ABSTRACT
Implication of autophagy in the downregulation of immune signaling pathways through the degradation of their components constitutes an emerging field of investigation. Our work showed that the selective interaction of Drosophila protein Kenny/IKKγ (CG16910) with the autophagic machinery is required for the degradation of the I-kappa B kinase complex. This regulatory mechanism is essential for the downregulation of the immune deficiency (IMD) pathway in response to commensal microbiota to prevent inflammation.
Autophagy is an evolutionary conserved catabolic process that is required for the recycling and degradation of deleterious or unnecessary cytosolic components such as proteins, lipids or organelles. It also constitutes an immunity effector through the elimination of intracellular pathogens and regulation of some immune signaling pathways.1 The specificity of autophagy relies on the presence of specialized receptor proteins able to bridge cargo to the autophagy machinery; notably through the interaction with autophagosomal membrane protein microtubule-associated protein 1 light chain 3 (MAP1LC3/LC3) family proteins, hereafter referred to as Atg8. The interaction with Atg8 proteins often depends on the presence of a short motif, known as LC3-interacting region (LIR) or Atg8-interacting motif (AIM).2 Most autophagy receptor proteins share a number of common features, including at least one LIR motif and often at least one ubiquitin-binding domain (UBD). There are 20 described families of UBDs which are structurally different and bind non-covalently to ubiquitin molecules and chains with a distinct mode of interaction.3 In Drosophila, the first described selective autophagy receptor Ref(2)P – homolog of mammalian Sequestosome-1 protein (SQSTM1, best known as p62) – is no exception as it possesses both an LIR motif and a UBD of the Ubiquitin-associated domain (UBA) family.4
To identify new selective autophagy in Drosophila, we screened the Drosophila proteome for UBD-containing proteins and screened them for LIR motifs. Only proteins containing at least one relaxed LIR (xLIR) within an intrinsically disordered region were considered.5
One of the hits we identified was the UBAN (ubiquitin binding in ABIN and NEMO) domain-containing protein Kenny/IKKγ (CG16910), homolog of mammalian NEMO (NF-kappa-B essential modulator).6 Together with ird5/IKKβ (Inhibitor of nuclear factor kappa-B kinase subunit beta), Kenny forms the I-kappa B kinase (IKK) complex, essential to the activation of the nuclear factor-kappa B (NF-κB)-like IMD (immune deficiency) signaling pathway in response to Diaminopimelic acid (DAP)-type peptidoglycan, usually carried by Gram-negative bacteria, in Drosophila.7
Using a combination of in vitro, in cellulo and in vivo experiments, we confirmed that Drosophila Kenny interacts with Atg8a in an LIR motif-dependent manner. This interaction is essential for targeting Kenny to the autophagosome and autolysosome for degradation. In its course to the lysosome, Kenny drags ird5 for the degradation of the whole IKK complex. Failure to degrade the IKK complex by autophagy leads to systemic and constitutive deregulation of the IMD immune pathway by the presence of commensal bacteria. Indeed, both Atg8a and Atg7-deficient flies display an upregulation of the expression of the antimicrobial peptide Diptericin that is lost in germ-free and in Kenny-deficient flies. The deregulation of the IMD pathway by commensal bacteria results in systemic inflammation. The Drosophila midgut host a broad spectrum of microbiota, which is controlled by the innate immune system. It was recently published that immune senescence associated with aging causes systemic inflammation in Drosophila, resulting in a depressed activity of the IMD pathway in the midgut and hyperplasia phenotype.8 We wondered whether the inflammation in autophagy-deficient flies also affects their gut integrity. We observed that the lack of autophagy is associated with an elevated number of phospho-histone 3 positive stem cells in the midgut, sign of an increased cell proliferation, a phenotype that has been associated with inflammation. The absence of commensal bacteria reduces the proportion of proliferative stem cells in the midgut. This observation correlates with the observation that diptericin upregulation in autophagy-deficient flies is lost when the flies are raised in germ-free conditions. The lining of the gut constitutes an efficient barrier, but can be damaged by pathogens and excessive inflammation, affecting the digestion. Using a dye to measure the number of deposits, we observed that flies with hyperplasia show an increased retention of ingested food.
Since Kenny is a functional ortholog to mammalian NEMO, we checked if its ability to be degraded by autophagy through direct interaction with member of the Atg8 family was conserved. Unlike Kenny, mammalian NEMO does not contain any xLIR motif and we confirmed that it does not interact with any Atg8 family proteins. Moreover, NEMO expressed in Drosophila is unable to localize to the autophagosome, suggesting it may require additional partners to be targeted for autophagic degradation. Using a deterministic mathematical model, we propose that the co-evolution between host and pathogen led to the loss of the LIR motif in NEMO. A hypothesis which is supported by independent studies showing that mammalian NEMO degradation by autophagy may be triggered by proteins expressed by pathogens. Indeed, the interaction of the murine cytomegalovirus M45 protein with NEMO induces its engulfment by the autophagosome.9 The bacterial effector geldanamycin can also trigger the lysosomal degradation of the IKK complex.10
Our study suggests that selective autophagic degradation of the IKK complex in Drosophila, mediated by the LIR motif-dependent interaction of Kenny with Atg8a, constitutes a negative regulator of the activation of the IMD pathway by commensal bacteria. Lack of autophagy due to mutation or aging causes Kenny to accumulate and aggregate, leading to the activation of NF-κB factor Relish and constitutive expression of antimicrobial peptides, which in turn induces inflammation and damages the intestinal epithelium (Figure 1).6
Figure 1.
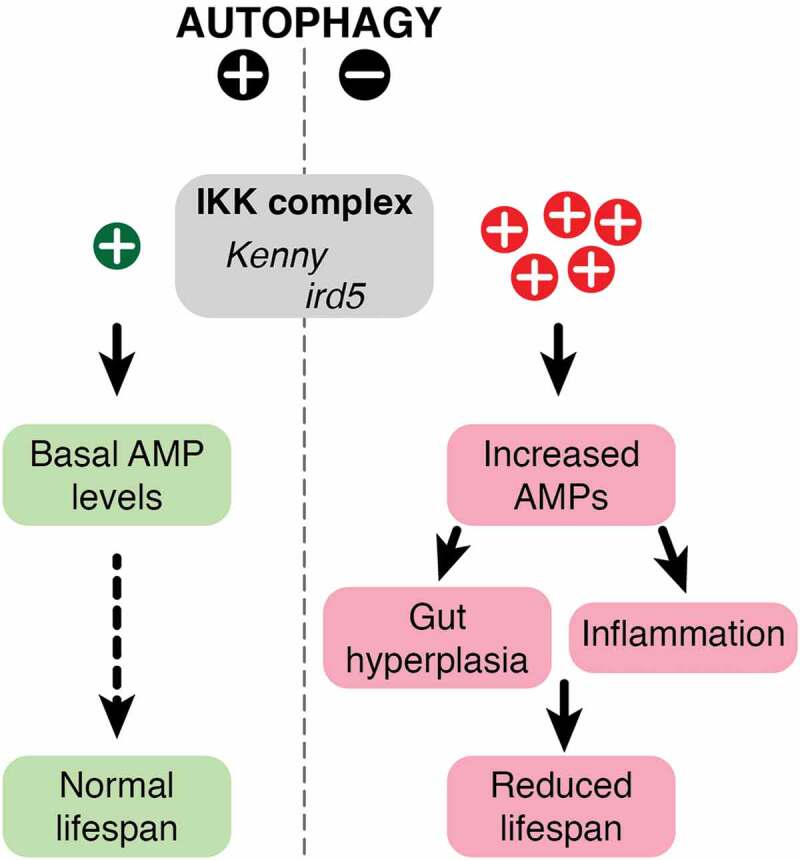
Maintenance of basal level of IKK complex by autophagy. In normal conditions, commensal microbiota trigger the production of AMPs but they are negatively regulated by autophagy and being kept at basal low levels through the selective autophagic degradation of the IKK complex components, Kenny and ird5. In autophagy-deficient conditions, Kenny is accumulated and activates the immune response that causes inflammation and gut hyperplasia and reduces lifespan. (Abbreviations: AMPs, antimicrobial peptides; IKK: I-kappa B kinase complex).
Funding Statement
This work was supported by the BBSRC grants BB/L006324/1 and BB/P007856/1 to IPN.
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the authors.
References
- 1.Gomes LC, Dikic I.. Autophagy in antimicrobial immunity. Molecular Cell. 2014;54:1–3. doi: 10.1016/j.molcel.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 2.Johansen T, Lamark T.. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J Mol Biol. 2019. doi: 10.1016/j.jmb.2019.07.016. [DOI] [PubMed] [Google Scholar]
- 3.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jain A, Rusten TE, Katheder N, Elvenes J, Bruun JA, Sjottem E, Lamark T, Johansen T. p62/Sequestosome-1, Autophagy-related Gene 8, and Autophagy in Drosophila Are Regulated by Nuclear Factor Erythroid 2-related Factor 2 (NRF2), Independent of Transcription Factor TFEB. The Journal of Biological Chemistry. 2015;290:14945–14962. doi: 10.1074/jbc.M115.656116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalvari I, Tsompanis S, Mulakkal NC, Osgood R, Johansen T, Nezis IP, Promponas V. iLIR: A web resource for prediction of Atg8-family interacting proteins. Autophagy. 2014;10:913–925. doi: 10.4161/auto.28260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tusco R, Jacomin AC, Jain A, Penman BS, Larsen KB, Johansen T, Nezis IP. Kenny mediates selective autophagic degradation of the IKK complex to control innate immune responses. Nature Communications. 2017;8:1264. doi: 10.1038/s41467-017-01287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silverman N, Zhou R, Stoven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes & Development. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Zheng X, Zheng Y. Age-associated loss of lamin-B leads to systemic inflammation and gut hyperplasia. Cell. 2014;159:829–843. doi: 10.1016/j.cell.2014.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fliss PM, Jowers TP, Brinkmann MM, Holstermann B, Mack C, Dickinson P, Hohenberg H, Ghazal P, Brune W. Viral mediated redirection of NEMO/IKKgamma to autophagosomes curtails the inflammatory cascade. PLoS Pathog. 2012;8:e1002517. doi: 10.1371/journal.ppat.1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qing G, Yan P, Xiao G. Hsp90 inhibition results in autophagy-mediated proteasome-independent degradation of IkappaB kinase (IKK). Cell Research. 2006;16:895–901. doi: 10.1038/sj.cr.7310109. [DOI] [PubMed] [Google Scholar]


