Abstract
Computations that require speed and temporal precision are implemented throughout the nervous system by neurons capable of firing at very high rates, rapidly encoding and transmitting a rich amount of information, but with substantial metabolic and physical costs. For economical fast spiking and high throughput information processing, neurons need to optimize multiple biophysical properties in parallel, but the mechanisms of this coordination remain unknown. We hypothesized that coordinated gene expression may underlie the coordinated tuning of the biophysical properties required for rapid firing and signal transmission. Taking advantage of the diversity of fast-spiking cell types in the medial vestibular nucleus of mice of both sexes, we examined the relationship between gene expression, ionic currents, and neuronal firing capacity. Across excitatory and inhibitory cell types, genes encoding voltage-gated ion channels responsible for depolarizing and repolarizing the action potential were tightly coexpressed, and their absolute expression levels increased with maximal firing rate. Remarkably, this coordinated gene expression extended to neurofilaments and specific presynaptic molecules, providing a mechanism for coregulating axon caliber and transmitter release to match firing capacity. These findings suggest the presence of a module of genes, which is coexpressed in a graded manner and jointly tunes multiple biophysical properties for economical differentiation of firing capacity. The graded tuning of fast-spiking capacity by the absolute expression levels of specific ion channels provides a counterexample to the widely held assumption that cell-type-specific firing patterns can be achieved via a vast combination of different ion channels.
SIGNIFICANCE STATEMENT Although essential roles of fast-spiking neurons in various neural circuits have been widely recognized, it remains unclear how neurons efficiently coordinate the multiple biophysical properties required to maintain high rates of action potential firing and transmitter release. Taking advantage of diverse fast-firing capacities among medial vestibular nucleus neurons of mice, we identify a group of ion channel, synaptic, and structural genes that exhibit mutually correlated expression levels, which covary with firing capacity. Coexpression of this fast-spiking gene module may be a basic strategy for neurons to efficiently and coordinately tune the speed of action potential generation and propagation and transmitter release at presynaptic terminals.
Keywords: flocculus, high-frequency firing, ion channel genes, Kv3, pre-cerebellar, sodium channel
Introduction
Fast-spiking neurons are essential components of auditory, vestibular, and proprioceptive sensory systems, inhibitory networks in the cerebral cortex and hippocampus, and basal ganglia, cerebellar, and brainstem components of motor systems. The ability to fire persistently at high rates enables neurons to rapidly and precisely convey timing information to downstream circuits and is critical for several aspects of brain function, including sensory encoding, population synchrony, and motor control. Maintaining high rates of firing and synaptic transmission, however, places several metabolic and physical demands on neurons. Restoring ionic gradients during action potential firing is energetically expensive (Carter and Bean, 2009; Sengupta et al., 2010). To remain sufficiently excitable during repetitive firing, neurons must express high levels of sodium and potassium channels (Carter and Bean, 2011; Gittis et al., 2010). To propagate signals rapidly, axons must be sufficiently thick (S. S. Wang et al., 2008; Perge et al., 2012), which consumes limited physical space in the CNS (Attwell and Laughlin, 2001). Finally, to sustain high rates of transmitter release, presynaptic terminals require multiple active zones that are morphologically and molecularly specialized (Taschenberger et al., 2002; Telgkamp et al., 2004; McElvain et al., 2015). How do neurons achieve coordinated expression of the multiple biophysical properties required to minimize the costs associated with high rates of firing and signal transmission?
Co-regulated expression of groups of functionally related genes via transcriptional mechanisms has been established as a fundamental strategy for cells to coordinate multiple properties required to achieve specific ends, such as the acquisition of neuronal identities (Kwan et al., 2012; Hobert and Kratsios, 2019) and responses to stress (Vihervaara et al., 2018). We hypothesized that tuned expression of a module of genes may coordinate multiple biophysical properties in fast-spiking neurons. Consistent with this idea, a specific set of genes associated with axonal function and transmitter release has been identified in several types of fast-spiking neurons (Saunders et al., 2018). It is unclear, however, whether expression levels of those genes are co-regulated to achieve specific fast-spiking capacities. To gain insights into transcriptional coordination underlying fast spiking, we took advantage of variations in firing capacities across distinct types of sensory-motor neurons in the medial vestibular nucleus (MVN) of mice. MVN neurons comprise multiple cell types (Bagnall et al., 2007; Shin et al., 2011; Kodama et al., 2012), which are functionally distinct with respect to transmitter types, projection targets, and cerebellar innervation, and collectively mediate both rapid and sustained eye movements (vestibulo-ocular reflex and optokinetic response). Although all MVN neurons are capable of firing at high rates (>50 spikes/s), the range of maximum sustainable firing rates varies widely, enabling us to examine how gene expression relates to firing capacity.
In this study, we first examine firing capacity and action potential waveforms in brainstem slice recordings from several functionally distinct types of MVN neurons. We then use single-cell quantitative PCR (qPCR) to identify ion channel genes in which expression levels covary with maximal firing rates within and across cell types. Using these ion channel genes as a proxy for fast-spiking capacity, we perform genome-wide screening to identify additional genes that exhibit graded coexpression with fast-spiking ion channel genes.
Materials and Methods
Slice preparation and current-clamp electrophysiology.
All experiments were performed under the approval of the Salk Institute Animal Care and Use Committee. Brainstem slices containing the vestibular nucleus were prepared from C57BL/6, YFP-16 (Feng et al., 2000), GlyT2-GFP (Zeilhofer et al., 2005), L7-Cre (Barski et al., 2000) crossed with Ai27 (Madisen et al., 2012; L7-Cre;Ai27), and GIN (Oliva et al., 2000) mouse lines of both sexes aged postnatal days 21–40 as described previously (Shin et al., 2011). Mice were anesthetized deeply with Nembutal, decapitated, and the hindbrain was rapidly removed from the skull and placed in ice-cold artificial CSF (ACSF; in mm: 124 NaCl, 5 KCl, 1.3 MgSO4, 26 NaHCO3, 2.5 CaCl2, 1 NaH2PO4, and 11 dextrose) aerated with 95% CO2/5% O2. Kynurenic acid (2 mm), picrotoxin (100 μm), and strychnine (10 μm) were added to the bath to block glutamatergic, GABAergic, and glycinergic synaptic transmission. Retrograde labeling of neurons was performed by stereotaxic injection of rhodamine dextran into target nuclei with subsequent slice physiology performed after a transport period of 2–6 d, as described by Kolkman et al. (2011). Pre-oculomotor neurons were identified as YFP+ in the YFP-16 lines that were retrogradely labeled from the contralateral abducens region. Commissural inhibitory neurons were identified as GFP+ neurons in the GlyT2 line that were retrogradely labeled from the contralateral MVN. Local inhibitory neurons were identified as GFP+ in the parvocellular region of the MVN in GIN mice that were consistently devoid of retrograde labeling from tracer in the contralateral MVN. Cerebellar target neurons were identified by the presence of a somatic innervation by fluorescent Purkinje cell terminals in L7-Cre;Ai27 mice. Action potential and firing data from pre-cerebellar neurons were derived from previously published recordings of neurons retrogradely labeled from the cerebellar flocculus (Kolkman et al., 2011). Whole-cell patch recordings with borosilicate glass pipettes were obtained from fluorescently labeled neurons or from neurons innervated by fluorescent Purkinje cell terminals. Recordings were performed with a MultiClamp 700B amplifier. Data were digitized at 40 kHz, filtered at 10 kHz, and collected with a Macintosh OS X with custom code written in Igor Pro (WaveMetrics).
Whole-cell recordings were performed in warmed (32°C) ACSF in current-clamp mode. Action potential analyses were standardized at firing rates of 10 Hz by adjusting intracellular DC current levels; holding currents were typically <200 nA. Bridge balance and capacitance neutralization were performed manually to minimize voltage artifacts and capacitative transients during current steps. Maximum firing rates were determined by injecting 1 s steps of intracellular current of increasing magnitude until the neuron exhibited depolarization block. Minimum sustained refractory period was measured as interspike interval at the maximum firing rate. Internal recording solutions were prepared with the following (in mm): 140 K-gluconate, 8 NaCl, 10 HEPES, 0.1 EGTA, 2 Mg-ATP, and 0.3 Na-GTP. ACSF was aerated with 95% O2, 5%CO2; and contained the following (in mm): 124 NaCl, 5 KCl, 1.3 MgSO4, 26 NaHCO3, 2.5 CaCl2, 1 NaH2PO4, and 11 dextrose. Kynurenic acid (2 mm), picrotoxin (100 μm), and strychnine (10 μm) were added to the bath to block glutamatergic, GABAergic, and glycinergic synaptic transmission, respectively.
Dissociated neuron preparation, voltage-clamp electrophysiology, and pharmacology.
Vestibular nucleus neurons were acutely dissociated from brainstem slices prepared from C57BL/6J wild-type, GIN (Oliva et al., 2000), or YFP-16 (Feng et al., 2000) mice of both sexes between 24 and 39 d of age. The data presented here are reanalyzed from recordings reported by Gittis and du Lac (2007). Vestibular nucleus neurons were enzymatically dissociated at 30°C for 10 min in a solution containing MEM powder (9.4 mg/ml; Invitrogen), HEPES (10 mm), cysteine (0.2 mm), and papain (40 U/ml; Worthington), pH 7.2. Excised vestibular nuclei were triturated with fire-polished Pasteur pipettes of decreasing diameter in 500 μl Tyrode's solution containing the following (in mm): 150 NaCl, 3.5 KCl, 2 CaCl, 1 MgCl2, 10 HEPES, 10 glucose to dissociate neurons. Following trituration, cells were plated on the glass slide of the recording chamber, allowed to settle for 10 min, and continuously perfused with oxygenated Tyrode's solution for the duration of the recording (2–3 h).
Whole-cell recordings from dissociated neurons were made at room temperature under continuous perfusion with oxygenated Tyrode's solution. After the formation of a gigaohm seal, cells were gently lifted from the bottom of the chamber and positioned directly in front of a small piece of tubing through which pharmacological solutions could be delivered. Borosilicate pipettes were filled with an intracellular solution containing the following (in mm): 140 KMeSO4, 8 NaCl, 10 HEPES, 0.02 EGTA, 2 Mg2-ATP, 0.3 Na2-GTP, and 14 Tris-creatine PO4. A measured junction potential of 15 mV was corrected off-line. Data were recorded with a MultiClamp 700B amplifier (Molecular Devices) and digitized with an ITC-16 interface board (InstruTECH). Stimulus delivery and data analysis were performed with Igor Pro.
Whole-cell currents were evoked with families of voltage steps, 150 ms in duration, from −55 to +15 mV from a holding potential of −65 mV. Individual components of whole-cell currents were isolated pharmacologically via rapid solution-exchange using a gravity-driven, VC-6 perfusion valve control system (Warner Instruments). Solutions were applied in the following order: (1) Tyrode's, (2) Tyrode's + 300 nm TTX, (3) 0 Ca Tyrode's (in which 2 mm CaCl2 was replaced with 1.7 mm MgCl2 and 0.3 mm CdCl2) + 300 nm TTX, and (4) 0 Ca Tyrode's + 300 nm TTX + 1 mm TEA. The transient Na current was measured by subtracting currents between solutions 1 and 2. The Ca-dependent K current was measured as the difference current between solutions 2 and 3. The 1 mm sensitive Kv3 current was measured as the difference between solutions 3 and 4. TTX was purchased from Tocris Bioscience; TEA and CdCl2 were purchased from Sigma-Aldrich. Stock solutions were diluted in water and stored at 4°C. The peak Na current was defined as the largest Na current evoked by the family of voltage steps and typically occurred at nominal −35 mV (−36.6 ± 3.9 mV). Peak Kv3 and KCa currents were defined as the average of the currents evoked by the largest applied voltage step (+15 mV).
Neurons included in the analyses fired spontaneously (13.4 ± 6.1 Hz) and had action potential amplitudes of >50 mV. Action potentials in dissociated neurons were recorded in current-clamp mode, filtered at 10 kHz, and digitized at 40 kHz. Following action potential recordings, the amplifier was switched to voltage-clamp mode. Whole-cell currents were filtered at 6 kHz and digitized at 20 kHz. Whole-cell capacitance was compensated via the amplifier and series resistance was compensated at 70%, average series resistance was 9 ± 3 MΩ; cells were excluded if they had a series resistance >20 MΩ. Uncompensated series resistance was 2.5 ± 1.0 MΩ. Recording stability in voltage-clamp was evaluated by comparing the sum of the integral of pharmacologically isolated current waveforms (evoked at nominal +15 mV) with that of outward current measured in TTX at the outset of pharmacological manipulations; the average error was 2%, and cells with >3% error were excluded.
Gene expression profiling of MVN neurons.
The single-cell cDNA samples used in this study (n = 167) were prepared from YFP-16, GlyT2-GFP, and GIN mouse lines and were used in a previous report (Kodama et al., 2012) to classify MVN neurons into molecularly and anatomically distinct cell types. On the basis of marker gene expression, connectivity patterns, and locations within the MVN, the excitatory (E1–E3) and inhibitory (I1–I3) cell types identified by Kodama et al. (2012) correspond with cell types analyzed in the present study as follows: E1(n = 21): pre-oculomotor, E2 (n = 20): pre-cerebellar, E3 (n = 6): unknown, I1 (n = 30): commissural inhibitory, I2 (n = 13): cerebellar target, and I3 (n = 77): local interneuron. Transcript levels were measured by real-time qPCR. For a subset of genes (Nefh, Nefm, Nefl, Vamp1, Cplx1, Cbln1), qPCR was performed in 102 single-cell cDNA samples, which covered each cell type (per-oculomotor, n = 20; pre-cerebellar, n = 19; commissural, n = 26; cerebellar target, n = 13; local interneuron, n = 18; unknown, n = 6).
For microarray analyses, single-cell cDNAs from each cell type were pooled, then subjected to an extra PCR to attach T7 promoter to the 3′ end of the cDNA fragments by which labeled cRNA was synthesized (Kurimoto et al., 2007). Commissural (I1) and local interneuron (I3) cell types were subdivided into two based on expression profiles of 94 genes as described later. In total, seven cell types were compared in the microarray analyses. Two technical replicates were made for each cell type. We followed the manufacturer's instruction to hybridize the labeled cRNAs with Affymetrix Genechip Mouse Genome 430 2.0. The microarray data have been published in NCBI GEO (GSE55656).
Experimental design and statistical analyses.
In the analyses of qPCR data, threshold cycle (Ct) values less than average Ct of 5-copy spike-in control (23.1) were regarded as significant expression. Note that noise in transcript quantification increases as transcript level decreases, which limits detection of correlative expression when expression levels are low. Pearson's correlation coefficient (r) between transcript levels were measured using log-transformed transcript copy numbers estimated from Ct values. Transcript copy number (N) was obtained by the equation: N = Nt/R∧Ct, where Nt is PCR product number at threshold cycle, and R is amplification rate in qPCR. Nt was determined by average Ct values for spike-in control corresponding to 1000-copy of transcripts. For 37 of 55 genes whose transcript levels are reported in this paper, the amplification rates were empirically measured by qPCR with a serial dilution series of cDNA prepared from mouse brain total RNA (Zyagen). The median value (1.936) of the experimentally measured amplification rates were used for genes whose amplification rates were not experimentally determined. Note that Pearson's r between log-transformed copy numbers is equal to r between corresponding original Ct values, because log-transformed copy number is a linearly transformed Ct value. Errors in the amplification rate therefore do not affect calculation of r. Correlations between transcript copy numbers were measured in logarithmic space, because they generally follow log-normal distribution (Bengtsson et al., 2005). For the same reason, mean of transcript copy numbers was calculated by geometric mean in Figures 3B and 6B. To compute the dendrogram shown in Figure 4A, Ward's hierarchical clustering method was used with 41 ion channels (see Table 3; note that Kcna5 was excluded because no cell expressed it), where “distance” between genes was defined by (1 − r).
Figure 3.

Expression of Scn8a and Kcnc1 correlates with fast-spiking capacity of MVN neurons. A, Scatter plots of transcript copy numbers for Kcnc1 and Scn8a genes, which encode Kv3.1 and Nav1.6 channel subunits, respectively. Each dot represents an individual neuron; colors indicate cell types. B, Average maximal firing rate of the MVN cell types plotted against population average transcript copy number for Scn8a and Kcnc1. Error bars: SEM.
Figure 6.
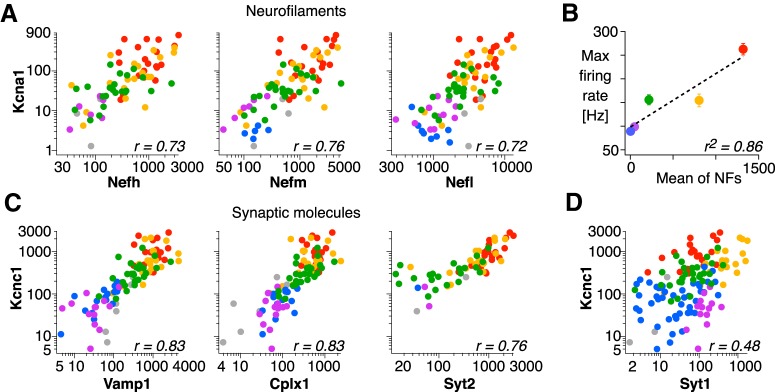
Confirmation of fast-spiking genes with single-cell qPCR. A, Transcript copy numbers of neurofilament genes (Nefh, Nefm, Nefl), plotted against Kcna1. B, Average maximal firing rate of the MVN cell types plotted against population average transcript copy number for the neurofilament genes. Error bars: SEM. C, Transcript copy numbers of synaptic molecule genes (Vamp1, Cplx1, Syt2), plotted against Kcnc1. D, Transcript copy numbers of Syt1 plotted against Kcnc1.
Figure 4.
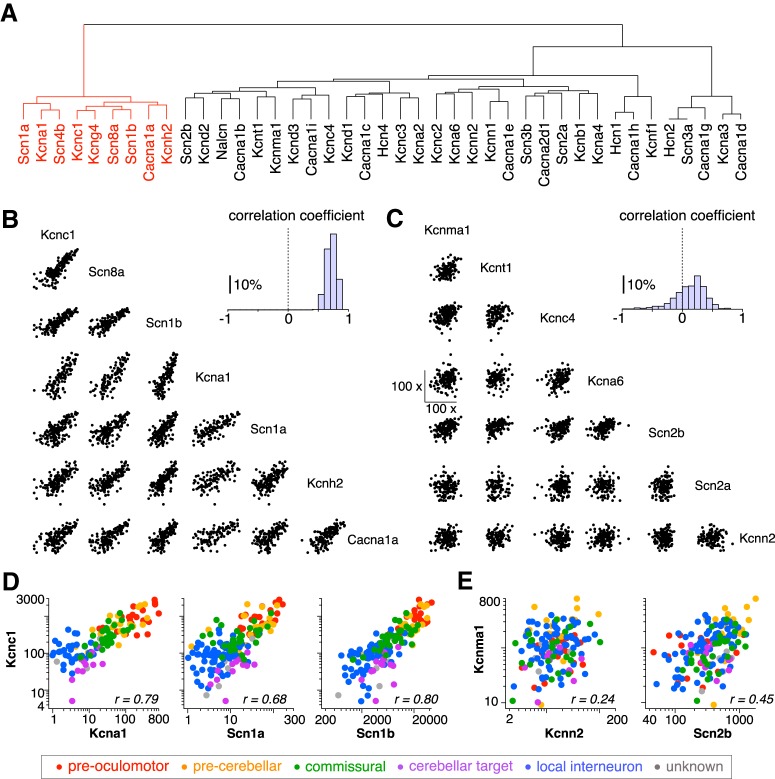
Correlated ion channel gene expression in MVN neurons. A, Dendrogram indicating correlations between transcript levels for 41 ion channels examined in MVN neurons (n = 167). B, C, Scatter plot matrix showing correlation of transcript levels in individual MVN neurons among the ion channels in the left (B) and right (C) branch of the dendrogram (left bottom). Histograms (right top) show distribution of Pearson's correlation coefficient (r). D, E, Representative scatter plots of transcript copy numbers of genes in the left (D) and right (E) branches of the dendrogram in which cell types are indicated by colors (see legend, bottom).
Table 3.
Ion channel genes (n = 42) profiled by qPCR with single-cell cDNA library prepared from MVN neurons (n = 167)
| Gene symbol | Gene name | No. of expressing cells |
|---|---|---|
| Scn1a | voltage-dependent sodium channel, type I, α subunit (NaV1.1) | 161 |
| Scn2a1 | voltage-dependent sodium channel, type II, α subunit (NaV1.2) | 136 |
| Scn3a | voltage-dependent sodium channel, type III, α subunit (NaV1.3) | 33 |
| Scn8a | voltage-dependent sodium channel, type VIII, α subunit (NaV1.6) | 160 |
| Cacna1a | voltage-dependent calcium channel, P/Q type, α 1A subunit (CaV2.1) | 153 |
| Cacna1b | voltage-dependent calcium channel, N type, α 1B subunit (CaV2.2) | 143 |
| Cacna1c | voltage-dependent calcium channel, L type, α 1C subunit (CaV1.2) | 61 |
| Cacna1d | voltage-dependent calcium channel, L type, α 1D subunit (CaV1.3) | 31 |
| Cacna1e | voltage-dependent calcium channel, R type, α 1E subunit (CaV2.3) | 50 |
| Cacna1g | voltage-dependent calcium channel, T type, α 1G subunit (CaV3.1) | 28 |
| Cacna1h | voltage-dependent calcium channel, T type, α 1H subunit (CaV3.2) | 45 |
| Cacna1i | voltage-dependent calcium channel, T type, α 1I subunit (CaV3.3) | 72 |
| Cacna2d1 | voltage-dependent calcium channel, α2/δ subunit 1 | 51 |
| Kcna1 | voltage-dependent potassium channel, shaker-related subfamily, member 1 (KV1.1) | 117 |
| Kcna2 | potassium voltage-gated channel, shaker-related subfamily, member 2 (KV1.2) | 88 |
| Kcna3 | potassium voltage-gated channel, shaker-related subfamily, member 3 (KV1.3) | 33 |
| Kcna4 | potassium voltage-gated channel, shaker-related subfamily, member 4 (KV1.4) | 108 |
| Kcna5 | potassium voltage-gated channel, shaker-related subfamily, member 5 (KV1.5) | 0 |
| Kcna6 | potassium voltage-gated channel, shaker-related, subfamily, member 6 (KV1.6) | 143 |
| Kcnb1 | potassium voltage gated channel, Shab-related subfamily, member 1 (KV2.1) | 69 |
| Kcnc1 | potassium voltage gated channel, Shaw-related subfamily, member 1 (KV3.1) | 166 |
| Kcnc2 | potassium voltage gated channel, Shaw-related subfamily, member 2 (KV3.2) | 71 |
| Kcnc3 | potassium voltage gated channel, Shaw-related subfamily, member 3 (KV3.3) | 44 |
| Kcnc4 | potassium voltage gated channel, Shaw-related subfamily, member 4 (KV3.4) | 157 |
| Kcnd1 | potassium voltage-gated channel, Shal-related family, member 1 (KV4.1) | 69 |
| Kcnd2 | potassium voltage-gated channel, Shal-related family, member 2 (KV4.2) | 155 |
| Kcnd3 | potassium voltage-gated channel, Shal-related family, member 3 (KV4.3) | 127 |
| Kcnh2 | potassium voltage-gated channel, subfamily H (eag-related), member 2 (ERG1) | 158 |
| Kcng4 | potassium voltage-gated channel, subfamily G, member 4 (KV6.4) | 92 |
| Kcnf1 | potassium voltage-gated channel, subfamily F, member 1 | 70 |
| Kcnn1 | calcium-dependent potassium channel, intermediate/small conductance, subfamily N, member 1 (SK1) | 30 |
| Kcnn2 | calcium-dependent potassium channel, intermediate/small conductance, subfamily N, member 2 (SK2) | 143 |
| Kcnma1 | calcium-dependent potassium channel, large conductance subfamily M, alpha member 1 (BK) | 163 |
| Kcnt1 | sodium-activated potassium channel, subfamily T, member 1 (Slack) | 120 |
| Hcn1 | hyperpolarization-activated, cyclic nucleotide-gated nonselective cation channel 1 | 13 |
| Hcn2 | hyperpolarization-activated, cyclic nucleotide-gated nonselective cation channel 2 | 13 |
| Hcn4 | hyperpolarization-activated, cyclic nucleotide-gated nonselective cation channel 4 | 67 |
| Scn1b | voltage-dependent sodium channel, type I, β subunit | 167 |
| Scn2b | voltage-dependent sodium channel, type II, β subunit | 163 |
| Scn3b | voltage-dependent sodium channel, type III, β subunit | 75 |
| Scn4b | voltage-dependent sodium channel, type IV, β subunit | 71 |
| Nalcn | sodium leak channel, non-selective | 162 |
No. of expressing cells indicates the number of MVN samples that showed Ct values less than a threshold corresponding to five copies of transcripts.
In the analyses of microarray data, GCRMA was used to normalize and compute expression levels (log2 transformed). The single-cell cDNAs used in the present study contain only 3′ end sequence of cDNAs [∼700 bp from the beginning of poly(A) tail]. From the analysis, we therefore excluded probe sets designed for cDNA sequences out of this range. We also excluded those showing difference in expression level >2 between technical duplicates, and those suspected of poor performance (Zhang et al., 2005). Total number of probe sets analyzed was 30843, including 18,267 annotated genes. To compute the dendrogram shown in Figure 5A, Ward's hierarchical clustering method was run exclusively with differentially expressed genes (difference between minimal and maximal expression level among cell types; >2.0, 1095 probe sets corresponding to 1004 annotated genes), where distance between genes was defined by (1 − Kendall's correlation coefficient τ). Kendall's τ was used for robust evaluation of correlation given the small number of samples (n = 7 cell types). The analyses were performed on R (r-project.org, v3.5.2) using gcrma and annaffy packages for expression level quantification.
Figure 5.
Genome-wide screening of genes that are coexpressed with fast-spiking ion channel genes in a graded manner. A, Dendrogram showing expression correlation of genes in MVN neurons. Gene expression was quantified by microarray using a pooled cDNA from each cell type. The branch indicated in red includes all of the ion channels that showed highly correlated expression with maximal firing rates of MVN neurons. Note that the microarray failed to detect Cacna1a even though its high expression was confirmed by qPCR (Fig. 4B, right). B, Transcript levels of neurofilaments plotted against Kcna1. Symbols in B indicate MVN cell types as shown in the legend on the right. C, Transcript levels of synaptic molecules (Vamp1, Cplx1, Syt2) plotted against Kcnc1. D, Transcript levels of Scn4b against Kcnc1. E, Transcript levels of representative genes that did not show positive correlation with fast-spiking ion channel genes, plotted against Kcnc1. For specifics of cell classification, see Figure 5-1, and for the list of probe sets included in the red branch of the dendogram in Figure 5A, see Figure 5-2.
Note that there is a minor difference between the cell classification used for the present study and the one previously reported; commissural cell type in this study includes one E3, one I2, and three I3 cells in the classification by Kodama et al., 2012 (the rightmost sub-branch of commissural cell type in Fig. 5-1, ). This is because the cell classification used in this study incorporated more genes (n = 94) than the previous report (Adcyap1, Cacna1a, Cacna1b, Cacna1c, Cacna1d, Cacna1e, Cacna1g, Cacna1h, Cacna1i, Cacna2d1, Cadps2, Cbln4, Cck, Cd24a, Clca1, Clca2, Clca3, Cnr1, Coch, Col5a1, Crh, Crhbp, Crhr1, Crhr2, Gad1, Gad2, Grin2c, Grin2d, Grin3a, Grin3b, Grm1, Grm4, Grm5, Grm7, Gsg1l, Hcn1, Hcn2, Hspb8, Kcna1, Kcna2, Kcna3, Kcna4, Kcna5, Kcna6, Kcnb1, Kcnc1, Kcnc2, Kcnc3, Kcnc4, Kcnd1, Kcnd2, Kcnd3, Kcne2, Kcnf1, Kcng4, Kcnh1, Kcnh2, Kcnma1, Kcnn1, Kcnn2, Kcnn3, Kcnn4, Lamb1–1, Nalcn, Nrn1, Oprl1, Otof, Phox2b, Ryr2, Ryr3, Scn11a, Scn1a, Scn1b, Scn2a1, Scn2b, Scn3a, Scn3b, Scn4b, Scn7a, Scn8a, Sema3a, Sema5a, Slc17a6, Slc17a7, Slc6a5, Spp1, Sst, Syt1, Syt2, Syt5, Syt9, Tacr3, Trpv1, Zic1).
Statistical analyses were conducted in R. Orthogonal regression was used for linear fit analyses to account for measurement errors in x- and y-axis variables. P values for Pearson's r were not adjusted for multiple comparison. In the text, A ± B reports mean (A) and SEM (B), unless otherwise stated.
Results
The MVN comprises a diverse population of excitatory and inhibitory neurons that differ in their intrinsic excitability, axonal projections, synaptic connections with the cerebellum, and functions in oculomotor behaviors (Fig. 1A). To compare firing properties across MVN neurons, we made whole-cell patch recordings in brainstem slices and targeted recordings to five distinct types of neurons (2 excitatory and 3 inhibitory) identified by their axonal projections, synaptic inputs, and reporter expression, as described in Materials and Methods. Excitatory pre-cerebellar mossy fiber neurons were retrogradely labeled from flocculus injections (Kolkman et al., 2011); excitatory premotor neurons were retrogradely labeled from the contralateral MVN. Inhibitory commissural neurons were identified via retrograde labeling from the contralateral MVN in GlyT2-EGFP reporter mice. Inhibitory cerebellar target neurons were identified by the presence of fluorescent Purkinje cell synapses in L7-Cre;Ai27 mice, and local GABAergic interneurons in the parvocellular MVN were targeted in GIN mice.
Figure 1.
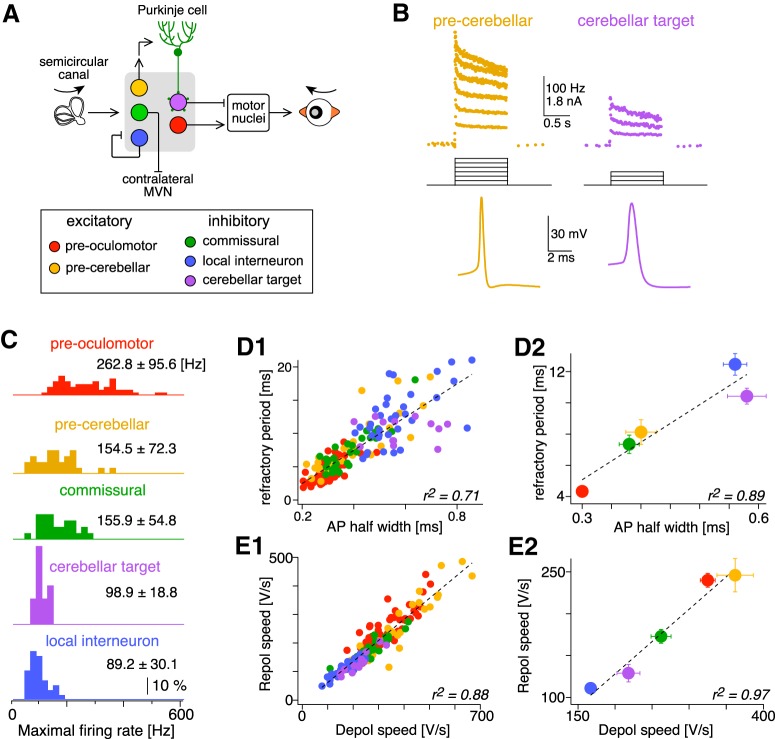
Graded firing capacity across MVN cell types. A, Schematic illustration of five MVN cell types and their connections in the VOR circuit. B, Action potential waveforms and instantaneous firing rates vs time of pre-cerebellar (left) and cerebellar target (right) neurons in the MVN evoked by depolarizing current steps. C, Normalized distribution of maximal firing rates in the five MVN cell types. The mean ± SD of maximal firing rates is indicated next to each histogram. The number of neurons analyzed was as follows: pre-oculomotor (n = 61), pre-cerebellar (n = 29), commissural (n = 27), cerebellar target neurons (n = 13), and local interneuron (n = 35). D, Minimum sustained refractory period (reciprocal of maximum firing rate) is plotted against action potential (AP) half-width for individual neurons (D1) and average of each cell type (D2). Coefficient of determination for linear regression (r2) is indicated in the plots. E, Repolarization (Repol) speed) of action potential is plotted against depolarization (Depol) speed), for individual cells (E1) and average of each cell type (E2). Error bars: SEM. Colors for the MVN cell types are consistent in this and all the following figures.
To examine firing capacities, neurons were depolarized intracellularly with incrementing steps of current for 1 s (Fig. 1B,C). All MVN neurons were capable of firing at high rates, but the maximum firing rates (peak average firing rates before entering depolarization block) varied widely between cell types, from 50 to >500 Hz (Fig. 1C). Excitatory premotor neurons exhibited both the highest maximum firing rates on average (262.8 Hz) and the widest distribution (114.6–531.3 Hz; SD 95.6 Hz). In contrast, local inhibitory interneurons had low maximum firing rates (average 89.2 Hz) that were relatively tightly clustered (range 47.5–160.7 Hz; SD 30.1 Hz). Firing capacities of MVN neurons skewed toward lower rates, consistent with the spared use of high firing rates to minimize energetic costs (Perge et al., 2012).
To be capable of sustaining high firing rates, neurons must overcome biophysical challenges imposed by voltage-gated sodium channels (NaV), which inactivate during the depolarization of each action potential and require tens to hundreds of milliseconds to become available for subsequent action potentials. As with other fast-spiking neurons (Rudy and McBain, 2001; Akemann and Knöpfel, 2006; Carter and Bean, 2009; Hu et al., 2014), MVN neurons meet these challenges by expressing Na currents, which are relatively resistant to inactivation (Gittis and du Lac, 2008), and KV3 currents, which are activated at high voltages and minimize NaV inactivation and neuronal refractory periods by rapidly repolarizing the action potential (Gittis and du Lac, 2008; Gittis et al., 2010). The net effect of these currents determines both the width of the action potential and the absolute refractory period; faster firing neurons with brief refractory periods have narrower action potentials than do slower firing neurons (Fig. 1C; Bagnall et al., 2007; Carter and Bean, 2009). The refractory period during sustained firing covaries linearly with action potential width (Fig. 1D), exhibiting a tight relationship across MVN neurons (r2 = 0.71 p < 0.00001, n = 167) and within each of the cell types (except cerebellar target neurons; Table 1). Interestingly, the speeds of action potential depolarization and repolarization also covary within and between cell types and are tightly and linearly correlated (Fig. 1E; Table 1), suggesting a constant ratio between NaV to and KV3 currents across diverse cell types. The balance between these currents is beneficial; insufficient KV3 current for a given NaV current leads to slower repolarization and accumulation of inactivated NaV (Gittis et al., 2010), limiting fast spiking. On the other hand, KV3 currents that exceed the requirements for NaV protection from inactivation would be a waste of metabolic resources. These results demonstrate that action potential shapes and firing speeds vary continuously both within and between MVN cell types and suggest the possibility of graded coexpression of the ion channels responsible for fast spiking.
Table 1.
Action potential and firing parameter correlations in MVN neurons
| Cell type | Refractory period vs APhw |
AP rise rate vs fall rate |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| r | r2 | Slope | n | p | r | r2 | Slope | n | p | |
| Pre-oculomotor | 0.65 | 0.43 | 0.019 | 61 | 1.2E-08 | 0.85 | 0.73 | 0.81 | 61 | 2.3E-18 |
| Pre-cerebellar | 0.83 | 0.68 | 0.026 | 29 | 3.1E-08 | 0.94 | 0.88 | 0.8 | 31 | 1.0E-14 |
| Commissural | 0.89 | 0.79 | 0.03 | 27 | 7.4E-10 | 0.93 | 0.87 | 0.6 | 27 | 1.5E-12 |
| Cerebellar target | 0 | 0 | 0 | 13 | 9.9E-01 | 0.94 | 0.88 | 0.67 | 13 | 2.3E-06 |
| Local interneuron | 0.63 | 0.4 | 0.022 | 35 | 4.8E-05 | 0.93 | 0.86 | 0.61 | 35 | 7.7E-16 |
| All | 0.84 | 0.71 | 0.025 | 165 | 1.3E-45 | 0.94 | 0.88 | 0.78 | 167 | 2.1E-77 |
| All (avg) | 0.94 | 0.89 | 0.024 | 5 | 1.5E-02 | 0.98 | 0.97 | 0.78 | 5 | 2.6E-03 |
Shown are results from linear regression of neuronal refractory period (inverse of maximum firing rate; see Materials and Methods) versus action potential half-width (APhw) and action potential (AP) rise versus fall rate for five types of MVN neurons, all of the MVN neurons (All), and averages of the five cell types [All (avg)]. The correlation coefficients r and r2, fit slope, number of neurons (n) and significance level (p) are indicated.
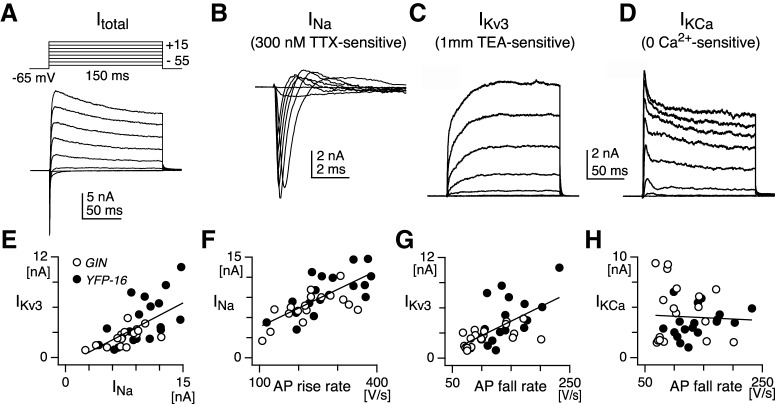
To determine whether Nav and Kv3 currents are coexpressed, we analyzed whole-cell voltage-clamp recordings from acutely dissociated vestibular nucleus neurons (Gittis and du Lac, 2007) obtained from mouse lines that express fluorescent proteins in specific neuronal subtypes (Bagnall et al., 2007). Cell types labeled in the YFP-16 line include pre-oculomotor, pre-cerebellar, commissural inhibitory (glycinergic), and unidentified excitatory neurons; the GIN line labels local and commissural inhibitory (GABAergic) neurons (Bagnall et al., 2007; Kodama et al., 2012). Following measurement of spontaneous action potentials in current-clamp mode, whole-cell currents were recorded in voltage-clamp in response to a series of voltage steps (Fig. 2A, top), and Na and K currents were pharmacologically isolated in each cell (see Materials and Methods). Voltage steps evoked a brief inward current which preceded sustained outward currents, as shown in Figure 2A for a representative YFP-16 neuron. The transient Na current (Fig. 2B) was identified as the large, fast inward current isolated by subtraction after applying TTX (300 nm). Kv3 and KCa currents (Figs. 2C,D) were identified by their sensitivity to TEA (1 mm) and replacement of extracellular Ca with Mg and Cd, respectively. The kinetics and voltage dependence of these currents were described previously (Gittis and du Lac, 2007).
Figure 2.
Na and Kv3 currents are positively correlated and scale with action potential speed. A, Whole-cell currents evoked by a series of voltage steps (−55 to 15 mV; 150 ms duration) from a holding potential of −65 mV in an acutely isolated MVN neuron. Individual components of whole-cell currents were isolated via pharmacological subtraction. B, Na currents evoked by the same voltage steps, revealed by subtracting currents in 300 nm TTX from whole-cell currents. C, Kv3 currents (1 mm TEA-sensitive). D, KCa currents (0 Ca-sensitive). E, Maximum Kv3 current (average current evoked at +15 mV) for 36 neurons correlates with the peak Na current for each neuron. F, Peak Na current measured in each neuron scales with action potential rise rate. G, Maximum Kv3 current scales with action potential fall rate. H, Maximum KCa current is not correlated with action potential fall rate.
Peak Kv3 and Na currents were well correlated in dissociated neurons (Fig. 2E; r = 0.64, p = 0.00028, n = 36 neurons total; r = 0.62, p = 0.0063, n = 18 for YFP-16 neurons; r = 0.68, p = 0.0017, n = 18 for GIN neurons), congruent with a global pattern of coexpression across MVN cell types. As expected from the respective roles of transient Na and Kv3 current in action potential generation in MVN neurons (Gittis and du Lac, 2007; Gittis et al., 2010), peak Na currents were well correlated with action potential rise rates (Fig. 2F; r = 0.71, p < 0.00001, n = 36 for all neurons; r = 0.70, p = 0.0012, n = 18 for YFP-16, r = 0.66, p = 0.0028, n = 18 for GIN) and peak Kv3 currents were well correlated with action potential fall rates (Fig. 2G; r = 0.63, p = 0.000045, n = 36 for all neurons; r = 0.65, p = 0.0036, n = 18 for YFP-16 neurons; r = 0.54, p = 0.54, n = 18 for GIN). In contrast, KCa currents, which are engaged predominantly during the interspike interval (Smith et al., 2002), are correlated with neither action potential rise rates (r = 0.10, p = 0.55, n = 36 for all neurons; r = 0.04, p = 0.87, n = 18 for YFP16 neurons; r = 0.06, p = 0.82, n = 18 for GIN neurons) nor fall rates (r = −0.05, p = 0.77, n = 36 for all neurons; r = 0.30, p = 022, n = 18 for YFP16; r = −0.098, p = 0.7, n = 18 GIN neurons). Given that action potential rise and fall rates are proxies of firing capacity and mutually correlated in dissociated neurons (Gittis et al., 2010) as in slice preparations (Fig. 1), these results indicate that variations in firing capacity across MVN neurons reflects an underlying pattern of graded coexpression of Na and Kv3 currents.
To assess whether the balanced expression of Nav and Kv3 currents was evident at the transcriptional level, we analyzed a single-cell cDNA library prepared from acutely dissociated MVN neurons, in which multiple transcripts for ion channels and marker genes were assessed quantitatively via real-time qPCR (single-cell qPCR) and neurons were classified via marker gene expression and retrograde labeling (Kodama et al., 2012). As shown in Figure 3A, expression levels of Kcnc1 and Scn8a, the genes encoding KV3.1 and NaV1.6 channels, which are primarily responsible for action potential generation in MVN neurons (Kodama et al., 2012), are well correlated over 100-fold range of transcript levels across the population of sampled neurons (r = 0.84, p < 0.00001, n = 161) and within each neuronal subtype except cerebellar target neurons, which expressed the lowest levels of transcripts (Table 2). Cell types that exhibited higher maximal firing rates expressed higher levels of Kcnc1 and Scn8a transcripts (Fig. 3B; r = 0.91, p = 0.034). The correspondence between ionic currents (Fig. 2E) and transcript levels (Fig. 3A) of Nav and Kv3 across individual MVN neurons suggests that the functional protein levels are primarily determined by transcript levels.
Table 2.
Correlations between Kcnc1 and Scn8a transcript levels within and between MVN cell types
| Cell type | r | n | p |
|---|---|---|---|
| Pre-oculomotor | 0.87 | 21 | 3.9E-07 |
| Pre-cerebellar | 0.88 | 20 | 4.2E-07 |
| Commissural | 0.70 | 30 | 1.5E-05 |
| Cerebellar target | 0.29 | 12 | 3.6E-01 |
| Local interneuron | 0.55 | 70 | 7.3E-07 |
| All | 0.84 | 153 | 1.0E-41 |
| All (avg) | 0.97 | 5 | 5.6E-03 |
Results from linear regression of transcript levels for Kcnc1 and Scn8a are indicated for five cell types individually, for the combined population of neurons (All), and for average values of the five cell types [All (avg)].
Several types of ion channels are engaged during repetitive firing, with distinct types of ion channels contributing differentially to the action potential and the interspike interval (Bean, 2007). To determine whether other ion channel genes exhibit similar expression patterns with Scn8a and Kcnc1, we assessed coexpression of 42 ion channel genes, which encode α subunits of voltage-gated and calcium/sodium-dependent channels and sodium channel β subunits (Table 3). Figure 4A shows the results of this analysis in the form of a dendrogram, in which closer nodes (genes) show higher correlation (measured across all the MVN neurons sampled). As shown in Figure 4, C and E, nine ion channels in one major branch of the dendrogram, including Scn8a and Kcnc1, were positively and significantly correlated across all neurons (Table 4) and within the cell types with the widest ranges of channel expression levels (pre-oculomotor and pre-cerebellar; Table 4). The ion channels in this branch are referred to as “fast-spiking ion channels”; the majority of them (Scn1a, Scn8a, Scn1b, Scn4b, Kcnc1, and Kcna1) are localized in the axon initial segment and nodes of Ranvier (H. Wang et al., 1993; Caldwell et al., 2000; Devaux et al., 2003; Duflocq et al., 2008; Buffington and Rasband, 2013; Wimmer et al., 2015; Rash et al., 2016) and contribute to the fast and reliable propagation of action potentials along the axon (see Discussion). In contrast, ion channels in the other major branch were significantly less correlated with each other (Fig. 4D,F; Table 5). These ion channels include calcium-dependent potassium channels (Kcnma1, Kcnn2) and A-type K channels (Kcnd2, Kcnd3, Kcnc4), which are active predominantly during the interspike interval and influence how neurons respond to synaptic inputs (Smith et al., 2002; Nelson et al., 2003; Bean, 2007).
Table 4.
Correlations among fast-spiking ion channel transcript levels within and across MVN cell types
| Cell type | r, avg | r, SD | p, avg | p, SD | No. sig; p < 0.05 | No. of valid comparisons | Significant, % | No. of cells, average | No. of cells, SD |
|---|---|---|---|---|---|---|---|---|---|
| Pre-oculomotor | 0.67 | 0.13 | 9.6E-03 | 2.4E-02 | 34 | 36 | 94.4 | 21.0 | 0.0 |
| Pre-cerebellar | 0.67 | 0.12 | 9.1E-03 | 1.6E-02 | 34 | 36 | 94.4 | 19.2 | 0.8 |
| Unknown | 0.84 | 0.10 | 5.9E-02 | 4.7E-02 | 6 | 15 | 40.0 | 5.7 | 0.5 |
| Commissural | 0.51 | 0.18 | 5.8E-02 | 1.4E-01 | 27 | 36 | 75.0 | 27.1 | 2.5 |
| Cerebellar target | 0.46 | 0.20 | 2.1E-01 | 2.3E-01 | 6 | 21 | 28.6 | 11.3 | 1.1 |
| Local interneuron | 0.25 | 0.24 | 3.2E-01 | 3.6E-01 | 17 | 33 | 51.5 | 42.2 | 25.5 |
| All | 0.71 | 0.08 | 1.1E-07 | 6.5E-07 | 36 | 36 | 100.0 | 116.3 | 36.4 |
Correlation coefficients and significance levels were evaluated for pairwise comparisons between ion channel genes in the left major branch of the dendrogram in Figure 4. Comparisons of transcript levels were considered valid when genes-of-interest expressed in more than three neurons. Table shows the average and SD of r and p values, the number of valid comparisons for each cell type, the percentage of correlations that were significant at the p < 0.05 level, and the mean and SD of the number of neurons in which correlations were evaluated in each group.
Table 5.
Correlations among non-fast-spiking ion channel transcript levels within and across MVN cell types
| Cell type | r, avg | r, SD | p, avg | p, SD | No. sig; p < 0.05 | No. of valid comparisons | Significant, % | No. of cells, average | No. of cells, SD |
|---|---|---|---|---|---|---|---|---|---|
| Pre-oculomotor | 0.17 | 0.39 | 4.3E-01 | 3.0E-01 | 33 | 292 | 11.3 | 11.0 | 4.8 |
| Pre-cerebellar | 0.06 | 0.41 | 4.3E-01 | 2.9E-01 | 33 | 389 | 8.5 | 10.7 | 5.1 |
| Unknown | 0.43 | 0.44 | 3.8E-01 | 2.9E-01 | 17 | 161 | 10.6 | 5.0 | 0.9 |
| Commissural | 0.21 | 0.39 | 3.8E-01 | 2.9E-01 | 42 | 315 | 13.3 | 11.6 | 6.7 |
| Cerebellar target | 0.24 | 0.42 | 4.3E-01 | 3.0E-01 | 10 | 170 | 5.9 | 7.6 | 2.9 |
| Local interneuron | 0.08 | 0.36 | 4.5E-01 | 3.0E-01 | 25 | 343 | 7.3 | 21.8 | 20.2 |
| All | 0.13 | 0.26 | 3.5E-01 | 3.1E-01 | 106 | 487 | 21.8 | 45.5 | 37.3 |
Correlation coefficients and significance levels were evaluated for pairwise comparisons between ion channel genes in the right major branch of the dendrogram in Figure 4. Table shows the average and SD of r and p values, the number of valid comparisons for each cell type, the percentage of correlations that were significant at the p < 0.05 level, and the mean and SD of the number of neurons in which correlations were evaluated in each group.
To identify additional genes that may be coexpressed with the fast-spiking ion channels, we pooled cDNA from each MVN cell type and assessed differentially expressed genes using microarrays. Figure 5A shows the dendrogram generated by hierarchical clustering of ion channel genes and all the other genes that exhibited differential expression across cell types (>4-fold difference between minimum and maximum expression levels), corresponding to 5.5% of genes on the array (1004/18266). The fast-spiking ion channel genes were clustered in a single branch of the dendrogram (red branch, termed “fast-spiking cluster”), which contains 134 genes in total (138 probe sets; Fig. 5-2). In contrast, genes encoding other types of ion channels are distributed in other branches of the dendrogram; these include Kcnn2, which encodes the SK2 type calcium activated potassium channel, which predominates during the interspike interval in MVN neurons, and Scn2a (NaV2.1), which mediates action potential backpropagation.
Cell classification used for microarray sample preparation in comparison with the one used in Kodama et al., 2012. Note that each node in the dendrogram corresponds to one MVN neuron. For details, see Materials and Methods. Download Figure 5-1, EPS file (248.1KB, eps)
Genes potentially co-regulated for fast-firing capacity across MVN neurons. Listed are 138 probe sets included in the red branch of the dendrogram shown in Figure 6A and corresponding gene symbols. Download Figure 5-2, XLS file (74.5KB, xls)
Notably, the fast-spiking cluster includes each of the four genes for neurofilament proteins expressed in the mammalian CNS (Nefh, Nefm, Nefl, and Ina, which encode neurofilament heavy, medium, and light chain and intermediate neurofilament protein, respectively). Expression levels of the neurofilament genes covaried with the fast-spiking ion channel genes (Fig. 5B; r = 0.75 ± 0.19 SD, 22/35 pairs with p < 0.05), consistent with a common transcription regulatory mechanism. The fast-spiking cluster also contained several genes for presynaptic proteins associated with fast, synchronous transmitter release, including Vamp1, Cplx1, and Syt2. As with the neurofilament genes, expression levels of these synaptic genes were highly correlated with the fast-spiking ion channel genes (Fig. 5C; r = 0.79 ± 0.19 SD, 18/28 pairs with p < 0.05).
Single-cell qPCR confirmed that transcript levels of neurofilament genes Nefh, Nefm, and Nefl were highly and significantly correlated with the fast-spiking ion channels across the population of MVN neurons (Fig. 6A; Table 6). Within each cell type, positive correlations were predominant in pre-cerebellar neurons and inhibitory commissural neurons (Table 6). Consistent with tight correspondence between fast-spiking ion channel gene expression and firing capacity across MVN cell types (Fig. 3B), average neurofilament expression levels were well correlated with maximal firing rates (Fig. 6B; r = 0.93, p = 0.023, n = 5 cell types). Given that expression levels of neurofilaments reflects the caliber of axons (Friede and Samorajski, 1970; Lee and Cleveland, 1996), the correlated expression of ion channel and neurofilament genes can account for the linear relationship between axon diameter and fast-spiking capacity, as has been reported in several cell types (Perge et al., 2012).
Table 6.
Correlations between fast-spiking ion channel gene and neurofilament transcript levels
| Cell type | r, avg | r, SD | p, avg | p, SD | No. sig; p < 0.05 | No. of valid comparisons | Significant, % | No. of cells, average | No. of cells, SD |
|---|---|---|---|---|---|---|---|---|---|
| Pre-oculomotor | 0.17 | 0.16 | 5.0E-01 | 3.1E-01 | 1 | 27 | 3.7 | 20.0 | 0.0 |
| Pre-cerebellar | 0.57 | 0.15 | 4.3E-02 | 7.2E-02 | 18 | 27 | 66.7 | 18.6 | 0.7 |
| Unknown | 0.55 | 0.31 | 3.3E-01 | 3.4E-01 | 3 | 18 | 16.7 | 5.8 | 0.4 |
| Commissural | 0.51 | 0.17 | 6.7E-02 | 1.7E-01 | 21 | 27 | 77.8 | 24.1 | 1.9 |
| Cerebellar target | 0.39 | 0.33 | 3.8E-01 | 2.8E-01 | 1 | 21 | 4.8 | 8.4 | 2.8 |
| Local interneuron | 0.35 | 0.36 | 3.4E-01 | 2.7E-01 | 3 | 23 | 13.0 | 10.7 | 4.9 |
| All | 0.67 | 0.08 | 2.8E-07 | 1.1E-06 | 27 | 27 | 100.0 | 83.1 | 12.4 |
Correlation coefficients and significance levels were evaluated for pairwise comparisons between fast-spiking ion channel genes and neurofilament genes (Nefh, Nefm, Nefl). Table shows the average and SD of r and p values, the number of valid comparisons for each cell type, the percentage of correlations that were significant at the p < 0.05 level, and the mean and SD of the number of neurons in which correlations were evaluated in each group.
Each of the three synaptic genes in the fast-spiking cluster (Vamp1, Cplx1 and Syt2) were confirmed with single-cell qPCR to be coexpressed with fast-spiking ion channels in a graded manner across neurons (Fig. 6C; Table 7). Within each cell type, significant positive correlations predominated in pre-cerebellar and inhibitory commissural neurons and were less evident in other cell types (Table 7). In contrast with these synaptic genes, Syt1, which was not in the fast-spiking cluster (Fig. 5A,E), exhibited a different pattern of cell-type-specific expression, which was not related to fast-firing capacity (Fig. 6D).
Table 7.
Correlations between fast-spiking ion channel gene and synaptic gene transcript levels within and across MVN cell types
| Cell type | Cell type | r, avg | r, SD | p, avg | p, SD | No. of valid comparisons | Significant, % | No. of cells, average | No. of cells, SD |
|---|---|---|---|---|---|---|---|---|---|
| Pre-oculomotor | 0.45 | 0.25 | 1.8E-01 | 2.4E-01 | 13 | 27 | 48.1 | 20.3 | 0.5 |
| Pre-cerebellar | 0.60 | 0.12 | 2.4E-02 | 4.3E-02 | 23 | 27 | 85.2 | 18.7 | 0.6 |
| Unknown | 0.76 | 0.14 | 1.0E-01 | 8.3E-02 | 5 | 12 | 41.7 | 5.8 | 0.4 |
| Commissural | 0.49 | 0.18 | 8.7E-02 | 1.5E-01 | 18 | 27 | 66.7 | 23.3 | 2.8 |
| Cerebellar target | 0.25 | 0.26 | 4.0E-01 | 2.9E-01 | 1 | 14 | 7.1 | 12.1 | 0.9 |
| Local interneuron | 0.38 | 0.31 | 2.0E-01 | 2.4E-01 | 6 | 16 | 37.5 | 14.8 | 4.4 |
| All | 0.70 | 0.10 | 1.6E-06 | 6.4E-06 | 27 | 27 | 100.0 | 81.9 | 17.3 |
Correlation coefficients and significance levels were evaluated for pairwise comparisons between fast-spiking ion channel genes and synaptic genes (Vamp1, Cplx1, Syt2). Table shows the average and SD of r and p values, the number of valid comparisons for each cell type, the percentage of correlations that were significant at the p < 0.05 level, and the mean and SD of the number of neurons in which correlations were evaluated in each group.
In total, we performed single-cell qPCR on 22 genes in the fast-spiking cluster identified by the microarray analysis. Of these, 15 exhibited graded coexpression with fast-spiking ion channel genes (>0.6 of average r to the fast-spiking ion channels; Table 8). Of note, some of the genes showing graded coexpression with fast-spiking channels were expressed sparsely in the cell types of low maximal firing rate (local interneurons, cerebellar target neurons; Table 8), suggesting that some genes associated with fast spiking are selectively expressed in the fastest firing cell types. The remaining seven genes were expressed in cell-type-specific patterns but did not covary with fast-spiking genes (<0.5 of average r to the fast-spiking ion channels across individual MVN neurons). These included Pvalb, which serves as a marker for fast-spiking inhibitory interneurons in the forebrain (Hu et al., 2014), Slc17a6 (Vglut2), Spp1 (osteopontin), Grm1 (metabotropic glutamate receptor), Gjd2 (connexin 36), Trhr (thyrotropin releasing hormone receptor), and Cbln1 (cerebellin 1).
Table 8.
Prevalence (%) of neurons with significant expression of genes that covary with fast-firing capacity in MVN cell types
| Pre-oculomotor | Pre-cerebellar | Unknown | Commissural | Cerebellar target | Local interneuron | |
|---|---|---|---|---|---|---|
| Cacna1a | 100 | 95 | 100 | 100 | 85 | 86 |
| Cplx1 | 100 | 100 | 100 | 100 | 100 | 100 |
| Kcna1 | 100 | 100 | 50 | 87 | 85 | 47 |
| Kcnc1 | 100 | 100 | 100 | 100 | 100 | 99 |
| Kcng4 | 100 | 90 | 0 | 90 | 15 | 31 |
| Kcnh2 | 100 | 95 | 100 | 100 | 92 | 91 |
| Nefh | 100 | 100 | 100 | 89 | 54 | 33 |
| Nefl | 100 | 100 | 100 | 100 | 100 | 100 |
| Nefm | 100 | 100 | 100 | 100 | 54 | 72 |
| Scn1a | 100 | 100 | 83 | 97 | 100 | 95 |
| Scn1b | 100 | 100 | 100 | 100 | 100 | 100 |
| Scn4b | 100 | 100 | 33 | 80 | 0 | 5 |
| Scn8a | 100 | 100 | 100 | 100 | 92 | 92 |
| Syt2 | 100 | 95 | 33 | 70 | 15 | 1 |
| Vamp1 | 100 | 100 | 100 | 100 | 100 | 100 |
Of 134 candidate genes the transcript levels of which potentially covary with fast-firing capacity of MVN neurons, 15 genes were confirmed for highly correlative expression (average r > 0.6) with the fast-firing ion channels across individual MVN samples. The percentage of cells showing significant expression of the genes are shown for each cell type. Cacna1a and Kcng4 are included in the list because, although the microarray failed to detect them, the high correlation of its expression to the fast-firing genes were confirmed at the single-cell level (Figure 4A,B). For Cplx1, Nefh, Nefl, Nefm, and Vamp1, 102 cells were examined (pre-oculomotor, n = 20; pre-cerebellar, n = 19; unknown, n = 6; commissural, n = 26; cerebellar target, n = 13; local interneuron, n = 18). For all the other genes, 167 cells were examined (pre-oculomotor, n = 21; pre-cerebellar, n = 20; unknown, n = 6; commissural, n = 30; cerebellar target, n = 13; local interneuron, n = 77).
Discussion
Fast and sustained firing of neurons consumes substantial metabolic and physical resources, giving rise to an evolutionary pressure for the minimal use of high firing rates in the CNS (Perge et al., 2012). However, several computational tasks do require neurons to encode and transmit signals as rapidly as possible. MVN neurons dynamically transform signals from the vestibular nerve axons (which fire tonically at 50–100 spikes/s) into appropriate premotor, pre-cerebellar, and local commands. The diversity of fast-spiking cell types in the MVN enabled us to study how neurons coordinate multiple biophysical properties to achieve specific firing capacities. Transcript profiling guided by electrophysiological analyses identified a group of genes in which expression levels covary with fast-spiking capacity in MVN neurons, enabling multiple biophysical properties associated with rapid firing to be tuned in parallel. Primary members of the identified gene group included voltage-gated ion channels, which maintain high excitability even during repetitive firing, neurofilaments, which reflect the caliber of the axons, and genes involved in neurotransmitter release, which may tune speed and sustainability of synaptic transmission. The covariation of transcript levels in association with fast-spiking capacity strongly suggests the presence of a “fast-spiking gene module” in which member genes are co-regulated for cost-efficient implementation of high throughput information processing.
The fast-spiking gene module that we propose comprises genes in which transcript levels are increased as the maximal number of spikes reliably discharged and transmitted through the axons and synapses in a given time (i.e., firing capacity) increases. A major function of the fast-spiking gene module is reliable generation and propagation of action potentials along the axon. NaV1.1 (Scn1a), NaV1.6 (Scn8a), NaVβ1 (Scn1b), NaVβ4 (Scn4b), and KV3.1 (Kcnc1) are densely localized in the axon initial segments and/or the nodes of Ranvier (H. Wang et al., 1993; Caldwell et al., 2000; Devaux et al., 2003; Duflocq et al., 2008; Buffington and Rasband, 2013; Wimmer et al., 2015; Rash et al., 2016). For neurons to sustain firing at high rates, it is essential to minimize inactivation of NaV during prolonged firing, for which Kcna1 and Kcnc1 play a critical role (Rudy and McBain, 2001; Kole et al., 2007; Carter and Bean, 2009; Foust et al., 2011; Kaczmarek and Zhang, 2017). Rapid repolarization of action potentials by KV3 is also critical for faster recovery of NaV from inactivation as well as cost-efficiency in action potential generation (Carter and Bean, 2011; Gittis et al., 2010; Hu et al., 2018). Higher expression of Scn1a and Scn8a would make neurons more resistant to accumulation of NaV inactivation, by increasing the fraction of inactivated NaV that neurons can accept without compromising firing capacity. Scn1b is likely to facilitate fast gating and localization of NaV (Isom et al., 1992; Brackenbury et al., 2008, 2010). The function of Kcnh2 (ERG channel) in fast spiking is unclear.
Genes encoding neurofilaments (Nefl, Nefm, Nefh, Ina) are enriched in the fast-spiking gene module. Correlated expression of the neurofilament genes across diverse neuronal populations and their link to fast-spiking capacity has been suggested (Saunders et al., 2018), for which we provide the first direct evidence (Fig. 6B). Given that the expression of neurofilaments is a proxy of the caliber of the axon (Friede and Samorajski, 1970; Lee and Cleveland, 1996), our findings also provide a molecular representation of the linear relationship between firing capacity of neurons and axon caliber (Perge et al., 2012). A thicker axon can accommodate more mitochondria (Perge et al., 2009), supplying more energy to maintain ionic gradients during repetitive firing. Larger caliber axons may also be beneficial to transport materials for forming more active zones in presynaptic terminals (Perge et al., 2012), as fast-spiking neurons tend to have highly arborized and/or enlarged terminals (Liberman, 1993; Goldberg, 2000; Carr and Soares, 2002; Sekirnjak et al., 2003; Telgkamp et al., 2004; Hu et al., 2014). Employing many active zones seems to be a common solution for fast and reliable transmitter release (Taschenberger et al., 2002; Telgkamp et al., 2004; McElvain et al., 2015; Delvendahl and Hallermann, 2016), consistent with the presence of active zone constituting molecules in the fast-spiking gene module (Cacnala, Vamp1, Cplx1, Syt2).
Some genes in the fast-spiking gene module were not detected with qPCR in MVN cell types with the lowest firing capacities. Syt2 (synaptotagmin2) and Scn4b (NaVβ4) are selectively expressed in pre-oculomotor, pre-cerebellar, and commissural neurons, but rarely in cerebellar target neurons and local interneurons. In contrast, synaptotagmin 1, which can be expressed in a complementary manner to synaptotagmin 2 (Fox and Sanes, 2007), was expressed in all MVN cell types but did not correlate with firing capacity (Fig. 6D). Synaptotagmin 2 has the fastest calcium-binding kinetics throughout the synaptotagmin family (Xu et al., 2007), and is known to facilitate fast synaptic signaling (Hu et al., 2014; Kochubey et al., 2016; Chen et al., 2017). NaVβ4 reduces the fraction of NaV entering the inactivated state after opening via an open channel blocking mechanism (Grieco et al., 2005). These genes would provide additional support for faster signaling.
Theoretical analyses (Bhalla and Bower, 1993; Goldman et al., 2001; Golowasch et al., 2002; Prinz et al., 2004; O'Leary et al., 2014) and experimental studies (Swensen and Bean, 2005; Schulz et al., 2006) have demonstrated that many different combinations of ionic currents can be used to produce the same temporal pattern of firing. These findings imply considerable degrees of freedom in ion channel gene expression. In contrast, our results suggest that absolute expression levels of a specific group of ion channel genes determine the firing capacities of fast-spiking neurons. Stringent biophysical requirements place tight constraints on the solution space for minimizing neuronal refractory periods while enabling sustained, high-frequency synaptic transmission. Our results suggest that, to this end, there is no other solution but to express specialized NaV channels at high levels, and sufficient amounts of KV3 for rapid repolarization of the action potential.
A variety of types of fast-spiking neurons have been identified in the CNS, including parvalbumin-expressing interneurons in the cerebral cortex and hippocampus and GABAergic neurons in the substantia nigra reticulata and globus pallidus. The cerebellum and brainstem are also populated by many types of fast-spiking neurons. Several lines of evidence indicate that the firing capacities of these diverse types of neurons are likely to be tuned by the fast-spiking gene module identified in MVN neurons, which are electrophysiologically representative of fast-spiking neurons in both the brainstem (Kolkman et al., 2011) and the cerebellar nuclei (Bagnall et al., 2009). As in MVN neurons, KV3 is essential for fast and sustained firing and synaptic transmission in auditory brainstem neurons, fast-spiking interneurons in the cerebral cortex and hippocampus, and cerebellar Purkinje cells (Rudy and McBain, 2001; Kaczmarek and Zhang, 2017). Expression of neurofilaments can distinguish neurons that presumably fire faster than neighboring neurons (White and Sillitoe, 2013; Ueta et al., 2014; Sun et al., 2018). The specific synaptic molecules we identified (Syt2, Cplx1, Vamp1) are also selectively expressed in fast-spiking neuronal cell types throughout the brain (Hu et al., 2014; Kochubey et al., 2016; Chen et al., 2017; Saunders et al., 2018). Given the consistently high representation of the same set of genes among diverse fast-spiking neuronal cell types, it is plausible that coordinated expression levels of these genes is a common mechanism for satisfying constraints on space and energy while optimally tuning the brain's need for speed.
Footnotes
This work was supported by NIH Grant EY11027 and HHMI.
The authors declare no competing financial interests.
References
- Akemann W, Knöpfel T (2006) Interaction of Kv3 potassium channels and resurgent sodium current influences the rate of spontaneous firing of Purkinje neurons. J Neurosci 26:4602–4612. 10.1523/JNEUROSCI.5204-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwell D, Laughlin SB (2001) An energy budget for signaling in the grey matter of the brain. J Cereb Blood Flow Metab 21:1133–1145. 10.1097/00004647-200110000-00001 [DOI] [PubMed] [Google Scholar]
- Bagnall MW, Stevens RJ, du Lac S (2007) Transgenic mouse lines subdivide medial vestibular nucleus neurons into discrete, neurochemically distinct populations. J Neurosci 27:2318–2330. 10.1523/JNEUROSCI.4322-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall MW, Zingg B, Sakatos A, Moghadam SH, Zeilhofer HU, de Lac S (2009) Glycinergic projection neurons of the cerebellum. J Neurosci 29:10104–10110. 10.1523/JNEUROSCI.2087-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski JJ, Dethleffsen K, Meyer M (2000) Cre recombinase expression in cerebellar Purkinje cells. Genesis 28:93–98. [DOI] [PubMed] [Google Scholar]
- Bean BP. (2007) The action potential in mammalian central neurons. Nat Rev Neurosci 8:451–465. 10.1038/nrn2148 [DOI] [PubMed] [Google Scholar]
- Bengtsson M, Ståhlberg A, Rorsman P, Kubista M (2005) Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res 15:1388–1392. 10.1101/gr.3820805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla US, Bower JM (1993) Exploring parameter space in detailed single neuron models: simulations of the mitral and granule cells of the olfactory bulb. J Neurophysiol 69:1948–1965. 10.1152/jn.1993.69.6.1948 [DOI] [PubMed] [Google Scholar]
- Brackenbury WJ, Davis TH, Chen C, Slat EA, Detrow MJ, Dickendesher TL, Ranscht B, Isom LL (2008) Voltage-gated Na+ channel β1 subunit-mediated neurite outgrowth requires fyn kinase and contributes to postnatal CNS development in vivo. J Neurosci 28:3246–3256. 10.1523/JNEUROSCI.5446-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackenbury WJ, Calhoun JD, Chen C, Miyazaki H, Nukina N, Oyama F, Ranscht B, Isom LL (2010) Functional reciprocity between Na+ channel Nav1.6 and β1 subunits in the coordinated regulation of excitability and neurite outgrowth. Proc Natl Acad Sci U S A 107:2283–2288. 10.1073/pnas.0909434107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffington SA, Rasband MN (2013) Na+ channel-dependent recruitment of Navβ4 to axon initial segments and nodes of Ranvier. J Neurosci 33:6191–6202. 10.1523/JNEUROSCI.4051-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell JH, Schaller KL, Lasher RS, Peles E, Levinson SR (2000) Sodium channel Nav1.6 is localized at nodes of Ranvier, dendrites, and synapses. Proc Natl Acad Sci U S A 97:5616–5620. 10.1073/pnas.090034797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr CE, Soares D (2002) Evolutionary convergence and shared computational principles in the auditory system. Brain Behav Evol 59:294–311. 10.1159/000063565 [DOI] [PubMed] [Google Scholar]
- Carter BC, Bean BP (2009) Sodium entry during action potentials of mammalian neurons: incomplete inactivation and reduced metabolic efficiency in fast-spiking neurons. Neuron 64:898–909. 10.1016/j.neuron.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter BC, Bean BP (2011) Incomplete inactivation and rapid recovery of voltage-dependent sodium channels during high-frequency firing in cerebellar Purkinje neurons. J Neurophysiol 105:860–871. 10.1152/jn.01056.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Arai I, Satterfield R, Young SM Jr, Jonas P (2017) Synaptotagmin 2 is the fast Ca2+ sensor at a central inhibitory synapse. Cell Rep 18:723–736. 10.1016/j.celrep.2016.12.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvendahl I, Hallermann S (2016) The cerebellar mossy fiber synapse as a model for high-frequency transmission in the mammalian CNS. Trends Neurosci 39:722–737. 10.1016/j.tins.2016.09.006 [DOI] [PubMed] [Google Scholar]
- Devaux J, Alcaraz G, Grinspan J, Bennett V, Joho R, Crest M, Scherer SS (2003) Kv3.1b is a novel component of CNS nodes. J Neurosci 23:4509–4518. 10.1523/JNEUROSCI.23-11-04509.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duflocq A, Le Bras B, Bullier E, Couraud F, Davenne M (2008) Nav1.1 is predominantly expressed in nodes of Ranvier and axon initial segments. Mol Cell Neurosci 39:180–192. 10.1016/j.mcn.2008.06.008 [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR (2000) Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28:41–51. 10.1016/S0896-6273(00)00084-2 [DOI] [PubMed] [Google Scholar]
- Foust AJ, Yu Y, Popovic M, Zecevic D, McCormick DA (2011) Somatic membrane potential and Kv1 channels control spike repolarization in cortical axon collaterals and presynaptic boutons. J Neurosci 31:15490–15498. 10.1523/JNEUROSCI.2752-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MA, Sanes JR (2007) Synaptotagmin I and II are present in distinct subsets of central synapses. J Comp Neurol 503:280–296. 10.1002/cne.21381 [DOI] [PubMed] [Google Scholar]
- Friede RL, Samorajski T (1970) Axon caliber related to neurofilaments and microtubules in sciatic nerve fibers of rats and mice. Anat Rec 167:379–387. 10.1002/ar.1091670402 [DOI] [PubMed] [Google Scholar]
- Gittis AH, du Lac S (2007) Firing properties of GABAergic versus non-GABAergic vestibular nucleus neurons conferred by a differential balance of potassium currents. J Neurophysiol 97:3986–3996. 10.1152/jn.00141.2007 [DOI] [PubMed] [Google Scholar]
- Gittis AH, du Lac S (2008) Similar properties of transient, persistent, and resurgent Na currents in GABAergic and non-GABAergic vestibular nucleus neurons. J Neurophysiol 99:2060–2065. 10.1152/jn.01389.2007 [DOI] [PubMed] [Google Scholar]
- Gittis AH, Moghadam SH, du Lac S (2010) Mechanisms of sustained high firing rates in two classes of vestibular nucleus neurons: differential contributions of resurgent na, Kv3, and BK currents. J Neurophysiol 104:1625–1634. 10.1152/jn.00378.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg JM. (2000) Afferent diversity and the organization of central vestibular pathways. Exp Brain Res 130:277–297. 10.1007/s002210050033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman MS, Golowasch J, Marder E, Abbott LF (2001) Global structure, robustness, and modulation of neuronal models. J Neurosci 21:5229–5238. 10.1523/JNEUROSCI.21-14-05229.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golowasch J, Goldman MS, Abbott LF, Marder E (2002) Failure of averaging in the construction of a conductance-based neuron model. J Neurophysiol 87:1129–1131. 10.1152/jn.00412.2001 [DOI] [PubMed] [Google Scholar]
- Grieco TM, Malhotra JD, Chen C, Isom LL, Raman IM (2005) Open-channel block by the cytoplasmic tail of sodium channel β4 as a mechanism for resurgent sodium current. Neuron 45:233–244. 10.1016/j.neuron.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Hobert O, Kratsios P (2019) Neuronal identity control by terminal selectors in worms, flies, and chordates. Curr Opin Neurobiol 56:97–105. 10.1016/j.conb.2018.12.006 [DOI] [PubMed] [Google Scholar]
- Hu H, Gan J, Jonas P (2014) Fast-spiking, parvalbumin+ GABAergic interneurons: from cellular design to microcircuit function. Science 345:1255263. 10.1126/science.1255263 [DOI] [PubMed] [Google Scholar]
- Hu H, Roth FC, Vandael D, Jonas P (2018) Complementary tuning of Na+ and K+ channel gating underlies fast and energy-efficient action potentials in GABAergic interneuron axons. Neuron 98:156–165.e6. 10.1016/j.neuron.2018.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom LL, De Jongh KS, Patton DE, Reber BF, Offord J, Charbonneau H, Walsh K, Goldin AL, Catterall WA (1992) Primary structure and functional expression of the beta 1 subunit of the rat brain sodium channel. Science 256:839–842. 10.1126/science.1375395 [DOI] [PubMed] [Google Scholar]
- Kaczmarek LK, Zhang Y (2017) Kv3 channels: enablers of rapid firing, neurotransmitter release, and neuronal endurance. Physiol Rev 97:1431–1468. 10.1152/physrev.00002.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochubey O, Babai N, Schneggenburger R (2016) A synaptotagmin isoform switch during the development of an identified CNS synapse. Neuron 90:984–999. 10.1016/j.neuron.2016.04.038 [DOI] [PubMed] [Google Scholar]
- Kodama T, Guerrero S, Shin M, Moghadam S, Faulstich M, du Lac S (2012) Neuronal classification and marker gene identification via single-cell expression profiling of brainstem vestibular neurons subserving cerebellar learning. J Neurosci 32:7819–7831. 10.1523/JNEUROSCI.0543-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole MH, Letzkus JJ, Stuart GJ (2007) Axon initial segment Kv1 channels control axonal action potential waveform and synaptic efficacy. Neuron 55:633–647. 10.1016/j.neuron.2007.07.031 [DOI] [PubMed] [Google Scholar]
- Kolkman KE, McElvain LE, du Lac S (2011) Diverse precerebellar neurons share similar intrinsic excitability. J Neurosci 31:16665–16674. 10.1523/JNEUROSCI.3314-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Saitou M (2007) Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc 2:739–752. 10.1038/nprot.2007.79 [DOI] [PubMed] [Google Scholar]
- Kwan KY, Sestan N, Anton ES (2012) Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development 139:1535–1546. 10.1242/dev.069963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Cleveland DW (1996) Neuronal intermediate filaments. Annu Rev Neurosci 19:187–217. 10.1146/annurev.ne.19.030196.001155 [DOI] [PubMed] [Google Scholar]
- Liberman MC. (1993) Central projections of auditory nerve fibers of differing spontaneous rate: II. Posteroventral and dorsal cochlear nuclei. J Comp Neurol 327:17–36. 10.1002/cne.903270103 [DOI] [PubMed] [Google Scholar]
- Madisen L, Mao T, Koch H, Zhuo JM, Berenyi A, Fujisawa S, Hsu YW, Garcia AJ 3rd, Gu X, Zanella S, Kidney J, Gu H, Mao Y, Hooks BM, Boyden ES, Buzsáki G, Ramirez JM, Jones AR, Svoboda K, Han X, et al. (2012) A toolbox of cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci 15:793–802. 10.1038/nn.3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElvain LE, Faulstich M, Jeanne JM, Moore JD, du Lac S (2015) Implementation of linear sensory signaling via multiple coordinated mechanisms at central vestibular nerve synapses. Neuron 85:1132–1144. 10.1016/j.neuron.2015.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AB, Krispel CM, Sekirnjak C, du Lac S (2003) Long-lasting increases in intrinsic excitability triggered by inhibition. Neuron 40:609–620. 10.1016/S0896-6273(03)00641-X [DOI] [PubMed] [Google Scholar]
- O'Leary T, Williams AH, Franci A, Marder E (2014) Cell types, network homeostasis, and pathological compensation from a biologically plausible ion channel expression model. Neuron 82:809–821. 10.1016/j.neuron.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva AA Jr, Jiang M, Lam T, Smith KL, Swann JW (2000) Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J Neurosci 20:3354–3368. 10.1523/JNEUROSCI.20-09-03354.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perge JA, Koch K, Miller R, Sterling P, Balasubramanian V (2009) How the optic nerve allocates space, energy capacity, and information. J Neurosci 29:7917–7928. 10.1523/JNEUROSCI.5200-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perge JA, Niven JE, Mugnaini E, Balasubramanian V, Sterling P (2012) Why do axons differ in caliber? J Neurosci 32:626–638. 10.1523/JNEUROSCI.4254-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz AA, Bucher D, Marder E (2004) Similar network activity from disparate circuit parameters. Nat Neurosci 7:1345–1352. 10.1038/nn1352 [DOI] [PubMed] [Google Scholar]
- Rash JE, Vanderpool KG, Yasumura T, Hickman J, Beatty JT, Nagy JI (2016) KV1 channels identified in rodent myelinated axons, linked to Cx29 in innermost myelin: support for electrically active myelin in mammalian saltatory conduction. J Neurophysiol 115:1836–1859. 10.1152/jn.01077.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B, McBain CJ (2001) Kv3 channels: voltage-gated K+ channels designed for high-frequency repetitive firing. Trends Neurosci 24:517–526. 10.1016/S0166-2236(00)01892-0 [DOI] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, Goeva A, Nemesh J, Kamitaki N, Brumbaugh S, Kulp D, McCarroll SA (2018) Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174:1015–1030.e16. 10.1016/j.cell.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz DJ, Goaillard JM, Marder E (2006) Variable channel expression in identified single and electrically coupled neurons in different animals. Nat Neurosci 9:356–362. 10.1038/nn1639 [DOI] [PubMed] [Google Scholar]
- Sekirnjak C, Vissel B, Bollinger J, Faulstich M, du Lac S (2003) Purkinje cell synapses target physiologically unique brainstem neurons. J Neurosci 23:6392–6398. 10.1523/JNEUROSCI.23-15-06392.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta B, Stemmler M, Laughlin SB, Niven JE (2010) Action potential energy efficiency varies among neuron types in vertebrates and invertebrates. PLoS Comput Biol 6:e1000840. 10.1371/journal.pcbi.1000840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M, Moghadam SH, Sekirnjak C, Bagnall MW, Kolkman KE, Jacobs R, Faulstich M, du Lac S (2011) Multiple types of cerebellar target neurons and their circuitry in the vestibulo-ocular reflex. J Neurosci 31:10776–10786. 10.1523/JNEUROSCI.0768-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MR, Nelson AB, du Lac S (2002) Regulation of firing response gain by calcium-dependent mechanisms in vestibular nucleus neurons. J Neurophysiol 87:2031–2042. 10.1152/jn.00821.2001 [DOI] [PubMed] [Google Scholar]
- Sun S, Babola T, Pregernig G, So KS, Nguyen M, Su SM, Palermo AT, Bergles DE, Burns JC, Müller U (2018) Hair cell mechanotransduction regulates spontaneous activity and spiral ganglion subtype specification in the auditory system. Cell 174:1247–1263.e15. 10.1016/j.cell.2018.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swensen AM, Bean BP (2005) Robustness of burst firing in dissociated Purkinje neurons with acute or long-term reductions in sodium conductance. J Neurosci 25:3509–3520. 10.1523/JNEUROSCI.3929-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschenberger H, Leão RM, Rowland KC, Spirou GA, von Gersdorff H (2002) Optimizing synaptic architecture and efficiency for high-frequency transmission. Neuron 36:1127–1143. 10.1016/S0896-6273(02)01137-6 [DOI] [PubMed] [Google Scholar]
- Telgkamp P, Padgett DE, Ledoux VA, Woolley CS, Raman IM (2004) Maintenance of high-frequency transmission at Purkinje to cerebellar nuclear synapses by spillover from boutons with multiple release sites. Neuron 41:113–126. 10.1016/S0896-6273(03)00802-X [DOI] [PubMed] [Google Scholar]
- Ueta Y, Otsuka T, Morishima M, Ushimaru M, Kawaguchi Y (2014) Multiple layer 5 pyramidal cell subtypes relay cortical feedback from secondary to primary motor areas in rats. Cereb Cortex 24:2362–2376. 10.1093/cercor/bht088 [DOI] [PubMed] [Google Scholar]
- Vihervaara A, Duarte FM, Lis JT (2018) Molecular mechanisms driving transcriptional stress responses. Nat Rev Genet 19:385–397. 10.1038/s41576-018-0001-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Kunkel DD, Martin TM, Schwartzkroin PA, Tempel BL (1993) Heteromultimeric K+ channels in terminal and juxtaparanodal regions of neurons. Nature 365:75–79. 10.1038/365075a0 [DOI] [PubMed] [Google Scholar]
- Wang SS, Shultz JR, Burish MJ, Harrison KH, Hof PR, Towns LC, Wagers MW, Wyatt KD (2008) Functional trade-offs in white matter axonal scaling. J Neurosci 28:4047–4056. 10.1523/JNEUROSCI.5559-05.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JJ, Sillitoe RV (2013) Postnatal development of cerebellar zones revealed by neurofilament heavy chain protein expression. Front Neuroanat 7:9. 10.3389/fnana.2013.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer VC, Harty RC, Richards KL, Phillips AM, Miyazaki H, Nukina N, Petrou S (2015) Sodium channel β1 subunit localizes to axon initial segments of excitatory and inhibitory neurons and shows regional heterogeneity in mouse brain. J Comp Neurol 523:814–830. 10.1002/cne.23715 [DOI] [PubMed] [Google Scholar]
- Xu J, Mashimo T, Südhof TC (2007) Synaptotagmin-1, -2, and -9: Ca2+ sensors for fast release that specify distinct presynaptic properties in subsets of neurons. Neuron 54:567–581. 10.1016/j.neuron.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Zeilhofer HU, Studler B, Arabadzisz D, Schweizer C, Ahmadi S, Layh B, Bösl MR, Fritschy JM (2005) Glycinergic neurons expressing enhanced green fluorescent protein in bacterial artificial chromosome transgenic mice. J Comp Neurol 482:123–141. 10.1002/cne.20349 [DOI] [PubMed] [Google Scholar]
- Zhang J, Finney RP, Clifford RJ, Derr LK, Buetow KH (2005) Detecting false expression signals in high-density oligonucleotide arrays by an in silico approach. Genomics 85:297–308. 10.1016/j.ygeno.2004.11.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell classification used for microarray sample preparation in comparison with the one used in Kodama et al., 2012. Note that each node in the dendrogram corresponds to one MVN neuron. For details, see Materials and Methods. Download Figure 5-1, EPS file (248.1KB, eps)
Genes potentially co-regulated for fast-firing capacity across MVN neurons. Listed are 138 probe sets included in the red branch of the dendrogram shown in Figure 6A and corresponding gene symbols. Download Figure 5-2, XLS file (74.5KB, xls)