Abstract
Successful fertilization is fundamental to sexual reproduction, yet little is known about the mechanisms that guide sperm to oocytes within the female reproductive tract. While in vitro studies suggest that sperm of internally fertilizing animals can respond to various cues from their surroundings, the inability to visualize their behavior inside the female reproductive tract creates a challenge for understanding sperm migration and mobility in its native environment. Here, we describe a method using C. elegans that overcomes this limitation and takes advantage of their transparent epidermis. C. elegans males stained with a mitochondrial dye are mated with adult hermaphrodites, which act as modified females, and deposit fluorescently labeled sperm into the hermaphrodite uterus. The migration and motility of the labeled sperm can then be directly tracked using an epi-fluorescence microscope in a live hermaphrodite. In wild-type animals, approximately 90% of the labeled sperm crawl through the uterus and reach the fertilization site, or spermatheca. Images of the uterus can be taken 1 h after mating to assess the distribution of the sperm within the uterus and the percentage of sperm that have reached the spermatheca. Alternatively, time-lapse images can be taken immediately after mating to assess sperm speed, directional velocity and reversal frequency. This method can be combined with other genetic and molecular tools available for the C.elegans to identify novel genetic and molecular mechanisms that are important in regulating sperm guidance and motility within the female reproductive tract.
Keywords: Developmental Biology, Issue 148, fertilization, sperm, chemotaxis, migration, oviduct, C. elegans, guidance, mating, prostaglandin
Introduction
The molecular mechanisms by which spermatozoa (referred to as sperm) navigate through the female reproductive tract toward the oocyte are not well understood, yet are fundamental to sexual reproduction. Sperm motility is highly dynamic and depends on robust communication signals that alter sperm velocity and directional motility1,2,3,4,5,6,7,8,9,10,11,12. C. elegans has become a powerful model for studying sperm movement in vivo because the hermaphrodite’s transparent epidermis permits the tracking of live sperm at single cell resolution2,3,8,10. The purpose of this paper is to provide methods for assessing sperm movement within the C. elegans hermaphrodite uterus.
In animal species where sperm and oocyte meet in the external environment (i.e., acquatic environments), sperm respond to chemotactic signals secreted by oocytes. These signals guide the direction of sperm movement, bringing them closer to the signal source4,6,11. However, much less is known about sperm movement in species that fertilize internally. A major challenge is the architecture of the female reproductive tract, which is inaccessible to microscopy in most species. In vitro studies in humans, mice, and pigs, for example, provide evidence that subpopulations of sperm can respond to chemoattractants, fluid flow, and thermal gradients1,5,7,9,12. With these systems, the inability to visualize and track sperm movement in vivo places serious limitations on strategies to discover the key mechanisms regulating these functions.
To overcome these limitations, we have developed methods using the nematode C. elegans to directly visualize sperm after insemination, to measure individual sperm migration parameters in vivo, and to measure the ability of a sperm population to target the fertilization site. These methods, together with the C. elegans molecular and genetic toolset, facilitate discovery of the chemical signaling molecules and molecular machinery that regulate sperm motility behaviors. For instance, genetic screens can be conducted in hermaphrodites or males to identify genes that are essential for efficient sperm movement in vivo13. Molecules can be injected into the hermaphrodite gonad to test for effects on sperm activation, migration velocity, and directional motility3. Additionally, the described methods can be used to monitor rogue sperm migration into ectopic body locations and to evaluate sperm competition10,14.
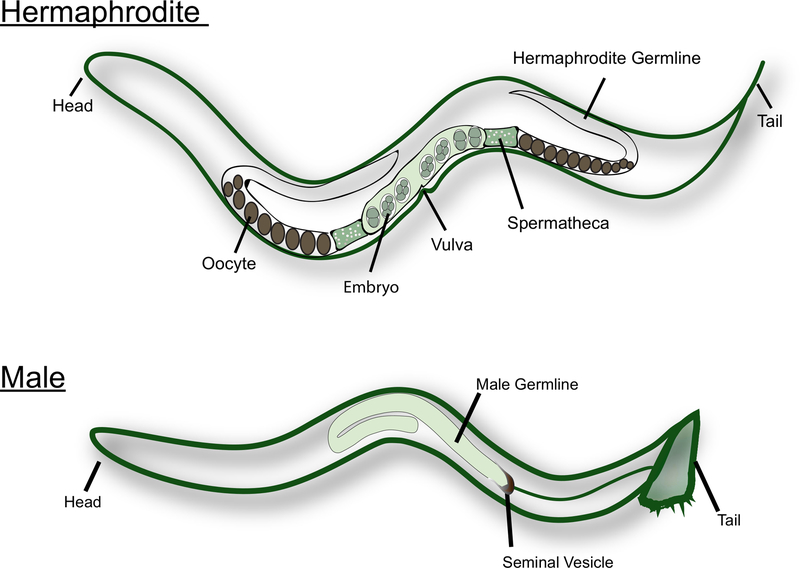
C. elegans exist in nature as hermaphrodites and males (see Figure 1). The hermaphrodite gonad has two U-shaped arms that are mirror images of each other. During the L4 larval stage, the most proximal germ cells (i.e., the cells near the spermatheca) undergo spermatogenesis. Each primary spermatocyte enters meiosis and produces four haploid spermatids. These spermatids are pushed into the spermatheca along with the first mature oocyte and undergo spermiogenesis15. Adult hermaphrodites switch from spermatogenesis to oogenesis. The oocytes mature in an assembly line fashion along the gonad, with the most mature oocyte at the proximal end of the gonad, next to the spermatheca. MSP signals from the sperm is needed to trigger meiotic maturation and ovulation16,17. Male C. elegans, on the other hand, have a J-shaped gonad that produce only sperm. The spermatids are stored in the seminal vesicle. Upon mating with the hermaphrodite or female, the male inserts the spicules near the tail into the vulva. Spermatids are activated during ejaculation, when they come in contact with the seminal fluid18. C. elegans sperm do not swim as they are not flagellated. Instead, they crawl through the reproductive tract, using the pseudopod for locomotion. It is well established that male sperm, which are larger in size, have a competitive advantage over hermaphrodite sperm14.
Figure 1: Cartoon of the adult C. elegans hermaphrodite and male.
Major reproductive structures are labeled in the figure.
In this method, male C. elegans act as the sperm donor and are mated to adult hermaprhodites. Adult males are stained with a fluorescent mitochondrial dye to produced labeled sperm. Once deposited through the hermaphrodite vulva, the sperm must crawl around the embryos in the uterus towards the spermatheca, or fertilization site. The transparent epidermis of the C. elegans model allows for the direct visualization of each individual sperm as it navigates through the female reproductive tract. In recent years, our lab has successfully used this method to demonstrate the importance of a class of F-series prostaglandins in guiding sperm from the vulva to the spermatheca19,20. The molecular mechansims governing its synthesis by the hermaphrodite and response by the sperm are still under investigation. However, this method for assessing sperm motility and migration greatly facilitates the identification of the key players that control sperm and oocyte communication in internally fertilizing animals. The following protocol describes step by step how to perform this assay.
Protocol
NOTE: All steps in this protocol are performed at room temperature (~20–22 °C) or in constant temperature incubators set to 16 °C or 20 °C. Male and hermaphrodite C. elegans are grown using standard culture conditions and NA22 or OP50 E. coli as a food source21,22. Wild-type N2 hermaphrodites and fog-2(q71) males are used in the procedure below.
1. Day 1: Picking L4 Stage Hermaphrodites for Mating
-
To obtain consistent results, all hermaphrodites should be synchronized as actively reproducing adults. Pick 20–30 L4 stage hermaphrodites to a 6 cm seeded nematode growth medium (NGM) plate. Incubate the hermaphrodites at 20 °C for 28–30 h.
NOTE: Only 12–15 hermaphrodites will be used for mating. The remaining hermaphrodites are surplus.
2. Day 1: Staining of Males with Fluorescent Mitochondrial Dye (Mito-dye)
Make a male staining plate by placing a dot of E. coli (food dot) at the center of an unseeded NGM plate. To make the food dot, use the end of a glass stirring rod to scrape E. coli from the bacteria lawn of a seeded plate and deposit it on the unseeded plate. The dot should be ~ 5–7 mm in diameter.
-
Mix together 2 μL of 1 mM mito-dye (see Table of Materials) in DMSO and 10 μL of M9 buffer (3 g of KH2PO4, 6 g of Na2HPO4, 5 g of NaCl, 1 mL of 1 M MgSO4, H2O to 1 L. Add MgSO4 after autoclaving). Pipette all of the mito-dye solution onto the food dot on the male staining plate. Let the plate dry in the dark (~30 min).
NOTE: Mito-dye is light sensitive. Shield all solution, plates, and worms containing mito-dye from light. Store the 1 mM stock at −20 °C.
-
Pick ~100 1–3 day old adult males23 to the mito-dye stained food dot on the male staining plate. Wrap the plate in aluminum foil and incubate overnight at 16 °C. For mating, use ~50–60 males per 12–15 hermaphrodites. If more than ~100 males are needed, make more staining plates to prevent overcrowding of males.
NOTE: Males can also be stained by incubating in a 10 μM mito-dye solution in M9 buffer for 3 h on a watch glass. Keep the worms covered to prevent evaporation and light exposure. After 3 h, use a Pasteur pipet to transfer males onto a 10 cm seeded NGM plate. Transfer as little of the mito-dye solution as possible. Wrap the plate in aluminum foil and incubate overnight in 16 °C.
Table of Materials
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description |
|---|---|---|---|
| Reagents and Material | |||
| 60mm × 15mm Petri Dish | Fisher | FB0875713A | |
| Agar | Fisher | BP1423–500 | |
| Sodium Chloride | Fisher | S671–3 | |
| Peptone | Fisher | BP1420–500 | |
| Cholesterol | Sigma-Aldrich | C8667 | |
| LB broth, Miller | Fisher | 1426–2 | |
| Escherichia coli strain NA22 | Caenorhabditis Genetics Center (CGC) | NA22 | Either this or OP50 E. coli can be used for C. elegans maintenance and assay. Both may be purchased at the CGC |
| N2 | CGC | N2 | |
| fog-2(q71) | CGC | CB4108 | |
| Platinum wire 0.25mm dia | Alfa Aesar | 10288 | |
| 5 3/4” Disposable Pasteur pipet | Fisher | 13-678-20A | |
| Watch glass | Fisher | 02–612A | |
| 5mm Dia. Glass rod | Fisher | 50-121-5269 | |
| MitoTracker CMXRos | Fisher | M7512 | Shield from light, store at −20℃ |
| Monopostassium phosphate | Fisher | P285-500 | |
| Disodium phosphate | Fisher | S374-1 | |
| Magnesium sulfate | Fisher | M63–500 | |
| Dimethyl sulfoxide | Fisher | BP231–1 | DMSO |
| Aluminum foil | Fisher | 01-213-102 | |
| Ethyl 3-aminobenzoate methanesulfonate | Sigma | E10521-10G | Tricaine is the common name |
| Tetramisole hydrochloride | Sigma | L9756-5G | |
| Agarose | Fisher | BP1356–100 | |
| Coverslips | Fisher | 12-548-A | 18 × 18-1 |
| Frosted microscope slides | Fisher | 12-552-3 | |
| Equipment | |||
| 16℃ and 20℃ incubators | Fisher | 97–990E | Same model, set at different temperatures. |
| Upright Microscope with epi-fluorescence illuminator, camera, and 10x and 60x objectives | Refer to the protocol section for specific model information | ||
| Software with image acquisition and tracking capabilities | Protocol given based on the NIS-Elements software | ||
| Stereo-microscope | Nikon | SMZ800N | Any stereo-microscope that can be used to visualize C. elegans may be used with this protocol |
3. Day 2: Mating
Pick the stained males from Day 1 onto a new, seeded NGM plate. Leave the plate in the dark until mating. This step ensures that excess mito-dye stained bacteria around the males are removed. Carryover of excessive mito-dye stained bacteria onto the mating plate can stain hermaphrodite tissue.
-
Make a mating plate by dropping 2 μL of a thick E. coli mixture on an unseeded NGM plate. Let the thick bacteria dry to make the mating dot. Males and hermaphrodites will be transferred onto this dot for mating. To make thick E. coli, spin down 3 mL of overnight E. coli and resuspend the bacteria pellet in 1 mL of M9. This mixture can be stored at 4 °C and reused for up to 6 months.
NOTE: The thickness of the E. coli solution may be adjusted. If the solution is too thin, males may crawl away from the mating dot instead of aggregating on it for mating. Mating dots made from an E. coli soluton that is too thick may decrease the mating efficiency.
-
While the mating plate from Step 3.2 is drying, mix together 300 μL of 1% (w/v) Tricaine (Tri), 300 μL of 0.1% (w/v) Tetramisole (Tet), and 900 μL of M9.
NOTE: Store 1% (w/v) Tricaine and 0.1% (w/v) Tetramisole as aliquotes at −20 °C. Avoid repeated freeze thaw.
Transfer 600 μL of the Tet/Tri solution to a watch glass.
-
Transfer 12–15 hermaphrodites picked on Day 1 to the Tet/Tri solution in the watch glass. Incubate for 30 min to immobilize the hermaphrodites. Keep the watch glass covered to prevent the Tet/Tri solution from evaporating.
NOTE: It is important that hermaphrodites are anesthetized for at least 30 min. Less time could result in a moving worm during image acquisition, which can interefere with imaging.
While hermaphrodites are incubating, pick 50–60 stained males from Step 3.1 onto the mating dot (Step 3.2). Store the plate in the dark until Step 3.8.
-
After the 30 min incubation in the Tet/Tri solution, use a glass Pasteur pipet to transfer the immobilized hermaphrodites from the watch glass onto an unseeded NGM plate. Remove as much liquid as possible and let the excess liquid dry.
NOTE: Do not let the hermaphrodites dry excessively. As soon as all visible liquid has evaporated, begin the next step.
Transfer the anesthetized hermaphrodites from the unseeded NGM plate onto the mating dot with the stained males. Incubate in the dark for 30 min to allow the males to mate with the hermaphrodites.
-
After mating for 30 min, mount the hermaphrodites immediately for time-lapse imaging or transfer the hermaphrodites onto a new, seeded NGM plate to rest for 1 h before imaging.
NOTE: Time-lapse videos of the hermaphrodite uterus are used to quantify sperm velocity and reversal frequency. Still images of the uterus taken 1 h after mating are used to quantify sperm distribution, or spem guidance.
4. Day 2: Mounting Worms for Visualization
-
Creating a mounting pad with 2% agarose in H2O
NOTE: 2% agarose can be made in bulk, aliquoted into glass test tubes, and stored at 4 °C. When needed, each aliquot can be microwaved before each use and stored in a heat block to prevent it from solidifying.- To make the mounting pad, align three glass microscope slides side by side with the long edges touching. Place two pieces of masking tape on top of each other on both of the outer slides. These outer glass slides with tape will act as the support so the thickness of the resulting agarose pad will be “two tape deep”.
- Place ~75 μL of melted 2% agarose on the center slide (this is the slide without tape). Immediately place a new glass microscope slide on top of the agarose. This top glass slide should be perpendicular to the other slides, with each end resting on the tape of the two support slides.
- Let the agarose harden (~30 s). Carefully remove the top glass slide by sliding it off the agarose pad.
Place 10–15 μL of the Tet/Tri solution onto the 2% agarose pad. Transfer the mated hermaphrodites onto the pad. Take care to transfer as little bacteria as possible.
Place a cover slip over the worms on the agarose pad.
5. Day 2: Image Acquisition Setup
NOTE: Any upright miscroscope equipped with epi-fluorescence, 10x and 60x objectives, and a digital camera can be used to acquire images for sperm distribution. Software capable of acquiring time-lapsed images are required for assessing sperm speed, directional velocity, and reversal frequency.
- Image acquisition 1 h after mating
-
Mount the slide onto the microscope stage. Look through the eye pieces to scan for worms on the agarose pad using the 10x objective with the red fluorescence emission filter (TRITC filter). Once a worm has been found, briefly turn on the fluorescence light to see if the worm has mated. If sperm is visible within the uterus, switch to the 60x objective.NOTE: The pressure created by the 60x objective on the coverslip may damage some fragile worms, causing the intestine or gonad to extrude from the animal. Scanning for successful mating using the 10x objective can minimize the worms’ exposure to the added pressure. Do not expose the mated worms to extended periods of fluorescent light.
-
Using Differential Interference Contrast Microscopy (DIC), position the worm so that both the vulva and one spermatheca are in view. Focus the image by focusing on the center of the spermatheca. Check the exposure for both DIC and TRITC channels. In DIC, internal worm structures should be clearly visible. In TRITC, individual sperm should be visible as distinct puncta.NOTE: Each image should capture the uterus from the vulva to one of the spermatheca. If the uterus is too long to fit on one image, two separate images can be taken. It is not necessary for all of the images to be taken at the same exposure level. However, it is important that individual sperm can be distinguished and quantified in the fluorescence images.
- Acquire DIC and fluorescence images for each uterus.
- Repeat Steps 5.1.1–5.1.3 until all mated hermaphrodites have been imaged.
-
- Capturing time-lapse videos
- Scan the agarose pad and locate hermaphrodites that contain labeled sperm within the uterus, as described in Step 5.1.1 and 5.1.2
- Configure the software to acquire time-lapse images in DIC and TRITC channels. Generally, time-lapse images are taken at 15–30 s intervals for 10–20 min per uterus.
6. Quantification
- Quantifying sperm distribution on uterus images taken 1 h after mating
- Starting with the vulva on one end and the spermatheca on the other, divide the uterus into thirds. These will represent the three zones. Zone 1 (Z1) contains the vulva and Zone 3 (Z3) contains the spermatheca.
-
Manually count the number of sperm within each third of the uterus, and report the number in each zone as a percent of the total sperm in the entire uterus. An example is provided below.NOTE: Sometimes, the signal intensity of the TRITC channel image needs to be adjusted so that every sperm that has been captured in the image can be visible and quantified.
-
Tracking sperm in time-lapse images
NOTE: In this paper, we used the NIS-Elements software for analysis. In the sections below, we give instructions for manually tracking sperm using this software (step 6.2.1) as well as the open source software ImageJ/Fiji (step 6.2.2).- Tracking sperm with NIS-Elements
- Open the .nd2 file with the time-lapse series to be tracked. To begin tracking, open up the Tracking panel by right clicking in the software and selecting Analysis Controls | Tracking.
- In the tracking panel, select Define New ROI. Define each region of interest (ROI) by clicking over each sperm that will be tracked. A colored mark will appear over the selected sperm. Click Finish when all ROIs have been selected.
-
Once the ROIs have been identified, move to the next frame in the time-lapse series. Drag the ROI marker to the new position of the sperm in the image. Continue doing this until the sperm can no longer be tracked. Dotted lines will appear connecting each of the locations the ROI marker has been placed through all the frames of the time-lapse image.NOTE: Only sperm in Zone 2 should be tracked as sperm in Zones 1 and 3 tend to move in a circular pattern even in wild-type animals.
- Export all quantifiable data (e.g., path length, time, XY position, etc.) from the tracked sperm to an Excel document by clicking Export in the Tracking panel.
- Tracking sperm with Fiji
- Convert the TRITC channel images in the time-lapse series to .tif files. Save all files from one series into one folder.
- Import the images to Fiji by using the BioFormats import function. Import images from one time-lapse series as one hyperstack.
- Open TrackMate in Fiji24 via Plugins | Tracking | Manual tracking with TrackMate. A diaglogue box will open.
- Select the TrackMate tool in the Fiji toolbar. Double click on the sperm that will be tracked. A green circle with dashed lines will appear. This circle may be repositioned by clicking inside the circle and dragging to the desired position. The size of this circle may be changed by simultaneously pressing the ALT key and scrolling the mouse.
- Once the size and position of the tracker has been set, click on the circle again. The dashed green lines will turn into a solid green line. Simultaneously hit the SHIFT and L keys to turn on tracking mode. This will be indicated in the Fiji toolbar.
- Move to the next frame in the time-lapse series. To set the new location of the tracked sperm in the new frame, hover the mouse over the new point and press the A key. The tracker will now appear at the new location, and a line will appear connecting the locations where the tracker has been placed in the previous frames.
- After the traces have been completed, click Analyze in the TrackMate dialogue box to generate the data needed.
- To calculate the speed, divide the total path length of the sperm by the elapsed time.
- To calculate the vectorial velocity, draw a line through the uterus starting from the vulva pointing toward the spermatheca. Measure the distance the sperm has migrated along this line from the beginning to the end of the trace. Divide this distance by the elapsed time. Negative values indicate the sperm has migrated away from the spermatheca.
- To record reversal frequency, count the number of times during which the sperm trace has generated an angle less than 90° during three consecutive time-lapse frames.
Representative Results
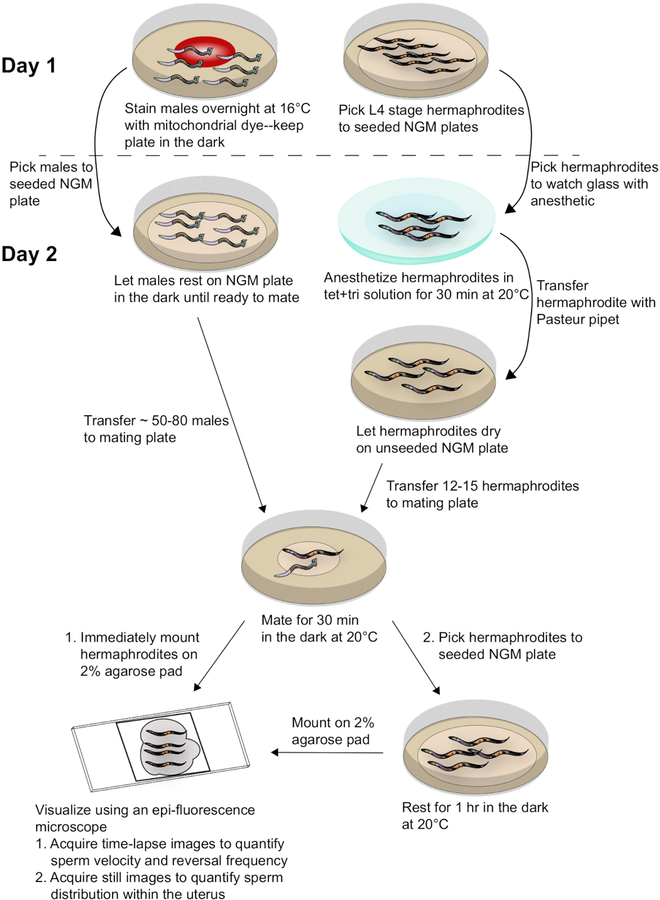
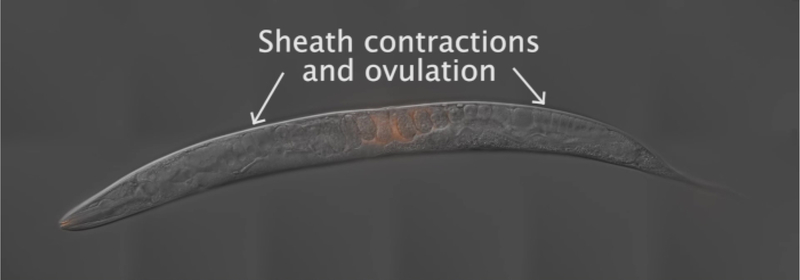
To generate the results depicted in this paper, fog-2(q71) males were stained with the mito-dye and mated to wild-type, N2 hermaphrodites. Figure 2 provides an overall scheme for the method, including worm preparation, mating, and analysis. As Movie 1 shows, the adult hermaphrodite reproductive tract has two arms that are mirror images of each other. Upon mating, labeled sperm are deposited in the hermaphrodite uterus through the vulva. The sperm move around the developing embryos within the uterus toward the spermatheca, where they are stored until fertilization. As germ cells in the adult hermaphrodite develop into oocytes, the proximal, most mature oocyte is pushed into the spermatheca via sheath cell contrations. Fertilization occurs while the oocyte is in the spermatheca.
Figure 2: Schematic diagram of sample preparation and data acquisition.
Males stained with the mito-dye are mated to synchronized adult hermaphrodites. Time-lapse images of mated hermaphrodites are taken immediately after mating to capture data for sperm velocity and reversal frequency. Still images of mated hermaphrodites are taken 1 h after mating to assess sperm distribution within the uterus. Refer to the text for more details.
Movie 1: Movie of sperm movement and migration.
A wildtype hermaphrodite was mated to fog-2(q71) males stained with mito-dye. The movie is a composite of time-lapse images taken at varied time intervals.
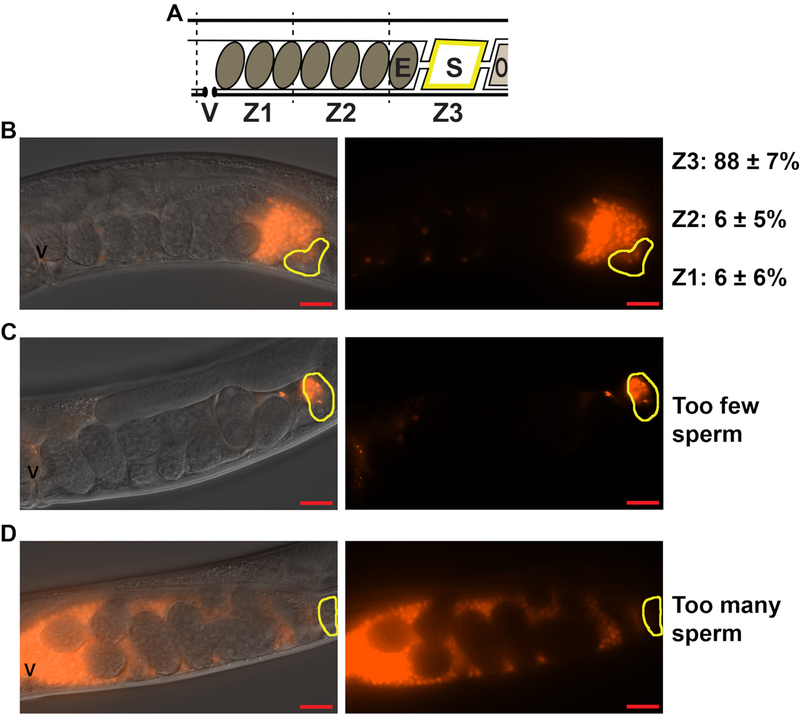
To quantify sperm distribution and migration through the female reproductive tract, the hermaphrodite uterus is divided into three zones (Movie 1, Figure 3A). Zone 1 spans the first third of the uterus, starting from the vulva. Zone 2 spans the middle third of the uterus, and Zone 3 spans the last third of the uterus and includes the spermatheca. Proper sperm guidance using wild-type, N2 hermaphrodites and fog-2(q71) males should result in approximately 90% of the labeled sperm reaching the spermatheca, or Zone 3 (Figure 3B). Matings that result in too few (Figure 3C, less than 10–15 sperm) or too many (Figure 3D, uterus filled with sperm) sperm in the uterus should not be counted. In matings that result in less than 10–15 sperm, 3–4 rogue sperm may heavily skew the data. Similarly, when the uterus is filled completely with sperm, the sperm cannot migrate appropriately. Sperm may seem scattered throughout the uteri of some mutants that display poor sperm guidance phenotype. However, in this case, sperm should not fill each crevice of the uterus, as seen in Figure 3D. Quantification of each arm of the gonad is considered one sample, or one n.
Figure 3: Quantifying sperm distribution within the hermaphrodite uterus.
(A). Schematic of the C. elegans hermaphrodite uterus. V = vulva, E = embryo, S = spermatheca, O = oocyte, Z1-Z3 = Zones 1–3 used to measure sperm distribution. (B-D). DIC+TRITC merged (left panels) and TRITC only (right panels) images of the wild-type hermaphrodite uteri 1 h after mating to fog-2(q71) males stained with the mito-dye. Sperm appear red. Yellow outlines indicate the location of the spermatheca. Scale bar: 20 μm. Z1, Z2, Z3 quantification in B represent the percent sperm in each zone ± standard deviation. Images in C and D represent matings that have resulted in too few (C) or too many (D) sperm for quantification.
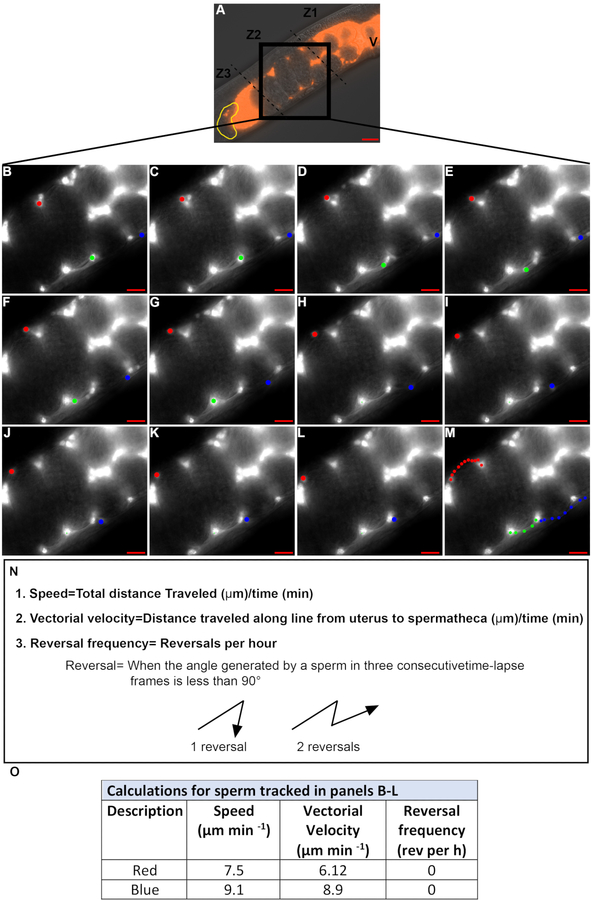
Time-lapse images are taken to quantify sperm velocity and reversal frequency. Only sperm in Zone 2 should be tracked (Figure 4A) because sperm in Zones 1 and 3 (Movie 1) tend to move in a circular pattern even within wild-type animals. Time-laspe images taken at 15–30 s intervals are usually used to track sperm. Only sperm that can be followed in consecutive frames for more than 2.5–3 min are quantified. In Figure 4B–M, the sperm marked by the red and blue dots satisfy this criterion, while the sperm marked by the green dot does not. Therefore, the values defined in Figure 4N are quantified for the sperm marked by the red and blue dots (Figure 4O), while those for the sperm marked by the green dot were not quantified.
Figure 4: Quantifying sperm velocity and reversal frequency during migration through the uterus.
(A). DIC+TRITC merged image of a hermaphrodite uterus containing fluorescent sperm (red). V = vulva, yellow = spermatheca, Z1–Z3 = three zones of the uterus, black box: zone 2. (B-M). Time-lapse TRITC channel images zoomed in on zone 2 (black box in A). Images were acquired at 20 s intervals. 3 individual sperm were tracked in each image (red, green, and blue dots). Colored dots in panel M represents the path of each sperm from B-L. Scale bar = 20μm. (N). Equations and definitions for sperm speed, vector velocity and reversal frequency. (O). Speed, vectorial velocity and reversal frequency of sperm tracked in panels B-L by the red and blue dots.
Discussion
The ability of sperm to navigate the convoluted female reproductive tract and find oocytes is critical for sexual reproduction. Recent studies using sperm of internally fertilizing animals suggest they actively respond to various environmental cues, including chemical signals, fluid flow, and temperature gradients1,5,7,9,12. However, these observations have largely resulted from in vitro experiments and little is known about sperm behavior and communication within the reproductive tract. One of the main barriers to acquiring in vivo data on sperm migration and motility is the lack of visibility within most female reproductive tracts. The method we have described here using C. elegans overcomes this limitation. As the representative results demonstrate, the transparent epidermis allows for direct visualization and tracking of each sperm at single cell resolution in a live, intact organism.
C. elegans has two sexes. The males, with an XO genotype, produce only sperm, and in this method, are used as the sperm donors. The hermaphrodite, with an XX genotype, are modified females. Their gonads first undergo spermatogenesis during the fourth larval stage and switch to oogenesis in adulthood25. This assay uses adult hermaphrodites, whose reproductive tissues provide a model for the female reproductive tract. The utilization of both sexes in this assay allows us to identify genetic and molecular pathways in both the male and female that may regulate sperm guidance and motility. Combined with the whole host of genetic and molecular techniques available for C. elegans, this method can lead to novel insights into sperm migration and motility as well as sperm and oocyte communication.
A few critical steps in this protocol warrant further consideration, in addition to the details provided in the protocol section.
Worms
fog-2(q71), him-5(e1490), or him-8(e1489) mutant males can be used in place of N2 males. These mutations increase the frequency of males in cultures, but do not affect male mating or sperm functions13. Females, such as fog-2(q71) females, may be used in place of hermaphrodites. However, females must be pre-mated with males to allow proper oocyte development. The presence of fertilized embryos from this pre-mating also ensures that the uterus is long enough to properly assess sperm distribution. If mutant or experimental hermaphrodites are being assessed, include a control group(s). For example, mutant hermaphrodites should be paired with wildtype N2 hermaphrodites as a control for other variables in the assay. Hermaphrodites that have been fed with bacteria containing plasmids for RNA interference assays should also be fed with bacteria containing empty vector control. The distance from the vulva, where sperm are inseminated, to the spermatheca, the fertilization site, can vary depending on the number of eggs in the uterus (i.e., the uterus expands with increasing egg number). If comparisons between genotypes are made, select hermaphrodites whose uteri contain similar numbers of eggs. Do not select hermaphrodites containing hatching embryos or moving larvae. A time course may be performed to identify the optimal age at which hermaphrodites should be assayed.
Picking hermaphrodites and males
It is important that the hermaphrodites picked for this assay are not from overgrown or starved plates. Food and pheromone cues modulate the expression of DAF-7, a TGFß homolog. The DAF-7 pathway has been shown regulate the synthesis of F-series prostaglandins that play important roles in guiding sperm to the spermatheca20. Picking hermaphrodites from overcrowded or starved plates may result in poor sperm guidance not related to the target of interest. The density of the plates do not seem to affect the male sperm. However, males that are too young or too old may result in decreased mating efficiency (i.e., the percent of hermaphrodites on the mating plate that have enough sperm in their uteri to quantify). 1–3 day old adult males are optimal for this assay23.
Anesthetizing hermaphrodites
In our hands, the Tetramisole and Tricaine combination ensures that worms are immobilized, and remain alive during mating and image acquisition. Other anesthetics, such as sodium azide, may also be used. However, sodium azide is highly toxic and conditions need to be standardized. Immobilization techniques using microbeads, agarose, and microfluidics chambers are not recommended as they interfere with mating.
Male staining
The mito-dye used in this manuscript, MitoTrackerCMXRos, has been widely used for labeling sperm, as well as other mitochondria, in C. elegans. The sperm labeled with this mito-dye is fully functional, retaining its ability to be activated, migrate, fertilize oocytes, and produce viable progeny26,27. Other mitochondrial dyes, such as rhodamine 6G and DiOC6 have been used to stain C. elegans mitochondria28,29. However, conditions for these dyes need to be standardized for labeling sperm in this assay. In addition to mitochrondrial labeling mechanisms, DNA stains, such as Syto17, may also be used to label sperm for migration assays30. While these labeling techniques are relatively easy and quick to perform, transgenic strategies may also be employed to generate sperm that express fluorescent tags under sperm specific promoters31,32.
Mating
Mating dots that are too thick may decrease mating efficiency. Care should be taken to transfer as little bacteria as possible when transferring males and anesthetized hermaphrodites onto the mating dot.
Making agarose pads and placing the cover slip
Air bubbles can be generated in the agarose pads. They may refract light during image acquisition or, when large enough, cause worms to fall through, making it impossible to acquire the image(s). Similarly, air bubbles may be created along the hermaphrodites when the cover slip is placed over them on the agarose pad. These bubbles refract light and lead to decreased image quality. Practice will help decrease the occurrence of air bubbles.
Quantification
When quantifying sperm distribution, different z-planes through the uterus of a worm may have slight differences in sperm distribution. We find that taking a single image focused on the spermatheca gives us reproducible results that are similar to results obtained by averaging multiple z-sections. We recommend focusing the image on the center of the spermatheca, but the focal planes can be altered slightly based on the experimenter’s needs. It is critical, however, that all images are taken in the same manner. Furthermore, it is important that only sperm that are in focus are counted. It is the counter’s discretion in determining the criteria for in-focus sperm. However, it is critical that the criteria is applied systematically to every worm that is quantified. For sperm tracking, many software offer automatic tracking capabilities. However, we find that manual tracking outperforms the software’s automatic tracking algorithm for two main reasons: 1) The abundance of similarly sized nuclei within the confined space makes it difficult for the software to distinguish between individual sperm and create defined ROIs for each sperm. 2) As sperm go in and out of focus, their intensities shift, making it difficult for the software to keep track of the sperm over extended periods of time.
Acknowledgments
We sincerely thank our late mentor, Dr. Michael Miller, for his inspiring and selfless mentorship and creation of this method as a tool to better understand sperm and oocyte communication. His sudden passing has been a tremendous loss for his family, his lab, and the scientific community. This study was supported by the NIH (R01GM085105 to MAM and F30HD094446 to MH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Video Link
The video component of this article can be found at https://www.jove.com/video/59783/
Disclosures
The authors have no conflicts of interest.
References
- 1.Boryshpolets S, Perez-Cerezales S, & Eisenbach M Behavioral mechanism of human sperm in thermotaxis: a role for hyperactivation. Human Reproduction. 30 (4), 884–892, (2015). [DOI] [PubMed] [Google Scholar]
- 2.Edmonds JW, McKinney SL, Prasain JK, & Miller MA The gap junctional protein INX-14 functions in oocyte precursors to promote C. elegans sperm guidance. Developmental Biology. 359 (1), 47–58, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edmonds JW et al. Insulin/FOXO signaling regulates ovarian prostaglandins critical for reproduction. Developmental Cell. 19 (6), 858–871, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espinal-Enriquez J, Priego-Espinosa DA, Darszon A, Beltran C, & Martinez-Mekler G Network model predicts that CatSper is the main Ca(2+) channel in the regulation of sea urchin sperm motility. Scientific Reports. 7 (1), 4236, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter RH, & Nichol R A preovulatory temperature gradient between the isthmus and ampulla of pig oviducts during the phase of sperm storage. Journal of Reproduction and Fertility. 77 (2), 599–606, (1986). [DOI] [PubMed] [Google Scholar]
- 6.Hussain YH, Guasto JS, Zimmer RK, Stocker R, & Riffell JA Sperm chemotaxis promotes individual fertilization success in sea urchins. Journal of Experimental Biology. 219 (Pt 10), 1458–1466, (2016). [DOI] [PubMed] [Google Scholar]
- 7.Kantsler V, Dunkel J, Blayney M, & Goldstein RE Rheotaxis facilitates upstream navigation of mammalian sperm cells. Elife. 3 e02403, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubagawa HM et al. Oocyte signals derived from polyunsaturated fatty acids control sperm recruitment in vivo. Nature Cell Biology. 8 (10), 1143–1148, (2006). [DOI] [PubMed] [Google Scholar]
- 9.Miki K, & Clapham DE Rheotaxis guides mammalian sperm. Current Biology. 23 (6), 443–452, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ting JJ, Tsai CN, Schalkowski R, & Cutter AD Genetic Contributions to Ectopic Sperm Cell Migration in Caenorhabditis Nematodes. G3 (Bethesda). 8 (12), 3891–3902, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanagimachi R et al. Chemical and physical guidance of fish spermatozoa into the egg through the micropyledagger,double dagger. Biology of Reproduction. 96 (4), 780–799, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y et al. Generation of Gradients on a Microfluidic Device: Toward a High-Throughput Investigation of Spermatozoa Chemotaxis. PloS One. 10 (11), e0142555, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoang HD, & Miller MA Chemosensory and hyperoxia circuits in C. elegans males influence sperm navigational capacity. PLoS Biology. 15 (6), e2002047, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen JM, Chavez DR, & Stanfield GM COMP-1 promotes competitive advantage of nematode sperm. Elife. 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.L’Hernault SW Spermatogenesis. WormBook: The Online Review of C. Elegans Biology. 1–14, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller MA et al. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 291 (5511), 2144–2147, (2001). [DOI] [PubMed] [Google Scholar]
- 17.Greenstein D Control of oocyte meiotic maturation and fertilization. WormBook: The Online Review of C. Elegans Biology. 1–12, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Hagan R, Wang J, & Barr MM Mating behavior, male sensory cilia, and polycystins in Caenorhabditis elegans. Seminars in Cell & Developmental Biology. 33 25–33, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoang HD, Prasain JK, Dorand D, & Miller MA A heterogeneous mixture of F-series prostaglandins promotes sperm guidance in the Caenorhabditis elegans reproductive tract. PLoS Genetics. 9 (1), e1003271, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKnight K et al. Neurosensory perception of environmental cues modulates sperm motility critical for fertilization. Science. 344 (6185), 754–757, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhuri J, Parihar M, & Pires-daSilva A An introduction to worm lab: from culturing worms to mutagenesis. Journal of Visualized Experiments. (47), (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stiernagle T Maintenance of C. elegans. WormBook: The Online Review of C. Elegans Biology. 1–11, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatterjee I et al. Dramatic fertility decline in aging C. elegans males is associated with mating execution deficits rather than diminished sperm quality. Experimental Gerontology. 48 (11), 1156–1166, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tinevez JY et al. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115 80–90, (2017). [DOI] [PubMed] [Google Scholar]
- 25.Corsi AK, Wightman B, & Chalfie M A Transparent Window into Biology: A Primer on Caenorhabditis elegans. Genetics. 200 (2), 387–407, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato M, & Sato K Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 334 (6059), 1141–1144, (2011). [DOI] [PubMed] [Google Scholar]
- 27.Wang Y et al. Kinetics and specificity of paternal mitochondrial elimination in Caenorhabditis elegans. Nature Communications. 7 12569, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolke U, Jezuit EA, & Priess JR Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development. 134 (12), 2227–2236, (2007). [DOI] [PubMed] [Google Scholar]
- 29.Mottram LF, Forbes S, Ackley BD, & Peterson BR Hydrophobic analogues of rhodamine B and rhodamine 101: potent fluorescent probes of mitochondria in living C. elegans. Beilstein Journal of Organic Chemistry. 8 2156–2165, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singson A, Hill KL, & L’Hernault SW Sperm competition in the absence of fertilization in Caenorhabditis elegans. Genetics. 152 (1), 201–208, (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu JC et al. Sperm development and motility are regulated by PP1 phosphatases in Caenorhabditis elegans. Genetics. 190 (1), 143–157, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seidel HS et al. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biology. 9 (7), e1001115, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]