Abstract
The off-target binding of aminoglycosides (AGs) to the A site of human mitochondrial ribosomes in addition to bacterial ribosomes causes ototoxicity and limits their potential as antibiotics. A fluorescence assay was employed to determine relative binding affinities of classical and improved AG compounds to synthetic RNA constructs representing the bacterial and mitochondrial A sites. Results compared well with previously reported in vitro translation assays with engineered ribosomes. Therefore, the minimal RNA motifs and fluorescence assay are shown here to be useful for assessing the selectivity of new compounds.
Keywords: Aminoglycosides, Helix 44, Ototoxicity, Mitochondrial A site, Fluorescence
1. Introduction
The bacterial ribosome is an ideal antibiotic drug target due to its central role in bacterial viability, cytosolic accessibility, lack of repair mechanisms, and structural differences with eukaryotic ribosomes [1,2]. Among the ribosome-targeting small molecules, aminoglycosides (AGs) are a class of potent broad-spectrum antibiotics [3–8]. The 4,5-deoxystreptamine class of AGs, as shown in Figure 1, contain protonated amines under physiological conditions, which is important for bacterial uptake as well as rRNA binding to bacterial ribosomes [8,9].
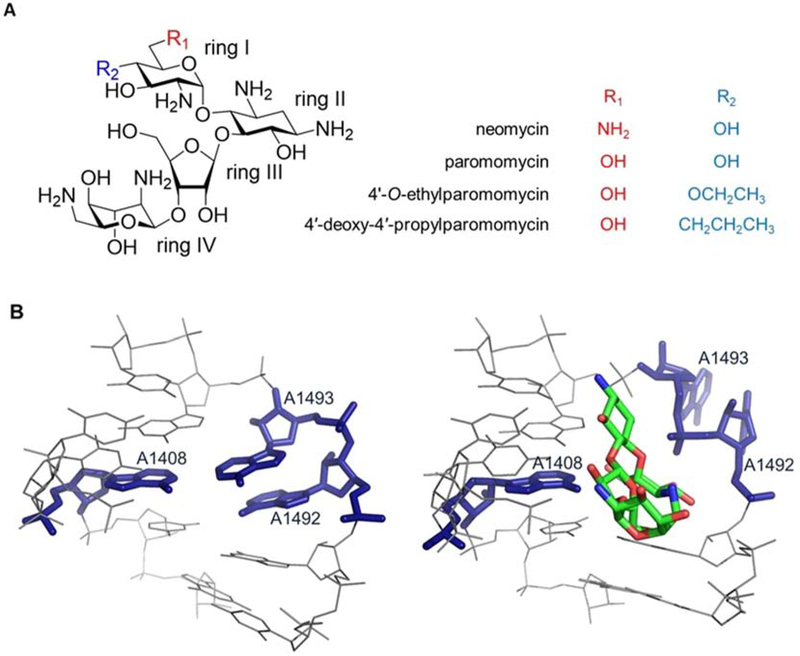
Figure 1. The structures of aminoglycosides and bacterial A site.
(A) Chemical structures of aminoglycosides used in this study are shown. (B) The crystal structure of the E. coli A site is shown with (right, PDB ID: 4V52) and without (left, PDB ID: 4V7A) paromomycin bound [3,7,8,9].
The AGs bind primarily to the aminoacyl-tRNA site (A site), or decoding region, of 16S rRNA of bacterial ribosomes [10,11]. Within the A site, AGs interact with the internal loop region of h44 [12–16], which consists of three conserved bases (A1408, A1492, and A1493) that participate in a non-Watson-Crick base pair (A1408•A1493) or remain unpaired (A1492) [10] (Figure 1B). These key residues sample two distinct conformational states referred to as ‘on’ and ‘off’ (flipped out vs. folded in, respectively), which plays an important role in translation [13,17,18]. Upon binding, AGs such as neomycin and paromomycin stabilize A1492 and A1493 in the ‘on’ state (Figure 1B) [13–16], which then interferes with discrimination of non-cognate versus cognate tRNAs and leads to accumulation of aberrant proteins and reduced bacterial growth [13,19–21].
Even though AGs are potent antibiotics, detrimental side effects on human health, including hearing impairment (ototoxicity), are a major limitation [22–24]. Binding of AGs to the mitochondrial (mt) ribosome (mitoribosome) in human cells correlates with induced ototoxicity [24–26]. Despite key differences in their ribosome sizes and protein content, considerable structural similarities exist between bacterial and mt rRNAs, specifically in the A site (Figure 2) [24–27]. The A site in mt 12S rRNA also contains an adenosine at position 1492 (1408 in E. coli), therefore AGs such as paromomycin can potentially bind to mitoribosomes, resulting in translation errors and miscoded mt proteins [28]. Consequently, perturbation in the cell respiratory pathway leads to apoptosis of inner-ear cells and ototoxicity [24–26]. In contrast, base mismatches (C1493•C1556; C1494•A1555) in the mt A site compared to Watson-Crick base pairs (C1409-G1491; A1410-U1490) in the corresponding E. coli RNA (Figure 2) provide unique opportunities to develop AGs with reduced affinity towards the mt rRNA.
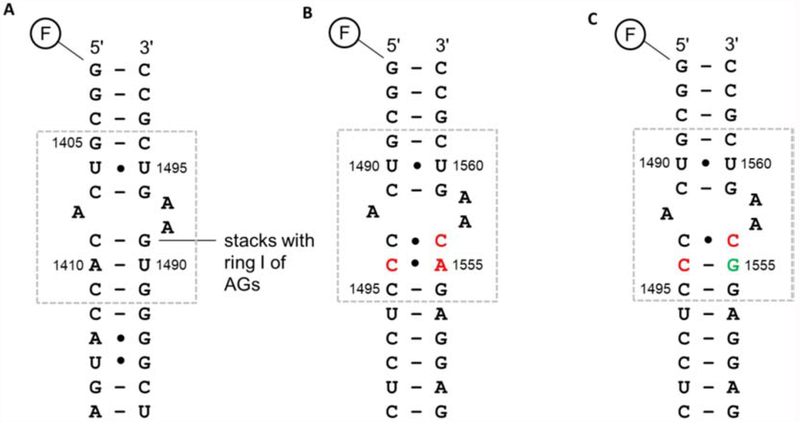
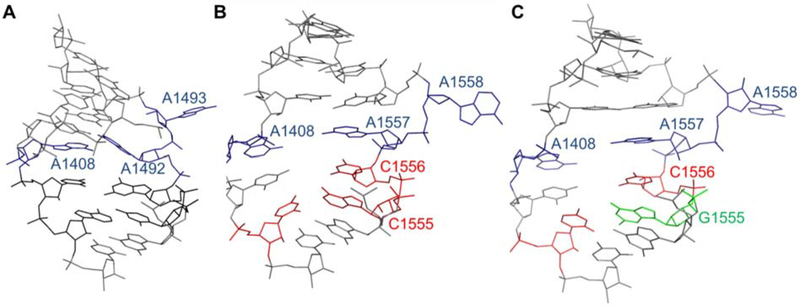
Figure 2. A-site RNA secondary structures.
RNA duplexes representing (A) E. coli (wt-bac), (B) wild-type human mitoribosomal (wt-mt), and (C) A1555G mutant human mitoribosomal (mut-mt) A-site sequences are shown (the A-site region is boxed). The nucleotides that are different in either wild-type or mutant mt sequences compared to the E. coli sequence are shown in red. The point mutation in the mut-mt sequence is shown in green. The rRNA numbering systems for E. coli and human mitochondria are used. The black dots (•) represent non-Watson-Crick base pairs and F is a 5′-fluorescein tag.
Clinical studies revealed that individuals with a mutation A1555G (U1490 in E. coli) in h44 of mitoribosomes have a higher chance of developing AG-induced ototoxicity [29]. Of note, restoration of a Watson-Crick base pair (C1494-G1555) in the mutant mt A site (Figure 2) could lead to a higher affinity of AGs (Figure 2) [28]. Therefore, it is important to compare AG affinity towards the A1555G mutant, as well as the mt A-site rRNA, when developing new compounds as potential antibiotics.
Modifying currently available AGs to retain the desired antibacterial activity but reducing anti-mitoribosomal activity is an approach for generating improved antibiotics [30–34]. Several modified AGs were identified based on screening of synthetic analogues of paromomycin in cell-free translation assays with bacterial and hybrid bacterial ribosomes with either wild-type or mutant human mt A sites [33,34,35]. Paromomycin was selected as the parent scaffold due to its considerable antibacterial activity [33,34], comparatively less ototoxic potential [33,34,36], and resistance against AG-modifying enzymes [34,37]. The paromomycin derivatives 4′-O-ethylparomomycin and 4′-deoxy-4′-propylparomomycin (propylamycin) (Figure 1A) showed reduced inhibition of hybrid ribosomes. In addition, propylamycin displayed increased antibacterial activity against Enterobacteriaceae and other Gram-negative pathogens [33,34].
The goal of this work was to use a fluorescence assay with synthetic RNAs to measure apparent dissociation constants (Kd values) of AG derivatives to bacterial and mt A sites. Conformational changes upon AG binding to short RNA duplexes representing the A site from E. coli (wt-bac) [14,16,42], wild-type human mitoribosomes (wt-mt), and A1555G mutant human mitoribosomes (mut-mt) (Figure 2) were monitored through an attached dye molecule (fluorescein, F) at the 5′ end of one strand [38–40]. This approach of F tagging the RNA has been used previously [38–41]. The main difference between the prior reports and this one is the use of a duplex instead of a hairpin construct [42]. The duplex construct represents the A site [16], but without the extra loop which has been shown previously to enhance non-specific binding [43]. The fluorescence changes associated with binding can be used to determine the relative affinities of the modified AGs to the model A-site RNA constructs, and to correlate with previously reported IC50S for translation inhibition with engineered ribosomes. Strong correlations with the previously reported antibacterial and anti-mitoribosomal activities suggest that the fluorescence method is a useful tool for the assessment of AG selectivity.
2. Materials and methods
2.1. Aminoglycoside compounds
Neomycin sulfate and paromomycin sulfate were purchased from Sigma (St. Louis, MO). Synthesis of the modified AGs 4′-O-ethylparomomycin pentaacetate and propylamycin pentaacetate was described previously [33,34] (Figure 1). The Dried AG compounds were dissolved in binding buffer (10 mM HEPES (pH 7.4), 150 mM NaCl, and 3 mM Na2EDTA) to generate 250 μM to 1 mM stocks.
2.2. RNA preparation
Chemically synthesized RNAs (Figure 2) were purchased from GE Healthcare Dharmacon, Inc. (Lafayette, Co) (5′ wt-bac: 5′-F-GGCGUCACACCAUGA; 5′ mt: 5′-F-GGCGUCACCCUC CUC; 3′ wt-bac: 5′-UCGGGGUGAAGUCGCC, 3′ wt-mt: 5′-GAGGAGACAAGUCGCC, and 3′ mut-mt: 5′-GAGGAGGCAAG UCGCC) and purified by using 20% denaturing polyacrylamide gel electrophoresis (PAGE) in 7 M urea. After PAGE, the RNAs were isolated by crush and soak, followed by ethanol precipitation. Briefly, a 3× volume of ice-cold 100% ethanol and 0.1× volume of 3 M NaOAc were added to each of the RNA samples. Then, the samples were kept at −80 °C for 45 min. Next, the RNAs were precipitated by centrifugation at 14,000 rpm for 15 min. The supernatants were removed and the RNA pellets were washed twice with ice-cold 70% ethanol. The RNA samples were centrifuged again at 14,000 rpm for 10 min. The supernatants were removed and the RNA pellets were dried briefly under vacuum. The purified RNAs were dissolved in binding buffer and stored at −20 °C. The concentrations were calculated by measuring the absorbance at 260 nm and using molar extinction coefficients of 168,100, 149,100, 153,700, 163,400, and 160,100 M−1cm−1 for 5′ wt-bac, 5′ mt, 3′ wt-bac, 3′ wt-mt, and 3′ mut-mt, respectively.
2.3. Fluorescence assay
The concentrations of the F-tagged RNAs (5′ wt-bac and 5′ mt) (Figure 2) were adjusted to 1 μM in binding buffer. The complementary RNAs (3′ wt-bac, 3′ wt-mt, and 3′ mut-mt) were annealed by incubating at 95 °C for 1 min followed by slow cooling to room temperature in a large (~2 L) water bath overnight. The fluorescence emission spectra were obtained using 300 μL of RNA and an excitation wavelength of 490 nm, emission wavelength range of 500–600 nm, and a slit width of 1.5 on a Varian Cary Eclipse fluorescence spectrophotometer. The AG compound (in binding buffer) was added sequentially (12 to 15 aliquots of 2 μL each) to the RNA. The samples were stirred manually (by gently pipetting the sample up and down four times) and incubated for 2 min after each addition. Fluorescence emission spectra were collected for each titration point until saturation was reached. Three controls were performed: 1) fluorescence measurements of RNA only (no AG) every 2 min for 1 h to check for photo-bleaching; 2) titration of RNA with titrated with 2 μL aliquots of buffer only (no AG) to determine background fluorescence; and 3) fluorescence measurements of AG compounds without RNA to determine AG background fluorescence.
Fluorescence intensities at 520 nm were corrected (Fi,corr) for the volume change by using Equation 1,
| (1) |
in which Fi,corr is the corrected fluorescence intensity for the ith point of titration, Fi,obs is the measured fluorescence intensity for the ith point of the titration, Vi is the volume after the ith addition, and Vo is the initial volume. The fraction of fluorescence intensity due to the bound species (Fr) was calculated using Equation 2,
| (2) |
in which F0,corr is the fluorescence intensity of the sample at the initial point (all free), Fi,corr is the fluorescence intensity of the sample at the ith point in the titration, and Ff,corr is the fluorescence intensity of the sample at the final titration point (all bound). The apparent dissociation constant, or Kd value, was determined by plotting Fr against the ligand concentration ([Lt]) and fitting the data to Equation 3,
| (3) |
in which [Lt] is the ligand concentration, [Rt] is the total RNA concentration, and C is a constant (109) [38]. Three independent trials were performed for each RNA and ligand combination, and error bars for each ligand concentration were derived from the standard deviations. The reported errors were obtained from the curve fitting analysis.
3. Results and discussion
3.1. Binding of aminoglycosides to A-site RNA constructs
In complete ribosomes, AGs such as paromomycin bind to h44 within the internal loop [12]. Similar binding interactions are observed in ribosome subunits or model RNA constructs of the A site [13–16,38,42]. The goal of this work was to determine the relative affinities of two semisynthetic AG compounds, 4′-O-ethylparomomycin and propylamycin, to the A sites from bacterial and mt ribosomes. The 5′-F-tagged RNAs employed were duplex versions of a previously used 27-nucleotide hairpin construct representing the bacterial A site [38]. Changes in fluorescence intensity at 520 nm of the F-tagged RNAs were monitored with increasing amounts of compound [38]. These changes are indicative of conformational changes of the RNA upon AG binding, which are propagated to the 5′ end of the construct where the fluorescein dye is located [38]. Control experiments were carried out to show that the changes in fluorescence upon AG addition were due to a binding event and not photobleaching or background fluorescence of the AGs. The fluorescence intensities of the F-tagged RNAs (no AG) did not change with repeated irradiations. Similarly, F-tagged RNAs were titrated with binding buffer and no changes in fluorescence intensity were observed. In addition, the AG compounds did not show any background fluorescence (data not shown), consistent with previous studies [38].
Neomycin, paromomycin, 4′-O-ethylparomomycin, and propylamycin binding to the F-tagged wt-bac duplex RNA was monitored by changes in the fluorescence emission intensity, starting at low nM concentrations until initial saturation was reached. As shown previously, further additions with higher concentrations of AGs led to non-specific binding and additional changes in fluorescence [38], but for this study titrations were not continued into the non-specific binding regime. When titrated with neomycin, the F-tagged wt-bac duplex RNA displayed a 20% decrease in fluorescence intensity, consistent with a conformational change in the bacterial A-site RNA upon AG binding. Assuming 1:1 complex formation, the data were fit as the fraction of fluorescence intensity (Fr) versus total neomycin concentration (Lt) (Equation 1) to give an apparent dissociation constant, or Kd value, of 0.001 ± 0.0002 μM (Figure S1 and Table 1). The Kd value with the F-tagged wt-bac duplex RNA is in good agreement with the previously reported value of 0.005 ± 0.002 μM for neomycin binding to a similar hairpin construct representing the bacterial A site [38].
Table 1.
Apparent dissociation constants of aminoglycosides for A-site RNA duplexes
| Aminoglycosides | Apparent dissociation constants, Kd values (μM) | ||
|---|---|---|---|
| bacterial A site (wt-bac) | mito. A site (wt-mt) | mutant mito. A site (mut-mt) | |
| neomycin | 0.001 ± 0.0002 | 1.1 ± 0.5 | 0.1 ± 0.02 |
| Paromomycin | 0.005 ± 0.001 | 6.5 ± 0.9 | 2.8 ± 0.5 |
| 4′-O-ethylparomomycin | 0.03 ± 0.01 | 12.8 ± 2.8 | 8.6 ± 2.1 |
| Propylamycin | 0.002 ± 0.001 | 26.9 ± 4.7 | 5.8 ± 1.8 |
Titrations with paromomycin, 4′-O-ethylparomomycin, and propylamycin with the F-tagged wt-bac duplex RNA yielded apparent Kd values of 0.005 ± 0.001, 0.03 ± 0.01, and 0.002 ± 0.001 μM, respectively (Figure 3, Figure S1, and Table 1). Of note, addition of neomycin to the F-tagged wt-bac induced quenching of fluorescence, whereas paromomycin and the modified AGs caused an increase in fluorescence (11 to 20% change in all cases). Previous computational studies showed that upon AG binding to the F-tagged RNA, a conformation change occurs along with ligand interactions with the F tag [38]. Thus, the AG functional groups may interact with the dye molecule differently, leading to different fluorescence changes.
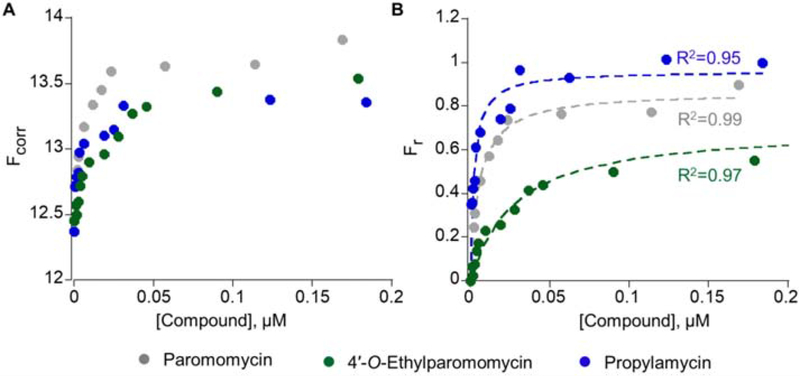
Figure 3. Propylamycin binding to bacterial A-site RNA construct.
(A) Changes in volume-corrected fluorescence intensity (Fcorr) upon titration with paromomycin, 4′-O-ethylparomomycin, and propylamycin are shown. (B) Representative binding curves of fraction bound (Fr) as a function of paromomycin, 4′-O-ethylparomomycin, and propylamycin concentration fitted to Equation 3 are overlaid (R2 = 0.99, 0.97, and 0.95, respectively).
Next, we compared AG interactions with the mt A-site rRNA due to its close resemblance to the bacterial A site. The F-tagged wt-mt and mut-mt duplex RNA constructs have similar sequences as the wt-bac RNA construct, particularly in the internal loop region, which is the observed binding site in X-ray crystal structures [13–16]. Titrations of neomycin, paromomycin, 4′-O-ethylparomomycin, and propylamycin against the wt-mt duplex RNAs gave apparent Kd values of 1.1 ± 0.5, 6.5 ± 0.9, 12.8 ± 2.8, and 26.9 ± 4.7 μM, respectively (Figure S2 and Table 1). With the mut-mt duplex RNA, neomycin, paromomycin, 4′-O-ethylparomomycin, and propylamycin gave apparent Kd values of 0.1 ± 0.02, 2.8 ± 0.5, 8.6 ± 2.1, and 5.8 ± 1.8 μM, respectively (Figure S3 and Table 1). These changes in binding affinities are consistent with altered contacts with the A-site residues of the wt-mt and mut-mt duplex RNAs, which have different residues at A1410 (C1494), G1491 (C1556), and U1490 (A/G1555) (Figure 2).
Neomycin showed the highest affinity for all three RNA constructs, with low μM to high nM binding to the mt RNAs. Compared to neomycin, paromomycin has a 5-, 6-, and 28-fold lower affinity for the wt-bac, wt-mt, and mut-mt RNAs, respectively. Despite these lower affinities, paromomycin has better selectivity for bacterial over human mt A sites, which is consistent with previous studies [33,34]. Although 4′-O-ethylparomomycin had the weakest affinity for wt-bac RNA (6-fold reduction in binding affinity compared to paromomycin), this derivative showed a moderate level of discrimination between wt-bac and mt RNAs (relative to paromomycin, 2- and 3-fold reduced affinity towards wt-mt and mut-mt RNAs, respectively). In contrast, propylamycin showed a 2.5-fold enhancement in binding to wt-bac compared to paromomycin, in addition to a 4- and 2-fold reduction in affinity towards the wt-mt and mut-mt RNAs, respectively. Thus, results with F-tagged RNAs are consistent with the 4′-deoxy-4′-propyl modification to paromomycin providing enhanced binding to the wt-bac and significant reduction in affinity to the undesired mt targets.
3.2. Correlation between in vitro binding affinities and in vivo activity of aminoglycosides
Modifying currently available AGs has been used to overcome AG-induced ototoxicity [33,34]. Quantifying the interactions between modified AGs and small RNA constructs resembling the target A site is therefore useful for making correlations between in vitro A-site RNA binding and in vivo activity (i.e., ribosome binding, translation inhibition, and antibacterial activity). Minimum inhibitory concentration (MIC) values of 4, 16, and 2 mg/L have been reported for paromomycin, 4′-O-ethylparomomycin, and propylamycin, respectively [33,34]. In addition, translation inhibition of bacterial ribosomes gave IC50 values of 0.03, 0.08, and 0.03 mg/L for paromomycin, 4′-O-ethylparomomycin, and propylamycin, respectively [33,34]. The fluorescence binding data for paromomycin, 4′-O-ethylparomomycin, and propylamycin with the F-tagged wt-bac duplex RNA construct therefore follow the same trend as the MIC and IC50 values, showing a good correlation between A-site target binding and biological effects of the AGs.
The IC50 values with hybrid ribosomes containing a human mitochondrial A site were reported to be 51, 97, and 167 mg/L for paromomycin, 4′-O-ethylparomomycin, and propylamycin, respectively [33–35]. The relative AG binding affinities obtained with the F-tagged wt-mt duplex RNA construct are consistent with those trends, in which propylamycin had the lowest affinity for the wt-mt A site and biggest loss in activity against the humanized ribosomes. Similarly, the IC50 values obtained for paromomycin, 4′-O-ethylparomomycin, and propylamycin with engineered ribosomes resembling the mutant mt A site (A1555G) were 6, 120, and 51 mg/L for paromomycin, 4′-O-ethylparomomycin, and propylamycin, respectively [33,34], which follows the same trend as apparent Kd values obtained by using the F-tagged mut-mt duplex RNA. In this case, 4′-O-ethylparomycin had the lowest affinity for the mut-mt A site and biggest loss in activity against the corresponding mut-mt ribosomes. Table 2 summarizes the trends in AG selectivity for the bacterial over mt A-site RNAs in terms of previously determined in vitro translation inhibition (IC50) [33,34] and the apparent Kd values obtained from fluorescence-based assays with the smaller RNA constructs, which are consistent for both the wt-mt and mut-mt constructs and the propylamycin derivative. In comparing the Kd values, 4′-O-ethylparomomycin is less selective than paromomycin for the wt-bac construct. In contrast, 4′-O-ethylparomomycin is more selective than paromomycin in the case of mut-mt ribosomes if the IC50 values are compared. Such discrepancies are not surprising, however, when comparing model and biological systems. The compound could have secondary ribosome binding sites that impact the IC50 values, but are not observed in the simplified model systems.
Table 2.
Selectivity of AG translation inhibition potential and binding affinity of A-site RNAs
| Aminoglycosides | Selectivity for bacterial over mt A site | |||
|---|---|---|---|---|
| ratio (wt-mt/wt-bac) | ratio (mut-mt/wt-bac) | |||
| IC50 | Kd | IC50 | Kd | |
| neomycin | 200 | 1100 | 20 | 100 |
| paromomycin | 1700 | 1300 | 200 | 560 |
| 4′-O-ethylparomomycin | 1212 | 427 | 1500 | 287 |
| propylamycin | 5566 | 13450 | 1733 | 2900 |
Both the chemical structure of AGs and the secondary structures of different A-site RNA constructs dictate the binding affinities of AGs. As observed in crystal structures and supported by solution and computational studies with RNA constructs resembling the bacterial A site, ring I of paromomycin forms a pseudo-Watson-Crick base pair with A1408, along with a stacking or CH/π interaction with G1491 [45–48]. In addition, chemical probing studies on ribosomes showed interactions of A1408 and G1491 among other residues with paromomycin [34,49]. Therefore, both ring I, which was modified in the two paromomycin derivatives, and the guanosine at position 1491 of the bacterial A-site RNA, which is a cytidine (C1556) in the mt RNA, are important for paromomycin class AG binding.
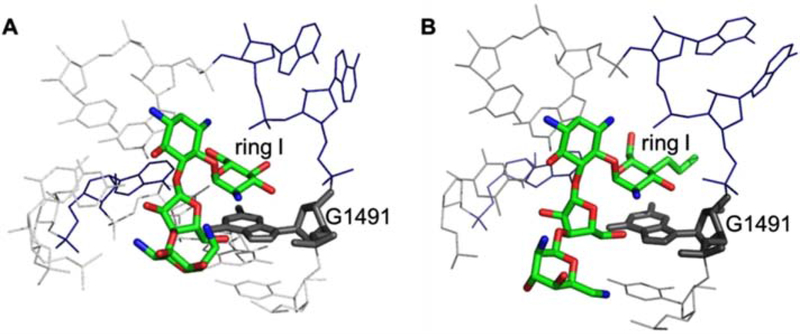
Compared to paromomycin, the 4′-O-ethyl and 4′-deoxy-4′-propyl modifications lead to either reduced or increased binding affinity, respectively, for the wt-bac RNA. As observed in a crystal structure of T. thermophilus 70S ribosomes, the interactions of propylamycin ring I with the A site also involve A1408 and G1491 (Figure 4) [34]. The ring I 6′-hydroxyl and 5′-oxygen form hydrogen bonds with the N1 and N6 of At 408, respectively. Therefore, it is not surprising that modifications to ring I impact affinity for the A site.
Figure 4. AG interactions with the bacterial A site.
The interactions of paromomycin (PDB ID:1IBK) [42] (A) and propylamycin (PDB ID:6097) [34] (B) with A-site rRNA are compared. The G1491 residue is highlighted to show the stacking interactions with ring I of AGs.
The three paromomycin compounds tested here display 400- to 13,000-fold tighter binding towards the F-tagged wt-bac duplex RNA over the wt-mt construct, and 300- to 3000-fold enhanced binding to the mut-mt RNA, supporting previous biological studies on engineered ribosomes with the same sequence variations in the A-site internal loop [33,34]. The internal loop of the mt constructs is expected to be wider than that of the bacterial RNA, particularly in the wt-mt A site, because the distances between bases in C1493•C1556 and C1494•A1555 non-canonical base pairs are typically longer than those in canonical Watson-Crick base pairs (Figure 2 and 5) [50]. These alterations in the internal loop structure may be less favorable for AG interactions [28]. In contrast, the presence of a canonical Watson-Crick base pair (C1492-G1555) in the mut-mt A site (Figure 2 and 5) may be responsible for the increased binding affinity of AGs compared to the wt-mt A site and therefore compromised selectivity for the bacterial sequence [28].
Figure 5. Secondary structures of the h44 internal loop.
The secondary structures of bacterial A-site rRNA (PDB ID: 4V4I) [10] (A) and RNA constructs resembling wild-type mitochondrial (PDB ID: 3BNN) (B) and mutant mitochondrial (A1555G) (PDB ID: 3BNP) A site are shown [51]. The nucleotides that are different in both wild-type and mutant mitochondrial sequences compared to the E. coli sequence are shown in red. The point mutation in mutant mitochondrial sequence is shown in green.
4. Conclusions
We have employed a fluorescence assay using 5′-fluorescein-labeled duplex RNA constructs resembling the bacterial and human mt A site to determine binding affinities of four AGs, neomycin, paromomycin, 4′-O-ethylparomomycin, and propylamycin. All AGs tested showed tighter binding towards the wt-bac duplex RNA construct over mt duplex RNA constructs. When considering the minimal mt constructs, neomycin has the tightest binding while 4′-O-ethylparomomycin and propylamycin have the weakest binding to mut-mt and wt-mt duplex RNAs, which can be correlated to the observed anti-mitoribosomal activity [33,34]. In particular, propylamycin showed significantly weaker binding to the mt duplex RNAs.
In comparing the mt RNA constructs, all four AGs showed increased binding to the mut-mt relative to the wt-mt RNA, supporting the previously reported correlation between AG binding to the mt A site and increased AG-induced ototoxicity potential in individuals carrying the mutation [29]. Nonetheless, the two modified paromomycin derivatives, 4′-O-ethylparomomycin and propylamycin, still displayed weaker binding to mut-mt constructs compared to paromomycin, which is also consistent with the translation inhibition activities [33,34].
In comparison, in vitro translation assays provide valuable information regarding AG activity, but require engineering of the ribosomes to generate the wild-type and mutant mitochondrial rRNA sequences, which can be challenging and time consuming. In contrast, the fluorescence assay takes advantage of small synthetic RNA constructs to obtain relative binding affinities of AGs to target (wt-bac) and off-target (wt-mt and mut-mt) RNAs, and can be performed in a relatively short period of time. Although possible ligand-dye interactions are a limitation, the assay can be utilized to obtain relative binding affinities of a particular ligand to related RNA sequences that are labeled with the same or different dyes. Previous studies have shown that the Kd values derived from the fluorescence assay compare well with those obtained by other methods such as mass spectrometry [42] and isothermal titration calorimetry [52]. Therefore, our studies have shown that the fluorescence assay with minimal RNA motifs is a useful tool to predict the selectivity of modified AGs for both wild-type and mutant forms of mitoribosomes.
Supplementary Material
Acknowledgments
This work is financially supported by the National Institutes of Health (AI123352 to DC) and a Schaap Faculty Fellowship (to CSC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare the following competing financial interest(s): David Crich is a cofounder and has an equity interest in Juvabis AG, a biotech start-up.
References
- 1.Poehlsgaard J; Douthwaite S Nat. Rev. Microbiol 2005, 3, 870–881. [DOI] [PubMed] [Google Scholar]
- 2.Böttger EC Trends Biotechnol. 2006, 24, 145–147. [DOI] [PubMed] [Google Scholar]
- 3.Davis BD. Microbiol. Rev 1987. 51, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begg EJ; Barclay ML Br. J. Clin. Pharmacol 1995, 39, 597–603. [PMC free article] [PubMed] [Google Scholar]
- 5.Wong CH; Hendrix M; Priestley ES; Greenberg WA. Chem. Biol 1998. 5, 397–406. [DOI] [PubMed] [Google Scholar]
- 6.Lynch SR; Puglisi JD. J. Mol. Biol 2001. 306, 1037–1058. [DOI] [PubMed] [Google Scholar]
- 7.Vicens Q; Westhof E Chembiochem. 2003, 6, 1018–1023. [DOI] [PubMed] [Google Scholar]
- 8.Magnet S; Blanchard JS. Chem. Rev 2005, 105, 477–4979. [DOI] [PubMed] [Google Scholar]
- 9.Hancock RE Annu. Rev. Microbiol 1984, 38, 237–264. [DOI] [PubMed] [Google Scholar]
- 10.Korostelev A; Trakhanov S.; Laurberg M.; Noller HF. Cell. 2006, 126, 1065–1077. [DOI] [PubMed] [Google Scholar]
- 11.Ramakrishnan V. Cell, 2002, 108, 557–572. [DOI] [PubMed] [Google Scholar]
- 12.Borovinskaya MA; Pai RD; Zhang W; Schuwirth BS; Holton JM; Hirokawa G; Kaji H; Kaji A; Cate JH Nat. Struct. Mol. Biol 2007, 14, 727–732. [DOI] [PubMed] [Google Scholar]
- 13.Ogle JM; Brodersen DE; Clemons WM Jr.; Tarry MJ; Carter AP; Ramakrishnan V Science. 2001, 292, 897–902. [DOI] [PubMed] [Google Scholar]
- 14.Fourmy D; Recht MI; Blanchard SC; Puglisi JD Science. 1996, 274, 1367–1371. [DOI] [PubMed] [Google Scholar]
- 15.Carter AP; Clemons WM; Brodersen DE; Morgan-Warren RJ; Wimberly BT; Ramakrishnan V Nature, 2000, 407, 340–348. [DOI] [PubMed] [Google Scholar]
- 16.François B; Russell RJM; Murray JB; Aboul-ela F; Masquida B; Vicens Q; Westhof E. Nucleic Acids. Res 2005, 33, 5677–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meroueh SO; Mobashery S Chem. Biol. Drug Des 2007, 69, 291–297. [DOI] [PubMed] [Google Scholar]
- 18.Dethoff EA; Petzold K; Chugh J; Casiano-Negroni A; Al-Hashimi HM Nature. 2012, 491, 724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaher HS; Green R. Cell. 2009, 136, 746–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pape T; Wintermeyer W; Rodnina MV Nat. Struct. Biol 2000, 7, 104–107. [DOI] [PubMed] [Google Scholar]
- 21.Kohanski MA; Dwyer DJ; Wierzbowski J; Cottarel G; Collins JJ. Cell. 2008, 135, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brammett RE; Fox KE Antimicrob. Agents Chemother 1989, 33, 797–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schacht J Otolaryngol. Clin. North. Am 1993, 5, 845–856. [PubMed] [Google Scholar]
- 24.Böttger EC; Schacht J. Hear. Res 2013. 303, 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobbie SN; Akshay S; Kalapala SK; Braell CM; Shcherbakov D; Böttger EC. Proc. Natl. Acad. Sci. USA. 2008105, 20888–20893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francis SP; Katz J; Fanning KD; Harris KA; Nicholas BD; Lacy M; Pagana J; Agris PF; Shin JB, J. Neurosci 2013, 33, 3079–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amunts A; Brown A; Toots J; Scheres SHW; Ramakrishnan V Science, 2015, 348, 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prokhorova I; Altman RB; Djumagulov M; Shrestha JP; Urzhumtsev A; Ferguson A; Chang CT; Yusupov M; Blanchard SC; Yusupova G. Proc Natl Acad Sci USA. 2017, 114, E10899–E10908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prezant TR; Agapian JV; Bohlman MC; Bu X; Oztas S; Qiu WQ; Arnos KS; Cortopassi GA; Jaber L; Rotter JI; Shohat M; Fischel-Ghodsian N Nat. Genet 1993, 4, 289–294. [DOI] [PubMed] [Google Scholar]
- 30.Mandhapati AR; Yang G; Kato T; Shcherbakov D; Hobbie SN; Vasella A; Böttger EC; Crich D J. Am. Chem. Soc 2017,139, 14611–14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sati GC; Shcherbakov D; Hobbie SN; Vasella A; Böttger EC; Crich D. ACS Infect. Dis 2017. 3, 368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Fernandez D; Shcherbakov D; Matt T; Leong NC; Kudyba I; Duscha S; Boukari H; Patak R; Dubbaka SR; Lang K; Meyer M; Akbergenov R; Freihofer P; Vaddi S; Thommes P; Ramakrishnan V; Vasella A; Böttger EC Nat. Commun 2014, 5, 3112–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duscha S; Boukari H; Shcherbakov D; Salian S; Silva S; Kendall A; Kato T; Akbergenov R; Perez-Fernandez D; Bernet B; Vaddi S; Thommes P; Schacht J; Crich D; Vasella A; Böttger EC mBio. 2014, 5, e01827–14; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsushita T; Sati GC; Kondasinghe N; Pirrone MG; Kato T; Waduge P; Kumar HS; Sanchon AC; Dobosz-Bartoszek M; Shcherbakov D; Juhas M; Hobbie SN; Schrepfer T; Chow CS; Polikanov YS; Schacht J; Vasella A; Böttger EC; Crich D J. Am. Chem. Soc. 2019, 141, 5051–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hobbie SN; Bruell CM; Akshay S; Kalapala SK; Shcherbakov D; Böttger EC. Proc. Natl. Acad. Sci. USA. 2008, 105, 3244–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikawa M; García-Mateo N; Čusak A; López-Hernández I; Fernández-Martínez M; Müller M; Rüttiger L; Singer W; Löwenheim H; Kosec G; Fujs S; Martínez-Martínez L; Schimmang T; Petković H; Knipper M; Durán-Alonso MB Sci. Rep 2019, 9, 2410–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bacot-Davis VR; Bassendenac AV; Berghuis AM Med. Chem. Commun 2016, 7, 103–113. [Google Scholar]
- 38.Llano-Sotelo B; Azucena EF Jr.; Kotra LP; Mobashery S; Chow CS Chem. Biol 2002, 9, 455–463. [DOI] [PubMed] [Google Scholar]
- 39.Haddad J; Kotra LP; Llano-Sotelo B; Kim C; Azucena EF; Liu M; Vakulenko SB; Chow CS; Mobashery S J. Am. Chem. Soc 2002, 124, 133229–1333237. [DOI] [PubMed] [Google Scholar]
- 40.Yan Z; Baranger AM Bioorg. Med. Chem. Lett 2004,14, 5889–5893. [DOI] [PubMed] [Google Scholar]
- 41.Wang NR; Hergenrother PJ Anal. Biochem 2007, 371, 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asare-Okai PN; Chow CS. Anal. Biochem 2011. 408, 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S; Huber PW; Cui M; Czarnik AW; Mei HY Biochemistry 1998, 37, 5549–5557. [DOI] [PubMed] [Google Scholar]
- 44.Sakakibara Y; Abeysirigunawardena SC; Due A-C; Dremann DN; Chow CS Angew. Chem. Int. Ed. Engl 2012, 51, 12095–12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vicens Q; Westhof E Structure, 2001, 9, 647–658. [DOI] [PubMed] [Google Scholar]
- 46.Asensio JL; Ardá A; Cañada FJ; Jiménez-Barbero J Acc. Chem. Res 2013, 46, 946–54. [DOI] [PubMed] [Google Scholar]
- 47.Vacas T; Corzana F; Jiménez-Osés G; González C; Gómez AM; Bastida A; Revuelta J; Luis Asensio J J. Am. Chem. Soc 2010, 132, 12074–12090. [DOI] [PubMed] [Google Scholar]
- 48.Moazed D; Noller HF. Nature. 1987. 327, 389–394. [DOI] [PubMed] [Google Scholar]
- 49.Purohit P; Stern S Nature. 1994, 370, 659–662. [DOI] [PubMed] [Google Scholar]
- 50.Roy A; Panigrahi S; Bhattacharyya M; Bhattacharyya D J. Phys. Chem. B 2008,112, 3786–3796. [DOI] [PubMed] [Google Scholar]
- 51.Kondo J; Westhof E. Nucleic Acids Res. 2008. 36, 2654–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaul M; Barbieri CM; Pilch DS. J. Mol. Biol 2005, 346, 119–134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.