Abstract
Introduction
Inflammatory response plays an important role in the disease progress or therapeutic effect. In this context, it is highly required to develop a technology to visualize the inflammatory response. In this study, macrophages and their microRNA (miRNA) which are involved in the inflammatory response, were focused while a system of molecular beacon (MB) to detect the miRNA of macrophages was designed and prepared.
Methods
Gelatin nanospheres were prepared by the conventional coacervation method. An antibody with an affinity for the surface receptor of macrophages was immobilized onto the gelatin nanospheres by several methods. A nucleic acid-based MB for a pro-inflammatory miRNA 155–5p was designed and incorporated into the antibody-immobilized gelatin nanospheres (MB-gelatin NS). Macrophages before and after the polarization into pro-inflammatory or anti-inflammatory phenotypes were cultured with the MB-gelatin NS and change in the intracellular fluorescence was observed.
Results
The antibody-immobilized gelatin nanospheres prepared by a coupling between the amino groups of gelatin and the sugar chains of antibody with NaIO4 showed the highest affinity for cellular receptor. MB complexed with the cell-penetrating (CP) peptide was successfully incorporated into the antibody-immobilized gelatin nanospheres. When cultured with pro-inflammatory macrophages, MB-gelatin NS efficiently detected the miRNA 155–5p to emit fluorescence.
Conclusions
By the NaIO4 method, the antibody was immobilized onto gelatin nanospheres with a high affinity remaining while the MB was incorporated into the antibody-immobilized gelatin nanospheres. The MB incorporated allowed mRNA to visualize the pro-inflammatory nature of macrophages.
Keywords: Gelatin nanospheres, Antibody immobilization, Molecular beacon, microRNA, Macrophages, Inflammatory response
Abbreviations: BCA, bicinchoninic acid; BHQ, black hole quencher; BSA, bovine serum albumin; CP, cell-penetrating; DDW, double-distilled water; DLS, dynamic light scattering; DSS, disuccinimidyl suberate; FCS, fetal calf serum; GA, glutaraldehyde; Ig, immunoglobulin; IL, interleukin; KPB, potassium phosphate-buffered; MB, molecular beacon; miRNA, microRNA; PBS, phosphate buffered-saline; qRT-PCR, quantitative real time-polymerase chain reaction; WST-8, 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium
Highlights
-
•
Antibody could be immobilized onto gelatin nanospheres with the affinity remaining.
-
•
MB for a pro-inflammatory miRNA was incorporated into gelatin nanospheres.
-
•
MB incorporated emitted the fluorescence in the pro-inflammatory macrophages.
1. Introduction
Inflammation is a biological response to an injury or damage caused by endogenous or exogenous stimuli, such as a pathogen invasion or disease. Recently, it has been revealed that the inflammatory response affects on the disease progress or therapeutic effect [1,2]. Based on that, there are several researches on the therapeutic approaches by the modulation of inflammatory responses [[2], [3], [4], [5], [6], [7], [8]]. On the other hand, for the sophistication of therapy, it is also highly required to develop the materials and technologies to non-invasively visualize the inflammatory response. In the visualization of inflammatory response, it is indispensable to design the imaging agent considering the cellular and molecular mechanism of inflammation. Several researches have been performed on the visualization of inflammatory cells [[9], [10], [11]] as well as inflammatory cytokine [[12], [13], [14], [15]].
In this study, macrophages were selected as a cellular target for the visualization of inflammatory response. Macrophages involve in all stages of inflammation by changing their phenotypes [16]. After injury, the classically activated (M1) macrophages, infiltrate into the injured site and produce pro-inflammatory substances, such as tissue necrosis factor-α, interleukin (IL)-1β, nitric oxide etc. and phagocyte the cell debris and pathogen. On the other hand, in the late stage of inflammation, the alternatively activated (M2) macrophages terminate the inflammation by producing anti-inflammatory substances, such as IL-10, transforming growth factor, arginase-1 etc. In addition, several types of microRNA (miRNA) have been reported to regulate the biological functions of macrophages [17,18]. The detection of these molecules by imaging agents enables to visualize the biological functions of macrophages. There are several researches on the development of imaging agents to visualize the biological function of macrophages [[19], [20], [21], [22]]. In this study, a miRNA was selected as a target molecule to visualize the biological functions of macrophages. Several methods have been used to detect the miRNA, such as northern blotting [23], microarray [24], and quantitative real time-polymerase chain reaction (qRT-PCR) [25]. However, the cell destruction is required in these methods, which hampers the non-invasive visualization.
Molecular beacon (MB) is a versatile activatable imaging agents to detect the nucleic acid. The MB has a stem-loop structured nucleic acid with the fluorophore and quencher at both the ends and emits the fluorescence only in the presence of complementary nucleic acid chain. The intracellular detection of miRNA can be achieved by MB without the cellular destruction. However, the efficiency of cellular internalization of naked MB is quite low due to the repulsion force between the MB and cell surface both with negative charges. Therefore, it is important to develop a delivery carrier into cells for the intracellular detection of miRNA.
Nanoparticles are promising carriers to incorporate and deliver drugs to the target tissues and cells. Drugs include therapeutic, diagnostic or preventive agents, dyes, and every agent with a biological activity. Based on this idea, there have been several researches on the development of nanoparticle-based carrier systems for the intracellular delivery of MB [[26], [27], [28], [29], [30], [31], [32], [33], [34]]. Targetable nanoparticles by the immobilization of ligands, such as peptides, oligosaccharides, and antibodies, enable drugs to more specifically deliver to the target site. Since the targetability depends on the affinity for receptors expressed on the surface of target cell, it is indispensable to develop the ligand immobilization method with the high affinity remaining.
In this study, antibody-immobilized gelatin nanospheres incorporating MB are attempted to be designed and prepared. Gelatin has been widely used for the preparation of nanoparticles because the biosafety and biocompatibility have been proven through long history of practical applications. Several methods were carried out to chemically immobilize an antibody against CD11b on the gelatin nanospheres in terms of affinity optimization for the receptor expressed on the macrophage. The MB for miRNA 155–5p was selected because its expression level become high in M1 macrophages. We examine the efficacy of the antibody-immobilized gelatin nanospheres incorporating MB to visualize the pro-inflammatory nature of macrophages.
2. Materials and methods
2.1. Materials
Gelatin with an isoelectric point of 5.0 and the weight-averaged molecular weight of 100,000 was kindly supplied by Nitta Gelatin Inc., Osaka, Japan. Acetone, glutaraldehyde (GA), Glycine, and sodium sulfite (Na2SO3) were purchased from Nacalai Tesque Inc., Kyoto, Japan. Sodium periodate (NaIO4), sodium cyanotrihydroborate (NaBH3CN), and 2-aminoethanol were purchased from FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan. Disuccinimidyl suberate (DSS) was obtained from Thermo Fisher Scientific Inc., Rockford, IL. Mouse anti-CD11b/Integrin alpha M (Anti-CD11b) antibody was purchased from R&D Systems, Inc. Minneapolis, MN. Anti-bovine serum albumin (BSA) antibody was obtained from Bethyl Laboratories, Inc. Montgomery, TX. Rat immunoglobulin (Ig) G (r-IgG) as an isotype control for anti-CD11b and anti-BSA antibodies was purchased from Jackson ImmunoResearch Inc. West Grove, PA. Molecular beacon for miRNA 155–5p was synthesized by Eurogentec S.A. Seraing, Belgium. The sequence of MB is 5’-[6-FAM]-CTGGTACCCCTATCACAATTAGCATTAAACCAG-[BHQ-1®]-3’ (BHQ: black hole quencher, underline: stem structure). Cell penetrating (CP) peptide with the sequence of GRRRRRRRRRPPQY was synthesized by Hokkaido System Science Co., Ltd., Hokkaido, Japan. The mimic of miRNA 155–5p (5′-UUAAUGCUAAUUGUGAUAGGGGU-3′) was purchased from QIAGEN K.K., Tokyo, Japan. RAW264 (RCB0535) cells of a mouse macrophage line were provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT/AMED, Japan. The cells were cultured in RPMI 1640 medium (Thermo Fisher Scientific Inc., Rockford, IL) supplemented with 10 vol% fetal calf serum (FCS, GE healthcare Life Sciences Hyclone Laboratories, Logan, UT), penicillin and streptomycin (50 U/ml each, Nacalai Tesque Inc., Kyoto, Japan) at 37 °C in a 5% CO2 –95% air atmospheric condition.
2.2. Preparation of gelatin nanospheres
Gelatin nanospheres were prepared by the conventional coacervation method with slight modifications [35]. Briefly, 10 ml of gelatin aqueous solution (25 mg/ml) was heated up to 40 °C. Next, 30 ml of acetone was added to the solution to form the coacervate. Then, 20 μl of GA was added to chemically crosslink the coacervate, followed by the stirring at 40 °C for 8 h to evaporate the residual acetone. To block the aldehyde groups unreacted, 15 mg of glycine was added. The gelatin nanospheres were collected by the centrifugation of 200,000 g for 30 min at 25 °C and resuspended in double-distilled water (DDW). The centrifugation and resuspension were repeated 3 times. The concentration of gelatin nanospheres was determined by measuring the weight after drying 100 μl of nanospheres suspension. When measured by dynamic light scattering (DLS) using Zetasizer Nano-ZS (Malvern Instruments Ltd., Worcestershire, UK), the size of nanospheres resuspended in 10 mM phosphate buffered-saline (PBS, pH7.4) was 161 ± 54.9 nm with the polydispersity index of 0.103.
2.3. Immobilization of antibody onto gelatin nanospheres
In this study, the antibody was immobilized onto gelatin nanospheres by three methods; one physical adsorption and two chemical immobilization methods using DSS (DSS method) and NaIO4 (NaIO4 method). In the physical adsorption method, 500 μl of gelatin nanospheres (1 mg/ml) and different concentrations of antibody (500 μl) were mixed in the PBS at room temperature for 2 h. Then, the gelatin nanospheres mixed with antibody were collected by the centrifugation of 20,800 g for 30 min at 25 °C and resuspended in DDW. The centrifugation and resuspension were repeated at 3 times to obtain the gelatin nanospheres immobilized with antibody by the physical adsorption method. In the DSS method, 500 μl of gelatin nanospheres (1 mg/ml) were mixed with 500 μl of DSS (0.608 mg/ml dissolved in dimethylsulfoxide) at room temperature for 30 min, followed by washing with PBS. The gelatin nanospheres activated with DSS were mixed with the antibody of different concentrations at room temperature for 2 h. The reaction suspension was washed by the centrifugation and resuspension at 3 times as described above to obtain the gelatin nanospheres immobilized with antibody by the DSS method. In the NaIO4 method [36], 17.2 μl of NaIO4 aqueous solution (0.1 M) was added to 172 μl of antibody solution (11.6 mg/ml) in PBS, followed by the incubation at room temperature for 15 min in the dark condition to proceed the oxidation reaction. After adding 17.2 μl of Na2SO3 (0.2 M) to quench the reaction, the solution was filtered with PD-10 column (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) to desalt and obtain the antibody with the oxidized aldehyde groups in the sugar chain. The gelatin nanospheres (1 mg/ml) were mixed with different concentrations of oxidized antibodies at room temperature for 2 h. To reduce the Schiff base obtained by the reaction of gelatin nanospheres and oxidized sugar chain in the antibody, 5 μl of NaBH3CN (5 M dissolved in 1 M NaOH) was added and incubated at room temperature for 30 min. After that, 25 μl of ethanolamine (1 M) was added at room temperature for 30 min to terminate the production of Schiff base. The reaction suspension was washed by the centrifugation and resuspension at three times as described above to obtain the gelatin nanospheres immobilized with antibody by the NaIO4 method.
The antibody was labeled with radioactive iodine (125I) by the conventional chloramine T method [36]. Briefly, to 50 μl of antibody solution (1 mg/ml) dissolved in 0.5 M potassium phosphate-buffered (KPB) solution (pH7.5) containing 0.5 M NaCl, 5 μl of Na125I (740 MBq/ml in 0.1 M NaOH aqueous solution, PerkinElmer Inc., Waltham, MA) solution and 100 μl of chloramine T (Nacalai Tesque Inc., Kyoto, Japan, 0.2 mg/ml) in 0.5 M KPB solution (pH7.2) containing 0.5 M NaCl were added. After the agitation at room temperature for 2 min, the aqueous solution (100 μl) of sodium pyrosulfite (Nacalai Tesque Inc, Kyoto, Japan, 4 mg/ml) was added to stop the radioiodination. The resulting mixture was passed through a PD-10 column to remove uncoupled, free 125I molecules to obtain an aqueous solution of 125I-labeled antibody. The concentration and radioactivity of 125I-labeled antibody were measured using a bicinchoninic acid (BCA) assay kit (Thermo Fisher Scientific, Rockford, IL) and gamma counter (Auto Well Gamma System ARC-380 CL, Aloka Co., Ltd, Tokyo, Japan), respectively. The amount of antibody immobilized onto gelatin nanospheres by each method was calculated based on the radioactivity of 125I-labeled antibody used for the immobilization.
2.4. Affinity evaluation of antibody immobilized onto gelatin nanospheres
The anti-BSA antibody or its isotype (r-IgG) was immobilized onto gelatin nanospheres by the DSS or NaIO4 method as described above. The BSA labeled with 125I by the conventional chloramine T method was added to the gelatin nanospheres immobilized with the same amount of antibody and the suspension was incubated for 3 h at 37 °C. After removing the 125I-labeled BSA unbound by three times centrifugation of 20,800 g for 10 min at 25 °C and resuspension in DDW, the radioactivity of nanospheres was measured using the gamma counter to calculate the amount of BSA bound to the gelatin nanospheres immobilized with antibody.
RAW 264 cells were seeded on each well of 24 multi-well culture plates (Corning Inc., Kennebunk, ME) at a density of 1 × 105 cells/well. The gelatin nanospheres immobilized with 125I-labeled anti-CD11b antibody or r-IgG by the DSS or NaIO4 method was added to RAW 264 cells and incubated for 2 h. After washing with cold PBS three times, the cells were collected by the trypsinization. The radioactivity of cells collected was measured using the gamma counter to evaluate the amount of antibody bound to the cells.
2.5. Incorporation of MB into antibody-immobilized gelatin nanospheres
MB (1 nmole) was mixed with CP peptide of different concentrations to obtain MB-CP complexes at different N/P ratios, which are defined as molar ratios of amino groups from CP peptide to phosphate groups from MB. To the complex, 100 μg of gelatin nanospheres immobilized with antibody by the NaIO4 method was added and incubated for 1 h at room temperature. The suspension was centrifuged with 20,800 g for 10 min at 25 °C and the supernatant was removed and the fresh PBS was added to obtain the antibody-immobilized gelatin nanospheres incorporating MB (MB-gelatin NS).
The absorbance of supernatant at the wavelength of 260 nm was measured on the spectrophotometer (NanoDrop 2000c, Thermo Fisher Scientific, Rockford, IL) to calculate the amount of MB incorporated into the antibody-immobilized gelatin nanospheres.
2.6. Sensitivity of MB incorporated into antibody-immobilized gelatin nanospheres
MB (20 nM) with or without incorporation into antibody-immobilized gelatin nanospheres was mixed with different concentrations of miRNA. After the incubation for 1 h at room temperature, the fluorescent intensity was measured by Multi-mode Microplate Reader (SpectraMax i3x, Molecular Devices Japan Co., Ltd., Tokyo, Japan) where the wavelengths of excitation and emission were 480 and 530 nm, respectively.
2.7. Cell viability of antibody-immobilized gelatin nanospheres
RAW 264 cells were seeded on each well of 96 multi-well culture plates (Corning Inc., Kennebunk, ME) at a density of 1 × 104 cells/well. After the incubation for 24 h at 37 °C, various concentrations of MB-gelatin NS, obtained from MB–CP complex at the N/P ratio of 5, were added to RAW 264 cells cultured. After further incubation for 24 h at 37 °C, 10 μl of 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium (WST-8, Nacalai Tesque. Inc., Kyoto, Japan) was added. After 4 h, the absorbance at the wavelength of 450 nm was measured by Multi-mode Microplate Reader. The viability of cells was expressed 100% for cells without addition of MB-gelatin NS.
2.8. miRNA expression for RAW264 cells stimulated with pro-inflammatory or anti-inflammatory agents
RAW 264 cells were seeded on each well of 6 multi-well culture plates (Corning Inc., Kennebunk, ME) at a density of 2 × 105 cells/well. After the incubation for 24 h at 37 °C, the cells were cultured for 24 h with the medium containing lipopolysaccharide (Sigma–Aldrich, Co., St. Louis, MO., 100 ng/ml) and interferon-γ (Pepro Tech, Inc., Rocky Hill, NJ., 20 ng/ml) or IL-4 (Pepro Tech, Inc., Rocky Hill, NJ., 20 ng/ml) to polarize the RAW 264 cells (M0) into pro-inflammatory (M1) or anti-inflammatory (M2) macrophages.
The total RNA was extracted from macrophages of different polarities by using mirVana™ miRNA Isolation Kit (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer's instructions. The reverse transcription was carried out using TaqMan® MicroRNA RT Kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The RT-qPCR was performed on 7500 Real Time PCR System (Applied Biosystems, Foster City, CA) using the solution containing TaqMan® Micro RNA Assay (Applied Biosystems, Foster City, CA) for miRNA 155–5p (ID: 001093) or RNU6B (ID: 464539), complementary DNA obtained above, TaqMan® Universal PCR Master Mix (Applied Biosystems, Foster City, CA), and Nuclease-free water. The reaction solution was incubated for the enzyme activation at 95 °C for 10 min, followed by 40 PCR cycles with 2 steps to denature at 95 °C for 15 s and anneal at 60 °C for 60 s. The level of miRNA 155–5p expression was normalized by that of RNU6B as an internal control.
2.9. Fluorescent detection of miRNA in RAW 264 by the MB-gelatin NS
RAW 264 cells were seeded on the glass bottom dish (Matsunami Glass Ind., Ltd., Osaka, Japan) at a density of 5 × 104 cells/dish. After the incubation for 24 h at 37 °C, the cells were stimulated as described above. After the stimulation for 24 h to obtain each cell population (M0, M1 or M2 macrophages), the medium was changed to the serum-free medium, followed by the culture for 3 h with the MB-gelatin NS (30 μg/ml) obtained from the addition of MB–CP complex at the N/P ratio of 5. Then, the medium was changed to the growth medium and the cells were incubated further for 24 h. After the fixation of cells with 4% paraformaldehyde, the cell nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific, Rockford, IL) and the fluorescent images were taken by the fluorescent microscopy (BZ-X700, KEYENCE Co., Ltd., Osaka, Japan). On the other hand, the cells cultured with the MB-gelatin NS were collected by the trypsinization. In each cell population (M0, M1 or M2 macrophages), the percentage of MB-derived fluorescent cells in all of cells collected by the trypsinization was independently analyzed and calculated by FACSCanto II flow cytometer through the BD FACSDiva software (BD Biosciences Ltd., Rockville, MD).
2.10. Statistical analysis
Data were analyzed using Tukey–Kramer paired comparison test, while the significance was accepted at p < 0.05. They were expressed as the means ± standard deviations.
3. Results
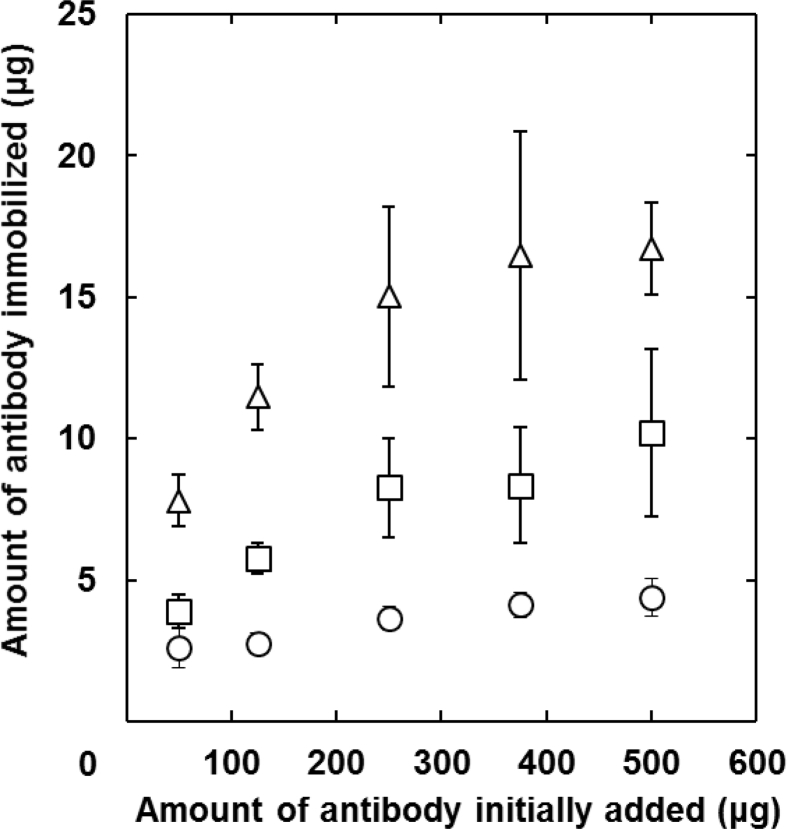
Fig. 1 shows the amount of antibody immobilized onto gelatin nanospheres by various methods. In every immobilization method, the amount of antibody immobilized onto gelatin nanospheres increased with an increase in the amount of antibody initially added. The amount depended on the type of immobilization method and it became smaller in the order of physical adsorption, NaIO4, and DSS methods. The gelatin nanospheres with antibody immobilized by NaIO4 and DSS methods were used in the following experiments. Because the amount of antibody physically immobilized onto gelatin nanospheres was too low to technically compare the antibody affinity.
Fig. 1.
The amount of antibody immobilized onto gelatin nanospheres as a function of antibody addition amount. The antibody is physically adsorbed (○) or chemically immobilized onto the gelatin nanospheres by the DSS (△) and NaIO4 methods (□).
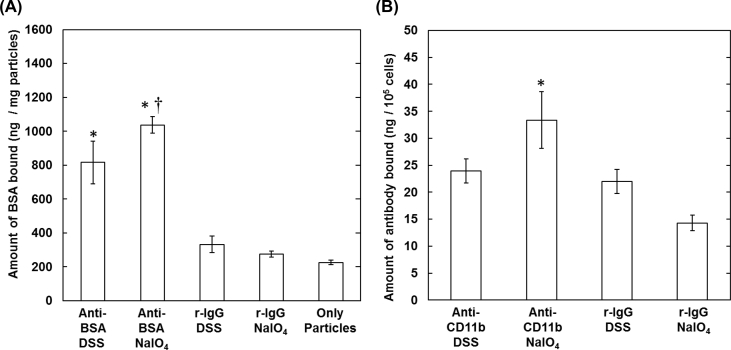
The affinity of antibody immobilized onto gelatin nanospheres was evaluated by two methods. First, the effect of antibody immobilization method on the extent of antigen molecule bound to antibody was investigated (Fig. 2A). In this experiment, BSA was used as an antigen and the antibody targeted to BSA or the isotype (r-IgG) was used for the immobilization onto gelatin nanospheres. The amount of BSA bound onto the gelatin nanospheres immobilized with the isotype antibody by the DSS and NaIO4 methods was low and similar to that of gelatin nanospheres without antibody immobilization. On the other hand, the amount of BSA bound onto the gelatin nanospheres immobilized with anti-BSA antibody by the NaIO4 method was significantly higher than that by the DSS method. Next, the affinity of antibody immobilized onto gelatin nanospheres was evaluated by the culture of macrophages, RAW 264 cells with the CD11b expressed on the surface (Fig. 2B). The binding amount to RAW 264 cells incubated with anti-CD11b antibody immobilized onto gelatin nanospheres by the NaIO4 method was significantly higher than that of both the isotype antibody-immobilized and the anti-CD11b antibody-immobilized nanospheres by the DSS method.
Fig. 2.
(A) The amount of BSA bound onto the gelatin nanospheres immobilized with antibody by the DSS and NaIO4 methods. The antibody used for the immobilization is the anti-BSA antibody or its isotype (r-IgG). The antibody is chemically immobilized onto the gelatin nanospheres by the DSS and NaIO4 methods. *, p < 0.05; significant against the nanospheres with or without the r-IgG immobilized. †, p < 0.05; significant against the nanospheres with the anti-BSA antibody immobilized by the DSS method (B) The amount of antibody on gelatin nanospheres bound to RAW264 cells. The antibody used for the immobilization is the anti-CD11b antibody or its isotype (r-IgG). The antibody is chemically immobilized onto the gelatin nanospheres by the DSS and NaIO4 methods. *, p < 0.05; significant against other groups.
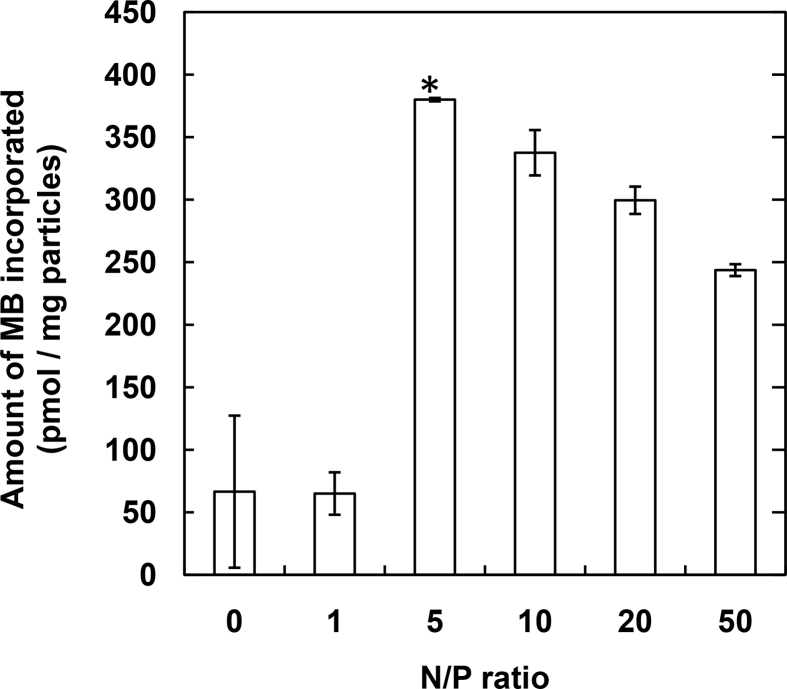
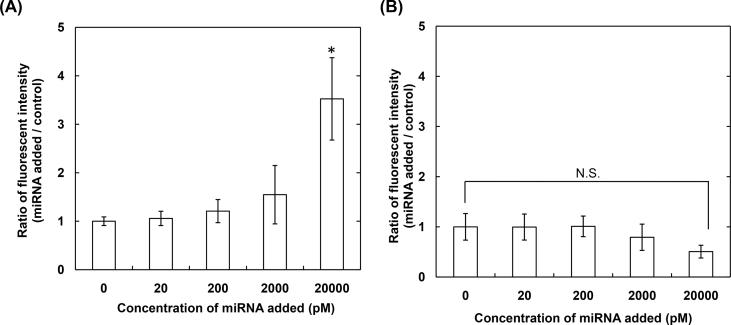
Fig. 3 shows the amount of MB incorporated into gelatin nanospheres immobilized with antibody by the NaIO4 method as a function of N/P ratios in the MB–CP complex. The significantly high amount of MB was observed when complexed with CP peptide at the N/P ratio of 5. The relative fluorescent intensity of MB after the addition of miRNA with different concentrations was evaluated (Fig. 4). The fluorescence of naked MB increased with an increase in the concentration of target miRNA added. On the contrary, the fluorescent of MB incorporated into gelatin nanospheres immobilized with antibody by the NaIO4 method was not changed, irrespective of the miRNA concentration.
Fig. 3.
The amount of MB incorporated into gelatin nanospheres immobilized with antibody by the NaIO4 method. The MB is complexed with CP peptide at various N/P ratios. *, p < 0.05; significant against other groups.
Fig. 4.
The relative fluorescent intensity of MB after the addition of miRNA with different concentrations. MB is used as the naked form (A) or the incorporated form (MB-gelatin NS, B). The MB-gelatin NS was obtained by the incorporation of MB complexed with CP peptide at the N/P ratio of 5 into gelatin nanospheres immobilized with antibody by the NaIO4 method. *, p < 0.05; significant against other concentrations.
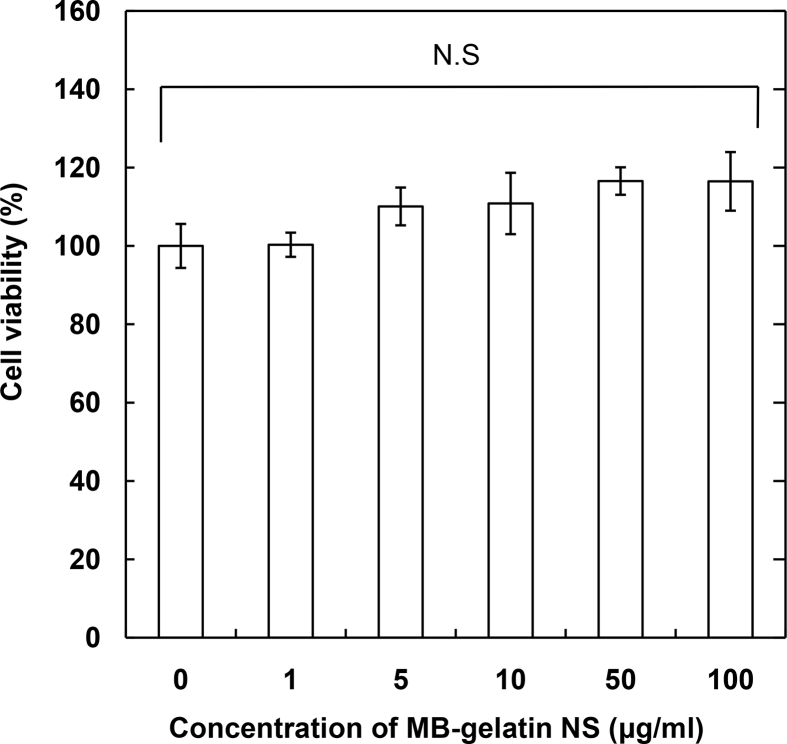
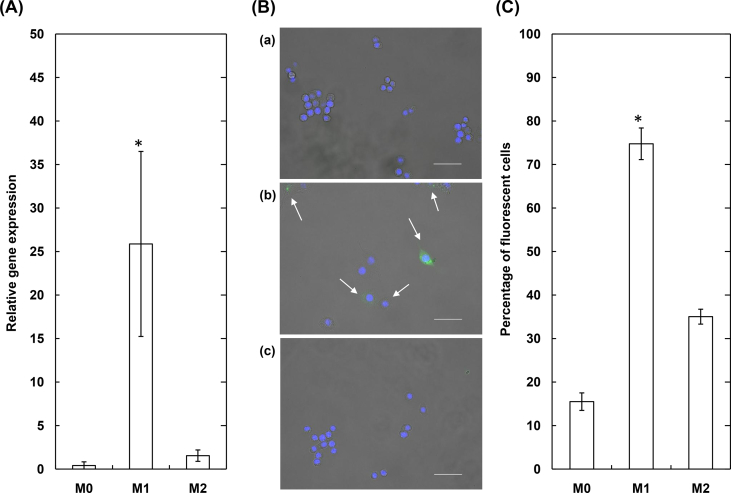
To investigate the feasibility of MB incorporated into gelatin nanospheres immobilized with antibody by the NaIO4 method (MB-gelatin NS) for the cell culture, first the relative viability of RAW 264 cells was evaluated at different concentrations of MB-gelatin NS (Fig. 5). No significant change in the cell viability was observed for every concentration of MB-gelatin NS up to 100 μg/ml. After the confirmation of significantly enhanced expression of miRNA 155–5p for RAW264 cells stimulated with the pro-inflammatory (M1) agent (Fig. 6A), the fluorescent behavior was investigated for RAW 264 cells cultured with miRNA 155–5p targeted MB-gelatin NS. In the fluorescent microscopy, the fluorescence emitted by MB was observed only for RAW264 cells stimulated with the M1 agent (Fig. 6B). When counted by the FACS, the percentage of MB-derived fluorescent cells after stimulation with the M1 agent was significantly higher than that before (M0) or after stimulation with the anti-inflammatory (M2) agent (Fig. 6C).
Fig. 5.
The relative viability of RAW 264 cultured with MB-gelatin NS of different concentrations for 24 h. The MB-gelatin NS was obtained by the incorporation of MB complexed with CP peptide at the N/P ratio of 5 into gelatin nanospheres immobilized with antibody by the NaIO4 method. The viability of RAW264 without MB-gelatin NS is 100%.
Fig. 6.
(A) The relative miRNA 155–5p expression of RAW264 before (M0) and after the stimulation with pro-inflammatory (M1) or anti-inflammatory agents (M2) (B) The representative fluorescent images of RAW264 before (a) and after the stimulation with pro-inflammatory (b) or anti-inflammatory agents (c). The RAW264 were cultured for 3 h with MB-gelatin NS (30 μg/ml) 24 h after the stimulation. The nucleus are stained in blue, while the MB interacted with miRNA 155–5p emits in green. The arrows indicate the cells which emit the fluorescence derived from MB. The size of scale bar is 50 μm (C) Percentage of MB-derived fluorescent cells which are not stimulated (M0) or stimulated with pro-inflammatory (M1) or anti-inflammatory agents (M2). The RAW264 were cultured for 3 h with MB-gelatin NS (30 μg/ml) 24 h after the stimulation. *, p < 0.05; significant against other groups.
4. Discussion
The present study demonstrates that a pro-inflammatory nature of macrophages could be visualized by the miRNA 155–5p targeted MB-gelatin NS. The antibody immobilization onto the gelatin nanospheres allowed MB to specifically internalize into macrophages and the MB internalized detected miRNA 155–5p. To our knowledge, this is the first report to visualize the pro-inflammatory nature of macrophages by the intracellular delivery of MB with nanoparticles.
Although the MB is a powerful nucleic acid to detect miRNA and mRNA, an intracellular delivery system is required for the live imaging. Various types of delivery procedures have been proposed so far [37], which include micro- or nano-injection, electroporation, liposome, and nanoparticles.
Gelatin nanospheres with the size of around 160 nm were used in this study. It was found from the preliminary experiment that the size of nanospheres increased with an increase in the gelatin concentration initially used for the coacervation formation (data not shown). In terms of productivity, much amount of gelatin nanospheres can be obtained from the gelatin aqueous solution with high concentration. On the other hand, it is well known that the internalization efficiency of nanoparticles depends on their sizes. Considering the balance of productivity and internalization efficiency, the gelatin nanospheres with the size of around 160 nm were selected in this study.
The amount of antibody immobilized onto the gelatin nanospheres was changed by the immobilization method (Fig. 1). The low amount by the physical adsorption method may be due to both the lack of adsorption site in the gelatin nanosphere and the interaction strength between the antibody and gelatin chains. Difference of amount between the DSS method and NaIO4 method would be ascribed to the number of functional groups contributing to the immobilization. It is likely that the number of amino groups in antibody used by the DSS method is higher than that of aldehyde group from the sugar chain by the NaIO4 method [36]. As a result, in the same amount of antibody initially added, the amount of antibody immobilized by the DSS method was higher than that by the NaIO4 method. The affinity of antibody immobilized onto gelatin nanospheres was evaluated in terms of binding to the antigen molecule (Fig. 2A) and cellular receptor (Fig. 2B). It is well known that the antigen is recognized by the epitope region of antibody. In the DSS method, the amino groups present even in the epitope region are used for the immobilization. In other words, it is possible that the DSS immobilization causes the hindrance of antibody recognition to the antigen. In contrast, in the NaIO4 method, the sugar chains present in the Fc portion, not in the epitope region, are used to react with gelatin amino groups. It was demonstrated from both evaluations that the highest amounts of antigen molecule and cellular receptor were observed for the gelatin nanospheres immobilized with antibody by the NaIO4 method. It is highly conceivable that the NaIO4 method enabled antibody to immobilize in the appropriate direction for the recognition, resulting in an increased binding amount of antigen molecule or cellular receptor. These results agree with other researches where the conjugation by the NaIO4 method enabled antibody to maintain the affinity for antigen [38,39].
The increased amount of MB incorporated into gelatin nanospheres by the complexation with CP peptide (Fig. 3) is due to the increased interaction strength of MB with gelatin chains. The reason why the amount slightly decreased with an increase in the amount of CP peptide for the complexation may be that the excess CP peptide which is not contributed to the complexation, hinders the incorporation of MB complex into the gelatin nanospheres. No fluorescence emission of MB incorporated into gelatin nanospheres was observed even in the presence of target miRNA (Fig. 4B). This is because the miRNA cannot interact with MB incorporated into gelatin nanospheres. In other words, this result demonstrated the successful incorporation of MB into the gelatin nanospheres.
The intracellular fluorescence was observed only for the pro-inflammatory macrophages cultured with the MB-gelatin NS (Fig. 6). The gelatin nanospheres immobilized with antibody by the NaIO4 method enabled to bind the receptor of macropahges (Fig. 2B). In addition, the MB was successfully incorporated into the gelatin nanospheres immobilized with antibody by the NaIO4 method (Fig. 4B). It has been demonstrated that the MB incorporated is released from gelatin nanospheres with time according to their degradation extents [34]. In the present study, it is possible that MB internalized via receptor-mediated endocytosis was released in the form of complex with CP peptide. The complexation with CP peptide may contribute to the easy transport from the endosome to the cytosol [40]. Taken together, it is conceivable that the MB-gelatin NS was internalized into macrophages and the MB released from the nanospheres in the cytosol was interacted with the miRNA 155–5p, resulting in the strong emission of fluorescence through the structural change of MB.
There are several issues to be solved for the improvement of inflammation imaging by the nanoparticle-based intracellular delivery of MB. First, the MB is a versatile activatable probe, that is, MB for every miRNA or mRNA can be theoretically designed and synthesized. In addition, nanoparticles enable to simultaneously encapsulate multiple types of agents. Taken together, more reliable evaluation of inflammation based on the biological functions of macrophages will be achieved by the nanoparticle incorporating multiple types of MB to detect multiple inflammation-related miRNAs. Next, the biological function is changed from time to time. The long-term visualization is highly required to detect the variable biological function. Intracellular controlled release is one of the most feasible methodologies and technologies to meet the requirement. Nanoparticles prepared from biodegradable polymers, such as gelatin and poly(lactic-co-glycolic acid) etc., are carriers for the intracellular controlled release. Actually, there are several researches on the nanoparticle-based intracellular controlled release of MB for the long-term detection of messenger RNA [34,41]. Therefore, the combination with the intracellular controlled release technology will enable MB to continuously detect the miRNA, resulting in the long-term visualization of biological function in macrophages.
5. Conclusions
The antibody was immobilized onto gelatin nanospheres with the high affinity remaining by the NaIO4 method while the MB was incorporated into antibody-immobilized gelatin nanospheres. The visualization of pro-inflammatory nature in macrophages through the detection of miRNA was achieved by the antibody-immobilized gelatin nanospheres incorporating MB.
Acknowledgements
This work was partly supported by Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Young Scientists (A) (JP17H04736).
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
References
- 1.Fullerton J.N., Gilroy D.W. Resolution of inflammation: a new therapeutic frontier. Nat Rev Drug Discov. 2016;15(8):551–567. doi: 10.1038/nrd.2016.39. [DOI] [PubMed] [Google Scholar]
- 2.Crupi A., Costa A., Tarnok A., Melzer S., Teodori L. Inflammation in tissue engineering: the Janus between engraftment and rejection. Eur J Immunol. 2015;45(12):3222–3236. doi: 10.1002/eji.201545818. [DOI] [PubMed] [Google Scholar]
- 3.Kim Y.H., Tabata Y. Enhancement of wound closure by modifying dual release patterns of stromal-derived cell factor-1 and a macrophage recruitment agent from gelatin hydrogels. J Tissue Eng Regenerat Med. 2017;11(11):2999–3013. doi: 10.1002/term.2202. [DOI] [PubMed] [Google Scholar]
- 4.Ritter B., Greten F.R. Modulating inflammation for cancer therapy. J Exp Med. 2019;216(6):1234–1243. doi: 10.1084/jem.20181739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung L., Maestas D.R., Jr., Housseau F., Elisseeff J.H. Key players in the immune response to biomaterial scaffolds for regenerative medicine. Adv Drug Deliv Rev. 2017;114:184–192. doi: 10.1016/j.addr.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Julier Z., Park A.J., Briquez P.S., Martino M.M. Promoting tissue regeneration by modulating the immune system. Acta Biomater. 2017;53:13–28. doi: 10.1016/j.actbio.2017.01.056. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez M.M., Liu J.C., Trujillo-de Santiago G., Cha B.H., Vishwakarma A., Ghaemmaghami A.M. Delivery strategies to control inflammatory response: modulating M1-M2 polarization in tissue engineering applications. J Control Release. 2016;240:349–363. doi: 10.1016/j.jconrel.2016.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagberg H., Mallard C., Ferriero D.M., Vannucci S.J., Levison S.W., Vexler Z.S. The role of inflammation in perinatal brain injury. Nat Rev Neurol. 2015;11(4):192–208. doi: 10.1038/nrneurol.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahrens E.T., Bulte J.W. Tracking immune cells in vivo using magnetic resonance imaging. Nat Rev Immunol. 2013;13(10):755–763. doi: 10.1038/nri3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malviya G., Galli F., Sonni I., Signore A. Imaging T-lymphocytes in inflammatory diseases: a nuclear medicine approach. Q J Nucl Med Mol Imaging. 2014;58(3):237–257. [PubMed] [Google Scholar]
- 11.Shirai T., Kohara H., Tabata Y. Inflammation imaging by silica nanoparticles with antibodies orientedly immobilized. J Drug Target. 2012;20(6):535–543. doi: 10.3109/1061186X.2012.693500. [DOI] [PubMed] [Google Scholar]
- 12.Tarkin J.M., Joshi F.R., Rudd J.H. PET imaging of inflammation in atherosclerosis. Nat Rev Cardiol. 2014;11(8):443–457. doi: 10.1038/nrcardio.2014.80. [DOI] [PubMed] [Google Scholar]
- 13.Schillaci O., Scimeca M., Trivigno D., Chiaravalloti A., Facchetti S., Anemona L. Prostate cancer and inflammation: a new molecular imaging challenge in the era of personalized medicine. Nucl Med Biol. 2019;68–69:66–79. doi: 10.1016/j.nucmedbio.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Lee H.J., Ehlerding E.B., Cai W. Antibody-based tracers for PET/SPECT imaging of chronic inflammatory diseases. Chembiochem. 2019;20(4):422–436. doi: 10.1002/cbic.201800429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anzola L.K., Galli F., Dierckx R.A. SPECT radiopharmaceuticals for imaging chronic inflammatory diseases in the last decade. Q J Nucl Med Mol Imaging. 2015;59(2):197–213. [PubMed] [Google Scholar]
- 16.Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graff J.W., Dickson A.M., Clay G., McCaffrey A.P., Wilson M.E. Identifying functional microRNAs in macrophages with polarized phenotypes. J Biol Chem. 2012;287(26):21816–21825. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Essandoh K., Li Y., Huo J., Fan G.C. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock. 2016;46(2):122–131. doi: 10.1097/SHK.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weissleder R., Nahrendorf M., Pittet M.J. Imaging macrophages with nanoparticles. Nat Mater. 2014;13(2):125–138. doi: 10.1038/nmat3780. [DOI] [PubMed] [Google Scholar]
- 20.Yang R., Sarkar S., Yong V.W., Dunn J.F. In vivo MR imaging of tumor-associated macrophages: the next frontier in cancer imaging. Magn Reson Insights. 2018;11 doi: 10.1177/1178623X18771974. 1178623X18771974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiemy W.F., Heeringa P., Kamps J., van der Laken C.J., Slart R., Brouwer E. Positron emission tomography (PET) and single photon emission computed tomography (SPECT) imaging of macrophages in large vessel vasculitis: current status and future prospects. Autoimmun Rev. 2018;17(7):715–726. doi: 10.1016/j.autrev.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Foss C.A., Sanchez-Bautista J., Jain S.K. Imaging macrophage-associated inflammation. Semin Nucl Med. 2018;48(3):242–245. doi: 10.1053/j.semnuclmed.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pall G.S., Hamilton A.J. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3(6):1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 24.Duan D., Zheng K.X., Shen Y., Cao R., Jiang L., Lu Z. Label-free high-throughput microRNA expression profiling from total RNA. Nucleic Acids Res. 2011;39(22):e154. doi: 10.1093/nar/gkr774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arefian E., Kiani J., Soleimani M., Shariati S.A., Aghaee-Bakhtiari S.H., Atashi A. Analysis of microRNA signatures using size-coded ligation-mediated PCR. Nucleic Acids Res. 2011;39(12):e80. doi: 10.1093/nar/gkr214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J.K., Choi K.J., Lee M., Jo M.H., Kim S. Molecular imaging of a cancer-targeting theragnostics probe using a nucleolin aptamer- and microRNA-221 molecular beacon-conjugated nanoparticle. Biomaterials. 2012;33(1):207–217. doi: 10.1016/j.biomaterials.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 27.Pan W., Yang H., Zhang T., Li Y., Li N., Tang B. Dual-targeted nanocarrier based on cell surface receptor and intracellular mRNA: an effective strategy for cancer cell imaging and therapy. Anal Chem. 2013;85(14):6930–6935. doi: 10.1021/ac401405n. [DOI] [PubMed] [Google Scholar]
- 28.Li H., Mu Y., Lu J., Wei W., Wan Y., Liu S. Target-cell-specific fluorescence silica nanoprobes for imaging and theranostics of cancer cells. Anal Chem. 2014;86(7):3602–3609. doi: 10.1021/ac500173d. [DOI] [PubMed] [Google Scholar]
- 29.Li H., Mu Y., Qian S., Lu J., Wan Y., Fu G. Synthesis of fluorescent dye-doped silica nanoparticles for target-cell-specific delivery and intracellular microRNA imaging. Analyst. 2015;140(2):567–573. doi: 10.1039/c4an01706d. [DOI] [PubMed] [Google Scholar]
- 30.Zhou W., Li D., Xiong C., Yuan R., Xiang Y. Multicolor-encoded reconfigurable DNA nanostructures enable multiplexed sensing of intracellular MicroRNAs in living cells. ACS Appl Mater Interfaces. 2016;8(21):13303–13308. doi: 10.1021/acsami.6b03165. [DOI] [PubMed] [Google Scholar]
- 31.Adinolfi B., Pellegrino M., Giannetti A., Tombelli S., Trono C., Sotgiu G. Molecular beacon-decorated polymethylmethacrylate core-shell fluorescent nanoparticles for the detection of survivin mRNA in human cancer cells. Biosens Bioelectron. 2017;88:15–24. doi: 10.1016/j.bios.2016.05.102. [DOI] [PubMed] [Google Scholar]
- 32.Adinolfi B., Pellegrino M., Tombelli S., Trono C., Giannetti A., Domenici C. Polymeric nanoparticles promote endocytosis of a survivin molecular beacon: localization and fate of nanoparticles and beacon in human A549 cells. Life Sci. 2018;215:106–112. doi: 10.1016/j.lfs.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Murata Y., Jo J.I., Tabata Y. Preparation of cationized gelatin nanospheres incorporating molecular beacon to visualize cell apoptosis. Sci Rep. 2018;8(1):14839. doi: 10.1038/s41598-018-33231-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murata Y., Jo J.I., Tabata Y. Intracellular controlled release of molecular beacon prolongs the time period of mRNA visualization. Tissue Eng A. 2019;25(21–22):1527–1537. doi: 10.1089/ten.TEA.2019.0017. [DOI] [PubMed] [Google Scholar]
- 35.Coester C.J., Langer K., van Briesen H., Kreuter J. Gelatin nanoparticles by two step desolvation--a new preparation method, surface modifications and cell uptake. J Microencapsul. 2000;17(2):187–193. doi: 10.1080/026520400288427. [DOI] [PubMed] [Google Scholar]
- 36.Hermanson G.T. Academic Press; London, UK: 2008. Bioconjugate techniques. [Google Scholar]
- 37.Kuang T., Chang L., Peng X., Hu X., Gallego-Perez D. Molecular beacon nano-sensors for probing living cancer cells. Trends Biotechnol. 2017;35(4):347–359. doi: 10.1016/j.tibtech.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Tresca J.P., Ricoux R., Pontet M., Engler R. Comparative activity of peroxidase-antibody conjugates with periodate and glutaraldehyde coupling according to an enzyme immunoassay. Ann Biol Clin. 1995;53(4):227–231. [PubMed] [Google Scholar]
- 39.Zhou Q., Stefano J.E., Manning C., Kyazike J., Chen B., Gianolio D.A. Site-specific antibody-drug conjugation through glycoengineering. Bioconjug Chem. 2014;25(3):510–520. doi: 10.1021/bc400505q. [DOI] [PubMed] [Google Scholar]
- 40.Najjar K., Erazo-Oliveras A., Mosior J.W., Whitlock M.J., Rostane I., Cinclair J.M. Unlocking endosomal entrapment with supercharged Arginine-rich peptides. Bioconjug Chem. 2017;28(12):2932–2941. doi: 10.1021/acs.bioconjchem.7b00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiraja C., Yeo D.C., Chong M.S., Xu C. Nanosensors for continuous and noninvasive monitoring of mesenchymal stem cell osteogenic differentiation. Small. 2016;12(10):1342–1350. doi: 10.1002/smll.201502047. [DOI] [PubMed] [Google Scholar]