Abstract
High-throughput screens in Drosophila cell lines have led to discovery of conserved gene functions related to signal transduction, host-pathogen interactions, ion transport, and more. CRISPR/Cas9 technology has opened the door to new types of large-scale cell-based screens. Whereas arrayed-format screens require liquid handling automation and assay miniaturization, pooled-format screens, in which reagents are introduced at random and in bulk, can be done in a standard lab setting. We provide a detailed protocol for conducting and evaluating genome-wide CRISPR single guide RNA (sgRNA) pooled screens in Drosophila S2R+ cultured cells. Specifically, we provide step-by-step instruction for library design and production, optimization of cytotoxin-based selection assays, genome-scale screening, and data analysis. This type of project takes about three months to complete. Results obtained in this type of screen can be used in follow-up studies performed in vivo in Drosophila, mammalian cells, and/or other systems.
Keywords: CRISPR, Drosophila, functional genomics, high-throughput screening, pooled-format screening
INTRODUCTION
Drosophila melanogaster is a well-established model for large-scale in vivo and cell-based screens. The relatively low-redundancy Drosophila genome, uniformity of cultured cells, and availability of transcriptomics and other large-scale data make fly cultured cells particularly useful for large-scale functional genomics studies. Impactful findings from large-scale screens in Drosophila cells done using RNAi include identification of a mediator of hypercapnic immune suppression (Helenius et al., 2016), new drug targets for development of therapeutic treatments for the proliferative disorder tuberous sclerosis complex (Housden et al., 2015), genes relevant to nucleolar size (Neumuller et al., 2013), genes relevant to host-pathogen interactions (e.g. Akimana et al., 2010; Sessions et al., 2009), and conserved ion transporters e.g. LETM1 (Jiang et al., 2009) and ORAI1/CRACM (Feske et al., 2006; Vig et al., 2006; Zhang et al., 2006) (additional examples at https://fgr.hms.harvard.edu/publications-data). These findings underscore the power of the Drosophila cultured cell system for gene function discovery. Nevertheless, the arrayed-format RNAi screening approach has limitations, including a need for specialized equipment and the potential for reagent-specific off-target effects.
Pooled-format screens using CRISPR technology provide an attractive alternative. Pooled-format approaches, in which reagents are introduced in bulk and at random, eliminate the need for liquid handling automation equipment. Moreover, CRISPR-Cas9 activity can result in knock out, rather than knockdown of a gene, and is associated with lower off-target potential. Pooled-format CRISPR screens was reported in 2014 as an approach to mammalian cell screening (Shalem et al., 2014; Wang et al., 2014). With CRISPR pooled-format cell screening technology, tens of thousands of CRISPR reagents are delivered to millions of isogenic cells simultaneously such that each cell receives a single reagent targeting a single locus. This is followed by outgrowth of the cells in the presence or absence of selection, such as with a drug (protocols include Joung et al., 2017; Piccioni et al., 2018). Technical comparisons of CRISPR and RNAi-based perturbations in mammalian cells have concluded that the approaches are complementary in large-scale screens or that CRISPR outperforms RNAi (Evers et al., 2016; Morgens et al., 2016).
We have established CRISPR pooled-format screening as an additional functional genomics approach for interrogation of gene function and drug target discover using Drosophila cultured cells (Viswanatha et al., 2018). In mammalian cells, introduction of the sgRNAs relies on lentiviral transduction and integration, a step that makes it possible to later track what sgRNAs are associated with positive results. We used bacterial site-specific recombination-based plasmid transfection to deliver comparably large CRISPR reagent libraries to Drosophila S2R+ cells (Fig. 1). We showed that pooled screening resulted in highly accurate identification of ~1300 cell-essential genes. Further, we applied the procedure to gene-drug interaction screening (Viswanatha et al., 2018). This method has also been used to identify new components of the insect molting hormone pathway (Okamoto et al., 2018). The approach is extensible to other Drosophila cell lines. The Drosophila Genomics Resource Center (DGRC) in Bloomington, IN, maintains a large catalog of Drosophila cell lines (https://dgrc.bio.indiana.edu).
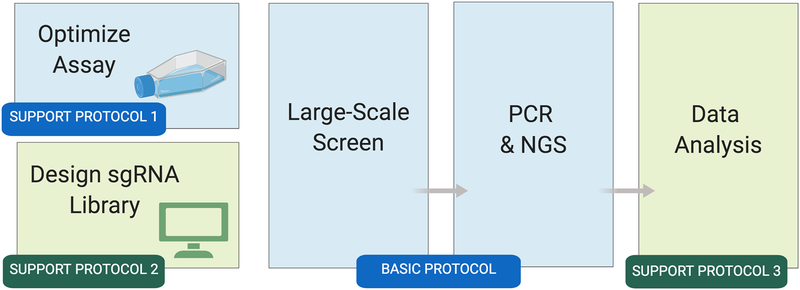
Figure 1: Overview of the Drosophila cell CRISPR pooled screen workflow.
Wet-bench activities are shown in blue; bioinformatics steps are shown in green. Once the cell-based assay is optimized (Support Protocol 1) and a library is obtained or built (Support Protocol 2), the large-scale screen can begin. The Basic Protocol takes about 45 days to complete. After the screen, the resulting next-generation sequencing data are analyzed to identify screen ‘hits’ (positive results) at the reagent and gene levels (Support Protocol 3).
Here, we provide a detailed protocol for conducting and evaluating Drosophila pooled CRISPR screens performed in the presence of a cytotoxin (i.e., cytotoxin sensitivity and resistance screen). An overview is shown in Fig. 1. Prior to screening, an appropriate Cas9-positive cell line should be obtained from the DGRC or established, the cell assay should be optimized (Support Protocol 1) and a large-scale sgRNA library in an appropriate vector should be obtained or generated (Support Protocol 2). A cell-line tailored for the assay (Viswanatha et al., 2018) is available at the DGRC (catalog #268, https://dgrc.bio.indiana.edu). The screen itself is comprised of three main phases: library transfection, library integration, and selection (Basic Protocol). Completion of these phases takes about 45 days. At the end of the screen period, sgRNA reagents present at the start and end of the assay are amplified, the resulting PCR fragments are purified, and the PCR fragments are subjected to next-generation sequencing (NGS). Screen data analyses include identification of sgRNAs that are enriched or depleted in cytotoxin treatment conditions vs. control conditions, followed by gene-level analyses (Support Protocol 3).
The screen can be done in the absence or presence of a cytotoxin. In the absence of a cytotoxin, a simple outgrowth assay is used to separate sgRNAs that confer cell lethality when knocked out (i.e., sgRNAs that target essential genes) from sgRNAs targeting non-essential genes. Such a screen can be performed to establish the set of essential genes in a given cell line (Viswanatha et al., 2018) or can be performed in parallel in wild-type and mutant cells, for example to identify genes that are synthetic lethal with knockout of a given gene. In the presence of a cytotoxin, CRISPR pooled screening can be used to identify sgRNAs that target genes for which knockout confers sensitivity or resistance to the cytotoxic treatment, which could be a cytotoxic drug such as a chemotherapeutic (e.g. see Viswanatha et al., 2018), a biomolecule that induces cell death (e.g. see Okamoto et al., 2018), an environmental toxin, an infectious pathogen, etc. Additional types of assays are compatible with the CRISPR pooled format, as discussed in the Commentary section.
STRATEGIC PLANNING
Getting Started
The Basic Protocol takes about 45 days to complete. Prior to this, the cells must be expanded to ensure that there are enough cells for the screen. A variety of simple tests can be used to make sure that the cells have the appropriate attributes. For example, integration can be tested using the empty pLib6.4 vector (as demonstrated in Fig. 1 of Viswanatha et al. (2018)). In addition, Cas9 activity can be confirmed, such as using by transfection with an sgRNA targeting mCherry followed by FACS (if using the mCherry-positive S2R+-MT::Cas9 cell line) or using a T7 endonuclease activity to assess cutting at a given sgRNA-targeted locus.
Set-Up for sgRNA Library Transfection into Cas9-Expressing S2R+ Cells
The amount of CRISPR library to transfect depends on the number of sgRNAs in the library, the desired coverage, and the integration efficiency. You should transfect the library in units of 6-well dish wells to contain potential contamination. Determine the number of wells needed using the following formula:
At least 400X coverage of the library is recommended in the final integrated pool. Integration efficiency is usually greater than 30%. The optimal density of S2R+ cells at the time of transfection is 3×106 per well. Using these parameters, for the genome wide Dsg_group1+2+3 library containing 86,364 sgRNAs (Viswanatha et al., 2018), ~40 wells must be transfected to cover the library, while in the Dsg_group1+2 library containing 26,566 sgRNAs, (Okamoto et al., 2018), ~11 wells must be transfected to cover the library.
Integration efficiency depends on transfection efficiency, which varies between experiments. It is therefore necessary to empirically evaluate integration efficiency within each experiment. In our hands, transfection efficiency (with Effectene and measured by flow cytometry) following transfection with pLib6.4 is >50% and integration efficiency is ~30%. Evaluating integration efficiency can be done by including the following controls during library transfection:
| pBS130 amount | CRISPR library amount | Passage in Puro? | |
|---|---|---|---|
| Well 1 | 0.2 g | 0.2 μg | No |
| Well 2 | - | 0.4 μg | No |
| Well 3-Well n | 0.2 μg | 0.2 μg | Yes |
Integration efficiency is approximately equal to the number of stable GFP-positive cells derived from Well 1 minus Well 2. Note that this value can only be determined after transiently delivered (non-integrated) plasmids have been effectively diluted out of the population, which takes approximately 3 weeks. We recommend evaluating after 3 weeks of passaging using a BD LSR II flow cytometer. If integration efficiency is below 30%, discard CRISPR cell pool and repeat procedure.
Thinking Ahead
Large-scale screens typically result in identification of a large number of ‘hits’ or positive results. Often, these can be subcategorized using secondary cell-based or in vivo assays (a) to determine detailed Drosophila cell phenotypes, (b) to explore in vivo relevance in Drosophila, either in general or in a specific tissue, stage or cell type, and/or (c) to understand the relevance of the finding to mammalian orthologs, e.g., by asking if a similar phenotype is observed for perturbation of orthologous genes in mammalian cultured cells. Secondary assays should be considered and developed prior to the large-scale cell-based screen. Knowing what follow-up assays you will do after the screen will be relevant to screen data analysis, as analysis can be adjusted with end goals in mind. If the follow-up assay is particularly time- or resource-intensive, for example, you might only want to test high-confidence hits. Thus, you would set a high statistical cut-off and/or look for other types of supporting evidence in the published literature or other datasets at in a screen data integration stage. If, conversely, you have one or more relatively easy and inexpensive secondary assay that can be applied to a large number of primary screen hits, you might ‘cast a broad net’ by setting a lower statistical threshold, and plan to test not only high-confidence hits but also moderate and low-confidence hits from the primary screen in the secondary assay.
BASIC PROTOCOL: Pooled-format screening with Cas9-expressing Drosophila S2R+ cells in the presence of a cytotoxin
INTRODUCTION
The basic protocol (Fig. 2) assumes that the goal of the screen is to identify genes for which knockout confers sensitivity or resistance to a cytotoxic treatment. Support Protocol 1 describes how to identify an optimal concentration of a cytotoxin for screening. Additional potential alternative protocols include a “drop-out” screen to identify genes that are essential in a given cell line, or separation via flow cytometry-based cell sorting based on a fluorescent marker or antibody. In addition, the basic protocol can be performed in parallel with both wild-type and mutant cells, e.g., to use a drop-out approach to identify synthetic lethal interactions detected in mutant but not wild-type cells (see Commentary for additional discussion on assay types).
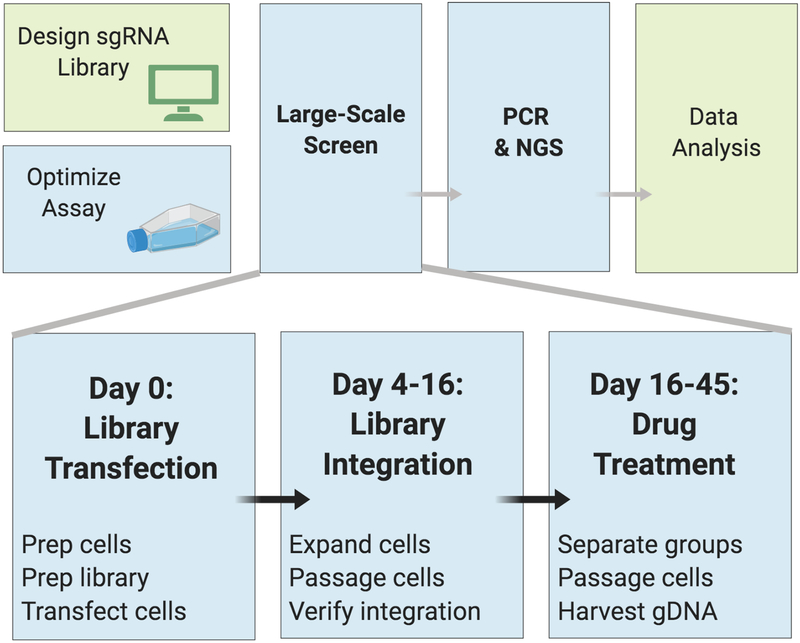
Figure 2: Overview of the Basic Protocol for Drosophila cell CRISPR pooled screening.
Workflow shown for a selection screen using a cytotoxic treatment at day 16. For a ‘drop out’ screen to identify genes that are essential, leave out drug selection and passage the cells without drug.
This basic protocol also assumes use of a Cas9+ S2R+ cell line with an attP integration site together with an existing sgRNA library, such as the library described in Viswanatha et al. (2018) and available from Addgene Viswanatha et al. (2018) and available from Addgene (catalog #’s 134582–134584). The specific strategy for integration-based insertion of sgRNAs in the library into cell genomes is shown in Fig. 3. New libraries can be developed as described in Support Protocol 2. Depending on the vector and library design, some modifications might be necessary to the protocol for data analysis, which is presented as Support Protocol 3.
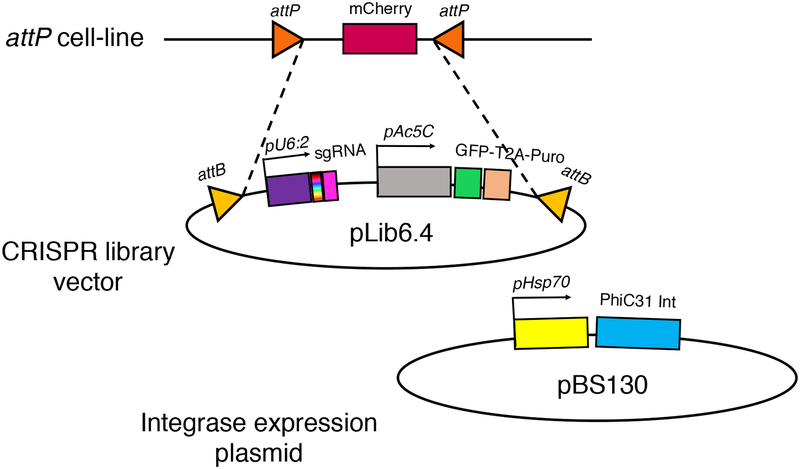
Figure 3. Plasmid-based library integration strategy for pooled CRISPR screening in Drosophila S2+ cells.
Co-transfection of cells with pLib6.4 mixture containing diverse sgRNAs and pBS130 PhiC13 Integrase vector causes a portion of transfected cells to integrate the sgRNA, GFP, and puromycin-resistance expression cassette.
MATERIALS LIST
S2R+-MT::Cas9 cultured cells, which are available from the Drosophila Genome Resource Center (DGRC cell stock #268)
Screen-specific cytotoxin (Support Protocol 1)
CRISPR plasmid sgRNA library for Drosophila gene knockout, such as the library described in Viswanatha et al. (2018) (Addgene catalog #’s 134582–134584), or developed as described in Support Protocol 2
Drosophila cell culture medium (Reagents and Solutions)
Qiagen Effectene transfection reagent (catalog #301427)
pBS130, pHSP70-PhiC31 integrase (Addgene #26290)
6-well tissue-culture treated dishes (Corning # 3516)
10-cm tissue-culture treated dishes (Corning # 430164)
Qiagen Gel Extraction Kit (Catalog # 28704)
Corning Cell-Lifter 3008 (Catalog # 3008)
250 mL media bottle (Nalgene # 2019–0250)
Plastic wrap, such as Reynolds #900 Food Service Film (Careforde Scientific # G1682075)
Zymo® gDNA Miniprep Kit (D3025)
PCR1 and PCR2 primers for sgRNA detection from cells (Table 1)
Table 1:
PCR primers used in the basic and support protocols.
| Primer name | Primer sequence |
|---|---|
|
Example
PCR1fwd* |
5’-CTTTCCCTACACGACGCTCTTCCGATCTNNNTAGCTTgttttcctcaatacttcgttcg-3’ Partial Read1 Sequence Fingerprint Barcode Drosophila U6:2 promoter sequence |
| PCR1rev | 5’-tttgtgtttttagaatatagaattgcatgctgggtacctc-3’ Complementary to pLib6.4 vector |
| PCR2fwd | 5’- AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3’ P5 sequence Spacer, required for NextSeq but not for HiSeq platforms Complete Read1 sequence |
| PCR2rev | 5’-CAAGCAGAAGACGGCATACGAGATtttgtgtttttagaatatagaattgcatgctgggtacctc-3’ Reverse complement of P7 Complementary to PCR1 |
Note: each PCR1 primer varies based on the length of the random nucleotide fingerprint region and the identity of the 6-bp barcode. In a typical experiment, it is necessary to order at least 4 distinguishable PCR1 primers.
Qubit fluorometer
Kapa Biosystems Library Quantification kit (#KK4835)
Illumina Next Seq 500 instrument (e.g. as accessed through a core facility)
Illumina Next Seq 500 v2 75-cycle High Output cartridge
Transfection of Library – Day 0
-
1
Determine the number of cells needed to obtain appropriate integration level for a given CRISRP plasmid library (see Strategic Planning).
-
2
Grow cells in a T-75 flask and verify they are doubling approximately once per day prior to transfection.
-
3
Dislodge by tapping flask and pipetting. To eliminate clumps and generate a single-cell suspension, withdraw medium and cells into a Pasteur pipette and press the tip of the pipette against a flat surface, such as the side of the flask, to narrow the opening. Dispense the medium with cells through narrow opening to generate a spray of cells. Repeat once or twice and confirm that most cells are now in a single-cell suspension.
-
4
Count cells with a Hemocytometer and adjust concentration to 1.5×106 cells per mL.
-
5
Dispense 2 mL of cell suspension per well into a 6-well dish (Corning # 3516) and allow to attach.
-
6
Prepare transfection mixtures according to the Qiagen Effectene Handbook, using the base protocol. Briefly, first prepare 0.4 μg DNA per well and then add 3.2 μL Enhancer solution per well. Vortex and spin down. Incubate 5 min. Add 10 μL Effectene reagent. Vortex. Incubate 15 min. Add complexes dropwise to cells.
-
7
Wrap stacks of 6-well dishes in plastic wrap to prevent evaporation and incubate at 25°C for 4 days.
Library Expansion and Maintenance – Day 4–16
-
8
Expand the transfected cells from 6-well dishes to 10cm plates. First, dislodge the cells from the 6-well plates by lifting up cells from each well individually using a Corning Cell-Lifter 3008 with the flat-side facing up. Eliminate clumps (see step 3) Transfer cells and medium together into a 10 cm dish containing 10 mL of Puromycin selection media. Stack and completely seal 10 cm dishes in plastic wrap and incubate at 25°C for 4 days.
-
9
CRISPR screen pool maintenance. Determine the number of cells needed in each passage to adequately maintain library diversity (see Strategic Planning). Dislodge cells from each dish using cell lifter. Dislodge cells from all 10 cm dishes and combine cell suspension into a single 250 mL media bottle. Eliminate clumps (see step 3). Invert bottle containing combined cell suspension 2–4 times to mix. Count cells and dispense the number of cells needed at a maximum density of 1×107 cells per 10 cm dish. Puromycin selection media.
-
10
Repeat passaging procedure every 4 days until Day 16.
Cytotoxin Treatment and Outgrowth – Day 16-end of screen (~Day 45)
-
11
From the remaining cell suspension, distribute integrated cell population into a minimum of 4 groups: 2 groups as untreated controls; 2 groups treated with the cytotoxin. We recommend continuing screen at 400X coverage (at least 400 cells per sgRNA per replicate). Note that the number of cells per replicate is critical to prevent bottlenecking the population during passages (see Strategic Planning). For example, for a library containing ~100,000 sgRNAs, at least four 10 cm dishes are needed per replicate. Plate 1×107 cells per 10 cm dish and allow to grow for 4–5 days.
-
12
Dislodge cells and combine together cells from the same replicates in one bottle. Invert 2–4 times to mix. Distribute 1×107 cells per dish to the same number of 10 cm dishes. This procedure maintains 400X coverage throughout the screen. Record cell density after each passage for each group, as this data can be used to verify that the cytotoxin caused expected reduction of cell proliferation.
-
13
Continue high-coverage passages until you reach the planned end-date for exposure to the cytotoxin. It is critical to maintain as many cells as possible during the passages to avoid bottlenecking the population (at least 400X coverage, see above). We recommend passaging cells for at least 30 days in selective medium for the initial screen.
-
14
Once the desired cytotoxin exposure period is reached, pellet cells in 50 mL aliquots. Aspirate supernatant and store pellets at −80°C to await genomic DNA (gDNA) extraction.
Sample Preparation for Next-Generation Sequencing (NGS)
The goal of this step is to extract the genomic DNA (gDNA) from the cells, then amplify the sgRNAs that have integrated into the gDNA. The specific sgRNAs present in gDNA from experimental and control cells (or experimental and time 0) will later be identified using NGS and then compared to identify positive results. A barcode strategy is applied during amplification of sgRNAs and used in analysis steps (Fig. 4).
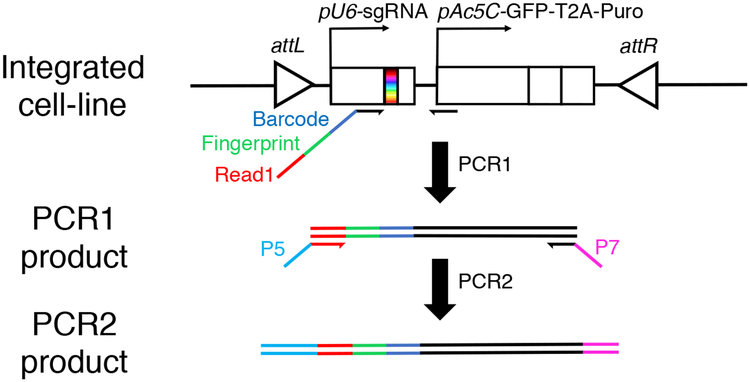
Figure 4. PCR strategy for amplifying sgRNA sequences from cell pools.
To amplify sgRNAs expression cassette from S2R+ cells, a 2-step PCR strategy is employed. In PCR1, barcoded primers are used to attach sample-barcodes to sgRNA sequences in each cell pool. The PCR1 forward primer also contains a fingerprint (or stagger) sequence that allows for cluster distinction during Illumina sequencing and a primer binding site for PCR2, which is a portion of the Illumina Read1 primer binding sequence. In PCR2, a second round of tailored PCR adds Illumina P5 and P7 sites for Illumina Flow Cell attachment.
gDNA Extraction
-
15
Thaw pelleted cells from the final passage (Day 45) on ice for about 10–15 minutes or until the pellet liquifies. To reduce thawing time, remove cells from ice and sit at room temperature.
-
16
Using Zymo® gDNA Miniprep Kit (D3025) set up 6 collection tubes with collection columns per pellet.
-
17
When pellet is thawed, resuspend in 5mL of gDNA Lysis Buffer by triturating and letting sit for 5 minutes.
-
18
Dispense in each of the 6 collection columns and then centrifuge at room temperature for 30 seconds at 10,000 × g.
-
19
Follow the rest of the manufacturer’s protocol
-
20
Pool each of the 6 Eppendorf tubes containing the DNA eluted in Elution Buffer coming from a single pellet.
PCR1: Amplification of sgRNAs from S2R+ cell-line genome
PCR mix and amplification reaction conditions appear below.
| PCR1 mix | |
|---|---|
| Component | Volume (uL) |
| Nuclease-free water | 4 |
| 5X Phusion HF Buffer | 4 |
| 2.5 mM dNTPs | 1.6 |
| 100 μM PCR1fwd | 0.1 |
| 100 μM PCR1rev | 0.1 |
| Template gDNA | 10 |
| Phusion DNA Polymerase | 0.2 |
| TOTAL VOLUME | 20 |
| Step | Temperature | Time |
|---|---|---|
| Initial Denaturation | 98°C | 2 min |
| 72°C | 30 s | |
| Elongation | 72°C | 10 min |
| Hold | 4°C |
-
21
Verify gDNA integrity and determine amount of PCR needed to provide appropriate coverage. Prepare 2-fold serial dilutions of gDNA preps and perform test amplifications using 2X Phusion PCR mastermix. Resolve each reaction on a 2% agarose gel (example appears below, Fig. 5). Identify the highest concentration of gDNA that still permits the reaction to yield a single band around 280–300 bp. Using a Qubit fluorometer, determine the concentration of DNA in this sample. You now have enough information to calculate the minimum required PCR volume for this sample. Based on the assumption that the diploid mammalian genome weighs 6.6 pg (Chen et al., 2015), the tetraploid S2R+ genome, which is ~11 times smaller, should weigh 0.60 pg. PCRs should contain at least 400 X coverage (Wang et al., 2014). For instance, using these parameters, for the genome wide Dsg_group1+2+3 library containing 86,364 sgRNAs (Viswanatha et al., 2018), a minimum of 21 μg of gDNA is needed. Typically, >700 μL of PCR are needed per sample.
-
22
Using a multichannel pipette, aliquot the pooled reaction in 20 μL aliquots into a 96-well plate and perform scaled up PCR to optimize coverage as required. After cycling, pool the reaction together and run approximately 50 μL of the pooled reaction on a 2% agarose SYBR Safe DNA gel.
-
23
Cleanly excise the 280–300 bp band.
-
24
Gel-purify DNA using Qiagen Gel Extraction Kit and proceed to PCR2.
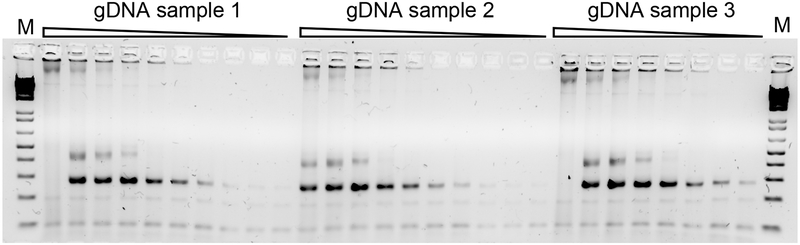
Figure 5. Small-scale verification of genomic DNA (gDNA) integrity.
After gDNA extraction, we perform 2-fold serial dilutions of gDNA to determine highest concentration of gDNA that will still allow detection of sgRNA cassette by PCR. In the example shown, PCRs containing indicated gDNA prep are serially diluted and then each added to 2X Phusion PCR mastermix and subjected to 23-cycles of PCR before being resolved on 2% agarose SYBR Safe DNA gel (image inverted). Expect a 280–300-bp band depending on PCR1 primer used. Marker, 1kb plus DNA Ladder (Thermo #10787018). Result indicates that gDNA samples 1 and 3 must be 2-fold diluted prior to PCR to detect the sgRNA cassette, whereas dilution is not necessary for gDNA sample 2.
PCR2: Addition of P5, Read1, and P7 to PCR1
PCR mix and amplification reaction conditions appear below.
| PCR2 mix | |
|---|---|
| Component | Volume (μL) |
| Nuclease-free water | (variable) |
| 5X Phusion HF or GC Buffer | 40 |
| 2.5 mM dNTPs | 16 |
| 100 μM PCR2fwd | 1 |
| 100 μM PCR2rev | 1 |
| Template DNA | (variable, 40 ng) |
| Phusion DNA Polymerase | 2 |
| TOTAL VOLUME | 200 |
| Step | Temperature | Time |
|---|---|---|
| Initial Denaturation | 98°C | 30 s |
| 72°C | 1 min | |
| Elongation | 72°C | 1 min |
| Hold | 4°C |
-
25
Attach the barcodes to each reaction individually by performing a second PCR using primers that attach P5, Read1, and P7 sequences required for downstream next-generation sequencing (NGS). To maintain representation, use 40 ng of purified PCR1 as template in each reaction as indicated. After cycling, pool the reaction together and run approximately half on a 2% agarose SYBR™ Safe™ gel.
-
26
Cleanly excise the ~350 bp band.
-
27
Gel-purify DNA using Qiagen Gel Extraction Kit and proceed to sample pooling for NGS.
Pooling Samples for Next-Generation Sequencing (NGS)
-
28
Measure the concentration of each PCR2 product using Qubit fluorometer. Using starting library diversity, derive the sgRNA concentration and normalize all samples by sgRNA concentration.
-
29
Pool together equal volumes of all samples.
-
30
Use Kapa Biosystems Library Quantification kit (#KK4835) or equivalent to more accurately determine library concentration.
-
31
Load pooled PCR2 directly on an Illumina Next Seq 500 v2 75-cycle High Output cartridge. Request 91 cycles from manufacturer-provided Read1 primer without Phi-X spike-in and without additional indexing reads.
-
32
Collect data as raw FASTQ file and for analysis, proceed to Support Protocol 3.
SUPPORT PROTOCOL 1: Optimization of the concentration of a cytotoxin for Drosophila cell screening
INTRODUCTION
The Basic Protocol describes a method for screening with a specific and predefined concentration of a cytotoxin. Optimization of the cytotoxic treatment is aimed at providing a robust assay (e.g., concentration is not too low) while at the same time giving cells a chance to survive (e.g., concentration is not too high). The protocol uses a readout of total ATP levels as a proxy for cell number/viability. Tests are first performed in a microwell plate to determine an optimal range, then performed in flasks to more closely mimic screen conditions.
MATERIALS LIST
S2R+-MT::Cas9, catalog #268 at DGRC (https://dgrc.bio.indiana.edu/)
200 ng/mL Hygromycin medium
96-well plate (Corning # 3799)
Cell Titer-Glo (Promega # G7573)
Cytotoxin; example cytotoxins: Rapamycin (LC Laboratories # R-5000), Trametinib (Selleck Chemicals # S2673), 20-Hydroxecdysone (Sigma # H5142)
Broad-Range Optimization Using 96-Well Plates
-
1
Obtain S2R+-MT::Cas9, Stock #268 from DGRC. Upon arrival, follow DGRC recommendations for thawing cells (see https://dgrc.bio.indiana.edu/include/file/ThawingCells.pdf), and passage for ~ 2 weeks in 200 ng/mL Hygromycin medium until cells are actively growing and doubling approximately once per day.
-
2
Put cells in media in all wells but use only non-edge wells as experimental wells to avoid the potential impact of ‘edge effects’ due to differences in well micro-environments (see example, Fig. 6). Prepare 10-fold serial dilutions of desired cytotoxin(s) in a total volume of 50 μL in experimental wells (or 50 μL vehicle control) and add to cells. Perform the assay in a flat-bottom 96-well plate.
-
3
Dislodge actively growing S2R+-MT::Cas9 cells from a T75 flask and adjust concentration to 600,000/mL. Serially dispense 50 μL of cell suspension to each well (30,000 cells/well) bringing the total volume in every well to 100 μL.
-
4
Wrap plate in plastic wrap and incubate at 25°C for 5 days.
-
5
Add 50 μL of Cell Titer Glo assay reagent (Promega) to each well, mix, and measure luciferase activity using a luminometer (plate reader).
-
6
Compare treated samples to vehicle controls and determine a cytotoxin concentration range for which cell growth is impeded but not completely halted. Typically, luminescence readings should be 10–50% of untreated value, indicating partial perturbation in cell growth.
-
7
To further narrow the concentration range, repeat Steps 1–4 with a two-fold dilution around the effective concentration range.
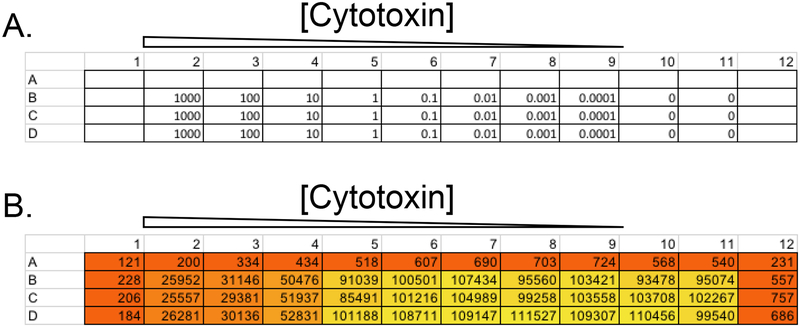
Figure 6. Determining an optimal concentration of a cytotoxin for a selection-based screen.
Shown is an example layout for a 96-well plate of cells used to determine the optimal concentration range of a cytotoxin. (A) Trametinib at indicated concentration (nM) was first added to the plate in 50 μL of growth medium. The same number of cells (50 μL of a cell suspension in fresh growth medium at 2 × 105 cells/mL, or 10,000 cells per well) was then added to each well. (B) Cells were grown for 5 days and then subjected to Cell-Titer Glo assay. Arbitraty luminometry values from corresponding wells shown. 50 nM was ultimately chosen as an optimal concentration for the screen.
Narrow-Range Optimization in T-12.5 Flasks
-
8
In a laminar flow tissue culture hood, dilute cytotoxin to concentrations identified from the 96-well assay 10 mL of growth media. Filter sterilize the cytotoxin-containing media by attaching a 0.22 μm filter and dispensing into a new 15 mL Falcon tube. Store at 4°C for no more than 2 weeks.
-
9
Add 500,000 cells to 2.5 mL of media with cytotoxin per T-12.5 flask and passage 3–5 days depending on growth rate. Count cells and seed 1×107 every passage.
-
10
Identify a cytotoxin concentration that consistently gives ~50% fewer cells on every passage. This should be useful for screening.
SUPPORT PROTOCOL 2: CRISPR sgRNA library design and production for Drosophila cell screening
INTRODUCTION
One option for screening is to use readymade CRISPR sgRNA libraries for Drosophila cell screening as reported in Viswanatha et al. (2018), which are available from Addgene (catalog #’s 134582–134584). Another option is to generate a new custom library with genome-wide or more limited coverage, using the plasmid vector pLib. This support protocol is based on use of Find CRISPR3.
Typically, there are many potential sgRNA target sites per gene. It is important to select the sgRNA designs with high specificity as well as high efficiency. The Find CRISPR tool implemented by DRSC allows you to find sgRNA sites and information such as predicted efficiency score, predicted likelihood of frame-shift mutation, and potential off-target sites (Housden et al., 2015; Ren et al., 2013). An improved version, Find CRISPR3 (https://www.flyrnai.org/crispr3/web/), provides efficiency scores predicted using different algorithms and includes an efficiency score calculated using a machine learning approach (unpublished) based on the genome-wide pooled screen reported in (Viswanatha et al., 2018), and a specificity score calculated based on potential off-target sites and annotation of protein domains. The sgRNAs reported in Find CRISPR3 (v3.0) were designed using the FlyBase reference genome. The tool also provides annotation of genome variants detected in the S2R+ cell line and protein domain annotations. Designs targeting within protein domains can be prioritized, even the in-frame mutations in these regions are more likely to impact the function of the protein (Shi et al., 2015). An overview of a workflow for choosing sgRNAs for inclusion in a library is provided in Mohr et al. (2016).
Find CRISPR3 provides a batch search option. You can search all sgRNA designs for a set of genes, download the list of sgRNAs, then select designs for inclusion in your library based on efficiency scores, specificity scores, SNP annotations, and/or protein domain annotations.
MATERIALS LIST
pLib6.4 plasmid vector (Addgene # 133783)
Phusion polymerase kit (polymerase, dNTPs, buffer)
Plastic pestle (VWR #KT749521–1590)
Qubit fluorometer (Thermo Q33238)
Qubit dsDNA HS Assay Kit (Thermo Q32854)
20% TBE Novex Gel (Thermo EC63152BOX)
O’Range 10 bp marker (Thermo SM1313)
SYBR Safe DNA gel stain (Thermo S33102)
Dark Reader (Bioexpress U-2235–18)
BpiI restriction enzyme (Thermo ER1012)
T4 DNA ligase buffer (New England Biolabs B0202S)
T7 DNA ligase (Enzymatics L6020L)
Plastic pestle (VWR #KT749521–1590)
109-mer oligo pools from Genscript (see protocol for design information)
Spin-X column (Corning CLS8161)
Electrocompetent E. coli such as Ecloni 10GF’ Electrocompetent cells (Novagen)
LB/agar plates with ampicillin
LB/broth with ampicillin
PCR Purification Kit (Qiagen 28106)
Custom sgRNA library design and synthesis
-
1
Use the “Find CRISPR3” batch design tool from DRSC/TRiP Functional Genomics Resources (flyrnai.org/crispr3) to obtain CRISPR sgRNA designs targeting a set of genes that are custom input by the user. You should note the Find CRISPR3 version number and FlyBase release number (Find CRISPR version 3.0 is based on FlyBase genome release r6.24). In choosing sgRNAs, exclude from the library sgRNA designs that target a region with genome variants in S2R+ compared with this reference genome. SNPs for S2R+-pMT::Cas9 cells are indicated in the results output at Find CRISPR (version 3; https://www.flyrnai.org/crispr3/web/). In addition, you can choose to prioritize designs targeting within protein domains, as this may increase knockout efficiency (Shi et al., 2015) (also indicated at Find CRISPR version 3).
-
2
Eliminate and manually replace any sgRNAs containing the U6 terminator (TTTT) or a BbsI or BpiI site (GAAGAC or TGCTTC).
-
3“Dialout” primers are used to retrieve classes of oligos from oligo pools. Append dialout primers, BpiI/BbsI recognition sites, U6:2 promoter sequence, and tracrRNA backbone sequence to each sgRNA sequence as follows:
- 5’-ctataatataggccagctcagctctggggggtGAAGACgcGTCGACTGCCGATTCGATTCGAACGTTTcgGTCTTCgttttatccaggcgaggggctgacaggggaatt-3’
- Dialout 5’
- BpiI/BbsI site
- sgRNA sequence
- Reverse BpiI/BbsI site
- Dialout 3’ (order reverse complement)
-
4
Order the resulting list of sequences as 109-mer oligo pools from a gene synthesis company such as GenScript (www.genescript.com).
Amplification and cloning of sgRNAs into the vector
-
5Use Phusion HF polymerase to amplify the library using the following PCR parameters. Note that you should divide the reaction into ~10 × 20 μL reactions.
Component Volume (uL) ddH20 138 5X Buffer 40 2.5 mM dNTPs 16 100 μm primer Dialout Fwd 1 100 μm primer Dialout Rev 1 Library@50ng/uL 2 Phusion 2 Step Temperature Time Initial Denaturation 98°C 30 s 72°C 1 min Elongation 72°C 1 min Hold 4°C -
6
Gel-purify 109-mer fragments from a 2% agarose gel.
-
7
Use 2 Qiagen Gel Extraction Kit columns to purify 109-mer amplicons. Elute each in 20 μL and pool. Add 10 μL Buffer G and 5 μL BpiI. Digest overnight at 37°C.
-
8
Run a 20% TBE Novex Gel for ~1 hour at 180V along with the O’Range 10 bp marker. Post-stain with Sybrsafe. Visualize in dark reader and cut the 24-nt band representing the double-stranded sgRNA fragments and transfer to a clean Eppendorf tube.
-
9
Using a plastic pestle, crush band into powder and reconstitute in 100 μL of nuclease-free water. Soak pulverized gel fragments with gentle shaking or end-over-end rotation at 37°C overnight.
-
10
Resuspend slurry and transfer to a Spin-X column.
-
11
Spin at maximum speed in a benchtop centrifuge.
-
12
Use Qubit or equivalent fluorometer to determine DNA concentration.
-
13
Obtain the pLib6.4 empty vector (Addgene # 133783) and linearize with BpiI (Thermo) in FastDigest Buffer for 2 hours (5 μg plasmid + 3 μL FastAP + 3 μL BpiI in a 60 μL reaction).
-
14
Resolve restriction digest reaction on a 1% agarose gel and purify the ~8 kb band containing linearized pLib6.4.
-
15
Mix pLib6.4 with all eluted DNA from Step 10 at a 1:2 molar ratio. Add 1/9 volume 10X T4 DNA ligase buffer. Add 1/20 volume T7 DNA ligase. Incubate overnight at 16°C.
-
16
Use a Qiagen PCR Cleanup Kit column to purify ligation reaction and elute in 20 μL ddH2O.
-
17
Perform test electroporation from 1 μL of resulting reaction into 25 μL of highly competent E. coli cells (such as Ecloni 10GF’ Electrocompetent cells from Novagen) and use provided recovery medium to recover reaction at 30°C for 1 hour with shaking. Plate 1 μL of bacterial culture onto Ampicillin LB/agar plate. Count resulting colonies.
-
18
Scale electroporation to cover expected library diversity. Specifically, perform enough 25 μL electroporation reactions that colony number = 100 times sgRNA number. Plate resulting bacterial cells onto enough 150 mm Ampicillin LB/agar plates to cover the library. To ensure proper separation of colonies, do not exceed 2E5 CFU/plate. Thus, a ~100,000-guide library should result in 1E7 CFU and be plated onto at least 50 plates. Grow plates at 30°C overnight. As soon as visible colonies are detected, proceed to next step.
-
19
Harvest plates into LB medium by pipetting ~10 mL LB medium directly into middle of each plate and then brushing plate with light downward pressure using a Cell Lifter 3008. This procedure will avoid scratching the plate causing loss of LB to the plate bottom.
-
20
Pool LB resulting from all plate harvests and extensively vortex to mix colonies. Pass cell suspension through a 0.45 μm cell strainer to clear chunks of agar.
-
21
Mix with an equal volume of 50% glycerol and freeze library in 1 mL aliquots at −80°C.
-
22
To prep library from an aliquot: quick thaw at 37°C and spin at max speed to harvest cell pellet.
-
23
Remove media and wash with 1 mL PBS. Vortex and spin at max speed.
-
24
Aspirate and discard PBS and replace with 250 μL Qiagen Buffer P1. Proceed with Qiagen Plasmid Miniprep.
-
25
Library validation: perform 12, 20 μL, 15-cycle Phusion High Fidelity PCRs using PCR1fwd and PCR1rev primers and ~10 ng of CRISPR plasmid library in each reaction.
-
26
Submit sample for Next Generation Sequencing.
-
27
Proceed to Support Protocol 3 to determine sgRNA distribution in plasmid library.
-
28
Proceed to Basic Protocol to introduce sgRNA library into S2R+-MT::Cas9 cells.
SUPPORT PROTOCOL 3: Barcode deconvolution and analysis of screen data
INTRODUCTION
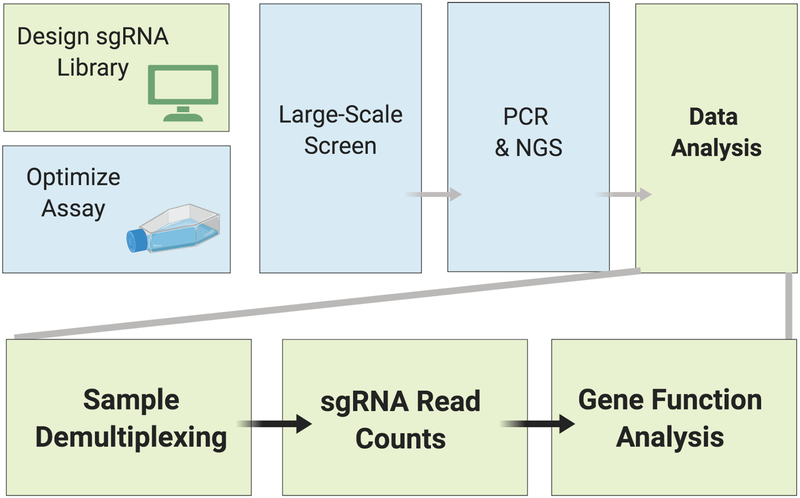
Analysis of the NGS data sets resulting from a screen can be divided into three sections: de-multiplexing the sample, determining the number of counts obtained for each unique sgRNA, and gene-level analysis of screen results (Fig. 7). Note that in addition to analysis of sgRNAs amplified from genomic DNA in a screen (Basic Protocol), this can also be applied to sgRNAs amplified from a newly generated library (Support Protocol 3) as a quality analysis step.
Figure 7: Data analysis for Drosophila cell CRISPR pooled screening.
Analysis of NGS data from a screen can be divided into three sections: de-multiplexing the sample, determining the number of counts obtained for each unique sgRNA, and gene-level analysis of screen results.
MATERIALS LIST
Tagdust software (Lassmann et al., 2009), http://genome.gsc.riken.jp/osc/english/dataresource/
MAGeCK software (Li et al., 2014), https://sourceforge.net/p/MAGeCK/wiki/Home/
1. Sample demultiplexing:
Process FASTQ file using TagDust (Lassmann et al., 2009) to demultiplex inline barcodes. Below are example commands to demultiplex 3 samples from the same sequencing output file:
tagdust file.fastq −1 F:NNN −2 B: TAGCTT −3 R:N -o Sample1 tagdust file.fastq −1 F:NNNN −2 B: CAGTCC −3 R:N -o Sample2 tagdust file.fastq −1 F:NNNNN −2 B: ACATGA −3 R:N -o Sample3
Successful TagDust demultiplexing results in a new file that contains the sample name followed by the barcode (e.g. sample1_BC_TAGCTT.fq…).
2. sgRNA counting in each sample.
To count reads matching to each sgRNA sequence, we use MAGeCK Count (Li et al., 2014). Since each PCR1 primer has 22 bp-homology to the U6:2 promoter, trimming the first 22 bp will arrive at the sgRNA sequence. Example demultiplexed sample files can be found at https://sharehost.hms.harvard.edu/genetics/?perrimon/CRISPR_fitness_screen_reads/reads/. Library files which relate the sgRNA sequences to the targeted gene must be provided for each library. Example MAGeCK-compatible library files can be downloaded at: https://sharehost.hms.harvard.edu/genetics/?perrimon/CRISPR_fitness_screen_reads/library_files/. Use the following commands to obtain readcount files for each demultiplexed .fq file:
count -l library_file.txt -n sample1 --sample-label sample1 --trim-5 22 --fastq sample1_BC_TAGCTT.fq count -l library_file.txt -n sample2 --sample-label sample2 --trim-5 22 --fastq sample2_BC_CAGTCC.fq count -l library_file.txt -n sample3 --sample-label sample3 --trim-5 22 --fastq sample3_BC_ACATGA.fq
Successful read counting will result in a new file that contains the sample name followed by “.count.txt” (e.g. …sample1.count.txt…). For our sample data, read counts can be found at https://sharehost.hms.harvard.edu/genetics/?perrimon/CRISPR_fitness_screen_reads/read_counts/.
3. Gene-level statistics.
To determine Z-scores at the gene-level, at least two sets of readcount files containing the same sgRNA sequences must be compared. Statistical significance occurs when multiple sgRNAs targeting the same gene show significant deviations between samples. For this analysis, we use MAGeCK MLE (using the maximum likelihood estimate statistical approach) (Li et al., 2014). Note that numerous analysis tools exist for CRISPR screening data each employing different statistical approaches, and no consensus yet exists regarding best practices (Schuster et al., 2019). To use MAGeCK MLE, first use standard spreadsheet software to reorganize sample readcounts into a single tab-delineated text file. In the example above, this can be done by sorting each file by sgRNA sequence (column 1) and then saving. Next, readcount column (column 3) from sample2.count.txt and sample 3.count.txt are copied and pasted to create new columns 4 and 5 within sample1.count.txt, and the new file is saved and renamed to “merged.count.txt”. Finally, a so-called “design matrix file” must be created to instruct the program to test every desired pairwise experimental combination of readcount files. For instance, the design matrix file below tests sample1 against sample 2 and sample1 against sample3. The contents of this file are then saved as a new text file called “design_matrix.txt”.
Samples baseline HL60 KBM7 sample1 1 0 0 sample2 1 1 0 sample3 1 0 1Now run MAGeCK MLE as follows: mageck mle --count-table merged.count.txt --design-matrix design_matrix.txt --permutation-round 100
For the example data set, a successful analysis using MAGeCK MLE to compare readcount files generated from the plasmid library vs cells outgrown for 45 days is available at https://sharehost.hms.harvard.edu/genetics/?perrimon/CRISPR_fitness_screen_reads/results_mageck_mle/.
REAGENTS & SOLUTIONS
Drosophila cell culture media (final concentrations)
Schneider’s media (Thermo 21720)
1X Pen/Strep (Thermo 15070063)
10% fetal bovine serum (Thermo 16140071)
Hygromycin selection media
Drosophila cell culture media (above)
200 ng/mL Hygromycin B (Calbiochem 400051, 1:2300 dilution of stock solution)
Puromycin selection media
Drosophila cell culture media (above)
5 μg/mL Puromycin (Calbiochem 540411)
Media preparation
To make the media, add Pen/Strep, FBS, and if relevant, additional antibiotics to achieve final concentrations shown above into Schneider’s media. Pass the media through a 0.2 μm bottle-top filter (Nalgene 566–0020) or equivalent to sterilize. Keep refrigerated until use.
COMMENTARY
Background information
This protocol presents a method for identification of genes that when knocked down confer sensitivity or resistance to a cytotoxin. CRISPR pooled-format screening is not limited to this type of assay; several other assay types are feasible and would allow for exploration of diverse gene functions (Table 2). Moreover, as noted in the introduction this protocol is extensible to other cell lines. Notably, Drosophila cell lines have been derived from different stages or tissue, have different transcriptomes, and have different growth requirements (reviewed in Luhur et al. (2019)). The same or similar assays could be applied to different cell lines to help uncover context-specific findings.
Table 2:
Example pooled screen assays for gene function discovery in Drosophila cells.
| Cells | Assay | Outcome(s) | Example |
|---|---|---|---|
| wild-type | treatment with a cytotoxic drug or compound |
|
Viswanatha et al. (2018) |
| wild-type | treatment with a ligand or hormone that induces lethality |
|
Okamoto et al. (2018) |
| wild-type | outgrowth |
|
Viswanatha et al. (2018) |
| wild-type (control) mutant (experimental) |
outgrowth |
|
|
| wild-type | FACS, TransWell, other method for physical separation of cell populations |
|
Critical Parameters
The following parameters are particularly critical for success: (1) optimization of the concentration of cytotoxin (see Support Protocol 1), (2) library design (see Support Protocol 2), (3) efficient integration of the library into cells (see Basic Protocol) and (4) avoiding bottlenecking the cell population during the screen (see Strategic Planning). Common issues with these steps are further addressed in Troubleshooting.
Troubleshooting
Common problems, causes, and solutions are presented in Table 3 below.
Table 3:
Common problems encountered, potential causes, and proposed solutions
| Problem | Potential cause | Solution |
|---|---|---|
| Low integration efficiency. |
|
|
| Cell doubling rate does not consistently decrease following cytotoxin treatment. | Cytotoxin may be losing activity. | Use Support Protocol 1 to confirm cytotoxin effect on naïve cells periodically during cytotoxin exposure to the CRISPR library to ensure that it has not lost activity; replace as needed. |
| Cell-lines crash during expansion |
|
|
| A few sgRNAs targeting different genes dominate reads |
|
|
Understanding Results
At the end of a screen, the outcome is a set of log-fold changes for sgRNAs. But of course, the overall goal is to identify the relationship between the phenotype and specific genes or gene sets. To accomplish this, we used mean and variance modeling to identify positive hits at the gene level (see Support Protocol 3 and (Viswanatha et al., 2018)).
Ideally, this screening approach can be used both to identify factors conferring growth advantage (e.g. resistance to a cytotoxin) or growth disadvantage (i.e. genes essential in a particular context such as genotype or treatment with a cytotoxin). For the former, it is reasonable to expect a relatively small number of genes. These should be further tested experimentally. For the latter, which we refer to as ‘fitness genes’ or essential genes, you can expect a larger number. As a validation of screen results, the set of fitness genes can be compared to RNAseq as we reported, as a gene in this category should be expressed. Moreover, these data can be used to determine a false discovery rate, as described in Viswanatha et al. (2018). A single replicate of a genome-wide screen yields approximately 1000 essential genes at a false-discovery rate of 5%. For positive selection screens, the predictions made by the screens must be validated experimentally.
To assist with the data interpretation, data from Viswanatha et al. (2018) have been made publicly available for download and reanalysis at: https://sharehost.hms.harvard.edu/genetics/?perrimon/CRISPR_fitness_screen_reads/
Time Considerations
Obtain or build sgRNA library, 0–2 weeks; thaw and revive S2R+-MT::Cas9 cells, 2 weeks; determine effective concentration of cytotoxin, 1–4 weeks; perform screen: 6–8 weeks.
The total time for a large-scale pooled format screen is ~9–14 weeks.
ACKNOWLEDGEMENTS
We thank Shannon Knight for helpful feedback on the manuscript. We thank the DNA Resource Core of the Dana-Farber/Harvard Cancer Center for Sanger sequencing services, as well as thank the Harvard Biopolymers Facility (Harvard Medical School) and Center for Computational and Integrative Biology (Massachusetts General Hospital) for next-generation sequencing services. Relevant support includes NIH NIGMS R01 GM084947 and P41 GM132087 (PI: N.P., Co-I: S.E.M.). S.E.M. is additionally supported in part by the Dana-Farber/Harvard Cancer Center, which is supported in part by an NCI Cancer Center Support Grant # NIH 5 P30 CA06516 (PI: L. Glimcher). R.V. is supported by NIH Training Grant # 2T32GM007748–40. N.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
INTERNET RESOURCES
- Find CRISPR3 sgRNA design resource; https://www.flyrnai.org/crispr3/web/
- Screen data from Viswanatha et al. (2018); https://sharehost.hms.harvard.edu/genetics/?perrimon/CRISPR_fitness_screen_reads.
- Tagdust software (Lassmann et al., 2009); http://genome.gsc.riken.jp/osc/english/dataresource/
- MAGeCK software (Li et al., 2014); https://sourceforge.net/p/MAGeCK/wiki/Home/
LITERATURE CITED
- Akimana C, Al-Khodor S, & Abu Kwaik Y (2010). Host factors required for modulation of phagosome biogenesis and proliferation of Francisella tularensis within the cytosol. PLoS One, 5(6), e11025. doi: 10.1371/journal.pone.0011025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, … Sharp PA (2015). Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell, 160(6), 1246–1260. doi: 10.1016/j.cell.2015.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers B, Jastrzebski K, Heijmans JP, Grernrum W, Beijersbergen RL, & Bernards R (2016). CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol, 34(6), 631–633. doi: 10.1038/nbt.3536 [DOI] [PubMed] [Google Scholar]
- Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, … Rao A (2006). A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature, 441(7090), 179–185. doi: 10.1038/nature04702 [DOI] [PubMed] [Google Scholar]
- Helenius IT, Haake RJ, Kwon YJ, Hu JA, Krupinski T, Casalino-Matsuda SM, … Beitel GJ (2016). Identification of Drosophila Zfh2 as a Mediator of Hypercapnic Immune Regulation by a Genome-Wide RNA Interference Screen. J Immunol, 196(2), 655–667. doi: 10.4049/jimmunol.1501708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housden BE, Valvezan AJ, Kelley C, Sopko R, Hu Y, Roesel C, … Perrimon N (2015). Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi. Sci Signal, 8(393), rs9. doi: 10.1126/scisignal.aab3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Zhao L, & Clapham DE (2009). Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science, 326(5949), 144–147. doi: 10.1126/science.1175145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, … Zhang F (2017). Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc, 12(4), 828–863. doi: 10.1038/nprot.2017.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Hayashizaki Y, & Daub CO (2009). TagDust--a program to eliminate artifacts from next generation sequencing data. Bioinformatics, 25(21), 2839–2840. doi: 10.1093/bioinformatics/btp527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, … Liu XS (2014). MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol, 15(12), 554. doi: 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhur A, Klueg KM, & Zelhof AC (2019). Generating and working with Drosophila cell cultures: Current challenges and opportunities. Wiley Interdiscip Rev Dev Biol, 8(3), e339. doi: 10.1002/wdev.339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr SE, Hu Y, Ewen-Campen B, Housden BE, Viswanatha R, & Perrimon N (2016). CRISPR guide RNA design for research applications. FEBS J, 283(17), 3232–3238. doi: 10.1111/febs.13777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens DW, Deans RM, Li A, & Bassik MC (2016). Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol, 34(6), 634–636. doi: 10.1038/nbt.3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Gross T, Samsonova AA, Vinayagam A, Buckner M, Founk K, … Perrimon N (2013). Conserved regulators of nucleolar size revealed by global phenotypic analyses. Sci Signal, 6(289), ra70. doi: 10.1126/scisignal.2004145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Viswanatha R, Bittar R, Li Z, Haga-Yamanaka S, Perrimon N, & Yamanaka N (2018). A Membrane Transporter Is Required for Steroid Hormone Uptake in Drosophila. Dev Cell, 47(3), 294–305 e297. doi: 10.1016/j.devcel.2018.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccioni F, Younger ST, & Root DE (2018). Pooled Lentiviral-Delivery Genetic Screens. Curr Protoc Mol Biol, 121, 32 31 31–32 31 21. doi: 10.1002/cpmb.52 [DOI] [PubMed] [Google Scholar]
- Ren X, Sun J, Housden BE, Hu Y, Roesel C, Lin S, … Ni JQ (2013). Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci U S A, 110(47), 19012–19017. doi: 10.1073/pnas.1318481110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster A, Erasimus H, Fritah S, Nazarov PV, van Dyck E, Niclou SP, & Golebiewska A (2019). RNAi/CRISPR Screens: from a Pool to a Valid Hit. Trends Biotechnol, 37(1), 38–55. doi: 10.1016/j.tibtech.2018.08.002 [DOI] [PubMed] [Google Scholar]
- Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, … Garcia-Blanco MA (2009). Discovery of insect and human dengue virus host factors. Nature, 458(7241), 1047–1050. doi: 10.1038/nature07967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, … Zhang F (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science, 343(6166), 84–87. doi: 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, & Vakoc CR (2015). Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol, 33(6), 661–667. doi: 10.1038/nbt.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, … Kinet JP (2006). CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science, 312(5777), 1220–1223. doi: 10.1126/science.1127883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanatha R, Li Z, Hu Y, & Perrimon N (2018). Pooled genome-wide CRISPR screening for basal and context-specific fitness gene essentiality in Drosophila cells. Elife, 7. doi: 10.7554/eLife.36333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, & Lander ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science, 343(6166), 80–84. doi: 10.1126/science.1246981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SL, Yeromin AV, Zhang XH, Yu Y, Safrina O, Penna A, … Cahalan MD (2006). Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proc Natl Acad Sci U S A, 103(24), 9357–9362. doi: 10.1073/pnas.0603161103 [DOI] [PMC free article] [PubMed] [Google Scholar]