Abstract
The MEK1/2–ERK1/2 pathway has been implicated in regulating the inflammatory response to lung injury and infection, and pharmacologic MEK1/2 inhibitor compounds are reported to reduce detrimental inflammation in multiple animal models of disease, in part through modulation of leukocyte responses. However, the specific contribution of myeloid MEK1 in regulating acute lung injury (ALI) and its resolution remain unknown. Here, the role of myeloid Mek1 was investigated in a murine model of LPS-induced ALI (LPS-ALI) by genetic deletion using the Cre-floxed system (LysMCre × Mekfl), and human alveolar macrophages from healthy volunteers and patients with acute respiratory distress syndrome (ARDS) were obtained to assess activation of the MEK1/2–ERK1/2 pathway. Myeloid Mek1 deletion results in a failure to resolve LPS-ALI, and alveolar macrophages lacking MEK1 had increased activation of MEK2 and the downstream target ERK1/2 on day 4 of LPS-ALI. The clinical significance of these findings is supported by increased activation of the MEK1/2–ERK1/2 pathway in alveolar macrophages from patients with ARDS compared with alveolar macrophages from healthy volunteers. This study reveals a critical role for myeloid MEK1 in promoting resolution of LPS-ALI and controlling the duration of macrophage proinflammatory responses.
Keywords: Inflammation, Pulmonology
Keywords: Innate immunity, Macrophages, Protein kinases
Myeloid Mek1 deletion in mice results in increased severity of LPS-ALI and failure to resolve the proinflammatory response.
Introduction
Acute respiratory distress syndrome (ARDS) is a severe form of acute lung injury (ALI), with associated significant morbidity and mortality, that affects 10% of patients hospitalized in intensive care units worldwide (1). Clinical treatments for ARDS are limited, mostly relying on improved supportive mechanical ventilation strategies, and mortality remains over 30% (1). These poor clinical outcomes reflect the need for a better understanding of the cellular and molecular pathogenesis of ALI and the basis for delayed resolution of the associated inflammatory response.
Macrophages are key effector cells of the pulmonary immune system, and they contribute to lung injury and its resolution by their unique functional heterogeneity and plasticity (2, 3). Macrophage plasticity allows these cells to adopt different polarized phenotypes that may evolve over time based on cell origin and environmental stimuli (4). In a murine model of experimental Pseudomonas aeruginosa pneumonia, resident alveolar macrophages are the predominant proinflammatory (M1-polarized) cells during the initiation of lung injury. During resolution of lung injury, these cells repolarize, and express markers associated with reparative phenotypes (M2-polarization) (5). In human ARDS, persistence of a proinflammatory M1-like M(IFN-γ) gene expression signature in macrophages obtained by bronchoalveolar lavage (BAL) was associated with worse clinical outcome, whereas presence of an M2-like M(IL-4) signature was associated with better outcome (6). In another study, failure to upregulate select macrophage surface markers, including the M2-polarization marker CD71, on alveolar macrophages was observed in a subset of ARDS patients with worse outcome (7). These results suggest that duration of macrophage proinflammatory function may be important in nonresolving ALI, and highlight the clinical relevance of understanding novel determinants of macrophage polarization.
In an effort to understand signaling pathways regulating macrophage activation, we identified the mitogen-activated protein kinases MEK1 (Map2k1 or Mek1) and MEK2 (Map2k2 or Mek2) as suppressors of macrophage reparative function (8). Targeting both MEK1 and MEK2 (MEK1/2) using commercially available inhibitor compounds enhanced the ability of macrophages to clear apoptotic polymorphonuclear leukocytes (PMNs) and respond to IL-4/IL-13 (8). Importantly, there was a therapeutic benefit of MEK1/2 inhibition in 2 murine lung injury models: sterile LPS–induced ALI (LPS-ALI) and bacterial pneumonia due to Pseudomonas aeruginosa infection (8, 9). In these ALI models, mice treated with a MEK1/2 inhibitor compound between 24 and 72 hours after LPS or 48 and 72 hours after bacterial infection experienced improved activity, faster recovery of body weight, reduced pulmonary neutrophilia, and increased macrophage M2 polarization (8, 9). Additional studies have demonstrated therapeutic potential of MEK1/2 inhibitors in other murine models of inflammation and infection. For example, MEK1/2 inhibitor pretreatment protected mice from LPS-ALI (10), MEK1/2 inhibitor coadministration with LPS protected mice in a lethal endotoxin shock model (11), and a MEK1/2 inhibitor was demonstrated to have a protective effect in the cecal ligation and puncture (CLP) model of sepsis (12). However, these approaches result in broad inhibition of MEK1/2 pathways in many cell types, and do not demonstrate specific roles for myeloid MEK1 in inflammatory conditions.
MEK1 and MEK2 participate in intracellular signaling networks and exert control on the downstream effector molecules, ERK1 and ERK2, via MEK1/2–dependent serine and tyrosine phosphorylation (13). The MEK1/2–ERK1/2 pathway can be stimulated by extracellular stimuli, such as growth factors and cytokines, and signal downstream of Ras and Raf (13). Abnormal regulation of these pathways has been reported across diseases, including cancer, cardiovascular disease, and pulmonary diseases, such as asthma and emphysema. Studies comparing rodent and human lung injury gene expression signatures revealed conserved pathways, including the MEK1/2–ERK1/2 pathway, as potential targets to counteract injury pathways (14, 15). MEK1 and MEK2 share 80% amino acid homology, suggesting that they may be functionally redundant, and in certain cases, deletion of both MEK1 and MEK2 is required for phenotypes to emerge (16). However, Mek2–/– mice are phenotypically normal, whereas Mek1 homozygous deletion is embryonically lethal, suggesting that MAPK cascade signaling is dependent on select isoforms in specific settings (17). Interestingly, several sites on MEK1, such as T292, have been described to impart an inhibitory effect on phosphorylation and function to reduce activation of the MEK1/2–ERK1/2 pathway (18). This regulatory site is absent in MEK2, further suggesting that there may be important and unique functions of MEK1 and MEK2 in different cellular contexts.
To gain a better understanding of the mechanism and cell source by which the MEK1/2 pathway regulates ALI and its resolution and to determine if MEK1 has distinct functions in regulating these responses, we generated mice deficient in MEK1 in myeloid cells using Lyz2Cre+/+ (LysMCre) deletion of Mek1fl/fl (herein called Mek1flLysMCre mice). Mek1flLysMCre mice have no apparent abnormal phenotype in unchallenged, naive conditions, but experience nonresolving LPS-ALI using a moderate LPS dose from which all Mek1fl mice recover. Despite similar neutrophil recruitment at early time points after LPS-ALI, Mek1flLysMCre mice have sustained and elevated inflammation at later time points that is associated with increased activation of the MEK1/2–ERK1/2 pathway in alveolar macrophages. Comparing alveolar macrophages obtained from healthy human subjects and patients with ARDS, we found elevated activation of the MEK1/2–ERK1/2 pathway in ARDS individuals, indicating that aberrant signaling of the macrophage MEK1/2–ERK1/2 pathway may have relevance to the pathogenesis of human ALI and could serve as a therapeutic target for clinical intervention.
Results
Mek1flLysMCre deletion significantly reduces alveolar macrophage MEK1 and decreases macrophage early proinflammatory responses to ex vivo stimulation with E.
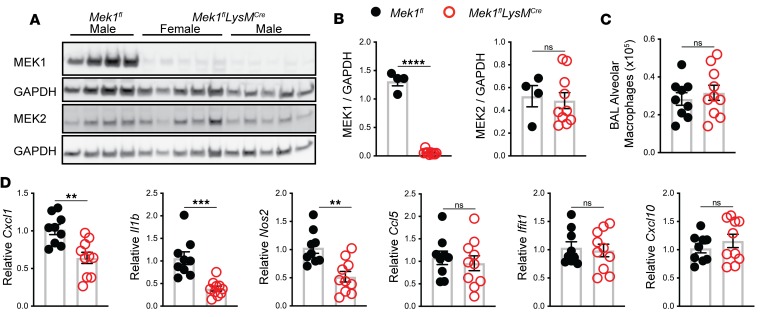
coli LPS. Mek1fl mice were crossed with LysMCre mice to generate Mek1flLysMCre and Mek1fl control mice (8). To confirm loss of MEK1 in alveolar macrophages, we performed BAL on naive mice to recover alveolar macrophages for protein analysis by Western blot. As expected, protein lysates and quantification of band densitometry revealed significant reduction of MEK1 (>95%, P < 0.001) (Figure 1, A and B) in Mek1flLysMCre compared with Mek1fl mice. There was no significant difference in the levels of MEK2 between the groups (Figure 1, A and B), and there was no significant difference in the total number of alveolar macrophages recovered from Mek1fl and Mek1flLysMCre mice (Figure 1C). We predicted that loss of MEK1 would decrease the macrophage proinflammatory response to LPS stimulation, which would be consistent with results demonstrated by use of pharmacological MEK1/2 inhibitor compounds (19). To test this hypothesis, we stimulated alveolar macrophages ex vivo with LPS for 4 hours and measured proinflammatory gene expression. Consistent with our hypothesis, relative expression of Cxcl1, Il1b, and Nos2 was significantly decreased in macrophages with loss of MEK1; however, relative expression of the type I IFN–responsive genes Ifit1 and Cxcl10, or of Ccl5 did not differ at this time point (Figure 1D). To determine if loss of MEK1 altered basal levels of proteins involved in recognition of LPS, we examined surface expression of CD14, TLR4, and the TLR4/MD2 complex on naive alveolar macrophages by flow cytometry (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.132377DS1). No significant differences were detected between Mek1fl and Mek1flLysMCre mice, suggesting that decreased proinflammatory gene expression following LPS stimulation is not due to reduced LPS detection. Further examination of mRNA levels of Cd14, Tlr4, and Ly96 (MD2) in unstimulated and 4-hour LPS-stimulated alveolar macrophages demonstrated LPS-induced upregulation of Cd14 and Ly96 and downregulation of Tlr4 (Supplemental Figure 1C) in both Mek1fl and Mek1flLysMCre cells, although the magnitude of change following LPS stimulation may be blunted due to MEK1 loss. Overall, these results suggest that loss of MEK1 alters the cellular response to LPS, likely through altered intracellular signaling, and not through the altered ability to detect LPS.
Figure 1. Mek1flLysMCre reduces alveolar macrophage MEK1 and decreases proinflammatory gene expression following LPS stimulation.
(A) Protein lysates from alveolar macrophages were recovered from naive female and male mice by bronchoalveolar lavage (BAL) and were used to detect total MEK1, MEK2, and GAPDH levels by Western blot. (B) Quantification by band densitometry revealed significant reduction in MEK1 without changes in MEK2 in Mek1flLysMCre mice compared with Mek1fl mice. Data points show values derived from an individual mouse (n = 4 for Mek1fl and n = 10 for Mek1flLysMCre), and the bars represent the mean ± SEM. (C) Quantitation of alveolar macrophages recovered from BAL of naive Mek1fl mice (n = 4 female, n = 5 male) and Mek1flLysMCre (n = 5 female, n = 5 male) reveal no differences in the total number of cells recovered. (D) Alveolar macrophages collected from the same mice as in C were allowed to attached to tissue culture plates and were stimulated with 50 ng/mL E. coli LPS for 4 hours. RNA was isolated and cDNA used as the template in qPCR reactions to measure proinflammatory gene expression. Relative Cxcl1, Il1b, Nos2, Ccl5, Ifit1, and Cxcl10 were compared to Hprt, and data are normalized to Mek1fl samples. Data points show values derived from an individual mouse and the bars represent the mean ± SEM. In A–D, statistical analysis used an unpaired t test. **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant.
Mek1flLysMCre deletion impairs resolution of LPS-ALI.
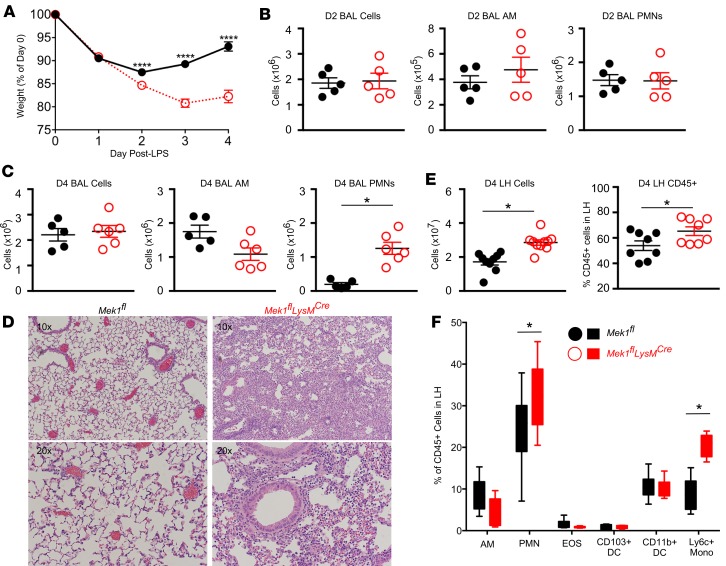
To evaluate the contribution of myeloid MEK1 in ALI and its resolution, we utilized a sterile LPS-ALI model in Mek1flLysMCre and Mek1fl mice. Using weight change as a surrogate marker for injury, both genotypes had a similar initial weight loss on day 1 following LPS-ALI (Figure 2A). However, weight loss was significantly increased in Mek1flLysMCre versus Mek1fl mice on day 2. Furthermore, recovery of weight, which was seen in Mek1fl mice from days 2 to 4, was markedly impaired in Mek1flLysMCre mice (Figure 2A). Quantification of BAL cells on days 1 (data not shown) and 2 revealed no significant differences in the total numbers of BAL cells, BAL alveolar macrophages, or BAL PMNs (Figure 2B). However, by day 4 of LPS-ALI, there was a marked increase in BAL PMNs in Mek1flLysMCre mice and no difference in alveolar macrophage numbers (Figure 2C). Histologic assessment of H&E–stained lung tissue on day 4 revealed a marked increase in alveolar and interstitial inflammation in Mek1flLysMCre mice, with minimal residual inflammation in Mek1fl mice (Figure 2D). Because day 4 BAL cell counts appeared to underestimate the amount of lung inflammation, we also quantified the cellularity and leukocyte composition of lung digests using flow cytometry (Figure 2, E and F; gating strategy represented in Supplemental Figure 2). Comparing day 4 of LPS-ALI Mek1fl versus Mek1flLysMCre mice, we found increased total lung homogenate cell counts, with a greater proportion of leukocytes (CD45+ cells) in Mek1flLysMCre mice (Figure 2E), and a majority of the increased inflammatory cells represented Ly6chi monocytes and PMNs (Figure 2F).
Figure 2. Mek1flLysMCre impairs resolution of LPS-ALI.
Mek1fl and Mek1flLysMCre mice (male and female) were subjected to LPS-induced acute lung injury (LPS-ALI) by oropharyngeal aspiration and (A) weight was monitored over 4 days. Data are the mean ± SEM from n = 35 Mek1fl and n = 34 Mek1flLysMCre mice (day 1), n = 30 Mek1fl and n = 29 Mek1flLysMCre mice (day 2), n = 25 Mek1fl and n = 24 Mek1flLysMCre mice (day 3), and n = 20 Mek1fl and n = 20 Mek1flLysMCre mice (day 4). Statistical analysis used an unpaired t test with correction for multiple comparisons using the Holm-Sidak method to compare values between each genotype on each day. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant. (B and C) Total bronchoalveolar lavage (BAL) cells were quantified and differential analyses on Diff-quick–stained cytospins distinguished between mononuclear (AM) and polymorphonuclear cells (PMNs) on day 2 (D2) (B) and day 4 (D4) (C). Representative H&E staining on a paraffin-embedded lung section from day 4 LPS-ALI (D) and (E and F) total lung homogenate (LH) digest cell counts and FACS analysis of the LH inflammatory cell populations (for gating strategy see Supplemental Figure 2) on day 4 of LPS-ALI confirm increased inflammation in Mek1flLysMCre mice. Data are the mean ± SEM of n = 8–14 for each genotype. In B, C, E, and F, an unpaired t test was used for statistical analysis to compare genotypes. *P < 0.05.
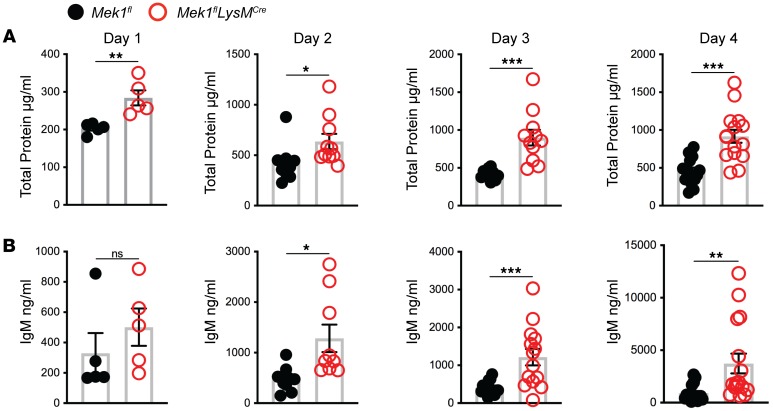
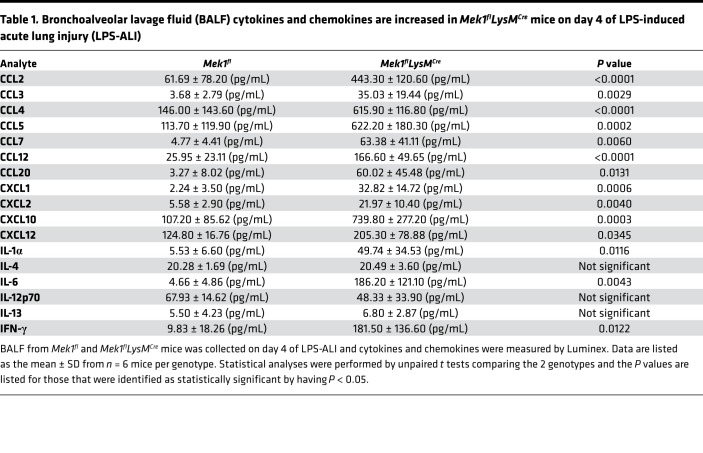
Since vascular leak is another important indicator of overall lung injury, we assessed markers of endothelial injury by measuring BAL fluid (BALF) total protein and IgM from days 1–4 after LPS. In addition to the differences in weight, Mek1flLysMCre mice displayed increased markers of inflammation and injury in the BALF compared with Mek1fl mice (Figure 3, A and B). There was no difference in either marker in naive mice (data not shown). However, on day 1 for total protein and day 2 for IgM, there were increased markers of vascular leak in Mek1flLysMCre compared with Mek1fl (Figure 3). Whereas IgM peaked on day 2 in Mek1fl mice and remained elevated, levels in Mek1flLysMCre mice continued to increase, consistent with nonresolving and more severe lung injury. Given the increase in injury and change in leukocyte influx, we assessed BALF for differences in chemokines and cytokines on day 4 following LPS-ALI (Table 1), which may be driving increased leukocyte recruitment. These studies revealed significantly increased proinflammatory mediators in Mek1flLysMCre mice compared with Mek1fl mice, including neutrophil chemokines, CXCL1 and CXCL2 and monocyte/macrophage chemokines, CCL2, CCL3, CCL4, and CCL5, among others. Together, these results demonstrate that Mek1flLysMCre mice have impaired resolution of LPS-ALI that corresponds to a significantly increased and sustained inflammatory response.
Figure 3. Vascular injury is increased in Mek1flLysMCre mice following LPS-ALI.
Cell-free bronchoalveolar lavage fluid (BALF) was used to measure (A) total protein and (B) IgM from Mek1fl and Mek1flLysMCre mice on days 1–4 following LPS-induced acute lung injury (LPS-ALI). Data show values from an individual mouse and the bars are the mean ± SEM. On day 1 total protein and IgM have n = 5 for both Mek1fl and Mek1flLysMCre mice. Day 2 total protein is n = 10 both groups and n = 9 for both groups for IgM. Day 3 total protein is n = 10 for Mek1fl and n = 11 for Mek1flLysMCre and IgM is n = 15 for Mek1fl and n = 14 for Mek1flLysMCre. Day 4 total protein is n = 16 for Mek1fl and n = 15 for Mek1flLysMCre and IgM is n = 16 for Mek1fl and n = 16 for Mek1flLysMCre. Statistical analyses used an unpaired t test to compare values between each genotype on each day. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant.
Table 1. Bronchoalveolar lavage fluid (BALF) cytokines and chemokines are increased in Mek1flLysMCre mice on day 4 of LPS-induced acute lung injury (LPS-ALI).
Mek1flLysMCre deletion alters macrophage proinflammatory responses following LPS-ALI.
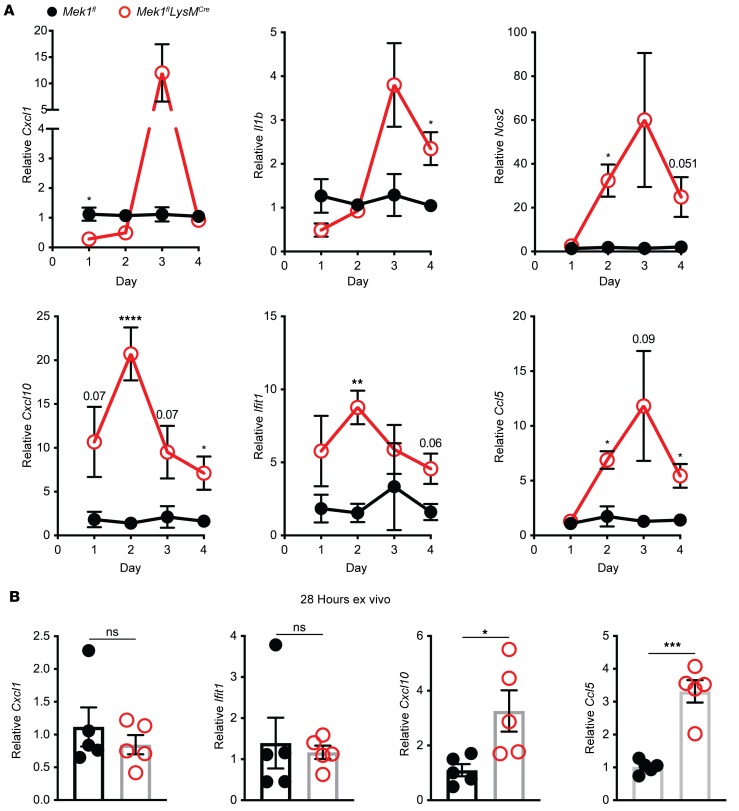
To determine the contribution of Mek1flLysMCre pulmonary macrophages to the increased proinflammatory cytokine and chemokine levels observed in the BAL from LPS-treated Mek1flLysMCre mice, we isolated the BAL macrophages from Mek1flLysMCre and Mek1fl mice on days 1–4 of LPS-ALI. On day 1 of LPS-ALI, Mek1flLysMCre mice had significantly decreased Cxcl1 compared with Mek1fl mice (Figure 4A), consistent with our ex vivo observation in Figure 1C. However, by day 3, Cxcl1 expression was elevated in Mek1flLysMCre mice compared with Mek1fl mice. Similarly, Il1b, which trended lower on day 1 in Mek1flLysMCre mice, was markedly higher in Mek1flLysMCre mice on days 3 and 4. Other genes, including the IFN-responsive genes Cxcl10 and Ifit1, or Nos2 and Ccl5, had significantly increased expression in Mek1flLysMCre mice on day 2 with sustained increases on day 4 (Figure 4A). These results indicate that loss of MEK1 results in abnormal macrophage gene expression that corresponds to the impaired resolution in Mek1flLysMCre mice following LPS-ALI. To determine if there is an inherent prolonged inflammatory profile in alveolar macrophages lacking MEK1, we isolated alveolar macrophages from naive mice and stimulated these cells ex vivo with E. coli LPS to assess proinflammatory gene expression at 28 hours. In contrast with the suppressed inflammatory responses seen at 4 hours (Figure 1D), at 28 hours we found increased expression of Cxcl10 and Ccl5 (Figure 4B and Supplemental Figure 3C), indicating that loss of MEK1 in alveolar macrophages results in prolonged proinflammatory gene expression following LPS stimulation, recapitulating the finding observed in the LPS-ALI in vivo model.
Figure 4. Aberrant alveolar macrophage proinflammatory gene expression in Mek1flLysMCre mice during LPS-ALI.
Mek1fl and Mek1flLysMCre mice were subjected to LPS-induced acute lung injury (LPS-ALI) and (A) RNA was collected from macrophages isolated by adherence from bronchoalveolar lavage (BAL) on days 1–4. Expression of Cxcl1, Il1b, Nos2, Cxcl10, Ifit1, and Ccl5 relative to Hprt was normalized to Mek1fl samples. Data are the mean ± SEM of n = 3–9 per genotype on each day. Statistical analysis used an unpaired t test with correction for multiple comparisons using the Holm-Sidak method. (B) Alveolar macrophages isolated from naive mice by BAL were stimulated with 50 ng/mL E. coli LPS for 28 hours. Expression of Cxcl1, Ifit1, Cxcl10, and Ccl5 relative to Hprt was normalized to Mek1fl samples. Data are the mean ± SEM of n = 5 for each genotype. Statistical analysis used an unpaired t test to compare genotypes. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant.
Mek1flLysMCre deletion increases MEK1/2–ERK1/2 pathway activation in alveolar macrophages.
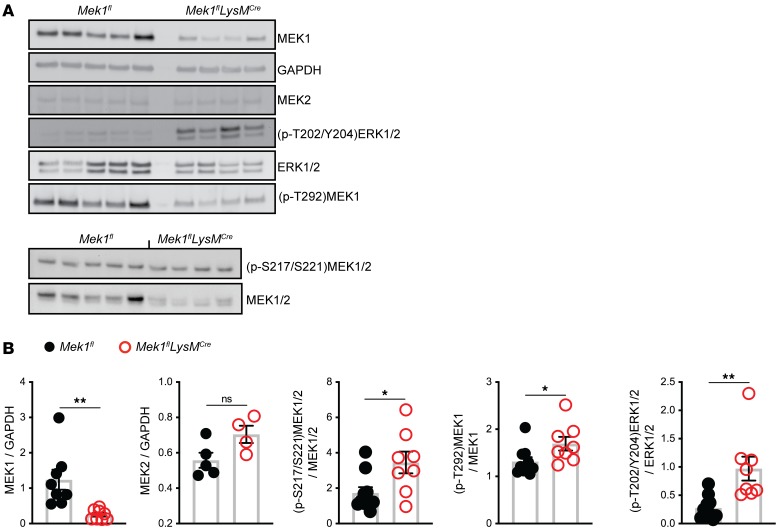
Our initial data demonstrating that Mek1 deletion reduces early proinflammatory responses to LPS stimulation (Figure 1C) are consistent with those observed with use of a MEK1/2 inhibitor, as described previously (19). However, the sustained inflammatory response of Mek1-deleted alveolar macrophages was unexpected and not observed with MEK1/2 inhibition. Although MEK1 and MEK2 are 80% homologous proteins that form heterodimers, MEK1 but not MEK2 contains a unique phosphorylation site (T292) that can be phosphorylated by ERK1/2 in a negative feedback loop to inactivate the MEK1/2 complex (20, 21). Therefore, we hypothesized that alveolar macrophages in Mek1flLysMCre mice would display increased MEK1/2–ERK1/2 pathway activity compared with Mek1fl mice following LPS-ALI due to loss of the MEK1-mediated negative feedback loop. To test this hypothesis, protein lysates from total BAL cells recovered on day 4 of LPS-ALI were obtained and used in Western blots to measure MEK1/2–ERK1/2 pathway activity with quantification by band densitometry (Figure 5, A and B). Blots and quantification demonstrated significantly decreased MEK1 levels without significant changes in MEK2 in Mek1flLysMCre mice compared with Mek1fl mice. In addition, quantification of the levels of phosphorylated (p-T202/Y204)ERK1/2 and total ERK1/2 and phosphorylated (p-S217/S221)MEK1/2 and total MEK1/2 revealed significantly increased ratios of phosphorylated protein for both ERK1/2 and MEK1/2 in Mek1flLysMCre mice compared with Mek1fl mice, confirming increased activation of the MEK1/2–ERK1/2 pathway in the absence of myeloid MEK1 (Figure 5B). Based on the significantly reduced levels of MEK1, these findings most likely represent increased activation of MEK2. We also examined the levels of phosphorylation of the inhibitor site of MEK1, (p-T292)MEK1, and the ratio of (p-T292)MEK1 to MEK1. In Mek1flLysMCre mice, the overall level of (p-T292)MEK1 is reduced, corresponding to the reduced total MEK1 levels. In the context of reduced MEK1 levels, the ratio of (p-T292)MEK1 to total MEK1 was slightly increased in Mek1flLysMCre (Figure 5, A and B). Overall, these data demonstrate that Mek1flLysMCre mice have reduced MEK1 levels but an increased ability to activate the downstream target ERK1/2.
Figure 5. Activation of the MEK1/2–ERK1/2 pathway is increased in total BAL cells in Mek1flLysMCre mice following LPS-ALI.
On day 4 following LPS-induced acute lung injury (LPS-ALI), protein lysates from total bronchoalveolar lavage (BAL) cells were collected from Mek1fl and Mek1flLysMCre mice and used in Western blots to detect levels of MEK1, MEK2, GAPDH, (p-T202/Y204)ERK1/2, ERK1/2, (p-T292)MEK1, (p-S217/221)MEK1/2, and MEK1/2. (A) Representative blots of n = 5 Mek1fl and n = 4 Mek1flLysMCre samples. (B) Densitometry quantitation show samples from an individual mouse and the bars represent the mean ± SEM. Statistical analyses were performed with unpaired t tests. *P < 0.05, **P < 0.01; ns, not significant.
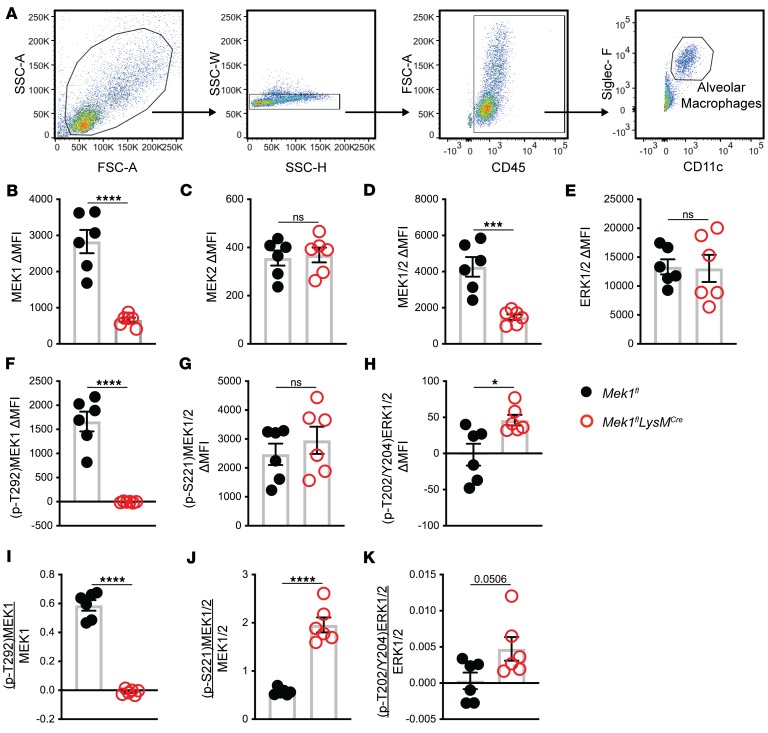
While the Western blot data support the hypothesis that loss of alveolar macrophage MEK1 results in increased MEK1/2–ERK1/2 pathway activity, a limitation of BAL cell lysates is the inclusion of other cells in the alveolar compartment in addition to alveolar macrophages that are collected during lavage. Since our data indicate that MEK1 regulates inflammatory programming of macrophages, we utilized flow cytometry to analyze activation of the MEK1/2–ERK1/2 pathway specifically in alveolar macrophages (CD45+CD11c+SigF+ cells) on day 4 of LPS-ALI (Figure 6A). As expected, alveolar macrophages had significantly reduced MEK1 (Figure 6B) without changes in MEK2 (Figure 6C). Total MEK1/2 was significantly reduced as a result of reduced MEK1 (Figure 6D), but total ERK1/2 was not significantly altered (Figure 6E). While (p-T292)MEK1 was detected in Mek1fl alveolar macrophages, it was undetectable in Mek1flLysMCre alveolar macrophages, consistent with the deletion of MEK1 in these cells (Figure 6F), and demonstrates a lack of this inhibitory pathway in alveolar macrophages (Figure 6I). The level of (p-S221)MEK1/2 was not significantly different (Figure 6G); however, when normalized to total MEK1/2, the ratio of activated MEK1/2 was increased in Mek1flLysMCre alveolar macrophages (Figure 6J), which supports the hypothesis of increased MEK2 activation. Consistent with increased MEK2 activation, activation of ERK1/2 as determined by (p-T202/Y204)ERK1/2 was increased in the absence of MEK1 (Figure 6, H and K). In addition to examining alveolar macrophages, we also performed the same analyses on Ly6G+ PMNs obtained from the BAL (Supplemental Figure 4). As expected, LysMCre mediated Mek1 deletion significantly reduced PMN MEK1 (Supplemental Figure 4B) without affecting total MEK2 (Supplemental Figure 4C) or ERK1/2 (Supplemental Figure 4E) levels. While the overall levels of all proteins examined (Supplemental Figure 4, B–H) were lower in PMNs than in alveolar macrophages, decreased (p-T292)MEK1 (Supplemental Figure 4I) and increased activation of MEK2 and ERK1/2 (Supplemental Figure 4, J and K) were observed in PMNs from Mek1flLysMCre mice compared with Mek1fl mice. Further, we were able to analyze Ly6G–Ly6C+CD11b+ monocytes that had infiltrated into the BAL (Supplemental Figure 5, A–K), and determined that MEK1 levels were also significantly reduced in these cells without changes in MEK2. However, we did not detect a positive signal for ERK1/2 phosphorylation (Supplemental Figure 5, H and K), while there was an indication that MEK2 activation was increased (Supplemental Figure 5J), similar to that seen in alveolar macrophages and PMNs. Overall, these changes in the MEK1/2–ERK1/2 pathway within the alveolar macrophages, PMNs, and monocytes match those seen by in total BAL cells observed by Western blot (Figure 5), and indicate a defect in a negative feedback loop involving MEK1, resulting in increased MEK1/2–ERK1/2 pathway activation.
Figure 6. Alveolar macrophage MEK1/2–ERK1/2 pathway activation is increased in Mek1flLysMCre mice following LPS-ALI.
Mek1fl and Mek1flLysMCre were subjected to LPS-induced acute lung injury (LPS-ALI) and alveolar cells were collected by bronchoalveolar lavage (BAL) on day 4. BAL cells were and fixed and permeabilized and stained for analysis by flow cytometry. (A) Representative gating strategy to identify total and single cells, CD45-positive cells, and alveolar macrophages as Siglec-F and CD11c double-positive cells. (B–H) The change in mean fluorescence intensity (ΔMFI) of alveolar macrophages for intracellular total proteins (B) MEK1, (C) MEK2, (D) MEK1/2, (E) ERK1/2, (F) (p-T292)MEK1, (G) (p-S221)MEK1/2, and (H) (p-T202/Y204)ERK1/2 was calculated by subtracting background signal using isotype-stained control samples. (I–K) The ratio of the ΔMFI of phosphorylated to total protein was calculated for (I) (p-T292)MEK1 to MEK1, (J) (p-S221)MEK1/2 to MEK1/2, and (K) (p-T202/Y204)ERK1/2 to ERK1/2. Dots represent individual mice and the bars are the mean ± SEM. Statistical analyses were performed by unpaired t tests comparing the 2 genotypes. *P < 0.05, ****P < 0.0001; ns, not significant.
To test if these findings can be recapitulated ex vivo, we analyzed the activation of ERK1/2 using cultured Mek1fl and Mek1flLysMCre alveolar macrophages. At 4 hours following LPS stimulation, there was a robust increase in (p-T202/Y204)ERK1/2 in Mek1fl cells that was blunted in LPS-treated Mek1flLysMCre cells (Supplemental Figure 3, A and B). At 28 hours, (p-T202/Y204)ERK1/2 levels approached those seen in unstimulated cells in Mek1fl. While the (p-T202/Y204)ERK1/2 levels between the two genotypes were similar at 28 hours, the activation of ERK1/2 tended to be slightly increased at 28 hours relative to 4 hours in Mek1flLysMCre mice. In addition, while LPS increased chemokine expression at 4 hours and 28 hours (Supplemental Figure 3C), there was greater expression of Ccl5 in Mek1flLysMCre compared with Mek1fl mice at 28 hours. Overall, these results indicate that loss of MEK1 alters alveolar macrophage responses to LPS stimulation; however, the sustained activation of the ERK1/2 pathway observed in vivo could not be recapitulated ex vivo.
MEK1/2–ERK1/2 pathway activation in alveolar macrophages from healthy human subjects and ARDS patients.
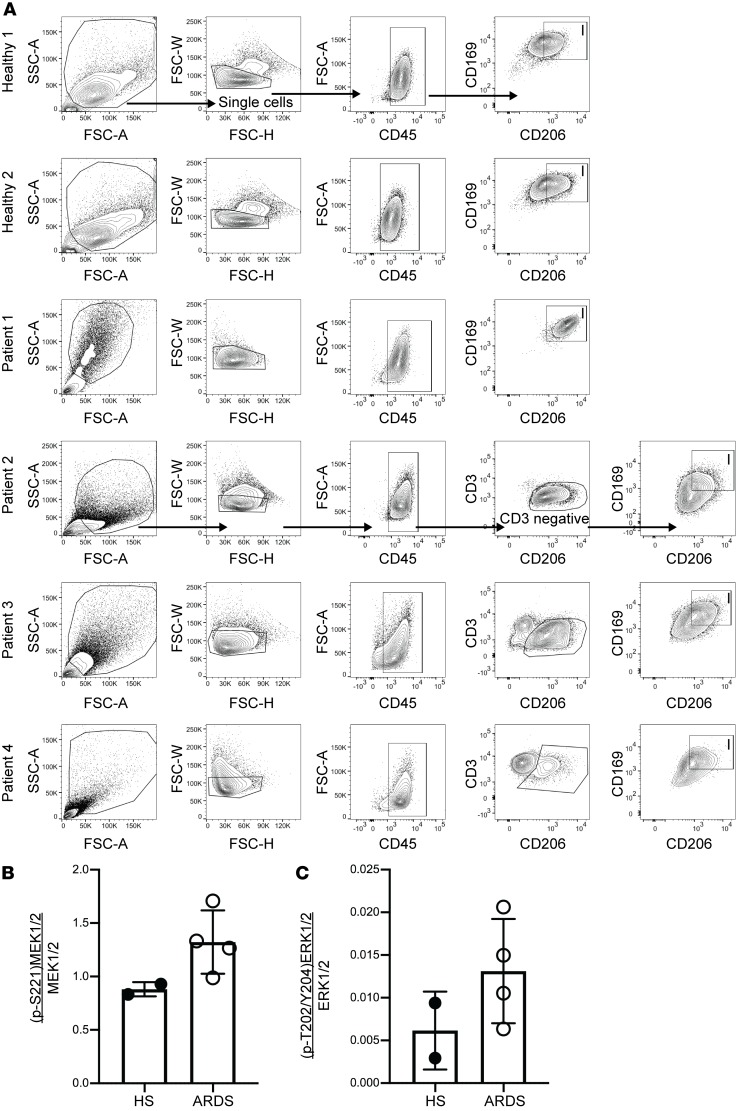
The data from this murine model support a key role of the macrophage MEK1/2–ERK1/2 pathway in regulating the severity and resolution of inflammation following ALI, and is consistent with our previous findings and results from other groups demonstrating that application of pharmacological inhibitors of the MEK1/2–ERK1/2 pathway can reduce detrimental proinflammatory responses in murine models of lung injury (8, 10), infection (9, 22, 23), asthma (24), sepsis (11, 12), and malaria (25). Based on these findings and results from our current studies, we hypothesized that subjects with ARDS would have increased activation of the MEK1/2–ERK1/2 pathway in alveolar macrophages compared with healthy subjects without ARDS. Lavage cells were obtained from spontaneously breathing healthy subjects or patients with ARDS who were supported on mechanical ventilation. We performed flow cytometry to assess the activation of the MEK1/2–ERK1/2 pathway in CD45+CD206+CD169+ alveolar macrophages (26) from these samples (Figure 7A). MEK1, MEK2, and ERK1/2 proteins were detected in alveolar macrophages from both healthy subjects and ARDS patients (data not shown), and there was a numeric increase in MEK1/2 and ERK1/2 activation in ARDS patients compared with healthy individuals (Figure 7, B and C).
Figure 7. MEK1/2–ERK1/2 pathway activation in alveolar macrophages from healthy human subjects and ARDS patients.
Human bronchoalveolar lavage (BAL) cells from 2 healthy subjects (HS) and 4 acute respiratory distress syndrome (ARDS) patients were fixed and stained to detect activation of the MEK1/2–ERK1/2 pathway. Gating strategy for each individual sample is shown (A) to identify CD45+ myeloid cells, and CD206+CD169+ alveolar macrophages designated as box I. In patients 2, 3, and 4, CD3+ cells were excluded in the gating strategy, whereas they were not present in healthy subjects 1 and 2 and patient 1 (data not shown). The (B) ratio of phosphorylated (p-S221)MEK1/2 to total MEK1/2 and the (C) ratio of phosphorylated (p-T202/Y204)ERK1/2 to total ERK1/2. Data shown are individual samples and the bars are the mean ± SEM.
Discussion
Understanding regulators of macrophage activation is important to advance development of new tools to regulate ALI and its resolution, which lack targeted pharmacologic interventions that result in improved clinical outcomes. In previous studies, we established the therapeutic potential of a MEK1/2 inhibitor compound to promote resolution of ALI in mice and a role of the MEK1/2–ERK1/2 pathway in control of reparative macrophage phenotypes (8, 9). Additional reports have demonstrated beneficial responses to therapeutic use of MEK1/2 inhibitor compounds in other murine models of lung injury (10), lung infection (22, 27), lung fibrosis (28, 29), asthma (24), sepsis (11, 12), atherosclerosis (30, 31), and cerebral malaria (25). Therefore, while there is a large interest in use of MEK1/2 inhibitors as an antiinflammatory therapy, the molecular mechanisms by which MEK1 regulates immune cell functions in disease states are not well described.
In the current study we expand our understanding of how the MEK1/2 pathway regulates macrophage responses during ALI and establish a key role for myeloid MEK1 in modulating the severity and recovery from ALI. In the absence of myeloid MEK1, mice experience similar early neutrophil recruitment, but at later time points, have prolonged and increased inflammation and vascular permeability, with a marked increase in monocytes and neutrophils in the lung. In an attempt to localize the source of increased proinflammatory mediators, we first examined BAL macrophages, which includes both resident and recruited cells. On day 1 of LPS-ALI, inflammatory gene expression was decreased or similar at early time points in MEK1-deficient cells compared with LPS-treated control mice. However, for most cytokines examined, there was greater expression in MEK1-deficient cells on days 2–4 of LPS-ALI. Although this finding could be the consequence of the altered inflammatory composition within LPS-injured Mek1flLysMCre mice, we performed an ex vivo LPS stimulation for 4 hours and 28 hours and observed similar proinflammatory gene expression patterns compared to those observed in vivo. Although ex vivo stimulation of alveolar macrophages from Mek1flLysMCre did not reproduce day 4 LPS-ALI observations of a substantial increase in ERK1/2 activation at 28 hours, a combination of different timing of the observation in addition to the in vivo setting being a more complex stimulatory environment than ex vivo LPS stimulation are likely factors playing a role in these different observations. Overall, these findings, coupled with greater responsiveness to IL-4/IL-13 with MEK1/2 inhibitor–treated macrophages and bone marrow–derived macrophages (BMDMs) from Mek1flLysMCre mice (8), indicate that MEK1 may serve complementary roles in macrophage biology — driving a proinflammatory response but also turning off that response and allowing cells to become more responsive to antiinflammatory cues. Hence, MEK1 may serve as a critical link in balancing macrophage proinflammatory and reparative responses over time.
One potential mechanism by which MEK1 controls the duration of proinflammatory responses is through a negative feedback loop that has been described to require phosphorylation of MEK1 T292 by ERK1/2 (32). In Mek1–/– fibroblasts, disabling this negative feedback loop stabilizes the phosphorylation of MEK2 and alters chemotactic ability of these cells (32). In further support of an inhibitory role of MEK1, mice with epiblast-restricted deletion of MEK1 have higher ERK1/2 activation levels in embryos, epidermis, and brain (33). Despite the homology between MEK1 and MEK2, the T292 inhibitory domain is not present in MEK2; therefore, MEK1 is required for additional regulation of MEK2 activity by this feedback circuit (20, 32). In this report, we found that loss of MEK1 resulted in increased activation of MEK2 and ERK1/2 in alveolar macrophages on day 4 of LPS-ALI. Together, these findings indicate an important regulatory role of MEK1 in modulating the duration of MEK1/2–ERK1/2 pathway activation and controlling macrophage inflammatory programming. Future studies aimed at dissecting the downstream transcriptional response in macrophages controlled by MEK1/2–ERK1/2 would yield a better understanding of how these pathways are involved in regulating the inflammatory response. As previously reviewed (34, 35), ERK1 and -2 have been described to regulate a large number of nuclear proteins and transcription factors involved in both chromatin remodeling and the regulation of the expression of specific genes (36–38). Interestingly, MEK1 was recently described to have important roles in T cell transcriptional responses through regulation of the nuclear receptor corepressor silencing mediator of retinoid and thyroid hormone (SMRT) at the promoter of c-Fos (39). In addition, there have been reports describing the interactions between PPAR-γ and the activation of MEK1/2–ERK1/2 (40), and MEK1 itself can play a role in regulating nuclear localization and activation of PPAR-γ (41–43). PPARs have important roles in regulating macrophage responses to inflammation (44, 45), and represents one of likely many potential mechanisms by which MEK1/2–ERK1/2 regulates macrophage responses.
Other studies investigating the function of Spred-2, a negative regulator of the Raf/MEK/ERK signaling pathway (46), highlight the importance that regulation of MEK1/2–ERK1/2 signaling has in controlling the inflammatory response. For example, Spred2–/– mice have increased phosphorylated ERK1/2, increased cytokine and chemokine production, and increased leukocyte infiltration following LPS-ALI, and Spred2–/– alveolar macrophages and BMDMs have increased proinflammatory cytokine and chemokine secretion following stimulation with 100 ng/mL LPS (19). While these studies demonstrate an importance of Spred-2 regulation of MEK1/2–ERK1/2 in macrophages (19), in influenza infection in mice, regulation of this pathway in the epithelium is critical for viral clearance and the ensuing inflammatory response (47). However, the importance of Spred-2 regulation of the MEK1/2–ERK1/2 pathway in epithelial cells is likely context specific, as it has been well described that inhibition of the MEK1/2–ERK1/2 pathway will disrupt viral replication (27, 48, 49). Our results further support a key role of the MEK1/2–ERK1/2 pathway in regulating the pulmonary inflammatory response.
One limitation of our in vivo studies is the use of Cre recombinase under the control of the LysM promoter, as LysM is also expressed in a minority of pulmonary epithelial cells (50). Hence, the contribution of Mek1 deletion in nonmyeloid cell types cannot be fully excluded, and the contribution of alveolar macrophages to the phenotypes reported here will need to be tested utilizing additional Cre recombinase breeding strategies. For example, we report that PMNs also have increased activation of the MEK1/2–ERK1/2 pathway due to Mek1 deletion, which could contribute to the increased proinflammatory responses and increased vascular injury observed in the Mek1flLysMCre mice. In addition, monocytes also have decreased MEK1 in Mek1flLysMCre mice, which also may contribute to impaired resolution of LPS-ALI. Future development of additional more specific myeloid cell deletion of Mek1, such as PMN-specific Mek1 deletion models, would help to determine the specific role of PMN MEK1 in regulating pulmonary injury. However, the ex vivo studies using alveolar macrophages do support a role for MEK1 in regulating macrophage polarization, independent of other cell types, and flow cytometry analyses also highlight sustained MEK1/2–ERK1/2 activation in Mek1-deleted alveolar macrophage from LPS-ALI mice. The studies with human cells, while limited by low sample numbers and heterogeneity among ARDS patients, revealed increased MEK1/2 activation in human ARDS alveolar macrophages, suggesting the murine model detailed here may be relevant in human disease.
In summary, this report details a role for MEK1 in regulating both the degree and duration of macrophage proinflammatory responses in vitro and in vivo, and the critical role of MEK1 to deactivate proinflammatory signals necessary for resolution of lung inflammation in LPS-ALI. Loss of a MEK1 negative feedback circuit leads to sustained MEK2–ERK1/2 activation. Hence, these data further support the relevance of the MEK1/2–ERK1/2 pathway for regulating the inflammatory response, and suggest that selective inhibition of this pathway in macrophages may be a therapeutic strategy for clinical intervention in human ALI/ARDS.
Methods
Antibodies and reagents.
All antibodies used in these studies are listed in Supplemental Tables 1 and 2. Mouse IgM ELISA Quantitation Set was purchased from Bethyl Laboratories. Total protein was quantified by BCA Assay (Pierce). E. coli LPS O111:B4 was purchased from Sigma-Aldrich.
Mice.
Animals were used in an SPF facility at the University of Washington utilizing a 14-hour light and 10-hour dark cycle. Mek1fl/fl mice were crossed with Lyz2Cre+/– (LysMCre) mice purchased from Jackson Laboratories, as previously reported (8) to generate Mek1fl/flLysMCre+/+ (myeloid MEK1 deletion, herein called Mek1flLysMCre) and Mek1fl/flLysMCre–/– (wild-type MEK1, herein called Mek1fl) mice for these studies. Male and female mice between the ages of 6 and 12 weeks were used in these studies.
Alveolar macrophage isolation, culture, and stimulation.
Naive mice were sacrificed by i.p. injection of Beuthanasia-D (Merck) and alveolar macrophages were collected from BAL and allowed to adhere to 24-well tissue culture plates for 20 minutes before nonadherent cells were removed by PBS rinses and RNA was collected. In some experiments, alveolar macrophages from multiple mice were collected, pooled, counted, and equal numbers of cells were seeded into 96-well plates with triplicate wells for each sample, or were seeded into 48-well plates for later collection of protein. Alveolar macrophages were allowed to adhere to plates for 1 hour and rinsed once with warm PBS prior to stimulation. Cells were stimulated for 4 hours or 28 hours with 50 ng/mL E. coli LPS in HEPES-buffered RPMI-1640 containing L-glutamine, 10% heat-inactivated FBS, and 1% penicillin/streptomycin.
LPS-induced experimental ALI and tissue processing.
Mice were anesthetized using isoflurane and PBS containing 1.5 mg/kg E. coli LPS was instilled into the lung by oropharyngeal aspiration. Mice were monitored for recovery from injury and anesthesia, and weighed daily. BAL was performed on mice that were euthanized by i.p. delivery of Beuthanasia-D. The trachea was surgically exposed and cannulated with a 20-gauge catheter (BD) and 3 serial lavages using 0.8 mL, 0.9 mL, and 0.9 mL of PBS without calcium or magnesium containing 2 mM EDTA were performed. BALF was centrifuged at 325 g for 10 minutes at 4°C to pellet BAL cells. BAL cells were resuspended in ice-cold PBS for further processing. BALF was collected and stored in aliquots at –80°C until further processing. The lungs were perfused with PBS through the right ventricle after severing the hepatic vein and cleared lungs were removed and stored on ice. Lung tissue was manually dissociated with a scissors and enzymatically digested using Liberase (1 mg/mL; Roche) and DNase I (1 mg/mL; Sigma-Aldrich) for 20 minutes at 37°C. Digests were passed through a 70-μm filter and washed. RBC Lysis Buffer (eBioscience) was used to lyse RBCs at room temperature and cell counts were performed on the cell suspensions from the lung digests. In some experiments, the lavaged lung tissues were inflated with 10% formalin, removed, and fixed in 10% formalin prior to paraffin embedding. Sections were stained with H&E. Cell counts were obtained on a Cellometer Auto 2000 using the ViaStain AOPI staining solution (Nexcelom Bioscience), and differential cell counts from the BAL were obtained from cytospin preparations subjected to Diff-Quick staining (Siemens).
Quantitative real-time PCR.
RNA isolations were performed using the NucleoSpin RNA isolation kit (Clontech Laboratories). RNA concentrations were determined using a NanoDrop 1000 (Thermo Fisher Scientific) and cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). cDNA was used as the template in quantitative real-time PCR (qPCR) reactions using the SensiMix II Probe Hi-ROX Kit (Bioline). The primer probes were purchased from Life Technologies and are listed in Supplemental Table 3.
Protein isolation and Western blots.
Protein lysates were collected in RIPA lysis buffer containing Halt Protease and Phosphatase Inhibitor (Invitrogen). Lysates were centrifuged for 5 minutes at 14,000 g to clear lysates and the supernatant was collected. Total protein was quantified by BCA assay following manufacturers protocols and 5–10 μg of total protein was used for each sample. Samples were prepared with Bolt 10× Reducing Agent and 4× LDS Sample Buffer and heated at 70°C for 10 minutes. Bolt Bis-Tris gradient gels (4%–12%) were used for electrophoresis and proteins were transferred onto 0.2-μm PVDF at 20 V for 75 minutes using Bolt transfer buffer containing 10% methanol. Wash buffer was TBS containing 0.05% Tween 20, and 5% BSA was added for blocking and incubation steps in primary and secondary antibodies. Bands were detected by chemiluminescence using Supersignal West Femto (Pierce) on an Omega Ultra-Lum imaging system. Images were cropped using Adobe Photoshop and quantitative densitometry was performed using ImageJ software (NIH). See complete unedited blots in the supplemental material.
Flow cytometry.
Staining of cells for flow cytometry was performed in 96-well V-bottom plates using between 1 × 105 and 1 × 106 cells per well. For phospho-flow staining, BAL cells were pelleted and resuspended in ice-cold PBS and immediately an equal volume of prewarmed BioLegend fixation buffer (catalog 420801) was added and samples were incubated at 37°C for 15 minutes. The BioLegend intracellular staining with True-Phos Perm Buffer (catalog 425401) protocol was followed and all washes were performed with BioLegend Cell Staining Buffer (catalog 425401). BioLegend Trustain FcX for human (catalog 422302) or mouse (clone 93, catalog 101320) were used to block samples before staining. Phospho-flow experiments were collected on a BD LSRII and lung homogenate analyses were collected using a BD FACSCanto RUO. FlowJo software (BD) was used for analysis of flow cytometry data.
Human studies.
Healthy subjects were recruited from the Seattle metropolitan area with the following exclusion criteria: (a) age < 18 or > 50; (b) active tobacco use; (c) weight < 100 or > 350 pounds; (d) any history of chronic illness; (e) pregnant or had given birth within the preceding 9 months; (f) symptoms of infection within the past 2 weeks; (g) prescription medication use (other than oral contraceptives); and (h) vaccinated in prior 6 weeks. Subjects were asked to refrain from any heavy exercise or alcohol use for 24 hours and to fast overnight. Bronchoscopy was performed using a flexible fiberoptic bronchoscope following the administration of topical anesthesia, most commonly via the nasal route. The bronchoscope was wedged in a lingular airway and five 30-mL aliquots of normal saline were serially instilled into the airway. After instillation of each aliquot, the BALF was retrieved by negative pressure. The process was repeated in the right middle lobe. Subjects provided written informed consent to participate.
ARDS subjects were identified from a cohort of intubated and mechanically ventilated patients undergoing BAL for clinical suspicion of ventilator-associated pneumonia (VAP) at Harborview Medical Center (HMC) in Seattle, Washington, USA as previously described (51). The indications and protocols for performing VAP BALs are standardized at HMC (52). Bronchoscopy with BAL is performed to evaluate for VAP if a patient meets the following 3 criteria: (a) radiographic abnormalities; (b) 1 or more of the following clinical signs: fever, purulent endotracheal secretions, or leukocytosis; and (c) no new antimicrobial drugs for ≥ 72 hours. All samples were obtained from the excess total BAL fluid not needed for clinical care and processed within 1 hour of the procedure. ARDS was defined using the 2012 Berlin Definition for ARDS (53). Characteristics of the human subjects participating in this study are located in Supplemental Table 4.
We filtered BAL fluid collected from both healthy and ARDS subjects using a 70-μm cell strainer (Corning) to remove mucous and debris. The collected flow-through from healthy subjects was centrifuged at 300 g for 10 minutes at 4°C and then washed twice with cold Hanks’ balanced salt solution. The collected flow-through from ARDS subjects was centrifuged at 300 g for 5 minutes and the supernatant was removed and stored. We eliminated RBCs in samples from ARDS subjects using a lysis buffer (BioLegend) and then the cells were washed once with PBS. Alveolar leukocytes from both healthy and ARDS subjects were resuspended in 2% FBS and an aliquot was processed for a cytospin. Cells were washed again and then cryopreserved in 7% DMSO (Thermo Fisher Scientific). For flow analyses, cells were thawed in media (HEPES-buffered RPMI-1640) containing DNase I, pelleted, and resuspended in media. An aliquot was taken for total cell enumeration and the remaining cells were fixed, permeabilized, and stained following the protocol described in the Flow cytometry section above.
Statistics.
GraphPad Prism 8 software was used to perform statistical analyses and specific statistical tests used are listed in individual figure legends. Multiple t tests were performed with corrections for multiple comparisons using the Holm-Sidak method, while 2-tailed unpaired t tests were used where indicated in figure legends. P < 0.05 was considered statistically significant and specific P-value identifiers are listed in each figure legend. Some data sets were checked for statistical outliers using the GraphPad Prism outlier calculator with an α of 0.05; if a data point was determined to be a significant outlier it was not included in the graphs or when calculating statistical significance.
Study approval.
The Institutional Animal Care and Use Committee at the University of Washington reviewed and approved all animal procedures described in these studies. All human studies were approved by the University of Washington Human Subjects Division Institutional Review Board (UW IRBs: STUDY00002504, STUDY00001438, and STUDY0004670). Written informed consent was received from participants prior to inclusion of the study or samples were obtained under a waiver of consent that was approved by the University of Washington Human Subjects Division.
Author contributions
MEL designed research studies, conducted experiments, acquired and analyzed data, and wrote and edited the manuscript. KQG and WEE assisted in design of research studies, conducted experiments, and acquired analyzed data. JSV conducted experiments and acquired and analyzed data. EDM, CM, TEW, and SJS designed research studies, acquired samples for analysis, and wrote and edited the manuscript. JC provided mice used in the study and edited the manuscript. WCL and AMM designed research studies, analyzed data, and wrote and edited the manuscript.
Supplementary Material
Acknowledgments
The work in this manuscript was supported by the Cystic Fibrosis Foundation through the University of Washington Research Development Program SINGH15R0 (to MEL), Postdoctoral Fellowship LONG18F0 (to MEL), and Postdoc-to-Faculty Transition Award LONG19F5 (to MEL); and the NIH grants R01 HL144656 (to AMM), R01 AI093646 (to SJS), R01 HL113382 (to TEW), R21 AI130797 (to TEW), F32 HL138746 (to EDM), and K23 HL120896 (to CM). We would like to thank Siriphan Boonsilp, Victoria Dmyterko, Helen Haile, and Brian Lee for assistance in recruiting subjects and sample handling, and Emily Hemann for additional technical assistance.
Version 1. 12/05/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(23):e132377.https://doi.org/10.1172/jci.insight.132377.
Contributor Information
Ke-Qin Gong, Email: kqgong@uw.edu.
Carmen Mikacenic, Email: cmikacen@uw.edu.
T. Eoin West, Email: tewest@uw.edu.
Jean Charron, Email: Jean.charron@crhdq.ulaval.ca.
Anne M. Manicone, Email: manicone@uw.edu.
References
- 1.Matthay MA, et al. Acute respiratory distress syndrome. Nat Rev Dis Primers. 2019;5(1):18. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3(1):23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 3.Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol. 2008;181(6):3733–3739. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- 4.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnston LK, Rims CR, Gill SE, McGuire JK, Manicone AM. Pulmonary macrophage subpopulations in the induction and resolution of acute lung injury. Am J Respir Cell Mol Biol. 2012;47(4):417–426. doi: 10.1165/rcmb.2012-0090OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrell ED, et al. Alveolar macrophage transcriptional programs are associated with outcomes in acute respiratory distress syndrome. Am J Respir Crit Care Med. 2019;200(6):732–741. doi: 10.1164/rccm.201807-1381OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosseau S, et al. Phenotypic characterization of alveolar monocyte recruitment in acute respiratory distress syndrome. Am J Physiol Lung Cell Mol Physiol. 2000;279(1):L25–L35. doi: 10.1152/ajplung.2000.279.1.L25. [DOI] [PubMed] [Google Scholar]
- 8.Long ME, et al. MEK1/2 inhibition promotes macrophage reparative properties. J Immunol. 2017;198(2):862–872. doi: 10.4049/jimmunol.1601059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long ME, Gong KQ, Eddy WE, Liles WC, Manicone AM. Pharmacologic inhibition of MEK1/2 reduces lung inflammation without impairing bacterial clearance in experimental Pseudomonas aeruginosa pneumonia. Pneumonia (Nathan) 2017;9:13. doi: 10.1186/s41479-017-0037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuh K, Pahl A. Inhibition of the MAP kinase ERK protects from lipopolysaccharide-induced lung injury. Biochem Pharmacol. 2009;77(12):1827–1834. doi: 10.1016/j.bcp.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Shi-Lin D, Yuan X, Zhan S, Luo-Jia T, Chao-Yang T. Trametinib, a novel MEK kinase inhibitor, suppresses lipopolysaccharide-induced tumor necrosis factor (TNF)-α production and endotoxin shock. Biochem Biophys Res Commun. 2015;458(3):667–673. doi: 10.1016/j.bbrc.2015.01.160. [DOI] [PubMed] [Google Scholar]
- 12.Smith JA, Mayeux PR, Schnellmann RG. Delayed Mitogen-activated protein kinase/extracellular signal-regulated kinase inhibition by trametinib attenuates systemic inflammatory responses and multiple organ injury in murine sepsis. Crit Care Med. 2016;44(8):e711–e720. doi: 10.1097/CCM.0000000000001672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer. 2015;15(10):577–592. doi: 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- 14.Moore BB. Groundhog day for rodent models of acute lung injury: clear relevance or renewed debate? Am J Respir Cell Mol Biol. 2017;57(2):141–142. doi: 10.1165/rcmb.2017-0119ED. [DOI] [PubMed] [Google Scholar]
- 15.Sweeney TE, Lofgren S, Khatri P, Rogers AJ. Gene expression analysis to assess the relevance of rodent models to human lung injury. Am J Respir Cell Mol Biol. 2017;57(2):184–192. doi: 10.1165/rcmb.2016-0395OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholl FA, et al. Mek1/2 MAPK kinases are essential for Mammalian development, homeostasis, and Raf-induced hyperplasia. Dev Cell. 2007;12(4):615–629. doi: 10.1016/j.devcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Bissonauth V, Roy S, Gravel M, Guillemette S, Charron J. Requirement for Map2k1 (Mek1) in extra-embryonic ectoderm during placentogenesis. Development. 2006;133(17):3429–3440. doi: 10.1242/dev.02526. [DOI] [PubMed] [Google Scholar]
- 18.Price S. Putative allosteric MEK1 and MEK2 inhbitors. Expert Opin Ther Pat. 2008;18(6):603–627. doi: 10.1517/13543776.18.6.603. [DOI] [Google Scholar]
- 19.Xu Y, et al. Spred-2 deficiency exacerbates lipopolysaccharide-induced acute lung inflammation in mice. PLoS One. 2014;9(9):e108914. doi: 10.1371/journal.pone.0108914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roskoski R. MEK1/2 dual-specificity protein kinases: structure and regulation. Biochem Biophys Res Commun. 2012;417(1):5–10. doi: 10.1016/j.bbrc.2011.11.145. [DOI] [PubMed] [Google Scholar]
- 21.Roskoski R. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 2012;66(2):105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Saint RJ, D’Elia RV, Bryant C, Clark GC, Atkins HS. Mitogen-activated protein kinases (MAPKs) are modulated during Francisella tularensis infection, but inhibition of extracellular-signal-regulated kinases (ERKs) is of limited therapeutic benefit. Eur J Clin Microbiol Infect Dis. 2016;35(12):2015–2024. doi: 10.1007/s10096-016-2754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruchhagen C, et al. Metabolic conversion of CI-1040 turns a cellular MEK-inhibitor into an antibacterial compound. Sci Rep. 2018;8(1):9114. doi: 10.1038/s41598-018-27445-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan W, Chan JH, Wong CH, Leung BP, Wong WS. Anti-inflammatory effects of mitogen-activated protein kinase kinase inhibitor U0126 in an asthma mouse model. J Immunol. 2004;172(11):7053–7059. doi: 10.4049/jimmunol.172.11.7053. [DOI] [PubMed] [Google Scholar]
- 25.Wu X, Dayanand KK, Thylur RP, Norbury CC, Gowda DC. Small molecule-based inhibition of MEK1/2 proteins dampens inflammatory responses to malaria, reduces parasite load, and mitigates pathogenic outcomes. J Biol Chem. 2017;292(33):13615–13634. doi: 10.1074/jbc.M116.770313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu YR, et al. Flow cytometric analysis of myeloid cells in human blood, bronchoalveolar lavage, and lung tissues. Am J Respir Cell Mol Biol. 2016;54(1):13–24. doi: 10.1165/rcmb.2015-0146OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Droebner K, Pleschka S, Ludwig S, Planz O. Antiviral activity of the MEK-inhibitor U0126 against pandemic H1N1v and highly pathogenic avian influenza virus in vitro and in vivo. Antiviral Res. 2011;92(2):195–203. doi: 10.1016/j.antiviral.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Galuppo M, et al. MEK inhibition suppresses the development of lung fibrosis in the bleomycin model. Naunyn Schmiedebergs Arch Pharmacol. 2011;384(1):21–37. doi: 10.1007/s00210-011-0637-7. [DOI] [PubMed] [Google Scholar]
- 29.Madala SK, Schmidt S, Davidson C, Ikegami M, Wert S, Hardie WD. MEK-ERK pathway modulation ameliorates pulmonary fibrosis associated with epidermal growth factor receptor activation. Am J Respir Cell Mol Biol. 2012;46(3):380–388. doi: 10.1165/rcmb.2011-0237OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, et al. MEK1/2 inhibitors activate macrophage ABCG1 expression and reverse cholesterol transport-An anti-atherogenic function of ERK1/2 inhibition. Biochim Biophys Acta. 2016;1861(9 Pt A):1180–1191. doi: 10.1016/j.bbalip.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, et al. Inhibition of ERK1/2 and activation of LXR synergistically reduce atherosclerotic lesions in ApoE-deficient mice. Arterioscler Thromb Vasc Biol. 2015;35(4):948–959. doi: 10.1161/ATVBAHA.114.305116. [DOI] [PubMed] [Google Scholar]
- 32.Catalanotti F, et al. A Mek1-Mek2 heterodimer determines the strength and duration of the Erk signal. Nat Struct Mol Biol. 2009;16(3):294–303. doi: 10.1038/nsmb.1564. [DOI] [PubMed] [Google Scholar]
- 33.Zmajkovicova K, Jesenberger V, Catalanotti F, Baumgartner C, Reyes G, Baccarini M. MEK1 is required for PTEN membrane recruitment, AKT regulation, and the maintenance of peripheral tolerance. Mol Cell. 2013;50(1):43–55. doi: 10.1016/j.molcel.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lake D, Corrêa SA, Müller J. Negative feedback regulation of the ERK1/2 MAPK pathway. Cell Mol Life Sci. 2016;73(23):4397–4413. doi: 10.1007/s00018-016-2297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wortzel I, Seger R. The ERK cascade: distinct functions within various subcellular organelles. Genes Cancer. 2011;2(3):195–209. doi: 10.1177/1947601911407328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikula M, et al. Genome-wide co-localization of active EGFR and downstream ERK pathway kinases mirrors mitogen-inducible RNA polymerase 2 genomic occupancy. Nucleic Acids Res. 2016;44(21):10150–10164. doi: 10.1093/nar/gkw763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikula M, Bomsztyk K. Direct recruitment of ERK cascade components to inducible genes is regulated by heterogeneous nuclear ribonucleoprotein (hnRNP) K. J Biol Chem. 2011;286(11):9763–9775. doi: 10.1074/jbc.M110.213330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson JD, LeBoeuf RC, Bomsztyk K. Direct recruitment of insulin receptor and ERK signaling cascade to insulin-inducible gene loci. Diabetes. 2011;60(1):127–137. doi: 10.2337/db09-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo L, Chen C, Liang Q, Karim MZ, Gorska MM, Alam R. Nuclear translocation of MEK1 triggers a complex T cell response through the corepressor silencing mediator of retinoid and thyroid hormone receptor. J Immunol. 2013;190(1):159–167. doi: 10.4049/jimmunol.1201657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolden A, Bernard L, Jones D, Akinyeke T, Stewart LV. The PPAR gamma agonist troglitazone regulates Erk 1/2 phosphorylation via a PPARγ-independent, MEK-dependent pathway in human prostate cancer cells. PPAR Res. 2012;2012:929052. doi: 10.1155/2012/929052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dave S, Nanduri R, Dkhar HK, Bhagyaraj E, Rao A, Gupta P. Nuclear MEK1 sequesters PPARγ and bisects MEK1/ERK signaling: a non-canonical pathway of retinoic acid inhibition of adipocyte differentiation. PLoS ONE. 2014;9(6):e100862. doi: 10.1371/journal.pone.0100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgermeister E, Chuderland D, Hanoch T, Meyer M, Liscovitch M, Seger R. Interaction with MEK causes nuclear export and downregulation of peroxisome proliferator-activated receptor gamma. Mol Cell Biol. 2007;27(3):803–817. doi: 10.1128/MCB.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng H, et al. Skp2 regulates subcellular localization of PPARγ by MEK signaling pathways in human breast cancer. Int J Mol Sci. 2013;14(8):16554–16569. doi: 10.3390/ijms140816554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chawla A. Control of macrophage activation and function by PPARs. Circ Res. 2010;106(10):1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heming M, et al. Peroxisome proliferator-activated receptor-γ modulates the response of macrophages to lipopolysaccharide and glucocorticoids. Front Immunol. 2018;9:893. doi: 10.3389/fimmu.2018.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakioka T, et al. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412(6847):647–651. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- 47.Ito T, et al. Sprouty-related Ena/vasodilator-stimulated phosphoprotein homology 1-domain-containing protein-2 critically regulates influenza A virus-induced pneumonia. Crit Care Med. 2016;44(7):e530–e543. doi: 10.1097/CCM.0000000000001562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haasbach E, et al. The MEK-inhibitor CI-1040 displays a broad anti-influenza virus activity in vitro and provides a prolonged treatment window compared to standard of care in vivo. Antiviral Res. 2017;142:178–184. doi: 10.1016/j.antiviral.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 49.Pleschka S, et al. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat Cell Biol. 2001;3(3):301–305. doi: 10.1038/35060098. [DOI] [PubMed] [Google Scholar]
- 50.McCubbrey AL, Allison KC, Lee-Sherick AB, Jakubzick CV, Janssen WJ. Promoter specificity and efficacy in conditional and inducible transgenic targeting of lung macrophages. Front Immunol. 2017;8:1618. doi: 10.3389/fimmu.2017.01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morrell ED, et al. Cytometry TOF identifies alveolar macrophage subtypes in acute respiratory distress syndrome. JCI Insight. 2018;3(10):99281. doi: 10.1172/jci.insight.99281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dellit TH, Chan JD, Skerrett SJ, Nathens AB. Development of a guideline for the management of ventilator-associated pneumonia based on local microbiologic findings and impact of the guideline on antimicrobial use practices. Infect Control Hosp Epidemiol. 2008;29(6):525–533. doi: 10.1086/588160. [DOI] [PubMed] [Google Scholar]
- 53.ARDS Definition Task Force, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307(23):2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.