Abstract
Glioblastoma multiforme is the most common and aggressive malignant primary brain tumor. As implied by its name, the disease displays impressive intrinsic heterogeneity. Among other complications, inter- and intratumoral diversity hamper glioblastoma research and therapy, typically leaving patients with little hope for long-term survival. Extensive genetic analyses, including omics, characterize several recurrent mutations. However, confounding factors mask crucial aspects of the pathology to conventional bulk approaches. In recent years, single-cell omics have made their first appearance in cancer research, and the methodology is about to reach its full potential for glioblastoma too. Here, recent glioblastoma single-cell omics investigations are reviewed, and most promising routes toward less grim prognoses and more efficient therapeutics are discussed.
Keywords: glioblastoma, glioblastoma multiforme heterogeneity, omics, single cells
1. Introduction
Brain malignancies are relatively rare. Prostate or breast cancers are respectively 60 and 50 times more prevalent in the United States.[1,2] Among other aspects, cancer development is hampered in the central nervous system (CNS) by a reduced tissue turnover as well as by the presence of microglia and brain barriers. The same factors, however, pose special challenges when brain cancer does occur.[3,4] With a median survival rate between 12 and 15 months and a 5-year survival percentage of roughly 5%,[5,6] glioblastoma multiforme (GBM) is one of the deadliest CNS neoplasms. Its especially poor prognosis causes it to be a serious economic and social concern[7]; in fact, GBM is among the most studied types of cancer.[8] The average annual age-adjusted incidence of GBM in the United States is around 3 per 100 000.[1]
The tumor owes the appellative multiforme to its variegated gross appearance. It features an inner core of necrotic tissue surrounded by a periphery of strongly anaplastic cells, with a variable degree of vascularization and hemorrhages. Edema and finger-like projections constitute the outermost attributes of this cancer. At times, and depending on etiology, smaller foci might form around the main one; these are termed secondary GBMs. Macrophages and microglia infiltrate the mass in high numbers. The GBM is a type of glioma, glialike tumors of the CNS.[9] Gliomas are traditionally classified and graded according to their histopathological characteristics, that is, they are given names according to the type of tissue they are most reminiscent of, as well as to their expected aggressiveness. The World Health Organization adopts a four-tier grading scheme of increasing malignancy (I–IV); a GBM is a grade-IV astrocytoma, meaning the most severe among those gliomas that resemble astrocytes.
The real cytological origin of GBM is not entirely understood. Cells giving rise to a GBM are named glioblastoma stem cells (GSCs), or more generally tumor- or glioma-initiating cells. In spite of its glial histological presentation, there is sound evidence supporting the notion that tumor-initiating cells in GBM might be other than astrocytes, and even not belong to glia at all.[2,10,11] The disease may originate in oligodendrocytes or in neural cell progenitors, precisely from a specific population of neural stem cells and progenitor cells of the subventricular zone. The fact that GBM is found in multiple brain locations would be a consequence of the early motility of cancer cells. GSCs too generally resemble astrocytes in appearance. They display a distinct metabolic plasticity, being able to shift from a quiescent and resistant-to-treatment state, to a proliferative and therapeutically targetable one. Instances of genetic markers for GSCs are cluster of differentiation (CD) 15 (CD15), CD44, CD133, and integrin alpha 6 (ITGA6).[2,11]
Irrespective of morphological features, grade-III and grade-IV astrocytomas can be categorized in genetic types that are highly predictive in terms of prognosis. The integration of molecular information into the histopathological classification is therefore now recommended to guide therapeutic approach. Biopsies are routinely screened for mutations in the loci of IDH1 or IDH2 (respectively, coding for cytosolic or mitochondrial NADP+ isocitrate dehydrogenases), and for hyper-methylation on the promoter of MGMT, the O6-alkylguanine DNA alkyltransferase.[12,13] The first aberration seems to mark a slightly milder kind of astrocytoma with a distinct onset. The methylation status of MGMT promoter predicts tumor response to alkylating chemotherapy, most relevantly to temozolomide (TMZ), the gold-standard chemotherapeutic agent for GBM.[14] Hypermethylation ablates MGMT function, preventing tumor cells to repair DNA damage caused by TMZ. Best results are obtained when TMZ is associated to radiotherapy, a practice known as Stupp regimen.[6]
The intratumoral variability of GBM is remarkable, both at the somatic and the genetic level. Understanding how patients differ with respect to the molecular paths that lead to the disease is an unavoidable step toward the development of effective treatments. However, such tumor-to-tumor personalization will likely not solve the problem alone. Even a single mass possesses, in fact, a strong intrinsic mosaicism: tumor clones arise, drift, and branch following evolutionary dynamics that are even fostered by generalized therapies. Secondary mutations generate genetic variability among proliferating malignant cells, and the frequency of each of such aberrations within a tumor changes in time, either simply by chance or because selective environmental forces are in place. For instance, an antitumoral drug may preferentially or exclusively kill cells that present a particular allele, therefore promoting the expansion of cancer clones that do not present that gene variant. Intratumoral diversity will ultimately constitute a major obstacle to a definitive cure for GBM. In order to prevent recurrences, toxicity must be achieved in all and only tumor cell types of a given individual.
In the last few years, single-cell (SC) workflows made their appearance in the field of omics. Due to their “atomic” resolution, SC approaches hold big promises for the treatment of GBM, and research dedicated to the subject is just about to blossom. Here we review most recent advances in SC research focused on GBM.
2. Single-Cell Omics Provide New Tools against Glioblastoma
2.1. Main Steps for Single-Cell Omics and Scope of the Current Work
A SC approach is in principle possible for most omics. The technique is limited by the amount of initial material that a single cell might yield, with some classes of molecules being more common than others. For instance, mRNA and proteins are more represented than DNA. In turn, some molecules present special advantages: nucleic acids, for example, can be amplified, while proteins cannot.
For SC omics, initial samples (that might be biopsies or cultures) are mechanically and/or chemically dispersed into SC suspensions. These may be selected by microfluidics, cell sorting, immunopanning or even manually, and end in SC wells, microchambers, or droplets. There the cell is lysed, and the molecular class of interest is analyzed by different methods. Bioinformatics is vastly used to handle resulting data, from initial raw signals to the most abstract semantic associations between sets of molecules, cells, and even studies. Due to the relatively lower sensitivity of SC versus bulk pipelines, high-order analyses are especially relevant in SC omics.[15,16] Also depending on the type of molecule studied, it is important that all laboratory procedures progress fast; namely, perishable cell types and molecules like RNA should be handled carefully and as quickly as possible.[15,16]
Considering a given framework as SC omics or not is, to some extent, a matter of definitions. SC omics are multistep processes aimed at isolating and analyzing cells with high-throughput modalities. There are a lot of layers, as a consequence, between the most big-data-oriented pipelines and mere qualitative evaluations on a few individual cells. An example of such mid-zone might be the investigation of Meyer and colleagues.[17] By a combination of conventional assays including bulk DNA microarrays, they characterized in vitro-expanded GBM clones, and found some that were resistant to TMZ even prior to treatment. They also screened potential drugs to target specific lineages. Similarly, Tome-Garcia et al.[18] performed bulk transcriptomic analyses on GBM or healthy stem-cell spheroids (3D colonies) obtained through culturing single cells isolated on the basis of their epidermal growth factor receptor (EGFR) phenotype, known to present aberrations in a relevant portion of GBM cases. Other groups took a step further: Piccirillo and colleagues[19] studied genomic alterations on single culture-derived GBM cells, while Liau et al.[20] investigated cell-cycle plasticity of cultured GSCs upon treatment. Likewise, Chen and co-authors[21] performed SC RNA sequencing (sc-RNA-seq) on cells obtained via growing slices from an MGMT-hyper-methylated, IDH-wild-type GBM biopsy.
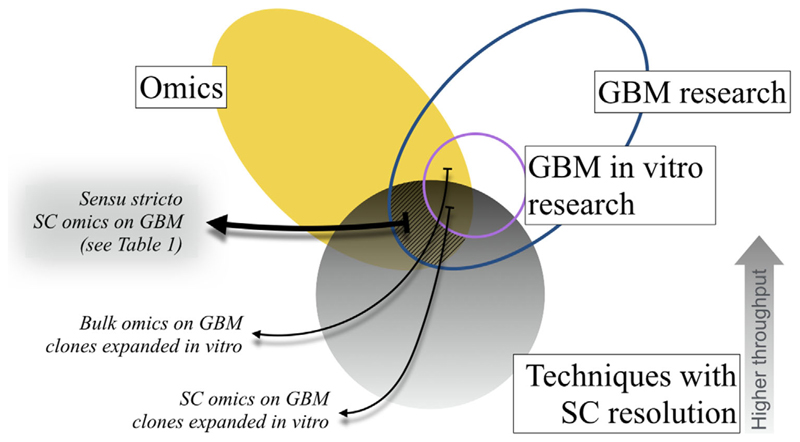
Here we focus on what we regarded as pure SC omic works. SC transcriptomics have been profitably used to study gliomas other than GBM, for example, oligodendroglioma[22] or histone H3 Lys27Met (H3K27M) glioma.[23] However, for the sake of brevity, we also restrict our attention to GBM research. Figure 1 illustrates inclusion and exclusion criteria defining the fundamental boundaries of our work.
Figure 1.
Logical relationships between research in glioblastoma, omics, and techniques with single-cell resolution. An Euler diagram shows criteria setting the requirements for a study to be considered of primary interest for the review. GBM, glioblastoma multiforme; SC, single cell.
In the light of the latest progress, the current review updates and integrates to various degree existing discussion[15,24–28] about this fast-evolving discipline. For more technical information about SC omics methods in cancer or specifically in GBM we suggest, respectively, Tsoucas et al.[29] and Sen et al.[30]
2.2. Recent Advances in Single-Cell Omics Research against Glioblastoma
Studies on clonal cell populations expanded in vitro from a single cell yielded valuable insight into the biology of GBM. However, evolutionary dynamics still occur during the growth of a laboratory clone, often introducing unwanted artefacts. In addition, the approach can practically survey only a limited number of founder cells.
About 5 years ago, sensu stricto SC omics made their first entrance in GBM research.[24] In 2014, two genomics investigations pioneered the field: Stieber and co-authors[31] studied variations in ploidy among tumor clones also by means of single nucleus (SN) comparative genomic hybridization, and Francis et al.[32] utilized an SN genomics pipeline to assess, in a number of GBM cells, the mutational status of EGFR. SN sequencing has advantages over SC sequencing in genomics, because it avoids bias in the cell dissociation steps. Complex cells like neurons might be preferentially lost during dissociation.[15] Shortly after, Patel et al.[33] performed sc-RNA-seq on patient-derived GBM tumors. These works represented a leap forward for the understanding of GBM heterogeneity, because direct, non-averaged measurements of transcript levels or locus copy number of individual cells could be acquired.
Authors were able to conclusively prove that intertumoral variability goes well beyond the conventional GBM classification from The Cancer Genome Atlas Research Network (TCGARN),[12,13] and that this has likely important prognostic implications. TCGARN divides GBMs into four histopathological subtypes: classical, proneural, neural, and mesenchymal. Patel and colleagues found that, while all examined tumors fell into a defined class when studied as a bulk, SC analyses clearly highlight that each mass is rather a combination of cells belonging to different categories. Given the variability in terms of anaplasia, proliferation potential, and response to treatment displayed by the different GBM subtypes, it becomes trivial to note that such internal heterogeneity cannot be overlooked. For each single case, they also detected variable expression of a number of membrane receptor genes that are relevant to the pathology, including the aforementioned proto-oncogene EGFR. Such mosaicism may well explain part of the long-term resistance of GBM to therapy. They also identified a few meta-signatures, that is, particular subsets of genes the expression of which consistently varies across single cells from either one or multiple tumors. These relate to specific processes, for instance, to cell cycle or hypoxia.
Müller et al.[34] combined bulk exome sequencing with sc-RNA-seq, and developed a procedure to reconstruct phylogenetic trees of clonal lineages for individual GBMs. The method produced detailed phylogenies of either EGF-driven or platelet-derived growth factor (PDGF)-driven GBM samples. While the first kind promoted increasingly aggressive, anaplastic cell types, the latter evolved toward a milder, oligodendrocyte progenitor-enriched phenotype. As those before them, authors stress that intratumoral variability is a major problem in GBM treatment. For instance, receptor tyrosine kinases (RTKs) display mosaic mutations, so that a cocktail of RTK inhibitors is necessary to achieve complete tumor regression in vitro. This is in agreement with Patel et al. as well.
SC omics can be used to derive information about the phylogenetic relationships among cells of the same tumor. Based on EGFR genotype, Francis and colleagues inferred intratumoral clonal evolution. The technique renewed interest toward those cells that are virtually at the base of a GBM evolutionary tree, the GSCs. Patel et al. confirmed that GSCs represent a minor population within the whole cancer, and that they display stemness-related features as a continuum, rather than being of a homogeneous type. Such characteristic constitutes another difference with respect to their in vitro counterpart, the spheroids. Furthermore, only a few tumor cells are actively cycling, ranging from around 1 to about 20%. This is markedly in contrast with cultured GBM models, which appear to be entirely proliferating.
The discussion around GSCs is still vivid in state-of-the-art SC omics. Two studies (published as non-peer reviewed preprints at the time of writing) redefined the transcriptional hallmarks of GSCs, and demonstrated that GSCs are the main causal factor of GBM heterogeneity.[35,36] Müller and co-authors used sc-RNA-seq on GBM samples, matching them with bulk exome sequencing. They too found a gradient of diversity in GSCs, which spans from an exquisitely proneural status to a more mesenchymal one. Immunohistochemical evidence suggested that mesenchymal GSCs are mainly located within the core of the cancer, are mostly quiescent, accumulate a comparatively high number of mutations, and mediate the chemotaxis of myeloid-derived suppressor cells. In turn, proneural ones appear to reside along the periphery of the tumor mass, and are characterized by a higher turnover. The ratio between proneural and mesenchymal GSCs positively correlates with survival. Further recent work[37] corroborated both the idea of a “transcriptional continuum” and the impression that classical categorization in subtypes does not fully grasp the real intricacy of GBM.
Cells can be tracked not only according to their type. Depending on pipelines, single cells can be classified as homogeneous with their surroundings or not. Disseminated cancer cells can be identified in otherwise healthy tissue, and vice versa infiltrating cells can be detected in tumor masses.[15,24] A mainstay of SC omics is the possibility to assess the contribution to the pathology from tumor microenvironment (TME), a major obstacle to immune therapy for GBM.[38] Tumor-associated macrophages (TAMs), highly present at cancer location, may act as local immune suppressors. However, the anatomical origin of TAMs has long remained obscure. Müller et al.[39] aimed to identify transcriptional signals that could distinguish blood-derived macrophages from microglial cells. For this purpose, they isolated TAMs from untreated GBMs or low-grade gliomas, and performed sc-RNA-seq. Experiments unveiled different signatures for the two TAM populations, pointing to a correlation between cancer type and TAM composition. Blood-derived TAMs profusely infiltrate pre-treatment gliomas, and appear to be especially relevant for immune suppression. The work profited new markers and more general information about TME.
Owing to SC omics, the role of TME can in summary be quantitatively approached with major benefits. Tumor niche is relevant for cancer characterization because it can complicate bulk analyses, and because of its crosstalk and interactions with tumor cells. Venteicher et al. discovered that bulk transcriptional differences between IDH-mutant astrocytoma (IDH-A) and IDH-mutant oligodendroglioma (IDH-O) are not observed with SC approach. Rather, transcriptome variations between the two may mostly account to their specific brain settings (more cortical for IDH-O). Much of the residual variation could simply be explained by their different genetic alterations. Hence, contrary to the traditional view, IDH-A and IDH-O might have similar etiology.[22,40] While Venteicher and colleagues[40] observed a continuum in infiltrating immune cells ranging from macrophage-like to microglia-like for IDH-A and IDH-O, Muller et al.[39] found their two TAM types to constitute discrete populations. Zong notes that more SC research is needed to clarify why low-grade gliomas differ from GBM in the distribution of diversity of TAMs.[27]
mRNA has been the main focus of SC omics, and GBM research is no exception. Depending on protocols, non-coding RNAs may still be included in sc-RNA-seq datasets. Hu et al.[41] mapped data from Patel and colleagues[33] to known mouse and human long non-coding RNAs (lncRNAs). Considering that lncRNAs are more cell-type dependent than mRNAs, they defined lncRNAs expressed by a quarter or more tumor cells as GBM-specific, and found lncRNA intertumoral diversity to be greater than lncRNA intratumoral diversity, so that different cases only show a minor overlapping of lncRNA transcriptomes. lncRNA-coding loci appear to display highly dynamic expression across cancer cells, to the point that each cell seemingly possesses a unique lncRNA transcriptional status. They concluded that stably expressed lncRNA genes are likely the most evolutionarily conserved ones.
While transcriptomics certainly dominates the panorama of GBM SC omics, a first SC proteomics work entered the field. The technique may be comparatively less mature, yet the approach is both promising and noteworthy. Wei and co-authors[42] grafted GBM cells into mice, then treated them with mammalian target of rapamycin (mTOR) kinase inhibitors and observed the tumor mass diminish (mTOR regulates proliferation). After this initial response, however, resistance to mTOR kinase inhibition typically occurs, and cancer grows back. They identified phosphorylation paths involved in adaptation to therapy by means of SC phosphoproteomics. Using a combination therapy, they could achieve long-term in vivo tumor regression. The study shows how adaptive, rather than genetic, mechanisms can sustain cancer progression, and how a deep understanding of population dynamics is crucial for the outcome of a treatment. A second investigation in SC phosphoproteomics for GBM is preliminarily divulgated as a non-peer reviewed preprint. Leelatian et al. propose an unbiased, unsupervised methodology to determine clinical risk in GBM cases as a continuum, by evaluating a group of 34 proteins (including phosphoproteins).[43]
The advent of SC omics refreshed the discussion about current challenges in developing a cure for GBM. With part of the tumor complexity sorted out, it appears increasingly evident that some tumor cells display key features that render GBM resistant to treatment, such as self-renewal, transient quiescence, resistance to DNA damage induced by therapy, and tolerance to hypoxia.[33] Specifically, GBM owes much of its aggressiveness to GSCs.[35] The long-held interest for GSCs is in short well placed: despite their low counts, these cells can sustain the development of new cancer tissue, and are not targeted by current therapies. Tumor biopsies are a limited resource so, as far as possible, it is recommendable to maximize their use by quantifying and localizing such cells in space and time, ideally even in relationship to treatment.[34] Cell-level information from tumor surroundings may also represent a decisive weapon for research and therapy. Darmanis et al.[44] performed an sc-RNA-seq of GBMs and stromal cells around them. By serially sorting different cell types, they identified GBM cells within the peritumoral tissue. The study has broad implications for the understanding of long-term tumor recurrence. In fact, authors discovered a tumor-supporting role for myeloid cells infiltrating the mass, possibly even participating in cancer dissemination. They also highlighted great intratumoral variability in GBM, but found that different masses adopt a convergent dissemination strategy. Importantly, they note that a criticality of GBM is its very diffuse nature, so that the existence of infiltrating cells with a predictable transcriptome signature could be relevant for a cure. Results are also potentially applicable to immunotherapy, for example, because they highlight highly variable expression of human leukocyte antigen (HLA) genes among different cases.
As SC omics workflows improve, more pervasive studies become possible.[35,37,39,40] The overall tendency is certainly to screen more cells and to probe each of them in greater depth. The knowledge gathered about GBM cells and lineages in the last few years is already profound; however, we should probably regard this a good start rather than an end point. Table 1 lists, to the best of our knowledge, all published and preprint articles strictly dedicated to SC GBM omics.
Table 1. Summary of single-cell omics studies about glioblastoma multiforme.
| Reference | Species | Sample(s) | Field | Technique | Notes |
|---|---|---|---|---|---|
| 2014, Francis et al.[32] | Human | BT325 and BT340 cells bearing EGFR alterations | Genomics | SN CNV-seq | — |
| 2014, Patel et al.[33] | Human | Five primary IDH1/2-wild-type GBMs, three of which EGFR-amplified; GBM6 and GBM8 cell lines | Transcriptomics | Smart-seq | — |
| 2014, Stieber et al.[31] | Human and human GBM cells in mouse hosts | 36 GBMs; some used for mouse xenografts | Genomics | SN array CGH | — |
| 2015, Hu et al.[41] | Human | Datasets from 2014, Patel et al. [2014, Patel] | Transcriptomics (bioinformatics) | sc-RNA-seq data mapped to known lncRNAs | — |
| 2016, Müller et al.[34] | Human | Three primary untreated GBMs, including an EGF-driven tumor and an PDGF-driven tumor | Transcriptomics | Fluidigm C1 IFC | — |
| 2016, Wei et al.[42] | Human GBM cells in mouse hosts | GBM39 mouse xenografts treated with mTOR kinase inhibitor CC214-2 | Phosphoproteomics | SCBC technology | — |
| 2017, Darmanis et al.[44] | Human | Four IDH1-negative GBMs, each with a second biopsy from peritumoral brain tissue | Transcriptomics | Smart-seq2 | Publishes a user-friendly website to consult dataset |
| 2017, Müller et al.[39] | Human | Purified TAMs from 13 untreated primary gliomas (11 GBMs, 2 LGGs) | Transcriptomics | Multiple sc-RNA-seq pipelines | — |
| 2017, Venteicher et al.[40] | Human | 16 IDH-mutant gliomas (ten astrocytomas with grades spanning from II to IV, six grade-II oligodendrogliomas) | Transcriptomics | Smart-seq2 | — |
| 2018, Couturier et al.[35] | Human; GSC-enriched samples proven tumorigenic by xenotransplantation in mouse | IDH-wild-type GBMs; isolated cells from four fetal telencephala (13–21 weeks of gestation) | Transcriptomics | Drop-seq | Currently a non-peer-reviewed preprint |
| 2018, Müller et al.5[36] | Human | Eight primary untreated GBMs | Transcriptomics | 10x genomics-based sc-RNA-seq | Currently a non-peer-reviewed preprint |
| 2018, Yuan et al.[37] | Eight HGGs, including GBMs | Transcriptomics | Own sc-RNA-seq method based on a high-density microwell platform | — | |
| 2019, Leelatian[43] | Human | 28 IDH-wild-type GBMs from adult patients | Phosphoproteomics | SC mass cytometry | Currently a non-peer-reviewed preprint |
| 2019, Peng[38] | Human | Three GBMs with their TME | Transcriptomics | Multiple sc-RNA-seq pipelines | — |
Only strictly single-cell (SC), strictly glioblastoma (GBM) articles are included.CGH, comparative genomic hybridization; CNV, copy-number variation; HGG, high-grade glioma; IFC, integrated fluidic circuit; LGG, low-grade glioma; lncRNA, long non-coding RNA; SCBC, SC barcode chip; sc-RNA-seq, SC RNA sequencing; SN, single nucleus; TAM, tumor-associated macrophage; TME, tumor microenvironment.
2.3. Ontogeny, Phylogeny, and Tumorigenesis. Single-Cell Omics Offer New Opportunities for Scientific Reflection
Ontogeny recapitulates phylogeny, goes an old adage often thought in embryology classes and formulated as such by Ernst Haeckel in the early nineteenth century. While the saying might appear simplistic nowadays, there are profound similarities between evolution and development, to the point that—even in modern biology—a dedicated branch exists for the study of the relationships between the two, called evolutionary developmental biology (in short, evo-devo). The morphological shaping of a heart, together with the underlying genetic programs, somehow resembles its evolutionary history. Similarly, the transition of a human fetus from the maternal womb to the external world is the archetype of the passage from an aquatic to a terrestrial lifestyle.
Tumorigenesis might recapitulate phylogeny too. At least, a growing body of scientific literature states that neoplasms share features with embryonic tissue, and some went as far as theorizing a cancer evo-devo.[45,46] From its first application to GBM, SC omics helped deciphering the connections between oncogenesis and CNS development. Different authors suggest that the cell complexity of GBM is somewhat reminiscent of brain ontogenesis.[33,35] Couturier et al., for instance, performed sc-RNA-seq on GBM cells and fetal brain cells, then compared the two groups. They concluded that GBM recapitulates normal brain development, with IDH-wild-type GBMs being organized into three lineages just like normal neural lineages, namely astrocytic, neuronal, and oligodendrocytic. Conversely, IDH-mutant GBMs would be subdivided in just two lineages, that is astrocytic and oligodendrocytic.[23,40]
While technology progresses, it becomes easier to appreciate that cancer exploits metabolic circuits that belong to developmental processes. Also at a more macroscopic scale, tumor progression and shaping closely resemble morphogenetic events. But to what extent carcinogenesis mocks developmental dynamics? What happens to a cancer cell when it is expanded in vitro? And how much overlap exists between tumor formation and evolution, or even evolutionary history?
SC omics just started to peer inside the spatiotemporal patterning of tumor diversity. With its impressive resolution, the approach offers unprecedented opportunities to quantitatively address fundamental questions that would otherwise largely remain in the realm of speculation. It allows tracing the formation, the expansion, and the extinction of clones within a neoplasm, as well as categorizing cells with a precision that almost defeats the classical conceptualization of cell types.
There are few doubts that oncological treatment, GBM included, will largely benefit from SC omics. However, it should be borne in mind that a similarly major impact is also expected in basic scientific discussion about cancer.
3. Conclusion
SC transcriptomes are becoming wider and more accurate. To sum up, some key advances against GBM brought about by SC transcriptomics were the conclusive proof of remarkable intratumoral diversity, the discovery of clinically significant genetic mosaicism associated to distinct lineage phenotype and metabolic signatures, and in general a better understanding of the GBM ecosystem, including important notions about GSCs and TME. With so few GBM investigations dedicated to SC genomics or proteomics, the non-transcriptomic SC omics of GBM are mostly untouched. Examples are surely about to come, and we can expect a lot from them.
Technical advance will hopefully lead to a reduction of costs per cell, wider options to study and combine different molecular classes, as well as neater and more standardized procedures. This in turn would bring a more widespread use of the approach, larger datasets in different contexts and, above all, its routine use in clinical practice.[15,28]
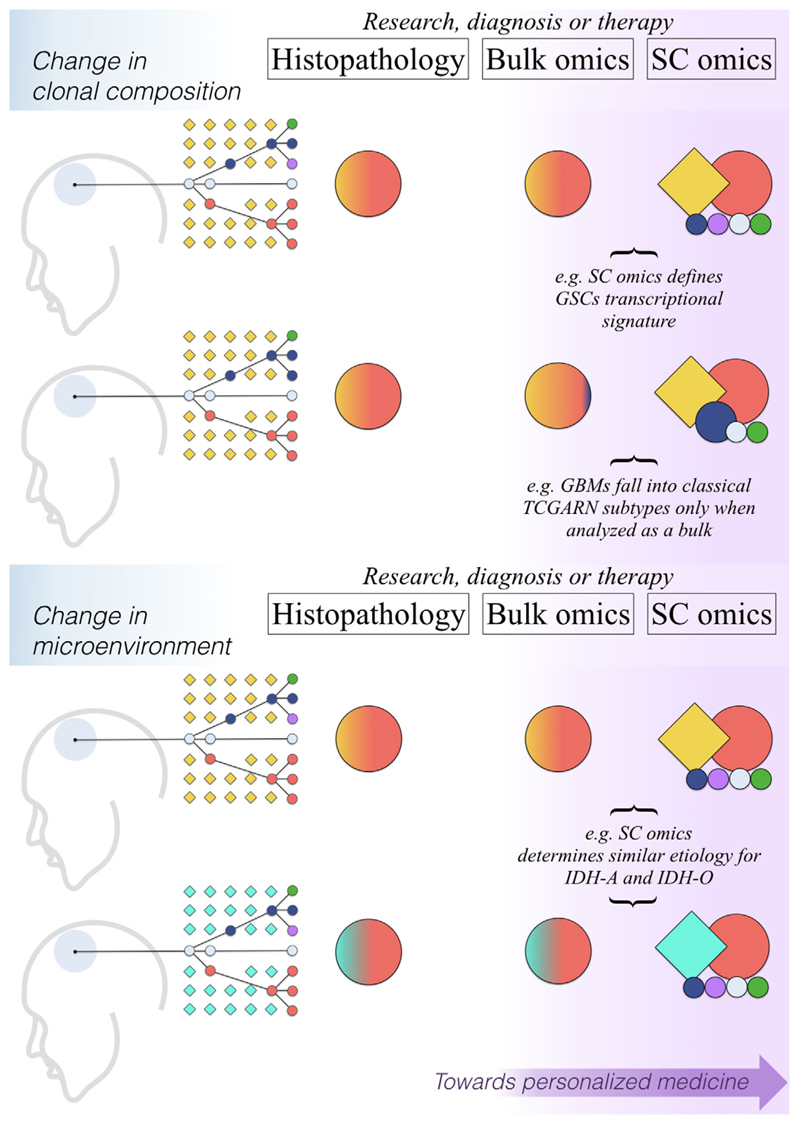
With time, SC omics will likely become indispensable tools for precision medicine, the more so for a currently intractable pathology such as GBM. SC methods will surely keep refining biomarkers, the appropriate identification and use of which are essential for, respectively, a punctual glioma classification as well as an early and accurate diagnosis. The technique will expectedly revisit detection targets, namely individual cells that, for either their molecular identity or localization, become critical for diagnosis. Eventually, it could guide therapies for the selective destruction of ideally all clinically relevant lineages—including rare cell types—in a timely and spatially controlled fashion.[24,28] Figure 2 provides a simplified overview of current and potential benefits of SC omics for GBM research and therapy, particularly in terms of improved capabilities of recognizing intratumoural complexity as well as of telling apart cancer cells from their non-cancerous surroundings.
Figure 2.
Single-cell omics push a shift in paradigm for glioblastoma study and treatment. Each of the four sketches on the left depicts a glioblastoma (GBM) site (pale borderless head area) with a representation of cell lineages found in it, circles representing cancer cells, squares symbolizing surrounding non-tumor cells (colors mark individual lineages, connecting lines indicate hypothetical phylogenetic relationships between GBM cells). “Research, diagnosis or therapy” tables report a simplification of typical outcomes for each of the three exemplified approaches to research, diagnosis, or treatment for GBM. Object colors indicate cell lines dictating results; their size correlates with the magnitude of the distinguishable signal they generate, and their shape indicates whether they can exquisitely associate a given signal to either tumor (circles) or surrounding (squares) cells. IDH-A, IDH-mutant astrocytoma; IDH-O, IDH-mutant oligodendroglioma; GSC, glioblastoma stem cell; SC, single cell; TCGARN, The Cancer Genome Atlas Research Network.
SC omics are a “double omics” in a sense. Namely, they possess a twofold complexity, because they study a whole class of molecules at a time, and because they encompass a large number of cells within a biological sample. The latter feature can potentially render electrophysiology, imaging, etc. “somewhat omic” too. Increased diversification and output can make it difficult to discern what is relevant from what it is not, a phenomenon that was aptly dubbed “data struggle.”[47] For these reasons, another desirable objective for the field is a growing attention toward responsible data handling, sharing, and integration. Commendable initiatives pointing in this direction are CancerSEA,[48] a user-friendly SC omics resource database for cancer that features some GBM results, and a simple website that allows browsing results from Darmanis et al. Others, such as the Ivy Glioblastoma Atlas, are integrating histopathological GBM data with molecular information, including SC omics.[49,50]
4. Search Strategy and Selection Criteria
Literature searches were mainly performed through three academic search engines, that is, NCBI Entrez (database PubMed), Google Scholar, and Scopus. In the following, we list the principal search strings used to query the three websites, mainly visited between December 2018 and February 2019. Strings are delimited by semicolons. Comments about individual strings are reported in braces.
For NCBI Entrez: “Single-Cell Analysis”[Mesh] AND “Glioblastoma”[Mesh]; “Neoplasms”[Mesh]) AND “Clonal Evolution”[Mesh] {only reviews, starting from 2009}; (“Glioblastoma”[Mesh]) AND (“Metabolism/cytology”[Mesh] OR “Metabolism/drug effects”[Mesh] OR “Metabolism/drug therapy”[Mesh] OR “Metabolism/genetics”[Mesh] OR “Metabolism/metabolism”[Mesh] OR “Metabolism/mortality”[Mesh] OR “Metabolism/pathology”[Mesh] OR “Metabolism/pharmacology”[Mesh] OR “Metabolism/physiology”[Mesh] OR “Metabolism/physiopathology”[Mesh] OR “Metabolism/therapy” [Mesh]) {only reviews, starting from 2009}; glioblastoma {only reviews, starting from 2014}.
For Google Scholar: single-cell glioblastoma; single-cell glioblastoma {starting from 2017}; (tumor OR cancer) AND evolution {starting from 2009}; (glioblastoma OR gbm) AND (metabolism OR pathways OR genetics); (glioblastoma OR gbm) AND heterogeneity; glioblastoma {starting from 2014}; cancer evolution with single cell sequencing.
For Scopus: single-cell glioblastoma {starting from 2017}; tumor evolution {only reviews, starting from 2009}; cancer evolution {only reviews, starting from 2009}; glioblastoma heterogeneity {only reviews, starting from 2009}; glioblastoma {only reviews, starting from 2014}.
Additional relevant references were retrieved after consultation of the initially chosen literature. Finally, we thoughtfully utilized regular search engines to further ensure we did not miss any crucial contribution. For the main body of the review, we were stringent in selecting narrowly speaking SC omics GBM studies. Only articles published in English were considered.
Acknowledgements
This work has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement N°709613, SLaMM).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
The ORCID identification number(s) for the author(s) of this article can be found under https://doi.org/10.1002/adtp.201900152
Contributor Information
Dr. Andrea Degl’Innocenti, Istituto Italiano di Tecnologia, Smart Bio-Interfaces, Viale Rinaldo Piaggio 34, 56025 Pontedera, Pisa, Italy.
Nicoletta di Leo, Istituto Italiano di Tecnologia, Smart Bio-Interfaces, Viale Rinaldo Piaggio 34, 56025 Pontedera, Pisa, Italy; Scuola Superiore Sant’Anna, The Biorobotics Institute, Viale Rinaldo Piaggio 34, 56025 Pontedera, Pisa, Italy.
Prof. Gianni Ciofani, Istituto Italiano di Tecnologia, Smart Bio-Interfaces, Viale Rinaldo Piaggio 34, 56025 Pontedera, Pisa, Italy; Politecnico di Torino, Department of Mechanical and Aerospace Engineering, Corso Duca degli Abruzzi 24, 10129 Torino, Italy.
References
- [1].Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, Wolinsky Y, Kruchko C, Barnholtz-Sloan J. Neuro-Oncology. 2014;16:iv1. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wirsching H-G, Galanis E, Weller M. In: Handbook of Clinical Neurology. 1st ed. Berger MS, Weller M, editors. Elsevier; Amsterdam: 2016. pp. 381–397. [DOI] [PubMed] [Google Scholar]
- [3].Wilhelm I, Molnár J, Fazakas C, Haskó J, Krizbai IA. Int J Mol Sci. 2013;14:1383. doi: 10.3390/ijms14011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wu S-Y, Watabe K. Front Biosci. 2017;22:1805. doi: 10.2741/4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Abbruzzese C, Matteoni S, Signore M, Cardone L, Nath K, Glickson JD, Paggi MG. J Exp Clin Cancer Res. 2017;36:169. doi: 10.1186/s13046-017-0642-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Stupp R, Mason WP, Van Den Bent MJ, Weller M, Fisher B, Taphoorn MJB, Belanger K, Brandes AA, Marosi C, Bogdahn U. N Engl J Med. 2005;352:987. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- [7].Raizer JJ, Fitzner KA, Jacobs DI, Bennett CL, Liebling DB, Luu TH, Trifilio SM, Grimm SA, Fisher MJ, Haleem MS. J Oncol Pract. 2014;11:e59. doi: 10.1200/JOP.2012.000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Geraldo LHM, Garcia C, da Fonseca ACC, Dubois LGF, Matias D, de Camargo Magalhães ES, do Amaral RF, da Rosa BG, Grimaldi I, Leser FS. Trends Cancer. 2018;5:46. [Google Scholar]
- [9].Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. Acta Neuropathol. 2007;114:97. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yao M, Li S, Wu X, Diao S, Zhang G, He H, Bian L, Lu Y. Cell Mol Immunol. 2018;15:737. doi: 10.1038/cmi.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Libby CJ, Tran AN, Scott SE, Griguer C, Hjelmeland AB. Biochim Biophys Acta,– Rev Cancer. 2018;1869:175. doi: 10.1016/j.bbcan.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].C. G. A. R. Network. Nature. 2008;455:1061. [Google Scholar]
- [13].Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP. Cancer Cell. 2010;17:98. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nam JY, de Groot JF. J Oncol Pract. 2017;13:629. doi: 10.1200/JOP.2017.025536. [DOI] [PubMed] [Google Scholar]
- [15].Tirosh I, Suvà ML. Neuro-Oncology. 2018;20:37. doi: 10.1093/neuonc/nox126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mincarelli L, Lister A, Lipscombe J, Macaulay IC. Proteomics. 2018;18 doi: 10.1002/pmic.201700312. 1700312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Meyer M, Reimand J, Lan X, Head R, Zhu X, Kushida M, Bayani J, Pressey C, Lionel AC, Clarke ID. Proc Natl Acad Sci USA. 2015;112:851. doi: 10.1073/pnas.1320611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tome-Garcia J, Tejero R, Nudelman G, Yong RL, Sebra R, Wang H, Fowkes M, Magid M, Walsh M, Silva-Vargas V. Stem Cell Rep. 2017;8:1421. doi: 10.1016/j.stemcr.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Piccirillo SGM, Colman S, Potter NE, Van Delft FW, Lillis S, Carnicer MJ, Kearney L, Watts C, Greaves M. Stem Cell Rep. 2015;4:7. doi: 10.1016/j.stemcr.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liau BB, Sievers C, Donohue LK, Gillespie SM, Flavahan WA, Miller TE, Venteicher AS, Hebert CH, Carey CD, Rodig SJ. Cell Stem Cell. 2017;20:233. doi: 10.1016/j.stem.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen X, Wen Q, Stucky A, Zeng Y, Gao S, Loudon WG, Ho HW, Kabeer MH, Li SC, Zhang X, Zhong JF. Carcinogenesis. 2018;39:931. doi: 10.1093/carcin/bgy052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G. Science. 2016;352:189. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, Mathewson ND, Neftel C, Frank N, Pelton K, Hebert CM. Science. 2018;360:331. doi: 10.1126/science.aao4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Qian M, Wang DC, Chen H, Cheng Y. Semin Cell Dev Biol. 2017;64:143. doi: 10.1016/j.semcdb.2016.09.003. [DOI] [PubMed] [Google Scholar]
- [25].Idbaih A. Ann Oncol. 2017;28:1415. doi: 10.1093/annonc/mdx217. [DOI] [PubMed] [Google Scholar]
- [26].Qazi MA, Vora P, Venugopal C, Sidhu SS, Moffat J, Swanton C, Singh SK. Ann Oncol. 2017;28:1448. doi: 10.1093/annonc/mdx169. [DOI] [PubMed] [Google Scholar]
- [27].Zong CC. Genome Biol. 2017;18:17. doi: 10.1186/s13059-017-1375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Johnson E, Dickerson KL, Connolly ID, Gephart MH. 2018;20:42. doi: 10.1007/s11912-018-0673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tsoucas D, Yuan G-C. Curr Opin Genet Dev. 2017;42:22. doi: 10.1016/j.gde.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sen R, Dolgalev I, Bayin NS, Heguy A, Tsirigos A, Placantonakis DG. In: Glioblastoma. Placantonakis DG, editor. Springer; New York: 2018. pp. 151–170. [DOI] [PubMed] [Google Scholar]
- [31].Stieber D, Golebiewska A, Evers L, Lenkiewicz E, Brons NHC, Nicot N, Oudin A, Bougnaud S, Hertel F, Bjerkvig R, Vallar L, et al. Acta Neuropathol. 2014;127:203. doi: 10.1007/s00401-013-1196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Francis JM, Zhang C-Z, Maire CL, Jung J, Manzo VE, Adalsteinsson VA, Homer H, Haidar S, Blumenstiel B, Pedamallu CS. Cancer Discovery. 2014;4:956. doi: 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, et al. Science. 2014;344:1396. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Müller S, Liu SJ, Di Lullo E, Malatesta M, Pollen AA, Nowakowski TJ, Kohanbash G, Aghi M, Kriegstein AR, Lim DA. Mol Syst Biol. 2016;12:889. doi: 10.15252/msb.20166969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Couturier CP, Ayyadhury S, Le PU, Monlong J, Riva G, Allache R, Baig S, Yan X, Bourgey M, Lee C, Wang YCD, et al. bioRxiv, Cancer Biol. 2018 449439. [Google Scholar]
- [36].Müller S, Di Lullo E, Bhaduri A, Alvarado B, Yagnik G, Kohanbash G, Aghi M, Diaz A. bioRxiv, Cancer Biol. 2018 377606. [Google Scholar]
- [37].Yuan J, Levitin HM, Frattini V, Bush EC, Boyett DM, Samanamud J, Ceccarelli M, Dovas A, Zanazzi G, Canoll P, Bruce JN, et al. Genome Med. 2018;10:57. doi: 10.1186/s13073-018-0567-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Peng S, Rath S, Vuong C, Bollam S, Eschbacher J. bioRxiv 2019. 2019 775197. [Google Scholar]
- [39].Müller S, Kohanbash G, Liu SJ, Alvarado B, Carrera D, Bhaduri A, Watchmaker PB, Yagnik G, Di Lullo E, Malatesta M, Amankulor NM, et al. Genome Biol. 2017;18:234. doi: 10.1186/s13059-017-1362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG, Hovestadt V, Escalante LE, Shaw ML, Rodman C. Science. 2017;355 doi: 10.1126/science.aai8478. eaai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hu W, Wang T, Yang Y, Zheng S. Cancer Genet. 2015;208:581. doi: 10.1016/j.cancergen.2015.09.005. [DOI] [PubMed] [Google Scholar]
- [42].Wei W, Shin YS, Xue M, Matsutani T, Masui K, Yang H, Ikegami S, Gu Y, Herrmann K, Johnson D. Cancer Cell. 2016;29:563. doi: 10.1016/j.ccell.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Leelatian N, Sinnaeve J, Mistry AM, Barone SM, Diggins KE, Greenplate AR, Weaver KD, Thompson RC, Chambless LB, Mobley BC. bioRxiv, Cancer Biol. 2019 632208. [Google Scholar]
- [44].Darmanis S, Sloan SA, Croote D, Mignardi M, Chernikova S, Samghababi P, Zhang Y, Neff N, Kowarsky M, Caneda C. Cell Rep. 2017;21:1399. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Anderson ARA, Quaranta V. Nat Rev Cancer. 2008;8:227. doi: 10.1038/nrc2329. [DOI] [PubMed] [Google Scholar]
- [46].Pearson RD. Med Hypotheses. 2009;72:629. doi: 10.1016/j.mehy.2009.01.014. [DOI] [PubMed] [Google Scholar]
- [47].D’Argenio V. High-Throughput. 2018;7:5. [Google Scholar]
- [48].Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G, Xu L, Luo T, Yan H, Long Z. Nucleic Acids Res. 2018;47:D900. doi: 10.1093/nar/gky939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Puchalski RB, Shah N, Miller J, Dalley R, Nomura SR, Yoon J-G, Smith KA, Lankerovich M, Bertagnolli D, Bickley K. Science. 2018;360:660. doi: 10.1126/science.aaf2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wick W, Kessler T. Nat Rev Neurol. 2018;14:453. doi: 10.1038/s41582-018-0038-3. [DOI] [PubMed] [Google Scholar]