cells (Figure 3(b)). On the other hand, the luciferase activity of the 3ʹUTR of WT FOXP1 was reduced in SKOV3/DDP cells co-transfected with the miR-29c-3p mimic, and the expression of FOXP1 was inhibited in these cells (Figure 3(c)).
Figure 3.
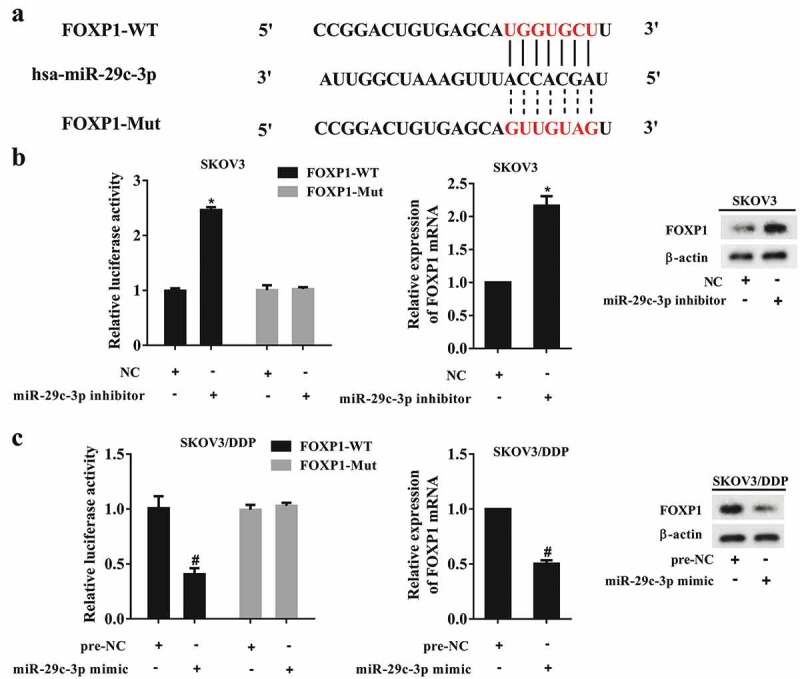
miR-29c-3p directly regulates FOXP1 expression. (a) The binding site between miR-29c-3p and the 3ʹUTR of FOXP1. (b) The luciferase activity of the 3ʹUTR of FOXP1 in SKOV3 cells. (c) The luciferase activity of the 3ʹUTR of FOXP1 in SKOV3/DDP cells. Three independent experiments with biological repeats. *P < 0.05, vs. NC, #P < 0.05, vs. pre-NC.
3.4. Effect of FOXP1 on ATG14 expression
The ChIP assay showed that FOXP1 could bind to the promoter of ATG14 (Figure 4(a)). Thus, we examined the effect of FOXP1 on ATG14 expression in SKOV3 cells and SKOV3/DDP cells. The pcDNA-FOXP1 was used to overexpress FOXP1, and siRNA-FOXP1 was used to silence FOXP1. The results demonstrated that overexpression of FOXP1 promoted the mRNA and protein levels of ATG14 in SKOV3 cells (Figure 4(b)). Silencing of FOXP1 reduced ATG14 expression in SKOV3/DDP cells (Figure 4(c)). These data indicated that ATG14 expression was transactivated by FOXP1.
Figure 4.
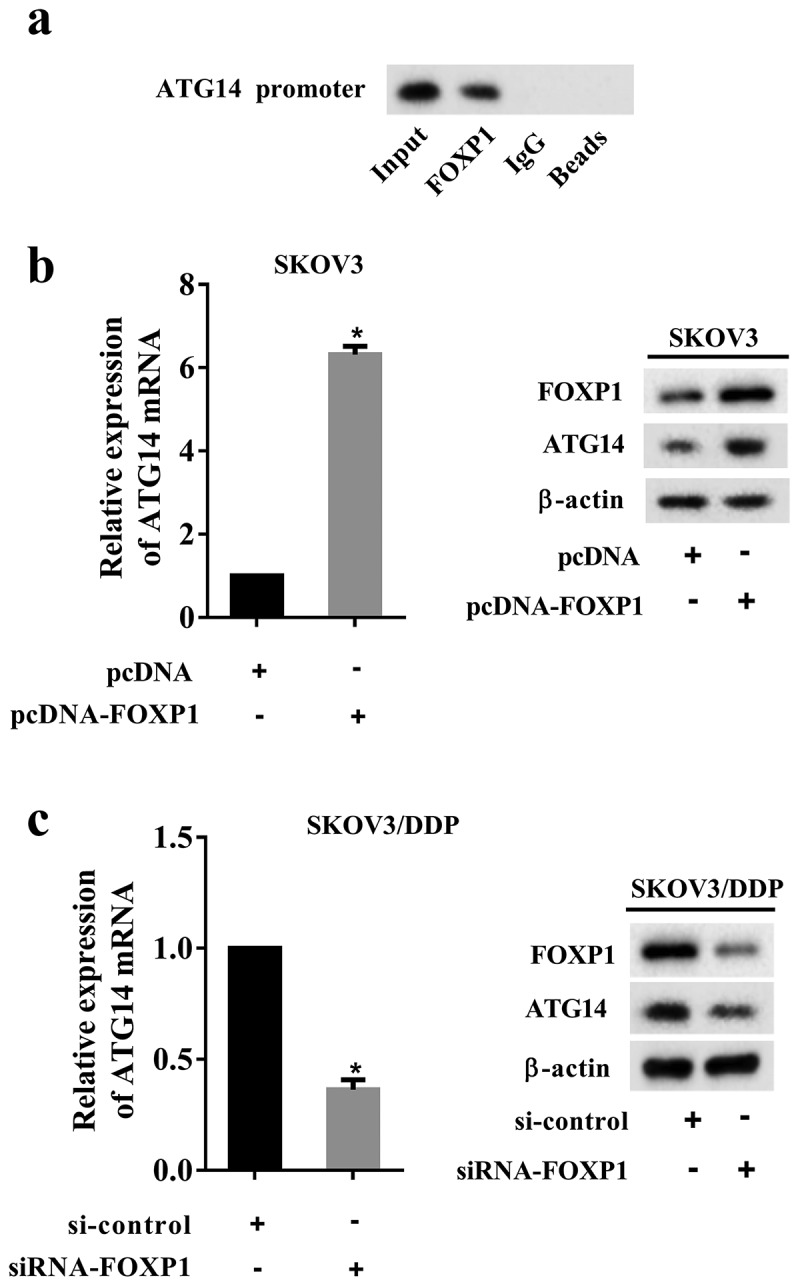
Effect of FOXP1 on ATG14 expression. (a) The interaction of FOXP1 and ATG14 promoter was measured using a ChIP assay. (b) The SKOV3 cells were transfected with pcDNA-FOXP1, and the mRNA and protein levels of ATG14 were detected using qRT-PCR and Western blot assay. (c) The SKOV3/DDP cells were transfected with siRNA-FOXP1, and the mRNA and protein levels of ATG14 were detected using qRT-PCR and Western blot assay. Three independent experiments with biological repeats. *P < 0.05, vs. pcDNA or si-control.
3.5. miR-29c-3p inhibits ATG14 expression via downregulating FOXP1
To investigate whether miR-29c-3p regulates ATG14 via FOXP1, the DDP-sensitive cell lines (SKOV3 and A2780) were divided into the NC, miR-29c-3p inhibitor, miR-29c-3p inhibitor + si-control, and miR-29c-3p inhibitor + siRNA-FOXP1 groups. The results showed that the miR-29c-3p inhibitor increased the POXP1 protein and the mRNA and protein levels of ATG14, whereas such response was negated by the siRNA-FOXP1 co-transfection (Figure 5(a)). The DDP-resistant cell lines (SKOV3/DDP and A2780/DDP) were divided into the pre-NC, miR-29c-3p mimic, miR-29c-3p mimic + pcDNA, and miR-29c-3p mimic + pcDNA-FOXP1 groups. The results showed that the miR-29c-3p mimic significantly reduced the FOXP1 protein and the mRNA and protein levels of ATG14, while such response was negated by the pcDNA-FOXP1 co-transfection (Figure 5(b)). Taken together, these findings indicated that miR-29c-3p inhibited ATG14 expression via downregulating FOXP1.
Figure 5.
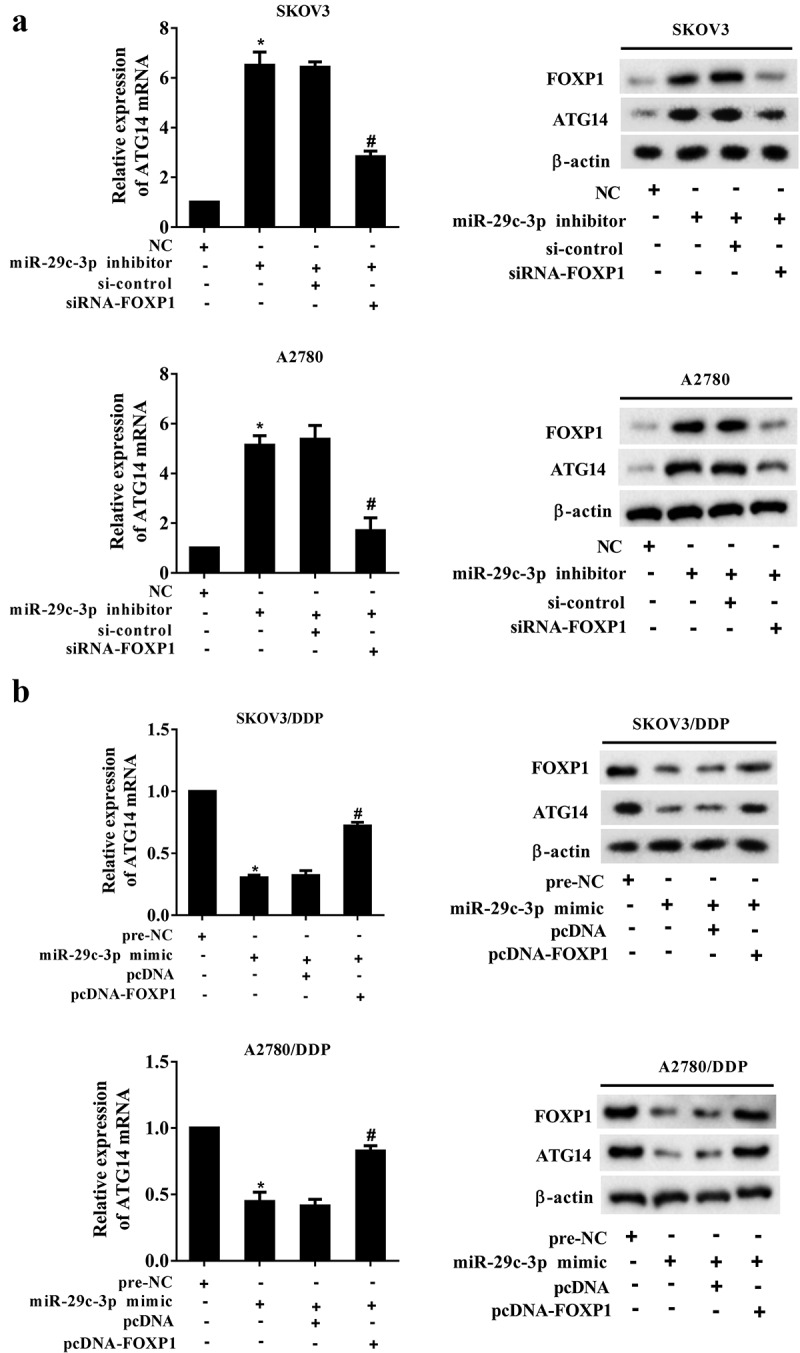
miR-29c-3p inhibits ATG14 expression by downregulating FOXP1. (a) The DDP-sensitive cell lines (SKOV3 and A2780) were divided into the NC, miR-29c-3p inhibitor, miR-29c-3p inhibitor + si-control, and miR-29c-3p inhibitor + siRNA-FOXP1 groups. The mRNA expression of ATG14 was detected using qRT-PCR, and the protein levels of FOXP1 and ATG14 were detected using Western blot assay. (b) The DDP-resistant cell lines (SKOV3/DDP and A2780/DDP) were divided into the pre-NC, miR-29c-3p mimic, miR-29c-3p mimic + pcDNA, and miR-29c-3p mimic + pcDNA-FOXP1 groups. The mRNA expression of ATG14 was detected using qRT-PCR, and the protein levels of FOXP1 and ATG14 were detected using Western blot assay. Three independent experiments with biological repeats. *P < 0.05, vs. NC or pre-NC, #P < 0.05, vs. miR-29c-3p inhibitor + si-control or miR-29c-3p mimic + pcDNA.
3.6. miR-29c-3p controls autophagy and DDP resistance by regulating FOXP1/ATG14 pathway
We next examined the effect of miR-29c-3p/FOXP1/ATG14 on autophagy of ovarian cells. The DDP-sensitive cell lines (SKOV3 and A2780) were transfected with miR-29c-3p inhibitor or co-transfected with siRNA-FOXP1 and pcDNA-ATG4, followed by the stimulation of rapamycin, which is an agonist of autophagy. The autophagy was promoted by miR-29c-3p inhibitor, and inhibited by siRNA-FOXP1, then it was promoted again by pcDNA-ATG14 (Figure 6(a)). The DDP-resistant cell lines (SKOV3/DDP and A2780/DDP) were transfected with miR-29c-3p mimic or co-transfected with pcDNA-FOXP1 and siRNA-ATG4, followed by the treatment of 3-MA, which is an antagonist of autophagy. The autophagy was inhibited by miR-29c-3p mimic, and promoted by pcDNA-FOXP1, then it was inhibited again by siRNA-ATG14 (Figure 6(b)).
Figure 6.
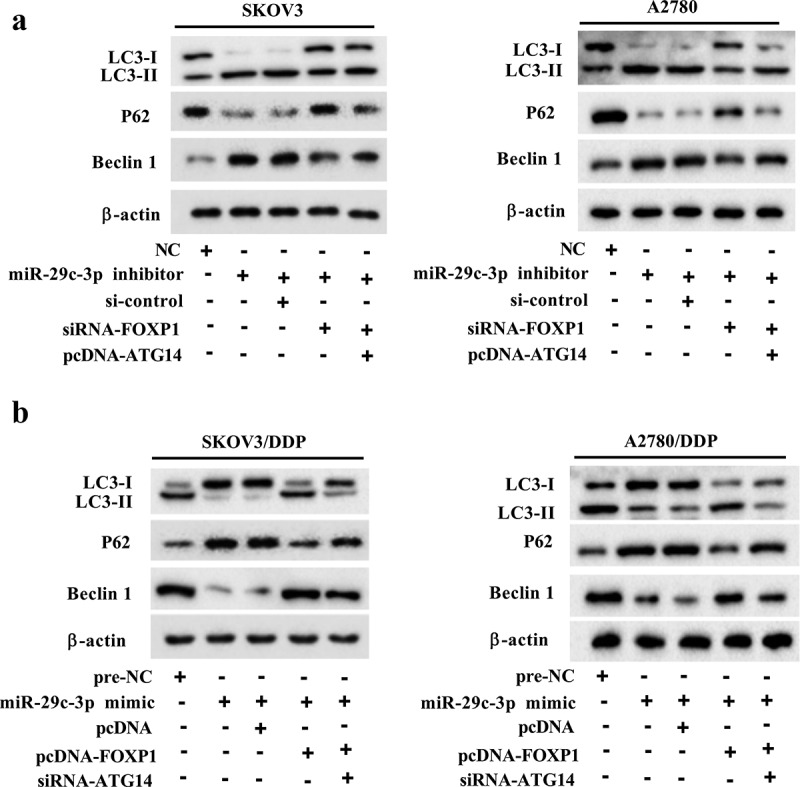
miR-29c-3p controls autophagy by regulating FOXP1/ATG14 pathway. (a) The DDP-sensitive cell lines (SKOV3 and A2780) were divided into the NC, miR-29c-3p inhibitor, miR-29c-3p inhibitor + si-control, miR-29c-3p inhibitor + si-FOXP1, and miR-29c-3p inhibitor + si-FOXP1 + pcDNA-ATG14 groups. Then the cells were treated with rapamycin, which is an agonist of autophagy. The expression of autophagy-related proteins was detected using Western blot assay. (b) The DDP-resistant cell lines (SKOV3/DDP and A2780/DDP) were divided into the pre-NC, miR-29c-3p mimic, miR-29c-3p mimic + pcDNA, miR-29c-3p mimic + pcDNA-FOXP1, and miR-29c-3p mimic + pcDNA-FOXP1 + si-ATG14 groups. Then the cells were treated with 3-MA, which is an antagonist of autophagy. The expression of autophagy-related proteins was detected using Western blot assay. Three independent experiments with biological repeats.
Cell viability was also detected in the DDP-sensitive cell lines (SKOV3 and A2780) and the DDP-resistant cell lines (SKOV3/DDP and A2780/DDP), which were transfected as described above. After the transfection, cells were stimulated by 5 μg/mL DPP for 24 h before the MTT assay. In the DDP-sensitive cell lines, cell viability and the protein expression of ATG14 and MDR-1 were promoted by miR-29c-3p inhibitor, and inhibited by siRNA-FOXP1, then it was promoted again by pcDNA-ATG14 (Figure 7(a and c)). In the DDP-resistant cell lines, cell viability and the protein expression of ATG14 and MDR-1 was inhibited by miR-29c-3p mimic, and promoted by pcDNA-FOXP1, then it was inhibited again by siRNA-ATG14 (Figure 7(b and d)).
Figure 7.
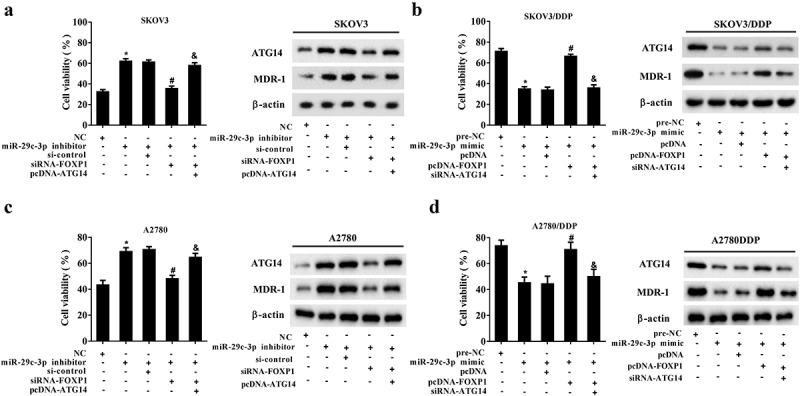
miR-29c-3p controls DDP resistance by regulating FOXP1/ATG14 pathway. (a and c) The DDP-sensitive cell lines (SKOV3 and A2780) were divided into the NC, miR-29c-3p inhibitor, miR-29c-3p inhibitor + si-control, miR-29c-3p inhibitor + si-FOXP1, and miR-29c-3p inhibitor + si-FOXP1 + pcDNA-ATG14 groups. Cell viability was detected using the MTT assay, and protein levels of ATG4 and MDR-1 were detected using Western blot assay. (b and d) The DDP-resistant cell lines (SKOV3/DDP and A2780/DDP) were divided into the pre-NC, miR-29c-3p mimic, miR-29c-3p mimic + pcDNA, miR-29c-3p mimic + pcDNA-FOXP1, and miR-29c-3p mimic + pcDNA-FOXP1 + si-ATG14 groups. Cell viability was detected using the MTT assay, and protein levels of ATG4 and MDR-1 were detected using Western blot assay. Three independent experiments with biological repeats. *P < 0.05, vs. NC or pre-NC; #P < 0.05, vs. miR-29c-3p inhibitor + si-control or miR-29c-3p mimic + pcDNA; P < 0.05, vs. miR-29c-3p inhibitor + siRNA-FOXP1, or miR-29c-3p mimic + pcDNA-FOXP1.
3.7. miR-29c-3p overexpression overcomes DDP resistance in vivo
To verify the effect of miR-29c-3p overexpression on DDP resistance in ovarian cancer, nude mice were subcutaneously injected with SKOV3/DDP cells which were transfected with miR-29c-3p mimic. The tumor volume was measured every 3 days. Compared with the control group, the miR-29c-3p overexpression in DDP-resistant cells significantly reduced the tumor volume (Figure 8(a)). Moreover, overexpression of miR-29c-3p reduced the expression of FOXP1, ATG14, MDR-1, LC3-Ⅱ/LC3-Ⅰ ratio and Beclin 1, while it increased P62 protein expression (Figure 8(b)), suggesting miR-29c-3p overexpression reduced autophagy and overcame DDP resistance in ovarian cancer. Meanwhile, the protein level of FOXP1 in nucleus was also reduced after miR-29c-3p overexpression, indicating a reduction of FOXP1 transportation from cytoplasm to nucleus, thus reducing the transcription and expression of ATG14 (Figure 8(b)).
Figure 8.
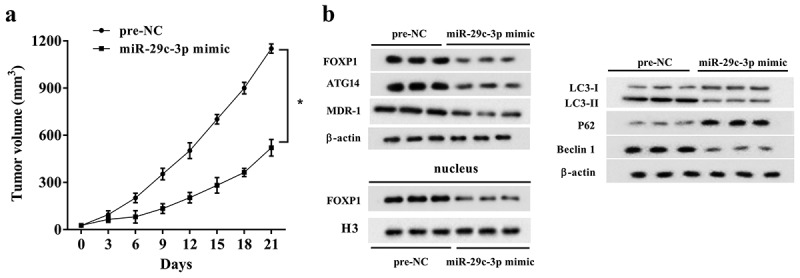
miR-29c-3p overexpression overcomes DDP resistance in vivo. Nude mice were subcutaneously injected with SKOV3/DDP cells, which were transfected with miR-29c-3p mimic (n = 5) or pre-NC (n = 5). DDP (4 mg/kg) was injected into nude mice every 3 days when the diameter of tumors reached 5 mm. (a) The sizes of the tumors were measured every 3 days. (b) After 21 d, the mice were killed, and the protein expression of FOXP1, ATG14, LC3-I, LC3-II, P62, Beclin 1, and MDR-1, and FOXP1 in nucleus was measured using Western blot assay. Three independent experiments with biological repeats of each mouse. *P < 0.05, vs. pre-NC.
4. Discussion
Ovarian cancer poses a serious threat to the health of women. A large number of studies confirmed that the induction of tumor cell apoptosis was the main mechanism of antitumor drugs. Recent studies demonstrated that some antitumor drugs enhanced autophagy of tumor cells [27] and that autophagy was the primary cause of a poor cancer prognosis [8]. Beclin 1 was the first mammalian autophagy gene to be identified, and is considered a key factor in autophagy in mammals [28]. LC3 is a homologue of yeast autophagy-related gene 8 in mammalian cells. Newly synthesized LC3 is processed into soluble LC3-I in cytoplasm and then binds to phosphatidyl ethanolamine on the surface of autophagic vacuoles to form LC3-Ⅱ [29]. The LC3 protein is a specific marker of autophagy, and the LC3-Ⅱ content and LC-II to LC3-I ratio reflect the quantity and extent of autophagy [30]. P62, a multifunctional protein, shows abnormal expression in most tumors [31], and it plays an important role in cell proliferation, differentiation, and apoptosis [32]. Increasing evidence suggests that P62 may serve as a substrate for selective autophagy [33]. MDR-1 encodes P-glycoprotein, a plasma membrane glycoprotein that acts as an efflux pump and causes multidrug resistance in cancer cells [34]. A previous study detected increased expression of MDR-1 in cancer cells after treatment with chemotherapeutic drugs [35]. In the present study, the results revealed an increased ratio of LC3-Ⅱ/LC3-I, upregulated expression of Beclin 1and MDR-1, and decreased expression of P62 in DDP-resistant cells, indicating that DDP resistance was associated with increased autophagy in ovarian cancer. Moreover, the results revealed differential expression of miR-29c-3p and FOXP1 in DDP-sensitive cells and DDP-resistant cells, suggesting that the two molecules (miR-29c-3p and FOXP1) probably participate in chemoresistance of ovarian cancer.
A number of recent studies demonstrated that miRNAs played an important role in ovarian cancer. For example, Liu et al. [36] showed that miR-1271 inhibited the growth of ovarian cancer cells by targeting cyclin G1. Guo et al. [37] demonstrated that miR-302a suppressed the proliferation of ovarian cancer cells by downregulating SDC1. Other research reported that miRNAs ware associated with DDP resistance in ovarian cancer. For example, Echevarríavargas et al. [38] found that miR-21 promoted DDP resistance in ovarian cancer cells via the JNK-1/c-Jun pathway. Another study showed that miR-489 exerted an inhibitory effect on DDP resistance of human ovarian cancer cells by repressing Akt3 [39]. The current study revealed that miR-29c-3p regulated DDP resistance in ovarian cancer cells by targeting FOXP1, which is widely expressed in both healthy and tumor tissues in humans and may be involved in the progression of cancers.
FOXP1 belongs to the subfamily P of the forkhead box transcription factor family. It has been reported that FOXP1 expression is closely related to the degree of malignancy of ovarian cancer and may be a reliable index for the prognosis [13]. Previous study revealed that FOXP1 expression can be regulated by several miRNAs, such as miR-152 [40] and miR-374b-5p [41]. In the present study, we demonstrated that FOXP1 expression was also regulated by miR-29c-3p, highlighting the oncogenic effect of FOXP1 in ovarian cancer. However, whether there are interactions among FOXP1 upstream regulators deserve further investigations. In addition, our findings also indicated that FOXP1 targeted ATG14 and participated in DDP resistance in ovarian cancer. Analysis of the relationship between miR-29c-3p/FOXP1 and ATG14 in ovarian cancer cells demonstrated that miR-29c-3p downregulated ATG14 via the regulation of FOXP1, resulting in inhibition of autophagy and DDP resistance in ovarian cancer cells. The regulatory mechanism was confirmed in vivo.
Taken together, the findings of the current study demonstrated that miR-29c-3p repressed autophagy and chemoresistance in ovarian cancer by reducing FOXP1/ATG14. These results indicate that miR-29c-3p may be a valuable target to overcome DDP resistance in ovarian cancer.
Funding Statement
This work is supported by the National Natural Science Foundation of China (No. 31670844); Sponsored by Program for Science and Technology Innovation Talents of Henan Province (No. 2018 JR0004); ZhongYuan thousand talents program-the Zhongyuan eminent doctor in Henan province (No. ZYQR201810107).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
References
- [1].Tawde SA, Chablani L, Akalkotkar A, et al. Evaluation of microparticulate ovarian cancer vaccine via transdermal route of delivery. J Control Release. 2016;235:147–154. [DOI] [PubMed] [Google Scholar]
- [2].Borley J, Brown R.. Epigenetic mechanisms and therapeutic targets of chemotherapy resistance in epithelial ovarian cancer. Ann Med. 2015;47:359–369. [DOI] [PubMed] [Google Scholar]
- [3].Zeller C, Brown R. Therapeutic modulation of epigenetic drivers of drug resistance in ovarian cancer. Ther Adv Med Oncol. 2010;2:319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jing K, Lim K. Why is autophagy important in human diseases? Exp Mol Med. 2012;44:69–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wu KK, Coffelt SB, Cho CH, et al. The autophagic paradox in cancer therapy. Oncogene. 2012;31:939–953. [DOI] [PubMed] [Google Scholar]
- [6].Du H, Yang W, Chen L, et al. Role of autophagy in resistance to oxaliplatin in hepatocellular carcinoma cells. Oncol Rep. 2012;27:143–150. [DOI] [PubMed] [Google Scholar]
- [7].Zhang SF, Wang XY, Fu ZQ, et al. TXNDC17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy. 2015;11:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang J, Wu GS. Role of autophagy in cisplatin resistance in ovarian cancer cells. J Biol Chem. 2014;289:17163–17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Katoh M, Igarashi M, Fukuda H, et al. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328:198–206. [DOI] [PubMed] [Google Scholar]
- [10].Koon HB, Ippolito GC, Banham AH, et al. FOXP1: a potential therapeutic target in cancer. Expert Opin Ther Targets. 2007;11:955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang X, Sun J, Cui M, et al. Downregulation of FOXP1 inhibits cell proliferation in hepatocellular carcinoma by inducing G1/S phase cell cycle arrest. Int J Mol Sci. 2016;17:1501–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Takayama K, Suzuki T, Tsutsumi S, et al. Integrative analysis of FOXP1 function reveals a tumor-suppressive effect in prostate cancer. Mol Endocrinol. 2014;28:2012–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hu Z, Zhu L, Gao J, et al. Expression of FOXP1 in Epithelial Ovarian Cancer (EOC) and its correlation with chemotherapy resistance and prognosis. Tumor Biol. 2015;36:7269–7275. [DOI] [PubMed] [Google Scholar]
- [14].Choi EJ, Seo EJ, Kim DK, et al. FOXP1 functions as an oncogene in promoting cancer stem cell-like characteristics in ovarian cancer cells. Oncotarget. 2016;7:3506–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chan SH, Wang LH. Regulation of cancer metastasis by microRNAs. Trends Cell Biol. 2014;24:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xia H, Hui KM. Mechanism of cancer drug resistance and the involvement of noncoding RNAs. Curr Med Chem. 2014;21:3029–3041. [DOI] [PubMed] [Google Scholar]
- [17].Iwagami Y, Eguchi H, Nagano H, et al. miR-320c regulates gemcitabine-resistance in pancreatic cancer via SMARCC1. Br J Cancer. 2013;109:502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhou Y, Wan G, Spizzo R, et al. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol Oncol. 2014;8:83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yin Y, Zhang B, Wang W, et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res. 2014;20:6187–6199. [DOI] [PubMed] [Google Scholar]
- [20].Xu S, Fu GB, Zhen T, et al. MiR-497 decreases cisplatin resistance in ovarian cancer cells by targeting mTOR/P70S6K1. Oncotarget. 2015;6:26457–26471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shuang T, Wang M, Shi C, et al. Down-regulated expression of miR-134 contributes to paclitaxel resistance in human ovarian cancer cells. FEBS Lett. 2015;589:3154–3164. [DOI] [PubMed] [Google Scholar]
- [22].Li N, Yang L, Wang H, et al. MiR-130a and MiR-374a function as novel regulators of cisplatin resistance in human ovarian cancer A2780 Cells. Plos One. 2015;10:e0128886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xu W, Li Z, Zhu X, et al. miR-29 family inhibits resistance to methotrexate and promotes cell apoptosis by targeting COL3A1 and MCL1 in osteosarcoma. Med Sci Monit. 2018;24:8812–8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Muluhngwi P, Alizadeh-Rad N, Vittitow SL, et al. The miR-29 transcriptome in endocrine-sensitive and resistant breast cancer cells. Sci Rep. 2017;7:5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yu PN, Yan MD, Lai HC, et al. Downregulation of miR-29 contributes to cisplatin resistance of ovarian cancer cells. Int J Cancer. 2014;134:542–551. [DOI] [PubMed] [Google Scholar]
- [26].Li X, Zhuang X, Xu T, et al. Expression analysis of microRNAs and mRNAs in ovarian granulosa cells after microcystin-LR exposure. Toxicon. 2017;129:11–19. [DOI] [PubMed] [Google Scholar]
- [27].Yang ZJ, Chee CE, Huang S, et al. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Valente G, Morani F, Nicotra G, et al. Expression and clinical significance of the autophagy proteins BECLIN 1 and LC3 in ovarian cancer. Biomed Res Int. 2015;2014:462658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tanida I, Minematsuikeguchi N, Ueno T, et al. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. [DOI] [PubMed] [Google Scholar]
- [30].Aparicio IM, Martin MP, Salido GM, et al. The autophagy-related protein LC3 is processed in stallion spermatozoa during short-and long-term storage and the related stressful conditions. Animal. 2016;10:1182–1191. [DOI] [PubMed] [Google Scholar]
- [31].Moscat J, Diaz-Meco MT. p62: a versatile multitasker takes on cancer. Trends Biochem Sci. 2012;37:230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Geetha T, Zheng C, Vishwaprakash N, et al. Sequestosome 1/p62, a scaffolding protein, is a newly identified partner of IRS-1 protein. J Biol Chem. 2012;287:29672–29678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ichimura Y, Kumanomidou T, Sou YS, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. [DOI] [PubMed] [Google Scholar]
- [34].Wu Q, Yang Z, Nie Y, et al. Multi-drug resistance in cancer chemotherapeutics: mechanisms and lab approaches. Cancer Lett. 2014;347:159–166. [DOI] [PubMed] [Google Scholar]
- [35].Santos SA, Paulo A. Small molecule inhibitors of multidrug resistance gene (MDR1) expression: preclinical evaluation and mechanisms of action. Curr Cancer Drug Targets. 2013;13:814. [DOI] [PubMed] [Google Scholar]
- [36].Liu X, Ma L, Rao Q, et al. MiR-1271 inhibits ovarian cancer growth by targeting cyclin G1. Med Sci Monit. 2015;21:3152–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Guo T, Wei Y, Lv S, et al. MiR-302a inhibits the tumorigenicity of ovarian cancer cells by suppression of SDC1. Int J Clin Exp Pathol. 2015;8:4869–4880. [PMC free article] [PubMed] [Google Scholar]
- [38].Echevarríavargas IM, Valiyeva F, Vivasmejía PE. Upregulation of miR-21 in Cisplatin Resistant Ovarian Cancer via JNK-1/c-Jun Pathway. Plos One. 2014;9:e97094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wu H, Xiao Z, Zhang H, et al. MiR-489 modulates cisplatin resistance in human ovarian cancer cells by targeting Akt3. Anticancer Drugs. 2014;25:799–809. [DOI] [PubMed] [Google Scholar]
- [40].Qin W, Xie W, He Q, et al. MicroRNA-152 inhibits ovarian cancer cell proliferation and migration and may infer improved outcomes in ovarian cancer through targeting FOXP1. Exp Ther Med. 2018;15:1672–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li H, Liang J, Qin F, et al. MiR-374b-5p-FOXP1 feedback loop regulates cell migration, epithelial-mesenchymal transition and chemosensitivity in ovarian cancer. Biochem Biophys Res Commun. 2018;505:554–560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


