ABSTRACT
Background: Homeobox B5 (HOXB5) is associated with the poor prognosis of various cancer types. However, the specific mechanism by which HOXB5 promotes the malignant progression of pancreatic cancer (PC) remains to be determined.
Methods: The Cancer Genome Atlas database indicated HOXB5 expression level correlated to PC prognosis. The biological functions of HOXB5 was confirmed by colony formation, migration, and invasion assays. The effects of HOXB5 on the expression of cancer stem cell and epithelial–mesenchymal transition markers were evaluated. The downstream target of HOXB5 was miR-6723, which was detected by transcriptional assay. A xenograft tumor model was established in nude mice for the assessment of the role of HOXB5 in tumor growth and metastasis.
Results: PC tissues had higher HOXB5 expression levels than noncancerous tissues, and high HOXB5 expression was significantly associated with poor PC prognosis. HOXB5 knockdown suppressed clone formation and the proliferation, invasion, and migration of PC cells in vitro. Conversely, these activities were enhanced by HOXB5 overexpression. The HOXB5 that bound two synergy motifs regulated miR-6723 expression and contributed to PC malignant progression. The role of HOXB5 in promoting tumor growth and metastasis was verified in vivo. Further investigation revealed that Twist1 and Zeb1 expression levels were increased by HOXB5.
Conclusions: HOXB5 overexpression was significantly correlated with poor PC prognosis. HOXB5 accelerated the malignant progression of PC by up-regulating miR-6723, which afforded PC cells stem-like properties and facilitated the epithelial–mesenchymal transition of PC cells.
KEYWORDS: Pancreatic cancer, homeobox B5, stemness, epithelial-mesenchymal transition, miR-6723
Introduction
Pancreatic cancer (PC) is the fourth leading cause of cancer-associated mortality worldwide and the second deadliest type of cancer. PC has poor prognosis, with a low 5 year survival rate and a medium overall survival time of <1 year [1,2]. Many target drugs that have been applied to PC therapy have no specific curative effects compared with chemotherapy drugs, and the mortality rate increases every year [3,4]. This serious situation may be caused by several reasons. First, owing to the lack of specific tumor targets and effective detecting methods, patients are generally diagnosed at the late stages. Second, PC shows resistance to chemotherapy, radiotherapy, and molecular targeted therapy. Complex epigenetic changes pose a major challenge to PC therapy [5–7].
Epithelial–mesenchymal transition (EMT) is a major malignant progression mechanism driving tumor cell metastasis and invasion [8–10]. During EMT, epithelial cells gain mesenchymal traits, which are characterized by the down-regulation of E-cadherin and up-regulation of vimentin and other mesoderm markers [11]. EMT plays a critical role in PC progression [9], and involves many transcriptional factors, including Twist1, Zeb1, Snail, and Slug [12–15]. Discovering the transcriptional factor that drives EMT is important because it may lead to the identification of the malignant progression of PC. The HOXB family involved in tumor EMT has been widely studied, but EMT HOXBs with PC are rarely reported.
Homeobox genes encode a transcriptional family that contain 39 members, and the aberrant expression of homeobox genes is frequently detected in various cancer types. Homeobox B5 (HOXB5), as a member of the homeobox gene family, is involved in several cancer types, including lung, breast, prostate, and bladder cancer [16–18]. As an important transcription factor, HOXB5 regulates many cancer cell functions, including proliferation, invasion, and migration. It enhances proliferation and Wnt/β-catenin pathways [19] and facilitates the malignant of non-small-cell lung cancer cells by miR455-3p [20]. HOXB5 can modulate cell proliferation and migration, which is correlated with the poor prognosis of patients with PC. HOXB5 overexpression is significantly correlated with cancer progression and poor prognosis. However, the specific mechanism by which HOXB5 regulates the malignant progression of PC remains unclear.
In this study, we aimed to analyze the potential mechanism of HOXB5 in the malignant progression of PC. Using the Cancer Genome Atlas (TCGA) database, we confirmed that HOXB5 is highly expressed in PC and high HOXB5 expression is associated with poor outcomes in patients with PC. We also detected the role of HOXB5 in several cancer-related processes, including cell proliferation, stemness, invasion, and migration. The results indicated the specific mechanism of HOXB5 in PC progression and may provide a new therapeutic target in PC treatment.
Methods
Tissue samples and TCGA database from patients with PC
A total of 196 cases of the preexisting paraffin-embedded PC tissue samples were collected from patients who underwent curative resection at the TCGA database. A total of 20 cases fresh tissue samples with paired adjacent noncancerous tissue were obtained from surgery. All patients were informed according to the protocols approved by the TCGA database (TCGA Program, National Cancer Institute, https://www. cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) and the ethical standards of Sichuan Academy of Medical Sciences & Sichuan Provincial People’s Hospital. This study complied with the ethical guidelines of the Helsinki Declaration.
Cell culture
Human PC cells ASPC-1, SW1990, DAN-G, capan-2, HPAC, and PANC-1 were purchased from American Type Culture Collection (Rockefeller, MD, USA) and the Cell Bank of Shanghai Institute of Cell Biology (Shanghai, China). All cells were cultured in Dulbecco’s modified eagle medium (DMEM; Gibco, Grand Island, NY, USA) and supplemented with 10% fetal bovine serum (FBS, Gibco) at 37°C in a humidified incubator containing 5% CO2.
Knockdown using shRNAs
shRNAs targeting HOXB5 were purchased from OriGene Biotechnology Company (Beijing, China). SKU TR312357, which is the sequence after experimental verification and can effectively target the coding sequence of HOXB5 mRNA, was inserted into a pRS vector. The efficacy of each shRNA was assessed by performing Western blot analysis on endogenous proteins in cells infected with viruses upon plating. The cells were then cultured for 3 days. The shRNA with the strongest knockdown efficiency was selected for further experiments.
Tumor growth in nude mice
The study was approved by the ethics committee of Sichuan Academy of Medical Sciences & Sichuan Provincial People’s Hospital and complied with the guidelines for Animal Experiments of Laboratory Animals. A total of 24 six-week-old mice were housed under specific pathogen-free conditions. capan-2/nc, capan-2/HOXB5, and capan-2/miR-6723 (1 × 106) cells were subcutaneously injected into the mice. The tail of each mouse was intravenously injected with or without miR-6723-aso. Tumor growth was monitored by measuring tumor diameter each week, and tumor weights were calculated at the end of the study as follows: tumor volume = length × width2/2. After the mice were sacrificed, tumors were taken out and fixed in paraffin for further analysis.
Histology and immunohistochemistry
Paraffin-embedded tissue samples from the nude mice were cut into 4 μm-thick sections, deparaffinized with xylene, and rehydrated through graded alcohol washes. Antigen retrieval was performed on all sections by heating in a microwave oven, and endogenous peroxidase activity was blocked with 3% H2O2 solution. Then, the sections were incubated using an anti-HOXB5 antibody (1:500; Abcam, Cambridge, MA, USA) or any of the following: an anti-MMP2, MMP9, E-cadherin, Vimentin, Twist1, or Zeb1 antibody (1:500; Abcam) at 4°C overnight. Lung sections from the mice in metastasis were stained with hematoxylin and eosin (H&E). Immunohistochemistry assays were carried out using a DAKO EnVision Detection System.
The immunohistochemistry staining scores of HOXB5 in the tissues were assessed by two pathologists using a semiquantitative method. They were blinded with the clinical data. The staining score was assessed as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). High expression was defined as a staining score of ≥2 with at least 50% of malignant cells showing positive HOXB5 staining, and a low expression level was defined as <50% of malignant cells showing nuclear staining or a staining score of <2.
Clone formation assay
Briefly, capan-2/NC, capan-2/HOXB5, PANC-1/NC, and PANC-1/shHOXB5 cells were seeded in a 6-well plate at a density of 100 cells/well. The medium was replaced every 3 days, and the cells were cultured for 14 days. The clones were fixed with 4% formaldehyde for 10 min and stained with 0.5% crystal violet for 3 min (Sigma-Aldrich). A clone was defined as having >50 cells, and its formation rate was calculated as follows: clone formation rate (%) = (number of clones/number of seeded cells) × 100%.
Cell migration and invasion assay
A 24-well plate with Transwell inserts having 8 μm pore size (Millipore, Billerica, MA, USA) was used in the evaluation of cell motility. In the invasion assay, the membranes were precoated with Matrigel (2 μg/well; BD Biosciences, San Jose, CA, USA) to simulate a matrix barrier. PC cells (8.0 × 104) were suspended in 300 μL of serum-free DMEM medium and added to the upper part of the insert chamber, and 800 μL of DMEM with 20% serum was added to the lower chamber. No migrated cells were cultured for 48 h and then removed from the upper chamber. The cells were used in the invasion assay. Cells that passed through the membrane were fixed with 4% formaldehyde and stained with 0.1% crystal violet. Three visual fields were randomly selected from each membrane, and cell were counted using a light microscope at 100× magnification. All experiments were performed in triplicate.
Wound healing assay
Capan-2 and PANC-1 cells were plated in a 24-well plate and grown to confluence. Then, the cell layers were scratched for the wound healing assay. A linear wound was created by dragging a 1 μL pipette tip through the monolayer. Cells were washed with PBS. Then, complete medium was added. Photographs were taken under 100× magnification at 0 and 48 h post-wounding, and the cells were evaluated under an Olympus inverted microscope.
Western blot analysis
Total protein was extracted from the tumor samples or cells. The protein samples were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred into nitrocellulose membranes. After blocking by using 5% skimmed milk, the membranes were incubated overnight at 4°C with the following specific primary antibodies: anti-HOXB5 antibody, anti-E-cadherin, vimentin, Twist1, and Zeb1 (Abcam). After washing, the membranes were incubated for 2 h with secondary HRP antibodies (Sigma-Aldrich). Protein intensity was determined and measured using Image Lab (5.2.1 Version, Bio-Rad Laboratories Co., Ltd., CA, USA). After normalization to GAPDH protein units for each sample, semiquantitative results for either the tumor or adjacent samples were obtained as ratios.
RNA isolation and real-time quantitative reverse transcription PCR
The total RNA of the capan-2 cells was extracted and purified using TRIzol (Takara, Japan) according to the manufacturer’s instructions. Then, cDNA was synthesized using 1 μg RNA and a reverse transcriptional kit following the manufacturer’s protocol. Quantitative PCR was performed using a SYBR premix ExTaq (Takara) on a CFX96 real-time PCR detection system (Bio-Rad). GAPDH as an internal control was used to normalize the mRNA expression of each gene. All reactions were detected in triplicate, and each experiment was performed at least three independent times. Related primer sequences were purchased from OriGene Biotechnology Company (Beijing, China).
Reporter gene assays
The miR-6723 motifs were PCR amplified from human genomic DNA and cloned into a pGL4.3 luciferase reporter vector (Promega). Transactivation assays were performed with a dual-luciferase reporter assay system (Promega). PANC-1 cells were cotransfected by culturing for 72 h. Luciferase activities were measured using a Synergy 2 microplate reader system (Gene).
Zymography assays
All media were collected and subjected to SDS-PAGE by using 0.01% wt/vol gelatine containing 10% polyacrylamide gel. After electrophoresis, the gels were equilibrated in 2.5% Triton X-100 and incubated in 50 mM Tris-HCl (pH of 7.5), 10 mM CaCl2, 150 mM NaCl, 1 mM ZnCl2, and 0.02% NaN3 for 40 h at 37°C. The gels were stained with Coomassie R250 and destained until the wash became clear with apparent cleared zones that are associated with MMP activity.
Statistical analysis
The data were expressed as the means and standard deviations from a minimum of three separate experiments. The statistical significance of the differences was examined using Student’s t-test. The correlations of the clinical characteristics in patients with PC were analyzed using Chi-squared test. Survival data were used in drawing the Kaplan–Meier curves, and the differences among the groups were analyzed using the log-rank assay. Statistical analyzes were performed using SPSS 21.0 (SPSS Inc., Chicago, IL, USA). P < 0.05 indicated statistically significant difference.
Results
HOXB5 expression was correlated with poor PC prognosis
To examine the correlation between HOXB5 and PC prognosis, we detected the expression of HOXB5 in 196 PC specimens from TCGA and 20 fresh PC tissues. We found that HOXB5 expression level was higher in PC tissues than in the paired noncancerous tissues (Figure 1(a)). Further exploration by Western blot revealed that the protein level of HOXB5 was higher in the PC specimens (P < 0.001, Figure 1(b)). The TPM of RNA-seq demonstrated the variable expression level of HOXB5 in the PC tissues (Figure 1(c)). The TCGA database suggested that a high HOXB5 expression level in PC tissue is correlated with short overall survival time and PFS. We also found that the high HOXB5 expression level in PC tissue was positively associated with pathological grade (Figure 1(f)), tumor mutational burden (Figure 1(g)), and PC nodes (Figure 1(h)). However, the relationship between the degree of HOXB5 staining and age or clinical stage was nonsignificant. In summary, our data indicated that the aberrant HOXB5 expression in PC tissue was significantly correlated with poor PC prognosis.
Figure 1.
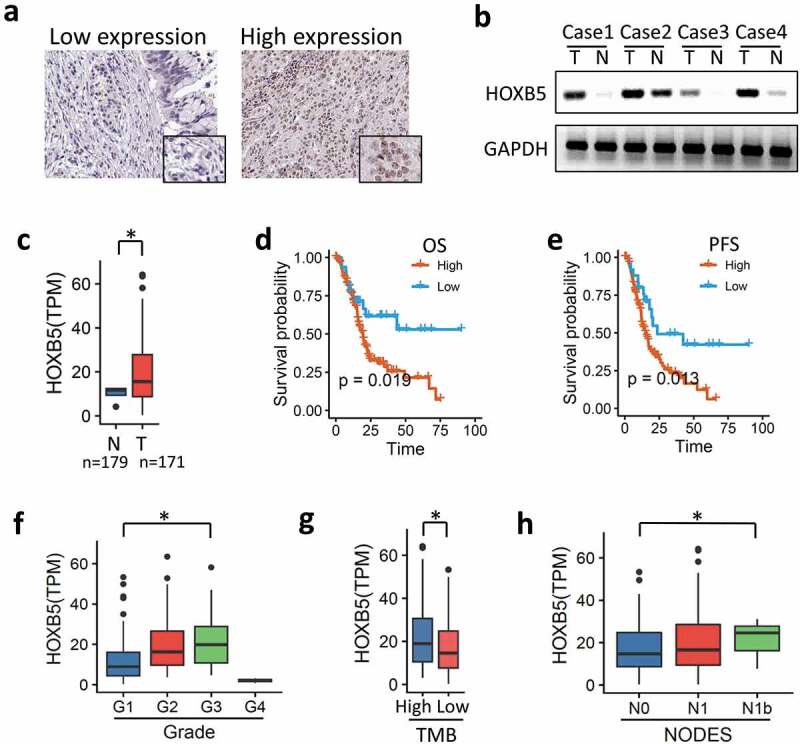
HOXB5 up-regulation in PC tissues correlates with poor prognosis. (a) IHC staining showed HOXB5 expression in the PC tissue with low and high cases. (b) Representative WB showing HOXB5 expression level in HCC and paired adjacent noncancerous tissues. (c) HOXB5 was negative in adjacent noncancerous tissues and positive in the PC tissues. (d) Kaplan–Meier survival analysis of OS in patients with PC. High HOXB5 level resulted in decreased survival time in patients with PC; low HOXB5 level was correlated with increased survival time (P < 0.05). (e) PFS in PC showed similar results with OS. (f) HOXB5 was positive in high grades in PC (P < 0.05). (g) HOXB5 has a positive correlation to tumor mutational burden (TMB), (P < 0.05). (h) HOXB5 has a positive correlation to lymph node metastasis (P < 0.05).
HOXB5 drove malignant progression in PC by bioinformatics analysis
According to the TCGA database, TPM was extracted from each RNA-seq for the analysis of the HOXB5 high-expression group as compared with the low-expression group. The GO enrichment results showed that embryonic-development-related pathways were effectively enriched (Figure 2(a)). Using a precise Gene Set Enrichment Analysis, the results showed that multicancer invasiveness signature cluster (P = 0.0028) and embryonic stem cell core cluster (P = 0.0056) were the indicators (Figure 2(b)). GO and GSEA indicated that HOXB5 is involved in embryonic difference and cell plasticity. WGCNA analysis by using MetaScape indicated that between the HOXB5 high and low expression groups, embryonic development associated pathway was enriched; epithelial cell differentiation (Figure 2(c)). The Molecular Complex Detection (MCODE) algorithm has been applied to identify densely connected network components. The MCODE networks identified for individual gene lists were determined, and the three best-scoring MCODE components by P value were retained, as shown in Figure 2(d). The function of MCODE-3 is chromatin assembly, and the function of MCODE-4 is muscle filament sliding. These data indicated that HOXB5 is involve in cell differentiation and stemness and potentially play malignant driving roles in PC progression.
Figure 2.
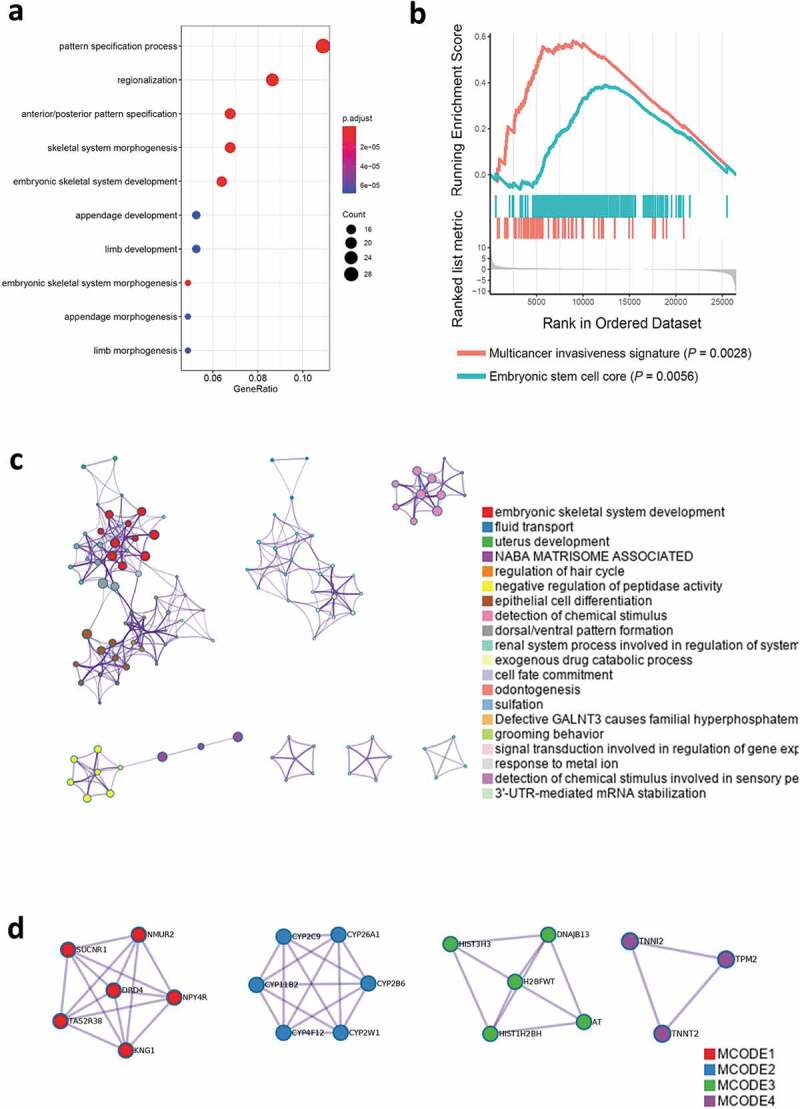
In the bioinformatics analysis, patients with HOXB5 high expression and low expression showed the differences among the spectra. (a) GO analysis showed that the embryo morphogenesis-associated pathways were enriched. (b) GSEA analysis showed that multicancer invasiveness and embryonic stem cells core were enriched. (c) Metascape analysis enriched embryonic-associated pathways. (d) Three best-scoring terms of MCODE components identified in HOXB5 upregulated genes.
HOXB5 increased PC cell malignant progression in vitro
To further validate HOXB5 expression in PC, we performed Western blot analysis to compare the HOXB5 protein expression levels in various PC cell lines. Capan-2 had a low HOXB5 expression level in contrast to PANC-1 (Figure 3(a)). Capan-2 cells were treated with HOXB5 expression plasmid pcDNA3.1-HOXB5. PANC-1 cells were knocked down by shRNA. These cells were detected through Western blot analysis (Figure 3(b)). When HOXB5 expression was up-regulated, the morphology of the capan-2 cells changed from tightly packed colonies to structures with scattered growth (Figure 3(c)). We tested the clone formation, migration, and invasion abilities of capan-2 and PANC-1 after treatment. As shown in Figure 3(d), a significant colony formation ability difference was found between highly and lowly expressed HOXB5 cells. Similarly, in the wound healing assay, significant difference in the speed of wound healing between the transfection and control groups was observed (Figure 3(e)). In the invasion assay result presented in Figure 3(f), cell invasion in HOXB5-transfected capan-2 cell line increased approximately tenfold compared with that in the negative vector control (P < 0.01). We also observed a tentfold decrease in HOXB5-shRNA-transfected PANC-1 cells compared with the negative vector control (P < 0.01). MMPs, which acted as effector molecules, are important in cell plasticity and EMT. According to zymographic assays, the MMP2 and MMP9 activities were significantly higher in HOXB5 up-regulated group in capan-2 compared with the control group. Given that MMP activity in PANC-1 cells decreased after HOXB5 was knocked down (Figure 3(g)). Through this process, tumor cells enhanced their expression and active MMPs through up-regulated HOXB5 expression, which was ultimately important to malignant progression. For EMT markers, high HOXB5 level was observed accompanied by vimentin, Twist1, and down-regulated E-cadherin in capan-2 and PANC-1 cells (Figure 3(h)).
Figure 3.
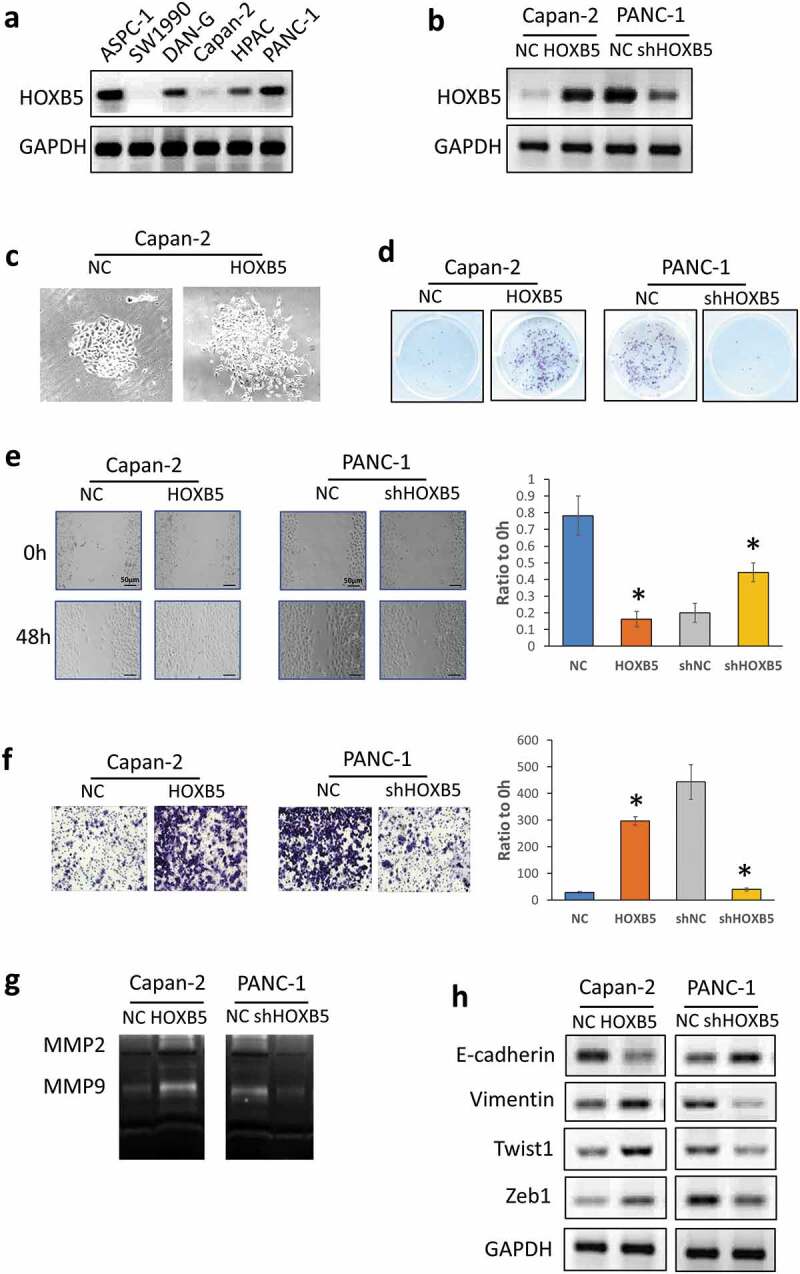
HOXB5 facilitated PC cell proliferation by promoting progression. (a) Protein HOXB5 level in different PC lines. (b) Efficiency of HOXB5 knockdown and overexpression in PANC-1 and capan-2 cells, respectively. (c) HOXB5 clonal morphology in capan-2 and PANC-1 cells. (d) Cell colony formation assay was used to examine the effect of HOXB5 on PC cell proliferation. HOXB5 promoted PC cell proliferation (P < 0.05). (e) Wound healing assay in capan-2 and PANC-1 cells. HOXB5 promoted PC cell migration (P < 0.05). (f) Invasion analysis by Transwell assay in capan-2 and PANC-1 cells. HOXB5 accelerated PC cell invasion (P < 0.05). (g, h). Zymography assays and WB were used to detect invasion and EMT markers expression level after HOXB5 knockdown and overexpression.
HOXB5 promoted miR-6723 by binding to conserved motifs in promoter region
According to ChIP-seq in Cistrome Data (http://dc2.cistrome.org/#/), we referred to Yan J.’s data published in Cell in 2013, which Chip by HOXB5 in LoVo cell, which is a gastrointestinal tract adenocarcinoma cell-line [21]. In this data, the top 10 target genes are shown in Figure 4(a). We also screened the binding motif by HOXB5 for the top target gene, that is, miR-6723, by using Wash U analysis (http://epigenomegateway.wustl.edu/legacy/?genome=hg38& datahub =http://dc2.cistrome.org/api/datahub/42672&coordinate=chr1:532324-732413). Using motif analysis, we defined motifs by ChIP-seq peaks in the miR-6723 promoter region by using the ChIPseeker online software. Two individual motifs and their locations within the miR-6723 promoter were found. Motifs 1 and 2 possessed the relative conserved sequences of Chr1 629791-630040 and Chr1 633904-634132, respectively (Figure 4(b)). Then, we used the up-regulated HOXB5 expression in capan-2 cell and knockdown in PANC-1 cell for the detection miR-6723 expression levels. The results showed that miR-6723 expression was up-regulated following high HOXB5 expression levels (Figure 4(c)). The truncated luciferase report plasmids were used for motif identification, and the miR-6723 motifs were truncated into two regions, one of which covered its peak sequence. The luciferase assay data showed that HOXB5 transcriptionally activated motifs 1 and 2 and enhanced by the 1 + 2 sequences, which indicated that motif 1 may synergize motif 2 through transcriptional activation by HOXB5 regulation (Figure 4(d)). To confirm the transcriptional activation of miR-6723 by HOXB5, we analyzed the correlation between HOXB5 and miR-6723 expression levels in the PC samples in the TCGA database. In TCGA, PAAD data was downloaded. R pack showed that HOXB5 had a positive correlation with miR-6723 (R = 0.12, P = 0.051; Figure 4(e)). We also analyzed the tumor sizes and node numbers, which had significant relationships with miR-6723 level (Figure 4(f,g)). However, clinical stage and grade had insignificant relationship to miR-6723, which indicated that miR-6723 may participate in the complex process in PAAD progression through the downstream target gene.
Figure 4.
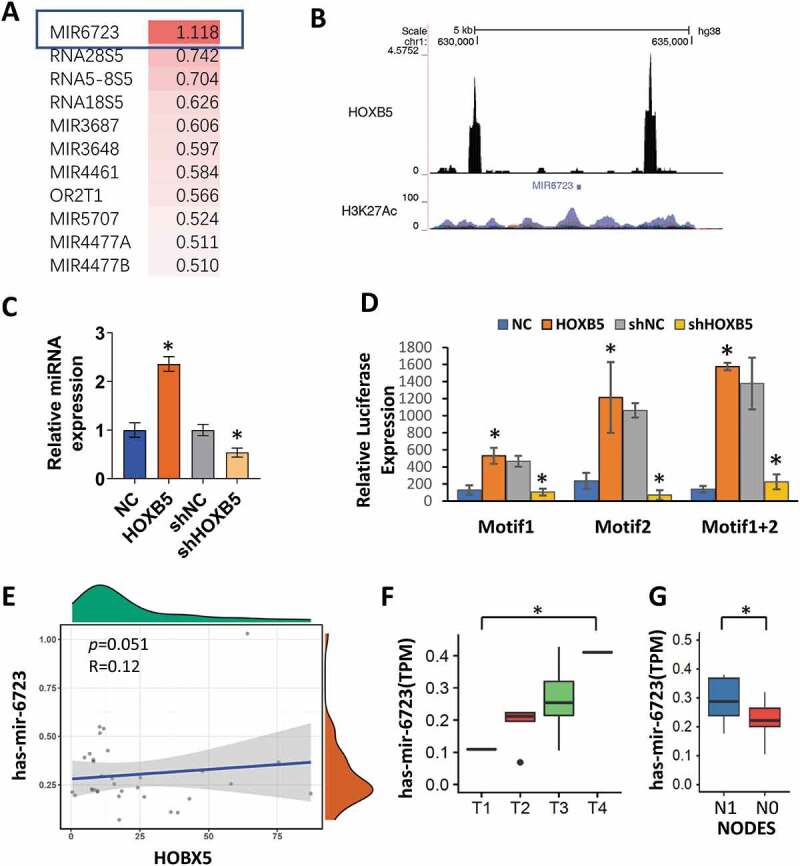
Motifs regulated by HOXB5 in its target miR-6723. (a) Top 10 target analysis by ChIP-seq in Cistrome data (http://dc2.cistrome.org/#/). (b) Motif analysis of the target gene, that is, miR-6723, in the promoter region by using the ChIPseeker. Two individual motifs and their locations within the promoters of miR-6723, Chr1 629791-630040 and Chr1 633904-634132. (c) qRT-PCR was used to detect the level of miR-6723 after HOXB5 knockdown and overexpression; HOXB5 promoted miR-6723 expression (P < 0.05). (d) Luciferase report assay showed that HOXB5 transcriptionally activated motifs 1 and 2 and enhanced by 1 + 2 sequences. (e) TCGA database showed a positive correlation between HOXB5 and miR-6723 expression in patients with PC. (f) HOXB5 had a positive correlation with miR-6723 (R = 0.12, p = 0.051). (g) Tumor sizes and node numbers showed a significant correlation with miR-6723 level (P < 0.05).
HOXB5 relied on miR-6723 to promote PC cell proliferation, migration, invasion, and EMT
RNA-seq GO and GSEA analysis indicated that malignant progression, such as proliferation, migration, invasion, and EMT ability, may be enhanced by HOXB5 in PC. Then, we identified the biological function of HOXB5 in PC cells. We used capan-2 and PANC-1 cells to evaluate the biological function of HOXB5 in PC process. To access the effect of HOXB5/miR-6723 on PC growth, we overexpressed HOXB5 and miR-6723 in capan-2 cells and knocked them down by shRNA in PANC-1 cells. For the recovery experiment, we overexpressed HOXB5 with miR-6723 knockdown by antisense oligonucleotide (ASO) in capan-2 or HOXB5 knockdown by shRNA with overexpressed miR-6723-mimic in PANC-1 cells. The results showed that HOXB5 overexpression or miR-6723-mimic promoted proliferation by colony formation assay in capan-2. Conversely, PANC-1 proliferation was inhibited due to the down-regulation HOXB5 by shRNA and miR-6723 ASO. Treatment with miR-6723 after knockdown of HOXB5 can reverse the effect of HOXB5 knockdown on PANC-1 cells (Figure 5(a)). Metastasis is vital for cancer progression. The effect of HOXB5 on the migration and invasion abilities of PC cells was examined. The wound healing assays showed that HOXB5 and miR-6723-mimic overexpression enhanced cell migration as presented by the narrowing gap (Figure 5(b)). Conversely, HOXB5 and miR-6723-ASO knockdown weakened cell migration as presented by the increased gap. Similar to proliferation data, when neutralizing the effect of HOXB5 by miR-6723-ASO, migration presented an offset. When the effect of shHOXB5 was neutralized by miR-6723-mimic, migration was restored. Transwell assays showed similar effect to wound healing assay, and HOXB5 and miR-6723 promoted invasion in PC cells. HOXB5 was knocked down and inhibited, and miR-6723 weakened the effect (Figure 5(c)). The results indicated that miR-6723 presented its effect on malignance progression dependent upon HOXB5 overexpression, which supported the transcriptional activation relationship between HOXB5 and miR-6723.
Figure 5.
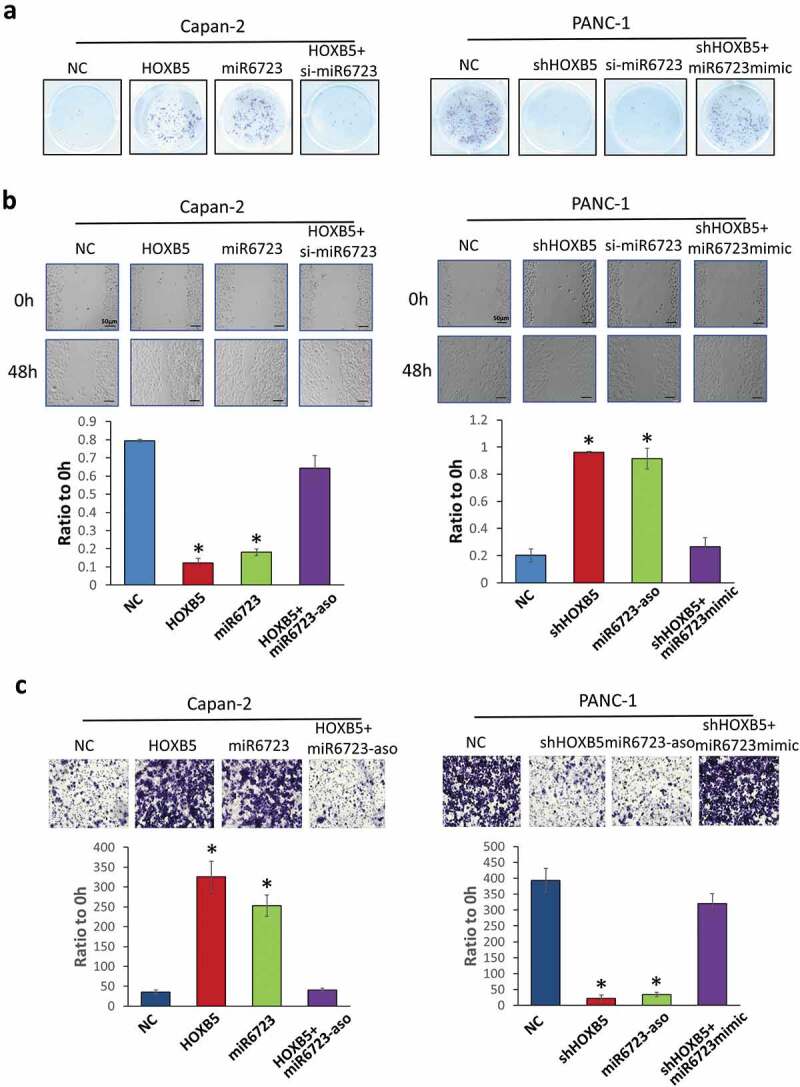
Cell function analysis of the effect of HOXB5/miR-6723 on PC. (a) Colony formation assay detected overexpressed HOXB5, promoted miR-6723 proliferation in Capan-2 cells and knockdown by shRNA suppressive proliferation in PANC-1 cells. Overexpressed HOXB5 with miR-6723 knocked down by antisense oligonucleotide (ASO) in capan-2 and HOXB5 knocked down by shRNA with overexpressed miR-6723-mimic in PANC-1 cells had neutralization effects. B. Wound healing assay. (c) Invasion assay had similar results with (a). HOXB5 transcriptional activation miR-6723 promoted cell progression.
HOXB5 facilitated PC progression in vivo
To further validate the role of HOXB5 in PC growth and metastasis, we established a capan-2 xenograft model by using BALB/c-nu/nu mice. Capan-2 stable transfection clone was obtained by G418 enrichment, which was then injected subcutaneously or in the tail vein of mice. When HOXB5 and miR-6723 were individually overexpression, tumor growth were significantly enhanced. Conversely, when repressed miR-6723, the growth curve of the tumor was weakened (Figure 6(a,b)). The quantification of lung metastasis node numbers showed that HOXB5 and miR-6723 indicated additional colonization in the lungs (Figure 6(c)). Survival analysis revealed that HOXB5 overexpression in PC decreased the survival time, and miR-6723 up-regulation also led to a worse outcome (Figure6(d)). The IHC staining of tumor tissues demonstrated that the invasion and metastasis of EMT markers were up-regulated, HOXB5 and miR-6723 were overexpressed, and HOXB5 had a negative correlation with miR-6723-ASO groups (Figure6(e)). Thus, HOXB5 accelerates PC progression depending upon its target, that is, miR-6723.
Figure 6.
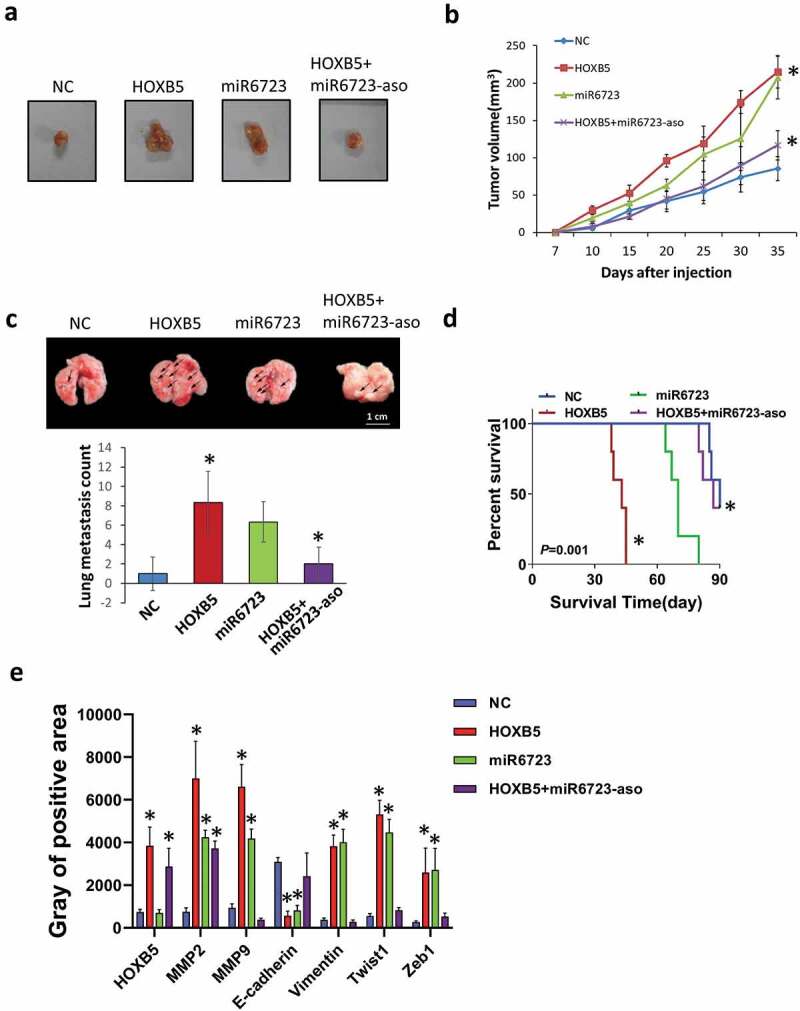
Effect of HOXB5 on PC cell growth and metastasis in vivo. (a) Representative tumor images in capan-2/scramble, capan-2/HOXB5, capan-2/miR-6723, and capan-2/HOXB5 + miR-6723-ASO groups (n = 6 per group). (b) Tumor volume follow days; HOXB5 and miR-6723 overexpression promote tumor growth. Quantification of fluorescence from metastatic tumors (P < 0.01). (c) H&E staining of metastatic tumors in lung tissues and node counts; HOXB5 and miR-6723 promoted lung metastasis. (d) Kaplan–Meier survival analysis of mice in different groups. (e) IHC staining of tumor samples and gray analysis of the positive area for the detection of progression markers for each group.
Discussion
HOXB5 as a member of the homeobox gene family that plays a crucial role in tumor progression, proliferation, and metastasis and angiogenesis [17,22]. Here, we found that HOXB5 was highly expressed in PC tissues and high HOXB5 expression was correlated with poor prognosis of patients with PC. HOXB5 enhanced the growth and metastasis in vitro and in vivo in PC cells. HOXB5 afforded stem-like properties and EMT and activated miR-6723 transcription. Our results suggested that HOXB5 promoted PC cell invasion and EMT and accelerated the malignant progression of PC by up-regulating miR-6723.
HOXB5 is dysregulated in several cancer types and associated with cancer progression. HOXB5 up-regulation has been demonstrated in gastric cancer and correlated with poor prognosis in patients [23]. The deregulation of HOXB5, colorectal, and ovarian cancer expression in breast cancer predicted poor outcomes in patients [17,24]. However, the specific mechanism by which HOXB5 regulates the malignant progression of PC remains unclear. Our study confirmed that HOXB5 was associated with poor PC prognosis. Our results also revealed that HOXB5 enhanced cancer stem-like properties and EMT in PC cells.
HOXB5 promotes long-term hematopoietic stem cell self-renewal and HOXB5 drives the progenitor’s behavior of cancer-associated fibroblast cells [25]. HOXB5 modulates cancer cell invasion and migration [20,26]. A recent study reported that HOXB5 promotes non-small-cell lung cancer migration and invasion, but the mechanism is unclear [16]. EMT plays a fundamental role in PC metastasis, and these cells acquire a mesenchymal and invasive/metastatic phenotype [27,28]. The deregulation of HOXB5 in breast cancer cells induces EMT and promotes migration and invasion [17]. HOXB5 knockdown results in migration and invasion depression with EMT alteration in lung cancer [20]. Our results revealed that HOXB5 had a substantial effect on the EMT phenotypes of PC cells, thereby enhancing PC cell motility and inducing EMT.
Although the role of HOXB5 in PC progression has been verified, the specific mechanism by which HOXB5 regulates PC remains unclear. Our results showed that the transcriptional activity of miR-6723 was up-regulated by HOXB5, thereby indicating that HOXB5 promoted the malignant progression of PC by up-regulating noncoding RNAs. Moreover, the HOXB5/miR-6723 axis was modulated by direct transcriptional regulation and afforded PC cells cancer stem-like and invasion properties in PC cells. The crosstalk between the AKT pathway and miR-6723 was involved in EMT and carcinogenesis via Twist1 and Zeb1 up-regulation. HOXB5 facilitated the metastasis of PC cells and up-regulated Twist1 and Zeb1 expression. This finding suggested that HOXB5 promoted PC cell metastasis by activating PI3K/AKT/Twist1 to promote EMT. In brief, HOXB5 promoted the malignant progression of PC by up-regulating the Twist1/Zeb1 pathway and vimentin, MMP2, and MMP9. Thus, HOXB5 can regulate several signaling pathways, but miR-6723 is the primary target activated by HOXB5 to modulate the malignant progression of PC.
Funding Statement
This study was funded by Special Fund of Sichuan Province People’s Hospital [grant number: 2018ZX01].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Froeling F, Tuveson D.. Pancreatic cancer foiled by a switch of tumour subtype. Nature. 2018;557(7706):500–501. [DOI] [PubMed] [Google Scholar]
- [2].Danai LV, Babic A, Rosenthal MH, et al. Altered exocrine function can drive adipose wasting in early pancreatic cancer. Nature. 2018;558(7711):600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mueller S, Engleitner T, Maresch R, et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature. 2018;554(7690):62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Olive KP, Jacobetz MA, Davidson CJ, et al. Inhibition of hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].van Kampen JG, Marijnissen-van Zanten MAJ, Simmer F, et al. Epigenetic targeting in pancreatic cancer. Cancer Treat Rev. 2014;40(5):656–664. [DOI] [PubMed] [Google Scholar]
- [6].Sato N, Goggins M. The role of epigenetic alterations in pancreatic cancer. J Hepatobiliary Pancreat Surg. 2006;13(4):286–295. [DOI] [PubMed] [Google Scholar]
- [7].Hessmann E, Johnsen SA, Siveke JT, et al. Epigenetic treatment of pancreatic cancer: is there a therapeutic perspective on the horizon?. Gut. 2017;66(1):168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Brabletz T, Kalluri R, Nieto MA, et al. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–134. [DOI] [PubMed] [Google Scholar]
- [9].Aiello NM, Brabletz T, Kang Y, et al. Upholding a role for EMT in pancreatic cancer metastasis. Nature. 2017;547(7661):E7–E8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Derynck R, Weinberg RA. EMT and cancer: more than meets the eye. Dev Cell. 2019;49(3):313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Meng J, Chen S, Han J-X, et al. Twist1 regulates vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 2018;78(15):4150–4162. [DOI] [PubMed] [Google Scholar]
- [13].Martin A, Cano A. Tumorigenesis: Twist1 links EMT to self-renewal. Nat Cell Biol. 2010;12(10):924–925. [DOI] [PubMed] [Google Scholar]
- [14].Krebs AM, Mitschke J, Lasierra Losada M, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19(5):518–529. [DOI] [PubMed] [Google Scholar]
- [15].Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12):1487–1495. [DOI] [PubMed] [Google Scholar]
- [16].Zhang B, Li N, Zhang H. Knockdown of homeobox B5 (HOXB5) inhibits cell proliferation, migration, and invasion in non-small cell lung cancer cells through inactivation of the Wnt/beta-catenin pathway. Oncol Res. 2018;26(1):37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lee JY, Hur H, Yun HJ, et al. HOXB5 promotes the proliferation and invasion of breast cancer cells. Int J Biol Sci. 2015;11(6):701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Luo J, Cai Q, Wang W, et al. A microRNA-7 binding site polymorphism in HOXB5 leads to differential gene expression in bladder cancer. PLoS One. 2012;7(6):e40127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tan X, Jiang L, Wu X, et al. MicroRNA-625 inhibits the progression of nonsmall cell lung cancer by directly targeting HOXB5 and deactivating the Wnt/beta-catenin pathway. Int J Mol Med. 2019;44(1):346–356. [DOI] [PubMed] [Google Scholar]
- [20].Gao X, Zhao H, Diao C, et al. miR-455-3p serves as prognostic factor and regulates the proliferation and migration of non-small cell lung cancer through targeting HOXB5. Biochem Biophys Res Commun. 2018;495(1):1074–1080. [DOI] [PubMed] [Google Scholar]
- [21].Yan J, Enge M, Whitington T, et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154(4):801–813. [DOI] [PubMed] [Google Scholar]
- [22].Lee JY, Kim JM, Jeong DS, et al. Transcriptional activation of EGFR by HOXB5 and its role in breast cancer cell invasion. Biochem Biophys Res Commun. 2018;503(4):2924–2930. [DOI] [PubMed] [Google Scholar]
- [23].Hong CS, Jeong O, Piao Z, et al. HOXB5 induces invasion and migration through direct transcriptional up-regulation of beta-catenin in human gastric carcinoma. Biochem J. 2015;472(3):393–403. [DOI] [PubMed] [Google Scholar]
- [24].Wu Q, Lothe RA, Ahlquist T, et al. DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol Cancer. 2007;6:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chen JY, Miyanishi M, Wang SK, et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature. 2016;530(7589):223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xu H, Zhao H, Yu J. HOXB5 promotes retinoblastoma cell migration and invasion via ERK1/2 pathway-mediated MMPs production. Am J Transl Res. 2018;10(6):1703–1712. [PMC free article] [PubMed] [Google Scholar]
- [27].Zhou P, Li B, Liu F, et al. The epithelial to mesenchymal transition (EMT) and cancer stem cells: implication for treatment resistance in pancreatic cancer. Mol Cancer. 2017;16(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang S, Huang S, Sun YL. Epithelial-mesenchymal transition in pancreatic cancer: a review. Biomed Res Int. 2017;2017:2646148. [DOI] [PMC free article] [PubMed] [Google Scholar]


