Abstract
Synthetic biologists are exploiting biofilms as an effective mechanism for producing various outputs. Metabolic optimization has become commonplace as a method of maximizing system output. In addition to production pathways, the biofilm itself contributes to the efficacy of production. The purpose of this review is to highlight opportunities that might be leveraged to further enhance production in preexisting biofilm production systems. These opportunities may be used with previously established production systems as a method of improving system efficiency further. This may be accomplished through the reduction in the cost of establishing and maintaining biofilms, and maintenance of the enhancement of product yield per unit of time, per unit of area, or per unit of required input.
Keywords: Synthetic biology, Tunable, Biofilm engineering
1.1. Introduction
1.1.1. What are biofilms?
Biofilms are three-dimensional structures of various bacteria that adhere to biotic or abiotic surfaces. Generally, biofilms are founded by single cells or small groups of cells that then divide and differentiate into complex communities [1] with extracellular matrices [2,3] water channels [3] embedded extracellular proteins [4], extracellular lipids [5] and embedded extracellular nucleic acids [6]. Many biofilms also include humic [7,8] and uronic [8,9] acids. Biofilms contribute to bacterial fitness by increasing adherence to various surfaces, protection from predation, desiccation, immune attack, antibiotics, and protection from starvation via carbon storage [10]. Biofilms can contribute to pathogenesis and environmental survival of bacteria [11,12]. Biofilms also can have significantly different structural elements; while Pseudomonas aeruginosa biofilms classically have tall mushroom-like structures, and well-defined water-channels, other bacteria such as Francisella tularensis can have relatively flat and undifferentiated biofilms [13,14]. Being able to regulate the production of biofilm through exogenously added components such as small molecules or anti-biofilm peptides is a topic of significant research in the field [13,15].
Currently, extracellular proteins, carbohydrates and nucleic acids are considered the principal components of biofilm. When these central extracellular components are enzymatically degraded biofilms size can be reduced considerably [[16], [17], [18]]. The substances surrounding the cells in a biofilm are often referred to as extracellular polymeric substances (EPS). While EPS includes all of the extracellular lipids, carbohydrates, protein, and acids associated within the biofilm [19], the majority of current EPS research focuses on the carbohydrate components as they are believed to generally constitute a majority of biofilm biomass [5,[20], [21], [22]]. The actual percent biomass contribution is likely dependent upon the nature of the biofilm being studied. The identity of each sugar component, the mechanism of linkage and the order in which the sugars are joined are highly variable across different species/conditions. These carbohydrate chains contribute considerably to the incredible diversity of biofilms found throughout nature.
These complex biofilm structures are associated with disease states [[23], [24], [25]], biocorrosion [26] and biofouling [27,28]. They are also associated with food production [29] and the maintenance of human health [30]. On a more functional level, biofilms have very different properties than planktonic cells such as increased resistance to antibiotics [31], antiseptics [32], disinfectants [33], protists [34], phages [34], shear force [35], heat [36], desiccation [37] and UV [38,39] as well as additional properties. While not every studied biofilm has each of the above qualities relative to planktonic form, biofilm has nonetheless been established as a unique state. Recently, multiple transcriptomics studies highlighting the difference between biofilm and planktonic cultures have been performed [[40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50]]. These studies have largely enabled synthetic biologists to understand what exactly constitutes a biofilm and how each of these constituents might be artificially manipulated to optimize the use of biofilms as a production unit.
1.1.2. What is synthetic biology?
Synthetic Biology has been defined by a variety of scientists and engineers from diverse fields [[51], [52], [53], [54]]. We have chosen to use the definition of “the design of life for useful purposes”. Those useful functions can generally be split into one of two categories: life designed in order to study life itself or life designed in order to produce a useful output. These are not necessarily exclusive categories as the latter generally involves pieces of the former in the design-build-test cycle. Important progress has been made in synthetic biology from the single-part standardization [55,56] and optimization [57,58], to the construction of modules/logic gates [[59], [60], [61], [62]], to the engineering of whole systems [[63], [64], [65], [66], [67]]. On the applied science side of synthetic biology, whole systems are being developed to effectively produce specific outputs [[68], [69], [70], [71]]. Since biofilms have additional properties compared to planktonic cells, synthetic biologists are interested in leveraging some of these properties to enhance preexisting biofilm production systems [[72], [73], [74], [75]].
1.1.3. Why monoculture?
Following the traditional reductionist approach, bacterial organisms are generally studied in monoculture. This has the major advantage of reducing the complexity of systems. While these monoculture experiments may not reflect the real world, they do allow understanding of basic principles that can then be applied and modified in a more complex system. Aristotle aptly pointed out a flaw in this approach “The whole is greater than the sum of its parts”. Some of the more complex phenomena of interspecies interaction in a mixed culture will not exist at all in a monoculture system [76], just as some additional properties may be observed in a single type of organism grown as a biofilm as opposed to planktonic cells.
In synthetic biology it is well known that different chassis (organisms) are suited to different production needs. The most important factors to consider are going to be highly dependent upon the users end goal. For protein production, folding and secretion abilities might take precedence in design, whereas for secondary metabolite production, resistance to the produced product may be a primary concern. In addition to inherent biological limitations, factors such as ease of genomic manipulation and culturing may be considered as additional contributors to experimental costs.
1.1.4. Why not monoculture?
While multispecies biofilms are more complicated to create and control, these biofilms have the potential for novel properties unavailable in mono-species biofilms [38] and may have the potential to streamline the production/detection of various products [74]. Multispecies biofilms may be metabolically more efficient as they separate different aspects of a process into unique compartments [77,78]. Mixed biofilms may also be better able to survive environmental challenges [[79], [80], [81], [82], [83]] which may be necessary for synthetic biofilms. Complimentary metabolic waste products might further enhance biofilm maintenance/growth [84]. Different species also have different abilities to retain/manufacture various chemical compounds such as iron containing magnetosomes [85] or various polyhydroxyalkanoates [86] each of which may be useful in a production scenario. Alternatively, additional species allow modular tuning opportunities such as quorum sensing pathways only present in some of the members of the biofilm [87]. Another reason for using mixed species biofilms is the potential for inclusion of difficult-to-culture microorganisms [[88], [89], [90]]. A multispecies biofilm may also be useful to culture vulnerable cells such as L-form cells [91] which may possess desirable qualities for production/secretion [[92], [93], [94]], but lack the ability to outcompete wild type cells [94]. Multispecies biofilms may also have potential to include chassis that normally do not from sizeable biofilms [95]. The addition of novel chassis to synthetic biology is welcome as each new organism contains a set of novel parts ready to be manipulated. Finally, multispecies biofilms have the potential to form novel complex three-dimensional structures, which may affect the system in a number of ways. Herein we will discuss the altering of biofilm architecture by synthetic biologists as potential mechanism for optimizing the output of biofilm.
1.1.5. Genetic manipulation of biofilm architecture
The current manipulation of biofilm genetics is done by editing the genome of planktonic bacteria, and allowing these cells to form a biofilm. There are methods of blocking the transcription of certain genes in biofilms directly [[96], [97], [98], [99]] or indirectly through the manipulation of quorum sensing [[100], [101], [102]]. These studies generally aim to eliminate or prevent biofilm completely. The ideal tool for genetic manipulation of transcription levels would allow for the fine tuning of a user specified gene. While there are a few promising new conjugation-based methods of multi-species biofilm genome editing [103,104], they have yet to mature into modular tools. For now, the best approach is editing the genome of individual planktonic species and letting those cells establish a biofilm.
To leverage a biofilms’ architecture to further enhance the efficiency of production, different aspects of biofilm architecture could be examined: the distribution quantity and relative function of the basic macromolecules, the distribution of cells across the biofilm as well as their spatial relationship to other species and the overall three-dimensional characteristics of the biofilm.
1.2. Macromolecules where and why
1.2.1. Protein
Embedded proteins are known to constitute a significant amount of biomass to biofilms [105]. These proteins are known to play roles in attachment of biofilm to both biotic and abiotic surfaces. New evidence points to the majority of proteins found in biofilm to not be secreted proteins or protein derived from lysed cells, but typically outer membrane proteins that may have been transported to the biofilm by outer membrane vesicles [106]. In contrast to embedded proteins, many of the secreted proteins bind specific biotic matrices such as fibronectin [107,108], chitin [[109], [110], [111]] or mannose [112,113] to mediate attachment. More general strategies for attachment involve the expression of pili [112,114,115], flagella [1,116] and the more recently uncovered fimbriae [[117], [118], [119], [120], [121]]. Both unique and generalist attachment strategies offer unique engineering opportunities. For example, a rhamnose-inducible pili would yield a greater concentration of biofilm near the rhamnose source. This serves two unique functions. First, this presents a relatively inexpensive way of structuring multispecies biofilms. Second, this serves as a potential find-the- leak type system, where biofilms preferentially form at higher concentrations of inducer identifying not just the presence but the sources of a contaminant. On the flip side, attachment can be directly selected against with the production of various biosurfactants, which can enhance motility and repress biofilm formation. These biosurfactants may be programmed to be species-specific and cell-surface attached [122] or they may be secreted [123].
A subtype of fimbria are called curli [124]. As curli form en-masse and overlap they are known as amyloid fibers [125]. Amyloid fibers are commonly found in biofilms [126]. These projections from the cell have been used for a variety of disparate functions. Some interested in nanoscale engineering have been using the fibers as nucleation points for metal deposit [127], essentially building a functional nano-wire with the reasonably electro-stable curli at the core. While this is an achievement a bit removed from biofilm, the subsequent achievement of tunable curli length [128] is of immediate value in more environments beyond tightly controlled laboratory setups. Interestingly, some bacterial species have the ability to elongate each other's amyloid fibers [129] presenting yet another avenue of potential regulation. As amyloid fibers are known to provide structural stability to biofilms [130], these tunable amyloid fibers can be harnessed to dictate how difficult it is to remove an artificial biofilm via physical force. Others have discovered that amyloid fibers have the potential to bind and retain certain chemical compounds [131]. As amyloid fibers with engineered binding sites are developed [132], a greater number of substrates may be captured. There are also now a greater number of proteins with useful functions which may be displayed in biofilms, such as proteins that fend off grazing protists [133] or proteins that entice cellular aggregation [134]. Biofilm can also act as a reservoir which holds potential for inducible release. This has utility beyond general biofilm dispersal, as rebuilding a biofilm is resource-intensive so a minimal loss of biomass associated with product release is often desirable. As designer proteases [[135], [136], [137], [138], [139], [140], [141]] become more common and dependable it is conceivable that a net full of amyloid fibers might be sheared off by a user by adding specific protease and thus releasing concentrated cargo within. This type of approach may be useful in setups where collection is costly.
Often, but not always, cells in a biofilm will also form an S-layer. The S-layer is closer to the cell membrane than amyloid fibers. The S-layer is found in most archaea and some bacteria; it is a layer of protein adorning the cell beyond any cell wall that may be present [142]. S-layer engineering has contributed immensely to the efficiency of many production systems. These engineered proteinaceous alterations tend to act as scaffolds for specific enzymatic reactions. S-layers are also known to help shield bacteria from adverse environmental conditions [143] as well as influence attachment [144]. S-layers can also be utilized as a sort of sieve by the cell [142] preventing larger molecules from approaching the peptidoglycan layer. A potential problem with S-layer and amyloid fibers is the potential for them to get clogged with compounds [145]. This leads to three overlapping downstream problems: diffusion to and from the cell of various materials may be hampered, the concentrated compound may inhibit local cells’ ordinary function and the compound may be blocked from reaching a final (beneficial to the system) location.
1.2.2. Lipids
Outer membrane vesicles (OMVs), essentially lipid bilayers spheres that bleb off of cells, are one of the cellular products that may have altered formation/diffusion patterns in systems with extensive S-layer/amyloid fiber engineering. While these OMVs are known to be important contributors to biofilm protein and biomass overall, their role in contributing biofilm lipids is unclear. It is known that the cells which form biofilms alter their lipid membranes relative to planktonic cells [146]. A study pointed to the enrichment of lipids in cell membranes that provided more structure and less membrane fluidity [147]. While some studies have demonstrated that knocking out [148] or effectively reducing fatty acids [149] prevents full biofilm formation, these studies must be interpreted carefully as changes in lipid membranes inherently alter the viability of all cells. As lipidomics is a field largely in its infancy these early results prevent synthetic biologists from effectively editing the lipid content of biofilms beyond the simple increase in lipid production for harvesting.
1.2.3. Nucleic acids
Extracellular DNA (eDNA) is DNA that is found in the biofilm matrix (EPS), outside of the cellular membrane. The tools and methods to isolate, manipulate, interpret and build nucleic acids are much farther along than their lipid counterparts. Originally, it was thought that the nucleic acids associated with biofilms were just remnants of lysed cells [150], but it became clear that in addition to lysis [151], many of these molecules were also released within OMVs [[152], [153], [154], [155], [156]]. The degree to which the outer membrane vesicle release of DNA and the quorum sensing regulated release of DNA (via lysis or OMV) are related is not yet clear [[157], [158], [159]]. OMVs are also known to be responsible for the release of many intracellular proteins. This highlights the ability for protein and/or eDNA release to be potentially tuned at a high level by regulating outer membrane vesicle release [160]. This may be useful as a method to lower signal-to-noise ratios in regards to extracellular molecule monitoring in biofilms. If this tunable circuit of outer membrane vesicle release is restricted to a single species this also allows for opportunities for ratioed protein contribution adjustment to biofilm.
eDNA has recently been shown to be important for the formation of biofilm [6,155,161,162]. This may be via a combination of enhanced attachment [161] and added structural stability [162]. It appears that eDNA is mostly randomly fragmented genomic DNA [163]; no studies have yet to investigate non B-form DNA structures (Z, G-quad, etc.) in relation to biofilm stability or attachment. These future studies might provide additional modular ways to use eDNA in a biofilm shaping manner, especially when combined with the idea that DNA fragment length (something that is easily artificially controlled) may be used to tune attachment. An experiment with Listeria monocytogenes showed that shorter pieces of DNA prevented attachment and that if added prior to long pieces of DNA, these short pieces could prevent subsequent long DNA-assisted biofilm attachment [161]. In mixed species biofilms, the release of eDNA from biofilm is more complex, as different organism have different propensities for releasing eDNA [163,164] and those propensities may be changed based on the other species in the biofilm [163,165]. The same study showed that both eDNA and peptidoglycan were codependent for attachment, further illustrating the opportunities for cooperation and involvement of eDNA in biofilm formation. Studies have shown that the lattice or the mechanism of cross-linking of eDNA contributes to the strength and resilience of the resulting biofilm [166].
eDNA presents unique control opportunities for biofilm attachment and structural stability. Its release might be modified in a species specific manner via specific protein mechanisms [156] or phage-based lysis [162] or quorum-sensing based OMV release [158] or antibiotic-based OMV release [152] that may be applied to alter mechanical biofilms properties. eDNA can also play additional roles in biofilm beyond basic establishment and maintenance.
eDNA can be taken up by living cells via a process known as competence. Sometimes this can just be a useful background mechanism for cells to survive during stationary phase using the DNA of previously lysed cells as a carbon source in order to survive longer [167]. Competence is also known to contribute to the effective transfer of genes and plasmids between bacteria [[168], [169], [170]]. This has been demonstrated to be a method of antibiotic resistance marker transfer. A distinct mechanism of antibiotic resistance caused by eDNA has also been uncovered in which neighboring cells detect eDNA with two component systems and upregulate various genes in response [171]. eDNA can also lead to antibiotic resistance via a structural mechanism. As DNA (as well as most bacterial membranes) has a negative charge, a layer of eDNA outside of biofilm cells can help shield biofilm cells from aminoglycosides and cationic antimicrobial peptides which are positively charged [172]. Releasing eDNA in a temporal manner might be ideal for generating a biofilm which can be made more resistant to positively charged molecule attack upon demand. This may be ideal if the user does not know exactly which cationic antimicrobials the biofilm will need to resist. As the release of DNA has tunability via quorum sensing regulation or user-controlled lysis, any of these mechanisms of antibiotic resistance might be considered.
eDNA also presents a novel opportunity to the synthetic biologist, one that is largely absent from other fields of engineering. Once the machine gets destroyed and the integrated circuit chip gets broken in half, that circuit is dead. Barring elaborate unprecedented artificial surgical reconstruction, that physical circuit chip will never contribute to a circuit again. In synthetic biology, even if your machine (biofilm) gets destroyed there is a reasonable chance parts of its code (eDNA) might be integrated into a whole new system. This is an interesting opportunity that has yet to be explored.
Much as eDNA can help physically stabilize biofilm, exopolysaccharides are also known to have this effect from both antibiotic based destabilization [173] and immune system based attack [[174], [175], [176]]. Constitutive antibiotic resistance engineered into biofilm may also be useful as a mechanism to thwart competing biofilms from getting established. An often overlooked facet of biofilm is that some cells that facultatively replicate intracellularly might form biofilm in an effort to be more efficiently recognized and phagocytosed [177]. Thus, it may be important to form a larger biofilm if the end goal is to insert your organism into another.
This point of view offers interesting potential to the engineer. Now by using biofilm as a sort of entry vector (which is modifiable in a myriad of ways), organisms that may be phagocytosed present a new opportunity to edit other chassis that may be more resistant to genetic engineering This may provide an interesting backdoor to genetically engineering unculturable protists [178,179] as a prerequisite to genetic engineering is traditionally the ability to culture the recipient organism.
1.2.4. Carbohydrates
Along with antibacterial agents, various exopolysaccharide agents can also trap micronutrients [180,181] which is essential for biofilms grown under flow conditions. Extracellular carbohydrates in EPS are also known for their ability to enhance attachment and therefore enhance biofilm formation [[182], [183], [184]]. Optimizing nutrient intake in biofilms grown in flow systems is a balancing act as ideally the biofilm will capture what it was designed to capture yet will not inhibit the flow of fluid (which more exopolysaccharide will contribute to).
Many complex polysaccharides are produced within biofilms in the EPS. Exopolysaccharide production in biofilms can be manipulated at the transcriptional level with repressors [185], at the post transcriptional level via withheld precursors which can therefore not be operated on by downstream enzymes [186] or via degradation of previously secreted sugars [16]. Manipulation of these processes can be utilized for synthesis of desired molecules such as the production of Glucosamine from Chitin [187]. Many of the carbohydrate-control-loci (in addition to expression of proteins and of eDNA) are influenced by intercellular small-molecule messages called quorum signals [188], meaning that quorum signals often regulate sugar production.
1.3. Interspecies biofilm relationships
Quorum signals are chemical species that get produced by prokaryotes and some eukaryotes when the population has amassed to a certain density. These signals are generally sensed by two component systems, where a membrane bound receptor binds the extracellular signal. This receptor then interacts with a response regulator which alters transcriptional patterns. While quorum signals could potentially regulate any transcriptional levels, the majority of established research has focused on the effects of quorum sensing on antibiotic resistance, virulence, and biofilm formation [189].
Quorum sensing logic gates provide fantastic opportunities for modular tunable control. Building artificial quorum sensing systems as a means for regulating biofilm continues to be the most prevalent way of genetically manipulating artificial biofilms. For a full review on artificial quorum systems see Hennig et al. [190]. The utility of quorum sensing systems is largely based on two facets, modularity and tunability. Quorum systems are ubiquitous in nature. Often distantly related organisms can communicate through the same molecule. While the actual production of certain molecules may prove difficult in certain chassis, the expression of a selected two-component system is usually a much easier feat. The great variety of quorum signals even within the same families offers researchers potential to tune response by using a more or less potent signal as small structural changes often have sizeable effects [191]. In addition, promoter sequences may be altered to change affinity of the sequence to the response regulator. This may prove to be a more effective, simpler mechanism of fine tuning an organism's response. The relationship between promoter sequence and the binding of response regulators was highlighted by He and Wang [192] where they demonstrated how the binding efficiency of PhoP, a response regulator found in Mycobacterium tuberculosis, changed with small changes to the promoter sequence.
Beyond creating or editing a quorum system to control one's own biofilm, signal production also has the potential to prevent competing biofilms. This is true for two reasons. First, not all quorum signals that get produced are necessarily responded to by the producer which was shown with Pseudomonas aeruginosa [193] and second, the same quorum signal may have the opposite effect on different species [194]. This strategy of fending of rivaling biofilms is problematic as the efficacy of any secreted signal against other cells would be entirely dependent upon their identity. Thus, this approach has little value beyond a system where the main competitor is known. Engineering quorum system can be problematic because of system noise and lack of true orthogonality with various quorum systems influencing each other in spaghetti code fashion [[195], [196], [197], [198]]. Other physical parameters also alter the efficacy of using quorum signals to control a system. These can be broken into two categories. The first being factors that prevent the signal from reaching its destination such as being bound by off target species or being removed by current. The second being factors that destroy the signal abiotically by either physical/chemical methods biotically via quorum quenching enzymes such as AHL-lactonase [199]. While some of these issues like fluid velocity may be beyond control in certain circumstances, macromolecular distribution of the biofilm can usually be manipulated via any number of mechanisms discussed herein.
The spatial orientation of bacteria relative to one another within the biofilm is important to consider when optimizing a biofilm for production. This is key for both inputs like external quorum signal application and nutrients as well as outputs such as metabolic waste and product secretion. The packing of bacteria in relation to one another can be controlled by specific protein interactions. For example Pseudomonas and Staphylococcus will comingle in biofilms if Pseudomonas expresses mucA and RpoN [83]. RpoN is a gene important involved in flagella and pili production. Interestingly, when pilH, a negative regulator of pilin, is knocked out of Pseudomonas causing more pili per unit of surface area, microcolonies of Pseudomonas and Staphylococcus grow bigger [83]. This may be associated with the binding of eDNA to the Pseduomonas pili [200] which then mediates an interaction with Staphylococcus. This mechanism is speculative and has yet to be confirmed. Evidence for pili-like cell-to-cell attachment was also shown in Acinetobacter baumanii monoculture biofilms [114]. In a separate study it was shown that Pseudomonas attaches to already formed Acinetobacter microcolonies before forming biofilm [201]. These studies highlight how monoculture biofilm formation may be a distinct unusual process for many microbes in nature.
Even in monoculture biofilms, specific bacterial packing appears to be a selected-for trait with the microcolonies (post attachment, pre-biofilm) of some species showing remarkable spacing regularity across surfaces [201] indicating that under certain circumstances it might benefit populations to organize their distribution before building biofilms. Packing can also be influenced by carbohydrate production with strains which do not produce as much EPS being packed significantly closer to one another [183]. The regulation of EPS has potential to change single cell toleration values of waste as more space between cells means greater room for diffusion away from the cell, assuming the EPS in question does not bind the waste product increasing local concentration by preventing diffusion into the medium. A high local concentration of metabolic waste is generally believed to reduce metabolic activity of cells [[202], [203], [204]], therefore limiting the production of output. Alternatively, EPS may be thought of as a modular spacer which better enables single cell resolution. This may be useful if the user output is something akin to fluorescence within a mixed species biofilm.
1.4. Biofilm shape
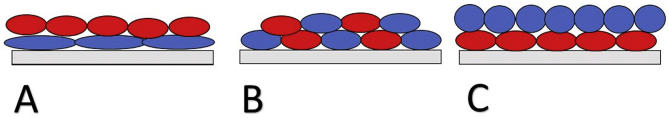
There is a non-trivial relationship between cell shape and biofilm organization. While coccoid cells have the advantage of a high surface area to volume ratio allowing them to exchange more easily than bacilli the cocci are generally at a disadvantage when it comes to attachment area. Cell shape in isolation is not a powerful enough factor to completely dictate attachment rate in nature. Under artificial conditions though, cell shape can absolutely be applied to altering biofilm arrangement. This was cleverly demonstrated by Smith et al. [205]. They made single amino acid changes to MreB (cell partitioning protein) in Escherichia coli resulting in substrains that replicated at nearly identical rates. They then observed these substrains when cultured together seeing a clear gradient from bacillus at the basal surface towards more coccus like cells near the top of the colony. This relationship is depicted in Fig. 1. This study highlights a unique single protein tunable structure that has profound effect on how cells relate structurally to one another. This is a potentially useful way to optimize metabolic consortia or to shield vulnerable population from external forces. For example, one might engineer coccus cells normally found on the periphery of the biofilm to be more bacillus like which would likely sequester them closer to the basal surface of the biofilm which would partially shield these cells from changing environmental conditions such as flow rate. Alternatively cell shape may be altered not to change cells local positioning but to alter the vertical height of a biofilm [206]. A useful review by Caccamo and Brun [207] highlights other potential proteins that might be manipulated to alter cell shape and therefore arrangement in biofilm.
Fig. 1.
Effect of cell shape on cell arrangement. A. The wild type population is red and coccobacillus in shape, whereas the blue population is genetically identical except for a single amino acid substitution rendering the cells more bacillus-like. B. Both the red and blue populations are genetically identical coccobacillus. C. The red population is wild-type coccobacillus which is genetically identical to the blue population except for a single amino acid substitution which renders cells coccus shaped.
In order to understand and control the effects of different inputs leading to varying outputs in biofilm systems, it would be useful to have computational models of biofilm formation and organization. Recent studies with non-typeable Haemophilus influenza have revealed difference between in vitro biofilm architecture versus in vivo biofilm architecture, with clusters being 1/10 the size in vivo. This was attributed to the removal of planktonic cells from the biofilm through the host response, leading to a similarly organized but structurally different biofilm [208]. An in silico model of this biofilm formation was then generated, which will allow the study and isolation of different parts of the biofilm production process, to potentially identify the parts of the system responsible for the differences between the morphologies of in vitro compared to in vivo biofilms. This is the first report that we have identified of a computational model that can integrate various inputs and predict the biofilm output.
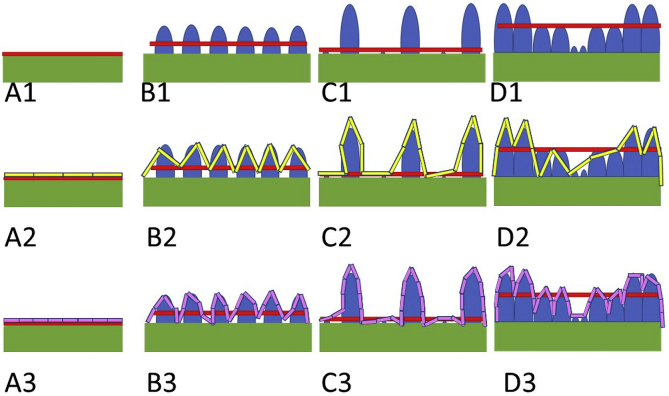
Surface area analysis can also be applied to apical surfaces. The apical surface area has potential engineerable factors including surface roughness and fractal dimensionality. Surface roughness is essentially a measure of the distance from a predefined smooth surface (having consistent curvature). This measure can be contrasted with fractal dimensionality, a metric of the degree to which an answer changes when the scale at which the object is measured is varied. At first glance fractal dimensionality may appear to be merely a measure of surface roughness standard deviation. In actuality, fractal dimensionality is a combination of the deviation from average surface and a measure of how regularly deviations occur over distances. The relationship between surface roughness and fractal dimensionality can be seen in Fig. 2. While applying fractal dimensionality to biofilm height and length is certainly useful, comparing that fractality to other dimensions such as height and width provides more advanced measurements for understanding architecture in response to environmental stimuli especially that of flow [209]. As biofilms are not rectangular prisms, but complex non-euclidian shapes there are a theoretically infinite amount of semi overlapping two-dimensional dimension comparisons a user could study. The limit to which these relationships are useful is likely defined by the regularity of flow with a pipe model being a simple model with fluid flow coming from one direction and a lattice of interconnected tubes being a complicated model with more complex dynamics. These deeper understandings of biofilm shape are necessary to understand to establish ideal biomass to productivity ratios. The idea is the more input your biofilm/factory needs to provide output, the less efficient a factory it is. This mechanical relationship is further complicated by the fact that biofilms can be extremely ephemeral in nature. So one must also ensure that the biofilm will both stay where it needs to be (attachment) and not get destroyed regularly (resisting shear force).
Fig. 2.
Relationship Between Surface Roughness and Fractal Dimensionality. The green rectangle represents underlying biofilm, the red line indicates average biofilm height, the blue ovals represent protrusions from the underlying biofilm, the yellow and pink rectangles represents rulers with the pink being exactly half of the yellow. Projections 1 through 3 are the same biofilms with either no ruler [1] yellow rulers [2] or pink rulers [3] A. Smooth biofilm with minimal surface roughness and minimal fractal dimensionality (4 yellow, 8 pink). B. Biofilm with medium roughness and low fractal dimensionality (12 yellow, 25 pink). C. Biofilm with high roughness and low fractal dimensionality (14 yellow, 31 pink). D. Biofilm with medium roughness and high fractal dimensionality (15 yellow, 36 pink).
Surface roughness and fractal dimensionality are important as they influence both the kinetics of chemical reactions and fluid dynamics. This can be more broadly thought of with the truism shape dictates function. As fractal dimensionality and surface roughness are properties of shapes they are part of defining this function. This was aptly demonstrated by Dewey who was studying the kinetics of proton exchange within lysozymes. Dewey showed using tritium that as reaction space got smaller relative to the unit that causes the change (in this case proton movement) the reaction sped up [210]. Beyond speeding up or slowing down the rate of reaction, optimizing the fractal dimensionality may also be useful for obtaining a more consistent rate of reaction which may be desirable if user input is required at discrete steps during biofilm establishment or maintenance. A more elaborate discussion of fractal dimensionality and how that affects the rates of chemical reactions in many different biological contexts may be found in a review by Gabriele Angelo Losa [211]. When looking at multispecies biofilms exchanging many different kinds of chemical species, as opposed to sub-cellular structures and singular chemical species, these relationships become more complex but, on the whole, it still appears that the more irregular the surface the faster reactions will proceed [212].
Two biofilms with equal surface roughness but unequal fractal dimensionalities have different ideal uses. The biofilm with low fractal dimensionality (more regularly patterned) is more suited for situations where the consistency of flow is of utmost importance as this biofilm will be easier to model. The biofilm with higher fractal dimensionality it more suited to applications where the exchange of diverse chemical species or biological entities is prioritized over regulated flow, as greater fractal dimensionality means greater opportunity for multiple surface niches. It is important to appreciate the influence of biofilm on flow and visa versa. These relationships are also dictated by apical elasticity and buoyancy. As both of these factors will affect deformation of biofilm in response to flow which has potential downstream effects on biofilm shear and nutrient acquisition [213]. Fractal dimensionality has been shown to have important effects in biofilms designed for electron transfer [214], where a more regular biofilm is better suited for uniform transfer. Fractal dimensionality has also been appreciated as important when addressing biofilms where flocculation and post-fragmentation are key characteristics [215]. Similar works has also pointed to fractal dimensionality as being important when biofilms are upstream of mandatory filtration steps [216] (smaller more irregular pieces plug up filters faster). Roughness can also be seen as having a non-trivial effect on cell populations. A study focusing on Legionella biofilms highlighted the importance of biofilm surface roughness with the maintenance of biofilm. They showed that asperities had a significant reduction in cell sloughing compared to smoother portions of the biofilm. This phenomenon is believed to be due to the change in local hydrodynamics [217]. A subsequent study measuring mixed species demonstrated that biofilm grown in turbulent flow are larger than those generated in shear flow [218]. This demonstrates how hydrodynamics can limit biofilm growth and why biological mechanisms for decreasing shear flow may have been selected for.
Interestingly, surface roughness also has a notable influence on microcolony growth. Sometimes edges of microcolonies are associated with mutant subpopulations. The phenomenon dubbed “surfing” involves mutants with a growth advantage having a much greater chance of spreading the mutation when the original mutant is on a promontory on the leading edge of the colony with ample room to spread [219]. This work has yet to be applied to biofilms but may very well be used to selective evolution biofilm experiments in the future in order to better fix mutants within the population. Another group showed how exposure to carbenicillin, an antibiotic which causes cell elongation can actually lead to the selection of carbenicillin sensitive cells, as this elongation helps these cells better reach the leading edge of a growing population [220].
As fractal dimensionality and surface roughness are already standard metrics for biofilms [221] it seems likely that these will be a greater focus in the future of biofilm optimization projects. Engineering surface roughness and fractal dimensionality may be accomplished via a variety of mechanisms depending on the starting composition of biofilm. Initial colonizers may be employed to change the underlying architecture which may influence apical surfaces on cells, or the periphery of the biofilm may be engineered, either constitutively or in response to a gradient, to produce more or less of a desired protein, carbohydrate etc. While biofilm surfaces are composed of a great many things (discussed above) the relative contribution of different types of molecules is not necessarily equal. A group studying Bacillus showed that at one point on the biofilm cycle surface roughness was largely attributable to a single protein BslA [222]. This highlights the feasibility of surface roughness engineering in biofilms. Alternatively, an external agent may be added which may bind the biofilm and essentially alter its surface topography.
The great challenge is altering roughness or fractal dimensionality in an orthogonal manner. Generally surface roughness and fractal dimensionality are thought of as consequences as other more central phenomena (charge, attachment ability, etc.). At present there is not a consistent, predictable way to alter roughness or fractal dimensionality without changing other perhaps more important characteristics of a biofilm. More empirical testing is needed in this area to enhance the viability of this option. It may be the case that surface roughness and fractal dimensionality will be targeted for manipulated in response to these future studies especially when guided by principal component analysis [[214], [215], [216]].
1.5. Conclusion
Biofilms are the new factories of tomorrow. As more and more design teams seek to harness biofilms abilities, a greater need for system efficiency is demanded. While metabolic engineering-type approaches are absolutely essential in directing metabolic flux, the underlying biofilm architecture should not be ignored. Each of the components of biofilm and the shape of the biofilm itself significantly affects the efficiency of any system. Efforts by biologists to optimize biofilms are summarized in Table 1 whereas future potential targets are summarized in Table 2. While not all modulations are possible in all biofilms, it is important to appreciate the plasticity of each chassis or group of chassis to best optimize performance to a certain task.
Table 1.
Current biofilm engineering targets.
| Molecular Target | Species | Goal | Mechanism | Reference |
|---|---|---|---|---|
| PqsE/PqsC (Quorum Sensing) | Pseudomonas aeruginosa | Enhanced Electric Production | Elimination of anaerobic repression leading to enhanced microbial metabolism | [223] |
| PtsG/PtsM/Glk/Gcd (Carbon utilization) | Escherichia coli | Enhanced Biomass | Creation of a second population which was unable to use the primary carbon source and thus used waste products of the primary strain to grow enhancing total growth | [224] |
| Diguanylate cyclase (regulator of intracellular signal c-di-GMP) | Pseudomonas putida | Enhanced Biomass/Haloalkane degredation | Raising the levels of cyclic di-GMP which allowed large biofilms which sped the biodegradation of haloalkanes | [225] |
| degQ (regulator of extracellular poly glutamate, capsule) | Bacillus subtilis | Enhanced Biomass/Antibiotic production | Increasing extracellular glutamate lead to a thicker capsule which lead to a thicker biofilm enhancing production | [226] |
| OmpR (regulator of curli production) | Escherichia coli | Enhanced Bimoass Production/Curli Fiber production/L-Halotryptophan production | Upregulating the amount of curli lead to a larger biofilm which lead to a greater L-Halotryptophan yield | [227] |
| BifA like protein (c-di-GMP phosphodiesterase) | Pseudomonas taiwanensis | Enhanced Biomass/Enhanced Attachment/Altered Surface Charge/styrene-oxide | A differing surface charge leads to enhanced attachment and then larger biofilms to produce styrene oxide | [228] |
Table 2.
Future biofilm engineering targets.
| Molecular Target | Net Effect | Mechanism |
|---|---|---|
| BslA | Alter surface roughness | Greater expression of BslA yielding rougher biofilms |
| MreB | Alter surface roughness, fractal dimensionality, cell stacking patterns | Altering the MreB protein causing cell so become more or less coccoid changing where they reside in mixed specie biofilms and potentially how the apical surface is shaped |
| MucB/RpoN/PilH | Alter surface roughness, fractal dimensionality, cell stacking patterns, cellular distribution | Increasing or decreasing pillin expression causing cells to coaggregate changing biofilm shape and cell distribution |
| EPS regulating protein (Glycosyl hydrolase) | Alter surface roughness, fractal dimensionality, cellular distribution | Increasing glycosyl hydrolases to decrease the amount of maintained EPS forcing cells closer together |
| AHL-lactonase | Deter establishment of rival biofilms | Constituent expression of modular quorum quenching enzymes to retard the establishment of rival systems |
Declaration of competing interest
None.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Stephen J. Kassinger, Email: skassin2@gmu.edu.
Monique L. van Hoek, Email: mvanhoek@gmu.edu.
References
- 1.O'toole G.A., Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 2.Branda S.S., Vik Å., Friedman L., Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Stoodley P., Boyle J.D., DeBeer D., Lappin‐Scott H.M. Evolving perspectives of biofilm structure. Biofouling. 1999;14:75–90. [Google Scholar]
- 4.Zhang X., Bishop P.L., Kupferle M.J. Measurement of polysaccharides and proteins in biofilm extracellular polymers. Water Sci Technol. 1998;37:345–348. [Google Scholar]
- 5.Gehrke T., Telegdi J., Thierry D., Sand W. Importance of extracellular polymeric substances from Thiobacillus ferrooxidans for bioleaching. Appl Environ Microbiol. 1998;64:2743–2747. doi: 10.1128/aem.64.7.2743-2747.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitchurch C.B., Tolker-Nielsen T., Ragas P.C., Mattick J.S. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295 doi: 10.1126/science.295.5559.1487. 1487-1487. [DOI] [PubMed] [Google Scholar]
- 7.Kroll A., Behra R., Kaegi R., Sigg L. Extracellular polymeric substances (EPS) of freshwater biofilms stabilize and modify CeO2 and Ag nanoparticles. PLoS One. 2014;9 doi: 10.1371/journal.pone.0110709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen P.H., Frølund B., Keiding K. Changes in the composition of extracellular polymeric substances in activated sludge during anaerobic storage. Appl Microbiol Biotechnol. 1996;44:823–830. doi: 10.1007/BF00178625. [DOI] [PubMed] [Google Scholar]
- 9.Mojica K., Elsey D., Cooney M.J. Quantitative analysis of biofilm EPS uronic acid content. J Microbiol Methods. 2007;71:61–65. doi: 10.1016/j.mimet.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 10.O'Toole G.A. To build a biofilm. J Bacteriol. 2003;185:2687–2689. doi: 10.1128/JB.185.9.2687-2689.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong G.C., O'Toole G.A. All together now: integrating biofilm research across disciplines. MRS Bull. 2011;36:339–342. doi: 10.1557/mrs.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Hoek M.L. Biofilms: an advancement in our understanding of Francisella species. Virulence. 2013;4:833–846. doi: 10.4161/viru.27023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean S.N., Chung M.C., van Hoek M.L. Burkholderia diffusible signal factor signals to Francisella novicida to disperse biofilm and increase siderophore production. Appl Environ Microbiol. 2015;81:7057–7066. doi: 10.1128/AEM.02165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durham-Colleran M.W., Verhoeven A.B., van Hoek M.L. Francisella novicida forms in vitro biofilms mediated by an orphan response regulator. Microb Ecol. 2010;59:457–465. doi: 10.1007/s00248-009-9586-9. [DOI] [PubMed] [Google Scholar]
- 15.de la Fuente-Nunez C., Reffuveille F., Haney E.F., Straus S.K., Hancock R.E. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung M.-C., Dean S., Marakasova E.S., Nwabueze A.O., van Hoek M.L. Chitinases are negative regulators of Francisella novicida biofilms. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan J.B., Ragunath C., Velliyagounder K., Fine D.H., Ramasubbu N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother. 2004;48:2633–2636. doi: 10.1128/AAC.48.7.2633-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu S. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 2015;25:1352. doi: 10.1038/cr.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xavier J.B., Picioreanu C., Rani S.A., van Loosdrecht M.C., Stewart P.S. Biofilm-control strategies based on enzymic disruption of the extracellular polymeric substance matrix–a modelling study. Microbiology. 2005;151:3817–3832. doi: 10.1099/mic.0.28165-0. [DOI] [PubMed] [Google Scholar]
- 20.Christensen B., Characklis W. Physical and chemical properties of biofilms. Biofilms. 1990;93:130. [Google Scholar]
- 21.Nielsen P.H., Jahn A., Palmgren R. Conceptual model for production and composition of exopolymers in biofilms. Water Sci Technol. 1997;36:11–19. [Google Scholar]
- 22.Jahn A., Nielsen P.H. Cell biomass and exopolymer composition in sewer biofilms. Water Sci Technol. 1998;37:17–24. [Google Scholar]
- 23.Pihlstrom B.L., Michalowicz B.S., Johnson N.W. Periodontal diseases. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 24.Costerton J.W. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 2001;9:50–52. doi: 10.1016/s0966-842x(00)01918-1. [DOI] [PubMed] [Google Scholar]
- 25.Potera C. Am Assoc Adv Sci. 1999 [Google Scholar]
- 26.Beech I.B., Sunner J. Biocorrosion: towards understanding interactions between biofilms and metals. Curr Opin Biotechnol. 2004;15:181–186. doi: 10.1016/j.copbio.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Baker J., Dudley L. Biofouling in membrane systems—a review. Desalination. 1998;118:81–89. [Google Scholar]
- 28.Geesey G.G. CRC Press; 1994. Biofouling and biocorrosion in industrial water systems. [DOI] [PubMed] [Google Scholar]
- 29.Holah J., Kearney L. Springer; 1992. Biofilms—science and technology; pp. 35–41. [Google Scholar]
- 30.Macfarlane S., Dillon J. Microbial biofilms in the human gastrointestinal tract. J Appl Microbiol. 2007;102:1187–1196. doi: 10.1111/j.1365-2672.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- 31.Nickel J., Ruseska I., Wright J., Costerton J. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother. 1985;27:619–624. doi: 10.1128/aac.27.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cargill K.L., Pyle B.H., Sauer R.L., McFeters G.A. Effects of culture conditions and biofilm formation on the iodine susceptibility of Legionella pneumophila. Can J Microbiol. 1992;38:423–429. doi: 10.1139/m92-071. [DOI] [PubMed] [Google Scholar]
- 33.Scher K., Romling U., Yaron S. Effect of heat, acidification, and chlorination on Salmonella enterica serovar Typhimurium cells in a biofilm formed at the air-liquid interface. Appl Environ Microbiol. 2005;71:1163–1168. doi: 10.1128/AEM.71.3.1163-1168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costerton J.W., Lewandowski Z., Caldwell D.E., Korber D.R., Lappin-Scott H.M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 35.Rittman B.E. The effect of shear stress on biofilm loss rate. Biotechnol Bioeng. 1982;24:501–506. doi: 10.1002/bit.260240219. [DOI] [PubMed] [Google Scholar]
- 36.Oh D.-H., Marshall D.L. Destruction of Listeria monocytogenes biofilms on stainless steel using monolaurin and heat. J Food Prot. 1995;58:251–255. doi: 10.4315/0362-028X-58.3.251. [DOI] [PubMed] [Google Scholar]
- 37.Espinal P., Marti S., Vila J. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J Hosp Infect. 2012;80:56–60. doi: 10.1016/j.jhin.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 38.Pozos N., Scow K., Wuertz S., Darby J. UV disinfection in a model distribution system:: biofilm growth and microbial community. Water Res. 2004;38:3083–3091. doi: 10.1016/j.watres.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 39.Elasri M.O., Miller R.V. Study of the response of a biofilm bacterial community to UV radiation. Appl Environ Microbiol. 1999;65:2025–2031. doi: 10.1128/aem.65.5.2025-2031.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo A.W. Comparative transcriptomic analysis of Porphyromonas gingivalis biofilm and planktonic cells. BMC Microbiol. 2009;9:18. doi: 10.1186/1471-2180-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castro J. Comparative transcriptomic analysis of Gardnerella vaginalis biofilms vs. planktonic cultures using RNA-seq. NPJ Biofilms Microbiome. 2017;3:3. doi: 10.1038/s41522-017-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu D. Comparative transcriptomic analysis of Clostridium acetobutylicum biofilm and planktonic cells. J Biotechnol. 2016;218:1–12. doi: 10.1016/j.jbiotec.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 43.Charlebois A., Jacques M., Archambault M. Comparative transcriptomic analysis of Clostridium perfringens biofilms and planktonic cells. Avian Pathol. 2016;45:593–601. doi: 10.1080/03079457.2016.1189512. [DOI] [PubMed] [Google Scholar]
- 44.Resch A. Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics. 2006;6:1867–1877. doi: 10.1002/pmic.200500531. [DOI] [PubMed] [Google Scholar]
- 45.Romero-Lastra P. Comparative gene expression analysis of Porphyromonas gingivalis ATCC 33277 in planktonic and biofilms states. PLoS One. 2017;12 doi: 10.1371/journal.pone.0174669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jahns A.C., Eilers H., Alexeyev O.A. Transcriptomic analysis of Propionibacterium acnes biofilms in vitro. Anaerobe. 2016;42:111–118. doi: 10.1016/j.anaerobe.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Manos J. Gene expression characteristics of a cystic fibrosis epidemic strain of Pseudomonas aeruginosa during biofilm and planktonic growth. FEMS Microbiol Lett. 2009;292:107–114. doi: 10.1111/j.1574-6968.2008.01472.x. [DOI] [PubMed] [Google Scholar]
- 48.Flemming L. 2010. Comparative proteomic and genomic analysis of Flavobacterium johnsoniae-like biofilm, planktonic and agar surface-associated cells. [Google Scholar]
- 49.Waite R.D., Papakonstantinopoulou A., Littler E., Curtis M.A. Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J Bacteriol. 2005;187:6571–6576. doi: 10.1128/JB.187.18.6571-6576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schembri M.A., Kjærgaard K., Klemm P. Global gene expression in Escherichia coli biofilms. Mol Microbiol. 2003;48:253–267. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- 51.Benner S.A., Sismour A.M. Synthetic biology. Nat Rev Genet. 2005;6:533. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khalil A.S., Collins J.J. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andrianantoandro E., Basu S., Karig D.K., Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwok R. Five hard truths for synthetic biology. Nature News. 2010;463:288–290. doi: 10.1038/463288a. [DOI] [PubMed] [Google Scholar]
- 55.Anderson J. BglBricks: a flexible standard for biological part assembly. J Biol Eng. 2010;4:1. doi: 10.1186/1754-1611-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Canton B., Labno A., Endy D. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol. 2008;26:787. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 57.Dueber J.E., Mirsky E.A., Lim W.A. Engineering synthetic signaling proteins with ultrasensitive input/output control. Nat Biotechnol. 2007;25:660. doi: 10.1038/nbt1308. [DOI] [PubMed] [Google Scholar]
- 58.Villalobos A., Ness J.E., Gustafsson C., Minshull J., Govindarajan S. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinf. 2006;7:285. doi: 10.1186/1471-2105-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Slusarczyk A.L., Lin A., Weiss R. Foundations for the design and implementation of synthetic genetic circuits. Nat Rev Genet. 2012;13:406. doi: 10.1038/nrg3227. [DOI] [PubMed] [Google Scholar]
- 60.Wang B., Kitney R.I., Joly N., Buck M. Engineering modular and orthogonal genetic logic gates for robust digital-like synthetic biology. Nat Commun. 2011;2:508. doi: 10.1038/ncomms1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siuti P., Yazbek J., Lu T.K. Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol. 2013;31:448. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- 62.Seelig G., Soloveichik D., Zhang D.Y., Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 63.Pósfai G. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 64.Bedau M.A. Artificial life: organization, adaptation and complexity from the bottom up. Trends Cogn Sci. 2003;7:505–512. doi: 10.1016/j.tics.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 65.Noireaux V., Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci. 2004;101:17669–17674. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gibson D.G. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 67.Toya Y., Shimizu H. Flux analysis and metabolomics for systematic metabolic engineering of microorganisms. Biotechnol Adv. 2013;31:818–826. doi: 10.1016/j.biotechadv.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 68.Hasunuma T. Metabolic pathway engineering based on metabolomics confers acetic and formic acid tolerance to a recombinant xylose-fermenting strain of Saccharomyces cerevisiae. Microb Cell Factories. 2011;10:2. doi: 10.1186/1475-2859-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McNeely K., Xu Y., Bennette N., Bryant D.A., Dismukes G.C. Redirecting reductant flux into hydrogen production via metabolic engineering of fermentative carbon metabolism in a cyanobacterium. Appl Environ Microbiol. 2010;76:5032–5038. doi: 10.1128/AEM.00862-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kizer L., Pitera D.J., Pfleger B.F., Keasling J.D. Application of functional genomics to pathway optimization for increased isoprenoid production. Appl Environ Microbiol. 2008;74:3229–3241. doi: 10.1128/AEM.02750-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pfleger B.F., Pitera D.J., Smolke C.D., Keasling J.D. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24:1027. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 72.Rudge T.J., Steiner P.J., Phillips A., Haseloff J. Computational modeling of synthetic microbial biofilms. ACS Synth Biol. 2012;1:345–352. doi: 10.1021/sb300031n. [DOI] [PubMed] [Google Scholar]
- 73.Hong S.H. Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device. Nat Commun. 2012;3:613. doi: 10.1038/ncomms1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brenner K., You L., Arnold F.H. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 2008;26:483–489. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 75.Nguyen P.Q. Synthetic biology engineering of biofilms as nanomaterials factories. Biochem Soc Trans. 2017;45:585–597. doi: 10.1042/BST20160348. [DOI] [PubMed] [Google Scholar]
- 76.Stoodley P., Lewandowski Z., Boyle J.D., Lappin‐Scott H.M. The formation of migratory ripples in a mixed species bacterial biofilm growing in turbulent flow. Environ Microbiol. 1999;1:447–455. doi: 10.1046/j.1462-2920.1999.00055.x. [DOI] [PubMed] [Google Scholar]
- 77.Eiteman M.A., Lee S.A., Altman E. A co-fermentation strategy to consume sugar mixtures effectively. J Biol Eng. 2008;2:3. doi: 10.1186/1754-1611-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jagmann N., von Rekowski K.S., Philipp B. Interactions of bacteria with different mechanisms for chitin degradation result in the formation of a mixed-species biofilm. FEMS Microbiol Lett. 2012;326:69–75. doi: 10.1111/j.1574-6968.2011.02435.x. [DOI] [PubMed] [Google Scholar]
- 79.Burmølle M. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol. 2006;72:3916–3923. doi: 10.1128/AEM.03022-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simoes M., Simoes L.C., Vieira M.J. Species association increases biofilm resistance to chemical and mechanical treatments. Water Res. 2009;43:229–237. doi: 10.1016/j.watres.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 81.Wang R., Kalchayanand N., Schmidt J.W., Harhay D.M. Mixed biofilm formation by Shiga toxin–producing Escherichia coli and Salmonella enterica serovar Typhimurium enhanced bacterial resistance to sanitization due to extracellular polymeric substances. J Food Prot. 2013;76:1513–1522. doi: 10.4315/0362-028X.JFP-13-077. [DOI] [PubMed] [Google Scholar]
- 82.Giaouris E., Chorianopoulos N., Doulgeraki A., Nychas G.-J. Co-culture with Listeria monocytogenes within a dual-species biofilm community strongly increases resistance of Pseudomonas putida to benzalkonium chloride. PLoS One. 2013;8 doi: 10.1371/journal.pone.0077276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang L. Pattern differentiation in co-culture biofilms formed by Staphylococcus aureus and Pseudomonas aeruginosa. FEMS Immunol Med Microbiol. 2011;62:339–347. doi: 10.1111/j.1574-695X.2011.00820.x. [DOI] [PubMed] [Google Scholar]
- 84.Kouzuma A., Kato S., Watanabe K. Microbial interspecies interactions: recent findings in syntrophic consortia. Front Microbiol. 2015;6:477. doi: 10.3389/fmicb.2015.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schüler D., Frankel R.B. Bacterial magnetosomes: microbiology, biomineralization and biotechnological applications. Appl Microbiol Biotechnol. 1999;52:464–473. doi: 10.1007/s002530051547. [DOI] [PubMed] [Google Scholar]
- 86.Anjum A. Microbial production of polyhydroxyalkanoates (PHAs) and its copolymers: a review of recent advancements. Int J Biol Macromol. 2016;89:161–174. doi: 10.1016/j.ijbiomac.2016.04.069. [DOI] [PubMed] [Google Scholar]
- 87.Tan C.H. Community quorum sensing signalling and quenching: microbial granular biofilm assembly. npj Biofilms Microbiome. 2015;1:15006. doi: 10.1038/npjbiofilms.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Giao M., Wilks S., Azevedo N., Vieira M., Keevil C. Incorporation of natural uncultivable Legionella pneumophila into potable water biofilms provides a protective niche against chlorination stress. Biofouling. 2009;25:345–351. doi: 10.1080/08927010902803305. [DOI] [PubMed] [Google Scholar]
- 89.Wade W. Unculturable bacteria—the uncharacterized organisms that cause oral infections. J R Soc Med. 2002;95:81–83. doi: 10.1258/jrsm.95.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Podolich O., Laschevskyy V., Ovcharenko L., Kozyrovska N., Pirttilä A. Methylobacterium sp. resides in unculturable state in potato tissues in vitro and becomes culturable after induction by Pseudomonas fluorescens IMGB163. J Appl Microbiol. 2009;106:728–737. doi: 10.1111/j.1365-2672.2008.03951.x. [DOI] [PubMed] [Google Scholar]
- 91.Klieneberger E. The natural occurrence of pleuropneumonia‐like organism in apparent symbiosis with Strrptobacillus moniliformis and other bacteria. J Pathol Bacteriol. 1935;40:93–105. [Google Scholar]
- 92.Sieben S., Hertle R., Gumpert J., Braun V. The Serratia marcescens hemolysin is secreted but not activated by stable protoplast-type L-forms of Proteus mirabilis. Arch Microbiol. 1998;170:236–242. doi: 10.1007/s002030050638. [DOI] [PubMed] [Google Scholar]
- 93.Rippmann J.F. Procaryotic expression of single-chain variable-fragment (scFv) antibodies: secretion in L-form cells of Proteus mirabilis leads to active product and overcomes the limitations of periplasmic expression in Escherichia coli. Appl Environ Microbiol. 1998;64:4862–4869. doi: 10.1128/aem.64.12.4862-4869.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gumpert J., Hoischen C. Use of cell wall-less bacteria (L-forms) for efficient expression and secretion of heterologous gene products. Curr Opin Biotechnol. 1998;9:506–509. doi: 10.1016/s0958-1669(98)80037-2. [DOI] [PubMed] [Google Scholar]
- 95.Liu W. Low‐abundant species facilitates specific spatial organization that promotes multispecies biofilm formation. Environ Microbiol. 2017;19:2893–2905. doi: 10.1111/1462-2920.13816. [DOI] [PubMed] [Google Scholar]
- 96.Xia Y., Xiong Y., Li X., Su X. Inhibition of biofilm formation by the antisense peptide nucleic acids targeted at the motA gene in Pseudomonas aeruginosa PAO1 strain. World J Microbiol Biotechnol. 2011;27:1981–1987. [Google Scholar]
- 97.Kalesinskas P., Kačergius T., Ambrozaitis A., Jimbo R., Ericson D. Streptococcus mutans biofilm inhibition using antisense oligonucleotide to glucosyltransferases B and C. Acta Med Litu. 2015;22 [Google Scholar]
- 98.Otsuka T. Antimicrobial activity of antisense peptide–peptide nucleic acid conjugates against non-typeable Haemophilus influenzae in planktonic and biofilm forms. J Antimicrob Chemother. 2016;72:137–144. doi: 10.1093/jac/dkw384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Howard J.J. Inhibition of Pseudomonas aeruginosa by peptide-conjugated phosphorodiamidate morpholino oligomers. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.01938-16. e01938-01916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Storz M.P. Validation of PqsD as an anti-biofilm target in Pseudomonas aeruginosa by development of small-molecule inhibitors. J Am Chem Soc. 2012;134:16143–16146. doi: 10.1021/ja3072397. [DOI] [PubMed] [Google Scholar]
- 101.Kim H.-S., Lee S.-H., Byun Y., Park H.-D. 6-Gingerol reduces Pseudomonas aeruginosa biofilm formation and virulence via quorum sensing inhibition. Sci Rep. 2015;5:8656. doi: 10.1038/srep08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hentzer M. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology. 2002;148:87–102. doi: 10.1099/00221287-148-1-87. [DOI] [PubMed] [Google Scholar]
- 103.Brophy J.A. Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. Nat. Microbiol. 2018;3:1043. doi: 10.1038/s41564-018-0216-5. [DOI] [PubMed] [Google Scholar]
- 104.Ronda C., Chen S.P., Cabral V., Yaung S.J., Wang H.H. Metagenomic engineering of the mammalian gut microbiome in situ. Nat Methods. 2019;16:167. doi: 10.1038/s41592-018-0301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jahn A., Griebe T., Nielsen P.H. Composition of Pseudomonas putida biofilms: accumulation of protein in the biofilm matrix. Biofouling. 1999;14:49–57. [Google Scholar]
- 106.Toyofuku M., Roschitzki B., Riedel K., Eberl L. Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J Proteome Res. 2012;11:4906–4915. doi: 10.1021/pr300395j. [DOI] [PubMed] [Google Scholar]
- 107.Christner M. The giant extracellular matrix‐binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol Microbiol. 2010;75:187–207. doi: 10.1111/j.1365-2958.2009.06981.x. [DOI] [PubMed] [Google Scholar]
- 108.O'Neill E. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol. 2008;190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brandl M.T. Salmonella biofilm formation on Aspergillus Niger involves cellulose–chitin interactions. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Margolis J.J. Contributions of Francisella tularensis subsp. novicida chitinases and Sec secretion system to biofilm formation on chitin. Appl Environ Microbiol. 2010;76:596–608. doi: 10.1128/AEM.02037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kirn T.J., Jude B.A., Taylor R.K. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature. 2005;438:863. doi: 10.1038/nature04249. [DOI] [PubMed] [Google Scholar]
- 112.Pratt L.A., Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 113.Jones G., RICHARDSON L.A. The attachment to, and invasion of HeLa cells by Salmonella typhimurium: the contribution of mannose-sensitive and mannoseresistant haemagglutinating activities. Microbiology. 1981;127:361–370. doi: 10.1099/00221287-127-2-361. [DOI] [PubMed] [Google Scholar]
- 114.Tomaras A.P., Dorsey C.W., Edelmann R.E., Actis L.A. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology. 2003;149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 115.Manetti A.G. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol Microbiol. 2007;64:968–983. doi: 10.1111/j.1365-2958.2007.05704.x. [DOI] [PubMed] [Google Scholar]
- 116.Kim T.-J., Young B.M., Young G.M. Effect of flagellar mutations on Yersinia enterocolitica biofilm formation. Appl Environ Microbiol. 2008;74:5466–5474. doi: 10.1128/AEM.00222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Park Y. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun. 2005;73:3983–3989. doi: 10.1128/IAI.73.7.3983-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wurpel D.J. Comparative proteomics of uropathogenic Escherichia coli during growth in human urine identify UCA-like (UCL) fimbriae as an adherence factor involved in biofilm formation and binding to uroepithelial cells. J proteom. 2016;131:177–189. doi: 10.1016/j.jprot.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 119.Vallet I., Olson J.W., Lory S., Lazdunski A., Filloux A. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci. 2001;98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Caserta R. Expression of Xylella fastidiosa fimbrial and afimbrial proteins during biofilm formation. Appl Environ Microbiol. 2010;76:4250–4259. doi: 10.1128/AEM.02114-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.De Jong W., Wösten H.A., Dijkhuizen L., Claessen D. Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Mol Microbiol. 2009;73:1128–1140. doi: 10.1111/j.1365-2958.2009.06838.x. [DOI] [PubMed] [Google Scholar]
- 122.Cárcamo-Oyarce G., Lumjiaktase P., Kümmerli R., Eberl L. Quorum sensing triggers the stochastic escape of individual cells from Pseudomonas putida biofilms. Nat Commun. 2015;6:5945. doi: 10.1038/ncomms6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xavier J.B., Kim W., Foster K.R. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol Microbiol. 2011;79:166–179. doi: 10.1111/j.1365-2958.2010.07436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cookson A.L., Cooley W.A., Woodward M.J. The role of type 1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. Int J Med Microbiol. 2002;292:195–205. doi: 10.1078/1438-4221-00203. [DOI] [PubMed] [Google Scholar]
- 125.Chapman M.R. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Larsen P. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 127.Scheibel T. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc Natl Acad Sci. 2003;100:4527–4532. doi: 10.1073/pnas.0431081100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Seker U.O.S., Chen A.Y., Citorik R.J., Lu T.K. Synthetic biogenesis of bacterial amyloid nanomaterials with tunable inorganic–organic interfaces and electrical conductivity. ACS Synth Biol. 2016;6:266–275. doi: 10.1021/acssynbio.6b00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou Y. Promiscuous cross-seeding between bacterial amyloids promotes interspecies biofilms. J Biol Chem. 2012;287:35092–35103. doi: 10.1074/jbc.M112.383737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Romero D., Aguilar C., Losick R., Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li D. Designed amyloid fibers as materials for selective carbon dioxide capture. Proc Natl Acad Sci. 2014;111:191–196. doi: 10.1073/pnas.1321797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nguyen P.Q., Botyanszki Z., Tay P.K.R., Joshi N.S. Programmable biofilm-based materials from engineered curli nanofibres. Nat Commun. 2014;5:4945. doi: 10.1038/ncomms5945. [DOI] [PubMed] [Google Scholar]
- 133.Koval S., Hynes S. Effect of paracrystalline protein surface layers on predation by Bdellovibrio bacteriovorus. J Bacteriol. 1991;173:2244–2249. doi: 10.1128/jb.173.7.2244-2249.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rickard A.H., Leach S.A., Buswell C.M., High N.J., Handley P.S. Coaggregation between aquatic bacteria is mediated by specific-growth-phase-dependent lectin-saccharide interactions. Appl Environ Microbiol. 2000;66:431–434. doi: 10.1128/aem.66.1.431-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.G. W. Duff, J. R. Sayers, S. Vitovski. (Google Patents, 2002).
- 136.E. Madison, J. Nguyen, S. Ruggles, C. Thanos. (Google Patents, 2007).
- 137.J. Nguyen, C. Thanos, S. W. Ruggles, C. S. Craik. (Google Patents, 2009).
- 138.S. Ruggles, J. Nguyen. (Google Patents, 2006).
- 139.E. L. Madison. (Google Patents, 2014).
- 140.S. W. Ruggles, J. Nguyen. (Google Patents, 2016).
- 141.C. C. Shone et al. (Google Patents, 2015).
- 142.Sleytr U.B., Beveridge T.J. Bacterial S-layers. Trends Microbiol. 1999;7:253–260. doi: 10.1016/s0966-842x(99)01513-9. [DOI] [PubMed] [Google Scholar]
- 143.Gerbino E., Carasi P., Mobili P., Serradell M., Gómez-Zavaglia A. Role of S-layer proteins in bacteria. World J Microbiol Biotechnol. 2015;31:1877–1887. doi: 10.1007/s11274-015-1952-9. [DOI] [PubMed] [Google Scholar]
- 144.Callegari M.L. The S-layer gene of Lactobacillus helveticus CNRZ 892: cloning, sequence and heterologous expression. Microbiology. 1998;144:719–726. doi: 10.1099/00221287-144-3-719. [DOI] [PubMed] [Google Scholar]
- 145.Moreau J.W. Extracellular proteins limit the dispersal of biogenic nanoparticles. Science. 2007;316:1600–1603. doi: 10.1126/science.1141064. [DOI] [PubMed] [Google Scholar]
- 146.Ciornei C.D. Biofilm-forming Pseudomonas aeruginosa bacteria undergo lipopolysaccharide structural modifications and induce enhanced inflammatory cytokine response in human monocytes. Innate Immun. 2010;16:288–301. doi: 10.1177/1753425909341807. [DOI] [PubMed] [Google Scholar]
- 147.Benamara H., Rihouey C., Jouenne T., Alexandre S. Impact of the biofilm mode of growth on the inner membrane phospholipid composition and lipid domains in Pseudomonas aeruginosa. Biochim Biophys Acta Biomembr. 2011;1808:98–105. doi: 10.1016/j.bbamem.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 148.Lattif A.A. Lipidomics of Candida albicans biofilms reveals phase-dependent production of phospholipid molecular classes and role for lipid rafts in biofilm formation. Microbiology. 2011;157:3232. doi: 10.1099/mic.0.051086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zara G. FLO11 expression and lipid biosynthesis are required for air–liquid biofilm formation in a Saccharomyces cerevisiae flor strain. FEMS Yeast Res. 2012;12:864–866. doi: 10.1111/j.1567-1364.2012.00831.x. [DOI] [PubMed] [Google Scholar]
- 150.Sutherland I.W. The biofilm matrix–an immobilized but dynamic microbial environment. Trends Microbiol. 2001;9:222–227. doi: 10.1016/s0966-842x(01)02012-1. [DOI] [PubMed] [Google Scholar]
- 151.Rice K.C. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kadurugamuwa J.L., Beveridge T.J. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Grande R. Helicobacter pylori ATCC 43629/NCTC 11639 outer membrane vesicles (OMVs) from biofilm and planktonic phase associated with extracellular DNA (eDNA) Front Microbiol. 2015;6:1369. doi: 10.3389/fmicb.2015.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hagemann S. DNA‐bearing membrane vesicles produced by Ahrensia kielensis and Pseudoalteromonas marina. J Basic Microbiol. 2014;54:1062–1072. doi: 10.1002/jobm.201300376. [DOI] [PubMed] [Google Scholar]
- 155.Sahu P.K., Iyer P.S., Oak A.M., Pardesi K.R., Chopade B.A. Characterization of eDNA from the clinical strain Acinetobacter baumannii AIIMS 7 and its role in biofilm formation. Sci World J. 2012;2012 doi: 10.1100/2012/973436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mann E.E. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Allesen‐Holm M. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 158.Thomas V.C. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol Microbiol. 2009;72:1022–1036. doi: 10.1111/j.1365-2958.2009.06703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.McMahon K.J., Castelli M.E., Vescovi E.G., Feldman M.F. Biogenesis of outer membrane vesicles in Serratia marcescens is thermoregulated and can be induced by activation of the Rcs phosphorelay system. J Bacteriol. 2012;194:3241–3249. doi: 10.1128/JB.00016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Song T. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol Microbiol. 2008;70:100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Harmsen M., Lappann M., Knøchel S., Molin S. Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Appl Environ Microbiol. 2010;76:2271–2279. doi: 10.1128/AEM.02361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gödeke J., Paul K., Lassak J., Thormann K.M. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J. 2011;5:613. doi: 10.1038/ismej.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Steinberger R., Holden P. Extracellular DNA in single-and multiple-species unsaturated biofilms. Appl Environ Microbiol. 2005;71:5404–5410. doi: 10.1128/AEM.71.9.5404-5410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dominiak D.M., Nielsen J.L., Nielsen P.H. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ Microbiol. 2011;13:710–721. doi: 10.1111/j.1462-2920.2010.02375.x. [DOI] [PubMed] [Google Scholar]
- 165.Pammi M., Liang R., Hicks J., Mistretta T.-A., Versalovic J. Biofilm extracellular DNA enhances mixed species biofilms of Staphylococcus epidermidis and Candida albicans. BMC Microbiol. 2013;13:257. doi: 10.1186/1471-2180-13-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Devaraj A. The extracellular DNA lattice of bacterial biofilms is structurally related to Holliday junction recombination intermediates. Proc Natl Acad Sci U S A. 2019;116:25068–25077. doi: 10.1073/pnas.1909017116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Finkel S.E., Kolter R. DNA as a nutrient: novel role for bacterial competence gene homologs. J Bacteriol. 2001;183:6288–6293. doi: 10.1128/JB.183.21.6288-6293.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Muto Y., Goto S. Transformation by extracellular DNA produced by Pseudomonas aeruginosa. Microbiol Immunol. 1986;30:621–628. doi: 10.1111/j.1348-0421.1986.tb02989.x. [DOI] [PubMed] [Google Scholar]
- 169.Hannan S. Transfer of antibiotic resistance by transformation with eDNA within oral biofilms. FEMS Immunol Med Microbiol. 2010;59:345–349. doi: 10.1111/j.1574-695X.2010.00661.x. [DOI] [PubMed] [Google Scholar]
- 170.Mao D. Persistence of extracellular DNA in river sediment facilitates antibiotic resistance gene propagation. Environ Sci Technol. 2013;48:71–78. doi: 10.1021/es404280v. [DOI] [PubMed] [Google Scholar]
- 171.Mulcahy H., Charron-Mazenod L., Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chiang W.-C. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2013;57:2352–2361. doi: 10.1128/AAC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Arciola C.R. Antibiotic resistance in exopolysaccharide-forming Staphylococcus epidermidis clinical isolates from orthopaedic implant infections. Biomaterials. 2005;26:6530–6535. doi: 10.1016/j.biomaterials.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 174.Pier G.B., Coleman F., Grout M., Franklin M., Ohman D.E. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect Immun. 2001;69:1895–1901. doi: 10.1128/IAI.69.3.1895-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Leid J.G. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-γ-mediated macrophage killing. J Immunol. 2005;175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- 176.Geisinger E., Isberg R.R. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.D. J. Hassett et al. (Taylor & Francis, 2003).
- 178.Andreoli I. Molecular phylogeny of unculturable Karyorelictea (Alveolata, Ciliophora) Zool Scr. 2009;38:651–662. [Google Scholar]
- 179.Xie L. Profiling the metatranscriptome of the protistan community in Coptotermes formosanus with emphasis on the lignocellulolytic system. Genomics. 2012;99:246–255. doi: 10.1016/j.ygeno.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 180.Costerton J.W. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 181.Harrison J.J., Ceri H., Turner R.J. Multimetal resistance and tolerance in microbial biofilms. Nat Rev Microbiol. 2007;5:928. doi: 10.1038/nrmicro1774. [DOI] [PubMed] [Google Scholar]
- 182.Frederix M. Mutation of praR in Rhizobium leguminosarum enhances root biofilms, improving nodulation competitiveness by increased expression of attachment proteins. Mol Microbiol. 2014;93:464–478. doi: 10.1111/mmi.12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nivens D.E., Ohman D.E., Williams J., Franklin M.J. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J Bacteriol. 2001;183:1047–1057. doi: 10.1128/JB.183.3.1047-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ma L., Jackson K.D., Landry R.M., Parsek M.R., Wozniak D.J. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol. 2006;188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chu F. A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;68:1117–1127. doi: 10.1111/j.1365-2958.2008.06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ma L. Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post‐transcriptionally regulated. Environ Microbiol. 2012;14:1995–2005. doi: 10.1111/j.1462-2920.2012.02753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bao J. Programming a biofilm-mediated multienzyme-assembly-cascade system for the biocatalytic production of glucosamine from chitin. J Agric Food Chem. 2018;66:8061–8068. doi: 10.1021/acs.jafc.8b02142. [DOI] [PubMed] [Google Scholar]
- 188.Miller M.B., Bassler B.L. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 189.Shih P.-C., Huang C.-T. Effects of quorum-sensing deficiency on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. J Antimicrob Chemother. 2002;49:309–314. doi: 10.1093/jac/49.2.309. [DOI] [PubMed] [Google Scholar]
- 190.Hennig S., Rödel G., Ostermann K. Artificial cell-cell communication as an emerging tool in synthetic biology applications. J Biol Eng. 2015;9:13. doi: 10.1186/s13036-015-0011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dean S.N., Chung M.-C., van Hoek M.L. Burkholderia diffusible signal factor signals to Francisella novicida to disperse biofilm and increase siderophore production. Appl Environ Microbiol. 2015;81:7057–7066. doi: 10.1128/AEM.02165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.He X., Wang S. DNA consensus sequence motif for binding response regulator PhoP, a virulence regulator of Mycobacterium tuberculosis. Biochemistry. 2014;53:8008–8020. doi: 10.1021/bi501019u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Allen R.C., McNally L., Popat R., Brown S.P. Quorum sensing protects bacterial co-operation from exploitation by cheats. ISME J. 2016;10:1706. doi: 10.1038/ismej.2015.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhou L., Zhang L.-H., Cámara M., He Y.-W. The DSF family of quorum sensing signals: diversity, biosynthesis, and turnover. Trends Microbiol. 2017;25:293–303. doi: 10.1016/j.tim.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 195.Liang H., Li L., Kong W., Shen L., Duan K. Identification of a novel regulator of the quorum-sensing systems in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2009;293:196–204. doi: 10.1111/j.1574-6968.2009.01544.x. [DOI] [PubMed] [Google Scholar]
- 196.McGrath S., Wade D.S., Pesci E.C. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS) FEMS Microbiol Lett. 2004;230:27–34. doi: 10.1016/S0378-1097(03)00849-8. [DOI] [PubMed] [Google Scholar]
- 197.McKnight S.L., Iglewski B.H., Pesci E.C. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 2000;182:2702–2708. doi: 10.1128/jb.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Schuster M., Greenberg E.P. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 199.Dong Y.-H. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature. 2001;411:813. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- 200.Van Schaik E.J. DNA binding: a novel function of Pseudomonas aeruginosa type IV pili. J Bacteriol. 2005;187:1455–1464. doi: 10.1128/JB.187.4.1455-1464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Haagensen J.A., Hansen S.K., Christensen B.B., Pamp S.J., Molin S. Development of spatial distribution patterns by biofilm cells. Appl Environ Microbiol. 2015;81:6120–6128. doi: 10.1128/AEM.01614-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Anwar H., Strap J., Costerton J. Establishment of aging biofilms: possible mechanism of bacterial resistance to antimicrobial therapy. Antimicrob Agents Chemother. 1992;36:1347. doi: 10.1128/aac.36.7.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.D'Acunto B., Frunzo L., Klapper I., Mattei M. Modeling multispecies biofilms including new bacterial species invasion. Math Biosci. 2015;259:20–26. doi: 10.1016/j.mbs.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 204.Chang W.-S., Halverson L.J. Reduced water availability influences the dynamics, development, and ultrastructural properties of Pseudomonas putida biofilms. J Bacteriol. 2003;185:6199–6204. doi: 10.1128/JB.185.20.6199-6204.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Smith W.P. Cell morphology drives spatial patterning in microbial communities. Proc Natl Acad Sci. 2017;114:E280–E286. doi: 10.1073/pnas.1613007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Beroz F. Verticalization of bacterial biofilms. Nat Phys. 2018;14:954. doi: 10.1038/s41567-018-0170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Caccamo P.D., Brun Y.V. The molecular basis of noncanonical bacterial morphology. Trends Microbiol. 2018;26:191–208. doi: 10.1016/j.tim.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Brown J.R. In silico modeling of biofilm formation by nontypeable Haemophilus influenzae in vivo. mSphere. 2019;4 doi: 10.1128/mSphere.00254-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hermanowicz S., Schindler U., Wilderer P. Fractal structure of biofilms: new tools for investigation of morphology. Water Sci Technol. 1995;32:99–105. [Google Scholar]
- 210.Dewey T.G. Fractal analysis of proton exchange kinetics in lysozyme. Proc Natl Acad Sci. 1994;91:12101–12104. doi: 10.1073/pnas.91.25.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Losa G.A. Fractals in biology and medicine. Rev Cell Biol Mol Med. 2006 [Google Scholar]
- 212.Zahid W., Ganczarczyk J. A technique for a characterization of RBC biofilm surface. Water Res. 1994;28:2229–2231. [Google Scholar]
- 213.Vo G.D., Heys J. Biofilm deformation in response to fluid flow in capillaries. Biotechnol Bioeng. 2011;108:1893–1899. doi: 10.1002/bit.23139. [DOI] [PubMed] [Google Scholar]
- 214.Artyushkova K. Relationship between surface chemistry, biofilm structure, and electron transfer in Shewanella anodes. Biointerphases. 2015;10 doi: 10.1116/1.4913783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Byrne E., Dzul S., Solomon M., Younger J., Bortz D.M. Postfragmentation density function for bacterial aggregates in laminar flow. Phys Rev E. 2011;83 doi: 10.1103/PhysRevE.83.041911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Van De Staey G. 2016. What is the role of activated sludge flocs in membrane fouling in membrane bioreactors (MBRs)? [Google Scholar]
- 217.Shen Y. Role of biofilm roughness and hydrodynamic conditions in Legionella pneumophila adhesion to and detachment from simulated drinking water biofilms. Environ Sci Technol. 2015;49:4274–4282. doi: 10.1021/es505842v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tsagkari E., Sloan W. Turbulence accelerates the growth of drinking water biofilms. Bioproc Biosyst Eng. 2018;41:757–770. doi: 10.1007/s00449-018-1909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Farrell F.D., Gralka M., Hallatschek O., Waclaw B. Mechanical interactions in bacterial colonies and the surfing probability of beneficial mutations. J R Soc Interface. 2017;14:20170073. doi: 10.1098/rsif.2017.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Frost I. Cooperation, competition and antibiotic resistance in bacterial colonies. ISME J. 2018;1 doi: 10.1038/s41396-018-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Heydorn A. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 222.Kesel S. Direct comparison of physical properties of Bacillus subtilis NCIB 3610 and B-1 biofilms. Appl Environ Microbiol. 2016;82:2424–2432. doi: 10.1128/AEM.03957-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang V.B. Engineering PQS biosynthesis pathway for enhancement of bioelectricity production in Pseudomonas aeruginosa microbial fuel cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bernstein H.C., Paulson S.D., Carlson R.P. Synthetic Escherichia coli consortia engineered for syntrophy demonstrate enhanced biomass productivity. J Biotechnol. 2012;157:159–166. doi: 10.1016/j.jbiotec.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Benedetti I., de Lorenzo V., Nikel P.I. Genetic programming of catalytic Pseudomonas putida biofilms for boosting biodegradation of haloalkanes. Metab Eng. 2016;33:109–118. doi: 10.1016/j.ymben.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 226.Rahman M.S., Ano T., Shoda M. Biofilm fermentation of iturin A by a recombinant strain of Bacillus subtilis 168. J Biotechnol. 2007;127:503–507. doi: 10.1016/j.jbiotec.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 227.Perni S., Hackett L., Goss R.J., Simmons M.J., Overton T.W. Optimisation of engineered Escherichia coli biofilms for enzymatic biosynthesis of L-halotryptophans. Amb Express. 2013;3:66. doi: 10.1186/2191-0855-3-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Schmutzler K., Kupitz K., Schmid A., Buehler K. Hyperadherence of Pseudomonas taiwanensis VLB120ΔC increases productivity of (S)‐styrene oxide formation. Microbial Biotechnol. 2017;10:735–744. doi: 10.1111/1751-7915.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]