ABSTRACT
A vast majority of the human genome encodes long non-coding RNAs (lncRNAs) as compared to protein-coding genes (PCGs). But most efforts to determine biomarkers of anticancer drug response have focused entirely on PCGs. Comprehensive investigation of lncRNAs and drug response demonstrates that lncRNAs are indeed crucial biomarkers of drug response.
KEYWORDS: Long non-coding RNA, pharmacogenomics, drug response prediction
A long-standing objective of cancer precision medicine is to select treatments that are tailored based on a tumor’s genetic profile with the goal to maximize the probability of clinical response. This requires careful determination of biomarkers that can predict response to each candidate drug. In order to find such biomarkers, researchers have successfully utilized in vitro models based on cancer cell lines treated with various anticancer agents. For example, the genomics of drug sensitivity in cancer (GDSC)1 and cancer therapeutics response portal (CTRP)2 studies screened nearly a thousand cell lines, along with detailed molecular profiles, to generate a comprehensive pharmacogenomic biomarker profile for hundreds of drugs. However, these large-scale drug screens suffered from a major caveat by focusing solely on protein-coding genes (PCGs). We now know that nearly 70% of the human genome encodes long non-coding RNAs (lncRNAs), as compared to about 2% encoding PCGs.3 With the emergence of lncRNAs as a key regulator of gene expression4 and drivers of malignant transformation,5 we believe it is critical that we investigate their potential contributions as biomarkers in cancer precision medicine.
To fill the gap in our understanding of lncRNAs as potential biomarkers, we performed a systematic analysis of the associations between the somatic lncRNA transcriptome and genome of about a thousand cell lines with detailed pharmacological profiles for hundreds of drugs6 (Figure 1). As lncRNA transcriptomes are notoriously difficult to profile, we first developed and implemented a novel computational tool to impute the lncRNA transcriptomes of the cell lines with missing lncRNA profiles.7 With a comprehensive picture of lncRNA transcriptome available at our disposal, we first evaluated the ability of the lncRNA transcriptome to predict response to all drugs as compared to PCGs. We found that lncRNAs were just as potent as the PCG transcriptome at drug response prediction, suggesting it is worthwhile digging deeper into the data to identify potential individual lncRNAs as biomarkers.
Figure 1.
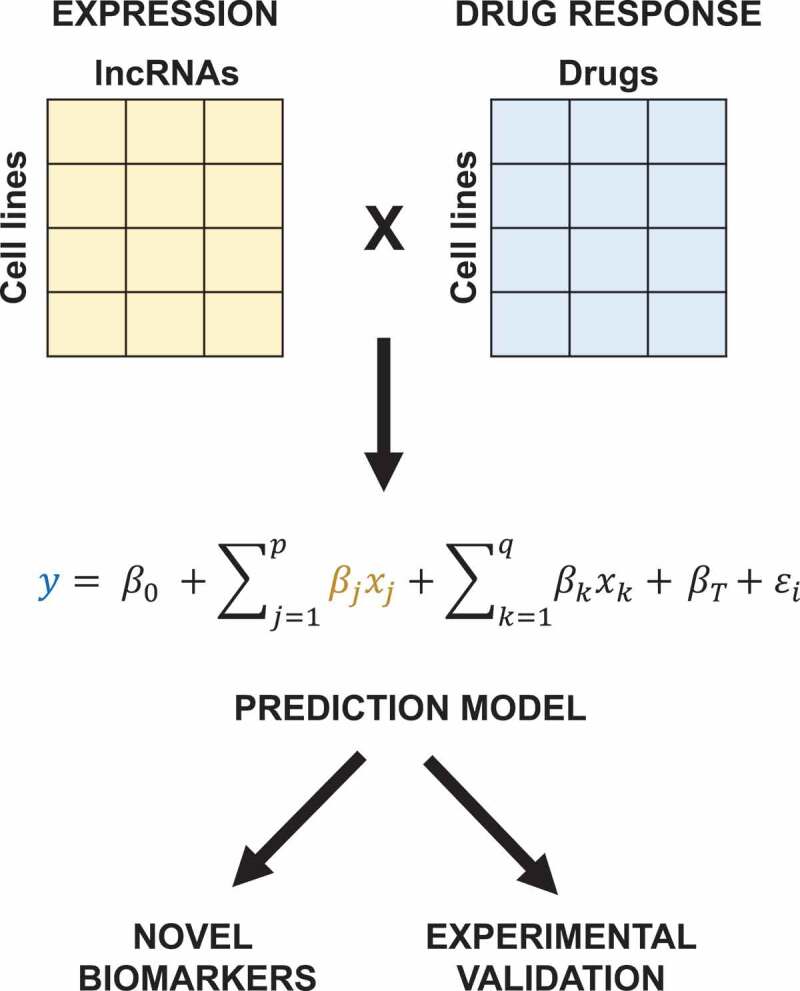
Pipeline to determine novel lncRNA biomarkers of drug response. Using the lncRNA transcriptome and drug response profiles of cell lines, we construct prediction models for each individual drug (y) as a function of lncRNA expression (βjxj) and other potential confounding variables, including cis-protein-coding genes (βkxk) and tissue-type (βT). Intercept and unexplained variance are denoted by β0 and ϵi respectively. Thus, we determined statistically significant novel biomarkers that could be candidates for further experimental validation.
However, identifying the potential of individual lncRNAs as novel biomarkers posed two critical challenges – (1) The close genomic proximity of many lncRNAs to PCGs means any statistical analysis correlating expression of lncRNAs with drug response maybe biased, or at the very least redundant, with neighboring PCGs; and (2) The top candidate lncRNAs biomarkers may not provide any additional predictive power beyond the well-established PCG clinical biomarkers. To address these issues, we carefully modified our predictions models to account for the possible confounding effects of these variables. First, we characterized the bias that may be introduced by the expression of neighboring cis-PCGs (within ±500Kb of the lncRNA), and found that nearly half of all the significant drug-lncRNA associations were in fact redundant from the associations with proximal cis-PCGs. Thus, by adjusting the prediction models for cis-PCGs, we were able to identified novel associations with known, cancer-associated, and uncharacterized lncRNAs. Moreover, these novel lncRNA predictors were located at distinct genomic loci compared to the top PCG biomarkers for most drugs, suggesting a potential association with drug response independent of the PCGs.
We next addressed the utility of these candidate lncRNAs in comparison with well-established clinical biomarkers by adjusting our models for these alterations. For example, we determined that two lncRNAs, EGFR-AS1 (epidermal growth factor receptor anti-sense 1) and MIR205HG (microRNA 205 host gene), could substantially improve upon the prediction of response to erlotinib and gefinitib over EGFR (epidermal growth factor receptor) somatic mutation and amplification status. In other words, our analysis suggested that there may be tumors that may respond to anti-EGFR therapy despite not carrying its established clinical biomarker.8 Using an in vitro model, we confirmed that sensitivity to erlotinib depends on the expression of EGFR-AS1 and MIR205HG in the NCI-H222 and HCC-827 lung cancer cell lines. Finally, we presented a statistical approach to determine lncRNA-specific somatic alterations undergoing positive selection and significantly associated with drug response.
Overall, we found that the lncRNAs generally outperformed established PCG biomarkers at predicting response to most drugs, suggesting a critical role of lncRNAs in cancer precision medicine. While we tested and validated the link between two lncRNAs and anti-EGFR response, our study revealed a plethora of new hypotheses that need to be studied in detail. In addition, future efforts must focus on further characterization of lncRNAs at both the functional level as well as in pre-clinical and clinical models for pharmacogenomic relevance.
Funding Statement
This work was supported by the National Cancer Institute [R01CA204856].
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, Aben N, Gonçalves E, Barthorpe S, Lightfoot H, et al. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166:1–3. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basu A, Bodycombe NE, Cheah JH, Price E, Liu K, Schaefer G, Ebright R, Stewart M, Ito D, Wang S, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattick JS, Rinn JL.. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 5.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 6.Nath A, Lau EYT, Lee AM, Geeleher P, Cho WCS, Huang RS. Discovering long noncoding RNA predictors of anticancer drug sensitivity beyond protein-coding genes. Proc Natl Acad Sci. 2019;201909998. doi: 10.1073/pnas.1909998116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nath A, Geeleher P, Huang RS. Long non-coding RNA transcriptome of uncharacterized samples can be accurately imputed using protein-coding genes. Brief Bioinform. 2019;bby129. doi:10.1093/bib/bby129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gazdar A. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28:S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]


