Phagocytosis level is an indicator of cancer aggressiveness since both are related to mechanical deformability of the cells.
Abstract
The malignancy potential is correlated with the mechanical deformability of the cancer cells. However, mechanical tests for clinical applications are limited. We present here a Triangular Correlation (TrC) between cell deformability, phagocytic capacity, and cancer aggressiveness, suggesting that phagocytic measurements can be a mechanical surrogate marker of malignancy. The TrC was proved in human prostate cancer cells with different malignancy potential, and in human bladder cancer and melanoma cells that were sorted into subpopulations based solely on their phagocytic capacity. The more phagocytic subpopulations showed elevated aggressiveness ex vivo and in vivo. The uptake potential was preserved, and differences in gene expression and in epigenetic signature were detected. In all cases, enhanced phagocytic and aggressiveness phenotypes were correlated with greater cell deformability and predicted by a computational model. Our multidisciplinary study provides the proof of concept that phagocytic measurements can be applied for cancer diagnostics and precision medicine.
INTRODUCTION
There is growing evidence correlating cancer aggressiveness and the mechanical deformability of tumor cells. Various processes of cancer development require mechanical adjustment of tumor cells to physical environments, such as the growth of high pressure tumors, the detachment of metastases-forming cells from the tumor, their efficient motion and passage through narrow confinements, and their establishment in a different location (1–5). During the epithelial-mesenchymal transition (EMT), epithelial cells lose their cell polarity and cell-cell adhesion and gain migratory and invasive properties to acquire a mesenchymal stem cell phenotype (6). The generation of force transmitted to the cell from the extracellular matrix (ECM) is associated with signal transduction and can control the cancerous level of cells (7). To date, mechanical measurements of cells are mostly done using single-cell methods and, in many cases, require microscopy-based image analysis. Common examples include atomic force microscopy (AFM), microplate stretcher, micropipette aspiration, magnetic twisting cytometry, and microscopy image analysis (8–11). While such measurements are elegant and precise, their main disadvantage is being low throughput in most cases, reaching a range of 100 tested cells in most optimal cases. Adamo et al. (12) have produced data of about 3000 cells using a designated software with a microfluidic device. For clinical applications, it is critically important to develop robust tests of cell deformability because of the large heterogeneity among cancer cells (13, 14).
In addition to the link between cell mechanics and cancer potential, in normal cells, it is known that cell deformability is related to the capacity of cells to uptake particles, mainly in engulfment-mediated internalization including phagocytosis and endocytosis (15–17). Given that cancer aggressiveness is correlated with cell deformability and that cell deformability can control uptake capacity, an outstanding question is whether the mechanical properties of cells can tie cancer aggressiveness with particle uptake. Proving such an aggressiveness-uptake-deformability triangular correlation (TrC) can revolutionize the fields of cancer diagnostics and drug delivery design. Differential particle uptake capacity can be considered in rational design of selective drug delivery systems used in precision schemes. Such mechanical-based specificity can potentially reduce the challenging need of allocating specific target molecules for selectivity (18–20).
Here, we found a clear link between cancer aggressiveness, cell uptake capability, and cell deformability. High-throughput uptake measurements with tens of thousands of cells were done using fluorescence-activated cell sorter (FACS), as well as microplate reader spectrometry followed by detailed microscopy to assess particle localization. Uptake capacity of cancer cells was markedly higher in skin cancer cells as compared to noncancerous skin cells. Additional experiments with eight different cancer cell lines demonstrated a nonmonotonic dependence on particle size, which can be explained by physical considerations. By comparing cells with varying malignancy originating from primary (prostate) or metastatic sites (21, 22), we demonstrated that particle uptake was tightly correlated with cell invasiveness ex vivo and also with elevated cell deformability measured by AFM, where higher levels of force compliance indicated greater deformability. To determine whether heterogeneity in particle uptake has biological consequences, we generated, using FACS sorting, “phagocytic” and “nonphagocytic” subpopulations of cells (“positive” and “negative,” respectively) originating from the same human cell origin: either melanoma or bladder carcinoma cells. Unexpectedly, the phagocytic phenotype was enhanced over generations, suggesting involvement of genetic effectors. Moreover, positive cells were found to be more malignant and invasive than the negative ones in ex vivo and in vivo experiments. In mice, we found notable differences in the rate and size of tumor growth between the phagocytic and nonphagocytic cells, associated with elevated histological markers of tumor aggressiveness. Genetic up-regulation in transcription of EMT and cytoskeleton reorganization genes was found, with differences also in chromatin level. AFM experiments showed that the phagocytic cells were more deformable compared with the nonphagocytic cells. Further mechanistic insights were provided by a physical model using the specific cell parameters measured here and which quantitatively predicted the uptake patterns found in the experiments.
The TrC established here can lead to the development of a simple diagnostic scheme using uptake measurements as an additional parameter for grading cancer. To date, cancer diagnostics has not accounted for mechanical properties of cells, although they have been proven to be biomarkers of the malignancy potential. Cell uptake assessments can provide a simple, robust, and accurate diagnostic method and add mechanical knowledge to the existing clinical tools. Moreover, finding specific cell uptake features may provide a mean to rationally design individual drug carriers with higher specificity.
RESULTS
Malignant cells uptake inert particles more readily in a size-dependent manner
We examined whether cancer cells have enhanced abilities of particle uptake as compared to normal cells from the same tissue origin (Fig. 1). Skin cells from a primary cancerous cell line (melanoma, A375-P) were compared to transformed skin cells originated from normal cells (HaCaT, keratinocyte cell line). Both types of cells were incubated overnight with fluorescent polystyrently labelled particles, ranging from tens of nanometers to several micrometers in size, and the normalized uptake (see eq. S1) was evaluated using flow cytometry, a microplate reader, and fluorescence microscopy, as shown respectively in Fig. 1, A to C. After incubation, we consistently found, using all three methods, that uptake was markedly higher in cancer cells compared with normal cells. In both cell types, vanishing uptake of small particles below 0.5 μm was observed (explained below).
Fig. 1. Cancer skin cells (A375-P) present an enhanced uptake compared with noncancer skin cells (HaCaT).
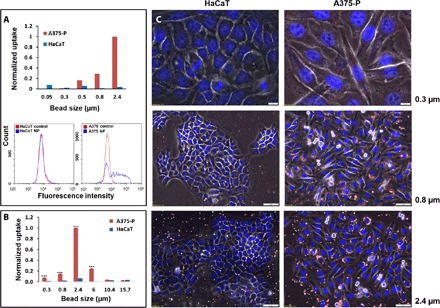
(A) FACS analysis is presented as column plot of normalized average uptake values of 0.05-, 0.3-, 0.5-, 0.8-, and 2.4-μm fluorescently labeled polystyrene beads. Representative histograms for 2.4-μm beads are shown at the bottom. (B) Microplate reader fluorescence analysis. Excitation, 530/25 and emission, 590/20, with 0.3-, 0.8-, 2.4-, 6-, 10.4-, and 15.7-μm beads. The unitless results in the column plots of (A) and (B) are normalized by the intensity of particle fluorescence to represent the relative mass of cell internalized particles and are scaled to a maximal value of 1 (see eq. S1). (C) Fluorescent microscopy images of cells incubated with 0.3-, 0.8-, and 2.4-μm beads. Blue, DAPI; red, beads. Scale bars, 10 μm (upper panels, 0.3 μm) or 50 μm (middle and lower panels, 0.8 and 2.4 μm).
To determine the relevant particle size for studying the TrC, eight different cell lines were tested from skin, prostate, bladder, and kidney cancers, which differed in their metastatic potential (see Materials and Methods). Experimental and theoretical results in Fig. 2 show a nonmonotonic dependence where the uptake increases with particle size, reaching a maximal value with 2.4-μm beads, and then declines. This was observed using FACS, the microplate reader (Fig. 2A), and confocal imaging (Fig. 2B), which also verified particle penetration into cells. The nonmonotonic dependence was predicted by a physical theory as illustrated in Fig. 2C. Engulfment and uptake of low-affinity particles by cells require a sufficiently large area of cell-particle adhesion to reduce the free energy, but minimal cell morphological changes as the latter increases the free energy. Hence, intermediate sized particles are optimal here for cell entrance, having a wide enough contact area with the cells and requiring small cell deformation during their engulfment.
Fig. 2. Uptake of beads by cancer cells has a nonmonotonic dependence on the bead size.
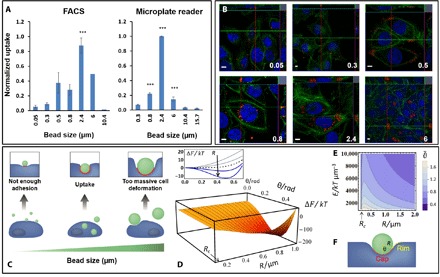
Cancer cell interaction with inert spherical fluorescent polystyrene beads increases with bead size, reaching a maximum around 2.4-μm bead diameter and then decreases. (A) Left: FACS analysis; average over normalized uptake values of eight cell lines: A375-p, A375-SM, PC3M-P, PC3M, LN4253J, 253 J-B5, SN12, and SN12-C. Right: Microplate fluorescent analysis (excitation, 530/25 and emission, 590/20), average over normalized uptake values of A375-P, A375-SM, PC3M-P, and PC3M-LN4. (B) Confocal fluorescent images (blue, DAPI; green, fluorescein isothiocyanate–phalloidin; red, polystyrene fluorescently labeled beads). Scale bars, 10 μm. (C) Physical model predicted the nonmonotonic dependence due to the interplay between adhesion and cell deformability. (D) The free energy of the wrapping model has a minimum as a function of both the bead radius and the wrapping angle, ϑ (bottom); however, for small beads of R < Rc, there is no stable adhesion [top; plots downward are for R = 0.05, 0.11, 0.14 (Rc, dashed), 0.18, and 0.23 μm]. (E) The stable adhesion angle, (rad) is shown versus Young’s modulus and R. (F) Model geometry. Parameters used in plots of (D) and (E) are based on AFM calculations in PC3M-LN4 cells (see table S1).
The physical theory proposed here is a combination of contact mechanics principles for three-dimensional (3D) cell deformations upon contact with a particle, as well as a 2D description of cell membrane morphology, providing the following change in free energy upon cell-particle contact:
| (1) |
ϑ is the reaction coordinate describing particle wrapping by the cell membrane. The particle is modeled as a rigid sphere of radius R. The first two terms describe the adhesion and 3D elastic cell deformation according to the Derjaguin-Muller-Toporov (DMT) model (23). Δγ, the work of adhesion per unit area, and E, the effective Young’s modulus of the cell, are calculated here per cell type from our AFM results (table S1). The third term reflects the elastic membrane curvature with bending modulus κ = 4.5kBT (24) in thermal energy units, where kB is the Boltzmann constant and T is the temperature. The model derivation and assumptions can be found in Materials and Methods.
The function ΔF(ϑ) has a minimum only when the particles are larger than a critical value
| (2) |
For small beads below the critical radius Rc, ΔF is a monotonically increasing function (box in Fig. 2D), and it is not expected that the cells will uptake the particles, as indeed observed in our experimental results. For each specific cell type, Rc values were measured here using AFM and are presented in table S1. We found an excellent match with the experiments in predicting realistic values of Rc, below which uptake capacity was vanishing. From Eq. 1, we can derive the stable (equilibrium) wrapping angle, ; the higher is, the more likely it is to have full engulfment and uptake. is maximal for intermediate values of R (Fig. 2E) in agreement with our experimental nonmonotonicity findings (Fig. 2, A and B), due to the energetic considerations mentioned above. From the dependence of on E, the stiffer the cell, the less bead uptake is probable, as indeed found in our experiments (see the next section). The model geometry is sketched in Fig. 2F.
TrC in parental versus metastatic cancer cells
We compared cells with varying malignancy that originated from either a primary site of prostate cancer (PC3M-P) or a highly metastatic subpopulation of these cells (PC3M-LN4) (21, 22). A comparison of the three factors—particle uptake, mechanical deformability, and malignant potential between these cell types—is presented in Fig. 3. Particle uptake was found to be higher in PC3M-LN4 cells as measured by FACS (Fig. 3A) and confocal microscopy (Fig. 3B) and as predicted by our model (Fig. 3C). In the physical model, we used specific measured cell parameters from table S1 and found that the free energy is lower and that is larger for the more metastatic cells, predicting a higher uptake, as indeed observed by the FACS and the confocal microscope. The highly phagocytic-like PC3M-LN4 cells were also found to be more invasive. Figure 3D shows an ex vivo 3D multicellular spheroid formation assay (25) in which the PC3M-LN4 cells were clearly more invasive in collagen as compared with PC3M-P cells. This was reflected in higher values of spheroid compactness (shown in red) that were calculated from image analysis–based photos (see fig. S1A). Examples of these photos, corresponding to the lowest microscope images, are presented at the bottom of Fig. 3D. Figure 3E presents AFM results. The Young’s modulus of the PC3M-LN4 cells was smaller than that of PC3M-P cells, and their maximal adhesion force was larger. This indicates a higher mechanical deformability of PC3M-LN4 compared with PC3M-P cells; the PC3M-LN4 cells were more compliant to shape changes induced by indentation or retraction of the AFM probe (see Materials and Methods). Figure 3F illustrates that the overall results of Fig. 3 (A to E) demonstrate the TrC, where PC3M-LN4 cells were clearly more invasive, deformable, and phagocytic than the PC3M-P cells.
Fig. 3. TrC in primary versus metastatic cancer cells.
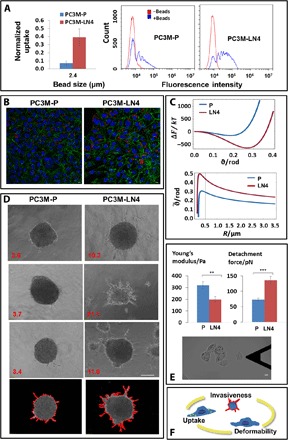
(A to C) Uptake comparison of 2.4-μm beads in primary versus metastatic cell lines of prostate cancer, PC3M-P and PC3M-LN4, respectively, showing elevated uptake by the metastatic cells: (A) FACS analysis, (B) confocal images (scale bars, 10 μm), and (C) results of the theoretical model with specific parameters of PC3M-P and PC3M-LN4: ΔF (ϑ) curves for R = 2.37 μm (top) as well as (R) (bottom) demonstrate that the metastatic cells internalize more particles. (D) Spheroid invasion test shows enhanced invasiveness of the metastatic compared to the primary cells. Compactness values are shown in red. At the bottom shown are examples of image analysis made for calculating the compactness of the microscope snapshots above them. Scale bar, 100 μm. (E) A comparison of the mechanical properties of the cells: Young’s modulus and the maximal adhesion force, using AFM, show that the metastatic cells are more elastic and more adhesive than the primary cells. A typical snapshot of a PC3M-LN4 cell from the AFM microscope is shown. Scale bar, 10 μm. The results presented in (A) to (E) support the TrC as shown in the cartoon (F). The PC3M-LN4 cells have higher uptake capacity (A and B), and they are more invasive (D) and are more deformable (E) in comparison with the PC3M-P cells, as also supported by the physical model in (C).
TrC in phagocytosis-based sorted cancer cells
We found an innate heterogeneity in uptake capacity among given cancer cell populations (Fig. 4A and movie S1). Hence, we sorted cancer cells from the same population by their phagocytic potential, aiming to determine whether such sorting can also subgroup cells according to their malignancy potential and their deformability. In our scheme (Fig. 4B), a FACS sorter was used for separating cells that did not take any particles (negative) from cells that had taken up any number of particles (positive). We used 2.4-μm beads, for which separate peaks of the FACS diagram corresponded to a discrete number of cell-inserted beads (fig. S2). This allowed us to control the number of beads taken by cells in the FACS sorting procedure. We started with two types of primary cancer cells: bladder (253 J) and melanoma (A375-P). Six cycles of FACS sorting were done, yielding very efficient enrichment patterns (Fig. 4C), indicating that uptake potential is a preserved characterization. The enrichment process resulted in four subpopulations: 253 J positive, 253 J negative, A375-P positive, and A375-P negative. Notably, the positive cells (both in 253 J and A375-P) showed a more invasive phenotype with higher compactness values as compared with the negative ones in a spheroid invasion test (Fig. 5A and fig. S1B). Proliferation rates, which can affect the extent of particle uptake (movies S2 and S3), were found to be identical in positive and negative subpopulations (fig. S3); therefore, they were not the cause for the differences in cell behavior. AFM measurements (Fig. 5B) confirmed TrC, showing that both of the positive subtypes were more adhesive than their negative counterparts. The A375-P–positive cells were also more elastic than the negative ones. The AFM results were used as input parameters in the physical model (Fig. 5C). In both cell pairs, the model predicted a higher extent of engulfment by positive as compared with negative cells, implying higher particle uptake by the positive cells. A complementary view was achieved by studying how stiffening of cancer cells affected the uptake of 0.8-μm beads. Figure S4 shows that the uptake by stiffened cells was notably lower than the uptake by nontreated cells. To shed light on the phagocytic phenotype and to pinpoint on relevant genes that may mediate this effect, we performed RNA sequencing of the four subpopulations. Gene set enrichment analysis (GSEA) was used on the hallmark gene set collection of the molecular signature database (mSigDB). In both cell lines, positive-enriched cells displayed up-regulation in the transcription of EMT and cytoskeleton-related gene sets (Fig. 5D). Top cytoskeleton-related up-regulated gene sets included GO_actin_cytoskeleton, GO_cytoskeletal_protein_binding, GO_actin_filament_based_process, KEGG_regulation_of_actin_cytoskeleton, and GO_structural_constituent_of_cytoskeleton. Figure 5E shows protein-protein interaction networks of the genes that contributed to the up-regulation. Next, we asked whether the differences in gene expression are reflected at the chromatin level as well. To that end, we performed chromatin immunoprecipitation sequencing (ChIP-seq) for the active histone mark, H3K4me2, on A375-P–positive and A375-P–negative cells. As expected, the vast majority of the peaks (i.e., 48,120 peaks) were shared between the positive and negative cells. However, we still observed 7327 unique peaks for the positive cells and 2940 peaks for the negative cells (Fig. 5F). The presence of a group-specific peak was correlated with the expression levels of the affected genes (Fig. 5G, two loci LINP1 and ADGRL2 are shown). Using Genomic Regions Enrichment of Annotations Tool (GREAT), we associated the specific peaks to their relevant genes and ran Gene ontology (GO) annotation. Similarly to the transcriptome analysis, the positive-specific peaks were enriched in genes that participate in cell polarity pathway and tyrosine kinase activity and the negative cells for epithelial structure maintenance. Together, our data suggest that the positive cell phenotype was associated with EMT.
Fig. 4. Sorting cancer cells based on uptake capacity of 2.4-μm beads.
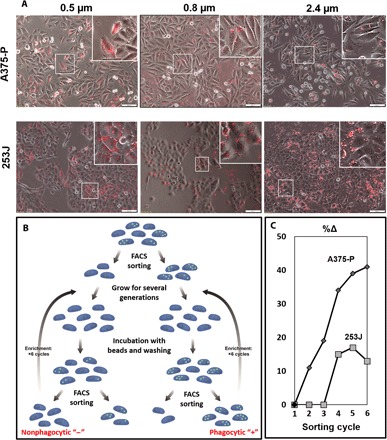
(A) Uptake is heterogenic among a cell population. Fluorescence microscope images of either A375-P or 253 J cell types after incubation with beads show that while some cells internalized many particles, others did not internalize any. Scale bars, 50 μm (upper images) and 100 μm (lower images). (B) Scheme of FACS sorting procedure for separating cells based on uptake capacity. Each sorted cycle resulted in two subpopulations of phagocytic (positive) cells and nonphagocytic (negative) cells. (C). Enrichment uptake graph. “Δ” is the percent of positive cells generated from a previously positive population, minus the percent of positive cells generated from a previously negative population (see Materials and Methods).
Fig. 5. TrC in cells that were sorted on the basis of their uptake capacity.
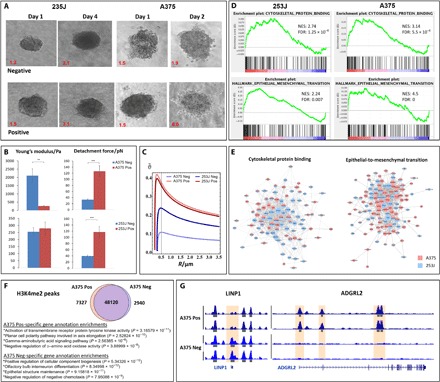
The cells with higher phagocytic capacity (positive cells) were more invasive and more deformable than the negative cells (A to C). (A) Spatial invasion assay with compactness values using spheroids of positive and negative A375-P and 253J cells showed that positive cells were more migratory and invasive than negative cells in collagen. (B) AFM results revealed that both of the positive subtypes were more adhesive than their negative counterparts. The A375-P positive cells were also more elastic than the negative ones. (C) The AFM results were used as parameters in the physical model. The stable wrapping angle was found to be larger in the positive cells compared with the negative cells (showing a higher effect in A375-P than in 253J, in agreement with the experimental results). The differences between positive and negative cells were further examined in bioinformatics assays (D and E). (D) GSEA plots showing up-regulation of cytoskeletal protein binding and EMT in A375-P and 253J cells. NES, normalized enrichment signal; FDR, false discovery rate. (E) Protein-protein interaction networks of genes are shown. Nodes represent proteins, and an edge connects proteins that interact [interactions taken from HIPPIE (2) and TSRING (3)], where the size of a node is proportional to the number of proteins that interact with it. Light blue, 253J; pink, A375; mixed light blue and pink, nodes that contributed to the enrichment signal in both cell lines. (F) Venn diagram of H3K4me3 ChIP-seq peaks in A375-positive and A375-negative cells. Overall, we have identified 58,387 peaks, of which 48,120 peaks (82%) are shared among the two conditions. Conversely, 7327 peaks (13%) are unique to A375-positive cells and 2940 peaks (5%) to A375 Neg. Analysis of gene annotation shows enrichment for EMT-related terms among genes associated with the A375 Pos peaks and maintenance of epithelial morphology–related terms among the A375 Neg–related genes. (G) Examples for A375 Pos and Neg differential peaks. An A375 Neg–specific H3K4me2 peak (left, highlighted box) is shown near the LINP1 promoter. Conversely, several A375 Pos–specific H3K4me2 ChIP-seq peaks were identified at the ADGRL2 gene (right, highlighted boxes). Both cases are in accordance with differential gene expression levels for those genes, as discussed above.
For a stronger level of validation of the differential malignancy in the uptake-based sorted cells, we performed an in vivo experiment. Three groups of nude mice were injected subcutaneously with cells originating from our sorting experiments: (i) original A375-P cells, (ii) positive A375-P cells, and (iii) negative A375-P cells. Tumor growth was monitored over 20 days in C57/BL mice. A notable (P = 0.033) difference in tumor volume was obtained after 9 days, where the negative cells’ tumors were 52 or 64% smaller in volume than the tumors formed in the positive or parental groups, respectively (Fig. 6A). Tumor images and weights are presented in Fig. 6B. It was expected that there would be a large divergence of the negative population and similarity between positive and parental cells, since the negative cells were formed by specifically isolating nonphagocytic cells from the rest of the population. As shown in Fig. 6C, histological examination of hematoxylin and eosin (H&E)–stained tissue sections revealed that positive cells induced dense and highly proliferative tumor tissues, while negative ones formed tissues with large areas of dead cells. To investigate the reason for the necrotic area, we performed immunohistological analysis using Ki67 as a marker for proliferation and cleaved caspase-3 to label apoptotic cells (Fig. 6D, top). The results suggest that the positive cell-originated cancers were more proliferative and showed reduced apoptosis as compared to the negative ones. Moreover, three EMT-related markers—Snail, E-cadherin, and smooth muscle actin (SMA) (Fig. 6D, bottom)—were tested to assess their level in the tumor tissues. The results revealed that Snail had higher levels in the positive tissues, while E-cadherin was lower, indicating on elevated EMT in the positive tumors, and that SMA levels were shown to be similar. The histological findings are summarized in Fig. 6E.
Fig. 6. TrC in uptake-based sorted cells was validated in a xenograft study.
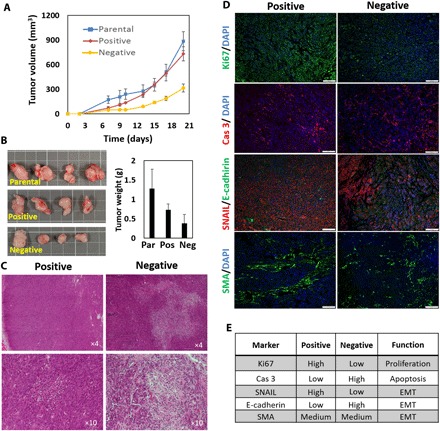
Parental, positive, and negative A375-P cells were injected subcutaneously to athymic nude mice. (A) Induced tumors were measured every other day from day 9 following the injection. The negative tumors were notably smaller in volume than the positive and the parental ones. (B) Extracted tumor tissues from the negative group were notably smaller compared with the positive and the parental tumors, as demonstrated visually and by weight measurements. Photo credit: O. Benny, HUJI. Square scale, 1 cm. H&E staining of the extracted tumors showed dense and highly proliferative tissues of the positive tumors (C) and, in contrast, large areas of necrosis in negative ones (D). Immunohistochemical staining for evaluating proliferation (anti-Ki67 in green), apoptosis [anti-cleaved caspase-3 (Cas 3) in red], and EMT (anti-SNAIL/SLUG in red versus anti–E-cadherin in green, as well as SMA in green). Positive tissues showed a higher level of proliferation (Ki67 staining), while negative tissues showed a higher level of apoptosis (cleaved caspase-3). SNAIL/SLUG versus E-cadherin show enhanced EMT in the positive tumors compared to the negative ones, while SMA values were similar. Scale bars, 100 μm. (E) A qualitative summary of histological immunohistological analysis as determined visually from captured images. Grading is based on positive-stained areas in a given tissue.
DISCUSSION
Growing knowledge about the high complexity of tumor cell biology has made it clear that an integrative approach is required in cancer research, including cell mechanics aspects. In the TrC proven here, mechanical deformability was found to be the mechanistic link that correlates cell uptake features with cancer aggressiveness. Correlations between cell deformability and cancer potential were previously studied. Various physical experiments were performed comparing the flexibility of normal and cancer cells with increasing malignancy. In numerous types of cancers, it was found that cell elasticity could be used as a biomarker for malignancy and for the metastatic potential (3, 8, 26, 27). Some studies found that a marked reduction of cell stiffness resulted in lower invasiveness potential, probably due to the difficulty in sensing and applying force (3, 28, 29). While cancer cells in vitro show enhanced elasticity, 3D tumor tissues are very stiff as compared with normal ones. This duality highlights the notable ability of cancer cells to adjust mechanically to different conditions such as the force exerted by the ECM (8, 30). It is well known that cell adhesiveness plays a major role in cancer progression. Turnover of specific cell adhesion molecules is required for enhanced dynamics of both cell-cell and cell-ECM adhesions during tumor growth and the spread of metastases (31). While most previous data focused on the relation between specific adhesions and malignancy, importantly, we found here that nonspecific adhesions were also correlated with cancer potential [supported by previous work (32)]. Namely, cell deformability, in terms of elasticity and nonspecific adhesiveness, is the mechanistic link between cancer aggressiveness and cell uptake capacity.
To date, information is sparse regarding the role of cell mechanics in particle-loading by cancer cells. A recent work by Ma et al. (33) showed that drug-resistant soft tumor-repopulating cells loaded cell-derived microparticles that contained drugs more efficiently than parental tumor cells that were stiffer and originally less resistant to drugs. Cancer cells often have enhanced abilities to internalize large micro-objects (34, 35), a property commonly referred to as a phagocytic ability (6, 28). Evidence of cancer cell cannibalism also implies on advanced capacities of internalizing large objects (36). In a work by Chandrasoma, published in 1980 (34), the author examined cells from a primary tumor of a patient with endometrial adenoacanthoma as well as from skin metastasis of the same patient. In the primary tumor, two cell populations were recognized; only one of them phagocytosed polymorphonuclear leucocytes. However, all metastasis cells phagocytosed polymorphs. It was suggested that the phagocytic abilities of primary tumor cells were related to their potential to form metastases. Since then, other reported studies outlined a phagocytic ability of cancer and metastases cells; however, direct comprehensive evidence of the aggressiveness-uptake link was not clear (6). Here, we found that cancer cells with higher metastatic potential had elevated capacities of particle uptake and that cell populations that were sorted on the basis of increased phagocytic properties were more aggressive. This provided a direct proof that the phagocytozing cells were more aggressive and vice versa.
Marked differences between particle uptake by keratinocyte cell line and cancer skin cells were found here, supporting the TrC. Moreover, we found that primary prostate cancer cells (PC3M-P) internalized particles fivefold less than a metastatic subpopulation of these cells (PC3M-LN4). The cells metastatic potential was validated using spheroid invasion tests showing higher invasiveness of the PC3M-LN4 cells, which were also found to be more elastic and adhesive in the AFM experiments. The AFM measurements were used as input parameters in our physical coarse-grained model, which qualitatively predicted uptake patterns of the primary and metastatic cell types using basic energetic arguments. The free energy terms of particle engulfment by cells include the interplay between the reduction of energy caused by cell-particle adhesion and an increase in energy resulting from cell deformation upon particle wrapping. We found here that cells of higher malignant and metastatic potential were more elastic and adhesive. Thus, the energetic penalty of their deformation is smaller and the gain in adhesion energy is larger, resulting in higher uptake capacity.
The size of particles was found to affect markedly the extent of uptake. A nonmonotonic dependence of uptake on particle size was found, with a maximum uptake for 2.4-μm beads. Other size dependencies reported in the literature [e.g., (35)], could have resulted from varying values of the physical adhesiveness and elasticity of cells or from biological factors such as the membrane surface brush (37), specific biochemical pathways such as clathrin- or caveolin-mediated internalization, and the formation of lipid rafts. Here, we tested beads of minimal cell affinity; thus, adhesion energy was insufficient to enable particle internalization in the case of small particles because of the small cell-particle contact area. Increased size of particles widens the contact area, and thus uptake becomes more favorable. However, a further increase in particle size requires massive cell deformation upon engulfment, and the uptake is reduced, resulting in a nonmonotonic dependence. Moreover, our model predicted the experimental observation of a critical radius, , that is specific for cell and particle type, below which particles do not penetrate cells.
A major challenge in cancer treatment results from internal heterogeneity and plasticity in terms of genetics, functionality, phenotypes, stem cell differentiation, microenvironment variability, and others (31, 38, 39). It should be recognized that mechanical heterogeneity is notable as well (13, 14) and that, ideally, all of these factors should be included in the profile of patients for optimizing their clinical therapy with precision schemes. We found a large variability in uptake capacity of cancer cells from the same population (Fig. 4A), raising two questions. Is uptake variability correlated with the cancer potential and with the mechanical properties of individual cells? And are these characteristics inherited from generation to generation and can therefore allocate potential targets for therapies? To address these questions, we sorted cells on the basis of uptake capacities using six enrichment cycles of FACS sorting. Differences between the sorted cells were notable and indicated conservation of the TrC. The fact that sorting of cells according to their particle uptake resulted in populations that were clearly distinguished from each other in their cancer potential after mice injection and in artificial 3D tumors (spheroids) may have an enormous impact on future drug design and diagnostics. Histologically, we found that positive cell populations had more aggressive phenotypes, enhanced proliferation, and an increase in EMT protein, Snail, and reduced E-cadherin expression, suggesting an EMT. Our gene expression analysis supported the mechanical link between uptake and cancer potential, suggesting the differential pattern of up-regulation in the transcription of EMT genes and of genes related to cytoskeleton reorganization. Differences between the sorted subpopulations persisted over generations with clear epigenetic differences. The differences in genes that were related to cell polarity pathway, tyrosine kinase activity, and epithelial structure maintenance, might explain the enrichment of the phagocytic phenotype with cell generations. Since EMT is associated with morphological cell changes, mobility, and cell polarity, it may be related to phagocytosis.
The significance of combining cell mechanics in the rational design of cancer therapies and in diagnostics is clear. However, robust, accurate, and simple mechanical tests for this aim are lacking. A consequence of the TrC is that uptake measurement can be used as a reliable high-throughput scheme of mechanical measurements in cancer. We anticipate that these findings will open up the possibility of practical simple procedures that use uptake tests as a routine in cancer diagnostics and in the rational design of selective therapies. Further development of our findings would involve focusing on clinical scenarios that can benefit from the TrC insights related to cell-particle interactions, including ex vivo diagnostics, or circulating cells. Of course, at the tissue level, additional effectors should be considered. Future research in this field can include investigations of different aspects of mechanical factors such as biochemical pathways and cytoskeleton remodeling. In addition, the efficacy of drug-loaded carriers can be examined for enhancing specificity on the basis of mechanical arguments.
MATERIALS AND METHODS
All institutional and national guidelines for the care and use of laboratory animals were followed, and protocols were approved by The Hebrew University of Jerusalem Ein Kearem Medical School Institutional Animal Care and Use Committee.
Cell culture
Human keratinocyte cell line, HaCaT cells (Thermo Fisher Scientific), was maintained in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific) supplemented with 10% (v/v) fetal bovine serum and 1% antibiotics [streptomycin (10,000 μg/ml) and penicillin (10,000 units/ml)] at 37°C with 5% CO2. A375-P (primary human melanoma cells), A375-SM (metastatic human melanoma cells), PC3M-P (primary prostate cancer cells), PC3M-LN4 (metastatic prostate cancer cells), 253 J (primary human bladder cancer cells), 253 J-B5 (metastatic human bladder cancer cells), SN12 (human renal primary cancer cells), and SN12-C SN12 (human renal metastatic cancer cells) were obtained by the Bielenberg laboratory at the Boston Children’s Hospital, USA and maintained as previously reported (22). Cells were serially passaged at 70 to 80% confluency, and then experiments were conducted with subconfluent cells. All cell lines were mycoplasma free (EZ-PCR Mycoplasma Test Kit Biological Industries, catalog number 2070020).
Materials and reagents
Unless otherwise stated, reagents were purchased form Sigma-Aldrich. Fluorescently labeled polystyrene spherical particles were purchased from Spherotech Inc. (USA). In most cases, purple fluorescence (excitation, 488 nm and emission, 545/60 nm) was used except for the 6-μm particles in Fig. 1B that were pink (excitation, 488 nm and emission, 615/30 nm). Antibodies for immunostaining were anti-Ki67 (1:100, Abcam, no. ab15580) for proliferation, anti–cleaved caspase-3 (1:100; Cell Signaling, no. MAB835) for detecting apoptosis, and markers for EMT: anti-SNAIL/SLUG (1:100, no. ab180714), anti–E-cadherin (1:100, Abcam, no. ab15148), and anti-SMA (1:100, Abcam, no. ab5694). Cells were blocked with 3% rabbit serum.
Measurement of particle uptake by cells
To measure the extent of particle internalization into cells, three methods were used: FACS analysis, spectrometry microplate reader analysis, and microscopy imaging. Measurements of uptake after 24 hours incubation with fluorescently labeled polystyrene particles were performed. Control experiments were done with 5-min incubation, ensuring low particle-cell affinity that does not depend on particle size (fig. S5).
For FACS analysis, cells were seeded in six-well plates for 4 days after overnight starvation in low (0.5%, v/v) serum media. Fluorescently labeled polystyrene particles were added and diluted either to final concentrations of 0.0007 to 0.003% (w/v) (Fig. 2A) or to fixed concentrations of 5% (v/v) otherwise, for O/N incubation in fresh full medium. Controls of cells without particles in the same conditions were done as well. The minimal number of cells in a sample was 10,000. Normalized uptake values were calculated using eq. S1. After the incubation with particles, the cells were washed with cold phosphate-buffered saline (PBS), detached using trypsin, washed again, and filtered through a 40- to 50-μm nylon mesh using a 50-ml conical tube to remove tissue debris mesh. Cells were then centrifuged and suspended in a FACS buffer containing 1% bovine serum albumin in PBS and 0.05% sodium azide. Analyses were performed using a Beckman Coulter CytoFLEX (USA) flow cytometer and analyzed using CytExpert software.
Further quantification of particle uptake by cells was done by fluorescence detection using a microplate reader (Synergy, HT, BioTek, USA). The tested cells were seeded in a final concentration of 20,000 cells per well in 96-well clear-bottom plates (Corning, Sigma-Aldrich) and cultured until cells reach 90% confluency. Polystyrene beads were added from 1% (w/v) stock to a final concentration of 5% (v/v). Normalized uptake values were calculated using eq. S1. Cells were incubated with beads for 24 hours, washed thoroughly with PBS, and read with the microplate reader in excitation: 530/25 and emission: 590/20. Cells without beads were used to obtain a baseline signal.
Cell imaging was used as a direct detection of particle uptake that we used, both with an inverted fluorescent microscope (model IX73, Olympus Corporation, Japan) and by confocal microscopy (Nikon’s A1 MP multiphoton confocal microscope equipped with a 639-nm diode). For fluorescence microscopy, cells were seeded in 24-well plates. After O/N starvation (serum-free medium), fluorescently labeled beads, in the size of 0.05 to 2.4 μm, were added for an O/N incubation. The cells were then washed repeatedly, fixed using 4% paraformaldehyde (PFA), and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Wells were then observed and photographed using fluorescent microscopy (Olympus IX73).
To further validate the extent of particle uptake, and differentiate between internalization and cell adhesion, a confocal microscope with optical sectioning was used. Cells were seeded in ibidi eight-well microplates for 3 days, during which cells were starved overnight and later incubated O/N with fluorescent-labeled beads ranging from 0.05 to 2.4 μm in fresh full medium. Cells were then washed thoroughly, fixated with 4% PFA, and stained with DAPI for nuclei staining with or without counterstaining of Alexa Fluor 488 phalloidin for actin staining. The wells were imaged and photographed using a ZEISS (Germany) confocal microscope. Z-stacks (~1 μm per slice) were performed for ortho and 3D analysis using ZEISS Zen software.
Time-lapse imaging of fluorescently tagged-polystyrene particle uptake
Real-time imaging of particle uptake was performed using the confocal microscope. Further details and movies are presented in the Supplementary Materials.
Atomic force microscopy
To evaluate the elasticity modulus of the cells and their adhesion to the silicon tip, indentation measurements were performed using an atomic force microscope. All measurements were performed using NanoWizard 3 (JPK Instruments, Germany). Indentation measurements were performed in a contact mode using silicon oxide colloidal AFM probes 6.2 μm in diameter (SQube, CP-PNPL-SiO-C, NanoAndMore GMBH, Germany). Each cell was probed at 50 points in two different locations not on cell nucleus with an area of 0.5 μm2 each. A force of 1 nN was used at a speed of 5 μm/s. The data were analyzed using JPK Instrument data analysis software. The Young’s modulus was determined using a DMT model, and the maximal adhesion force was calculated by the minimum point of the force curve. Cells that were more deformable had higher compliance to shape changes induced by indentation or retraction of the AFM probe. Higher deformability therefore corresponded to lower values of Young’s modulus and higher values of the maximal adhesion force. Cells that have small values of Young’s modulus are highly elastic and can greatly deform and allow massive indentation of the probe. High values of the maximal adhesion force represent cells that showed large deformation keeping their membrane adhered to the retracting probe.
Cell invasion assay
To assess the migratory and invasiveness potential of cells, we measured spatial cell invasion from multicellular cancer spheroids (3D culture) in an ECM. Briefly, cells were seeded in a multiwall 2% Agarose microwells array templated with a Master 3D Petri Dish 96-well arrays at 3000 cells per well (Microtissues Inc., USA) as previously described, 3000 cells per well (25). Following cell seeding, the microwells were incubated with fresh medium to allow spheroid formation for 48 hours. Forty-eight hours after spheroids incubation, the spheroids were harvested and embedded in a solution of 0.25% methyl cellulose in suitable media. Spheroids were then mixed with collagen (rat tail collagen type I, Corning, USA) in neutral pH, seeded in a 24-well plate, and incubated for 30 min at 37°C. After gel stabilization, 300 μl of fresh media was added to provide sufficient media supply. The gel containing the spheroids was gently detached from the well, and images of the spheroids were taken by an inverted fluorescent microscope (Olympus Corporation, Japan, model IX73) every few hours.
FACS sorting
Generation of two subcell populations originated from a single cell line, differing by their capacity to uptake 2.4-μm beads, was done by the FACS sorter. Each FACS sorting procedure separated cells that did not uptake any particles (negative, FL2Low) from the rest of the cells (positive, uptake of at least one bead, FLhigh). Therefore, after the first sorting, in each cycle, we obtained four different subsets of cells: positive-positive; positive-negative, negative-positive, and negative-negative. Only the positive-positive and negative-negative were kept for further sorting. In each cycle, the sorting enrichment, Δ (presented in Fig. 4C), was defined as , where was the percent of positive cells obtained in a sorting procedure on the positive-positive population of the previous cycle and was the percent of positive cells obtained in a sorting procedure on the negative-negative population of the previous cycle. An exception to this definition is the first cycle where both of the percent values of positive cells were calculated relative to the same origin population of cells, yielding . Each sorting included the flowing steps. Cells were cultured for five generations and incubated with 2.4-μm beads for 24 hours. Cells were then washed thoroughly with PBS and detached with trypsin, centrifuged, counted, and diluted to 1.0 × 107 cells/ml in PBS. Using a BD FACSAria III cell sorter, cells were analyzed and sorted on the basis of the fluorescence intensity distribution (FL-2). The levels of fluorescence intensity indicated accurately on the number of internalized particles, as shown in fig. S2, allowing for a highly sensitive uptake-based sorting.
Xenograft in vivo experiment
Following the fifth sorting cycle, A375 cells were first cultivated to dispose the sorting particles, harvested, and injected (5 × 106 cells in 100 μl of PBS containing 5% Matrigel) subcutaneously to male athymic nude–Foxn1 HSD 5- to 6-week-old mice, under isoflurane anesthesia. Mice were inspected and weighed every other day. From day 7 of the injection, tumors were measureable and their volume calculated through the ellipsoid equation (square of the tumor width and multiplied by 0.52). Twenty-two days after the injection, mice were euthanized by cervical dislocation under ketamine/xylazine sedation, and tumors were extracted, weighed, and photographed. Tumors were fixated in formalin and embedded in paraffin for histological examinations.
Histology
Tumors extracted from the xenograft experiment were embedded in paraffin and sectioned to 8-μm samples. Samples were stained with an H&E dye for detecting tissue structure using a standard procedure. For immunofluorescence, sections were fixed and 0.1% Triton was used for tissue permeability. After blocking with 3% normal goat serum, sections were incubated with primary antibodies for apoptosis and proliferation (cleaved caspase-3 and Ki67, respectively), and three antibodies for EMT: anti-SMA, anti–E-cadherin, and anti-Snail/Slug. Alexa Fluor 488 or Alexa Fluor 647 (Abcam) was used for secondary antibody labeling, and fluorescent mounting media was used with DAPI (VECTASHIELD, Vector Laboratories, USA) for imaging using a fluorescent microscope (Olympus Corporation, Japan).
RNA extraction
For RNA extraction, the standard QIAzol protocol was used. Tissue samples were homogenized in a QIAzol Lysis Reagent. After addition of chloroform, the homogenate was separated into aqueous and organic phases by centrifugation. RNA partitioned to the upper, aqueous phase, while DNA partitioned to the interphase and proteins to the lower, organic phase. RNA was precipitated from the aqueous phase by addition of isopropanol. The pellet was then washed with ethanol and redissolved in ribonuclease-free water.
RNA sequencing
Sequencing of positive and negative cell samples was performed in The Nancy and Stephen Grand Israel National Center for Personalized Medicine (G-INCPM)–Weizmann Institute of Science (Israel). Libraries were prepared using the INCPM-mRNA sequencing. Briefly, the polyA fraction (mRNA) was purified from 500 ng of total RNA following by fragmentation and the generation of double-stranded complementary DNA. Then, end repair, A base addition, adapter ligation, and polymerase chain reaction amplification steps were performed. Libraries were evaluated by Qubit (Thermo Fisher Scientific) and TapeStation (Agilent). Sequencing libraries were constructed with barcodes to allow multiplexing of 12 samples in one lane. Around 21 million single-end 60–base pair (bp) reads were sequenced per sample on an Illumina HiSeq 2500 V4 instrument.
Bioinformatics
Reads were trimmed using cutadapt (40) and mapped to the human genome GRCh38 using STAR (41) v2.4.2a (with End To End option and out Filter Mismatch NoverLmax set to 0.04). Counting proceeded over genes annotated in Ensembl release 88, using an HTSeq count (42) (intersection-strict mode). Differential expression analysis was performed using DESeq2 (43) with the betaPrior, cooksCutoff, and independent Filtering parameters set to false. Raw P values were adjusted for multiple testing using the procedure of Benjamini and Hochberg (44). The pipeline was constructed using Snakemake (45).
Statistical analysis
All experiments were performed and repeated three or more times. A parametric test and a two-tailed Student’s t test were used for calculating significant differences between two sets of results. An analysis of variance (ANOVA) test followed by a Tukey post hoc test were used for multiple comparisons. Error bars represent SEs. A small fraction of outliner points were discordant in the AFM data and were subtracted using a Z test with 95% confidence. P values of 0.0001 to 0.001 and 0.001 to 0.01, respectively, are represented by marks of “***” and “***” in the graphs.
Gene set enrichment analysis
Whole differential expression data were subjected to gene set enrichment analysis using GSEA (46). GSEA uses all differential expression data (cutoff independent) to determine whether a priori defined sets of genes show statistically significant, concordant differences between two biological states. Gene sets with cytoskeleton-related functions were extracted from the MSigDB (v6.1, October 2017).
Network analysis
Protein-protein interactions data were extracted from HIPPIE (47) and STRING (48), and images were generated using Cytoscape (49).
Chromatin immunoprecipitation
ChIP assay was performed as follows (50). Cells were fixed with 0.8% formaldehyde for 10 min, and then formaldehyde was quenched with 125 mM glycine. The cells were lysed for 20 min on ice, and DNA was fragmented by adding Micrococcal Nuclease (MNase, Thermo Fisher Scientific, 88216) for 20 min at 37°C. The chromatin fragments were then added to Protein A/G beads (Invitrogen, 10004D/10002D) that were attached to H3K4me2 antibody (Millipore, 07-030). Samples were washed twice with radioimmunoprecipitation assay (RIPA) buffer, twice with RIPA high-salt buffer, twice with LiCl wash buffer, and twice with 10 mM tris-HCl (pH 8). DNA was eluted and extracted using AMPure XP beads (A63881, Beckman Coulter Genomics). ChIP libraries were prepared according to the Illumina Genomic DNA protocol.
ChIP-seq analysis
ChIP-seq fragments were sequenced (in paired-end, read lengths of 42 bp), and then mapped to the human genome (hg19) using Burrows-Wheeler Aligner (BWA) (51) with default parameters. Binary Sequence Alignment/Map (BAM) files were then filtered for quality and proper mapping using SAMtools’s “-q 10 -F 1796” (52). We then calculated the ChIP-seq read coverage along the genome using deepTools bamCoverage (53), with the flags “-bs 50 -e 500 --normalize Using RPGC --effectiveGenomeSize 2451960000,” and presented the generated tracks using the University of California, Santa Cruz (UCSC) Genome Browser (54).
Peak calling and annotation enrichments
Peak calling was performed using MACS2 (55), with the parameters “--slocal=2000 --llocal=50000 --nomodel --extsize=1500 -q .0005 -g hs” on each replicate, and then a calculation of the two sets of peaks union was done. Peaks were then intersected using mergePeaks (HOMER, v4.10) (56), and differential peaks were assigned to nearby genes and analyzed for gene ontology enrichments using GREAT (57).
Theoretical model
The free energy change in the adhesive interaction between a rigid spherical particle and a cell is presented in Eq. 1. The mechanism of particle engulfment is provided in a coarse-grained manner (details of the cytoskeletal dynamics were beyond the scope of the present work). A single particle was considered, neglecting particle-particle interactions. An initial stable envelope of the bead particle by the cell membrane was hypothesized to be a necessary step for full engulfment and uptake, and therefore our model provides the upper limit of the extent of particle internalization. A cell was assumed to behave as a continuum 3D elastic material wrapped by an elastic 2D membrane. We used the DMT model for describing the free energy of the cohesion and the 3D cell deformation, consistent with our DMT-based calculations of parameters from the AFM measurements (23). The Helfrisch theory was used for describing membrane elasticity (58). The DMT model accounts for attractive interactions of adhesion and Van der Waals (VdW) that are not spatially limited to the cell-bead contact area. The interactions are assumed to be elastic, describing the case of low penetration. The bead was assumed to be small compared with the cell, and the spontaneous curvature of the cell was taken as zero so that the initial cell geometry was modeled as half a space. The bead and cell surfaces were assumed to be smooth and tightly adhering along their contact surface. Smaller factors of the free energy, such as entropic terms and membrane-stretching tension, were neglected in our coarse-grained model as well as coupling between the DMT and Helfrich terms. Despite the model’s simplicity, excellent quantitative agreement was found between the model predictions and our experimental data. This indicates that the model components were well suited for describing the interactions of cells with the wide size range of particles studied here. The derivation leading to Eq. 1 is as follows. According to the DMT model, the force χ upon particle wrapping is given by
| (3) |
providing the following energetic term
| (4) |
The effective Young’s modulus, E, in the case of a rigid bead is given by where E1 and ν1 are, respectively, the elastic modulus and Poisson ratio of the cell; z is the indentation depth.
Upon contact with a bead, the membrane geometry is approximated as a spherical cap surrounded by a torus sector rim (red and yellow lines, respectively, in Fig. 2F). Then, the bending energy can be written analytically as
| (5) |
The cap area, ac, can be expressed in terms of ϑ as 2πR2(1 − cosϑ) with the bead radius R being the radius of curvature. ar, the rim surface area, is
| (6) |
where Rr is the rim radius of curvature that is the radius of the torus tube. Scaling arguments imply that Rr ∝ R for the following simple reason. If we multiply the dimensions of the bead by a given factor, this is equivalent to scaling the whole-cell bead system by the same factor as long as we remain within the regime where the cell can be modeled as half a space. Thus, the rim radius of curvature should be multiplied by the same factor as well. A direct consequence is that the radius of rim curvature should be proportional to the bead size. In our simplified derivation, we assume a constant Rr equal to the bead radius
| (7) |
Thus, summing the membrane terms we obtain
| (8) |
From Eq. 7, we obtain z = 2R(1 − cosϑ), and therefore, the DMT term can be written as a function of ϑ, yielding the total change in free energy as written in Eq. 1 of the main text
| (9) |
Under small-angle approximations , we obtain
| (10) |
where ΔF(ϑ) has a minimum (for ϑ ≠ 0) when
| (11) |
This provides the following condition for stable adhesion of the particles to the cell (Eq. 2 in the main text)
| (12) |
This is a very important result. For small beads below Rc, ΔF is a monotonically increasing function, and therefore, it is not expected that particles would be wrapped and taken by cells, as indeed observed in our experimental results [Figs. 1, A and B, and 2, A and B]. Most intuitively, Rc increases with the membrane bending modulus, κ (as for small beads, 3D cell deformations are less relevant and the main energetic penalty comes from membrane deformation) and decreases with the adhesion gain (in terms of Δγ).
Supplementary Material
Acknowledgments
We thank The Crown Genomics institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine, Weizmann Institute of Science for RNA sequencing and G. Friedlander, Mantoux Bioinformatics Institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine, Weizmann Institute of Science, for analyzing the RNA sequence. We thank N. Melamed-Book from the Bio-Imaging Unit of The Hebrew University of Jerusalem for assistance with time-lapse confocal imaging. Funding: This project received funding from the European Research Council (ERC-StG) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 0305260). We thank the Israel Ministry of Science and Technology (MOST) (grant agreement no. 0394906) and the Israel Science Foundation (grant agreement no. 0394883) for funding and the Teacher-Scholars program of The Hebrew University of Jerusalem supported by the Jerusalem Municipality, JDA and the Trump Foundation. Author contributions: Y.B.-K.: Conception and experimental design and analysis, writing of the paper with point assistance of other authors, and physical model. D.D.: Experimental design and performance of in vitro and in vivo experiments, FACS, microscopy, and histology. T.M.: FACS experiments and analysis. Y.G.: Uptake experiments and statistical analysis. O.S.: Technical assistance, microscopy, uptake experiments, and RNA extraction. N.O.: FACS experiments and microscopy. T.S.: spheroid cell assays. T.D.-A. and M.R.: Design and performance of AFM studies. H.B.: Bioinformatics. D.B.: Experimental design of cell assays. M.J. and Y.B.: Design and performance of epigenetic analysis. M.V.-A.: FACS experiments. N.L. and T.K.: Bioinformatics analysis of epigenetic studies. O.B.: Conception and experimental design and analysis, writing of the paper with point assistance of other authors, and supervision of the entire work. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The RNA sequencing data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) (59) and are accessible through GEO Series accession number GSE135193 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135193). The DNA sequencing and epigenetic data have been deposited in NCBI’s GEO (59) and are accessible through GEO Series accession number GSE137316 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137316).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/3/eaax2861/DC1
Fig. S1. Quantification of cell invasiveness in spheroid tests.
Fig. S2. Separate peaks of the FACS diagram corresponded respectively to the number of 2.4-μm beads inside the cells, allowing for accurate FACS sorter cell separation.
Fig. S3. Cycle time of cells was not affected by the FACS sorting procedure.
Fig. S4. Cell stiffening resulted in a decrease in the uptake capacity of cancer cells.
Fig. S5. Zero time control in FACS uptake experiments.
Table S1. Cells parameters calculated from AFM measurements, as well as the critical radius for particle uptake based on the physical model results.
Table S2. Normalization factors of internal bead fluorescence.
Movie S1. An example demonstrating the innate heterogeneity in cancer cell populations in terms of their uptake capacity.
Movie S2. Time evolution of 2.4-μm beads uptake by A375 cells.
Movie S3. Time evolution of 2.4-μm beads uptake by 253J cells.
Equation S1. Normalization of uptake data.
REFERENCES AND NOTES
- 1.Chen J., Zhou W., Jia Q., Chen J., Zhang S., Yao W., Wei F., Zhang Y., Yang F., Huang W., Zhang Y., Zhang H., Zhang Y., Huang B., Zhang Z., Jia H., Wang N., Efficient extravasation of tumor-repopulating cells depends on cell deformability. Sci. Rep. 6, 19304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker E. L., Lu J., Yu D., Bonnecaze R. T., Zaman M. H., Cancer cell stiffness: Integrated roles of three-dimensional matrix stiffness and transforming potential. Biophys. J. 99, 2048–2057 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wirtz D., Konstantopoulos K., Searson P. C., The physics of cancer: The role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer 11, 512–522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedl P., Wolf K., Tumour-cell invasion and migration: Diversity and escape mechanisms. Nat. Rev. Cancer 3, 362–374 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Butcher D. T., Alliston T., Weaver V. M., A tense situation: Forcing tumour progression. Nat. Rev. Cancer 9, 108–122 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seyfried T. N., Huysentruyt L. C., On the origin of cancer metastasis. Crit. Rev. Oncog. 18, 43–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar S., Weaver V. M., Mechanics, malignancy, and metastasis: The force journey of a tumor cell. Cancer Metastasis Rev. 28, 113–127 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suresh S., Biomechanics and biophysics of cancer cells. Acta Biomater. 3, 413–438 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez-Elizondo M. B., Weihs D., Cell-Gel Mechanical Interactions as an Approach to Rapidly and Quantitatively Reveal Invasive Subpopulations of Metastatic Cancer Cells. Tissue Eng. Part C Methods 23, 180–187 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Du G., Ravetto A., Fang Q., den Toonder J. M. J., Cell types can be distinguished by measuring their viscoelastic recovery times using a micro-fluidic device. Biomed. Microdevices 13, 29–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu S., Liu G., Chen W., Li X., Lu W., Lam R. H. W., Fu J., Multiparametric biomechanical and biochemical phenotypic profiling of single cancer cells using an elasticity microcytometer. Small 12, 2300–2311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adamo A., Sharei A., Adamo L., Lee B., Mao S., Jensen K. F., Microfluidics-based assessment of cell deformability. Anal. Chem. 84, 6438–6443 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu T., Babaniyi O. A., Hall T. J., Barbone P. E., Oberai A. A., Noninvasive in-vivo quantification of mechanical heterogeneity of invasive breast carcinomas. PLOS ONE 10, e0130258 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malandrino A., Mak M., Kamm R. D., Moeendarbary E., Complex mechanics of the heterogeneous extracellular matrix in cancer. Extreme Mech. Lett. 21, 25–34 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S., Gao H., Bao G., Physical Principles of Nanoparticle Cellular Endocytosis. ACS Nano 9, 8655–8671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhdanov V. P., Physical aspects of the initial phase of endocytosis. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 88, 064701 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Richards D. M., Endres R. G., The mechanism of phagocytosis: Two stages of engulfment. Biophys. J. 107, 1542–1553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muro S., Challenges in design and characterization of ligand-targeted drug delivery systems. J. Control. Release 164, 125–137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torchilin V. P., Drug targeting. Eur. J. Pharm. Sci. 11 (Suppl 2), S81–S91 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Au J. L.-S., Yeung B. Z., Wientjes M. G., Lu Z., Wientjes M. G., Delivery of cancer therapeutics to extracellular and intracellular targets: Determinants, barriers, challenges and opportunities. Adv. Drug Deliv. Rev. 97, 280–301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinney C. P. N., Fishbeck R., Singh R. K., Eve B., Pathak S., Brown N., Xie B., Fan D., Bucana C. D., Fidler I. J., Killion J. J., Isolation and characterization of metastatic variants from human transitional cell carcinoma passaged by orthotopic implantation in athymic nude mice. J. Urol. 154, 1532–1538 (1995). [PubMed] [Google Scholar]
- 22.Coughlin M. F., Bielenberg D. R., Lenormand G., Marinkovic M., Waghorne C. G., Zetter B. R., Fredberg J. J., Cytoskeletal stiffness, friction, and fluidity of cancer cell lines with different metastatic potential. Clin. Exp. Metastasis 30, 237–250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roa J. J., Oncins G., Diaz J., Sanz F., Segarra M., Calculation of Young's modulus value by means of AFM. Recent Pat. Nanotechnol. 5, 27–36 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Händel C., Schmidt B. U. S., Schiller J., Dietrich U., Mohn T., Kießling T. R., Pawlizak S., Fritsch A. W., Horn L.-C., Briest S., Höckel M., Zink M., Käs J. A., Cell membrane softening in human breast and cervical cancer cells. New J. Phys. 17, 083008 (2015). [Google Scholar]
- 25.Shoval H., Karsch-Bluman A., Brill-Karniely Y., Stern T., Zamir G., Hubert A., Benny O., Tumor cells and their crosstalk with endothelial cells in 3D spheroids. Sci. Rep. 7, 10428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park S., Nano-mechanical Phenotype as a promising biomarker to evaluate cancer development, progression, and anti-cancer drug efficacy. J. Cancer Prev. 21, 73–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Yu J., Bomba H. N., Zhu Y., Gu Z., Mechanical Force-Triggered Drug Delivery. Chem. Rev. 116, 12536–12563 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Mierke C. T., Phagocytized beads reduce the α5β1 integrin facilitated invasiveness of cancer cells by regulating cellular stiffness. Cell Biochem. Biophys. 66, 599–622 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Grundy T. J., De Leon E., Griffin K. R., Stringer B. W., Day B. W., Fabry B., Cooper-White J., O’Neill G. M., Differential response of patient-derived primary glioblastoma cells to environmental stiffness. Sci. Rep. 6, 23353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mierke C. T., The fundamental role of mechanical properties in the progression of cancer disease and inflammation. Rep. Prog. Phys. 77, 076602 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Friedl P., Alexander S., Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 147, 992–1009 (2011). [DOI] [PubMed] [Google Scholar]
- 32.X. Tang, T. Cappa, T. Kuhlenschmidt, M. Kuhlenschmidt, T. Saif, in Mechanobiology of Cell-Cell and Cell-Matrix Interactions, A. Wagoner Johnson, B. A. C. Harley, Eds. (Springer US, 2011), pp. 105–122. [Google Scholar]
- 33.Ma J., Zhang Y., Tang K., Zhang H., Yin X., Li Y., Xu P., Sun Y., Ma R., Ji T., Chen J., Zhang S., Zhang T., Luo S., Jin Y., Luo X., Li C., Gong H., Long Z., Lu J., Hu Z., Cao X., Wang N., Yang X., Huang B., Reversing drug resistance of soft tumor-repopulating cells by tumor cell-derived chemotherapeutic microparticles. Cell Res. 26, 713–727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandrasoma P., Polymorph phagocytosis by cancer cells in an endometrial adenoacanthoma. Cancer 45, 2348–2351 (1980). [DOI] [PubMed] [Google Scholar]
- 35.Rejman J., Oberle V., Zuhorn I. S., Hoekstra D., Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 377, 159–169 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma N., Dey P., Cell cannibalism and cancer. Diagn. Cytopathol. 39, 229–233 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Iyer S., Woodworth C. D., Gaikwad R. M., Kievsky Y. Y., Sokolov I., Towards nonspecific detection of malignant cervical cells with fluorescent silica beads. Small 5, 2277–2284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Junttila M. R., de Sauvage F. J., Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501, 346–354 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Meacham C. E., Morrison S. J., Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 41.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjamini Y., Hochberg Y., Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995). [Google Scholar]
- 45.Koster J., Rahmann S., Snakemake--a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaefer M. H., Fontaine J. F., Vinayagam A., Porras P., Wanker E. E., Andrade-Navarro M. A., HIPPIE: Integrating protein interaction networks with experiment based quality scores. PLOS ONE 7, e31826 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., von Mering C., STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benchetrit H., Jaber M., Zayat V., Sebban S., Pushett A., Makedonski K., Zakheim Z., Radwan A., Maoz N., Lasry R., Renous N., Inbar M., Ram O., Kaplan T., Buganim Y., Direct induction of the three pre-implantation blastocyst cell types from fibroblasts. Cell Stem Cell 24, 983–994.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramirez F., Ryan D. P., Gruning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dundar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D., The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M., Bejerano G., GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helfrich W., Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. C 28, 693–703 (1973). [DOI] [PubMed] [Google Scholar]
- 59.Edgar R., Domrachev M., Lash A. E., Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raudenska M., Kratochvilova M., Vicar T., Gumulec J., Balvan J., Polanska H., Pribyl J., Masarik M., Cisplatin enhances cell stiffness and decreases invasiveness rate in prostate cancer cells by actin accumulation. Sci. Rep. 9, 1660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yashunsky V., Shimron S., Lirtsman V., Weiss A. M., Melamed-Book N., Golosovsky M., Davidov D., Aroeti B., Real-time monitoring of transferrin-induced endocytic vesicle formation by mid-infrared surface plasmon resonance. Biophys. J. 97, 1003–1012 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/3/eaax2861/DC1
Fig. S1. Quantification of cell invasiveness in spheroid tests.
Fig. S2. Separate peaks of the FACS diagram corresponded respectively to the number of 2.4-μm beads inside the cells, allowing for accurate FACS sorter cell separation.
Fig. S3. Cycle time of cells was not affected by the FACS sorting procedure.
Fig. S4. Cell stiffening resulted in a decrease in the uptake capacity of cancer cells.
Fig. S5. Zero time control in FACS uptake experiments.
Table S1. Cells parameters calculated from AFM measurements, as well as the critical radius for particle uptake based on the physical model results.
Table S2. Normalization factors of internal bead fluorescence.
Movie S1. An example demonstrating the innate heterogeneity in cancer cell populations in terms of their uptake capacity.
Movie S2. Time evolution of 2.4-μm beads uptake by A375 cells.
Movie S3. Time evolution of 2.4-μm beads uptake by 253J cells.
Equation S1. Normalization of uptake data.


