Abstract
DNA is a versatile biomaterial with well-defined mechanical and biochemical properties. It has been broadly adopted to synthesize tension sensors that calibrate and visualize cellular forces at the cell-matrix interface. Here we showed that DNA-based tension sensors are vulnerable to deoxyribonucleases (DNases) which cells may express on cell membrane or secret to the culture environment. These DNases can damage the sensors, lower signal-to-noise ratio or even produce false signal in cellular force imaging. To address this issue, we tested peptide nucleic acid (PNA), chemically modified RNA and their hybrids with DNA as alternative biomaterials for constructing tension sensors. Four duplexes: double-stranded DNA (dsDNA), PNA/DNA, dsRNA (modified RNA) and PNA/RNA, were tested and evaluated in terms of DNase resistance, cellular force imaging ability and material robustness. The results showed that all PNA/DNA, dsRNA and PNA/RNA exhibited strong resistance to both soluble DNase I and membrane-bound DNase on cells. However, PNA/RNA-based tension sensor had low signal-to-noise ratio in cellular force imaging, while dsRNA-based tension sensor exhibited strong non-specific signal unrelated to cellular forces. Only PNA/DNA-based tension sensor reported cellular forces with highest signal-to-noise ratio and specificity. Collectively, we confirmed that PNA/DNA hybrid is an accessible material for the synthesis of DNase-resistant tension sensor that retains the force-reporting capability and remains stable in DNase-expressing cells. This new class of tension sensors will broaden the application of tension sensors in the study of cell mechanobiology.
Keywords: Peptide nucleic acid, DNA biosensor, DNase resistance, integrin, tension sensor, cellular force imaging
1. Introduction
Integrin-mediated force transmission is important for many cellular functions, including cell survival (Aoudjit and Vuori, 2012; Giancotti, 1997; Illario et al., 2003), spreading (Cavalcanti-Adam et al., 2007; Price et al., 1998), migration (Hood and Cheresh, 2002; Huttenlocher and Horwitz, 2011), proliferation (Moreno-Layseca and Streuli, 2014; Shankar et al., 1993) and differentiation (Gomez-Lamarca et al., 2014; Wang et al., 2015a). A variety of methods have been developed to measure and map cellular force (Polacheck and Chen, 2016; Roca-Cusachs et al., 2017). To visualize the invisible cellular force at high resolution, one powerful strategy is to convert the molecular force transmitted by integrins to fluorescence using integrin tension sensors equipped with a dye-dye pair or a dye-quencher pair. Biomaterials such as spider silk peptide (Brenner et al., 2016), polyethylene glycol (Legant et al., 2010; Miller et al., 2010) and DNA (Blakely et al., 2014; Wang and Ha, 2013; Zhang et al., 2014) have been adopted for constructing the tension sensors. Among these materials, DNA attracted tremendous interest for its programmable tension sensing ability, matured chemistry for the conjugation with dyes and integrin ligands, and well-calibrated mechanical property (Cocco et al., 2001; Hatch et al., 2008). Both hairpin DNA (Blakely et al., 2014; Zhang et al., 2014) and dsDNA (Wang and Ha, 2013; Wang et al., 2018) were adopted for calibrating integrin tension and imaging cellular force. These tension sensors have been successfully applied to a broad scope of studies including platelet functions (Wang et al., 2018; Zhang et al., 2018), cell migration (Zhao et al., 2018) and immune cell activation (Liu et al., 2016), etc.
Despite its versatility as biomaterial for molecular engineering, DNA is susceptible to degradation by DNase, either soluble DNases released by cells (Fischer et al., 2011; Perry and Chalkley, 1981; Zhang et al., 2009) or membrane-bound DNases expressed by some cell types (Shiokawa et al., 2007; Wang et al., 2019). During cell culture and plating, some cells may rupture due to cell death, apoptosis or mechanical shear stress, releasing soluble DNases into the culture medium. These DNases may gradually deteriorate the tension sensors immobilized on the substrate. Moreover, many cancer cells tend to express substantial membrane-bound DNases which degrade surface-immobilized DNA (Wang et al., 2019). Collectively, these soluble and membrane-bound DNases can degrade DNA-based tension sensors or even produce false force signals, limiting the application of DNA-based tension sensors in the study of cells that have extracellular DNase activity.
Previously we developed dsDNA-based Integrative tension sensor (ITS) (Wang et al., 2018) and applied it to image cellular force in platelets, keratocytes and other cell types which have no or relatively low DNase activity. A typical ITS is a dsDNA with the upper strand conjugated with a quencher and a RGD (Arginine-glycine-aspartic acid) peptide ligand (Mondal et al., 2013) targeting integrins, and the bottom strand conjugated with a dye in the proximity of the quencher and a biotin for surface immobilization. During cellular force mapping, ITS is coated on a coverslip where cells are plated subsequently. Upon cell adhesion, integrins bind and transmit force to ITS and activate fluorescent signal by mechanically dissociating the dsDNA of the ITS and separating the dye-quencher pair, so that the force is reported by fluorescence. However, when applied to the force study in cancer cells such as MDA-MB-231 breast cancer cells, MTC (mouse thyroid carcinoma) cells, and macrophages, ITS suffered from degradation or even produced false force signal. We identified that the DNase activity on the cell membrane caused the ITS degradation and false signal generation. The DNase activity in these cell types would likely degrade other DNA-based tension sensors as well. Overall, the application of DNA-based tension sensors is hampered in the cellular force study of many DNase-active cell types. To address this issue, here we developed DNase-resistant tension sensors that tolerate both soluble and membrane-bound DNases while retaining the ability of cellular force imaging.
2. Materials and Methods
2.1. Synthesis of integrative tension sensors based on dsDNA, dsRNA, PNA/DNA and PNA/RNA hybrids
DNAs and chemically modified RNAs were customized and purchased from Integrated DNA Technologies. PNA was customized and purchased from PNA Bio Inc. The sequences of the nucleic acids are:
2.2. Immobilizing ITS on glass bottom petridishes
The detail of ITS coating can be found in articles (Wang et al., 2015b; Zhao et al., 2019). In brief, a glass bottom petri dish was incubated with 100 μg/ml BSA-biotin (Sigma-Aldrich, A8549) spiked with 5 μg/ml fibronectin in PBS for 30 min and washed with PBS 3 times. BSA-biotin coated the surface to prevent non-specific binding and provide biotin which was available for avidin protein binding. Afterward the surface was incubated with 50 μg/ml Neutravidin (Thermo Fisher Scientific, 31000) in PBS for 30 min and washed with PBS 3 times. Finally, the surface was incubated in 1μM ITS in PBS for 30min. It is important to make sure that the surface should not dry or the imaging background will be messy.
2.3. Cell culture and plating
All cells except platelets were cultured in culture media recommended by ATCC.org. To plate cells on ITS surface, the cells in culture flasks were washed with mild cell detaching solution (recipe: 100 mL 10× HBSS + 10 mL 1 M HEPES (PH7.6) + 10 mL 7.5% sodium bicarbonate + 2.4 mL 500 mM EDTA + 1 L H2O). The detaching solution was then added to treat the cells for 5–10 min. Detached cells were dispersed with a pipette, collected and centrifuged for 3 min with 300 RCF. Supernatant was discarded and serum-free culture medium was added to re-suspend the cells at a cell density around 1×106/mL. The cell solution was plated on the ITS surface and incubated in 5% CO2 and at 37°C for 2 hours before imaging or immunostaining.
The process of culturing platelets can be found in (Wang et al., 2018). Platelet-rich plasma (PRP) was provided by collaborators. After receiving PRP, it was centrifuged for 8 min with 800 RCF. Supernatant was discarded, the cell pellet was re-suspended in F-12 medium (HFL05, Caisson Laboratories) and 10 μM Adenosine diphosphate (ADP) was added to platelets media in order for the activation of platelets. The platelets were cultured onto ITS surface for 40 min before fixation and imaging.
2.4. Vinculin, actin immunostaining of cells on ITS surface
After 2h of culture of cells on ITS surface, cells were fixed with 4% paraformaldehyde (PFA) for 10 min and rinsed with PBS 3 times. Then the sample was permeabilized with 0.5% Triton X for 10 min and rinsed with PBS 3 times. Then the sample was incubated with 2μg/mL BSA solution at 4 °C to block non-specific absorption and washed with PBS 3 times. Afterwards, primary antibody (anti-Vinculin, from Sigma-Aldrich, FAK100) and secondary antibody with Alexa405 dye was respectively applied to the sample for 1–2h at 4 °C, with 3 wash with PBS + 0.05% Tween20 after each step. Finally, 5 units/mL of Phalloidin-Alexa 647 (Thermo Fisher Scientific, A22287) was applied to the sample for 20 min and the sample was rinsed with PBS 3 times.
2.5. Fluorescent imaging of Cy3 molecules conjugated on DNA on PNA
We used pegylated glass coverslips (preparation of PEG glass can be found in (Wang and Ha, 2013)) for imaging of Cy3 molecules. PEG glass was incubated with 50 μg/ml neutravidin for 30 min and washed with PBS once. Then the glass was incubated with 2 pM Cy3 molecules conjugated onto dsDNA or hybrid DNA/PNA without Quencher for 30 min. The glass was rinsed once with PBS and soaked in PBS during imaging.
2.6. Soluble Dnase I treatment on tension sensor
After the immobilization of ITS onto glass bottom petri dish, the dish was treated with 20 unit/mL DNase I (Thermo Fisher Scientific, 89836) in DMEM media (Corning, 10–013-CV). Time elapse imaging was performed immediately.
2.7. Imaging
The imaging of MDA-MB-231 cells, platelets and DNase I treatment was conducted with an epi-fluorescent microscope (Ti, Nikon) with a 40× lens (Plan Fluor 40×/0.75, Nikon) and TRITC or GFP-B filters (Nikon).
The imaging of all other samples was performed with a total internal reflection fluorescence microscope (TIRFM) (Ti-2, Nikon) with a TIRF objective lens (CFI Apo TIRF 100× Oil, Nikon). The laser (LU-N Laser Unit, Nikon) with 405 nm, 488 nm, 561 nm, 640 nm wavelengths was used as excitation light. Laser Quad Band Set (TRF89901-EMv2, CHROMA) was used as the universal optical filter for fluorescence imaging with all the four laser wavelengths.
3. Results
3.1. dsDNA-based tension sensor is susceptible to the degradation by membrane-bound DNases
Mammalian cells may express membrane-bound DNases (Los et al., 2000; Shiokawa et al., 2007) which may protect the cells from exogenous gene invasion. Many cancer cells, including MDA-MB-231 and MTC cells, express DNase X, a type of membrane-bound DNase (Wang et al., 2019). To test if DNase X disrupts the performance of DNA-based ITS, we prepared two dsDNA constructs, with one conjugated with integrin ligand RGD (ITS) and the other without RGD (RGD-null ITS) (Fig. 1A). In principle, the ITS can be activated to fluorescent state by either DNA dissociation or DNA degradation which both can free the dye from quenching (Fig. 1B). The two constructs were immobilized on glass surfaces along with fibronectin which facilitates cell adhesion. MDA-MB-231 cells were plated and cultured on the surfaces for 2 hours. Bright fluorescent signals were generated on both surfaces, regardless with or without RGD (Fig. 1C), suggesting that the ITS is not reporting integrin-transmitted cellular force. To confirm that the fluorescent signal was caused by DNase X, the cells on ITS surfaces were fixed and immunostained with DNase X antibody and Alexa 647 labeled secondary antibody. The immunostained DNase X was well co-localized with the fluorescent pattern of ITS (Fig. 1D), suggesting the ITS signal was mainly caused by DNase X. A previous study (Wang et al., 2019) also showed that knocking down DNase X expression in MDA-MB-231 cells with siRNA significantly reduced the degradation of surface-immobilized dsDNA. Therefore, DNA-based ITS was degraded by DNase X on the cell membrane, giving rise to false force signal. We also found that the false signal is generally much brighter than the fluorescent signal produced by cellular force, perhaps because DNases degrade DNA more efficiently than cellular forces dissociate DNA. It is likely that tension sensors based on other DNA structures such as DNA hairpins would also be degraded or produce false force signal when applied to the force study of cells expressing membrane-bound DNases.
Figure 1. Membrane-bound DNase disrupts the performance of DNA-based tension sensor.
(A) Structures of dsDNA-based tension sensor (ITS, shear configuration of dsDNA) and RGD-null ITS (without integrin ligand). (B) ITS can be activated to fluoresce either by integrin tension or by DNase cleavage. RGD-null ITS can be activated to fluorescence by DNase cleavage, but not by integrin tension. (C) Both ITS and RGD-null ITS showed similar fluorescence pattern, suggesting that ITS is activated by DNase instead of integrin tension. (D) MDA-MB-231 cells exhibited rich DNase activity on cell membrane, evidenced by the co-localization between immunostained DNase X and ITS. Scale bar: 20 μm
3.2. PNA/DNA, modified dsRNA and PNA/RNA resist both soluble and membrane-bound DNases
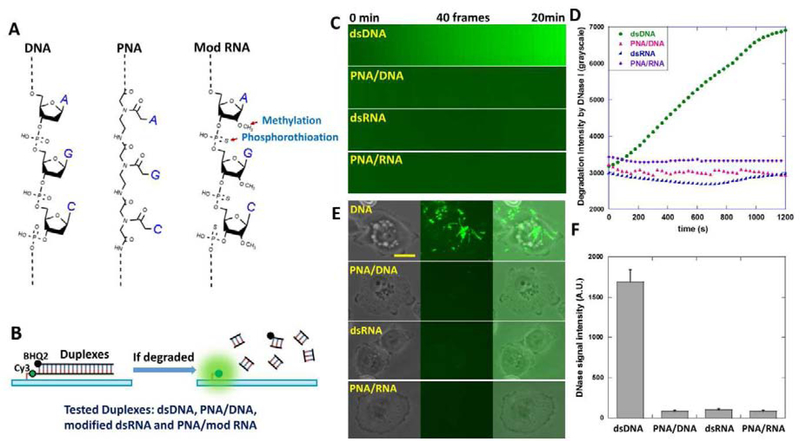
In order to overcome the susceptibility of DNA-based tension sensor to DNases, we tested PNA (peptide nucleic acid) and chemically modified RNA as alternative biomaterials for tension sensor construction. PNA is a nucleic acid with peptide N-(2-aminoethyl)-glycine as the backbone (Fig. 2A). It was previously reported that the PNA/DNA hybrid duplex has strong resistance to DNases and proteases (Pellestor and Paulasova, 2004). RNA as the other candidate is naturally resistant to DNase degradation. However, RNA is highly susceptible to RNases at even trace level. As a result, RNA is unstable in general lab setting and in cell culture unless extreme caution is taken. To increase the robustness of RNA material, we adopted modified RNA for tension sensor construction. The RNA in this paper was modified with 2’-O-Methylation at all of its nucleotides and phosphorothioation at the backbone (Fig. 2A). These two modifications were reported to increase RNA stability during in vivo nucleic acid experiments (Hernandez et al., 2012; Putney et al., 1981). In following context, “RNA” refers to the RNA with these two modifications.
Figure 2. Testing the resistance of dsDNA, PNA/DNA, modified dsRNA and PNA/RNA to both soluble DNase and membrane-bound DNase.
(A) DNA, PNA, RNA modified by methylation and phosphorothioation and their hybrid duplexes were prepared and tested as candidate DNase-resistant biomaterials. (B) dsDNA, PNA/DNA, dsRNA (modified) and PNA/RNA as duplexes (RGD-null ITS) were designed as in the figure. In both PNA/DNA and PNA/RNA constructs, PNA is the lower strand immobilized on the surface. Duplex degradation by DNase would de-quench Cy3 and be reported by local fluorescence. (C) Four surfaces coated with these duplexes, respectively, were treated with 20 unit/mL DNase I. Surface fluorescence intensities was monitored by 20 min time-lapse imaging. (D) Surface fluorescence intensity versus time on four surfaces. (E) MTC cells expressing membrane-bound DNase were plated on the four surfaces. (F) Fluorescence signal intensities caused by membrane-bound DNase on the four surfaces. Scale bar: 10 μm.
Four duplex constructs, dsDNA, PNA/DNA, dsRNA and PNA/RNA with the same nucleic acid sequences (except that the thymine was replaced with uracil in RNA constructs) were prepared according to Table 1. Their resistance to both soluble DNase and cell membrane-bound DNase were tested. The four duplex constructs were conjugated with Cy3-BHQ2 pairs and immobilized on glass surfaces through biotin-neutravidin interaction (Fig. 2B). No RGD ligand was conjugated to these duplexes. In both PNA/DNA and PNA/RNA hybrid duplexes, PNA is the lower strand that is labeled with Cy3 and directly immobilized on the surface. If the duplex construct is degraded, Cy3 will be left on the surface and freed from quenching, hence emitting fluorescence and reporting the duplex degradation. First, we tested their resistance to DNase I, a common type of soluble DNase. Glass-bottom petridishes coated with the duplexes were treated with 20 unit/mL DNase I in DMEM cell culture medium. Time-lapse imaging was conducted and the fluorescence intensity was recorded for each surface (Fig. 2C). As expected, during the 20 min treatment, fluorescence intensity of the dsDNA construct rapidly increased, reporting the dsDNA degradation by DNase I. In contrast, fluorescence intensities of other three duplexes did not have obvious increment (Fig. 2C–2D), suggesting that PNA/DNA, dsRNA and PNA/RNA are indeed strongly resistant to DNase I.
Table. 1.
Nucleic acids for the synthesis of RGD-null ITS that reports their DNA resistance to DNases. (Sequences are from 5‵ end to 3‵ end)
| 1 | DNA upper strand for RGD-null ITS | GGG CGG CGA CCT CAG CAT/3BHQ_2/ |
| 2 | DNA lower strand for RGD-null ITS | /5BiosG/T/iCy3/ATG CTG AGG TCG CCG CCC/ |
| 3 | RNA upper strand for RGD-null ITS | mG*mG*mG* mC*mG*mG* mC*mG*mA* mC*mC*mU* mC*mA*mG* mC*mA*mU*/3BHQ_2/ |
| 4 | RNA lower strand for RGD-null ITS | /5BiosG/mU*/iCy3/mA*mU*mG* mC*mU*mG* mA*mG*mG* mU*mC*mG* mC*mC*mG* mC*mC*mC*/ |
| 5 | PNA lower strand for RGD-null ITS | Biotin-OO-Lys(Cy3)-O-ATGCTGAGGTCGCCGCCC |
Notes:
Strands 1 and 2 were hybridized to dsDNA-based RGD-null ITS.
Strands 3 and 4 were hybridized to dsRNA-based RGD-null ITS.
Strands 1 and 5 were hybridized to PNA/DNA-based RGD-null ITS.
Strands 3 and 5 were hybridized to PNA/RNA-based RGD-null ITS.
/3BHQ_2/ represents a Black Hole Quencher 2 at the 3’ end.
/5BiosG/ represent a biotin tag at the 5’ end of a nucleic acid.
m_* in strands 5 and 6 represents a phosphorothioated 2’O-methyl base.
“O” in the PNA sequence represents an ethylene glycol that increases the PNA solubility in water.
We then tested the susceptibilities of these duplexes to membrane-bound DNase X in live cells. MTC cells were cultured on the surfaces separately coated with these duplexes and incubated for 2 hours. In contrast to the significant DNase signal shown by dsDNA construct, all other three constructs showed little fluorescent response to cells (Fig. 2E). The statistics also shown that the average fluorescence intensities produced by DNase of MTC cells on DNase-resistant constructs are less than 5% of the signal intensity produced on the dsDNA-based construct (Fig. 2F), indicating that the resistance of PNA/DNA, dsRNA and PNA/RNA duplexes to membrane-bound DNases are at least 20 times higher than dsDNA.
3.3. Cellular force imaging by tension sensors based on PNA/DNA, modified dsRNA and PNA/RNA
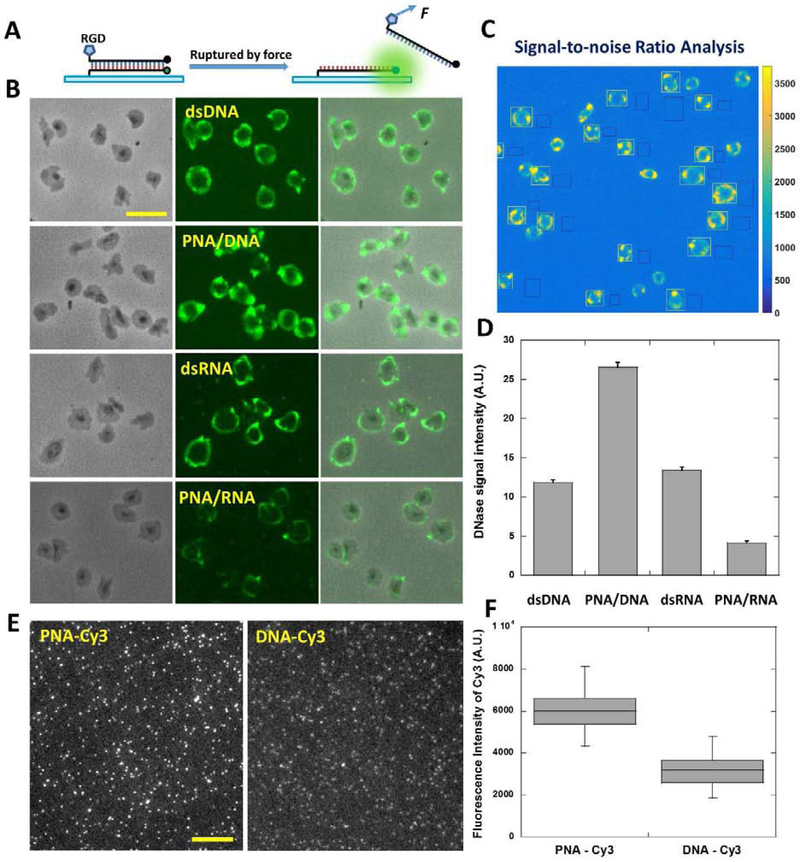
We confirmed the resistance of the alternative duplexes to DNases. Next we synthesized tension sensor ITSs based on these duplexes by conjugating integrin ligand RGD to them in the unzipping configuration, with the RGD and the biotin at the same end of the duplex (Fig. 3A). Refer to table 2 for the nucleic acid sequences and modifications. PNA is the lower strand in both PNA/DNA and PNA/RNA ITSs. To evaluate their force reporting abilities exclusively, we selected platelets as cell models as platelets reliably produce cellular force maps and exhibited little DNase activity in our previous force assays (Wang et al., 2018). Platelets were incubated on four surfaces coated with tension sensors based on dsDNA, PNA/DNA, dsRNA and PNA/RNA, respectively. After 30 min incubation, cell samples and the force maps were imaged with a fluorescence microscope (Fig. 3B) with the same optical settings (light source power and exposure time, etc.). The typical ring-shaped platelet force patterns were observed on all tension sensor surfaces. To evaluate the force mapping capability, the signal-to-noise ratios (SNR) of platelet force maps against the background were quantified (Fig. 3C). Among these four types of tension sensors, PNA/DNA ITS has the highest SNR, and dsRNA ITS has a SNR at a similar level to that of dsDNA-based ITS. This shows that tension sensors based on PNA/DNA and dsRNA duplexes have comparable or even better performance than dsDNA-based tension sensor. However, the SNR of PNA/RNA ITS is significantly lower, suggesting its inferior cellular force reporting ability, possibly because PNA/RNA has higher binding energy than other duplexes (Natsume et al., 2007) and therefore is more difficult to be ruptured by cellular forces. In addition, PNA/RNA is also the most costly one among the four duplexes in terms of sample preparation. Therefore PNA/RNA is not recommended for tension sensor synthesis and it was not used in the following tests.
Figure 3. Evaluating cellular force mapping abilities of tension sensors with DNase-free platelets.
(A) Tension sensors based on four types of duplexes, dsDNA, PNA/DNA, dsRNA (modified RNA) and PNA/RNA, were prepared in an unzipping configuration. For the two hybrid duplexes, PNA is the lower strand which is immobilized on substrates by biotin-neutravidin interaction. (B) Platelet force maps on the four tension sensor surfaces. Scale bar: 10 μm. (C) Signal-to-noise ratios of platelet force maps were evaluated with the division quotient of fluorescence intensity of platelets (regions circled by white rectangles) and pixel-to-pixel standard deviation of background fluorescence (regions circled by black rectangles). Fluorescence intensity of single platelets was obtained by subtracting background fluorescence intensity from fluorescence intensity in platelet regions. (D) Statistics of signal-to-noise ratios of platelet force maps on four tension sensor surfaces. (E) Fluorescence imaging of single Cy3s conjugated to PNA and DNA, respectively. Scale bar: 10 μm. (F) Fluorescence intensities of single PNA-Cy3s and single DNA-Cy3s under the same imaging settings.
Table. 2.
Nucleic acids for the synthesis of ITS reporting cellular forces. (Sequences are from 5‵ end to 3‵ end)
| 1 | DNA upper strand for ITS | /5ThioMC6-D/GGG CGG CGA CCT CAG CAT/3BHQ_2/ |
| 2 | DNA upper strand for ITS (Linked with RGD) | /RGD/GGG CGG CGA CCT CAG CAT/3BHQ_2/ |
| 3 | DNA lower strand for ITS | /5Cy3/ATG CTG AGG TCG CCG CCC/3Bio/ |
| 4 | RNA upper strand for ITS | /5ThioMC6-D/ mG*mG*mG* mC*mG*mG* mC*mG*mA* mC*mC*mU* mC*mA*mG* mC*mA*mU*/3BHQ_2/ |
| 5 | RNA upper strand for ITS (Linked with RGD) | /RGD/ mG*mG*mG* mC*mG*mG* mC*mG*mA* mC*mC*mU* mC*mA*mG* mC*mA*mU*/3BHQ_2/ |
| 6 | RNA lower strand | /5Cy5/mA*mU*mG* mC*mU*mG* mA*mG*mG* mU*mC*mG* mC*mC*mG* mC*mC*mC*/3Bio/ |
| 7 | PNA lower strand | Cy3-O-ATGCTGAGGTCGCCGCCC-KKKK(Biotin) |
Notes:
/5ThioMC6-D/ represents a thiol modification which is used to conjugate a RGD peptide to a DNA or a RNA. Protocol of conjugating RGD to DNA strand 1 or RNA strand 6 can be found in (Zhao et al., 2019).
/3Bio/ represent a biotin tag at the 3’ end of a nucleic acid.
“K” in the PNA sequence represents a lysine that increases the PNA solubility in water.
Strands 2 and 3 were hybridized to dsDNA-based RGD ITS.
Strands 5 and 6 were hybridized to dsRNA-based RGD ITS.
Strands 2 and 7 were hybridized to PNA/DNA-based RGD ITS.
Strands 5 and 7 were hybridized to PNA/RNA-based RGD ITS.
We investigated why PNA/DNA ITS exhibits higher SNR than other ITSs in cellular force mapping. One potential reason is that the fluorescence intensity of cyanine dyes is enhanced at the proximity (nanometer range) of a protein or a peptide, known as a phenomenon named PIFE (Protein induced fluorescence enhancement) (Hwang and Myong, 2014). We performed single molecule imaging on Cy3 conjugated to a DNA or a PNA and compared their fluorescence intensities (Fig. 3E–3F). Under the same imaging setting, Average fluorescence intensity of single Cy3s on PNA is two times of the fluorescence intensity of single Cy3s on DNA. The PIFE effect brings an additional advantage to the PNA-based tension sensor in terms of signal strength.
3.4. Modified dsRNA-based Tension sensor exhibits erratic activation in some cell types
Although dsRNA ITS has strong resistance to DNases and shows good performance in platelet force mapping, we found that this tension sensor exhibits erratic activation in some cell types. During cellular force imaging in NIH 3T3 cells, a type of fibroblasts, we repeatedly observed significant amount of fluorescent signal in a punctate pattern on dsRNA ITS surface (Fig. 4A). Immunostaining of focal adhesions showed little co-localization between these dots and focal adhesions, suggesting that these fluorescent dots are not force signal. In contrast, PNA/DNA ITS had normal performance in cellular force mapping of NIH 3T3 cells and reported cellular force in a typical streak pattern which is well co-localized with focal adhesions. Therefore, the dsRNA-based tension sensor even with the two RNA modifications is still not robust in cellular force mapping. The exact reason for this erratic fluorescence activation is unknown. A possibility is that NIH 3T3 cells may release RNases that degraded the RNA despite its methylation and phosphorothioation, producing local fluorescence signal that is not caused by cellular force. Therefore, after all these tests, PNA/DNA duplex is the most robust construct for the development of DNase-resistant tension sensor.
Figure 4. dsRNA ITS exhibits non-force signal in NIH 3T3 cells.

(A) dsRNA ITS produced fluorescent signal in a punctate pattern that has no apparent co-localization with focal adhesions (immunostained vinculin), suggesting that the fluorescent dots are not related with integrin-transmitted cellular force. (B) PNA/DNA ITS reports cellular force in a streak pattern which is spatially associated with focal adhesions. Scale bar: 10μm
3.5. Tests of PNA/DNA-based ITS in a variety of cell types
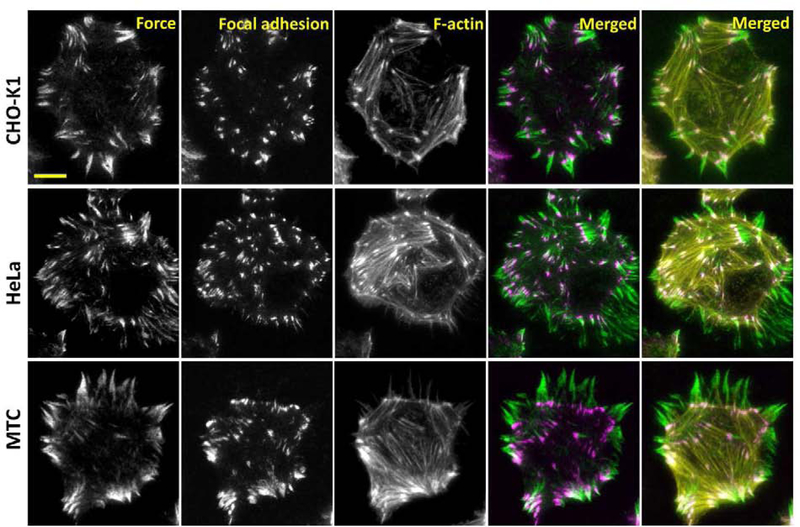
We have shown that PNA/DNA ITS has high DNase resistance and reliable force-reporting capability, making it an excellent alternative biomaterial replacing DNA for synthesizing tension sensors applicable to the study of cells exhibiting strong DNase activities. PNA/DNA ITS has successfully imaged cellular forces in platelets and NIH 3T3 cells. We further tested its force-reporting capability with three other cell lines: CHO-K1 cells, HeLa cells and MTC cells. All these cells were incubated on PNA/DNA ITS surfaces (co-coated with fibronectin) for 2 h, and then fixed and stained with antibody against vinculin and phalloidin. Cellular force maps reported by PNA/DNA ITS, focal adhesions marked by the antibody and F-actin marked by phalloidin were co-imaged in these cells (Fig. 5). The integrin-transmitted cellular force, focal adhesions and stress fibers showed excellent spatial associations, suggesting that PNA/DNA ITS reported cellular force properly in these cells. Moreover, owing to the PIFE effect, fluorescence intensity of cellular force maps reported by PNA/DNA ITS was enhanced to a level similar to that of cellular structural imaging, making cellular force as bright as cell structures. These experiments confirmed that PNA/DNA ITS can routinely and robustly report cellular forces in various cell types.
Figure 5. Co-imaging of cellular force and cell structures with PNA/DNA ITS.
PNA/DNA ITS was applied to cellular force imaging in CHO-K1, HeLa and MTC cells. Focal adhesions and F-actin were stained by antibody against Vinculin (secondary antibody is labeled with Alexa 405) and phalloidin labeled with Alexa 647, respectively. The fluorescence intensities are at comparable levels in three imaging channels, indicating that cellular force reported by PNA/DNA ITS is as visible as cell structures. Scale bar: 10 μm
4. Conclusion
DNase is ubiquitous in cells and their local environment. DNase I exists in serum (Kishi et al., 1990) and organs such as kidney, liver and pancreas. DNase II exists in urine (Yasuda et al., 1992) and spleen (Koerner and Sinsheimer, 1957), Membrane-bound DNase is expressed on cancer cells and muscle cells. As DNA-based tension sensors have been developed and applied to study a wide range of cellular functions, tension sensors with DNase resistance are desired to extend cellular force study to cells exhibiting elevated DNase activity.
In this work, we tested PNA/DNA, chemically modified dsRNA and PNA/RNA as alternative biomaterials to develop DNase-resistant tension sensors. These tension sensors were evaluated in terms of DNase resistance, cellular force imaging ability and general robustness in applications. We found that even with methylation and phosphorothioation, RNA is still not a robust biomaterial for tension sensor synthesis and application. In contrast, PNA/DNA hybrid duplex shows strong resistance to both soluble DNase and membrane-bound DNase in cells, and exhibits high sensitivity and specificity in cellular force imaging of all tested cells. Therefore, PNA/DNA provides a valuable and accessible biomaterial to replace DNA for the synthesis of tension sensor when its resistance to DNases is required.
Currently, DNA-based tension sensors are still applicable for a large number of cell types that have insignificant DNase activity on cell membrane and in culture environment. However, when applying the DNA-based tension sensor to a new type of cells, it is advised to confirm that cells do not degrade the sensor or produce false force signal using the RGD-null tension sensor. In our tests, we found that cancer cell-lines and macrophages generally have higher probability to express DNases on cell membrane. In such case, the PNA/DNA hybrid would be a good alternative material for the synthesis of the tension sensor to resist the DNase activity. Overall, PNA-based tension sensors greatly increase the robustness of molecular tension sensors and broaden their application in the study of cell mechanobiology.
Acknowledgement
The work was supported by National Institute of General Medical Sciences (1R35GM128747) and by National Science Foundation (1825724). We thank Dr. Dana LeVine for providing canine blood sample used in the platelet force assay.
Footnotes
Replacing DNA with PNA in constructing molecular tension sensors
PNA-based tension sensors resist DNase which is ubiquitous in cell environment
PNA enhances fluorescence, making force signal as bright as cell structural signal
Yuanchang Zhao: Methodology, Data acquisition, Data analysis, Original draft preparation.
Anwesha Sarkar: Platelet sample preparation.
Xuefeng Wang: Conceptualization, Methodology, Manuscript Writing.
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
□ The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Aoudjit F, Vuori K, 2012. Chemother Res Pract 2012, 283181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely BL, Dumelin CE, Trappmann B, McGregor LM, Choi CK, Anthony PC, Duesterberg V, Baker BM, Block SM, Liu DR, Chen CS, 2014. Nature Methods 11(12), 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner MD, Zhou RB, Conway DE, Lanzano L, Gratton E, Schwartz MA, Ha T, 2016. Nano Lett 16(3), 2096–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz JP, 2007. Biophys J 92(8), 2964–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco S, Monasson R, Marko JF, 2001. P Natl Acad Sci USA 98(15), 8608–8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H, Scherz J, Szabo S, Mildner M, Benarafa C, Torriglia A, Tschachler E, Eckhart L, 2011. Plos One 6(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti FG, 1997. Curr Opin Cell Biol 9(5), 691–700. [DOI] [PubMed] [Google Scholar]
- Gomez-Lamarca MJ, Cobreros-Reguera L, Ibanez-Jimenez B, Palacios IM, Martin-Bermudo MD, 2014. Journal of Cell Science 127(Pt 21), 4667–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch K, Danilowicz C, Coljee V, Prentiss M, 2008. Phys Rev E 78(1). [DOI] [PubMed] [Google Scholar]
- Hernandez FJ, Stockdale KR, Huang LY, Horswill AR, Behlke MA, McNamara JO, 2012. Nucleic Acid Ther 22(1), 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JD, Cheresh DA, 2002. Nat Rev Cancer 2(2), 91–100. [DOI] [PubMed] [Google Scholar]
- Huttenlocher A, Horwitz AR, 2011. Cold Spring Harb Perspect Biol 3(9), a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Myong S, 2014. Chem Soc Rev 43(4), 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illario M, Amideo V, Casamassima A, Andreucci M, di Matola T, Miele C, Rossi G, Fenzi G, Vitale M, 2003. J Clin Endocrinol Metab 88(1), 260–269. [DOI] [PubMed] [Google Scholar]
- Kishi K, Yasuda T, Ikehara Y, Sawazaki K, Sato W, Lida R, 1990. Am J Hum Genet 47(1), 121–126. [PMC free article] [PubMed] [Google Scholar]
- Koerner JF, Sinsheimer RL, 1957. J Biol Chem 228(2), 1039–1048. [PubMed] [Google Scholar]
- Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS, 2010. Nature Methods 7(12), 969–U113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Blanchfield L, Ma VPY, Andargachew R, Galior K, Liu Z, Evavold B, Salaita K, 2016. P Natl Acad Sci USA 113(20), 5610–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los M, Neubuser D, Coy JF, Mozoluk M, Poustka A, Schulze-Osthoff K, 2000. Biochemistry-Us 39(25), 7365–7373. [DOI] [PubMed] [Google Scholar]
- Miller JS, Shen CJ, Legant WR, Baranski JD, Blakely BL, Chen CS, 2010. Biomaterials 31(13), 3736–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal G, Barui S, Chaudhuri A, 2013. Biomaterials 34(26), 6249–6260. [DOI] [PubMed] [Google Scholar]
- Moreno-Layseca P, Streuli CH, 2014. Matrix Biol 34, 144–153. [DOI] [PubMed] [Google Scholar]
- Natsume T, Ishikawa Y, Dedachi K, Tsukamoto T, Kurita N, 2007. Chem Phys Lett 434(1–3), 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellestor F, Paulasova P, 2004. Eur J Hum Genet 12(9), 694–700. [DOI] [PubMed] [Google Scholar]
- Perry M, Chalkley R, 1981. J Biol Chem 256(7), 3313–3318. [PubMed] [Google Scholar]
- Polacheck WJ, Chen CS, 2016. Nature Methods 13(5), 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price LS, Leng J, Schwartz MA, Bokoch GM, 1998. Mol Biol Cell 9(7), 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney SD, Benkovic SJ, Schimmel PR, 1981. Proceedings of the National Academy of Sciences 78(12), 7350–7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P, Conte V, Trepat X, 2017. Nat Cell Biol 19(7), 742–751. [DOI] [PubMed] [Google Scholar]
- Shankar G, Davison I, Helfrich MH, Mason WT, Horton MA, 1993. Journal of Cell Science 105 ( Pt 1), 61–68. [DOI] [PubMed] [Google Scholar]
- Shiokawa D, Matsushita T, Shika Y, Shimizu M, Maeda M, Tanuma SI, 2007. J Biol Chem 282(23), 17132–17140. [DOI] [PubMed] [Google Scholar]
- Wang H, Luo X, Leighton J, 2015a. Biochem Insights 8(Suppl 2), 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XF, Ha T, 2013. Science 340(6135), 991–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XF, Sun J, Xu Q, Chowdhury F, Roein-Peikar M, Wang YX, Ha T, 2015b. Biophys J 109(11), 2259–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, LeVine DN, Gannon M, Zhao YC, Sarkar A, Hoch B, Wang XF, 2018. Biosens Bioelectron 100, 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Zhao YC, Sarkar A, Wang XF, 2019. J Biophotonics 12(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Nadano D, Awazu S, Kishi K, 1992. Biochim Biophys Acta 1119(2), 185–193. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ge CH, Zhu C, Salaita K, 2014. Nat Commun 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qiu Y, Blanchard AT, Chang Y, Brockman JM, Ma VP, Lam WA, Salaita K, 2018. Proc Natl Acad Sci U S A 115(2), 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang YY, Liu B, 2009. Anal Chem 81(10), 3731–3737. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Wetter NM, Wang X, 2019. JoVE (Journal of Visualized Experiments)(146), e59476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YC, Wang YL, Sarkar A, Wang XF, 2018. Iscience 9, 502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]