Abstract
At diagnosis, most people with type 1 diabetes (T1D) produce measurable levels of endogenous insulin, but the rate at which insulin secretion declines is heterogeneous. To explain this heterogeneity, we sought to identify a composite signature predictive of insulin secretion, using a collaborative assay evaluation and analysis pipeline that incorporated multiple cellular and serum measures reflecting β cell health and immune system activity. The ability to predict decline in insulin secretion would be useful for patient stratification for clinical trial enrollment or therapeutic selection. Analytes from 12 qualified assays were measured in shared samples from subjects newly diagnosed with T1D. We developed a computational tool (DIFAcTO, Data Integration Flexible to Account for different Types of data and Outcomes) to identify a composite panel associated with decline in insulin secretion over 2 years following diagnosis. DIFAcTO uses multiple filtering steps to reduce data dimensionality, incorporates error estimation techniques including cross-validation and sensitivity analysis, and is flexible to assay type, clinical outcome, and disease setting. Using this novel analytical tool, we identified a panel of immune markers that, in combination, are highly associated with loss of insulin secretion. The methods used here represent a potentially novel process for identifying combined immune signatures that predict outcomes relevant for complex and heterogeneous diseases like T1D.
Keywords: Autoimmunity
Keywords: Diabetes, Immunotherapy, Molecular pathology
A panel of immune markers that, in combination, are highly associated with loss of insulin secretion in type 1 diabetes was identified using a computational tool.
Introduction
Type 1 diabetes (T1D) is caused by immune destruction of pancreatic β cells, leading to an inability to produce sufficient insulin. At diagnosis, most people with T1D still produce some endogenous insulin, but both the level and rate of continued decline vary markedly among individuals. Age represents 1 component of this heterogeneity because subjects diagnosed at a younger age tend to have lower levels of insulin secretion at diagnosis and to lose insulin secretion more rapidly after onset (1–3). The other sources of heterogeneity in insulin secretion after diagnosis are not well understood. Addressing this gap could help enable patient selection for clinical trial enrollment, where enriching for subjects with a faster rate of decline could reduce the size or duration of efficacy trials (4, 5). Clinically, maintenance of insulin secretion after diagnosis can contribute to a reduction in the rate of disease complications (6). The rate of loss of insulin secretion is therefore considered a metric of postdiagnosis disease progression.
Immune system parameters are also hypothesized to contribute to varied rates of disease progression. Individuals with T1D are heterogeneous in regard to their immunobiology at and after diagnosis, as supported by the breadth and intersubject range of individual immunological features within this population (7–11). In recent studies of individual assays, this heterogeneity has been suggested to predict the rate of progression after diagnosis for some subjects (12–15). Better fundamental knowledge of the array of immune drivers of disease could help explain the differing rates of loss of β cells observed across subjects and potentially indicate immunotherapeutic targets.
Determining whether a combination of immune and β cell features can together define rate of disease progression across a range of subjects requires (a) sufficient samples from shared, well-annotated subjects and (b) appropriately qualified assays that broadly describe response in a given subject. Sample availability can be overcome by working collaboratively to obtain retrospective samples from clinical trials or similarly annotated longitudinal collections. Because samples from such collections are necessarily limited, ensuring their best use requires fit-for-purpose qualification of any biomarker assays used. Here, we planned to run each assay in a single batch. Thus, the key performance parameters for assay qualification were detectability (frequency present) and intra-assay precision of each analyte, when tested in the target patient population, with the sample type, and with available sample volume. Notably, sample volumes may disproportionately affect low-frequency analytes, such as low-abundance cell populations, because stochastic sampling error can result in false positive associations, especially when many measurements are made.
In this study, we aimed to determine whether multiple unique measures in aggregate correlate with rate of loss of insulin secretion. To this end, we established a stringent, well-defined, and collaborative assay evaluation and data analysis method. The assays selected for study included measures that were hypothesized to directly relate to T1D pathogenesis; these included antigen-specific CD8+ T cell frequencies (16), a Treg transcriptional signature previously associated with T1D (17), the ratio of proinsulin to C-peptide (18), and a measure of demethylated insulin DNA in serum (19). Other selected assays showed prior utility in understanding T1D pathogenesis. These included genome-scale technologies, included here for broad screening and hypothesis generation: whole-blood and cell subset RNA-Seq (15, 20) and an assay measuring the transcriptional response to T1D serum (13, 21, 22) were used. Assays also included screening assessments that were smaller scale, including immunophenotyping by flow cytometry (23) and measurements of serum miRNA (24). Using these assays, we conducted a proof-of-concept study in a cohort of recent-onset T1D subjects with variable rates of C-peptide decline, who were followed meticulously for both metabolic outcomes and ancillary sample collection over a 2-year period within the context of clinical trial monitoring. After filtering based on individual assay validity and consistency, data from all assays were integrated, their dimensionality was reduced to facilitate combined modeling, and features associated with C-peptide decline were identified. Model estimation error was assessed by cross-validation. Selected analytes were then subjected to sensitivity analysis. We found 12 analytes that, in combination, were prognostic for decline in C-peptide; these originated from 3 immune assays (signature of serum exposure, cell type–specific RNA-Seq, and flow cytometry) implicating at least 3 immune cell types. Attributes of immune activation, suggestive of an attempt to control the immune response, were positively associated with maintenance of insulin secretion. Immune trafficking and B cell activation, the latter of which was recently associated with poor response to therapy (20), were both associated with increased rate of disease progression.
Results
Study approach and heterogeneity in C-peptide decline.
We collaboratively defined an approach to identify a robust composite signature of decline in insulin secretion. This method involved 2 key steps. First, we conducted blinded replicate testing to measure the detectability and intra-assay precision of a panel of selected assays, as listed in Table 1. Next, we deployed each qualified assay on samples collected at T1D diagnosis to identify analytes that were prognostic for decline in insulin secretion over the following 2 years. PBMC, serum, and whole-blood RNA samples for this step were collected from control-arm subjects (n = 50) enrolled in 1 of 3 new-onset T1D trials conducted by the Immune Tolerance Network (25–27). Subjects were meticulously followed after diagnosis, with insulin secretion assessed by mixed-meal tolerance testing at least 5 times between diagnosis and 2 years after diagnosis. Clinical and demographic data for included subjects are listed in Table 2.
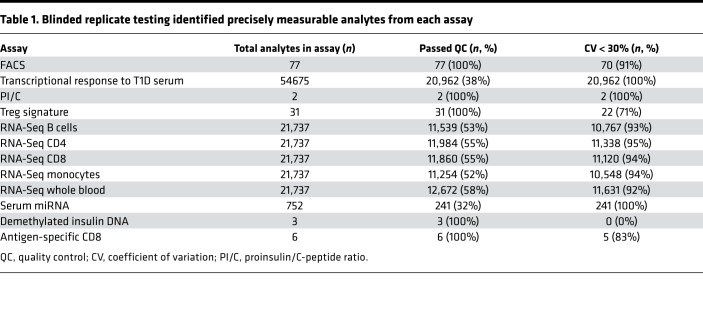
Table 1. Blinded replicate testing identified precisely measurable analytes from each assay.
Table 2. Clinical and demographic data for recent-onset cohort.
As previously described (28, 29), decline in insulin secretion (as measured by circulating C-peptide) in the years after diagnosis was highly heterogeneous. Figure 1 shows these data for all subjects whose samples were used in this study. Here, we used a log rate of decline calculated using a mixed model approximating all time points available for each subject (15, 30). Change in insulin secretion in this cohort most strongly approximated exponential decay (15, 30). In our data set, as in previous work (1–3), age is partially predictive of rate of decline. However, it is an imperfect predictor, particularly in subjects of younger ages, where variability in decline rates is highest (15). This variation is also present in subjects diagnosed at older ages. For example, the subjects highlighted in magenta and green (Figure 1) are aged 19 and 17, respectively, and show substantively different rates of decline.
Figure 1. C-peptide decay rate for each subject included in the recent-onset cohort.
C-peptide decay rate for each subject included in the recent-onset cohort (n = 50). Each line is 1 subject. Subjects highlighted in blue and green demonstrate slow and rapid decline, respectively. The subject highlighted in olive showed no detectable insulin secretion at a time point before the end of the study; decay rate calculations used throughout exclude the second time point at the limit of C-peptide detection. Some subjects (example in green/magenta) are similar in age but have disparate C-peptide decay rates.
Replicate testing identifies sufficiently precise arrays.
We selected a broad range of assays to test (Table 1). This included low-dimension assays selected to assess expected features of disease progression, including proinsulin/C-peptide ratio (18, 31), a marker of pancreatic islet β cell dysfunction; a demethylated insulin DNA assay measuring β cell death (19); antigen-specific CD8+ T cell frequency and phenotype as measured by qDot multimer assay (16); and a transcriptional signature of regulatory T cells that had previously been identified to discriminate between subjects with and without T1D (17). Higher-dimension (genome-scale) assays were also included to identify data-driven features of disease progression. These included RNA-Seq of whole blood (30) and sorted B cell, CD4+ and CD8+ T cell, and monocyte subsets (32); immunophenotyping by flow cytometry (33); transcriptional response to T1D serum (22) assessed using Affymetrix microarrays; and serum miRNAs measured by quantitative PCR (qPCR) (24).
Assays with poor precision were excluded to limit the effect of technical variation on eroding statistical power in the planned composite model. To assess assay precision in a blinded fashion, triplicate aliquot samples from 3–5 subjects with T1D were tested by each assay (per design in Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.126917DS1.) Over 160,000 individual analytes were measured across all assays (Table 1). Of these, 49% (80,852 analytes) met initial quality control or detection limits specific to each assay. Down-selection at this step was driven primarily by the higher dimension assays; for example, much of the genome is not expected to be transcribed in CD4+ T cells, and thus these unexpressed genes are filtered out from the RNA-Seq assays. Next, the CV (here expressed as a percentage) for each analyte was calculated per subject; any analyte with a mean CV less than 30% was eligible to be included in downstream analyses. The majority of analytes that met detection limits (91%) met the CV cutoff. Replicate testing data are available in Supplemental Figures 2–7. All analytes that did not meet this CV threshold were removed from further analysis.
The demethylated insulin assay was removed entirely from our pipeline at this phase because the CV in samples from subjects with T1D (regardless of duration) was consistently above our cutoff (Supplemental Figure 4). This may have been driven by sample quality because the serum samples for both replicate testing and the recent-onset cohort had been stored for many years and were not collected according to protocols optimized for this assay. The antigen-specific CD8+ T cell assay (qDot multimer, Supplemental Figure 5) was also removed at this phase. This assay had an acceptable CV but could be applied only to assay HLA-A2+ subjects, and the number of subjects in the recent-onset cohort bearing the HLA of interest was too low for the assay to have utility. The miRNA assay (Supplemental Figure 6) was attempted in the recent-onset cohort samples; however, it was removed before data analysis. Laboratory processing failures that occurred during miRNA extraction resulted in too few samples with sufficient available data for analysis. This stringent culling of assays and analytes reduced our total analyte count substantially, likely improving our statistical power based on reduced measurement error in the retained analytes. The process resulted in the removal of 3 assays (demethylated insulin, CD8 antigen–specific T cells, and serum miRNA). Although a considerable number of analytes were removed, we found that the vast majority of analytes tested in the 9 assays that were included in the rest of the study (95%) met our predefined precision cutoffs.
Development and tuning of an analysis pipeline incorporating multiple data types.
There are many tools optimized for single types of assay data but few that integrate multiple data types. Thus, we developed an analytical tool that was capable of incorporating the multiple data types tested here (Figure 2). The tool brings together established analytical methods (such as LASSO; ref. 34) with newly developed code to integrate varied data types. It was developed in the R programming language and is freely available. The tool is flexible to data types and to outcomes and thus is named DIFAcTO: Data Integration Flexible to Account for different Types of data and Outcomes; here, DIFAcTO was tested with 5 data types and a continuous clinical outcome. An optional dimension reduction step can be applied to whole-genome assays. In our study, each whole-genome assay was subjected to modular analysis by weighted gene co-expression network analysis (WGCNA) (35), and modules identified were treated as independent analytes, which were scaled and clustered as described below.
Figure 2. Schematic of analysis pipeline developed to integrate multiple data types.
Analytes are merged and scaled. Initial univariate filtering leaves only those analytes with at least modest correlations to the outcome for each assay. Subsequent filtering (clustering) identifies best correlated analyte in a given cluster (regardless of assay). From these, a composite model is generated using LASSO with cross-validation. The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range.
We used a machine learning approach that combines the following multistep analyte filtering. Step 1 is preprocessing by scaling and preliminary feature selection. Step 2 then identifies a multivariate model (using LASSO), which also implements internal feature selection. The goal of the first preprocessing step is to remove analytes (features) that are poorly associated with the dependent variable (rate of C-peptide loss) so that an acceptable number of features are included in the later LASSO step. The first step of preprocessing is itself done in 2 parts. The first is univariate filtering: within each assay, all individual associations between each analyte and the dependent variable are estimated. Only the top analytes are kept for further analysis, based on rank or significance thresholds (with correction for multiple comparisons). The second is clustering of associated analytes across multiple assays. Many of the selected analytes may strongly correlate with each other, even across different assays. For the final multivariate model, this may cause multicollinearity, which would complicate interpretation of associations. To avoid this, analytes are clustered by Pearson’s correlation using hierarchical clustering. The number of clusters is determined by a tuning parameter (described in detail below). A single representative analyte is then selected to represent the cluster; this analyte is most strongly associated with the dependent variable in each cluster. Lastly, a multivariate model (LASSO; refs. 34, 36) with predefined covariates (clinical, demographic) and the remaining analytes is performed. The LASSO also implements a regularized step that removes additional nonsignificant analytes. For this study, we incorporated baseline C-peptide, age, HLA, and BMI as known potential predictive covariates; these analytes were included in the LASSO analysis to ensure they were considered as potential components of the model.
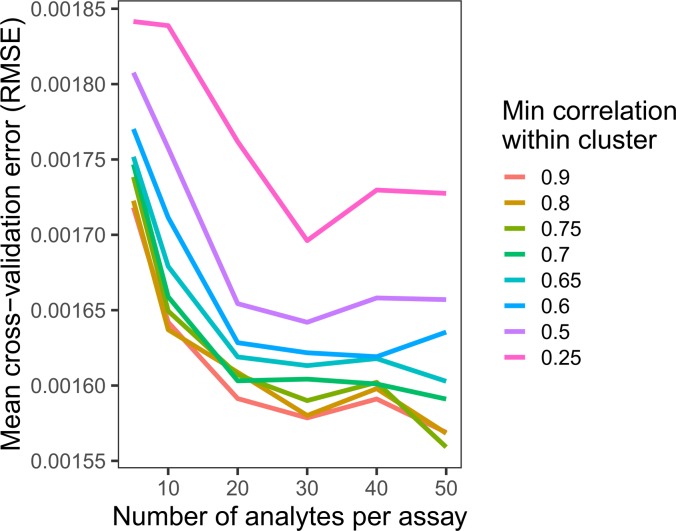
To allow a wide range of analytes and outcomes, DIFAcTO includes multiple user-adaptable parameters. The user can specify the number of analytes from each assay that enter the clustering step. The minimum within-cluster correlation used at the clustering step can also be tuned. Increasing this value results in a larger number of highly correlated clusters and thus a larger number of analytes entering the feature selection step; decreasing this value results in poorly correlated analytes within a cluster but fewer analytes entering the feature selection step. Both of these parameters (number of analytes per assay and minimum correlation per cluster) should be tested with each data set to identify optimal parameter settings. For this study, we evaluated a range of minimum correlation values and analyte counts per assay and assessed the cross-validation error rates after feature selection (Figure 3). Error rates reached a near minimum for this data set, with 30 analytes per assay included in the feature selection and within-cluster correlation of r = 0.7. In total, 201 clusters were identified, and a single analyte from each was used for LASSO feature selection.
Figure 3. Establishment of optimal parameter settings.
Changing settings within the analytical tool (DIFAcTO) identifies point of lowest cross-validation error associated with higher within-cluster correlation (r = 0.7) and a moderate number of analytes per assay (n = 30). Each line shows the cross-validation error (root mean squared error, RMSE) for a given analyte per assay setting (x axis) at a given minimum correlation within cluster (indicated by color).
Identification and sensitivity analysis of a composite analyte panel associated with insulin secretion.
We next applied DIFAcTO to our primary question: which immune markers measured at baseline (diagnosis) would, in a multivariate model, be prognostic for rate of C-peptide decline over the ensuing 2 years of disease? Using these optimized parameter settings, we identified a model composed of 17 immune analytes that, measured at diagnosis, were prognostic for rate of decline in insulin secretion. This rate was determined by using a standard clinical measure of C-peptide during mixed-meal tolerance testing (AUC C-peptide) at 5 time points over 2 years, as described previously (15, 30). Table 1 lists each individual analyte and the assay from which it was measured. C-peptide level at diagnosis is a known correlate of future C-peptide decline and was selected by the tool. We noted that age, HLA, and BMI were not selected as independent predictors in analysis of this cohort.
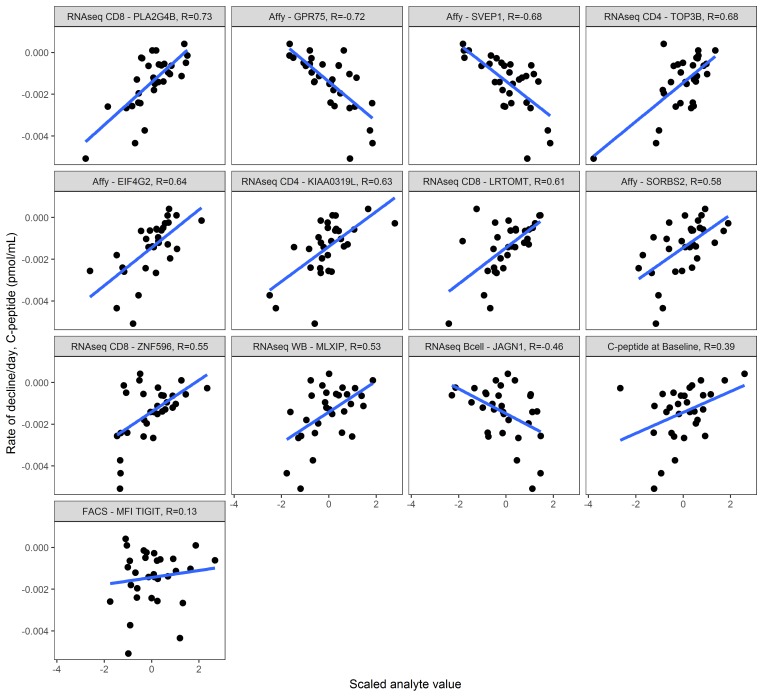
Although we did considerable filtering of this data set and performed cross-validation in feature selection, we recognized the continuing risk of identifying false positive associations given the number of analytes assessed. We therefore performed a preliminary sensitivity analysis on the remaining 17 analytes. We reasoned that an analyte should, at minimum, be robust to minor changes in settings of our own analytical tool; slight modifications to numbers of analytes per cluster or to clustering correlations should still result in a similar set of analytes. As expected, C-peptide at diagnosis (baseline) was consistently selected by DIFAcTO using all parameters tested (Figure 4). We identified 12 analytes that were robust to analytical tool settings (Table 3). Five analytes, however, were specific to only a few settings in DIFAcTO (annotated as “dropped” in Table 3); we predict that these analytes would likely not be associated with C-peptide decline in a separate validation data set because they were not robustly selected in this one. Individual correlations between C-peptide decline rate and each remaining analyte, as well as baseline C-peptide, are shown in Figure 5, ranked by correlation to C-peptide decline. Of the 12 analytes selected by the tool, 11 were more highly correlated with C-peptide decline than was baseline C-peptide. Because age was not selected by the tool, we also inspected the relationship between age and each individual analyte (Supplemental Figure 8). The analyte most highly correlated with age was the MFI of T cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domains (TIGIT) on naive CD8+ T cells; this was also the analyte with the lowest individual correlation with C-peptide decline. The remaining analytes and baseline C-peptide had limited correlations with age in this data set.
Figure 4. Characteristics of analytes selected by tool.
Sensitivity analysis shows that, even with cross-validation implemented in LASSO, 5/17 analytes are not robust to different parameter settings within the tool. These 5, indicated in grayscale, are unlikely to be validated in an independent cohort. Each individual plot’s x and y axes represent settings used to run the analytical tool; the number of analytes per assay setting is on the x axis, and minimum correlation per cluster is on the y axis of each miniplot. Darkest coloring indicates that the analyte was selected by the tool using that combination of x and y settings. Lighter coloring indicates that another analyte in that same cluster was selected by the tool. Light gray indicates that this analyte was not selected using that combination of x and y settings. Each miniplot is labeled by analyte and the assay from which it was originally measured. “Affy” indicates the transcriptional response to T1D serum assay as this is conducted on the Affymetrix platform.
Table 3. Analytes selected by the tool, assay from which they were derived, and status after sensitivity analysis.
Figure 5. Individual correlations between each selected analyte and insulin secretion.
Immune markers were measured at trial enrollment (within 90 days of diagnosis, n = 30 subjects), and y axis indicates C-peptide decay rate per day over the 2 years after diagnosis. Each miniplot uses the scaled value for the analyte on the x axis. Pearson’s correlation values are listed at the top of each miniplot; miniplots are ordered by absolute correlation value. Regression lines in blue. Note that in this data set, several immune parameters have higher correlation values with rate of C-peptide decay than does C-peptide level at diagnosis (Pearson’s r = 0.39). Assays and analyte names are truncated; full names can be found in Table 3. “Affy” indicates the transcriptional response to T1D serum assay as this is conducted on the Affymetrix platform.
Finally, we assessed the performance of our tool in predicting C-peptide decline in this data set and tested its ability to identify previously published analytes from an independent data set in a different disease context. C-peptide decline prediction was tested by fitting a linear model to the data using 3 sets of predictors: a baseline model using C-peptide at diagnosis as a known predictor, a “full” model using all 17 identified analytes, and a “maintained” model using only the 12 analytes that were found to be robust to tool settings. From these fits we calculated an adjusted R2 value that reflects how well the model fits the data. Additionally, we calculated a robust, cross-validated RMSE by separating the data into 5 folds and, for each fold, training on the other four-fifths of the data and predicting the held-out fold. We did this 1000 times to get a robust estimate of the RMSE along with a 90% confidence interval. The results are shown in Supplemental Table 1. Using our selected analytes improved prediction over baseline C-peptide in this data set, as reflected both in adjusted R2 and RMSE. Additionally, the improvement in performance of the maintained model over the full model supports our decision to remove those analytes that were not found to be robust to initial parameter settings. In an initial step toward comparison of this tool to elastic net, we tested an independent data set from a clinical trial of an HIV vaccine, RV144 (37). DIFAcTO successfully identified previously known immunological predictors in this high-dimensional data set (Supplemental Methods).
Discussion
Here, we describe a collaborative, generalizable method to identify robust, inclusive correlates of clinical outcomes and a proof-of-concept usage of that method to identify a panel of markers associated with decline in insulin secretion after diagnosis. In developing this method, we had 3 key goals: first, including data from as many assay types as possible and incorporating the expertise and perspective of as many investigators as was feasible; second, blind assessment of reproducibility for all analytes included in any analyses to increase likelihood of future success; and third, transferability to other studies and data sets.
One important component of our method was the collaborative identification of assays that might yield results of interest (38). We focused on choosing assays that were thought to be independent and mechanistically related to T1D pathogenesis, such as the proinsulin/C-peptide ratio, as well as assays that could generate more broad-based, hypothesis-generating results, such as RNA-Seq of multiple immune cell subsets. Nine investigators chose to participate in, and provide data for, this collaborative project; this resulted in a rich data set, generated from the same sets of subjects that are now being mined for other clinical outcomes and associations between immune markers. We partnered in this effort with a major autoimmunity clinical trial network (the Immune Tolerance Network; ITN), which furthered the visibility of this work in the T1D research community and may improve the possibilities for future clinical translation of our findings.
Our second focus was on assay quality. The technical precision of each individual analyte is essential to the reproducibility of the composite panel and thus to our ability to identify meaningful correlates of clinical outcomes. An early step in our process, therefore, was a preliminary assessment of immune marker reproducibility. Should these markers be of interest for future translation, many other assessments, including broader reproducibility measurements and multicenter validation, as recently described (39), would clearly be needed. Here, we applied a moderate level of rigor — requiring a mean CV less than 30% across a limited number of subjects with T1D for each assay. Still, this was sufficient to remove over 4000 analytes even after initial QC was applied. For the genome-scale assays, CV cutoffs were applied after detection thresholding processes; although a much larger number of analytes were removed because of lack of detection, technical imprecision resulted in the removal of hundreds to thousands of additional analytes for each RNA-Seq assay. This method of dimension reduction could be applied with relative ease and should reduce type 1 error resulting from detection of random associations between variables that can occur with imprecise measurements. One caveat to this substantial data reduction is that analytes associated with C-peptide decay may have been excluded because they did not fit our criteria; for example, there may be measurements that are more highly expressed and thus more reproducible in specific subjects or at specific disease stages. Future studies may reassess reproducibility in larger populations of T1D and other subjects. However, we accepted the trade-off between known reproducibility characteristics and the theoretical loss of important analytes. Separately, gene expression data tend to cluster into strongly correlated groups of genes (35), and groups of immune cell populations also show strongly correlated clusters (40). In some cases, the relationship to C-peptide that may be present for an analyte with low reproducibility will be represented by a more reproducible analyte in that or another assay.
Our third focus was on building a broadly useful analytical process. This method, from reproducibility testing through the use of our new analytical tool, is generalizable. DIFAcTO has been used to find analytes associated with both categorical and continuous outcomes. It has user-modifiable parameters that allow it to accommodate data sets of different sizes and data with different variability profiles. Indeed, in this study we used parameters that enabled us to prioritize selection of variables from multiple assays that may help understand T1D biology, rather than focusing solely on predictive performance. The tool can incorporate multiple data types, including genome-scale data sets. It has been developed in R/MLR (41), which is an advanced, generic, robust analytical framework for multivariate analysis modeling and feature selection methods. As mentioned, we are now mining this data set to identify robust predictors of other clinical outcomes in these subjects. However, DIFAcTO could easily be applied by those investigators and consortia with very large clinical and mechanistic data sets.
We have used this method to identify a diverse multivariate model that predicts C-peptide decline. Of course, to move toward clinical utility, this panel would need to be confirmed in an independent replication cohort. In addition, the panel would likely need to be transitioned to more focused assays, as opposed to the genome-scale data analyzed here, and therefore would need full, independent qualification using the focused assay methods, as has been considered in other studies (reviewed in refs. 5, 42). One might speculate that this panel could be informative at earlier stages of T1D, including the antibody-positive at-risk setting, or that it may be predictive of future disease-associated complications. This remains to be tested in other data sets.
Interestingly, this composite panel incorporates markers of multiple cell types and pathways. The expression level of each component immune feature in our signature differs across subjects. However, we can see common immunological themes. We found that markers associated with immune activation (TOP3B, LRTOMT), ER processing (ZNF596, EIF4G2), and regulation of activation (SORBS2, TIGIT on naive CD8+ T cells) were positively correlated with slower loss of insulin secretion. Additional markers were associated with viral or interferon responses (PLA2G4B, KIAA0319L). These positive correlations may initially be counterintuitive because islet β cell destruction is thought to be immune mediated. However, a similar association of immune activation and increased regulation has been observed in regulatory T cell studies in autoimmunity, where there is an ultimately failed attempt to control the immune response (43). Thus, in part, our data suggest that immune activation positively associated with slower disease progression, representing immune processes directed toward controlling autoimmunity. In contrast, in the same T1D population, we also found negative correlations with maintenance of cell trafficking and insulin secretion, respectively (SVEP1, GRP75), as well as functional markers of B cells (JAGN1). This is consistent with 2 previous findings: increased B cells in pancreatic sections (7) and a B cell transcriptional signature found to correlate with poor response to therapy and more rapid C-peptide decline (15, 20). Together, these data suggest that there may be common immune processes that associate with slower disease progression, but they likely differ in composition and predominance across subjects, highlighting the universal value of a composite signature.
Immune, β cell, and demographic data were all included as potential predictors in the LASSO analysis; we note that 12 immune features and baseline C-peptide were selected. Importantly, nearly all the selected markers (11 of 12) when analyzed independently were more highly associated with decline in insulin secretion than was baseline C-peptide, highlighting the possible relevance of these potentially novel immune markers. As would be expected from previous studies (1, 44, 45), the tool identified baseline C-peptide as an important predictor of insulin secretion. HLA, BMI, and age, however, were not selected to contribute to the composite model. Age and HLA have each been established to play a role in predicting risk of T1D development (46–48); a role for BMI has been investigated, but the relationship with disease risk varies by study (49–53). In agreement with our findings, HLA and BMI have not been consistently identified as predictors of insulin secretion after diagnosis (15, 44, 54). Our expectation was that age, however, would be selected by the analytical tool. One immune marker showed a moderate correlation with age (MFI of TIGIT on naive CD8+ T cells); we speculate that some of the role age plays in disease progression may be reflected in part by this marker. Intriguingly, baseline C-peptide was itself not strongly correlated with age at onset in this data set. Another potential reason that age was not selected is an enrichment in this data set of relatively older-onset T1D subjects (median age 19.25 years for subjects included in LASSO analysis), which is not unexpected based on the demographics of T1D development in many nations (55–57). Although this cohort may underrepresent early-onset cases, it is representative of subjects who qualify for immune intervention at disease onset.
In summary, we have developed a method to generate high-quality data across multiple assays and an analytical pipeline to combine and analyze disparate data types. This method identified a composite model associated with decline in insulin secretion that includes both expected and novel biological insights that could move toward replication in other cohorts and potentially assessment in other stages of T1D.
Methods
Subjects and clinical outcome data.
All recent-onset samples (serum, PBMC, purified RNA, and Tempus tubes) were provided by the ITN and were obtained from subjects with T1D randomized to the control arms of 3 new-onset T1D trials (25–27). PBMC and serum samples were processed at a central location as described in the original trials; RNA samples were processed as described previously (15). Clinical and demographic data were obtained from ITN TrialShare (58), a freely available source of results from ITN trials. Samples from the AbATE study were not received for the cell subset RNA-Seq assays and thus could not be included in the LASSO selection, leaving n = 30 subjects.
Rate of C-peptide decline was calculated as previously described (15, 30). In brief, C-peptide 2-hour AUC measurements from the baseline and 6-, 12-, 18-, and 24-month visits were log-transformed and fit to a linear model with participant as a random effect. Repeated C-peptide measurements at the lower limits of detection were removed from fitting. The C-peptide decay rate was extracted from the fit as the slope of the linear model. The fits were performed using the lme4 package.
Assay methods.
Each of these assays has been previously published individually. Whole-blood RNA-Seq data were generated from Tempus tubes (Applied Biosystems) using the process described previously (20, 30). Cell subsets were sorted and RNA-Seq data generated using the process described previously (59). All RNA-Seq data were prefiltered to remove all non–protein-coding genes. RNA-Seq sample identity was verified using kinship comparisons based on genomic variants called from the RNA-Seq reads (20) and clustering of sample data by source cell type; all samples matched their annotated subject and cell type. Flow immunophenotyping panels were those routinely implemented for ITN clinical trials, as described previously (26, 33). The transcriptional response to T1D serum assay was conducted as described previously (13). The miRNA assay was conducted using the Exiqon qPCR platform as described previously (24). The Treg transcript assay was performed using the NanoString platform as described previously (14, 17), with samples for the Treg assay sorted concomitantly with the cell subset RNA-Seq sample set. The proinsulin/C-peptide assay was conducted using a trefoil time-resolved fluorescence immunoassay (31), adapted to an AutoDelfia automatic system (PerkinElmer) (18). The demethylated insulin assay was conducted using the RainDance droplet digital PCR platform (RainDance Technologies). The antigen-specific CD8+ T cell assay was conducted using a multicolor quantum dot multimer assay described previously (16). For additional information on data processing for each genome-scale assay, please see the supplemental materials.
Replicate testing data.
The replicate testing cohort was used to determine the technical precision of each analyte by calculating the intrasubject CV for 3 biological replicate aliquots from the same blood draw from 5 subjects. Briefly, after data processing, the intrasubject CV was calculated from the 3 replicates. For each analyte, the mean CV across subjects was calculated. Analytes were retained for the analyte selection pipeline if the mean CV was below 30%. Replicate testing data for all RNA-Seq assays and the transcriptional response to serum assay are available at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database at GSE131528.
Analyte selection tool.
All analyses were conducted using the R programming language and software environment (60). Analytes were sequentially processed in the following steps: analyte scaling, univariate filtering, assay merging, hierarchical clustering, and multivariate modeling. Analyte scaling was performed for each assay separately by subtracting the mean and dividing by the standard deviation, such that the mean of all analyte values was equal to 0 and the standard deviation of all analyte values was equal to 1. Univariate filtering was performed by first applying a linear model to each analyte using the analytes as the predictor, C-peptide decay rate as the outcome, and sex, study, BMI, age, and baseline C-peptide as covariates. The P value of the analyte term was extracted from each generalized model and adjusted for multiple testing using the Benjamini-Hochberg algorithm. A number of the most significant analytes from each assay were retained for the subsequent steps. The number of analytes selected per assay is an adjustable parameter and is used for error estimation and sensitivity analysis. Next, the top significant analytes from each assay were merged into a single array by subject. After merging, all missing values were imputed with the k-nearest neighbors method (k = 20). The array containing analytes from all assays was then hierarchically clustered on analytes, with distance metric = 1 – cor, where cor is Pearson’s correlation. The distance metric value that defines which analytes are clustered together is 1 – cor*, where cor* is analogous to the minimum correlation within cluster. The minimum correlation within cluster is an adjustable parameter and is used for error estimation and sensitivity analysis. After clustering, the analytes with the lowest adjusted P value from each cluster were selected as the representative analytes and retained for the subsequent steps. The remaining analytes were included as covariates in a linear regression model with C-peptide decay rate as the outcome. Sex, study, age, BMI, and baseline C-peptide were included as additional covariates. The analytes and covariates were additionally included in a LASSO regression model for an optional last feature selection method. Linear models were run with the glmnet package. The value of the regularization parameter used in the LASSO models was calculated by glmnet.cv as lambda.min. The optimal values for the 2 adjustable pipeline parameters were selected as the parameter set that minimizes the resulting cross-validation RMSE, while maintaining a conservative number of analytes that are input into the linear model.
Cross-validation was performed by randomly subsetting the data set into a training set (75% of subjects) and a test set (25% of subjects), using the training set as input into the feature selection pipeline, and calculating the prediction error (RMSE) on the test set. This process was performed 1000 times for these sets of parameters: number of analytes per assay and minimum correlation within cluster. The mean RMSE of the 1000 iterations was calculated for each parameter set.
Sensitivity analysis.
Analytes that were selected when using the optimal pipeline parameters were then analyzed for parameter sensitivity. Results of the pipeline from all parameter sets were inspected for presence of the analytes of the optimal parameter set, either selected explicitly or clustered (i.e., highly correlated) with a selected analyte. Analytes from the optimal parameter that were sensitive to parameter choice (present in <50% of the parameter sets tested) were removed from the final list of analytes.
Data and code availability.
Code for the analytical tool is available on GitHub at https://github.com/FredHutch/JDRFCAV using branch name “master” and commit ID 905178c, along with data necessary to recreate the analyses presented here. Whole-genome data (cell subset RNA-Seq and transcriptional response to serum assays, as well as all replicate testing data) are available in the NCBI GEO database under accession number GSE131528. Whole-blood RNA-Seq data are available in NCBI GEO under accession number GSE124400.
Statistics.
All statistical analyses are described in the Analyte Selection Tool section of Methods.
Study approval.
All human studies were approved by appropriate institutional review boards. ITN studies were approved by independent IRBs at each participating clinical site, as described in the original clinical trial reports (25–27). Use of human samples for assay replicate testing was approved by the Benaroya Research Institute IRB. Written informed consent was received from all participants in all cohorts before inclusion in the study.
Author contributions
CS designed the study, conducted experiments, acquired data, analyzed data, and wrote the first draft of and edited the manuscript. SOS analyzed data, developed the analytical tool, designed analyses, wrote sections of the manuscript, and reviewed the manuscript. DB analyzed data, developed the analytical tool, and edited the manuscript. EW analyzed data, developed the analytical tool, and reviewed the manuscript. MJD analyzed data, calculated the clinical outcomes, and edited the manuscript. WCY analyzed data and edited the manuscript. AMP acquired and analyzed data, provided reagents and protocols, and edited the manuscript. JMO conceived and designed the study and edited the manuscript. FKG analyzed data, provided protocols, and edited the manuscript. EAJ analyzed data, provided reagents and protocols, and edited the manuscript. MKL analyzed data, provided reagents and protocols, and reviewed the manuscript. PSL analyzed and provided data and reviewed the manuscript. EMA analyzed and provided data and reviewed the manuscript. AP analyzed and provided data and reviewed the manuscript. MJH analyzed and provided data, provided reagents and protocols, and reviewed the manuscript. RG oversaw analytical tool development, conceived and designed the study, and reviewed the manuscript. GTN conceived and designed the study, reviewed data, and reviewed the manuscript. SAL was involved in study design, interpreted data, and assisted in writing of the manuscript.
Supplementary Material
Acknowledgments
We wish to acknowledge the clinical staff who diligently conducted each of the ITN trials here mentioned, as well as all participants in those trials. We also thank the many technical staff in each of the laboratories that contributed data, who are too numerous to name individually here. We appreciate fruitful discussions with the JDRF Biomarker Working Group and its individual members. Simi Ahmed of JDRF was a strong supporter of this work; we thank her for many helpful discussions. We thank Carla Greenbaum for useful manuscript comments; she and many Benaroya Research Institute (BRI) Diabetes Clinical Research Program members played critical roles in collecting samples distributed through the BRI sample registry and repository. Colin O’Rourke and Jordan Klaiman played key roles in data cleanup and figure editing for the final version of this text. This work was funded by JDRF under grant numbers 17-2013-316 toGTN, 3-SRA-2016-209-Q-R to SAL, 3-SRA-2019-791-S-B to CS, 2-PAR-2015-123-Q-R to GTN, 2-SRA-2015-107-Q-R to EAJ, 2-SRA-2015-122-Q-R to AP, and 1-PNF-2015-113-Q-R to MKL. PSL received funding from the NIH for this work under grant number DP3 DK104465-01. AMP was supported by fellowships from JDRF and the JDRF Canadian Clinical Trial Network. MKL receives a Scientist Salary Award from the BC Children’s Hospital Research Institute. Portions of the research reported in this publication were performed as projects of the ITN and supported by the National Institute of Diabetes and Digestive and Kidney Diseases and the National Institute of Allergy and Infectious Diseases of the NIH under award number UM1AI109565. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Version 1. 10/31/2019
In-Press Preview
Version 2. 12/05/2019
Electronic publication
Footnotes
EW’s present address is: Celgene, Inc., Seattle, Washington, USA.
JMO’s present address is: Gilead Sciences, Inc., Seattle, Washington, USA.
AMP’s present address is: University College London, London, United Kingdom.
Conflict of interest: EW receives a salary from Celgene, Inc. JMO receives a salary from Gilead Sciences, Inc. MKL has received research support from TxCell, Pfizer, and Bristol Myers Squibb and has patents pending related to alloantigen-specific chimeric antigen receptors (PCT/CA2018/051167 and PCT/CA2018/051174). PSL receives research support from Bristol Myers Squibb and is an inventor on patent US5844095A, “CTLA4Ig fusion proteins.” RG has received consulting income from Juno Therapeutics, Takeda, Infotech Soft, and Celgene, Inc., and has received research support from Janssen Pharmaceuticals and Juno Therapeutics.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(23):e126917.https://doi.org/10.1172/jci.insight.126917.
Contributor Information
Cate Speake, Email: CSpeake@benaroyaresearch.org.
Samuel O. Skinner, Email: samueloskinner@gmail.com.
Dror Berel, Email: dror.berel@gmail.com.
Elizabeth Whalen, Email: ewhalen@benaroyaresearch.org.
Matthew J. Dufort, Email: Mdufort@benaroyaresearch.org.
William Chad Young, Email: wyoung@scharp.org.
Jared M. Odegard, Email: jared.odegard@gmail.com.
Anne M. Pesenacker, Email: a.pesenacker@ucl.ac.uk.
Frans K. Gorus, Email: frans.gorus@az.vub.ac.be.
Eddie A. James, Email: ejames@benaroyaresearch.org.
Megan K. Levings, Email: megan.levings@ubc.ca.
Peter S. Linsley, Email: plinsley@benaroyaresearch.org.
Eitan M. Akirav, Email: akiravei@gmail.com.
Alberto Pugliese, Email: apuglies@med.miami.edu.
Martin J. Hessner, Email: mhessner@mcw.edu.
Gerald T. Nepom, Email: jnepom@benaroyaresearch.org.
Raphael Gottardo, Email: rgottard@fhcrc.org.
S. Alice Long, Email: along@benaroyaresearch.org.
References
- 1.Bundy BN, Krischer JP, Type 1 Diabetes TrialNet Study Group A model-based approach to sample size estimation in recent onset type 1 diabetes. Diabetes Metab Res Rev. 2016;32(8):827–834. doi: 10.1002/dmrr.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hao W, Gitelman S, DiMeglio LA, Boulware D, Greenbaum CJ, Type 1 Diabetes TrialNet Study Group Fall in C-peptide during first 4 years from diagnosis of type 1 diabetes: variable relation to age, HbA1c, and insulin dose. Diabetes Care. 2016;39(10):1664–1670. doi: 10.2337/dc16-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker A, et al. Age-dependent decline of β-cell function in type 1 diabetes after diagnosis: a multi-centre longitudinal study. Diabetes Obes Metab. 2014;16(3):262–267. doi: 10.1111/dom.12216. [DOI] [PubMed] [Google Scholar]
- 4.Mallone R, Roep BO. Biomarkers for immune intervention trials in type 1 diabetes. Clin Immunol. 2013;149(3):286–296. doi: 10.1016/j.clim.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Mathieu C, Lahesmaa R, Bonifacio E, Achenbach P, Tree T. Immunological biomarkers for the development and progression of type 1 diabetes. Diabetologia. 2018;61(11):2252–2258. doi: 10.1007/s00125-018-4726-8. [DOI] [PubMed] [Google Scholar]
- 6.Lachin JM, McGee P, Palmer JP, DCCT/EDIC Research Group Impact of C-peptide preservation on metabolic and clinical outcomes in the Diabetes Control and Complications Trial. Diabetes. 2014;63(2):739–748. doi: 10.2337/db13-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arif S, et al. Blood and islet phenotypes indicate immunological heterogeneity in type 1 diabetes. Diabetes. 2014;63(11):3835–3845. doi: 10.2337/db14-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Michels A, Gottlieb P. Pathogenesis of type 1A diabetes. In: De Groot LJ, et al, eds. Endotext. South Dartmouth, Massachusetts, USA: MDText.com; 2000. [Google Scholar]
- 9.Leete P, Mallone R, Richardson SJ, Sosenko JM, Redondo MJ, Evans-Molina C. The effect of age on the progression and severity of type 1 diabetes: potential effects on disease mechanisms. Curr Diab Rep. 2018;18(11):115. doi: 10.1007/s11892-018-1083-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laban S, et al. Heterogeneity of circulating CD8 T-cells specific to islet, neo-antigen and virus in patients with type 1 diabetes mellitus. PLoS ONE. 2018;13(8):e0200818. doi: 10.1371/journal.pone.0200818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bollyky JB, et al. Heterogeneity in recent-onset type 1 diabetes - a clinical trial perspective. Diabetes Metab Res Rev. 2015;31(6):588–594. doi: 10.1002/dmrr.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeo L, et al. Autoreactive T effector memory differentiation mirrors β cell function in type 1 diabetes. J Clin Invest. 2018;128(8):3460–3474. doi: 10.1172/JCI120555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabrera SM, et al. Innate immune activity as a predictor of persistent insulin secretion and association with responsiveness to CTLA4-Ig treatment in recent-onset type 1 diabetes. Diabetologia. 2018;61(11):2356–2370. doi: 10.1007/s00125-018-4708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pesenacker AM, et al. Treg gene signatures predict and measure type 1 diabetes trajectory. JCI Insight. 2019;4(6):123879. doi: 10.1172/jci.insight.123879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dufort MJ, Greenbaum CJ, Speake C, Linsley PS. Cell type-specific immune phenotypes predict loss of insulin secretion in new-onset type 1 diabetes. JCI Insight. 2019;4(4):125556. doi: 10.1172/jci.insight.125556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James EA, et al. Combinatorial detection of autoreactive CD8+ T cells with HLA-A2 multimers: a multi-centre study by the Immunology of Diabetes Society T Cell Workshop. Diabetologia. 2018;61(3):658–670. doi: 10.1007/s00125-017-4508-8. [DOI] [PubMed] [Google Scholar]
- 17.Pesenacker AM, et al. A regulatory T-cell gene signature is a specific and sensitive biomarker to identify children with new-onset type 1 diabetes. Diabetes. 2016;65(4):1031–1039. doi: 10.2337/db15-0572. [DOI] [PubMed] [Google Scholar]
- 18.Van Dalem A, et al. Prediction of impending type 1 diabetes through automated dual-label measurement of proinsulin:C-peptide ratio. PLoS ONE. 2016;11(12):e0166702. doi: 10.1371/journal.pone.0166702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akirav EM, et al. Detection of β cell death in diabetes using differentially methylated circulating DNA. Proc Natl Acad Sci USA. 2011;108(47):19018–19023. doi: 10.1073/pnas.1111008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linsley PS, Greenbaum CJ, Speake C, Long SA, Dufort MJ. B lymphocyte alterations accompany abatacept resistance in new-onset type 1 diabetes. JCI Insight. 2019;4(4):126136. doi: 10.1172/jci.insight.126136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabrera SM, et al. Interleukin-1 antagonism moderates the inflammatory state associated with Type 1 diabetes during clinical trials conducted at disease onset. Eur J Immunol. 2016;46(4):1030–1046. doi: 10.1002/eji.201546005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen YG, et al. Molecular signatures differentiate immune states in type 1 diabetic families. Diabetes. 2014;63(11):3960–3973. doi: 10.2337/db14-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long SA, et al. Partial exhaustion of CD8 T cells and clinical response to teplizumab in new-onset type 1 diabetes. Sci Immunol. 2016;1(5):eaai7793. doi: 10.1126/sciimmunol.aai7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snowhite, Allende G, Sosenko J, Pastori RL, Messinger Cayetano S, Pugliese A. Association of serum microRNAs with islet autoimmunity, disease progression and metabolic impairment in relatives at risk of type 1 diabetes. Diabetologia. 2017;60(8):1409–1422. doi: 10.1007/s00125-017-4294-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gitelman SE, et al. Antithymocyte globulin therapy for patients with recent-onset type 1 diabetes: 2 year results of a randomised trial. Diabetologia. 2016;59(6):1153–1161. doi: 10.1007/s00125-016-3917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rigby MR, et al. Alefacept provides sustained clinical and immunological effects in new-onset type 1 diabetes patients. J Clin Invest. 2015;125(8):3285–3296. doi: 10.1172/JCI81722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herold KC, et al. Teplizumab (anti-CD3 mAb) treatment preserves C-peptide responses in patients with new-onset type 1 diabetes in a randomized controlled trial: metabolic and immunologic features at baseline identify a subgroup of responders. Diabetes. 2013;62(11):3766–3774. doi: 10.2337/db13-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis AK, et al. Prevalence of detectable C-peptide according to age at diagnosis and duration of type 1 diabetes. Diabetes Care. 2015;38(3):476–481. doi: 10.2337/dc14-1952. [DOI] [PubMed] [Google Scholar]
- 29.Oram RA, et al. Most people with long-duration type 1 diabetes in a large population-based study are insulin microsecretors. Diabetes Care. 2015;38(2):323–328. doi: 10.2337/dc14-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linsley PS, Greenbaum CJ, Rosasco M, Presnell S, Herold KC, Dufort MJ. Elevated T cell levels in peripheral blood predict poor clinical response following rituximab treatment in new-onset type 1 diabetes. Genes Immun. 2019;20(4):293–307. doi: 10.1038/s41435-018-0032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Pauw PE, et al. Simultaneous measurement of plasma concentrations of proinsulin and C-peptide and their ratio with a trefoil-type time-resolved fluorescence immunoassay. Clin Chem. 2008;54(12):1990–1998. doi: 10.1373/clinchem.2008.109710. [DOI] [PubMed] [Google Scholar]
- 32.Linsley PS, Chaussabel D, Speake C. The relationship of immune cell signatures to patient survival varies within and between tumor types. PLoS ONE. 2015;10(9):e0138726. doi: 10.1371/journal.pone.0138726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long SA, et al. Remodeling T cell compartments during anti-CD3 immunotherapy of type 1 diabetes. Cell Immunol. 2017;319:3–9. doi: 10.1016/j.cellimm.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tibshirani R. Regression shrinkage and selection via the Lasso. Journal of the Royal Statistical Society, Series B (Methodological) 1996;58(1):267–288. doi: 10.1111/j.2517-6161.1996.tb02080.x. [DOI] [Google Scholar]
- 35.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 36.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 37.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366(14):1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Speake C, Odegard JM. Evaluation of candidate biomarkers of type 1 diabetes via the core for assay validation. Biomark Insights. 2015;10(Suppl 4):19–24. doi: 10.4137/BMI.S29697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivison S, et al. A standardized immune phenotyping and automated data analysis platform for multicenter biomarker studies. JCI Insight. 2018;3(23):121867. doi: 10.1172/jci.insight.121867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaczorowski KJ, et al. Continuous immunotypes describe human immune variation and predict diverse responses. Proc Natl Acad Sci USA. 2017;114(30):E6097–E6106. doi: 10.1073/pnas.1705065114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bischl B, et al. mlr: Machine Learning in R. Journal of Machine Learning Research. 2016;17(170):1–5. [Google Scholar]
- 42.Ahmed S, et al. Standardizing T-cell biomarkers in type 1 diabetes: challenges and recent advances. Diabetes. 2019;68(7):1366–1379. doi: 10.2337/db19-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long SA, Buckner JH. CD4+FOXP3+ T regulatory cells in human autoimmunity: more than a numbers game. J Immunol. 2011;187(5):2061–2066. doi: 10.4049/jimmunol.1003224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenbaum CJ, et al. Fall in C-peptide during first 2 years from diagnosis: evidence of at least 2 distinct phases from composite Type 1 Diabetes TrialNet data. Diabetes. 2012;61(8):2066–2073. doi: 10.2337/db11-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ludvigsson J, et al. Decline of C-peptide during the first year after diagnosis of Type 1 diabetes in children and adolescents. Diabetes Res Clin Pract. 2013;100(2):203–209. doi: 10.1016/j.diabres.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Ziegler AG, et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA. 2013;309(23):2473–2479. doi: 10.1001/jama.2013.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wherrett DK, et al. Defining pathways for development of disease-modifying therapies in children with type 1 diabetes: a consensus report. Diabetes Care. 2015;38(10):1975–1985. doi: 10.2337/dc15-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steck AK, et al. Predictors of progression from the appearance of islet autoantibodies to early childhood diabetes: The Environmental Determinants of Diabetes in the Young (TEDDY) Diabetes Care. 2015;38(5):808–813. doi: 10.2337/dc14-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meah FA, et al. The relationship between BMI and insulin resistance and progression from single to multiple autoantibody positivity and type 1 diabetes among TrialNet Pathway to Prevention participants. Diabetologia. 2016;59(6):1186–1195. doi: 10.1007/s00125-016-3924-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferrara CT, et al. The role of age and excess body mass index in progression to type 1 diabetes in at-risk adults. J Clin Endocrinol Metab. 2017;102(12):4596–4603. doi: 10.1210/jc.2017-01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferrara CT, et al. Excess BMI in childhood: a modifiable risk factor for type 1 diabetes development? Diabetes Care. 2017;40(5):698–701. doi: 10.2337/dc16-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J, et al. Prevalence of obesity was related to HLA-DQ in 2-4-year-old children at genetic risk for type 1 diabetes. Int J Obes (Lond) 2014;38(12):1491–1496. doi: 10.1038/ijo.2014.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winkler C, Marienfeld S, Zwilling M, Bonifacio E, Ziegler AG. Is islet autoimmunity related to insulin sensitivity or body weight in children of parents with type 1 diabetes? Diabetologia. 2009;52(10):2072–2078. doi: 10.1007/s00125-009-1461-1. [DOI] [PubMed] [Google Scholar]
- 54.Lauria A, et al. BMI is an important driver of β-cell loss in type 1 diabetes upon diagnosis in 10 to 18-year-old children. Eur J Endocrinol. 2015;172(2):107–113. doi: 10.1530/EJE-14-0522. [DOI] [PubMed] [Google Scholar]
- 55.Mølbak AG, Christau B, Marner B, Borch-Johnsen K, Nerup J. Incidence of insulin-dependent diabetes mellitus in age groups over 30 years in Denmark. Diabet Med. 1994;11(7):650–655. doi: 10.1111/j.1464-5491.1994.tb00327.x. [DOI] [PubMed] [Google Scholar]
- 56.Vandewalle CL, et al. Epidemiology, clinical aspects, and biology of IDDM patients under age 40 years. Comparison of data from Antwerp with complete ascertainment with data from Belgium with 40% ascertainment. The Belgian Diabetes Registry. Diabetes Care. 1997;20(10):1556–1561. doi: 10.2337/diacare.20.10.1556. [DOI] [PubMed] [Google Scholar]
- 57.Kyvik KO, et al. The epidemiology of Type 1 diabetes mellitus is not the same in young adults as in children. Diabetologia. 2004;47(3):377–384. doi: 10.1007/s00125-004-1331-9. [DOI] [PubMed] [Google Scholar]
- 58.Asare AL, Carey VJ, Rotrosen D, Nepom GT. Clinical trial data access: opening doors with TrialShare. J Allergy Clin Immunol. 2016;138(3):724–726. doi: 10.1016/j.jaci.2016.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Linsley PS, Speake C, Whalen E, Chaussabel D. Copy number loss of the interferon gene cluster in melanomas is linked to reduced T cell infiltrate and poor patient prognosis. PLoS ONE. 2014;9(10):e109760. doi: 10.1371/journal.pone.0109760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. R Core Team. R: A language and environment for statistical computing. http://www.R-project.org Accessed November 5, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code for the analytical tool is available on GitHub at https://github.com/FredHutch/JDRFCAV using branch name “master” and commit ID 905178c, along with data necessary to recreate the analyses presented here. Whole-genome data (cell subset RNA-Seq and transcriptional response to serum assays, as well as all replicate testing data) are available in the NCBI GEO database under accession number GSE131528. Whole-blood RNA-Seq data are available in NCBI GEO under accession number GSE124400.