Antibiotic production in Streptomyces coelicolor is coordinated by a division of labor through large genome deletions.
Abstract
One of the hallmark behaviors of social groups is division of labor, where different group members become specialized to carry out complementary tasks. By dividing labor, cooperative groups increase efficiency, thereby raising group fitness even if these behaviors reduce individual fitness. We find that antibiotic production in colonies of Streptomyces coelicolor is coordinated by a division of labor. We show that S. coelicolor colonies are genetically heterogeneous because of amplifications and deletions to the chromosome. Cells with chromosomal changes produce diversified secondary metabolites and secrete more antibiotics; however, these changes reduced individual fitness, providing evidence for a trade-off between antibiotic production and fitness. Last, we show that colonies containing mixtures of mutants and their parents produce significantly more antibiotics, while colony-wide spore production remains unchanged. By generating specialized mutants that hyper-produce antibiotics, streptomycetes reduce the fitness costs of secreted secondary metabolites while maximizing the yield and diversity of these products.
INTRODUCTION
Social insects provide some of the most compelling examples of divisions of labor, with extremes in morphological differentiation associated with highly specialized functions and reproductive sterility in all colony members, except the queen (1). However, conditions that select for division of labor are not limited to animals, and it has become increasingly clear that microbes offer unique opportunities to identify and study the mechanistic underpinnings of divisions of labor (2–8). First, microbes are typically clonal, which helps ensure that a division of labor is favored by kin selection (4). Second, microbial populations are highly social, often cooperating to carry out coordinated behaviors such as migration or biofilm formation that require the secretion of metabolically expensive public goods that can be shared among clonemates (9, 10). If these conditions are met, and investment in public good secretion trades off with fitness, divisions of labor are predicted to evolve (4, 11).
Here, we describe the cause and evolutionary benefits of a unique division of labor that has evolved in colonies of the filamentous actinomycete Streptomyces coelicolor. After germinating from unichromosomal spores, these bacteria establish multicellular networks of vegetative hyphae, reminiscent of fungal colonies (12–14). Vegetative hyphae secrete a broad variety of public goods, such as chitinases and cellulases that are used to acquire resources, as well as a chemically diverse suite of antibiotics that are used to kill or inhibit competing organisms (15–17). Streptomycetes are prolific producers of antibiotics and are responsible for producing more than 50% of our clinically relevant antibiotics (18). Although the terminal differentiation of Streptomyces colonies into vegetative hyphae (soma) and viable spores (germ) is well understood (19–21), no other divisions of labor in these multicellular bacteria are known. However, opportunities for phenotypic differentiation are possible, because although colonies begin clonally, they can become genetically heterogeneous because of unexplained high-frequency rearrangements and deletions in their large, ~9-Mb linear chromosome (22–25). The work we describe shows that these two phenomena are intertwined. Briefly, we find that genomic instability causes irreversible genetic differentiation within a subpopulation of growing cells. This differentiation, in turn, gives rise to a division of labor that increases the productivity and diversity of secreted antibiotics and increases colony-wide fitness.
RESULTS
Genomic instability and phenotypic heterogeneity are coupled
Genomic instability and phenotypic heterogeneity have been observed in several Streptomyces species (26–32), but there are no explanations for the evolution or functional consequences of this extreme mutability. To begin addressing this question, we quantified the phenotypic heterogeneity arising within 81 random single colonies of S. coelicolor M145 by harvesting the spores of each of these colonies and then replating the collected spores onto a new agar surface. Although most progeny are morphologically homogeneous and similar to the wild type (WT), notably aberrant colonies (Fig. 1A) arise at high frequencies (0.79 ± 0.06%, mean ± SEM, ranging from 0 to 2.15%, n = 81) (Fig. 1A). Similarly high rates were obtained on two minimal media (MM: 2.13 ± 0.14%; MM + casamino acids: 5.13 ± 0.37%; mean ± SEM; n = 30 and n = 40, respectively) (fig. S1), thereby ruling out the possibility that these mutations are an artifact of rapid growth on rich resources. The differences we observe on these two media types also suggest that the mutant frequencies we estimate based on spore counts may underestimate their values within growing colonies, given that mutants may be compromised in growth or sporulation (as we confirm below). This is supported by the nearly twofold difference in mutant frequencies on MM + casamino acids compared to unsupplemented MM, where auxotrophs arising by mutation would be unable to persist. To determine the heritability of these aberrant phenotypes, we restreaked 15 random colonies from different plates onto a new agar plate, which revealed remarkable variability in colony morphology (Fig. 1B). Rather than reverting to the WT morphology, as would be anticipated if the initial heterogeneity was due to phenotypic plasticity or another form of bistability, the colonies derived from mutant colonies are themselves hypervariable, giving rise to up to nine diverse phenotypes from any single colony. Thus, in the course of two cycles of colony outgrowth, an array of colony types that differ in size, shape, and color emerged (Fig. 1B). Because our ability to discern colony heterogeneity is limited to only a few visually distinct phenotypic characters, we assume that these estimates of diversity are lower than their true level of occurrence.
Fig. 1. Emergence of phenotypic heterogeneity in colonies of S. coelicolor.
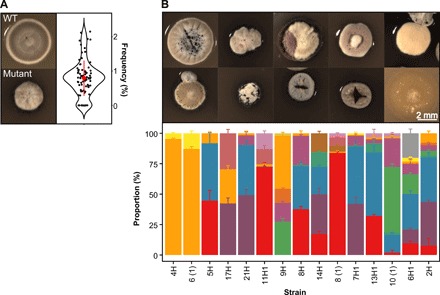
(A) WT (top) and mutant (bottom) colonies and the frequency that mutants emerge from WT colonies on SFM agar (right). (B) Phenotypically diverse progeny (top) emerges after restreaking mutant colonies that vary in size, shape, and pigmentation. Representative colonies are shown. The bottom graph depicts the range of distinct morphologies that emerge after restreaking 15 random colonies. Each color represents a distinct colony phenotype.
Using whole-genome sequencing of eight random mutants, we confirmed that these isolates contained profound chromosomal changes. As shown in Fig. 2A, large genome deletions were observed at the chromosome ends in all eight strains. In three cases, we found an ~297-kb amplification on the left chromosomal arm flanked by the Insertion Sequence IS1649, encoded by SCO0091 and SCO0368. Average sequence coverage of the amplified region suggests that it contains between 2 and 15 copies of this amplification (Fig. 2A and fig. S2). Sequencing results were expanded using pulsed-field gel electrophoresis (PFGE) analysis of 30 mutant isolates (Fig. 2B and fig. S3). Consistent with our sequencing results, this analysis revealed that mutants contained variably sized deletions of up to ~240 or ~872 kb on the left chromosome arm and up to 1.6 Mb on the right chromosome arm, deleting more than 1000 genes. In addition, 8 of 30 strains contained the same large amplification between copies of IS1649 as noted above. These strains are conspicuously yellow, which might be caused by the overproduction of carotenoids due to the amplification of the crt gene cluster (SCO0185-0191) (33–35). In addition to this and other phenotypic effects associated with these changes that are discussed below, deletions to the right chromosome arm cause the loss of two loci, argG (SCO7036) and cmlR1 (SCO7526)/cmlR2 (SCO7662), that result in two easily scorable phenotypes: arginine auxotrophy and chloramphenicol susceptibility, respectively. Scoring these phenotypes allows rapid determination of the minimal size of the deletion on the right chromosome arm in the absence of molecular characterization. Chloramphenicol susceptibility indicates a deletion of at least 322 kb, while the addition of arginine auxotrophy indicates a deletion of at least 843 kb (Fig. 2B).
Fig. 2. Genome diversity of mutant colonies determined from whole-genome sequencing and PFGE.
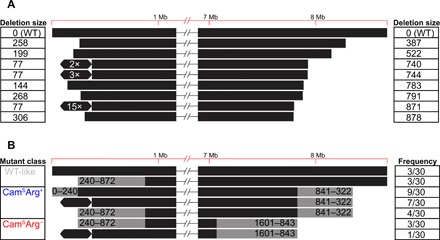
Values in (A) correspond to the size (in kilobases) of genome deletions, while the hexagons represent an ~297-kb genome amplification. Each line in (B) depicts the range of deletion sizes (gray) in each mutant class, together with their respective frequencies from 30 sampled mutant strains.
Mutants increase the production and diversity of antibiotics
Mutant strains were conspicuously pigmented when compared to their parental WT strains (Fig. 1). Because several antibiotics produced by S. coelicolor are pigmented, namely, actinorhodin, prodigines, and coelimycin P1, which are blue, red, and yellow, respectively, we tested whether mutant strains had altered secondary metabolite and inhibitory profiles. Secreted metabolites from mutant and WT strains grown on agar surfaces were analyzed using quantitative 1H nuclear magnetic resonance (NMR) profiling (36, 37). Principal components analysis (Fig. 3A) supports the partition of strains into three well-separated groups: WT and WT-like strains and then two clusters of mutant isolates. In each case, groupings corresponded to the size of genomic lesions mentioned above. More specifically, strains grouping in the WT and WT-like cluster were chloramphenicol resistant (CamR) and arginine prototrophic (Arg+), while those clustering within the blue ellipse were chloramphenicol susceptible (CamS) and arginine protrophic (Arg+), and those in the red ellipsed cluster were chloramphenicol susceptible (CamS) and arginine auxotrophic (Arg−). To assess whether genomic deletions affected antibiotic biosynthesis, we used mass spectrometry (MS)–based quantitative proteomics on five representative strains from the two mutant clusters. This analysis revealed that the biosynthetic pathways for actinorhodin, coelimycin P1, and calcium-dependent antibiotic were significantly up-regulated in all mutants (Fig. 3, B and C; fig. S4; and table S1). Because the expression level of biosynthetic enzymes directly correlates with antibiotic production (38), these MS results are consistent with increased antibiotic production in these strains (Fig. 3, B to D; fig. S4; and table S1). In addition to antibiotic biosynthesis clusters, pathways regulating arginine and pyrimidine biosynthesis were also increased in both arginine auxotrophic strains (fig. S4B and table S1) (39). No antibiotic-related proteins were down-regulated in this analysis.
Fig. 3. Secondary metabolite production in mutant strains determined by 1H NMR, quantitative proteomics, or zones of inhibition on B. subtilis or 40 different natural streptomycete isolates.

(A) Principal components (PC) analysis plot of 1H NMR data. Each cluster enclosed in a colored ellipse (with 95% confidence interval) corresponds to a mutant class with a different phenotype and degree of genomic instability: WT-like strains (gray), CamSArg+ strains (blue), and CamSArg− strains (red). (B and C) Volcano plots of MS-based quantitative proteomics of two representative strains 9H1A (CamSArg−) (B) and 9H1B (CamSArg+) (C). Protein level is indicated by the size of the dot, and genes with ≤2-fold change and/or P ≥ 0.05 are grayed out. (D) Zones of inhibition of each strain when grown with a B. subtilis soft agar overlay. Colors represent the same mutant classes as in (A). The large dot represents the mean of four replicates, while error bars represent the SE. (E) Partial least-squares (PLS) plot of 1H NMR data partitioned by the same clusters as in (A). The heat map indicates the size of the zone of inhibition on B. subtilis. (F) Zones of inhibition of four representative mutant strains with an overlay of 40 different natural streptomycetes, each represented by a different line. Statistics are given in the main text.
We next asked whether these different metabolic and proteomic profiles translated to differences in biological activity, specifically the ability to inhibit the growth of other bacteria. Thirty mutant strains were grown on agar plates and then covered with a soft agar overlay containing Bacillus subtilis. Inhibition was visualized as an absence of growth surrounding the mutant colony, and the extent of inhibition was determined from the size of the inhibition zone. As shown in Fig. 3D, all but three WT-like mutant strains produced significantly larger zones of inhibition than the WT strain (one-tailed t tests, all P < 0.05). In addition, we observed significant heterogeneity among mutant strains in halo size [one-way analysis of variance (ANOVA), F29,90 = 5.45, P < 0.001].
To test whether the increased inhibition we observed against B. subtilis was correlated with the 1H NMR profiles, we used a partial least-squares regression (Fig. 3E) (37). This showed that the separation into different groups significantly correlates with halo size (Q2 = 0.879), which was further validated by both permutation tests and ANOVA of cross-validated residuals (CV-ANOVA, F8,116 = 104.443, P < 0.001). To identify possible compounds that are overproduced in mutants compared to WT, we identified several 1H NMR signals that varied across strains and strongly correlated with the size of the zone of inhibition against B. subtilis (table S2); notable among these are several aromatic signals, which correspond to actinorhodin, consistent with our proteomic analyses (Fig. 3, B and C; fig. S4; and table S1).
Phenotypic results indicate that mutant strains produce more antibiotics than their WT parent when assayed against a single bacterial target, as anticipated given our NMR and proteomic results. However, they do not distinguish whether strains can be further partitioned on the basis of which other species they inhibit. Score plots of principal components analysis based on 1H NMR signals reveal clear separation between mutant clones within and between clusters (Fig. 3A), suggesting that their inhibitory spectra may vary. In addition, quantitative proteomic data show that different strains vary in their production of known antimicrobials. To test this, we measured the ability of four mutant clones to inhibit 48 recently isolated Streptomyces strains (40). Streptomyces targets were chosen because these are likely to represent important competitors for other streptomycetes in soil environments. At least one of the four mutant strains produced a significantly larger halo than the WT strain against 40 of 48 targets, indicating increased inhibition (Fig. 3F). For these 40 targets, we observed significant differences between the mutant strains themselves. We found differences in the size of the zone of inhibition on different target species (two-way ANOVA, F39,117 = 21.21, P < 0.001) as well as a significant interaction between mutant strain and the target species (two-way ANOVA, F117,320 = 5.83, P < 0.001), indicating that the inhibitory profile of each mutant strain is distinct from the others. Together, these results reveal that mutants arising within colonies not only are more effective at inhibiting other strains but also are diversified in who they can inhibit because their inhibition spectra do not overlap. They also suggest that the beneficiary of diversified antibiotic secretion is the parent strain, because competing bacteria are unlikely to be resistant to this broadened combination of secreted antimicrobials.
Antibiotic production is coordinated by a division of labor
Having shown that Streptomyces colonies differentiate into distinct subpopulations that vary in their antibiotic production, we next asked how this differentiation affects colony fitness. To answer this, we measured the fitness of each mutant strain by quantifying the number of spores they produce when grown in isolation. As shown in Fig. 4A, mutants produce significantly fewer spores than the WT strain (28 of 30; P < 0.01, two-sample t tests) and, in extreme cases, as much as 10,000-fold less, with significant heterogeneity among strains (one-way ANOVA, F29,59 = 132.57, P < 0.001). The reduction in spore production is significantly negatively correlated with antibiotic production (F1,29 = 26.58, r2 = 0.478, P < 0.001) (fig. S5A). This provides evidence that antibiotic production is costly to S. coelicolor and that there is a direct trade-off between antibiotic production and reproductive capacity, possibly because energy is redirected from development to antibiotic production (41). In addition, we observed a significant negative correlation between the size of the genome deletion and colony-forming unit (CFU) (F1,7 = 12.32, r2 = 0.638, P = 0.0099) and a positive correlation between deletion size and bioactivity against B. subtilis (F1,7 = 37.97, r2 = 0.844, P < 0.001), suggesting that these phenotypes scale with the magnitude of genomic changes (Fig. 4B).
Fig. 4. Fitness of mutant strains grown alone or during coculture with the WT strain and the effects of genome deletions on fitness and antibiotic production.
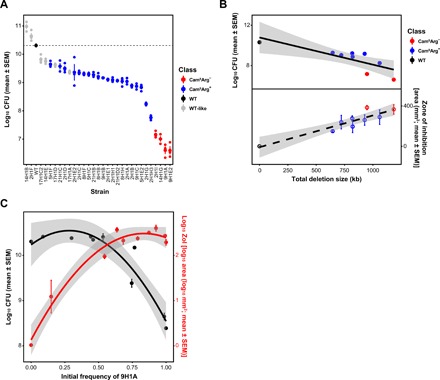
(A) Fitness [colony-forming unit (CFU)] of mutant strains. (B) Decreases in genome size negatively correlate with fitness (top) and positively correlate with antibiotic production (bottom). (C) Division of labor during coculture of the WT and strain 9H1A at different starting frequencies. Increasing frequencies of 9H1A cause increased antibiotic production (F2,7 = 107.7, r2 = 0.969, P < 0.001) (red) but only negatively affect colony fitness at frequencies >~50% (F2,7 = 37.95, r2 = 0.916, P < 0.001) (black). Quadratic regression lines include the 95% confidence interval.
To examine the effects of mutant strains on the colony as a whole, we mixed mutant strains with their WT parent at increasing frequencies and quantified colony-wide spore production and the ability of these mixtures to kill B. subtilis. We measured responses across a broad range of initial mutant frequencies to reflect the large variation in these values across media types and colonies and to also address uncertainties in their frequencies and spatial distribution during colony growth. Results of these experiments, shown in Fig. 4C, support two important conclusions: (i) Increasing fractions of mutants lead to increased antibiotic production, and (ii) although mutant strains have individually reduced fitness (fig. S5B), their presence within colonies has no effect on colony-wide spore production, until the mutant frequency exceeds >50% of the total. We carried out the same assay with three additional mutant clones, but at fewer frequencies, to estimate spore production and observed concordant results (fig. S6): Up to a frequency of ~50%, mutant strains have no effect on colony-wide spore production, while each incremental increase in the frequency of these strains enhances colony-wide antibiotic output. These results indicate that the benefits of producing cells with chromosomal lesions are evident across a broad range of frequencies, but that even with extremely high mutation rates, the costs to colony-wide fitness are minimal or entirely absent.
DISCUSSION
Streptomycetes are prolific producers of antibiotics, with genomes typically containing more than 20 secondary metabolite gene clusters that comprise more than 5% of their entire genome (34, 42, 43). They invest heavily in these products, and their biosynthesis and secretion are costly. Our results suggest that, by limiting antibiotic production to a fraction of the colony through division of labor, S. coelicolor can eliminate the overall costs of biosynthesis while maximizing both the magnitude and diversity of their secreted antibiotics. Although this comes at a large individual cost, it increases group fitness by improving the ability for S. coelicolor to inhibit their competitors. Moreover, our results reveal that the range of conditions that select for a division of labor are quite broad, because colony-wide fitness is unaffected, even if mutant strains are as frequent as ~50%.
Division of labor is predicted to be favored in this system for several reasons. First, Streptomyces colonies emerge from a single spore and are clonal (19). This fact, together with their filamentous mode of growth, ensures that costly individual traits can be maintained because of their indirect fitness benefits (4, 5, 13). In addition, because resistance to the diversified antibiotic profile of mutant strains is unlikely to be present in competing strains, only the parent strain stands to benefit from their sacrifice. Second, the costs of antibiotic production via large and dedicated multistep biosynthetic pathways, e.g., nonribosomal peptide or polyketide synthases, are likely to be highest at the initiation of antibiotic production but diminish thereafter, meaning that producing cells become more efficient at making antibiotics through time (11); furthermore, we show that antibiotic production trades off with reproduction. Last, many antibiotics are secreted, so the entire colony, but not susceptible competitors, can benefit from the protection they provide (44).
Even if conditions predispose to a division of labor, there must still be a process that generates phenotypic heterogeneity. Our results show that, in Streptomyces, this is caused by genomic instability that creates a subpopulation of cells within colonies that contain large deletions or amplifications at the termini (23, 25, 31). These mutations are severe and irreversible. Because strains, or portions of colonies, containing these deletions have significantly reduced fitness, they effectively behave like a sterile caste that provide direct benefits to the rest of the colony and receive little in return (1). Even when the initial frequency of mutants in mixed colonies approaches 80%, their final frequency declines to less than 1% after one cycle of colony growth (fig. S7). This suggests that the division of labor in S. coelicolor is reestablished independently and differently in each colony, a mechanism that may help to maximize the diversity of secreted antibiotics.
It remains unclear whether there are mechanisms regulating the size and frequency of chromosomal deletions and amplifications. One possibility is that these events are induced by external environmental conditions and that their rate is context dependent. Instability can be elevated by exposure to certain toxicants, e.g., mitomycin C or nitrous acid (45), although no explicit stress was added in the experiments we report, and it also varies with media type. It may also be increased during competition with other strains, a process that is known to alter the secretion of secondary metabolites (16, 46). Another possibility is that deletions and the benefits they bring for antibiotic production are a fortuitous by-product of the cell death that accompanies development (19). By this argument, chromosome degradation would be regulated but would not always be lethal. Although we have not confirmed this experimentally, it is likely that conserved amplifications result from the flanking copies of IS1649, which can facilitate intragenomic rearrangements (47). In either case, the expectation is that increased antibiotic production results from the deregulation of biosynthetic clusters following the deletion of hundreds of genes, many known to coordinate antibiotic biosynthesis (34). In addition, because deletions are stochastic, especially following the removal of protective telomeres at the ends of linear chromosomes, this would also cause antibiotic production to vary in different sections of the colony.
Our preliminary surveys have found similar levels of genomic instability in streptomycete strains that we have freshly isolated from soil, suggesting that the division of labor we describe here is general. We are limited, however, in our ability to detect polymorphisms; color changes are conspicuous and are invariably associated with changes to pigmented secondary metabolites, but other secreted public goods may also become modified in these multicellular bacteria. Understanding which, if any other, public goods vary in the ways shown here is crucial because it will help to identify conditions that lead to a genetically encoded division of labor as compared to other forms of regulation that allow complex multicellular microbial systems to coordinate their behaviors and maximize their fitness.
MATERIALS AND METHODS
Bacterial strains and growth conditions
S. coelicolor A3(2) M145 was obtained from the John Innes Centre strain collection. The strain was cultivated at 30°C on soy flour mannitol medium agar plates (SFM) for strain isolation and to quantify CFU (48). SFM contains mannitol (20 g liter−1), agar (20 g liter−1), and soya flour (20 g liter−1). To examine antibiotic production and to extract secondary metabolites, we used MM supplied with 0.5% mannitol and casamino acids (740 μg ml−1). MM contains asparagine (0.5 g liter−1), K2HPO4 (0.5 g liter−1), MgSO4·7H2O (0.2 g liter−1), FeSO4·7H20 (0.01 g liter−1), and agar (10 g liter−1). For DNA extraction, strains were grown in liquid flasks shaken at 200 rpm at 30°C in TSBS:YEME (1:1, v:v) supplemented with 0.5% glycine and 5 mM MgCl2. TSBS contains tryptic soya broth powder (30 g liter−1) and sucrose (100 g liter−1), and YEME contains 3 g of yeast extract, 5 g of peptone, 3 g of malt extract, 10 g of glucose, and 340 g of sucrose. Escherichia coli and B. subtilis were cultivated at 37°C in LB media with constant shaking or on LB agar plates.
All strains were derived from a single isolate of S. coelicolor A3(2) M145 (designed as WT). Briefly, samples from a frozen spore stock were diluted and plated onto SFM agar to obtain single colonies. After 5 days of growth, single colonies with WT morphology were diluted and plated onto another SFM plate. From each plate, single colonies with conspicuously mutant phenotypes were picked into sterile water and plated at appropriate dilutions onto SFM agar (n = 3 per colony), from which we estimated the frequency of different mutant phenotype classes. Each derived type was plated to confluence on SFM agar, and after 7 days of growth, spores were harvested to generate spore stocks, which were stored at −80°C in 20% glycerol. To quantify mutation frequency, single colonies were grown for 5 days on three different media, and then we picked the colonies with WT morphology, diluted, and plated them onto the corresponding media. Mutation frequency was scored on the basis of the phenotypes after 3 to 5 days. Table S3 provides strain designations and indicates which strains were examined in each set of assays.
Phenotypic scoring
Two phenotypes that are related to the loss of loci in the right arm were scored (n = 3 per strain). The arginine auxotrophs were identified by replicating 103 CFUs of each strain on MM supplied with 0.5% mannitol with and without arginine (37 μg ml−1) (45). After 5 days of growth, auxotrophs were identified as those strains that only grow on the media supplied with arginine. Chloramphenicol resistance was estimated by using the disk diffusion method. In detail, 2 × 105 spores were spread onto MM supplemented with casamino acids (740 μg ml−1) (45) in 12-mm square petri dishes, followed by placing a paper disk containing 25 μg of chloramphenicol on it. After 4 days, the radius of the inhibition zone around the disk was measured using ImageJ (49). Inhibition zones that were smaller than 5 mm were scored as resistant, while those that are larger than 5 mm were scored as susceptible.
Antibiotic production
Spores of each strain were diluted to 105 CFU ml−1 in Milli-Q water, and 1 μl was spotted onto MM + casamino acid agar plates (n = 4 per strain). After growth for 5 days at 30°C, plates were covered with 15 ml of LB soft agar (0.7%) containing 300 μl of a freshly grown indicator strain [optical density at 600 nm (OD600) = 0.4 to 0.6]. After overnight incubation at 30°C, zones of inhibition around producing colonies were measured using ImageJ.
The bioactivity against Streptomyces isolates was tested for four strains: 2H1A, 8H1B, 9H1B, and 9H1A. Three milliliters of SFM agar was poured onto each well of a 100-mm square petri dish (Thermo Fisher Scientific), after which we spotted 1 μl of each test strain containing ~106 total spores in the corner of each well. After 5-day growth, 500 μl of MM supplemented with 0.5% mannitol and casamino acids (740 μg ml−1) containing ~105 spores of the target strain was overlaid on top. Zones of inhibition were measured 2 days later using ImageJ.
1H NMR profiling and data analysis
Spores (2 × 105) were spread onto MM + casamino acids in 12-mm square petri dishes (n = 3 per strain, except n = 2 for one WT clone). After 5 days of incubation at 30°C, agar was chopped into small pieces using a sterile metal spatula and secreted compounds were extracted in 50 ml of ethyl acetate for 72 hours at room temperature. Next, the supernatant was poured off and evaporated at 37°C using a rotating evaporator. Pellets were obtained by drying at room temperature to remove extra solvents and then freeze-dried to remove remaining water. After adding 500 μl of methanol-d4 to the dried pellets, the mixtures were vortexed for 30 s followed by a 10-min centrifugation at 16,000 rpm. The supernatants were then loaded into a 3-mm NMR tube and analyzed using 60-MHz 1H NMR (Bruker, Karlsruhe, Germany) (36, 37).
Data bucketing of NMR profiles was performed using AMIX software (version 3.9.12, Bruker BioSpin GmbH) set to include the region from δ 10.02 to 0.2 with a bin of 0.04 parts per million scaled to total intensity, while the signal regions of residual H2O in methanol (δ 4.9 to 4.7) and methanol (δ 3.34 to 3.28) were excluded. Multivariate data analysis was performed with the SIMCA software (version 15, Umetrics, Sweden) (36).
MS-based quantitative proteomics
Spores (104) were spotted on SFM agar covered with cellophane and incubated for 5 days at 30°C. Colonies were scraped off and snap-frozen in liquid N2 in tubes and then lysed three times in a precooled TissueLyser (Qiagen, The Netherlands). Proteins were dissolved in lysis buffer [4% SDS, 100 mM tris-HCl (pH 7.6), 50 mM EDTA] and then precipitated using chloroform-methanol (50). The dried proteins were dissolved in 0.1% RapiGest SF surfactant (Waters, USA) at 95°C. Protein digestion steps were done according to van Rooden et al. (51). After digestion, trifluoroacetic acid was added for complete degradation and removal of RapiGest SF. Peptide solution containing 8 μg of peptide was then cleaned and desalted using the STAGE-Tipping technique (52). Final peptide concentration was adjusted to 40 ng μl−1 with 3% acetonitrile solution containing 0.5% formic acid. Two hundred nanograms of digested peptide was injected and analyzed by reversed-phase liquid chromatography on a nanoACQUITY UPLC system (Waters) equipped with HSS-T3 C18 1.8 μm, 75 μm × 250 mm column (Waters). A gradient from 1 to 40% acetonitrile in 110 min was applied, and [Glu1]-fibrinopeptide B was used as the lock mass compound and sampled every 30 s. Online MS/MS analysis was done using a Synapt G2-Si HDMS mass spectrometer (Waters) with an UDMSE method setup as described (51).
Mass spectrum data were generated using ProteinLynx Global SERVER (PLGS, version 3.0.3), with MSE processing parameters with charge 2 lock mass 785.8426. Reference protein database was downloaded from GenBank with the accession number NC_003888.3. The resulting data were imported to ISOQuant (53) for label-free quantification. TOP3 quantification result from ISOQuant was used in later data processing steps.
In total, of the 7767 proteins from the database, 2261 proteins were identified across all samples. For each sample, on average, 1435 proteins were identified. TOP3 quantification was filtered to remove identifications meeting both criteria: (i) identified in less than 70% of samples of each strain and (ii) the sum of TOP3 value less than 1 × 105. This led to the removal of 297 protein quantification results. Proteins were considered significantly altered in expression when log2 fold change ≥ 1 and P ≤ 0.05. Volcano plots were made from filtered data, with the four biosynthetic gene clusters color-coded.
CFU production
To quantify CFU, 104 spores of each strain were spread onto SFM agar (n = 3 per strain, except n = 2 for 9H1B) and left to grow for 7 days to confluence. After 7 days, spores were harvested by first adding 10 ml of Milli-Q water onto the plate and then using a cotton swab to remove spores and mycelial fragments from the plate surface. Next, the water suspension was filtered through an 18-gauge syringe plugged with cotton wool to remove mycelial fragments. After centrifuging the filtered suspension at 4000 rpm for 10 min, the supernatant was poured off and the spore pellet was dissolved in a total volume of 1 ml of 20% glycerol. CFU per plate was determined via serial dilution onto SFM agar.
DNA extraction and sequencing
Nine strains, including one WT and eight mutants, were selected for sequencing with the Sequel Systems from Pacific Biosciences (PacBio, USA). Roughly 108 spores were inoculated in 25 ml of TSBS:YEME (1:1, v:v) supplemented with 0.5% glycine and 5 mM MgCl2 and cultivated at 30°C with 200 rpm shaking speed overnight. The pellet was then collected after centrifugation and washed twice with 10.3% sucrose. Samples were then resuspended in DNA/RNA Shield (Zymo Research, USA) with 10× volume at room temperature and sent to be commercially sequenced at BaseClear (Leiden, The Netherlands).
Subreads of the sequenced results shorter than 50 base pairs (bp) were filtered and stored in BAM format. The reference alignments were performed against S. coelicolor A3(2) genome (NC_003888.3) using BLASR (v5.3.2) (54). Resulting BAM files were then sorted and indexed using SAMtools (v1.9) (55). For the calculation of genome rearrangements, the depths were called and exported through the depth function in SAMtools. The edges of genome were identified by manually checking the break point where the coverage drops to zero. The size of the amplified region was defined by the markedly higher coverage compared to the adjacent sequences. All results were further confirmed by visualizing them in IGV (v2.4.15) (56, 57).
Pulsed-field gel electrophoresis
Approximately 108 spores were inoculated into 25 ml of TSBS:YEME (1:1, v:v) supplemented with 0.5% glycine and 5 mM MgCl2 and cultivated overnight at 30°C at 200 rpm. After centrifuging the culture at 4000 rpm for 10 min, the pellet was resuspended in 400 μl of cell suspension buffer containing 100 mM tris:100 mM EDTA (pH 8.0) and lysozyme (1 mg ml−1) and mixed with the same volume of 1% SeaKem Gold Agarose (Lonza, USA) in TE buffer containing 10 mM tris:1 mM EDTA (pH 8.0) with 1% SDS. This mixture was immediately loaded into the PFGE plug mold (Bio-Rad, USA). Next, plugs were lysed in 5 ml of cell lysis buffer containing 50 mM tris:50 mM EDTA (pH 8.0), 1% N-lauroylsarcosine sodium salt, and lysozyme (4 mg ml−1) incubated for 4 hours at 37°C with gentle agitation. This was then followed by a 5-hour incubation in 5-ml cell lysis buffer containing proteinase K (0.1 mg ml−1; Qiagen, The Netherlands) at 56°C and 50 rpm. Last, the plug was washed twice in preheated Milli-Q water and four times in preheated TE buffer and incubated at 56°C for at least 15 min with gentle mixing. Plugs were sliced into 2-mm width pieces and presoaked in 200 μl of 1× NEBuffer 3.1 for at least 30 min. After replacing the buffer with 200 μl of 1× NEBuffer 3.1, 2 μl of the rare-cutter Ase I (New England Biolabs, UK) was added and incubated at 30°C overnight. Agarose (1%) was used for running fragments in 0.5× freshly prepared tris-borate-EDTA. S. cerevisiae chromosomal DNA (0.225 to 2.2 Mb; Bio-Rad, USA) and WT S. coelicolor DNA were used as size markers to estimate fragment sizes. Two electrophoresis conditions were applied to separate and visualize the smaller (<1016 kb) and larger (>1016 kb) fragments: (switch time: 2.2 to 75 s; voltage: 200 V; running time: 19 hours) and (switch time: 60 to 125 s; voltage: 200 V; running time: 20 hours), respectively.
PFGE results were compared to the Ase I restriction maps of the WT strain, which contains 17 fragments ranging from 26 bp to 1601 kb (fig. S3). Two fragments, 240 and 632 kb, can be easily resolved if they are deleted from the left arm, while one large 1601-kb fragment can be affected when deletions occur in the right arm.
Fitness estimates
The relative fitness of four fully sequenced mutant isolates—2H1A, 8H1B, 9H1B, and 9H1A—was estimated during pairwise competition with the WT parent. To distinguish strains, we first transformed mutant and WT strains with the integrating plasmids PIJ82 and pSET152, which confer hygromycin B and apramycin resistance, respectively. Potential fitness effects of the markers were determined by generating two WT variants that were transformed with either single marker. No effects were observed in these control experiments (one-tailed t test, t = 2.029, P = 0.082). Fitness assays were initiated by normalizing each strain to a density of 106 spores ml−1 and then mixing at different starting ratios of mutant:WT. One hundred microliters of this mixture, containing 105 spores, was plated as a lawn onto SFM agar and incubated at 30°C for 5 days, while another fraction of the sample was plated after serial dilution onto SFM containing either apramycin (50 μg ml−1) or hygromycin B (50 μg ml−1) to precisely quantify the densities of each strain. After 5 days of growth, bacteria were harvested as above and plated by serial dilution onto SFM containing either apramycin (50 μg ml−1) or hygromycin B (50 μg ml−1). Fitness was quantified, following Lenski et al. (58), by calculating the ratio of the Malthusian parameters of both strains. Values below 1 indicate that mutant strains have lower fitness than the WT strain. More detailed assays were carried out with strain 9H1A, where we simultaneously estimated the fitness of this strain at a broader range of frequencies from 10 to 99% and determined how the frequency of the mutant strain influenced antibiotic production, as measured by the size of the zone of inhibition against a B. subtilis indicator in a soft agar overlay.
Supplementary Material
Acknowledgments
We acknowledge the helpful comments of C. Kost, S. Shitut, and S. Westhoff on an earlier version of this manuscript and assistance with NMR analysis from L. Bayona and C. Wu. We acknowledge B. Florea of Leiden University for running and monitoring proteome measurements and the bio-organic synthesis group at Leiden University for providing access to instrumentation. Funding: Funding was provided by the China Scholarship Council (CSC) to Z.Z., JPI-AMR to A.L. and D.E.R., and NWO to D.C. Author contributions: Z.Z., D.C., and D.E.R. conceptualized and planned the study. Z.Z., C.D., and F.d.B. collected data. All authors analyzed and interpreted the results. Z.Z., D.C., and D.E.R. drafted the manuscript with input from the other authors. All authors approved the final submitted version of this paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD014413 and 10.6019/PXD014413. Raw data available at Dryad. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/3/eaay5781/DC1
Fig. S1. Mutant frequencies in different media.
Fig. S2. PacBio sequencing results of nine selected strains.
Fig. S3. PFGE results of all sampled strains.
Fig. S4. Volcano plots of proteomics from four mutant strain.
Fig. S5. Trade-off between fitness and antibiotic production.
Fig. S6. Extended evidence for division of labor during coculture of the WT and three mutant strains at different starting frequencies.
Fig. S7. Competition assays between WT and mutant strain 9H1A at different starting frequencies.
Table S1. Filtered proteomics data.
Table S2. 1H NMR signals ranked by X and Y weights (w* and c) for PLS component 1.
Table S3. Mutant strains used in this study.
REFERENCES AND NOTES
- 1.Robinson G. E., Regulation of division of labor in insect societies. Annu. Rev. Entomol. 37, 637–665 (1992). [DOI] [PubMed] [Google Scholar]
- 2.van Gestel J., Vlamakis H., Kolter R., Division of labor in biofilms: The ecology of cell differentiation. Microbiol. Spectr. 3, 1–24 (2015). [DOI] [PubMed] [Google Scholar]
- 3.van Gestel J., Vlamakis H., Kolter R., From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLOS Biol. 13, e1002141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West S. A., Cooper G. A., Division of labour in microorganisms: An evolutionary perspective. Nat. Rev. Microbiol. 14, 716–723 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z., Claessen D., Rozen D. E., Understanding microbial divisions of labor. Front. Microbiol. 7, 2070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim W., Levy S. B., Foster K. R., Rapid radiation in bacteria leads to a division of labour. Nat. Commun. 8, 10508 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dragoš A., Kiesewalter H., Martin M., Hsu C. Y., Hartmann R., Wechsler T., Eriksen C., Brix S., Drescher K., Stanley-Wall N., Kümmerli R., Kovács Á. T., Division of labor during biofilm matrix production. Curr. Biol. 28, 1903–1913.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dragoš A., Martin M., García C. F., Kricks L., Pausch P., Heimerl T., Bálint B., Maróti G., Bange G., López D., Lieleg O., Kovács Á. T., Collapse of genetic division of labour and evolution of autonomy in pellicle biofilms. Nat. Microbiol. 3, 1451–1460 (2018). [DOI] [PubMed] [Google Scholar]
- 9.West S. A., Diggle S. P., Buckling A., Gardner A., Griffin A. S., The social lives of microbes. Annu. Rev. Ecol. Evol. Syst. 38, 53–77 (2007). [Google Scholar]
- 10.Tarnita C. E., The ecology and evolution of social behavior in microbes. J. Exp. Biol. 220, 18–24 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Schiessl K. T., Ross-Gillespie A., Cornforth D. M., Weigert M., Bigosch C., Brown S. P., Ackermann M., Kümmerli R., Individual- versus group-optimality in the production of secreted bacterial compounds. Evolution 73, 675–688 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barka E. A., Vatsa P., Sanchez L., Gaveau-Vaillant N., Jacquard C., Klenk H.-P., Clément C., Ouhdouch Y., van Wezel G. P., Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claessen D., Rozen D. E., Kuipers O. P., Søgaard-Andersen L., van Wezel G. P., Bacterial solutions to multicellularity: A tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 12, 115–124 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Flärdh K., Buttner M. J., Streptomyces morphogenetics: Dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7, 36–49 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Chater K. F., Biró S., Lee K. J., Palmer T., Schrempf H., The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 34, 171–198 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Abrudan M. I., Smakman F., Grimbergen A. J., Westhoff S., Miller E. L., van Wezel G. P., Rozen D. E., Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc. Natl. Acad. Sci. U.S.A. 112, 11054–11059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D. A. Hopwood, Streptomyces in Nature and Medicine: The Antibiotic Makers (Oxford Univ. Press, 2007). [Google Scholar]
- 18.van der Heul H. U., Bilyk B. L., McDowall K. J., Seipke R. F., van Wezel G. P., Regulation of antibiotic production in Actinobacteria: New perspectives from the post-genomic era. Nat. Prod. Rep. 35, 575–604 (2018). [DOI] [PubMed] [Google Scholar]
- 19.McCormick J. R., Flärdh K., Signals and regulators that govern Streptomyces development. FEMS Microbiol. Rev. 36, 206–231 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bibb M. J., Kelemen G. H., Fernández-Abalos J. M., Sun J., Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145, 2221–2227 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Kelemen G. H., Viollier P. H., Tenor J., Marri L., Buttner M. J., Thompson C. J., A connection between stress and development in the multicellular prokaryote Streptomyces coelicolor A3(2). Mol. Microbiol. 40, 804–814 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Cullum J., Altenbuchner J., Flett F., Piendl W., DNA amplification and genetic instability in Streptomyces. Biotechnol. Genet. Eng. Rev. 4, 59–78 (1986). [DOI] [PubMed] [Google Scholar]
- 23.Birch A., Häusler A., Hütter R., Genome rearrangement and genetic instability in Streptomyces spp. J. Bacteriol. 172, 4138–4142 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leblond P., Decaris B., New insights into the genetic instability of streptomyces. FEMS Microbiol. Lett. 123, 225–232 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Volff J. N., Altenbuchner J., Genetic instability of the Streptomyces chromosome. Mol. Microbiol. 27, 239–246 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Freeman R. F., Bibb M. J., Hopwood D. A., Chloramphenicol acetyltransferase-independent chloramphenicol resistance in Streptomyces coelicolor A3(2). Microbiology 98, 453–465 (1977). [DOI] [PubMed] [Google Scholar]
- 27.Matsubara-Nakano M., Kataoka Y., Ogawara H., Unstable mutation of beta-lactamase production in Streptomyces lavendulae. Antimicrob. Agents Chemother. 17, 124–128 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coyne V. E., Usdin K., Kirby R., The effect of inhibitors of DNA repair on the genetic instability of Streptomyces cattleya. J. Gen. Microbiol. 130, 887–892 (1984). [DOI] [PubMed] [Google Scholar]
- 29.Altenbuchner J., Cullum J., DNA amplification and an unstable arginine gene in Streptomyces lividans 66. Mol. Gen. Genet. 195, 134–138 (1984). [DOI] [PubMed] [Google Scholar]
- 30.Dyson P., Schrempf H., Genetic instability and DNA amplification in Streptomyces lividans 66. J. Bacteriol. 169, 4796–4803 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.P. Leblond, P. Demuyter, L. Moutier, M. Laakel, B. Decaris, J. Simonet, Hypervariability, a new phenomenon of genetic instability, related to DNA amplification in Streptomyces ambofaciens. 171, 419–423 (1989). [DOI] [PMC free article] [PubMed]
- 32.Gravius B., Bezmalinović T., Hranueli D., Cullum J., Genetic instability and strain degeneration in Streptomyces rimosus. Appl. Environ. Microbiol. 59, 2220–2228 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krügel H., Krubasik P., Weber K., Saluz H. P., Sandmann G., Functional analysis of genes from Streptomyces griseus involved in the synthesis of isorenieratene, a carotenoid with aromatic end groups, revealed a novel type of carotenoid desaturase. Biochim. Biophys. Acta 1439, 57–64 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Bentley S. D., Chater K. F., Cerdeño-Tárraga A.-M., Challis G. L., Thomson N. R., James K. D., Harris D. E., Quail M. A., Kieser H., Harper D., Bateman A., Brown S., Chandra G., Chen C. W., Collins M., Cronin A., Fraser A., Goble A., Hidalgo J., Hornsby T., Howarth S., Huang C.-H., Kieser T., Larke L., Murphy L., Oliver K., O’Neil S., Rabbinowitsch E., Rajandream M.-A., Rutherford K., Rutter S., Seeger K., Saunders D., Sharp S., Squares R., Squares S., Taylor K., Warren T., Wietzorrek A., Woodward J., Barrell B. G., Parkhill J., Hopwood D. A., Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Takano K., Light-induced carotenogenesis in Streptomyces coelicolor A3(2): Identification of an extracytoplasmic function sigma factor that directs photodependent transcription of the carotenoid biosynthesis gene cluster. J. Bacteriol. 187, 1825–1832 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H. K., Choi Y. H., Verpoorte R., NMR-based metabolomic analysis of plants. Nat. Protoc. 5, 536–549 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Wu C., Du C., Gubbens J., Choi Y. H., van Wezel G. P., Metabolomics-driven discovery of a prenylated isatin antibiotic produced by Streptomyces species MBT28. J. Nat. Prod. 78, 2355–2363 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Gubbens J., Zhu H., Girard G., Song L., Florea B. I., Aston P., Ichinose K., Filippov D. V., Choi Y. H., Overkleeft H. S., Challis G. L., van Wezel G. P., Natural product proteomining, a quantitative proteomics platform, allows rapid discovery of biosynthetic gene clusters for different classes of natural products. Chem. Biol. 21, 707–718 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Pérez-Redondo R., Rodríguez-García A., Botas A., Santamarta I., Martín J. F., Liras P., ArgR of Streptomyces coelicolor is a versatile regulator. PLOS ONE 7, e32697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu H., Swierstra J., Wu C., Girard G., Choi Y. H., van Wamel W., Sandiford S. K., van Wezel G. P., Eliciting antibiotics active against the ESKAPE pathogens in a collection of actinomycetes isolated from mountain soils. Microbioly 160, 1714–1725 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Komatsu M., Uchiyama T., Omura S., Cane D. E., Ikeda H., Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 107, 2646–2651 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Challis G. L., Hopwood D. A., Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. U.S.A. 100, 14555–14561 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Craney A., Ahmed S., Nodwell J., Towards a new science of secondary metabolism. J. Antibiot. 66, 387–400 (2013). [DOI] [PubMed] [Google Scholar]
- 44.van der Meij A., Worsley S. F., Hutchings M. I., van Wezel G. P., Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 41, 392–416 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Volff J. N., Vandewiele D., Simonet J. M., Decaris B., Ultraviolet light, mitomycin C and nitrous acid induce genetic instability in Streptomyces ambofaciens ATCC23877. Mutat. Res. 287, 141–156 (1993). [DOI] [PubMed] [Google Scholar]
- 46.Traxler M. F., Watrous J. D., Alexandrov T., Dorrestein P. C., Kolter R., Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio 4, e00459-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Widenbrant E. M., Kao C. M., Introduction of the foreign transposon Tn4560 in Streptomyces coelicolor leads to genetic instability near the native insertion sequence IS1649. J. Bacteriol. 189, 9108–9116 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.T. Kieser, M. J. Bibb, M. J. Buttner, K. F. Chater, D. A. Hopwood, Practical Streptomyces genetics (John Innes Foundation, Norwich, 2000). [Google Scholar]
- 49.Rueden C. T., Schindelin J., Hiner M. C., DeZonia B. E., Walter A. E., Arena E. T., Eliceiri K. W., ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wessel D., Flügge U. I., A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 (1984). [DOI] [PubMed] [Google Scholar]
- 51.van Rooden E. J., Florea B. I., Deng H., Baggelaar M. P., van Esbroeck A. C. M., Zhou J., Overkleeft H. S., van der Stelt M., Mapping in vivo target interaction profiles of covalent inhibitors using chemical proteomics with label-free quantification. Nat. Protoc. 13, 752–767 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Rappsilber J., Ishihama Y., Mann M., Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Distler U., Kuharev J., Navarro P., Levin Y., Schild H., Tenzer S., Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat. Methods 11, 167–170 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Chaisson M. J., Tesler G., Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): Application and theory. BMC Bioinformatics 13, 238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lenski R., Rose M., Simpson S., Tadler S., Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138, 1315–1341 (1991). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/3/eaay5781/DC1
Fig. S1. Mutant frequencies in different media.
Fig. S2. PacBio sequencing results of nine selected strains.
Fig. S3. PFGE results of all sampled strains.
Fig. S4. Volcano plots of proteomics from four mutant strain.
Fig. S5. Trade-off between fitness and antibiotic production.
Fig. S6. Extended evidence for division of labor during coculture of the WT and three mutant strains at different starting frequencies.
Fig. S7. Competition assays between WT and mutant strain 9H1A at different starting frequencies.
Table S1. Filtered proteomics data.
Table S2. 1H NMR signals ranked by X and Y weights (w* and c) for PLS component 1.
Table S3. Mutant strains used in this study.


