Abstract
Dietary salt consumption leads to cutaneous Na+ storage and is associated with various disorders, including osteopenia. Here, we explore the impact of Na+ and the osmoprotective transcription factor nuclear factor of activated T cell 5 (NFAT5) on bone density and osteoclastogenesis. Compared with treatment of mice with high-salt diet, low-salt diet (LSD) increased bone density, decreased osteoclast numbers, and elevated Na+ content and Nfat5 levels in the BM. This response to LSD was dependent on NFAT5 expressed in myeloid cells. Simulating in vivo findings, we exposed osteoclast precursors and osteoblasts to elevated Na+ content (high-salt conditions; HS¢), resulting in increased NFAT5 binding to the promotor region of RANKL decoy receptor osteoprotegerin (OPG). These data not only demonstrate that NFAT5 in myeloid cells determines the Na+ content in BM, but that NFAT5 is able to govern the expression of the osteoprotective gene OPG. This provides insights into mechanisms of Na+-induced cessation of osteoclastogenesis and offers potentially new targets for treating salt-induced osteopenia.
Keywords: Bone Biology
Keywords: Bone development, Bone marrow
NFAT5 in myeloid cells determines the Na+ content in bone marrow and govern the expression of the osteoprotective gene osteoprotegerin.
Introduction
Bones are essential for moving, protecting organs, supporting hematopoiesis, sustaining brain and lung function, and storing minerals. Bone is a vascularized, living, ever-changing, mineralized connective tissue. To ensure bone stability and integrity, about 10% of the bone material is renewed every year (1–3). Bone remodeling is a complex process characterized by the interaction of bone-forming osteoblasts and bone-resorbing osteoclasts (1, 4). During physiological bone remodeling, bone formation and resorption are strictly coupled to avoid any change in bone quality or mass. Many pathological conditions, like osteoporosis, are associated with enhanced bone resorption compared with formation (5, 6). While osteoblasts are derived from mesenchymal stem cells (7), bone osteoclasts differentiate from hematopoietic stem cells (8) induced by the 2 essential factors macrophage CSF (M-CSF) and RANKL (8). RANKL acts as the primary factor that promotes the differentiation of osteoclasts precursor cells to active, bone-resorbing osteoclasts (9, 10). The activity of RANKL is controlled by its decoy receptor osteoprotegerin (OPG) (11–13). The interaction of RANKL and OPG is essential for the control of osteoclastogenesis (14–16).
Contemporary Western diets contain a superfluous amount of Na+ (17–19). Excessive dietary Na+ intake is, for example, linked with hypertension (19, 20) and osteopenia (21). Dietary increases in Na+ consumption induce various physiological responses (22). These include cutaneous Na+ storage and induction of a macrophage-driven cutaneous response, which facilitates Na+ mobilization from the skin to avoid excess increases in blood pressure (23–25). This critically involves the activity of the osmoprotective transcription factor nuclear factor of activated T cell 5 (NFAT5, also known as tonicity-dependent enhancer binding protein [TonEBP]), which can be induced by osmotic stress in a calcineurin-independent manner (26–28)
Surprisingly, both hyponatremia and Na+-rich diets are linked to osteopenia (21, 29–37). Osteoclasts, which are derived from mononuclear phagocytes, play a major role in bone resorption and remodeling (1, 2). Previous studies demonstrate that Na+ availability influences osteoclastogenesis (38, 39). The impact of Na+-rich diets on BM Na+ content and the role of NFAT5-driven osmoprotective responses in osteoclastogenesis is, however, unexplored.
Results
Myeloid cell–derived NFAT5 prevents bone loss upon a low-salt diet.
We fed myeloid cell–specific conditional Nfat5-KO mice (Nfat5Δmyel) and control mice either a low-salt diet (LSD) or a high salt diet (HSD) for 2 weeks. Control mice kept under LSD displayed increased bone volume/total volume ratio (BV/TV) compared with HSD-treated control mice, indicating enhanced bone density (Figure 1, A and B). Surprisingly, Nfat5Δmyel mice did not display an increased BV/TV ratio after LSD treatment (Figure 1, A and B). We detected no changes in osteoblast numbers neither in control nor in Nfat5Δmyel mice (Figure 1C), while the number of osteoclasts was reduced in control mice kept on LSD (Figure 1D). In contrast, osteoclast numbers did not change in Nfat5Δmyel mice upon LSD (Figure 1D). In line with that, we detected decreased serum levels of TRACP-5b (Figure 1E) and β-CrossLaps (β-CTx) (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.127868DS1) in control, but not in Nfat5Δmyel mice, with LSD. The RANKL-OPG axis plays a key role in osteoclastogenesis and bone density (16, 40). LSD did not affect RANKL expression in the BM of control and Nfat5Δmyel mice (Figure 1F). In contrast, LSD increased the expression of the RANKL decoy receptor Opg mRNA only in control mice (Figure 1G). As reported earlier (23), we noted an increased cutaneous Na+ accumulation upon HSD (Supplemental Figure 2). In order to obtain BM, we flushed the femora of mice with distilled water. Detailed assessment of electrolyte content in BM, however, revealed no changes in Cl– or K+ content in BM of control and Nfat5Δmyel mice (Supplemental Figure 3, A and B). In contrast, LSD enhanced Na+ content (Figure 1H) and osmolality (Supplemental Figure 3C) in the BM homogenate of control mice but not in Nfat5Δmyel mice. In line with these results, we detected an enhanced expression of the osmoprotective transcription factor Nfat5 only in control mice exposed to a LSD (Figure 1I). From these findings, we conclude that (A) HSD does not uniformly result in Na+ accumulation in all organs, (b) myeloid cell–derived NFAT5 is important for Na+ accumulation in BM, and (c) myeloid cell–derived NFAT5 is required for increased bone density and low osteoclast numbers in animals fed LSD.
Figure 1. Myeloid cell–derived NFAT5 prevents bone loss upon a low-salt diet.
(A) Representative pictures of μCT analysis of HSD- and LSD-fed mice. Analyzed region of interest is colored in red. (B) Analysis of bone to total volume ratio (BV/TV). (C and D) Osteoblast (C) and osteoclast (D) numbers per bone perimeter (B. Pm). (E) TRACP-5b levels in serum. (F and G) Rankl (F) and Opg (G) gene expression in BM. (H and I) Na+ content (H) and Nfat5 (I) mRNA expression in BM. n = 6 for each group. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Unpaired, 2-tailed Student’s t tests.
Increased osmolality due to high-salt conditions (HS¢) fully incapacitates osteoclastogenesis.
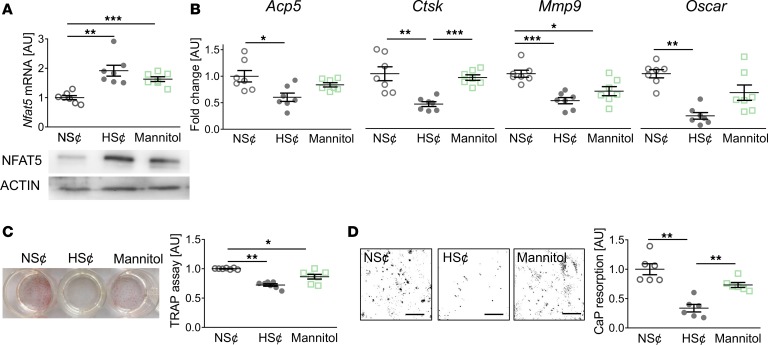
To further assess the role of increased osmolality and Na+ content on osteoclastogenesis, we exposed RANKL/M-CSF–treated WT RAW264.7 cells to either an increase of 40 mM NaCl (HS¢) or 80 mM mannitol. In contrast to NaCl, mannitol represents a nonionic osmolyte that is known to increase tonicity but does not penetrate the cell membrane (41, 42). Exposure to HS¢ or mannitol increased Nfat5 levels in RANKL/M-CSF–treated WT RAW264.7 cells (Figure 2A) and BM-derived macrophages (Supplemental Figure 4). Of note, LDH assays indicated that increases in Na+ are not cytotoxic (Supplemental Figure 5). HS¢ blunted the expression of various osteoclast-specific genes, such as acid phosphatase 5 (Acp5), cathepsin K (Ctsk), matrix metalloproteinase 9 (Mmp9), and osteoclast-associated immunoglobulin-like receptor (Oscar) (Figure 2B). Mannitol, in contrast, only affected Mmp9 gene expression significantly (Figure 2B). However, increases of osmolality by both addition of Na+ or mannitol reduced TRAP staining (Figure 2C). Nonetheless, calcium phosphate (CaP) resorption was only significantly impaired by HS¢ (Figure 2D). These data demonstrate that, although exposure to mannitol is able to blunt osteoclastogenesis, only increases in osmolality with Na+ fully incapacitate osteoclastogenesis.
Figure 2. Increased osmolality due to high salt (HS¢) fully incapacitates osteoclastogenesis.
(A) Nfat5 mRNA expression. Representative NFAT5 immunoblot. (B) Expression of osteoclast-specific genes. (C) Representative TRAP staining and TRAP assay of cell culture supernatants. (D) Representative images of CaP resorption assay. Resorbed CaP areas appear as black gaps. Quantification of CaP resorption assay using ImageJ. n = 6 for each group. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Welch-corrected ANOVA with Games-Howell post hoc tests. Scale bar: 100 µm.
Nfat5 overexpression prevented osteoclastogenesis of RANKL/M-CSF–treated RAW264.7 cells.
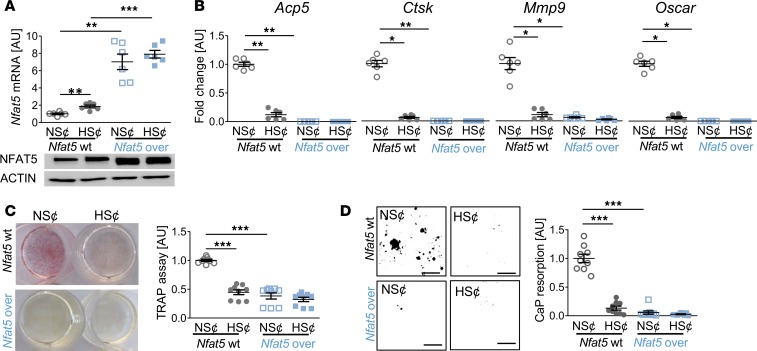
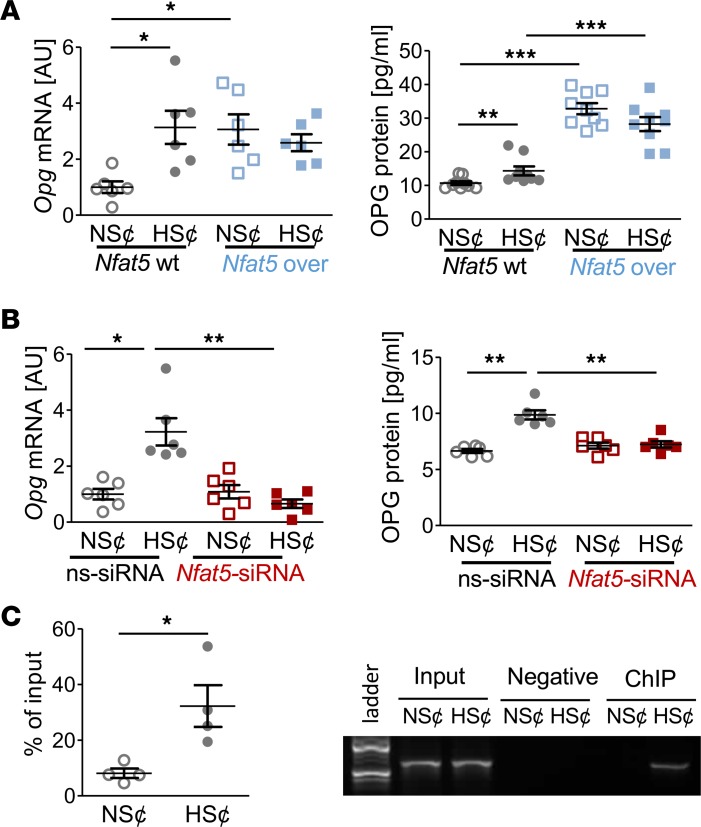
To further assess the role of NFAT5 in this state of affairs, we tested whether constitutive overexpression of Nfat5 in RAW264.7 cells (Nfat5-over) under normal salt conditions (NS¢) (Figure 3A) is sufficient to disturb osteoclast differentiation. We found that Nfat5 overexpression abolished the expression of osteoclast-specific genes (Figure 3B) and impaired TRAP staining (Figure 3C) and CaP resorption (Figure 3D) upon exposure to RANKL/M-CSF. Moreover, HS¢ did not further impair osteoclastogenesis in Nfat5-over cells (Figure 3, B–D). These data indicate that Nfat5 overexpression is sufficient to impair RANKL/M-CSF–driven osteoclastogenesis.
Figure 3. Nfat5 overexpression prevented osteoclastogenesis of RANKL/M-CSF–treated RAW264.7 cells.
(A) Nfat5 mRNA expression in RAW264.7 cells without (Nfat5-WT) or with Nfat5 overexpression (Nfat5-over, n = 6). Representative NFAT5 immunoblot. (B) Expression of osteoclast-specific genes in Nfat5-WT or Nfat5-over cells (n = 6). (C) Representative TRAP staining (red) of Nfat5-WT or Nfat5-over cells and TRAP assay of the supernatants (n = 9). (D) Representative images of CaP resorption assay. Resorbed CaP areas appear as black gaps. Quantification of CaP resorption assay using ImageJ (n = 9). AU, arbitrary units. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Welch-corrected ANOVA with Games-Howell post hoc tests. Scale bar: 100 µm.
Nfat5 siRNA treatment restored osteoclastogenesis of RANKL/M-CSF–treated RAW264.7 cells under HS¢.
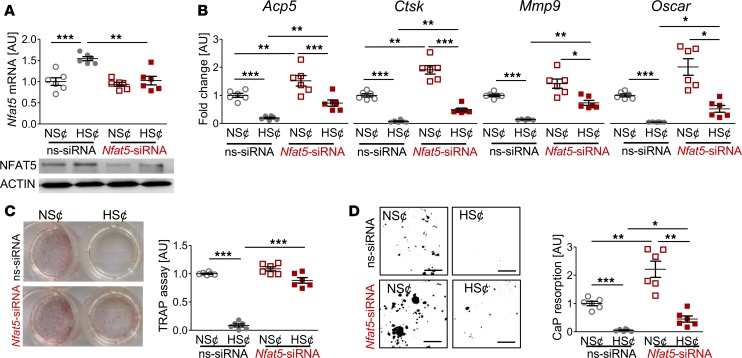
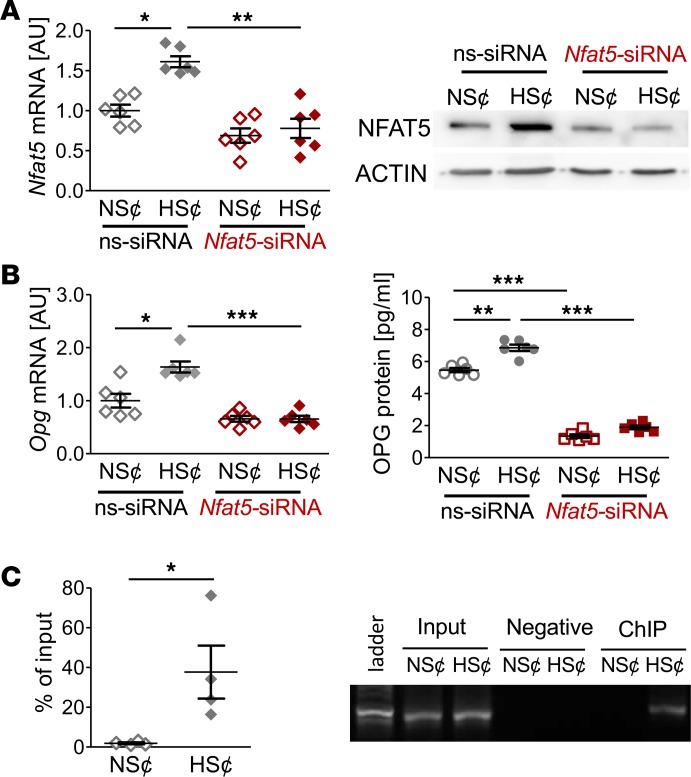
To test the contribution of Nfat5 in HS¢, we silenced Nfat5 expression in RAW264.7 cells (Figure 4A). This alleviated the HS-induced blockade of osteoclast-specific gene expression (Figure 4B). Furthermore, in Nfat5-silenced cells, the suppressive action of HS¢ on TRAP staining was largely abolished (Figure 4C). Moreover, Nfat5-deficient cells displayed higher CaP resorption than control cells upon incubation in HS¢ (Figure 4D). Nevertheless, even in the absence of Nfat5 in macrophages, HS¢ exerted substantial impairment on CaP resorption. Of note, Nfat5 siRNA transfection tended to increase the TRAP level under NS¢ (Figure 4D). More importantly, Nfat5 deficiency was accompanied by significantly enhanced CaP resorption, even in NS¢ in vitro (Figure 4D). These data demonstrate that NFAT5 is critically involved in impaired osteoclastogenesis under HS¢.
Figure 4. Nfat5 siRNA treatment restored osteoclastogenesis of RANKL/M-CSF–treated RAW264.7 cells under HS¢.
(A) Nfat5 mRNA expression in RAW264.7 cells treated with ns-siRNA or Nfat5 siRNA. Representative NFAT5 immunoblot of ns-siRNA– or Nfat5 siRNA–treated samples. (B) Expression of osteoclast-specific genes in ns-siRNA– or Nfat5 siRNA–treated RAW264.7 cells. (C) Representative TRAP staining of ns-siRNA– or Nfat5 siRNA–treated RAW264.7 cells and TRAP assay of the supernatants. (D) Representative pictures of CaP resorption assay. Resorbed CaP areas appear as black gaps. Quantification of CaP resorption assay using ImageJ. n = 6 for each group. AU, arbitrary units. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Welch-corrected ANOVA with Games-Howell post hoc tests. Scale bar: 100 µm.
NFAT5 governs OPG expression under HS¢ in RANKL/M-CSF–treated RAW264.7 cells.
Since diet-dependent Opg expression in BM hinged on Nfat5 expression in myeloid cells (Figure 1G), we quantified OPG levels in cell culture supernatants. HS¢ boosted OPG gene and protein expression in RANKL/M-CSF–treated RAW264.7 cells (Figure 5A). Likewise, Nfat5 overexpression enhanced OPG expression on mRNA and protein level in both NS¢ and HS¢ (Figure 5A), and Nfat5 silencing truncated HS¢-induced OPG on mRNA and protein level (Figure 5B). The Opg promotor region contains 3 putative NFAT5 binding sites (Supplemental Figure 6). ChIP analysis revealed binding of NFAT5 to these promoter regions upon exposure to HS¢ in RANKL/M-CSF–treated RAW264.7 cells (Figure 5C). These findings establish that OPG is an NFAT5 target gene in these cells.
Figure 5. NFAT5 governs OPG expression under HS¢ in RANKL/M-CSF–treated RAW264.7 cells.
(A) OPG mRNA (n = 6) and protein secretion (n = 8) in Nfat5-WT or Nfat5-overexpressing RAW264.7 cells (Nfat5-over). (B) OPG mRNA and protein expression in ns-siRNA– or Nfat5 siRNA–treated RAW264.7 cells (n = 6). (C) ChIP analysis of interaction between NFAT5 and Opg promotor in RAW264.7 cells under NS¢ or HS¢ (n = 4). AU, arbitrary units. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Welch-corrected ANOVA with Games-Howell post hoc tests, except C (unpaired, 2-tailed Student’s t test).
NFAT5 regulates OPG expression under HS¢ in murine osteoblasts.
Since osteoblasts are the main source of OPG, we also tested the impact of NFAT5. As we found in RANKL/M-CSF–treated RAW264.7 cells, murine osteoblasts subjected to HS¢ increased NFAT5 and OPG expression (Figure 6, A and B). Again, silencing of NFAT5 was associated with reduced OPG expression in osteoblasts exposed to HS¢ (Figure 6, A and B). ChIP analysis revealed that NFAT5 is able to bind to the OPG promoter upon exposure to HS¢ in osteoblasts, as well (Figure 6C). From these findings, we conclude that NFAT5 governs HS¢-triggered OPG expression in osteoblasts.
Figure 6. NFAT5 regulates OPG expression under HS¢ in murine osteoblasts.
NFAT5 influences OPG expression under high-salt conditions (HS¢) in murine osteoblasts. (A) NFAT5 mRNA and protein expression in ns-siRNA– or Nfat5 siRNA–treated osteoblasts under NS¢ or HS¢ (n = 6). Representative NFAT5 immunoblot of ns-siRNA– or Nfat5 siRNA–treated samples under NS¢ or HS¢. (B) OPG mRNA and protein expression in ns-siRNA– or Nfat5 siRNA–treated osteoblasts under NS¢ or HS¢ (n = 6 per group). (C) ChIP analysis of interaction between NFAT5 and Opg promotor in osteoblasts under NS¢ or HS¢ (n = 4). AU, arbitrary units. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Welch-corrected ANOVA with Games-Howell post hoc tests, except C (unpaired, 2-tailed Student’s t test).
Discussion
In accordance with earlier studies, we show that disturbances in the Na+ balance are linked with osteopenia and reduced bone density in various studies (21, 30–32, 37).
In line with previous studies, we found that HSD resulted in enhanced skin Na+ content (24, 25). Surprisingly, we found that LSD resulted in increased Na+ content in the BM. Of note, this response is confined to the BM, while total bone Na+ content remained unaffected, as demonstrated earlier (43). Our findings substantiate the notion that Na+ contents are not evenly distributed in the body and may vary locally, depending on dietary and other environmental challenges such as inflammation or infection (23–25, 44–46). In line with this idea, recent studies demonstrate that HSD not only induces Na+ storage in the skin, but results in a complete reorganization of body metabolism (18, 44).
Expression of the osmoprotective transcription NFAT5 in myeloid cells regulates local electrolyte content in the skin (23, 24). Here, we demonstrate that expression of this transcription factor in myeloid cells is required for Na+ accumulation in the BM. How LSD triggers increased Nfat5 expression in the BM is unclear. Moreover, the mechanism employed by NFAT5 in myeloid cells to increase the Na+ content in the BM upon LSD remains elusive. Both issues warrant further exploration. It is tempting to speculate that the renin/angiotensin/aldosterone axis might be involved, as well (47, 48).
Local Na+ contents are known to impact homeostatic and inflammatory innate myeloid cell function (24, 49). For instance, exposure of macrophages to HS¢ impaired the regulatory, antiinflammatory activity of macrophages (41), while increases in local Na+ enhanced their antimicrobial activity. Increases in Na+ have been demonstrated to help fight against the protozoan parasite Leishmania major (45) and against the bacterial pathogen E. coli (42, 50). Recent evidence also suggests that elevation in Na+ facilitates antiviral responses against vesicle stomatitis virus (51). In this report, we focused on the impact of this Na+ microenvironment on the cessation of osteoclastogenesis. It is conceivable that Na+ directly influences bone resorbing activity of fully differentiated osteoclasts, in addition, since Na+/H+ exchanger activity (52–55), Na+/Ca2+ exchanger activity (56–58), and Na+-dependent phosphate transport (59) are involved in the functionality of the resorptive hemivacuole. In addition, local Na+ content might affect the activity of the Na+/K+-ATPase (60, 61), which is required for secondary ion transport, for example by the Na+/Ca2+ and Na+/H+ exchangers.
Here, we observed that increases in local Na+ by 40 mM truncated osteoclast differentiation. This is in line with Wu et al., who found that excesses of 50 mM Na+ inhibited osteoclastogenesis of murine osteoclast progenitor cells (38). In contrast, when less than 50 mM of Na+ was added, Wu et al. noted enhanced osteoclastogenesis (38). Moreover, low Na+ content reportedly promote osteoclastogenesis (39). Therefore, it is conceivable that low Na+ content is able to facilitate osteoclastogenesis, whereas high Na+ content impairs osteoclastogenesis. The mechanisms linked to enhanced osteoclastogenesis upon low Na+ exposure remain, however, unknown.
We found that increases of Na+ by 40 mM were paralleled by increased expression of the RANKL decoy receptor (2, 62) OPG in osteoclast-precursor cells and osteoblasts upon exposure of cells to HS¢. With increased Na+ content, the osmoprotective transcription factor NFAT5 binds to the OPG promotor region in both cell types in vitro. Moreover, increased NFAT5 levels due to enhanced Na+ content or conditional NFAT5 expression resulted in OPG upregulation. These findings suggest that OPG is a potentially novel osmoprotective target gene and is able to regulate bone homeostasis, dependent on local Na+ content in the BM. This expands the regulatory repertoire governing OPG expression, which includes WNT- and IL-1–dependent signaling (63, 64). Although the contribution of OPG in this situation in vivo requires further detailed investigation, it is very likely that local increases in Na+ content in the BM trigger OPG expression primarily in osteoblasts, since these cells are known to be the major source of OPG in vivo (16, 40).
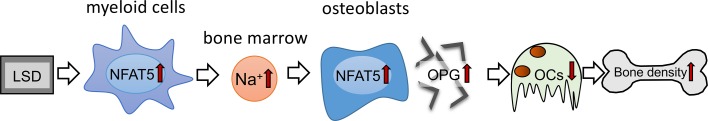
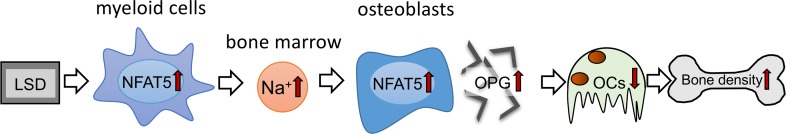
In summary, our data suggest that diet-dependent alterations of local Na+ content in BM impact on osteoclastogenesis and bone density. Surprisingly, LSD increased Na+ content, specifically in BM affected by myeloid cell–derived NFAT5. This local Na+ accumulation in BM of LSD-treated mice blocks RANKL-induced osteoclastogenesis by upregulating the expression of the RANKL decoy receptor OPG in an NFAT5-dependent manner (Figure 7). Our data suggest that favoring Na+ accumulation and OPG expression in BM via increasing NFAT5 activity is a potentially new method of fighting osteopenia. Our work provides insights into the mechanisms of salt-induced cessation of osteoclastogenesis and offers avenues to explain salt-induced osteopenia.
Figure 7. Schematic representation of the action of myeloid-derived NFAT5 in BM upon dietary challenges.
NFAT5 performs a dual function in bone homeostasis. On the one hand, myeloid cell–derived NFAT5 is critically involved in regulation of Na+ content in the BM, and on the other hand, NFAT5 directly controls osteoclastogenesis by binding to the OPG promotor and enhancing the expression of this osteoprotective gene.
Methods
Supplemental Methods are available online with this article.
Animal experiments.
A total of 12 male LysMWTNfat5fl/fl (control) and 12 LysMCreNfat5fl/fl (Nfat5Δmyel) mice were either kept on a LSD (<0.1% NaCl chow [ssniff-Spezialdiäten] and tap water) or a HSD (4% NaCl chow [ssniff-Spezialdiäten] plus 0.9% saline to drink) for 2 weeks and euthanized at the age of about 22 weeks (n = 6 of each group). We analyzed bone morphogenic parameters of tibia (n = 6 of each group) via μCT and histomorphometry. Additionally, we isolated RNA from BM and analyzed Nfat5, Rankl, and Opg gene expression (n = 6 of each group). We also measured TRACP-5b and β-CTx levels in serum and assessed electrolyte content in BM (n = 6 of each group).
Nfat5-WT and Nfat5-over cells.
RAW264.7 WT macrophages (Nfat5-WT) were obtained from Cell Lines Service (400319). We additionally used Nfat5 overexpressing RAW264.7 macrophages (Nfat5-over), as described earlier (24).
Nfat5-WT and Nfat5-over cells were seeded onto 12-well cell culture plates (10,000 cells) and cultured in 1 mL α-MEM (F0925, Biochrom), supplemented with 10% FBS (P30-3306, PAN-Biotech), 1% L-glutamine (SH30034.01, GE Healthcare), and 1% antibiotics/antimycotics (A5955, Sigma-Aldrich). Induction of osteoclastogenesis was performed by adding 30 ng/mL M-CSF (576404, BioLegend) and 50 ng/mL RANKL (577102, BioLegend) at day 0 under NS¢ or under HS¢ by adding 40 mM NaCl for at least 5 days. After that time, we analyzed mRNA, protein, TRAP, and CaP resorption.
Murine osteoblast-like MC3T3-E1 cells.
Murine osteoblast like MC3T3-E1 (Sigma-Aldrich) were seeded onto 12-well cell culture plates (100,000 cells) and cultured in 1 mL α-MEM (F0925, Biochrom), supplemented with 10% FBS (P30-3306, PAN-Biotech), 1% L-glutamine (SH30034.01, GE Healthcare), and 1% antibiotics/antimycotics (A5955, Sigma-Aldrich). After 24 preincubation cells, were either kept under NS¢ or HS¢ by adding 40 mM NaCl for at least another 24 hours. After that time, we analyzed mRNA and protein expression.
siRNA transfection.
Nonsilencing siRNA (ns-siRNA) oligonucleotides were purchased from Qiagen. siRNA nucleotides directed against Nfat5 were purchased from Dharmacon’s prevalidated siRNA database (L 058868). Transfer of siRNA duplexes was performed as described previously (65). Briefly, we transferred siRNA duplexes to a 4-mm cuvette (Molecular Bioproducts) and filled up to a volume of 50 μL. We added 50 μL of a cell suspension (containing 2 × 106 RAW264.7 macrophages) resolved in OPTI-MEM and pulsed in a GenePulser Xcell (Bio-Rad). Pulse conditions were 400 V, 150 μF, and 100 Ω. Using a fluorescein-labeled ns-siRNA, we routinely observed a transfection efficiency of over 90% (data not shown). After electroporation, we differentiated cells in α-MEM (supplemented as indicated in the section cell culture models) under NS¢ or HS¢ (by adding 40 mM NaCl) for 5 days. After that time, we analyzed RNA, protein, TRAP, and CaP resorption.
Isolation and purity assessment of total RNA.
We extracted total RNA by applying 0.5 mL peqGOLD TriFast (PEQLAB Biotechnology GmbH) per well and further processing according to the manufacturer’s instructions. We eluted the obtained RNA pellet in 20 μL nuclease-free water (T143, Bioscience-Grade, Carl Roth GmbH & Co.) and immediately cooled on ice. The used extraction protocol ensured good RNA integrity (RIN, 28S/18S ratio), as well as absence of genomic DNA and contamination, as shown before. For purity assessment and quantification of the eluted total RNA, OD was photometrically measured at 280 nm, 260 nm, and 230 nm (Implen).
Reverse transcription.
For cDNA synthesis, we transcribed a standardized amount of 100 ng RNA per sample using 0.1 nmol of an oligo(dT)18 primer (1 μL, SO131, Thermo Fisher Scientific), 0.1 nmol of random hexamer primers (1 μL, SO142, Invitrogen), 40 nmol dNTP mix (1 μL, 10 nmol/dNTP, Roti-Mix (PCR3, L785.2; Carl Roth), 4 μL 5× M-MLV buffer (M1705, Promega), 40 U (1 μL) of an RNase inhibitor (EO0381, Invitrogen), 200 U (1 μL) reverse transcriptase (M1705, Promega), and 20 μL nuclease-free H2O (T143, Carl Roth). We incubated the samples for 60 minutes at 37°C. After heat inactivation of reverse transcriptase (95°C, 2 minutes), the first-strand cDNA was stored until use at −20°C. cDNA synthesis was performed for all samples at the same time to minimize experimental variations. For quantitative PCR (qPCR), we diluted cDNA 1:5 with nuclease-free H2O (T143, Carl Roth GmbH & Co. KG).
qPCR.
We performed qPCR with the Mastercycler ep realplex-S thermocycler (Eppendorf AG) and 96-well PCR plates (TW-MT, 712282, Biozym Scientific GmbH), combined with BZO Seal Filmcover sheeting (712350, Biozym Scientific GmbH). Each reaction mix contained 7.5 μL SYBR Green JumpStart Taq ReadyMix (Sigma–Aldrich, S4438), as well as 7.5 pmol (0.75 μL) of the respective primer pair (3.75 pmol/primer), 1.5 μL of the respective cDNA solution (dilution 1:5), and 15 μL nuclease-free H2O (BioScience Grade T143, Carl Roth GmbH & Co. KG). To avoid technical errors due to manual pipetting, all components except the cDNA solution were prepared as a master mix. cDNA amplification was performed in 45 cycles (initial heat activation 95°C/5 minutes per cycle at 95°C/10-second denaturation, 60°C/8-second annealing, and 72°C/8-second extension) in duplets for each gene and biological sample. SYBR Green I fluorescence was quantified at 521 nm at the end of each extension step. Cycle quantification (Cq) values were determined as a second derivative maximum of the fluorescence signal curve using the software realplex (version 2.2, Eppendorf AG, CalqPlex algorithm, automatic baseline, drift correction on), and the arithmetic mean of each Cq duplet per gene and sample was used for further analysis. For normalization of target genes (relative gene expression), we used a set of 2 reference genes (Hprt/Tbp), which have been shown to be stably expressed in RAW264.7 cells under the conditions investigated. Relative gene expression was calculated as 2–ΔCq (66) with ΔCq = Cq (target gene) – Cq (mean Hprt/Tbp), divided by the respective arithmetic 2–ΔCq mean of the untreated controls at each time point to set their relative gene expression to 1 and used for statistical analysis.
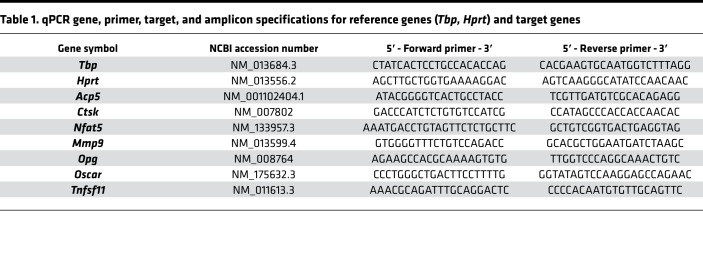
All intron-flanking, gene-specific primers (Table 1) were constructed according to MIQE quality guidelines (67) using NCBI PrimerBLAST and additional software (BeaconDesigner Free Edition [Premier BioSoft International] and UNAFold [Integrated DNA Technologies Inc.]), considering absence of dimers and secondary structures at annealing temperature. The unmodified primers were synthesized and purified by Eurofins MWG Operon LLC (High Purity Salt Free Purification HPSF). For each primer pair and qPCR run, a no-template control (NTC) without cDNA was tested to assess a possible bias in results by primer dimers or contaminating DNA.
Table 1. qPCR gene, primer, target, and amplicon specifications for reference genes (Tbp, Hprt) and target genes.
qPCR specificity was validated as described before using melting curve analysis and agarose gel electrophoresis (68).
Immunoblotting.
We washed cells 3 times with PBS and incubated in 8 M urea for 10 minutes on ice. Afterward, we removed the cells from the plate and centrifuged for 10 minutes at 16,200 g. We used the supernatant for immunoblot analysis. For immunoblotting, we separated equal amounts of total protein (20 μg) on 8% SDS-polyacrylamide gels under reducing conditions and electroblotted onto a polyvinylidene diflouride (PVDF) membrane. We blocked the blots with 5% nonfat milk in phosphate-buffered saline (PBS) and 0.1% Tween 20 (9127.1, Carl Roth), pH 7.5, for 1 hour at room temperature and then incubated overnight at 4°C with anti-NFAT5 (PA1-203, Thermo Fisher Scientific) diluted 1:1,000 or anti–β-actin (E1C602, EnoGene) diluted 1:3,000. After 3 washes in PBS with 0.1% Tween 20, pH7.5, we incubated the blots for 1 hour with horseradish peroxidase–conjugated anti-rabbit IgG (611-1302, Rockland) diluted 1:2,000 in blocking solution at room temperature. We visualized antibody binding using an enhanced chemiluminescence system (Pierce).
TRAP staining of cells and TRAP assay.
Histochemical TRAP staining was used to detect differentiated osteoclast-like cells after 5 days on NS or HS medium. We prepared fresh TRAP staining solution for each analysis by dissolving 0.3 mg Fast RED Violet LB (F-3381, Sigma Aldrich) in 1 mL TRAP buffer (50 mL, 0.1 M acetate buffer [35.2 mL 0.2 M sodium acetate solution], 14.8 mL 0.2 M acetic acid solution, 50 mL H2O, 10 mL 0.3 M sodium tartrate [S-8640, Sigma-Aldrich], 1 mL 10 mg/mL Naphtol AS-MX phosphate [N-5000, Sigma-Aldrich], 100 μL Triton X-100 [T-8787, Sigma Aldrich], and 38.9 mL H2Odd). We removed the medium and washed the cells with PBS. Then, these cells were fixed with 10% glutaraldehyde (G-5882, Sigma-Aldrich) for 15 minutes at 37°C. After washing the cells 2 times with prewarmed PBS, the cells were stained with 300 μL TRAP staining solution per 12 wells for 10 minutes at 37°C. We washed the cells with PBS and took pictures of 12-well plates to obtain an overview of stained or not-stained cells.
To receive quantitative data, we performed a TRAP assay using a TRAP staining kit (PMC-AK04F-COS, Cosmo Bio) with samples acquired from the supernatant. To this aim, we transferred 30 μL cell culture supernatant into a 96-well plate in duplets. Afterward, we added 50 μL chromogenic substrate including tartrate-containing buffer and incubated for 3 hours at 37°C. We measured staining efficiency at 540 nm with an ELISA reader (Multiscan GO, Thermo Fisher Scientific). Obtained data were normalized to Nfat5-WT NS or ns-siRNA NS controls.
CaP resorption assay.
For CaP resorption assay, we coated 12-well plates as previously described (69). Briefly, we prepared a simulated body fluid (SBF) by mixing 50% Tris buffer (50 mM Tris base [T1503, Sigma-Aldrich], pH 7.4, with 1 M HCl [X942.1, Carl Roth]), 25% calcium stock solution (25 mM CaCl2•H2O [C5080, Sigma-Aldrich], 1.37 M NaCl [3957.1, Carl Roth], 15 mM MgCl2•6H2O [M2670, Sigma-Aldrich] in Tris buffer, pH 7.4), and 25% phosphate stock solution (11.1 mM Na2HPO4 [P030.1, Carl Roth], 42 mM NaHCO3 [8551.1, Carl Roth] in Tris buffer, pH 7.4). CaP solution (CPS) was prepared by first adding 41 mL HCl (1 M) to 800 mL H2Odd and then dissolving 2.25 mM Na2PO4•H2O, 4 mM CaCl2•H2O, 0.14 M NaCl, and 50 mM Tris before adjusting the pH to 7.4 and the volume to 1 L. We sterilized the solution by filtration with a 0.22 μm MillexGV (MilliporeSigma). Twelve-well tissue culture plates were incubated with SBF (1 mL/well) for 3 days at room temperature. We aspirated SBF solution an added CPS (1 mL/well) for 1 day at room temperature. Then we aspirated CPS and added 70% ethanol. Afterward, CaP-coated plates were washed twice with distilled water and dried overnight at 37°C. Prior to cell plating, we incubated coated plates with FCS for 1 hour at 37°C. Then we plated 10,000 cells per 12-well and incubated for 5 days under NS or HS in α-MEM supplemented with RANKL and M-CSF, as indicated before. Activated osteoclasts resorbed this CaP coating, and the resulting gaps were quantified using ImageJ (NIH).
ChIP.
We performed ChIP using ChIP Assay Kit (17-295, MilliporeSigma) according to the manufacturer’s instructions. Briefly, about 2 × 106 RAW264.7 macrophages were left unstimulated (NS) or stimulated with 40 mM NaCl (HS) for 24 hours in DMEM, high glucose, 10% FCS. We performed crosslinking by adding formaldehyde to a final concentration of 1% directly to the medium and incubated for 10 minutes at 37°C. We then removed the medium and washed the cells twice with ice-cold PBS and proteinase inhibitors (1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg/mL aprotinin, and 1 μg/mL pepstatin A). The cells were scraped into a conical tube and pelleted for 4 minutes at 2,000 rpm at 4°C. We resuspended the cells in 400 μL SDS lysis buffer including proteinase inhibitors and divided them to 200 μL aliquots. Lysates were sonicated to lengths of 700 bp, and samples were centrifuged for 10 minutes at 16,200 g at 4°C. We diluted the supernatant 10-fold in ChIP dilution buffer plus protein inhibitors and kept about 20 μL aside for input control. To reduce nonspecific background, we incubated the samples with 75 μL Protein A Agarose/Salmon Sperm DNA (50% slurry) for 30 minutes at 4°C with agitation. We removed agarose by centrifugation with 100 g 1 minute at 4°C. We added about 1 μL NFAT5 antibody (PA1-203, Thermo Fisher Scientific) for immunoprecipitation and incubated overnight with agitation (for negative control, no antibody was added). We added about 60 μL Protein A Agarose/Salmon Sperm DNA (50% slurry) to all samples and incubated for 1 hour at 4°C with agitation. Agarose was pelleted by centrifugation at 100 g for 1 minute at 4°C. We removed the supernatant, and agarose was washed for 5 minutes, first with 1 mL low-salt immune complex wash buffer, then with 1 mL high-salt immune complex wash buffer, followed by 1 mL LiCl immune complex wash buffer, and twice with 1 mL TE Buffer with rotation. After washing, we eluted the histone complex from the antibody by adding 250 μL elution buffer (1% SDS, 0.1 M NaHCO3) to the pelleted agarose, followed by incubation at room temperature for 15 minutes with rotation. Agarose was spun down, and the supernatant was collected. We repeated this step, and the supernatant fractions were pooled (total volume 500 μL). About 20 μL 5 M NaCl was added to the eluates, and crosslinks were reversed by heating at 65°C for 4 hours. After that, we added 10 μL of 0.5 M EDTA, 20 μL 1 M Tris-HCl, pH 6.5 and 2 μL of 10 mg/mL Proteinase K to eluates and incubated for 1 hour at 45°C. We recovered the DNA by adding 500 μL phenol/chloroform to the samples. After centrifugation, this step was repeated with the supernatant. We centrifuged samples at 16,200 g for 10 minutes, and the supernatant was precipitated with isopropanol overnight at –20°C. Then, samples were centrifuged at 13,000 rpm for 30 minutes at 4°C. We washed the pellet twice with 70% ethanol. After that, pellets were dried and resuspended in 15 μL Tris-HCl, pH 8. We used about 5 μL of samples and 1 μL of input controls for PCR. The following PCR conditions were used for each sample: 1 μL 10× Puffer (12161567001, Roche), 0.25 μL fwd primer 5′ - TGTCTGCGTGTGGGATAGTT - 3′, 0.25 μL rev primer 5′ - TCCTGGGCTACGCTGTAAA - 3′, 0.2 μL dNTPs (11581295001, Roche), and 0.1 μL Taq-Polymerase (120329929001, Roche). H2Odd was added up to 10 μL. Amplification protocol included the following: (a) 95°C for 5 minutes; (b) 95°C for 20 seconds, (c) 60°C for 45 seconds, and (d) 4°C. Steps b and c were repeated 40×. We loaded samples on a 1% agarose gel and analyzed under UV light.
OPG ELISA.
OPG ELISA was obtained from Thermo Fisher Scientific (EMTNFRSF11B) and performed according to the manufacturer’s instructions.
TRACP-5b ELISA.
TRACP-5b Elisa was obtained from MyBiosource (MBS763504) and performed according to the manufacturer’s instructions.
μCT.
For μCT, we used the GE V-Tome-X S240 from GE Healthcare. Tibiae of 20-week-old mice were scanned using Fast-Scan protocol (33 minutes, nanofocus tube, voxel size 4.5 μm; magnification, 44.4×, picture number 2,000; timing 1,000 ms; voltage 35 kV; electricity 145). We obtained bone morphometric parameters in a region of interest (ROI) ranging from 0.2–2 mm below the growth boundary. We quantified BV/TV and trabecular thickness (Tb.Th) using VGL3.0 (Volume graphics GmbH).
Histological analysis.
Tibiae were fixed in 4% paraformaldehyde, stored in 70% ethanol until decalcification, embedded in paraffin, and sectioned at 5 μm. We deparaffinized slides overnight at 37°C and hydrogenated them. To assess bone histomorphometrically, we performed TRAP and toluidine blue stainings.
For TRAP staining, we placed the section in a freshly prepared TRAP buffer consisting of 1.64 g sodium acetate (6773.1, Carl Roth) and 23 g of disodium tartrate dihydrate (T110.1, Carl Roth) to 500 mL of H2Odd (pH 5) for 10 minutes at room temperature. Sections were then placed in a freshly prepared staining solution consisting of 40 mg Naphtol AS-MX Phosphate Disodium Salt (N5000, Sigma-Aldrich), 4 mL N,N-dimethylformamide (D4551, Sigma-Aldrich), 240 mg Fast Red Violet LB Salt (F3381, Sigma-Aldrich), 2 mL Triton X-100 (T9284, Sigma-Aldrich), and 200 mL previously prepared TRAP buffer. The sections were incubated at 37°C for 2 hours, rinsed in H2Odd, and counterstained for 3 minutes with filtered Hayer’s hematoxylin solution (51275, Sigma-Aldrich) at room temperature. Subsequently, the sections were covered immediately with Aquatex (1085620050, Merck).
For toluidine blue stainings, we put the deparaffinized and rehydrogenated slides in staining solution containing toluidine blue (89640, Fluka) and sodium-tetraborate (221732, Sigma-Aldrich) for 1 minute. After incubation for 10 minutes in H2Odd and for at least 20 minutes in xylene, the coverslips were applied with entellan (1.07961.0500, Merck).
Quantitative histomorphometry was performed on TRAP and toluidine blue–stained sections according to standard protocols (70) using the Osteomeasure histomorphometry system (Osteometrix). Experiments were performed in a blinded fashion.
Na+ measurements.
Femur BMs of mice were flushed with H2Odd. Wet weight of the flushed BM was determined and samples were stored at 4°C until analysis. Na+ was measured by atomic absorption spectrometry after appropriate dilution (Model 3100, Perkin Elmer).
Statistics.
Statistics were performed with the software application SPSS Statistics 24 (IBM). Descriptive statistics are given as mean ± SEM. Two-tailed, unpaired Student’s t test was used, where appropriate. Otherwise, the experimental groups were independently compared by 1-way ANOVAs, which were validated by applying Welch’s test, since homogeneity of variance was absent. Post hoc tests using the Games-Howell approach for heterogeneous variances were used for pairwise comparisons. μCT analysis and Na+ measurements consisted of comparison of means of data from animal experiments calculated by multivariate or univariate analysis using the General Linear Measurements (GLM) procedure. All differences were considered statistically significant at P ≤ 0.05.
Study approval.
All animal experiments were performed according to German law in compliance with the ARRIVE guidelines. The present study in animals was reviewed and approved by the Regierung von Unterfranken, Würzburg.
Author contributions
AS planned and performed experiments and wrote the manuscript. PN performed experiments. AB designed experiments and helped with analysis of μCT and bone morphology. WN provided critical material and contributed to manuscript preparation. JT and PP were involved in the planning of experiments, trial design, and manuscript editing. CK and JJ planned and supervised the experiments and contributed to manuscript preparation and revision.
Supplementary Material
Acknowledgments
The authors thank Kathrin Bauer and Eva Zaglauer for their technical support in performing the qPCR analyses and cell culture experiments, as well as Monika Nowottny for animal genotyping and Birgit Striegl for performing the μCT analysis (DFG-Nr.: INST 102/11-1 FUGG). AS received funding from the Faculty for Medicine Regensburg (ReForM A), CK from the German Orthodontic Society DGKFO (Kirschneck 08/2018), and JJ from the German Research Foundation DFG (JA1993/4-1).
Version 1. 12/05/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(23):e127868.https://doi.org/10.1172/jci.insight.127868.
Contributor Information
Agnes Schröder, Email: agnes.schroeder@ukr.de.
Patrick Neubert, Email: patrick.neubert@ukr.de.
Jens Titze, Email: jens.titze@duke-nus.edu.sg.
Aline Bozec, Email: aline.bozec@uk-erlangen.de.
Wolfgang Neuhofer, Email: wolfgang.neuhofer@helios-gesundheit.de.
Peter Proff, Email: peter.proff@ukr.de.
Christian Kirschneck, Email: christian.kirschneck@ukr.de.
Jonathan Jantsch, Email: jonathan.jantsch@ukr.de.
References
- 1.Feng X, Teitelbaum SL. Osteoclasts: New Insights. Bone Res. 2013;1(1):11–26. doi: 10.4248/BR201301003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Proff P, Römer P. The molecular mechanism behind bone remodelling: a review. Clin Oral Investig. 2009;13(4):355–362. doi: 10.1007/s00784-009-0268-2. [DOI] [PubMed] [Google Scholar]
- 3.Parfitt AM. Osteonal and hemi-osteonal remodeling: the spatial and temporal framework for signal traffic in adult human bone. J Cell Biochem. 1994;55(3):273–286. doi: 10.1002/jcb.240550303. [DOI] [PubMed] [Google Scholar]
- 4.Kong YY, Penninger JM. Molecular control of bone remodeling and osteoporosis. Exp Gerontol. 2000;35(8):947–956. doi: 10.1016/S0531-5565(00)00178-9. [DOI] [PubMed] [Google Scholar]
- 5.Raisz LG. Pathogenesis of osteoporosis: concepts, conflicts, and prospects. J Clin Invest. 2005;115(12):3318–3325. doi: 10.1172/JCI27071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schett G, Teitelbaum SL. Osteoclasts and Arthritis. J Bone Miner Res. 2009;24(7):1142–1146. doi: 10.1359/jbmr.090533. [DOI] [PubMed] [Google Scholar]
- 7.Ducy P, Schinke T, Karsenty G. The osteoblast: a sophisticated fibroblast under central surveillance. Science. 2000;289(5484):1501–1504. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- 8.Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289(5484):1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 9.Anderson DM, et al. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature. 1997;390(6656):175–179. doi: 10.1038/36593. [DOI] [PubMed] [Google Scholar]
- 10.Wong BR, et al. TRANCE is a novel ligand of the tumor necrosis factor receptor family that activates c-Jun N-terminal kinase in T cells. J Biol Chem. 1997;272(40):25190–25194. doi: 10.1074/jbc.272.40.25190. [DOI] [PubMed] [Google Scholar]
- 11.Bucay N, et al. osteoprotegerin-deficient mice develop early onset osteoporosis and arterial calcification. Genes Dev. 1998;12(9):1260–1268. doi: 10.1101/gad.12.9.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacey DL, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93(2):165–176. doi: 10.1016/S0092-8674(00)81569-X. [DOI] [PubMed] [Google Scholar]
- 13.Simonet WS, et al. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell. 1997;89(2):309–319. doi: 10.1016/S0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 14.Boyce BF, Xing L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch Biochem Biophys. 2008;473(2):139–146. doi: 10.1016/j.abb.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto K, Takayanagi H. Osteoimmunology. Cold Spring Harb Perspect Med. 2019;9(1):a031245. doi: 10.1101/cshperspect.a031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takayanagi H. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol. 2007;7(4):292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- 17.Kloss L, Meyer JD, Graeve L, Vetter W. Sodium intake and its reduction by food reformulation in the European Union – a review. NFS Journal. 2015;1:9–19. doi: 10.1016/j.nfs.2015.03.001. [DOI] [Google Scholar]
- 18.Rakova N, et al. Increased salt consumption induces body water conservation and decreases fluid intake. J Clin Invest. 2017;127(5):1932–1943. doi: 10.1172/JCI88530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rakova N, et al. Long-term space flight simulation reveals infradian rhythmicity in human Na(+) balance. Cell Metab. 2013;17(1):125–131. doi: 10.1016/j.cmet.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Kopp C, et al. 23Na magnetic resonance imaging-determined tissue sodium in healthy subjects and hypertensive patients. Hypertension. 2013;61(3):635–640. doi: 10.1161/HYPERTENSIONAHA.111.00566. [DOI] [PubMed] [Google Scholar]
- 21.Titze J, et al. Hypertension, sodium retention, calcium excretion and osteopenia in Dahl rats. J Hypertens. 2004;22(4):803–810. doi: 10.1097/00004872-200404000-00024. [DOI] [PubMed] [Google Scholar]
- 22.Titze J, et al. Spooky sodium balance. Kidney Int. 2014;85(4):759–767. doi: 10.1038/ki.2013.367. [DOI] [PubMed] [Google Scholar]
- 23.Wiig H, et al. Immune cells control skin lymphatic electrolyte homeostasis and blood pressure. J Clin Invest. 2013;123(7):2803–2815. doi: 10.1172/JCI60113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machnik A, et al. Macrophages regulate salt-dependent volume and blood pressure by a vascular endothelial growth factor-C-dependent buffering mechanism. Nat Med. 2009;15(5):545–552. doi: 10.1038/nm.1960. [DOI] [PubMed] [Google Scholar]
- 25.Machnik A, et al. Mononuclear phagocyte system depletion blocks interstitial tonicity-responsive enhancer binding protein/vascular endothelial growth factor C expression and induces salt-sensitive hypertension in rats. Hypertension. 2010;55(3):755–761. doi: 10.1161/HYPERTENSIONAHA.109.143339. [DOI] [PubMed] [Google Scholar]
- 26.López-Rodríguez C, Aramburu J, Jin L, Rakeman AS, Michino M, Rao A. Bridging the NFAT and NF-kappaB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress. Immunity. 2001;15(1):47–58. doi: 10.1016/S1074-7613(01)00165-0. [DOI] [PubMed] [Google Scholar]
- 27.Ko BC, Turck CW, Lee KW, Yang Y, Chung SS. Purification, identification, and characterization of an osmotic response element binding protein. Biochem Biophys Res Commun. 2000;270(1):52–61. doi: 10.1006/bbrc.2000.2376. [DOI] [PubMed] [Google Scholar]
- 28.Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM. Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci USA. 1999;96(5):2538–2542. doi: 10.1073/pnas.96.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordin BE, Need AG, Morris HA, Horowitz M. The nature and significance of the relationship between urinary sodium and urinary calcium in women. J Nutr. 1993;123(9):1615–1622. doi: 10.1093/jn/123.9.1615. [DOI] [PubMed] [Google Scholar]
- 30.Park SM, et al. High Dietary Sodium Intake Assessed by 24-hour Urine Specimen Increase Urinary Calcium Excretion and Bone Resorption Marker. J Bone Metab. 2014;21(3):189–194. doi: 10.11005/jbm.2014.21.3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frings-Meuthen P, Baecker N, Heer M. Low-grade metabolic acidosis may be the cause of sodium chloride-induced exaggerated bone resorption. J Bone Miner Res. 2008;23(4):517–524. doi: 10.1359/jbmr.071118. [DOI] [PubMed] [Google Scholar]
- 32.Hannon MJ, Verbalis JG. Sodium homeostasis and bone. Curr Opin Nephrol Hypertens. 2014;23(4):370–376. doi: 10.1097/01.mnh.0000447022.51722.f4. [DOI] [PubMed] [Google Scholar]
- 33.Usala RL, et al. Hyponatremia Is Associated With Increased Osteoporosis and Bone Fractures in a Large US Health System Population. J Clin Endocrinol Metab. 2015;100(8):3021–3031. doi: 10.1210/jc.2015-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verbalis JG, et al. Hyponatremia-induced osteoporosis. J Bone Miner Res. 2010;25(3):554–563. doi: 10.1359/jbmr.090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devine A, Criddle RA, Dick IM, Kerr DA, Prince RL. A longitudinal study of the effect of sodium and calcium intakes on regional bone density in postmenopausal women. Am J Clin Nutr. 1995;62(4):740–745. doi: 10.1093/ajcn/62.4.740. [DOI] [PubMed] [Google Scholar]
- 36.Harrington JT, et al. Risedronate rapidly reduces the risk for nonvertebral fractures in women with postmenopausal osteoporosis. Calcif Tissue Int. 2004;74(2):129–135. doi: 10.1007/s00223-003-0042-4. [DOI] [PubMed] [Google Scholar]
- 37.Goulding A, Campbell DR. Effects of oral loads of sodium chloride on bone composition in growing rats consuming ample dietary calcium. Miner Electrolyte Metab. 1984;10(1):58–62. [PubMed] [Google Scholar]
- 38.Wu L, et al. Increased levels of sodium chloride directly increase osteoclastic differentiation and resorption in mice and men. Osteoporos Int. 2017;28(11):3215–3228. doi: 10.1007/s00198-017-4163-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barsony J, Sugimura Y, Verbalis JG. Osteoclast response to low extracellular sodium and the mechanism of hyponatremia-induced bone loss. J Biol Chem. 2011;286(12):10864–10875. doi: 10.1074/jbc.M110.155002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsukasaki M, Takayanagi H. Osteoimmunology: evolving concepts in bone-immune interactions in health and disease. Nat Rev Immunol. 2019;19(10):626–642. doi: 10.1038/s41577-019-0178-8. [DOI] [PubMed] [Google Scholar]
- 41.Binger KJ, et al. High salt reduces the activation of IL-4- and IL-13-stimulated macrophages. J Clin Invest. 2015;125(11):4223–4238. doi: 10.1172/JCI80919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neubert P, et al. HIF1A and NFAT5 coordinate Na+-boosted antibacterial defense via enhanced autophagy and autolysosomal targeting. Autophagy. 2019;15(11):1899–1916. doi: 10.1080/15548627.2019.1596483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziomber A, et al. Sodium-, potassium-, chloride-, and bicarbonate-related effects on blood pressure and electrolyte homeostasis in deoxycorticosterone acetate-treated rats. Am J Physiol Renal Physiol. 2008;295(6):F1752–F1763. doi: 10.1152/ajprenal.00531.2007. [DOI] [PubMed] [Google Scholar]
- 44.Kitada K, et al. High salt intake reprioritizes osmolyte and energy metabolism for body fluid conservation. J Clin Invest. 2017;127(5):1944–1959. doi: 10.1172/JCI88532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jantsch J, et al. Cutaneous Na+ storage strengthens the antimicrobial barrier function of the skin and boosts macrophage-driven host defense. Cell Metab. 2015;21(3):493–501. doi: 10.1016/j.cmet.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matthias J, et al. Sodium chloride is an ionic checkpoint for human TH2 cells and shapes the atopic skin microenvironment. Sci Transl Med. 2019;11(480):eaau0683. doi: 10.1126/scitranslmed.aau0683. [DOI] [PubMed] [Google Scholar]
- 47.Carbone LD, et al. The renin-angiotensin aldosterone system and osteoporosis: findings from the Women’s Health Initiative. Osteoporos Int. 2019;30(10):2039–2056. doi: 10.1007/s00198-019-05041-3. [DOI] [PubMed] [Google Scholar]
- 48.Chen XF, et al. Inhibition on angiotensin-converting enzyme exerts beneficial effects on trabecular bone in orchidectomized mice. Pharmacol Rep. 2018;70(4):705–711. doi: 10.1016/j.pharep.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schatz V, et al. Elementary immunology: Na+ as a regulator of immunity. Pediatr Nephrol. 2017;32(2):201–210. doi: 10.1007/s00467-016-3349-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berry MR, et al. Renal Sodium Gradient Orchestrates a Dynamic Antibacterial Defense Zone. Cell. 2017;170(5):860–874.e19. doi: 10.1016/j.cell.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 51.Zhang WC, et al. Elevated sodium chloride drives type I interferon signaling in macrophages and increases antiviral resistance. J Biol Chem. 2018;293(3):1030–1039. doi: 10.1074/jbc.M117.805093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ravesloot JH, Eisen T, Baron R, Boron WF. Role of Na-H exchangers and vacuolar H+ pumps in intracellular pH regulation in neonatal rat osteoclasts. J Gen Physiol. 1995;105(2):177–208. doi: 10.1085/jgp.105.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta A, Miyauchi A, Fujimori A, Hruska KA. Phosphate transport in osteoclasts: a functional and immunochemical characterization. Kidney Int. 1996;49(4):968–974. doi: 10.1038/ki.1996.137. [DOI] [PubMed] [Google Scholar]
- 54.Nordström T, et al. Regulation of cytoplasmic pH in osteoclasts. Contribution of proton pumps and a proton-selective conductance. J Biol Chem. 1995;270(5):2203–2212. doi: 10.1074/jbc.270.5.2203. [DOI] [PubMed] [Google Scholar]
- 55.Hofstetter W, Siegrist M, Simonin A, Bonny O, Fuster DG. Sodium/hydrogen exchanger NHA2 in osteoclasts: subcellular localization and role in vitro and in vivo. Bone. 2010;47(2):331–340. doi: 10.1016/j.bone.2010.04.605. [DOI] [PubMed] [Google Scholar]
- 56.Moonga BS, et al. Identification and characterization of a sodium/calcium exchanger, NCX-1, in osteoclasts and its role in bone resorption. Biochem Biophys Res Commun. 2001;283(4):770–775. doi: 10.1006/bbrc.2001.4870. [DOI] [PubMed] [Google Scholar]
- 57.Li JP, Kajiya H, Okamoto F, Nakao A, Iwamoto T, Okabe K. Three Na+/Ca2+ exchanger (NCX) variants are expressed in mouse osteoclasts and mediate calcium transport during bone resorption. Endocrinology. 2007;148(5):2116–2125. doi: 10.1210/en.2006-1321. [DOI] [PubMed] [Google Scholar]
- 58.Albano G, et al. Increased bone resorption by osteoclast-specific deletion of the sodium/calcium exchanger isoform 1 (NCX1) Pflugers Arch. 2017;469(2):225–233. doi: 10.1007/s00424-016-1923-5. [DOI] [PubMed] [Google Scholar]
- 59.Gupta A, Guo XL, Alvarez UM, Hruska KA. Regulation of sodium-dependent phosphate transport in osteoclasts. J Clin Invest. 1997;100(3):538–549. doi: 10.1172/JCI119563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Makihira S, et al. Blocking of sodium and potassium ion-dependent adenosine triphosphatase-α1 with ouabain and vanadate suppresses cell-cell fusion during RANKL-mediated osteoclastogenesis. Eur J Pharmacol. 2011;670(2-3):409–418. doi: 10.1016/j.ejphar.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 61.Baron R, Neff L, Roy C, Boisvert A, Caplan M. Evidence for a high and specific concentration of (Na+,K+)ATPase in the plasma membrane of the osteoclast. Cell. 1986;46(2):311–320. doi: 10.1016/0092-8674(86)90748-8. [DOI] [PubMed] [Google Scholar]
- 62.Yasuda H, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc Natl Acad Sci USA. 1998;95(7):3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCarthy TL, Kallen CB, Centrella M. β-Catenin independent cross-control between the estradiol and Wnt pathways in osteoblasts. Gene. 2011;479(1-2):16–28. doi: 10.1016/j.gene.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vidal ON, Sjögren K, Eriksson BI, Ljunggren O, Ohlsson C. Osteoprotegerin mRNA is increased by interleukin-1 alpha in the human osteosarcoma cell line MG-63 and in human osteoblast-like cells. Biochem Biophys Res Commun. 1998;248(3):696–700. doi: 10.1006/bbrc.1998.9035. [DOI] [PubMed] [Google Scholar]
- 65.Siegert I, Schatz V, Prechtel AT, Steinkasserer A, Bogdan C, Jantsch J. Electroporation of siRNA into mouse BM-derived macrophages and dendritic cells. Methods Mol Biol. 2014;1121:111–119. doi: 10.1007/978-1-4614-9632-8_9. [DOI] [PubMed] [Google Scholar]
- 66.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 67.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 68.Kirschneck C, Batschkus S, Proff P, Köstler J, Spanier G, Schröder A. Valid gene expression normalization by RT-qPCR in studies on hPDL fibroblasts with focus on orthodontic tooth movement and periodontitis. Sci Rep. 2017;7(1):14751. doi: 10.1038/s41598-017-15281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maria SM, Prukner C, Sheikh Z, Mueller F, Barralet JE, Komarova SV. Reproducible quantification of osteoclastic activity: characterization of a biomimetic calcium phosphate assay. J Biomed Mater Res Part B Appl Biomater. 2014;102(5):903–912. doi: 10.1002/jbm.b.33071. [DOI] [PubMed] [Google Scholar]
- 70.Parfitt AM, et al. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 1987;2(6):595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.