Abstract
Recent studies of N-terminal acetylation have identified new N-terminal acetyltransferases (NATs) and expanded the known functions of these enzymes beyond their roles as ribosome-associated co-translational modifiers. For instance, the identification of Golgi- and chloroplast-associated NATs shows that acetylation of N-termini also happens post-translationally. In addition, we now appreciate that some NATs are highly specific: for example, a dedicated NAT responsible for post-translational N-terminal acetylation of actin was recently revealed. Other studies have extended NAT function beyond Nt-acetylation, including functions as lysine acetyltransferases (KATs) and non-catalytic roles. Finally, emerging studies emphasize the physiological relevance of N-terminal acetylation, including roles in calorie restriction-induced longevity and in pathological α-synuclein aggregation, relevant for Parkinson’s disease. Combined, the NATs rise as multifunctional proteins and N-terminal acetylation is gaining recognition as a major cellular regulator.
Keywords: Acetylation, actin, histones, KAT, lysine acetyltransferase, NAA10, NAA80, Nα-acetyltransferase, N-terminal acetyltransferase, NAT, N-terminal acetylation, protein folding, protein degradation, protein targeting
N-terminal acetylation – an abundant and regulated protein modification
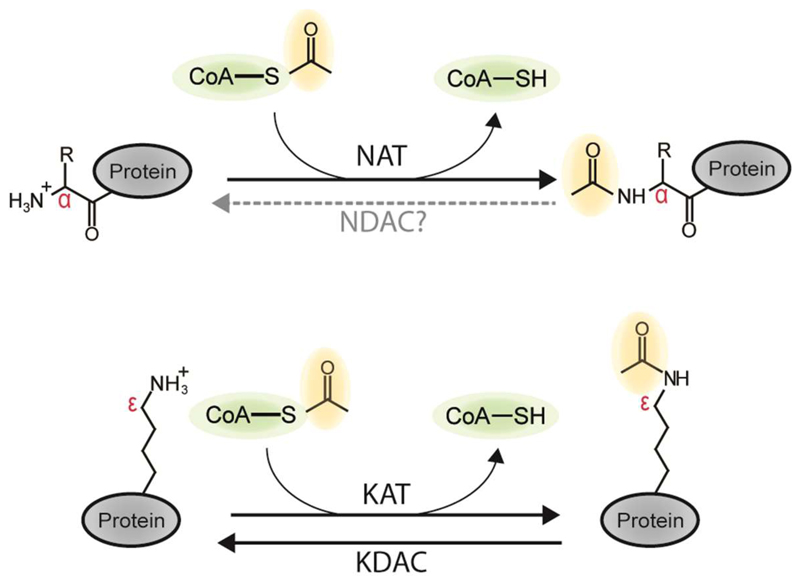
N-terminal (Nt) acetylation is among the most common protein modifications in eukaryotic cells. In fact, a majority of the proteome is Nt-acetylated (Arnesen et al., 2009; Bienvenut et al., 2012). This applies to both soluble cytosolic proteins as well as transmembrane proteins (Aksnes et al., 2015c). Nt-acetylation refers to the addition of an acetyl group to the N-terminus of a protein, i.e. the amino group on the α-carbon of the first amino acid in the protein (Figure 1). The source of the acetyl group is acetyl coenzyme A (Ac-CoA) and the transfer of acetyl is catalyzed by N-terminal acetyltransferases (NATs) (Aksnes et al., 2016). In contrast, the more extensively investigated acetylation of internal lysines is catalyzed by lysine acetyltransferases (KATs) and this reaction is reversible by the action of lysine deacetylases (KDACs) (Drazic et al., 2016). No deacetylase acting on acetylated N-termini has been identified, leading to the assumption that Nt-acetylation is both irreversible and completely static. However, recent studies suggest that it is probably more complex than that.
Figure 1. Protein Nε- and Nα-acetylation.
Protein N-acetyltransferases catalyze the transfer of an acetyl group from acetyl-CoA (green, acetyl group in yellow) onto a substrate protein. An N-terminal acetyltransferase (NAT, top panel) targets the α-amino group (red) of the N-terminal residue, whereas a lysine acetyltransferase (KAT, bottom panel) targets the ε-amine group (red) of an internal lysine. targets. Lysines can be deacetylated by a lysine deacetylase (KDAC), while currently, no N-terminal deacetylases (NDACs) are known, and N-terminal acetylation is thus considered irreversible.
This review provides a summary of these and other recent advances in our knowledge on N-terminal acetylation as well as point to new knowledge concerning the NATs and their cellular roles. The overall scope is a timely and complete overview of the cotranslational, posttranslational and non-catalytic roles of this enzyme family, including its most recent member, the actin Nt-acetyltransferase.
Regulation of N-terminal acetylation
Recent reports of cases in which N-terminal acetylation is reduced or increased as a response to certain factors, challenge the prevailing view of this modification as constitutive and static. In cancer cells, fluctuations in Ac-CoA was found to impact the level of Nt-acetylation and thereby apoptotic fate (Yi et al., 2011). On the contrary, in yeast cells exposed to huge variations in Ac-CoA concentration the bulk of the proteome was remarkably unaffected in its Nt-acetylation state, albeit some exceptions suggested a more specific regulation for a part of the proteome (Varland et al., 2018a). In plants, drought stress was found to potentially regulate Nt-acetylation levels (Linster et al., 2015). In this case, drought-induced phytohormone abscisic acid signaling downregulated the major NAT, NatA, with a concomitant increase in free N-termini.. Downregulation of NatA resulted in plant adaptation and thereby drought resistance. A perhaps similar recent finding is that in yeast, calorie restriction downregulates NatD as well as reduces the Nt-acetylation of a specific substrate, leading to increased stress resistance and longevity, further described below (Molina-Serrano et al., 2016). Another example is that NAA10, the catalytic subunit of NatA, is hydroxylated during normoxia therein changing its enzymatic properties (Kang et al., 2018), as discussed below. So, although still immature, current data suggest that Nt-acetylation may be regulated both via Ac-CoA-levels and NAT-levels/function to mediate organismal and cellular signaling.
The Nt-acetylome
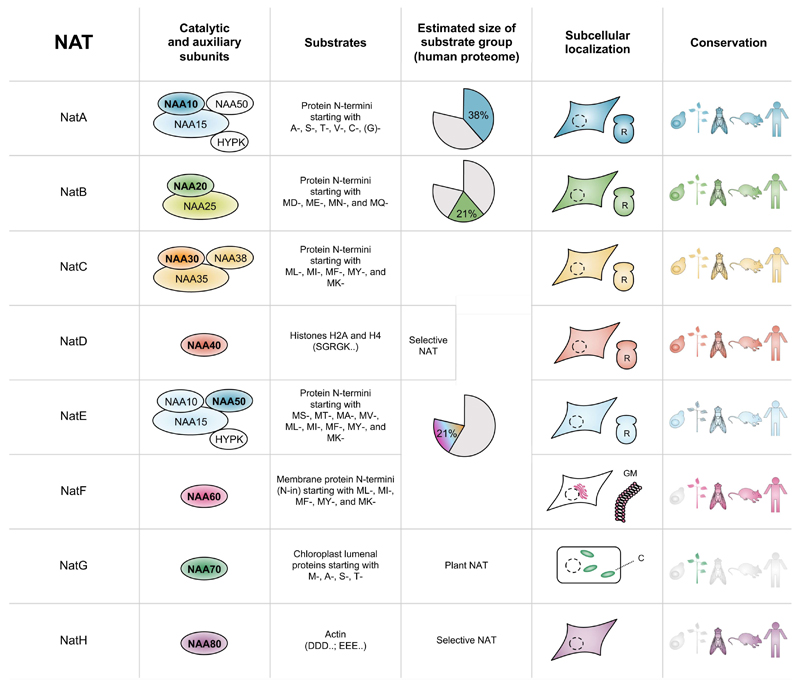
The currently known eight members of the NAT family all have distinctive features (summarized in Figure 2). Many NATs have a broad substrate specificity and together the NAT substrate classes make up the Nt-acetylome. Whether or not a protein is modified, and by which NAT, is mainly determined by the identity of the first two amino acids at the N-terminus (Figure 2, column 3). If the second amino acid is relatively small (G, A, V, S, T, C, P), the initiator methionine (iMet) is in most cases cleaved by methionine aminopeptidases (MetAPs) (Frottin et al., 2006), after which NatA may potentially acetylate the newly exposed N-terminus (Arnesen et al., 2009; Mullen et al., 1989; Polevoda et al., 1999). Among this group of substrates there are large differences: Ser- N-termini are for instance almost always Nt-acetylated while Gly- N-termini are rarely acetylated and Pro- N-termini are almost never acetylated (Arnesen et al., 2009; Goetze et al., 2009; Van Damme et al., 2011b). For suboptimal substrates, partial Nt-acetylation is often observed. With its broad specificity, NatA is the major NAT acetylating thousands of different proteins, representing ~40% of the human proteome (Figure 2, column 4). In contrast, NatD, which also acetylates Ser- starting N-termini, is highly specific for histones H2A and H4 (Hole et al., 2011; Song et al., 2003).
Figure 2. The eukaryotic N-terminal acetyltransferases.
Overview of the composition and characteristics of the eight currently known eukaryotic NATs, NatA to NatH (column 1). The catalytic subunits (NAA10 to NAA80) typically associate with up to two auxiliary subunits (NAA15, NAA25, NAA35, and NAA38) that contribute to activity of the NAT complex through ribosome anchoring and/or substrate specificity-modulation (column 2). NATs have specificity towards different proteins based on the identity of mainly the first two amino acids (column 3) (Aksnes et al., 2016). Taking into account the abundance of each N-termini subtype in the proteome, each NAT is estimated to acetylate a certain percentage of the proteome (column 4) (Aksnes et al., 2016; Aksnes et al., 2015c). NATs vary in their intracellular distribution from cytosolic and ribosome associated (NatA-NatE) to Golgi membrane (NatF), organelle lumen (NatG) and cytosolic but non-ribosomal (NatH) (column 5). In addition, some of the NAT catalytic subunits appear to additionally localize to the nucleus (e.g., NAA10, NAA40 and NAA50; not specifically denoted). The traditional ribosome-associated NATs are well conserved, whereas the more recently discovered NATs performing post-translational Nt-acetylation, are less conserved (column 6). R, Ribosome; GM, Golgi membrane; C, Chloroplast.
The iMet-retaining N-termini are modified by other enzymes. Met-“Asx/Glx”-type N termini (MD-, ME-, MN-, and MQ-starting) are almost 100% Nt-acetylated by NatB (Van Damme et al., 2012) whereas other Met- N-termini (like ML-, MI-, MF-, MY-, MK-, MT-starting) are more variably Nt-acetylated by NatC, NatF and NatE (Figure 2, column 3) (Aksnes et al., 2015c; Van Damme et al., 2011a; Van Damme et al., 2015; Van Damme et al., 2011b; Van Damme et al., 2016). While NatB alone acetylates ~20% of the human proteome, the combined actions of NatC, NatE and NatF accounts for the same number (Figure 2, column 4). The distinguishing factors between the substrates for each of these three NATs is less defined (Aksnes et al., 2016). However, NatF specifically Nt-acetylates membrane proteins.
Taking these three substrate classes together, the total fraction of the proteome that is Nt-acetylated, also called the Nt-acetylome, is ~80% in humans. In chloroplasts, proteins with various N-termini (M, A, S, T) are Nt-acetylated by NatG (Dinh et al., 2015). Finally, in the animal kingdom, actins are uniquely processed by a peptidase to generate mature acidic actin N-termini starting with several Asp or Glu residues (Redman and Rubenstein, 1981; Rubenstein and Martin, 1983). The actins are then Nt-acetylated by the recently identified NatH (Drazic et al., 2018).
Crosstalk with other N-terminal modifications
A major proportion of the eukaryotic proteome is Nt-acetylated. So what about the remaining fraction that is not Nt-acetylated? These protein N-termini are not necessarily unmodified. In fact, protein N-termini may undergo several different modifications. This embeds a layer of regulatory complexity to protein N-termini and opens for a functional interplay between different modifications at the N-terminus (Varland et al., 2015). For instance, Gly-starting N-termini (after iMet removal) are less frequently Nt-acetylated than many other types of N-termini (Arnesen et al., 2009) and a recent global investigation of Gly-N-termini not only revealed that many proteins were Nt-myristoylated instead of Nt-acetylated, but also that several specific proteins could dually exist in either Nt-acetylated or Nt-myristoylated states (Castrec et al., 2018).
Another example involves the Ser-starting myosin regulatory light chain 9 (MYL9) which is present in both Nt-acetylated and Nt-methylated states in vivo (Petkowski et al., 2012). Interestingly, these two modification states of MYL9 were recently described to be differentially modified also by internal PTMs, and to further have different binding properties, as well as promote different cellular roles (Nevitt et al., 2018). This work showed that Nt-acetylation of MYL9 is associated with increased phosphorylation at serine 19, as well as skewing towards MYL9’s cytoplasmic roles, whereas exclusive Nt-methylation is associated with increased DNA binding and increased nuclear function as a transcription factor while additionally also negatively impacting interactions between the MYL9 N-terminus and several cytoskeletal proteins.
Other types of interplay includes NatD mediated Histone H4 Nt-acetylation which was found to block H4Arg3 methylation in one case and H4Ser1 phosphorylation in another, impacting transcription from such promoters (Ju et al., 2017; Schiza et al., 2013). A recent study found an inverse relationship between protein Nt-ubiquitination and Nt-acetylation (Akimov et al., 2018). Among the here detected Nt-ubiquitinated proteins, there was an over-representation of Pro- and Val-starting proteins, i.e. types of N-termini known to rarely be Nt-acetylated (Aksnes et al., 2016). Interestingly, this work also suggested that Nt-ubiquitination likely has more roles than merely being a label for degradation.
The eukaryotic NAT machinery
The NAT machinery is classically thought to act co-translationally, and the five established ribosome associated NATs (NatA to NatE) are highly conserved among eukaryotes (Figure 2, column 5 and 6) (Aksnes et al., 2016). Each NAT has a catalytic subunit and all but one of these ribosomal NATs have a dedicated ribosomal anchor subunit (Polevoda et al., 2008).
NatA was first discovered in Saccharomyces cerevisiae and is composed of the catalytic subunit NAA10 and the auxiliary subunit NAA15 (Figure 2, column 2) (Arnesen et al., 2005a; Mullen et al., 1989; Park and Szostak, 1992). NAA15 is essential for NatA activity in two ways: it acts as a ribosomal anchor via contacts to the ribosome (Gautschi et al., 2003; Magin et al., 2017; Varland and Arnesen, 2018), and it modulates the substrate specificity of NAA10 (Liszczak et al., 2013). While monomeric NAA10 may target acidic amino acids (at least in vitro), the bulk of cellular NAA10 is bound to NAA15 and this complex prefers substrate N-termini starting with small amino acids like Ser and Thr (Liszczak et al., 2013; Van Damme et al., 2011a). The anchoring of NatA to the ribosome was recently revealed to involve binding to ribosomal RNA expansion segments thus positioning NatA dynamically underneath the polypeptide exit tunnel (Knorr et al., 2019). The Huntingtin interactor and chaperone-like protein HYPK (Raychaudhuri et al., 2008) was found to be a stably associated component of the human NatA complex and further to be important for proper NatA mediated Nt-acetylation of cellular proteins (Arnesen et al., 2010). The structures of the human and C. thermophilum NAA10-NAA15-HYPK complexes were recently revealed, and biochemical studies interestingly identified HYPK as a negative regulator of NatA activity (Gottlieb and Marmorstein, 2018; Weyer et al., 2017). Further investigations are required to define the overall impact of HYPK for NatA activity in cells and in vivo, but these data may imply that HYPK may act as an essential component of NatA ensuring quality control of this enzymatic process.
NatB is composed of the catalytic subunit NAA20 and the auxiliary subunit NAA25 (Polevoda et al., 2003; Starheim et al., 2008). NAA25 wraps around NAA20 in a similar way as observed for NAA15 and NAA10 (Hong et al., 2017), but the catalytic site of NAA20 is specialized to accommodate its large cellular pool of MD, ME, MN and MQ substrates.
The NatC complex acetylates a large variety of Met-hydrophobic N-termini and is the third of the major NATs (Polevoda et al., 1999; Tercero et al., 1993; Van Damme et al., 2016). NatC subunits NAA30 and NAA35 are analogues to the catalytic and ribosomal anchor subunits of the NatA and NatB complexes (Polevoda and Sherman, 2001; Starheim et al., 2009), but their structures remain to be solved. The small NAA38 subunit which has been identified in both S. cerevisiae and human NatC has not yet been functionally defined, but its Sm-like domain may suggest a role in RNA-binding.
NatD is the only human ribosomal NAT without a known ribosomal anchor subunit since it is solely composed of the catalytic NAA40 (Hole et al., 2011; Song et al., 2003). Its catalytic core is specifically designed to accommodate its only two known substrates, histones H2A and H4 (Magin et al., 2015). The N-termini of these histones are first processed by MetAP, similar to NatA substrates, before the Ser residues are Nt-acetylated by NatD. Thus, NatD is unique among these ribosomal NATs in terms of the lack of additional subunits as well as its highly selective substrate pool.
NatE is defined as the activity of the catalytic NAT-subunit NAA50 that may physically associate with NAA10 and NAA15 (Arnesen et al., 2006a; Gautschi et al., 2003; Williams et al., 2003). NAA50 may act alone towards Met-hydrophobic N-termini, but when in complex with NAA10-NAA15 it will be anchored to the ribosome and might then act on the rare NatA-type N-termini for which the iMet has not been removed by MetAPs (Met-Ser, Met-Thr etc.) (Evjenth et al., 2009; Van Damme et al., 2011a; Van Damme et al., 2015).
While these five ribosomal NATs may be considered as a part of the co-translational machinery common for all eukaryotes, the most recently identified NATs are mostly more specialized both in terms of defined post-translational actions and their presence in only some eukaryotic species.
NatF is composed of the catalytic NAA60 which is present in multicellular eukaryotes like animals and plants, but not in unicellular yeast (Van Damme et al., 2011b). It acetylates a variety of Met-starting N-termini and its natural substrates appear to be defined to the cytosolic N-termini of transmembrane proteins (Aksnes et al., 2015c). NAA60 has a C-terminal extension, not present in other NATs, that integrates into the cytosolic surface of the Golgi membrane (Aksnes et al., 2017). This points to a post-translational activity for NAA60.
NatG is defined as the plant kingdom specific NAA70 enzyme (Dinh et al., 2015). A variety of Arabidopsis thaliana proteins are imported into chloroplasts and are post-translationally Nt-acetylated following import. NAA70 is localized to the chloroplast stroma and displays a broad NAT substrate specificity, thus making it likely that it is responsible for a portion of these chloroplast lumenal Nt-acetylation events in addition to cotranslational Nt-acetylation of proteins encoded by the plastid genome (Dinh et al., 2015). Further, the authors found several additional putative chloroplast-localized NATs that may complete this picture in the near future, however a plastoribosomal anchoring subunit for AtNAA70 has not been described.
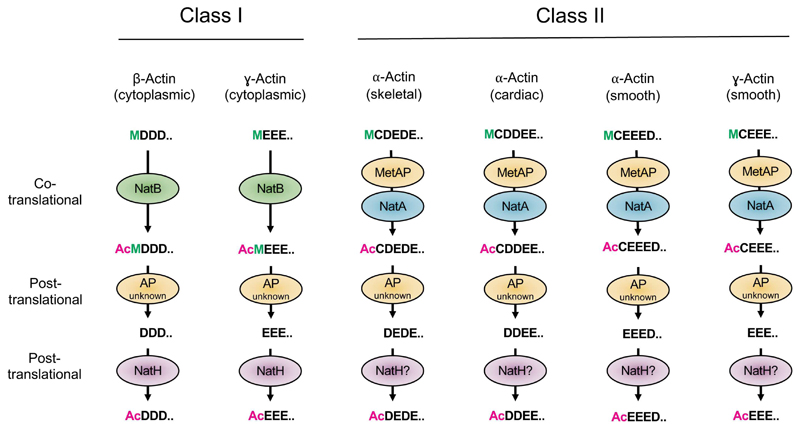
NatH/NAA80 is the most recently described member of the NAT family (Drazic et al., 2018). This NAT is neither ribosome associated, nor organelle localized, thus performing its Nt-acetylation post-translationally in the cytosol. Here, NAA80, which is a highly selective NAT, acts on one of the most abundant cytosolic proteins, namely actin. Both β- and γ-actin are proven substrates of NAA80 and it is likely that also the remaining four actins are NAA80 substrates (Drazic et al., 2018; Rubenstein and Martin, 1983; Wiame et al., 2018). All actins have a negatively-charged N-terminus and NAA80’s specificity for this type of N-terminus was explained as the structure of Drosophila NAA80 was solved in complex with a peptide-CoA bi-substrate analog mimicking the N-terminus of β-actin (Goris et al., 2018). NAA80-mediated actin Nt-acetylation is part of a unique animal-specific stepwise N-terminal processing in which Nt-acetylation actually occurs twice (Figure 3) (Berger et al., 1981; Redman and Rubenstein, 1981; Rubenstein and Martin, 1983). For animal β- and γ-actin (Class I actins), an initiator Methionine that is initially cotranslationally Nt-acetylated by NatB (Van Damme et al., 2012), is removed by an aminopeptidase in the proceeding step (Redman and Rubenstein, 1981; Rubenstein and Martin, 1983). The newly exposed N-terminus is highly acidic and NAA80-mediated Nt-acetylation further ensures acidity by removing the positive charge on the α-amino group in the final step. The first and cotranslational Nt-acetylation step in actin processing is common for all eukaryotes and depends on NatB/NAA20, whereas the posttranslational steps are specific for animals, which express both NatH/NAA80 and acidic actin N-termini, suggesting coevolution of this NAT with such actin N-termini (Drazic et al., 2018). The N-terminus of the Class II actins match the substrate specificity of NatA after iMet removal. After the initial acetylation these probably undergo the same post-translational processing as Class I actins.
Figure 3. N-terminal processing of actin.
In eukaryotes, the Class I actins, β- and γ-actin, match the substrate specificity of NatB and are co-translationally Nt-acetylated by NatB. This acetylated methionine is next cleaved off probably by the enzymatic action of an unidentified aminopeptidase. The newly exposed N-terminus is post-translationally Nt-acetylated by NatH/NAA80. These post-translational steps are specific for animals. The Class II actins, muscle α- and γ-actin, likely undergo co-translational NatA-mediated Nt-acetylation after iMet removal and then follow the same post-translational processing as the Class I actins.
To summarize, the NATs together provide Nt-acetylation for a majority of the proteome. In terms of substrate classes of N-termini, the major NATs appear to be accounted for. However, additional high-specificity NATs, like NatD and NatH may yet remain to be identified. Additionally, NATs acting posttranslationally on digested N-termini, as well as NATs localized to the organelle interior also possibly exist.
Impact of protein Nt-acetylation: from protein to organism
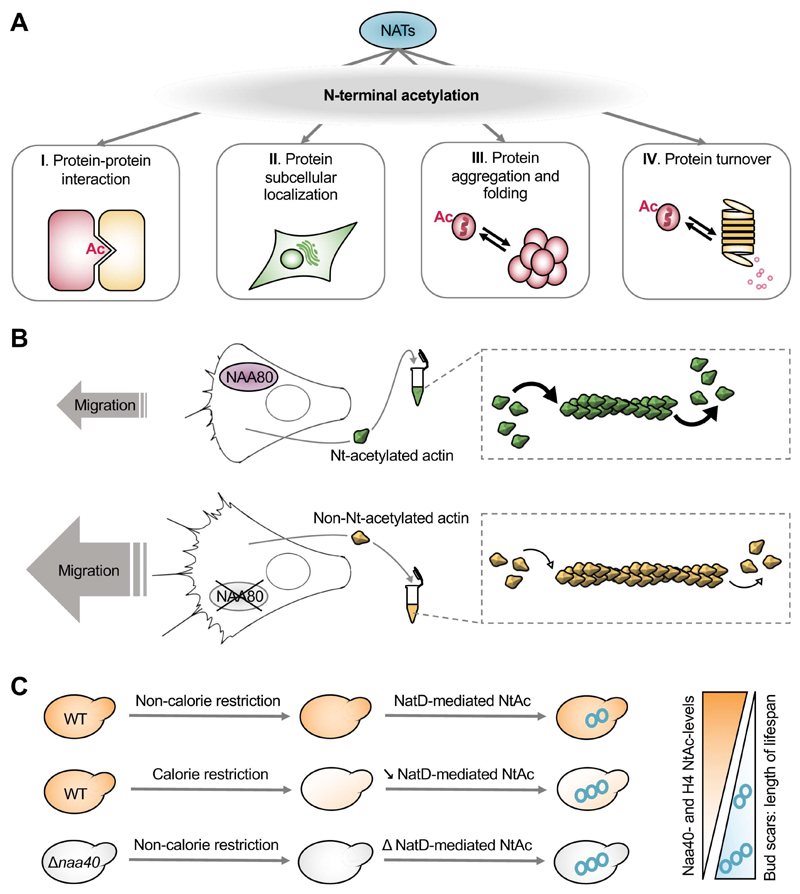
For the highly selective N-terminal acetyltranserase, NatH/NAA80, identification of the consequence of the Nt-acetyl group is measurable from protein level to cellular phenotypes. For the more typical NAT, that causes Nt-acetylation of many thousands of proteins, it can be challenging to connect effects at the protein level to specific cellular and organismal phenotypes. The studies describing impact of the Nt-acetyl group at the protein level show that Nt-acetylation can have quite diverse effects on the receiving proteins (Figure 4A).
Figure 4. Implications of the N-terminal acetyl group.
(A) A wide spectrum of protein functioning can be affected by Nt-acetylation. Nt-acetylation may be involved in protein-protein interactions. The interaction can directly involve the Nt-acetyl group’s insertion in a hydrophobic binding groove like depicted here or the modification can stabilize a binding region. Correct subcellular localization may be obscured in the absence of Nt-acetylation and in some cases the Nt-acetyl group may stabilize and take part in a membrane binding region. Absence of Nt-acetylation is also associated with aggregation, suggesting a role in global protein folding. In some cases the Nt-acetyl group may act as a proteasomal degradation signal or blocking such a signal, regulating protein lifetime or turnover rate.
(B) N-terminal acetylation of actin by NAA80/NatH affects cell morphology and migration rates as well as actin polymerization dynamics in vitro.
(C) NatD/Naa40 is downregulated during calorie restriction-induced longevity in yeast and the following reduced Nt-acetylation of histone H4 is suggested to mediate the increased replicative age seen in starved yeast. Deletion of yeast naa40 mimic the effect of calorie restriction.
Nt-acetylation impacts protein interactions and complex formation
Several studies show a direct involvement of the Nt-acetyl group in protein-protein interactions and protein complex formation (Figure 4A-I). The NatC Nt-acetylated Met-1 of the E2 enzyme UBC12 (human UBE2M) binds in a hydrophobic pocket in the E3 DCN1 (human DCUN1D1) (Scott et al., 2011), and this type of interaction is common for mammalian NEDD8 ligation enzymes (Monda et al., 2013). This direct involvement of the Nt-acetyl group in protein binding was recently antagonized by inhibitors with selectivity for the Nt-acetyl pockets of DCN1 and DCN2 (Scott et al., 2017). Nt-acetylation may also indirectly affect binding, such as for Sir3’s binding to the nucleosome in which the Nt-acetylation dependency is explained by stabilization of a binding loop (Arnaudo et al., 2013; Yang et al., 2013).
Nt-acetylation impacts protein subcellular targeting
Nt-acetylation can be crucial for correct subcellular localization. Several yeast proteins mislocalize in the absence of their respective NAT (Aksnes et al., 2015b; Behnia et al., 2004; Setty et al., 2004). However, despite attempts, Nt-acetylation could not thus far be defined as a global localization determinant in yeast (Aksnes et al., 2013; Caesar et al., 2006). In fact, some proteins that are localized by means of their Nt-acetyl group actually require it for protein-protein interaction at the localization site. In the plant Arabidopsis thaliana, Nt-acetylation by NatC is required for efficient photosynthesis and it was suggested that certain chloroplast precursor protein(s) were dependent on their Nt-acetyl group for chloroplast localization (Pesaresi et al., 2003).
An example of a human Nt-acetylated protein directly interacting with the membrane is α-synuclein, a protein that is implicated in Parkinson’s disease. Here, Nt-acetylation, together with copper binding to the Nt-region, stabilizes an N-terminal α-helix thereby increasing α-synuclein’s affinity for membranes (Dikiy and Eliezer, 2014; Miotto et al., 2015). Another subcellular targeting mechanism in which Nt-acetylation appears to play an important role, is post-translational translocation across the endoplasmic reticulum (ER) membrane. Here, the Nt-acetyl group was found to be part of an early targeting step inhibiting the signal recognition particle (SRP)-independent pathway for post-translational translocation through the Sec62 channel (Forte et al., 2011).
Nt-acetylation impacts protein aggregation and folding
Nt-acetylation has been connected to protein aggregation with both positive and negative correlation, the latter depicted in Figure 4A-III. We know of two proteins whose pathology-causing aggregation processes is somehow slowed down by their Nt-acetylation. Aggregation of α-synuclein (α-Syn) is a hallmark of Parkinson’s disease and other neuropathologies and in vivo this protein is fully Nt-acetylated. Although no Nt-acetylation-reducing disease variants have been characterized, in vitro experiments using the Nt-un-acetylated α-Syn has shown that this version has a considerable higher rate of aggregation (Kang et al., 2012). This indicates the biological importance of this modification. At the protein level, addition of an acetyl group at the N-terminus of α-Syn disrupts intermolecular hydrogen bonds that are important for α-Syn oligomerization, thereby decelerating the oligomerization process (Bu et al., 2017). Specifically, two H-bond donors at the N-terminus are absent in the presence of the Nt-acetyl group and this disrupts the intra molecular H-bond network (Bu et al., 2017). Thus, α-Syn is a very interesting case that demonstrates the importance of Nt-acetylation as the Nt-acetyl group is relevant for both aggregation propensity and membrane interaction as described above.
Another protein that becomes prone to aggregation in the absence of Nt-acetylation, is huntingtin (Htt), although the possible direct effect of an Nt-acetyl group on Htt is less defined since Htt’s aggregation is prevented by the chaperone function of Htt-interacting protein K (HYPK) (Raychaudhuri et al., 2008) which is also a stable interactor in the NatA complex (Figure 2), that is necessary for its Nt-acetylation activity (Arnesen et al., 2010). Nevertheless, it appears that the NatA-HYPK complex is essential in keeping Htt from aggregating, possibly through Nt-acetylation of Htt.
An example in which Nt-acetylation facilitates rather than prevents aggregation is the NatA substrate Sup35. This protein forms amyloids in prion [PSI+] yeast cells and deletion of NatA in these yeast decreases the stability of Sup35 amyloid thereby relieving the phenotype of the [PSI+] cells (Holmes et al., 2014). The N-terminus of Sup35 is outside of the amyloid core, suggesting a cooperative or conformational effect related to protein structure, analogous to the intra protein H-bond effect for α-Syn. Thus there are indications of a general effect of Nt-acetylation status on protein conformation or folding. Indeed, the same study (Holmes et al., 2014), proposed Nt-acetylation to have a global role in protein folding since Nt-acetylation deficiency in NatA-deleted yeast caused accumulation of misfolded proteins and increased levels of chaperones. However, indirect events may also come into play when studying global folding/aggregation effects resulting from depletion of NATs. For example, it was recently found that the yeast Hsp90 chaperone system is greatly impaired in cells depleted for NAA10, due to the unacetylated status of Hsp90 and several of its cochaperones (Oh et al., 2017).
Nt-acetylation impacts protein turnover through proteasomal degradation pathways
For several decades, Nt-acetylation was considered to contribute to protein stability in vivo (Hershko et al., 1984). In 2010, this concept was challenged by the discovery of the Ac/N-degron (Hwang et al., 2010). Nt-acetylated protein termini starting with M, S, A, T, or V were identified as potential Ac/N-degrons which may be recognized by the E3 ubiquitin ligases Doa10 or Not4 and targeted for degradation (Hwang et al., 2010; Shemorry et al., 2013). Degradation of Nt-acetylated proteins appears to be particularly important in the context of protein quality control. Improper folding or abnormal stoichiometries of protein complex subunits may render certain protein N-termini exposed and thus available for the degradation machinery (Shemorry et al., 2013). At the same time, for several proteins like Hyx in Drosophila (Goetze et al., 2009) or the human THOC7 (Myklebust et al., 2015), Nt-acetylation certainly confers increased protein half-lives.
An interesting example relevant for human physiology involves degradation of Rgs2 mediated through an Nt-acetylated as well as an unacetylated N-terminus. Rgs2 is a regulator of G-protein signaling and the wild type MQ-Rgs2 is fully Nt-acetylated by NatB and degraded via its Ac/N-degron. However, an Rgs2 N-terminal variant associated with hypertension, ML-Rgs2, is only partially Nt-acetylated by NatC and thus may be degraded via both its Ac/N-degron or via its unacetylated N-terminus which is a classical Arg/N-degron (Varshavsky, 2019). In sum, the ML-Rgs2 variant has a shorter half-life than the normal MQ-Rgs2 (degraded at least twice as rapidly) resulting in abnormal hypertension signaling in the patients expressing this variant (Aksnes et al., 2015a; Park et al., 2015). Thus both Nt-acetylated and unacetylated N-termini may act as degrons per se.
A recent global investigation on yeast N-terminal degrons suggested that a quarter of all nascent protein N-termini may potentially encode degrons (Kats et al., 2018). However, this study suggested that the E3 ligase Doa10 primarily targets hydrophobic and not Nt-acetylated N-termini. Further, Nt-acetylation was found to inhibit degradation via Ubr1 and Arg/N-degrons. So while there is agreement on Nt-acetylated proteins not being targeted via the classical Arg/N-degron pathway as shown for Ubr1, the proteome-wide extent of the Ac/N-degron pathway needs further elaboration. While the most recent investigation is comprehensive and elegantly designed, it is still a reporter based system, so global investigations based on endogenous proteins in different cellular conditions are required to clarify this. Also, the relative contributions of the E3 ligases Not4 and Doa10 in the Ac/N-degron pathway are interesting to investigate further (Kats et al., 2018; Zattas et al., 2013).
Nt-acetylation impacts cellular functions
Although loss of Nt-acetylation is associated with many cellular abnormalities, the molecular basis of these have only been defined in a few cases. The highly selective NAT, NAA80, offers a unique possibility to study effects of Nt-acetylation on substrate function. NAA80’s substrate, actin, was indeed demonstrated to be impacted by the Nt-acetyl group in both its chemical and cellular functioning as shown in Figure 4B (Drazic et al., 2018). NAA80 knockout cells were here described to have actin-related phenotypes, migrating more and with increased velocity compared to control cells holding Nt-acetylated actin. The cells with unacetylated actin also had an increased presence of protrusive structures characteristic of migrating cells and furthermore, actin purified from these cells had altered turnover of actin filaments. Actin has several binding partners that contribute to the regulation of its function and it is likely that actin’s Nt-acetyl group may contribute to some of its binding capacity. The hypermotility phenotype of the NAA80 knockout cells could suggest a role for NAA80 in keeping cellular migration at a healthy rate. In a cancer cell, the lack of proper NAA80 function could conceivably induce cancer metastasis, an idea that seems plausible due to the localization of the NAA80 gene in a tumor-suppressor region (Wei et al., 1996). Actin is a key protein in a broad range of cellular functions and functional roles of actin Nt-acetylation are likely to be further defined as NAA80 deletion is further studied in additional model systems.
Our knowledge on implications of Ntacetylation is growing as an increasing number of NAT knockout cells are characterized. A recent study using NAA20 and NAA25 knockout cells, found that Nt-acetylation by NatB is required for the shutoff activity of Influenza A virus and viral polymerase activity (Oishi et al., 2018). Influenza A virus suppresses, or shutoff, host gene expression through the viral endonuclease PA-X, that cleaves host mRNAs. An N-terminal endonuclease active site in PA-X, in addition to specific C-terminal basic amino acids, is important for its shutoff activity. Oishi and coworkers found that the shutoff activity of PA-X was suppressed in NatB-deficient cells and that Nt-mutants of PA-X not matching the NatB specificity had reduced shutoff activities (Oishi et al., 2018). This work indicated that Nt-acetylation alone is not sufficient for PA-X shutoff activity, since mutants still matching NatB specificity (PA-X E2D and PA-X E2N) showed somewhat reduced shutoff activity and mutants fully Nt-acetylated by NatA (PA-X E2A), showed reduced shutoff activity.
Yeast knockouts have had an important role in the early days of the Nt-acetylation field and now continue to contribute in expanding our understanding of how the Nt-acetyl group of certain proteins may be crucial for cellular function. A recent example is the deletion of Naa40 which causes prolonged lifespan under calorie restricted conditions, probably through decreased Nt-acetylation of the NatD substrate, histone H4 (Molina-Serrano et al., 2016). Here, histone H4 was found to be a regulator of yeast cellular lifespan in that it increases stress resistance and longevity as a response to calorie restriction. Interestingly, this mechanism seems to be regulated by the Nt-acetylation status of histone H4. Besides, NAA40 too seems to undergo regulation since calorie restriction caused its downregulation in addition to reduction in the levels of Nt-acetylated H4. The extended lifespan associated with calorie restriction was reduced upon overexpression of Naa40 associated with maintained levels of histone H4 Nt-acetylation. Similar to the previously shown effect of NatA depletion in plants (Linster et al., 2015), deletion of NAA40 in yeast mimics the effects of calorie restriction, here especially in the induction of stress-response genes. Hence this mutant is “primed” for scarce food supply, analogous to how the NatA depleted plants handle drought. This work also showed that an important regulator of calorie restriction-mediated longevity that prevents the accumulation of intracellular nicotinamide during stress, nicotinamidase Pnc1, is required for naa40Δ-mediated longevity.
Another recent paper also found a link between Nt-acetylation and NAD+/NADH metabolism as it described NatB as a NAD+ homeostasis factor in yeast (Croft et al., 2018). Here, NatB-deficient yeast flagged in a screen for mutants with increased release of the NAD+ intermediates NA and NAM. Also, the levels of NAD+ were significantly reduced in the NatB-deficient yeast and it was shown that decreased levels of the nicotinamide mononucleotide adenylyltransferases Nma1 and Nma2 likely caused these NAD+ defects. These two proteins were identified as NatB substrates, demonstrating that their Nt-acetylation status is essential for maintaining NAD+ levels.
In C. elegans, Nt-acetylation is critical for the function of the NatB substrate SYP-1 (Gao et al., 2016). This protein is a component of the synaptonemal complex, a proteinaceous structure ubiquitously present during meiosis. NatB-mediated Nt-acetylation of SYP-1 is necessary for the assembly of this structure, elucidating a role for Nt-acetylation during meiosis. Further findings in this study suggested the importance of Nt-acetylation in regulating biological functions in the nucleus as there was an enrichment for global protein Nt-acetylation in the nuclear fraction (82%) compared with whole-worm lysates (74%). Interestingly, underlying this higher amount of Nt-acetylated proteins in the nucleus is an increased presence of N-termini with a high degree of Nt-acetylation (S-, NatA, 99%; MD-, NatB, 100%; and ME-, NatB 99%, (Aksnes et al., 2016)); accompanied with a decreased presence of N-termini with a low degree of Nt-acetylation (G-, NatA, 33%; P-, 0%; T-, NatA, 82%; and V-, NatA, 20%) (Gao et al., 2016). Taking these data together, Gao et al. suggest that Nt-acetylated proteins might more readily form stable macromolecular structures in the nucleus, like the synaptonemal complex, perhaps through a facilitating function of Nt-acetylation in protein-protein interactions, as described above.
NAA10 as a multifunctional protein?
Defining the functional and biological roles is particularly challenging for one of the N-terminal acetyltransferases, NAA10. Not only does NatA have the highest number of cellular substrates, but also, the catalytic subunit NAA10 seems to have extensive moonlighting functions. We here consider three aspects of NAA10 function. First, the canonical role as the catalytic subunit of the NatA complex. Second, the role of NAA10 as a posttranslational KAT. Third, NAA10 may have non-catalytic functions outside of the NatA complex.
NAA10 in the NatA complex
Among the NATs, NAA10 is first in number of substrates, acetylating an estimated 40 % of the human proteome cotranslationally as part of the NatA complex (Arnesen et al., 2009). Binding to NAA15 ensures ribosomal anchoring and specificity for its large pool of cellular substrates (Liszczak et al., 2013). The result of these acetylation events are as diverse as the substrates, but generally the loss of NatA activity has been linked to diminished cell proliferation or increased cell death (Arnesen et al., 2006c; Fisher et al., 2005; Gromyko et al., 2010; Myklebust et al., 2015), lethality in Arabidopsis (Linster et al., 2015; Xu et al., 2015), as well as abnormal development in humans and in several model organisms (Ingram et al., 2000; Mullen et al., 1989; Ree et al., 2015; Rope et al., 2011; Sonnichsen et al., 2005; Wang et al., 2010) (Figure 5A).
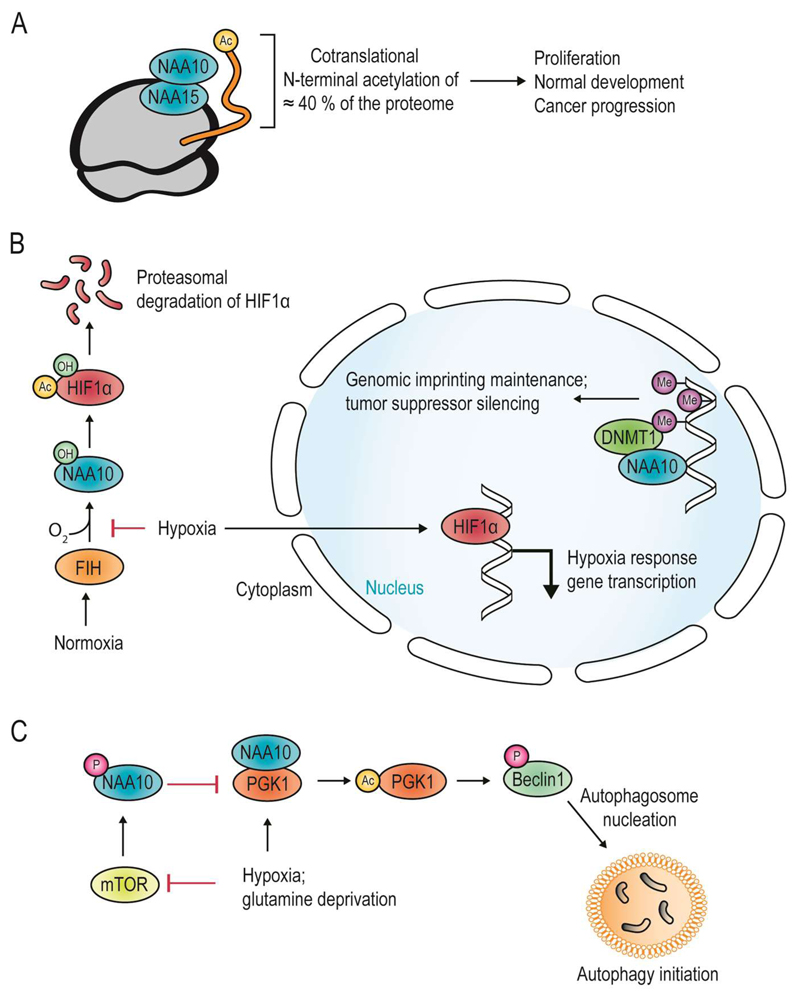
Figure 5. The diverse catalytic and non-catalytic roles of NAA10.
(A) The classical role of NAA10 as the catalytic subunit of the NatA complex. NatA is found on the ribosome and acetylates approximately 40 % of all nascent N-termini cotranslationally, contributing to diverse cellular functions, such as proliferation, normal organismal development, and as an oncoprotein and potentially a tumor suppressor, depending on the context and the function of the N-terminally acetylated protein.
(B) Examples of non-classical roles of NAA10 as a KAT (left) and a non-catalytic regulator (right). Left: NAA10 is hydroxylated during normoxia by factor inhibiting HIF-1α (FIH), which also hydroxylates HIF-1α. Upon hydroxylation, a loop in NAA10 shifts, enabling NAA10 to accommodate lysine substrates and Nε-acetylate HIF-1α. This contributes to the instability and subsequent proteasomal degradation of HIF1α. During hypoxia, neither NAA10 nor HIF-1α are hydroxylated, and HIF-1α is free to translocate to the nucleus and promote transcription of hypoxia response genes. Right: an acetyltransferase-independent function of NAA10. DNA methyltransferase 1 (DNMT1) is recruited to CpG islands by NAA10, which binds to nonmethylated DNA regions. DNMT1 binding to CpG islands furthers genomic imprinting and tumor suppressor silencing. Imprinting is required for normal murine development, but may also contribute to cancer progression.
(C) Example of non-classical role of NAA10 as a KAT. NAA10 phosphorylation by mammalian target of rapamycin (mTOR) is inhibited in states of hypoxia or glutamine deprivation. Loss of phosphorylation leads to NAA10 interaction with PGK1 and Nε-acetylation of a PGK1 lysine. Acetylated PGK1 is able to phosphorylate Beclin1 and Beclin1-mediated autophagosome nucleation and autophagy may occur.
NAA10 as a KAT
Several papers indicate that NAA10 has KAT activity, thus catalyzing the acetylation of ε-amino groups on internal lysines (Figure 1). Nε-acetylation of β-catenin is dependent on an interaction between NAA10 and β-catenin, as well as normoxia (Lim et al., 2008; Lim et al., 2006). NAA10 auto-Nε-acetylation enables NAA10 to Nε-acetylate β-catenin, turn on the activating protein 1-pathway, and stimulate cell proliferation (Seo et al., 2010). NAA10 has been implicated in the degradation of HIF-1α, by Nε-acetylating it and furthering its targeting by the proteasome by increasing the affinity of HIF-1α to the E3 ligase Von Hippel Lindau protein (Jeong et al., 2002). During hypoxia, HIF-1α is not hydroxylated and is free to translocate to the nucleus and activate the hypoxic response (Figure 5B, left). This claim has been disputed, as replication efforts failed to find neither an increase in hypoxia response genes, nor that the short half-life of HIF-1α under normoxia is increased by NAA10 depletion (Bilton et al., 2005; Fisher et al., 2005). Furthermore, NAA10 was not able to acetylate HIF-1α in vitro (Arnesen et al., 2005b). Nevertheless, a recent study shed light on how this acetylation may take place, showing that NAA10 is hydroxylated on C-2 of the indole ring of Trp38 by factor inhibiting HIF-1α (FIH) during normoxia and suggesting that this oxygen-dependent hydroxylation results in a conformational change in NAA10. Hydroxylation was suggested, by molecular dynamics simulations, to widen the substrate gate of NAA10, allowing it to accommodate the target lysine of HIF-1α (Kang et al., 2018), thus indicating a regulation of NAT versus KAT activity that is responsive to oxygen conditions (Figure 5B, left).
A number of additional KAT targets of NAA10 have been proposed. Nε-acetylation of the cell cycle regulator Cdc25A by NAA10 increases its stability, and this happens as a response to genotoxic stress. This acetylation, which can be reversed by HDAC11, is suggested to allow time for DNA repair after DNA damage (Lozada et al., 2016). NAA10 is also able to promote autophagosome nucleation through Nε-acetylation of phosphoglycerate kinase 1 (PGK1) (Figure 5C). Also in this case, oxygen conditions affect post-translational modification of NAA10. During normoxia, NAA10 is phosphorylated by mammalian target of rapamycin (mTOR), blocking its interaction with PGK1. Hypoxia or glutamine deprivation inhibits mTOR so that NAA10 is allowed to Nε-acetylate PGK1, which in turn may phosphorylate Beclin1 and start the process of autophagosome nucleation. NAA10 thus regulates nutrient availability and macromolecule recirculation through induction of autophagy under hypoxic or stress conditions. Heat shock protein 70 (Hsp70) works to promote protein refolding during the immediate time after chemical stress. NAA10 is also involved in stress response through Nε-acetylation of Hsp70, increasing the affinity of Hsp70 for a co-chaperone, thus promoting its refolding activity (Seo et al., 2016). Further, NAA10 interacts with and Nε-acetylates androgen receptor (AR), a mechanistic explanation for the fact that NAA10 is upregulated in prostate cancer. By acetylating AR, NAA10 promotes the expression of AR target genes (Wang et al., 2012). Finally, NAA10 is reported to Nε-acetylate the deoxynucleotide triphosphohydrolase SAM domain and HD domain containing protein 1 (SAMHD1). SAMHD1 acetylation leads to increased proliferation through promoting G1 to S-phase transition (Lee et al., 2017b).
From a structural point of view the KAT activity of NAA10 is controversial, as crystallographic structures indicate that there is not enough space in the groove between the α1-α2 and β6-β7 loops to accommodate a lysine substrate, only an N-terminal substrate (Figure 6A) (Gottlieb and Marmorstein, 2018; Liszczak et al., 2013). Several of the KAT substrates have so far failed to be replicated biochemically, and some results may stem from non-enzymatic acetylation (Magin et al., 2016). Indeed, non-enzymatic lysine acetylation occurs in a non-random manner and appears to be biased towards lysine sites with flanking basic residues, or where basic residues are found nearby in the 3D folding of a protein (Baeza et al., 2015). This also holds true for mitochondrial acetylation sites which are regulated by Sirt3 (Hebert et al., 2013) and are considered to be products of non-enzymatic acetylation (reviewed in Drazic et al., 2016). Two of the lysine sites discussed by Magin et al., Runx2 and MSRA, are found in basic patches of the protein 3D structure (Magin et al., 2016). This may explain why Magin found them to be readily non-enzymatically acetylated, and lends credence to the idea that they are not true NAA10 substrates. The molecular dynamics simulation of the consequence of a novel PTM on NAA10, Trp38 hydroxylation, may hold a clue as to how these opposing views may be reconciled (Kang et al., 2018). More research is needed to clarify under what conditions NAA10 can shift from a NAT to a KAT and to what degree such a change in structure adapts it to its different functions, and its implications for NAA10’s many physiological roles.
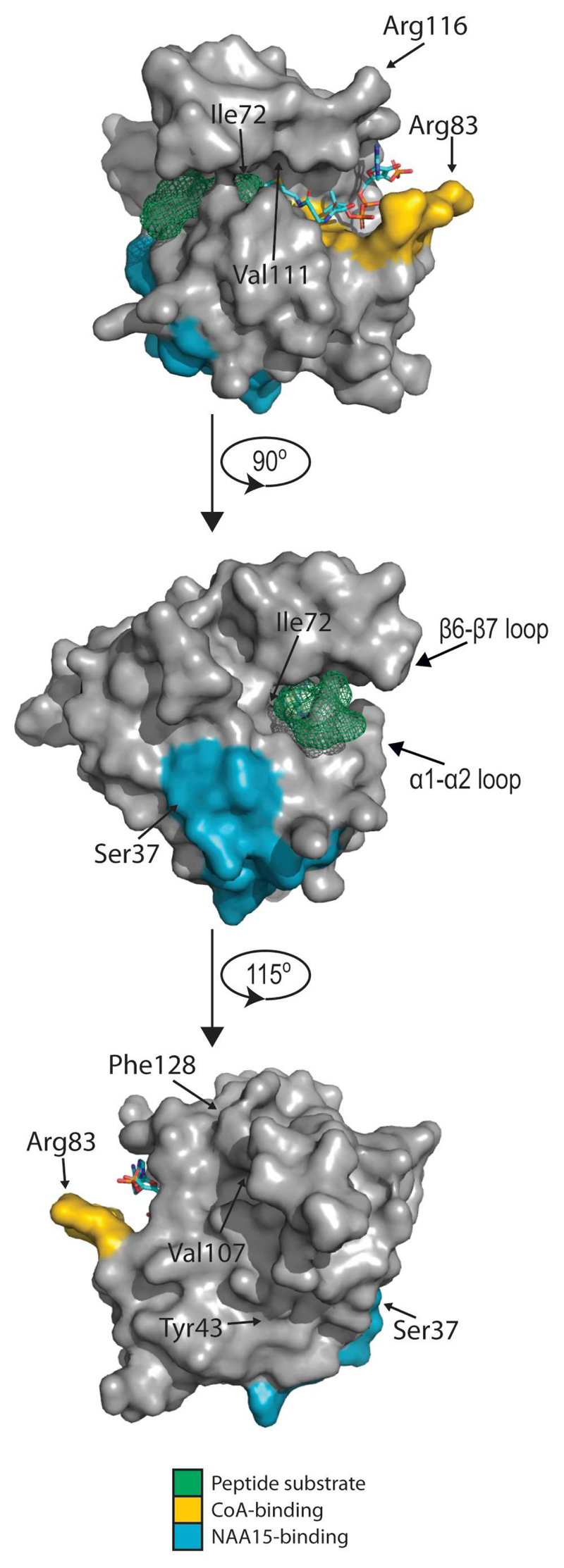
Figure 6. Structure of NAA10 and location of pathological mutation sites.
A structural model of NAA10 with SASE peptide substrate and CoA was made in PyMol by aligning the structure of human NAA10 in complex with NAA15 (not shown) (PDB ID: 6C9M; (Gottlieb and Marmorstein, 2018)) to the structure of Schizosaccharomyces pombe NAA10 solved in complex with NAA15 (not shown), CoA and substrate peptide (4KVM; (Liszczak et al., 2013)). CoA is shown as a licorice model colored according to element, SASE substrate peptide is represented in green, CoA-binding residues in yellow and NAA15-binding residues in blue. Mutation sites are indicated with arrows.
Non-catalytic roles of NAA10
NAA10 interacts with several proteins in a functionally meaningful manner. In addition to the reported substrate interactions described above (HIF-1α, Hsp70, SAMHD1, PGK1, Cdcd25A and AR), NAA10 can also affect binding partners in acetyltransferase-independent manners. In the case of signal transducer and activator of transcription 5a (STAT5a), NAA10 interaction decreases the expression of STAT5a target genes, in particular inhibitor of differentiation 1 (ID1). ID1 is considered to promote metastasis, and STAT5a is an essential driver of ID1 expression. The authors observed that NAA10 expression correlates inversely with metastasis, which may be explained in part by this inhibitory, acetyltransferase independent effect on STAT5a (Zeng et al., 2014).
Another described interactor of NAA10 is PIX. At the leading edge of a migrating cell, GTPases, guanine nucleotide exchange factors (GEFs), and extracellular matrix anchors cooperate at focal adhesions to drive the cell edge forward. One such complex consists of, among other proteins, the G protein-coupled receptor kinase-interacting protein (GIT) (a GTPase-activating protein), and p21-activated kinase interacting exchange factor (PIX, a GEF). NAA10 interacts with PIX and disrupts its binding to GIT, thus reducing Cdc42 and Rac1 activity at focal adhesions and decreasing the rate of cell migration (Hua et al., 2011). Interestingly, in both these cases, NAA10 appears to have an anti-metastatic function, inhibiting either the downstream effects of ID1 or cell migration capability through disruption of a migratory complex.
A completely different role of NAA10 was also presented, namely a direct binding to particular non-methylated DNA motifs and recruitment of DNA methyltransferase 1 (DNMT1) (Figure 5B, right). This maintains genomic imprinting, silencing genes that are not to be expressed, but may also silence tumor suppressor genes and contribute to cancer development (Lee et al., 2017a; Lee et al., 2010). Naa10 KO mice have several developmental defects, and 35 % male and 27 % female pups die prenatally, mainly as a result of placental deficiency. Surviving pups have retarded growth and hydrocephalus with dilated ventricles. Surviving heterozygous females are fertile, but have low body weight and decreased litter size. Heterozygous pups born of homozygous Naa10-/- mothers had a low perinatal survival rate, which was attributed to a maternal effect, given that heterozygous pups from heterozygous females and wildtype males survived for longer after birth. NAA10’s interaction with DNMT1 is acetyltransferase-independent (Lee et al., 2010), and when disrupted, leads to widespread dysregulation of genomic imprinting (Lee et al., 2017a). The paralog NAA11 has some functional redundancy with NAA10 (Arnesen et al., 2006b) and this might explain why the Naa10 KO mice are viable and fertile. The degree to which NAA11 or other paralogs or NATs can substitute for NAA10 in its acetyltransferase-dependent and –independent functions in KO models is an open question.
Emerging roles of NATs in human pathology
NatA variants cause developmental syndromes
The list of identified NAA10 mutations in patients has grown substantially since 2011, implicating Nt-acetylation in congenital syndromes and non-syndromic developmental delay (Figure 6, Table 1). There is considerable heterogeneity in the presentation and severity of cases of NAA10 deficiency, but broadly, mutations in conserved regions of NAA10 are associated with intellectual disability, developmental delay, growth failure, and specific organ development phenotypes, including skeletal, cardiac and ophthalmological abnormalities (Table 1). The genotype-phenotype relationship is complex, and does not depend directly on the loss of catalytic activity. Some variants, like Ser37Pro, which is the cause of Ogden syndrome, leads to a modest reduction in in vitro activity but has a very severe clinical presentation (Rope et al., 2011), while the Ile72Thr variant is at least as reduced for the monomeric activity, but nevertheless leads to a comparatively mild phenotype (Stove et al., 2018).
Table 1. Patient mutations in NAA10 and their consequences.
ID: intellectual disability; DD: developmental delay.
| Mutation | Patients | Symptoms | Functional effect | References |
|---|---|---|---|---|
| Ser37Pro | 8 male , 5 female carriers (mothers of affected boys) | Global DD; craniofacial abnormalities; aged appearance; cardiac arrhythmia; death in infancy | Decreased catalytic activity; impaired NAA15 and NAA50 interaction; decreased binding to ICRs; patient fibroblasts have decreased Nt-acetylation; abnormal cell migration and proliferation | (Lee et al., 2017a; Myklebust et al., 2015; Rope et al., 2011) |
| Tyr43Ser | 2 male (brothers), 1 heterozygous female with some symptoms (their mother) | ID; facial dysmorphism; scoliosis; long QT | Decreased catalytic activity; unstable | (Casey et al., 2015) |
| Ile72Thr | 3 male (2 brothers, 1 unrelated) | DD; ID; cardiac abnormalities | Decreased monomeric catalytic activity; unstable | (Stove et al., 2018) |
| Arg83Cys | 8 females (1 girl and 2 deceased boys are siblings; the rest de novo), 2 males | DD; ID; skeletal abnormalities; cardiac abnormalities | Decreased catalytic activity | (Saunier et al., 2016) |
| Val107Phe | 1 female | ID; abnormal muscle tone; growth failure; long QT; skeletal abnormalities | Decreased catalytic activity; decreased binding to ICRs | (Popp et al., 2015) |
| Val111Gly | 1 female | ID; delayed motor and language development | Decreased monomeric catalytic activity; unstable | (McTiernan et al., 2018) |
| Arg116Trp | 1 male, 1 female (unrelated) | ID; abnormal muscle tone; growth failure; skeletal abnormalities | Small reduction in catalytic activity; decreased binding to ICRs | (Lee et al., 2017a; Popp et al., 2015; Rauch et al., 2012; Saunier et al., 2016) |
| Phe128Ile | 1 female | ID; growth failure; abnormal muscle tone | Decreased stability; enzymatic activity not assessed | (Saunier et al., 2016; Thevenon et al., 2016) |
| Phe128Leu | 2 females (unrelated) | ID; growth failure; siezures; abnormal muscle tone | Decreased catalytic activity; decreased stability | (Saunier et al., 2016) |
| c.471+2T → A | 8 male (3 brothers, 4 others in same family) | ID (60% of cases); anophthalmia or microphthalmia; skeletal and genitourinary abnormalities | Truncated protein; enzymatic activity not assessed; patient fibroblasts have cell proliferation defects | (Esmailpour et al., 2014) |
NAA10 is located on the X chromosome, and hemizygous males would be expected to have graver symptoms of NAA10 deficiency than heterozygous females. However, in the case of very damaging mutations, female carriers may still exhibit serious symptoms, while male hemizygotes would not be viable and perish at an early age. One of the AcCoA binding mutations (Arg83Cys), which results in a very limited catalytic capacity, could be such a case. A mother who is germline mosaic for the variant gave birth to a boy who died at one week of age (Saunier et al., 2016).
Functional in vitro experimental work on NAA10 variants suggests that mutations may manifest themselves in three different ways at the protein level.
First, a mutation may cause the catalytic activity of NAA10 to diminish. This happens through abrogated interaction with a substrate – either AcCoA or the peptide substrate – or through missense mutation at a site directly involved in catalysis. Such mutations result in enzymes that are readily purified and stable, but have decreased catalytic capacity. Molecular dynamics simulations indicate that the Ser37Pro mutation impairs the flexibility of the substrate-binding region (Myklebust et al., 2015). The Arg83Cys mutation has a more direct impact as it affects the interaction of NAA10 with AcCoA (Saunier et al., 2016), and shows a heavily reduced catalytic activity. It would be presumed that all catalytic activities, NAT as well as KAT, would be impaired for such mutants.
Second, a structural or destabilizing mutant may cause NAA10 to be misfolded, and the misfolded protein to be rapidly degraded. Such mutations make the enzyme difficult to purify. Necessarily, they also cause a loss of in vitro and in vivo catalytic efficiency, and will have shorter half-lives. As Phe128 is located in a hydrophobic pocket at the core of NAA10, the Phe128Leu mutation was predicted to destabilize the fold and lead to a loss of enzymatic activity and stability, as was observed (Saunier et al., 2016). Some of these destabilizing variants like NAA10 V111G and I72T display interesting in vitro data suggesting that when bound to NAA15, the NatA complex NAT-activity is intact, while the monomeric catalytic activity is strongly impaired (McTiernan et al., 2018; Stove et al., 2018). in vivo evidence is missing, but this would support non-NatA impairment as causative for disease.
Third, a mutation located to the interface between NAA10 and another protein or macromolecule may lead to functional impairment. Mutations that affect association with NAA15 may interfere with NatA complex formation. In the case of such mutations, it is expected that cotranslational NatA activity is reduced, while the NAT and KAT activity of NAA10 towards posttranslational substrates may be unaffected or only mildly impaired. Although Ser37 does not directly interact with NAA15, mutation of Ser37 to a Pro shortens and shifts a helix which is involved in NAA15 binding (Myklebust et al., 2015). Recently, a novel role for NAA10 as a DNA-binding protein was described. Three patient mutations apparently disrupt NAA10 binding to imprinting control regions (ICRs) (Lee et al., 2017a) (Table 1). This suggests that some of the symptoms seen in these patients could be due to a disruption of NAA10 and its role in genomic imprinting maintenance.
Some mutations in the NAA15 gene, encoding the ribosome-binding subunit of NatA, have also been described, with a clinical picture overlapping with NAA10 mutations. NAA15 has been identified in screens for novel intellectual disability-associated genes (Zhao et al., 2018) and for variants associated with autism or developmental disability (Stessman et al., 2017). Comparing 38 cases of 25 different NAA15 variants, including splice-site, frameshift and missense mutations, a clinical picture with considerable heterogeneity, yet with some central common features, could be seen, Patients had neurodevelopmental disabilities including intellectual disability, autism spectrum disorder, impaired motor function, as well as delayed development. Some had cardiac anomalies and facial dysmorphia (Cheng et al., 2018). Seen in connection with the common feature of intellectual disability or neuromotor impairment in all cases of NAA10 and NAA15 mutation, this suggests that NatA has a crucial function in brain development, and further that at least some of these phenotypes are caused by an impaired NatA function (in contrast to NatA independent functions of NAA10 and NAA15).
NATs in cancer
Many of the NATs have an impact on cancer cell proliferation or survival and likely have a role as oncoproteins (Drazic et al., 2016; Kalvik and Arnesen, 2013). In particular NAA10 was found to play a key role in many cancer types. Several cancer tissues and cells display elevated levels of NAA10 as compared to non-cancer tissues including breast cancer, urinary bladder cancer, oral squamous cell carcinoma, hepatocellular carcinoma, colorectal cancer, lung cancer and prostate cancer (Lee et al., 2010; Midorikawa et al., 2002; Ren et al., 2008; Wang et al., 2012; Yu et al., 2009a; Yu et al., 2009b; Zeng et al., 2016). In several cases, increased NAA10 expression is correlated with increased tumor aggressiveness and low survival. However, also the opposite or more complex correlations may be found. For instance, in oral squamous cell carcinomas NAA10 is overexpressed in tumor tissue, but at the same time increased NAA10 is correlated with negative lymph node status and lower recurrence rates in patients (Zeng et al., 2016). Since NAA10 potentially has many modes of action as discussed earlier, the interpretation of the exact substrates and signaling pathways through which it impacts cancer cells is complicated. This ranges from its major role in the NatA complex as a NAT (Arnesen et al., 2006c; Gromyko et al., 2010) to various KAT activities towards specific substrates (Lee et al., 2017b; Lim et al., 2006; Qian et al., 2017; Seo et al., 2016; Shin et al., 2009; Shin et al., 2014; Vo et al., 2017; Wang et al., 2012; Yoon et al., 2014) and non-catalytic roles (Hua et al., 2011; Lee et al., 2017a). The cancer relevance of the KAT and non-catalytic roles of NAA10 are increasingly underpinned. Recent insights include two novel KAT targets of NAA10, SAMHD1 and Aurora kinase A, in which Nε-acetylation of both these proteins promote cancer cell proliferation. For the dNTPase SAMHD1, NAA10 mediated acetylation of Lys405 enhanced its catalytic activity (Lee et al., 2017b), while for Aurora kinase A acetylation of Lys75 and Lys125 increased the kinase activity and thus promoted cell cycle progression.
Autophagy is a process where proteins are broken down into constituent components for recycling during times of starvation or stress. However, cancer cells can also upregulate autophagy when subjected to nutrient stress or oxygen deprivation, allowing them to survive in tumor interiors with poor vascularization. NAA10 can induce autophagy by different mechanisms. Qian and colleagues subjected glioblastoma cells to glutamine deprivation or hypoxia, which inhibits mTOR signaling. They found that NAA10 interacts with phosphoglycerate kinase 1 (PGK1) and acetylates it on an internal lysine. This signals PGK1 to phosphorylate Beclin1 and induce the formation of an autophagosome, and increases the tumorigenicity of glioblastoma cells (Qian et al., 2017) (Figure 5C). In another mechanism, NAA10 indirectly acts as an inhibitor of mTOR. IκB kinase β (IKKβ), a kinase which acts downstream of tumor necrosis factor α, phosphorylates and targets for degradation two tumor suppressors, tuberous sclerosis complex 1 and forkhead box O3. In breast cancer cells, IKKβ phosphorylates NAA10 and targets it for degradation (Kuo et al., 2009). NAA10 acts on mTOR to inhibit its activity and promote autophagy. However, it also acetylates Hsp70, promoting autophagy through a distinct mechanism (Zhang et al., 2018).
A suggested role of NAA10 as a metastasis suppressor is based on several independent studies (Hua et al., 2011; Shin et al., 2009; Zeng et al., 2016). Mechanistically, this motility inhibition has two alternative explanations: a) NAA10 non-catalytically binds to p21-activated kinase (PAK)-interacting exchange factor (PIX) proteins and prevents formation of the GIT-PIX-Paxillin complex and subsequent activation of Rac1/Cdc42 (Hua et al., 2011), and b) NAA10 binds and acetylates Lys608 of MLCK thereby inhibiting the MLCK catalyzed phosphorylation of MLC (Shin et al., 2009).
Understanding the multifaceted roles of NAA10 in tumor progression and metastasis will remain a major challenge for future studies. With the numerous NAT, KAT, and non-catalytic targets of NAA10, and the huge variability in activated signaling pathways in different cancer cells, this is not straightforward.
NatB, NatC and NatD were all recently presented as vital for cancer cell survival and proliferation. NatB subunits are overexpressed in hepatocellular carcinoma and NatB mediated Nt-acetylation of tropomyosin and CDK2 contributes to cancer cell proliferation (Neri et al., 2017). NatC catalytic subunit NAA30 increases cancer cell viability when overexpressed (Varland et al., 2018b), while depletion of NAA30 disrupts mitochondrial function (Van Damme et al., 2016) and in glioblastoma initiating cells tumorigenic features are reduced (Mughal et al., 2015). NatD is essential for colon cancer cell survival (Pavlou and Kirmizis, 2016). In primary human lung cancer NatD was found to be upregulated and correlated with decreased survival rates (Ju et al., 2017). Cancer migration was mediated via Slug and repression of the epithelial to mesenchymal transition. Recently, also NatH/NAA80 mediated actin Nt-acetylation was revealed as a potential inhibitor of cancer cell motility (Drazic et al., 2018), as described above.
Nt acetylation of α-Synuclein and Parkinson’s disease
Parkinson’s disease (PD) is a neurodegenerative disease in which aggregation of the abundant presynaptic protein, α-Synuclein (α-Syn) is the major cause (Goedert, 2001). As described above, Nt-acetylation has a protective effect against α-Syn oligomerization. Formation of oligomers is the initial part of the PD pathogenesis as these may function as aggregation seeds. For mutated versions of α-Syn that are associated with early-onset Parkinson’s disease (A30P, E46K and A53T), the anti-aggregation effect of Nt-acetylation becomes impaired, suggesting a cooperative link between the regions of these mutations and the N-terminus (Ruzafa et al., 2017). PD specifically affects dopaminergic neurons and a neurotoxic effect on α-Syn caused by DOPAL (3,4-dihydroxyphenylacetaldehyde), a biproduct in the synthesis of dopamine, has been investigated by Lima and coworkers, amongst others (Lima et al., 2018). Very recently they found that wildtype Nt-acetylated α-Syn is less prone to form oligomers in the presence of DOPAL than the non-Nt-acetylated α-Syn, whereas the opposite was found for inherited mutations of α-Syn (A53T, E46K and H50Q) (Lima et al., 2018).
These recent studies on α-Syn underline the importance of using correctly modified (Nt-acetylated) α-synuclein in biochemical studies. Of particular interest in that regard, is a recently optimized recombinant expression system for rapid production of Nt-acetylated proteins that can be used for screening potential drugs targeting Nt-Ac-dependent α-Syn oligomerization (Eastwood et al., 2017).
Concluding remarks and future perspectives
A majority of eukaryotic proteins are Nt-acetylated and it is likely that we have now identified most of the components of the NAT machinery acting in the cytosol. At the same time, it is possible that there are yet to be discovered specific proteins, like actin, that require Nt-acetylation by a dedicated NAT. With NatF and NatG, the first organelle associated NATs have emerged (Aksnes et al., 2015c; Dinh et al., 2015), but in contrast to cytosolic NATs, it is quite likely that the collection of organellar NATs will expand in the future when the cellular compartments are further investigated.
The regulation of Nt-acetylation remains enigmatic, but may be on its way into the limelight. Transcriptional downregulation of NatA and an apparent downstream reduction of Nt-acetylation by drought-stress in plants represents a rare example (Linster et al., 2015), but was recently paralleled by NatD’s role in calorie restriction-induced longevity in yeast (Molina-Serrano et al., 2016). It would be interesting to explore this regulation in more detail and elucidate the role of Nt-acetylation in response to abiotic and biotic stresses as well as longevity studies in additional model organisms. Regulation could also occur through enzyme activation or inactivation via PTMs or similar, but such cases are yet limited to the examples regulating the KAT activity of NAA10. The cellular concentration of acetyl-CoA could impact Nt-acetylation rates, but this does not appear to be a general regulation mechanism (Varland et al., 2018a; Yi et al., 2011). Finally, the Nt-acetylation reaction could be reversible by the action of N-terminal deacetylases (NDACs) similar to KDACs acting on acetylated lysine residues. NDAC activity could fulfill a specific regulatory function by acting on particular targets. It could for example regulate protein half-lives or protein-protein or -lipid interactions as a response to cellular signaling. However, an NDAC enzyme is yet to be discovered.
Crosstalk and potential competition between Nt-acetylation and other N-terminal modifications could constitute a way for Nt-acetylation to indirectly pose regulation on a wide range of cellular functions. Nt-acetylation could for example counteract the effects of modifications such as Nt-myristoylation, Nt-methylation, or Nt-ubiquitination. Such crosstalk is thus far only at the beginning of its characterization and it would be important to explore the extent of its regulatory complexity at the molecular, cellular and organismal level.
Nt-acetylation may impact proteins in terms of complex formation, subcellular targeting, folding, aggregation, stabilization and degradation. However, for most proteins the impact has not been uncovered, and for some it is possible that Nt-acetylation does not have any major functional effect that is measurable on the single protein level. However, loss of Nt-acetylation is associated with many cellular phenotypes and given the number of substrates for a typical NAT, the underlying molecular mechanisms could be combinatorial. The recent global investigations are beginning to shed light on this perspective. Based on the important role of Nt-acetylation in development and in surviving conditions of stress and scarce food it could be hypothesized that subtle changes in the Nt-acetylation status of multiple proteins could in a way “set the stage” for various cellular and metabolic conditions. Such global combinatorial effects are less accessible to research, but could perhaps be addressed with large-scale proteomics and systems biology. For the high-selectivity NAT, NatH/NAA80, the substrate actin, which is the most abundant cellular protein, was recently shown to be affected by its Nt-acetyl group at both biochemical and cellular levels (Drazic et al., 2018). The animal kingdom processing and Nt-acetylation by NatH/NAA80 turns out to be a key factor in actin polymerization dynamics and cell motility. Future studies will unravel further molecular effects on actin function and not least why animal actins require this unique N-terminal processing.
A new direction in NAT research is undoubtedly the emerging moonlighting functions, in particular for NAA10. While the evolutionarily conserved role of NATs is catalyzing Nt-acetylation (Arnesen et al., 2009), many studies point to mammalian NAA10 as a KAT (Lee et al., 2017b; Lim et al., 2006; Qian et al., 2017; Seo et al., 2016; Shin et al., 2009; Shin et al., 2014; Wang et al., 2012; Yoon et al., 2014) or as a non-catalytic regulator (Hua et al., 2011; Lee et al., 2017a) of signaling pathways coupled to autophagy, cell stress, hypoxia, apoptosis, cell division and cell motility. A recently proposed mechanistic explanation for this NAA10 duality is the hydroxylation state of NAA10, where a non-hydroxylated pool of NAA10 has NAT activity while the hydroxylated NAA10 has KAT activity (Kang et al., 2018). This oxygen dependent hydroxylation would ensure NAA10 mediated KAT acetylation and degradation of HIF-1α, but the generality of this observation remains to be investigated.
In conclusion, the field is now entering an exciting phase where mechanisms of regulation and biological impact are likely to be key aspects.
Acknowledgements
The authors are supported by the Research Council of Norway grant 249843, the Norwegian Cancer Society, the Norwegian Health Authorities of Western Norway (Project 912176), Novo Nordisk Foundation grant NNF17OC0026694, and European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme under grant agreement No 772039.
References
- Akimov V, Barrio-Hernandez I, Hansen SVF, Hallenborg P, Pedersen AK, Bekker-Jensen DB, Puglia M, Christensen SDK, Vanselow JT, Nielsen MM, et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nature structural & molecular biology. 2018;25:631–640. doi: 10.1038/s41594-018-0084-y. [DOI] [PubMed] [Google Scholar]
- Aksnes H, Drazic A, Arnesen T. (Hyper)tension release by N-terminal acetylation. Trends in biochemical sciences. 2015a;40:422–424. doi: 10.1016/j.tibs.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Aksnes H, Drazic A, Marie M, Arnesen T. First Things First: Vital Protein Marks by N-Terminal Acetyltransferases. Trends in biochemical sciences. 2016;41:746–760. doi: 10.1016/j.tibs.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Aksnes H, Goris M, Stromland O, Drazic A, Waheed Q, Reuter N, Arnesen T. Molecular determinants of the N-terminal acetyltransferase Naa60 anchoring to the Golgi membrane. The Journal of biological chemistry. 2017;292:6821–6837. doi: 10.1074/jbc.M116.770362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksnes H, Hole K, Arnesen T. Molecular, cellular, and physiological significance of N-terminal acetylation. International review of cell and molecular biology. 2015b;316:267–305. doi: 10.1016/bs.ircmb.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Aksnes H, Osberg C, Arnesen T. N-terminal acetylation by NatC is not a general determinant for substrate subcellular localization in Saccharomyces cerevisiae. PloS one. 2013;8:e61012. doi: 10.1371/journal.pone.0061012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksnes H, Van Damme P, Goris M, Starheim KK, Marie M, Stove SI, Hoel C, Kalvik TV, Hole K, Glomnes N, et al. An organellar Nα-acetyltransferase, Naa60, acetylates cytosolic N termini of transmembrane proteins and maintains Golgi integrity. Cell reports. 2015c;10:1362–1374. doi: 10.1016/j.celrep.2015.01.053. [DOI] [PubMed] [Google Scholar]
- Arnaudo N, Fernandez IS, McLaughlin SH, Peak-Chew SY, Rhodes D, Martino F. The N-terminal acetylation of Sir3 stabilizes its binding to the nucleosome core particle. Nature structural & molecular biology. 2013;20:1119–1121. doi: 10.1038/nsmb.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesen T, Anderson D, Baldersheim C, Lanotte M, Varhaug JE, Lillehaug JR. Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. The Biochemical journal. 2005a;386:433–443. doi: 10.1042/BJ20041071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesen T, Anderson D, Torsvik J, Halseth HB, Varhaug JE, Lillehaug JR. Cloning and characterization of hNAT5/hSAN: an evolutionarily conserved component of the NatA protein N-alpha-acetyltransferase complex. Gene. 2006a;371:291–295. doi: 10.1016/j.gene.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Arnesen T, Betts MJ, Pendino F, Liberles DA, Anderson D, Caro J, Kong X, Varhaug JE, Lillehaug JR. Characterization of hARD2, a processed hARD1 gene duplicate, encoding a human protein N-alpha-acetyltransferase. BMC biochemistry. 2006b;7:13. doi: 10.1186/1471-2091-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesen T, Gromyko D, Pendino F, Ryningen A, Varhaug JE, Lillehaug JR. Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein N-alpha-acetyltransferase complex. Oncogene. 2006c;25:4350–4360. doi: 10.1038/sj.onc.1209469. [DOI] [PubMed] [Google Scholar]
- Arnesen T, Kong X, Evjenth R, Gromyko D, Varhaug JE, Lin Z, Sang N, Caro J, Lillehaug JR. Interaction between HIF-1 alpha (ODD) and hARD1 does not induce acetylation and destabilization of HIF-1 alpha. FEBS letters. 2005b;579:6428–6432. doi: 10.1016/j.febslet.2005.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesen T, Starheim KK, Van Damme P, Evjenth R, Dinh H, Betts MJ, Ryningen A, Vandekerckhove J, Gevaert K, Anderson D. The chaperone-like protein HYPK acts together with NatA in cotranslational N-terminal acetylation and prevention of Huntingtin aggregation. Molecular and cellular biology. 2010;30:1898–1909. doi: 10.1128/MCB.01199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesen T, Van Damme P, Polevoda B, Helsens K, Evjenth R, Colaert N, Varhaug JE, Vandekerckhove J, Lillehaug JR, Sherman F, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J, Smallegan MJ, Denu JM. Site-specific reactivity of nonenzymatic lysine acetylation. ACS chemical biology. 2015;10:122–128. doi: 10.1021/cb500848p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R, Panic B, Whyte JR, Munro S. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nature cell biology. 2004;6:405–413. doi: 10.1038/ncb1120. [DOI] [PubMed] [Google Scholar]
- Berger EM, Cox G, Weber L, Kenney JS. Actin acetylation in Drosophila tissue culture cells. Biochem Genet. 1981;19:321–331. doi: 10.1007/BF00504277. [DOI] [PubMed] [Google Scholar]
- Bienvenut WV, Sumpton D, Martinez A, Lilla S, Espagne C, Meinnel T, Giglione C. Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-alpha-acetylation features. Molecular & cellular proteomics : MCP. 2012;11 doi: 10.1074/mcp.M111.015131. M111 015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilton R, Mazure N, Trottier E, Hattab M, Dery MA, Richard DE, Pouyssegur J, Brahimi-Horn MC. Arrest-defective-1 protein, an acetyltransferase, does not alter stability of hypoxia-inducible factor (HIF)-1alpha and is not induced by hypoxia or HIF. The Journal of biological chemistry. 2005;280:31132–31140. doi: 10.1074/jbc.M504482200. [DOI] [PubMed] [Google Scholar]
- Bu B, Tong X, Li D, Hu Y, He W, Zhao C, Hu R, Li X, Shao Y, Liu C, et al. N-Terminal Acetylation Preserves alpha-Synuclein from Oligomerization by Blocking Intermolecular Hydrogen Bonds. ACS Chem Neurosci. 2017;8:2145–2151. doi: 10.1021/acschemneuro.7b00250. [DOI] [PubMed] [Google Scholar]
- Caesar R, Warringer J, Blomberg A. Physiological importance and identification of novel targets for the N-terminal acetyltransferase NatB. Eukaryotic cell. 2006;5:368–378. doi: 10.1128/EC.5.2.368-378.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JP, Stove SI, McGorrian C, Galvin J, Blenski M, Dunne A, Ennis S, Brett F, King MD, Arnesen T, et al. NAA10 mutation causing a novel intellectual disability syndrome with Long QT due to N-terminal acetyltransferase impairment. Scientific reports. 2015;5 doi: 10.1038/srep16022. 16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrec B, Dian C, Ciccone S, Ebert CL, Bienvenut WV, Le Caer JP, Steyaert JM, Giglione C, Meinnel T. Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern. Nat Chem Biol. 2018;14:671–679. doi: 10.1038/s41589-018-0077-5. [DOI] [PubMed] [Google Scholar]
- Cheng H, Dharmadhikari AV, Varland S, Ma N, Domingo D, Kleyner R, Rope AF, Yoon M, Stray-Pedersen A, Posey JE, et al. Truncating Variants in NAA15 Are Associated with Variable Levels of Intellectual Disability, Autism Spectrum Disorder, and Congenital Anomalies. American journal of human genetics. 2018;102:985–994. doi: 10.1016/j.ajhg.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft T, James Theoga Raj C, Salemi M, Phinney BS, Lin SJ. A functional link between NAD(+) homeostasis and N-terminal protein acetylation in Saccharomyces cerevisiae. The Journal of biological chemistry. 2018;293:2927–2938. doi: 10.1074/jbc.M117.807214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikiy I, Eliezer D. N-terminal acetylation stabilizes N-terminal helicity in lipid- and micelle-bound alpha-synuclein and increases its affinity for physiological membranes. The Journal of biological chemistry. 2014;289:3652–3665. doi: 10.1074/jbc.M113.512459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh TV, Bienvenut WV, Linster E, Feldman-Salit A, Jung VA, Meinnel T, Hell R, Giglione C, Wirtz M. Molecular identification and functional characterization of the first Nalpha-acetyltransferase in plastids by global acetylome profiling. Proteomics. 2015;15:2426–2435. doi: 10.1002/pmic.201500025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazic A, Aksnes H, Marie M, Boczkowska M, Varland S, Timmerman E, Foyn H, Glomnes N, Rebowski G, Impens F, et al. NAA80 is actin's N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:4399–4404. doi: 10.1073/pnas.1718336115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazic A, Myklebust LM, Ree R, Arnesen T. The world of protein acetylation. Biochimica et biophysica acta. 2016;1864:1372–1401. doi: 10.1016/j.bbapap.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Eastwood TA, Baker K, Brooker HR, Frank S, Mulvihill DP. An enhanced recombinant amino-terminal acetylation system and novel in vivo high-throughput screen for molecules affecting alpha-synuclein oligomerisation. FEBS letters. 2017;591:833–841. doi: 10.1002/1873-3468.12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmailpour T, Riazifar H, Liu L, Donkervoort S, Huang VH, Madaan S, Shoucri BM, Busch A, Wu J, Towbin A, et al. A splice donor mutation in NAA10 results in the dysregulation of the retinoic acid signalling pathway and causes Lenz microphthalmia syndrome. J Med Genet. 2014;51:185–196. doi: 10.1136/jmedgenet-2013-101660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evjenth R, Hole K, Karlsen OA, Ziegler M, Arnesen T, Lillehaug JR. Human Naa50p (Nat5/San) displays both protein N alpha- and N epsilon-acetyltransferase activity. The Journal of biological chemistry. 2009;284:31122–31129. doi: 10.1074/jbc.M109.001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher TS, Etages SD, Hayes L, Crimin K, Li B. Analysis of ARD1 function in hypoxia response using retroviral RNA interference. The Journal of biological chemistry. 2005;280:17749–17757. doi: 10.1074/jbc.M412055200. [DOI] [PubMed] [Google Scholar]
- Forte GM, Pool MR, Stirling CJ. N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS biology. 2011;9:e1001073. doi: 10.1371/journal.pbio.1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T. The proteomics of N-terminal methionine cleavage. Molecular & cellular proteomics : MCP. 2006;5:2336–2349. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- Gao J, Barroso C, Zhang P, Kim HM, Li S, Labrador L, Lightfoot J, Gerashchenko MV, Labunskyy VM, Dong MQ, et al. N-terminal acetylation promotes synaptonemal complex assembly in C. elegans. Genes & development. 2016;30:2404–2416. doi: 10.1101/gad.277350.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Just S, Mun A, Ross S, Rucknagel P, Dubaquie Y, Ehrenhofer-Murray A, Rospert S. The yeast N(alpha)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Molecular and cellular biology. 2003;23:7403–7414. doi: 10.1128/MCB.23.20.7403-7414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M. Alpha-synuclein and neurodegenerative diseases. Nat Rev Neurosci. 2001;2:492–501. doi: 10.1038/35081564. [DOI] [PubMed] [Google Scholar]
- Goetze S, Qeli E, Mosimann C, Staes A, Gerrits B, Roschitzki B, Mohanty S, Niederer EM, Laczko E, Timmerman E, et al. Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLoS biology. 2009;7:e1000236. doi: 10.1371/journal.pbio.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris M, Magin RS, Foyn H, Myklebust LM, Varland S, Ree R, Drazic A, Bhambra P, Stove SI, Baumann M, et al. Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:4405–4410. doi: 10.1073/pnas.1719251115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb L, Marmorstein R. Structure of Human NatA and Its Regulation by the Huntingtin Interacting Protein HYPK. Structure. 2018;26:925–935 e928. doi: 10.1016/j.str.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromyko D, Arnesen T, Ryningen A, Varhaug JE, Lillehaug JR. Depletion of the human Nalpha-terminal acetyltransferase A induces p53-dependent apoptosis and p53-independent growth inhibition. Int J Cancer. 2010;127:2777–2789. doi: 10.1002/ijc.25275. [DOI] [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Molecular cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Heller H, Eytan E, Kaklij G, Rose IA. Role of the alpha-amino group of protein in ubiquitin-mediated protein breakdown. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:7021–7025. doi: 10.1073/pnas.81.22.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hole K, Van Damme P, Dalva M, Aksnes H, Glomnes N, Varhaug JE, Lillehaug JR, Gevaert K, Arnesen T. The human N-alpha-acetyltransferase 40 (hNaa40p/hNatD) is conserved from yeast and N-terminally acetylates histones H2A and H4. PloS one. 2011;6:e24713. doi: 10.1371/journal.pone.0024713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes WM, Mannakee BK, Gutenkunst RN, Serio TR. Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding. Nature communications. 2014;5 doi: 10.1038/ncomms5383. 4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Cai Y, Zhang S, Ding H, Wang H, Han A. Molecular Basis of Substrate Specific Acetylation by N-Terminal Acetyltransferase NatB. Structure. 2017;25:641–649 e643. doi: 10.1016/j.str.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Hua KT, Tan CT, Johansson G, Lee JM, Yang PW, Lu HY, Chen CK, Su JL, Chen PB, Wu YL, et al. N-alpha-acetyltransferase 10 protein suppresses cancer cell metastasis by binding PIX proteins and inhibiting Cdc42/Rac1 activity. Cancer Cell. 2011;19:218–231. doi: 10.1016/j.ccr.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram AK, Cross GA, Horn D. Genetic manipulation indicates that ARD1 is an essential N(alpha)-acetyltransferase in Trypanosoma brucei. Mol Biochem Parasitol. 2000;111:309–317. doi: 10.1016/s0166-6851(00)00322-4. [DOI] [PubMed] [Google Scholar]
- Jeong JW, Bae MK, Ahn MY, Kim SH, Sohn TK, Bae MH, Yoo MA, Song EJ, Lee KJ, Kim KW. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell. 2002;111:709–720. doi: 10.1016/s0092-8674(02)01085-1. [DOI] [PubMed] [Google Scholar]
- Ju J, Chen A, Deng Y, Liu M, Wang Y, Wang Y, Nie M, Wang C, Ding H, Yao B, et al. NatD promotes lung cancer progression by preventing histone H4 serine phosphorylation to activate Slug expression. Nature communications. 2017;8:928. doi: 10.1038/s41467-017-00988-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvik TV, Arnesen T. Protein N-terminal acetyltransferases in cancer. Oncogene. 2013;32:269–276. doi: 10.1038/onc.2012.82. [DOI] [PubMed] [Google Scholar]
- Kang J, Chun YS, Huh J, and Park JW. FIH permits NAA10 to catalyze the oxygen-dependent lysyl-acetylation of HIF-1alpha. Redox Biol. 2018;19:364–374. doi: 10.1016/j.redox.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Moriarty GM, Woods LA, Ashcroft AE, Radford SE, Baum J. N-terminal acetylation of alpha-synuclein induces increased transient helical propensity and decreased aggregation rates in the intrinsically disordered monomer. Protein science : a publication of the Protein Society. 2012;21:911–917. doi: 10.1002/pro.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kats I, Khmelinskii A, Kschonsak M, Huber F, Kniess RA, Bartosik A, Knop M. Mapping Degradation Signals and Pathways in a Eukaryotic N-terminome. Molecular cell. 2018;70:488–501 e485. doi: 10.1016/j.molcel.2018.03.033. [DOI] [PubMed] [Google Scholar]
- Knorr AG, Schmidt C, Tesina P, Berninghausen O, Becker T, Beatrix B, Beckmann R. Ribosome-NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation. Nature structural & molecular biology. 2019;26:35–39. doi: 10.1038/s41594-018-0165-y. [DOI] [PubMed] [Google Scholar]
- Kuo HP, Lee DF, Xia W, Lai CC, Li LY, Hung MC. Phosphorylation of ARD1 by IKKbeta contributes to its destabilization and degradation. Biochemical and biophysical research communications. 2009;389:156–161. doi: 10.1016/j.bbrc.2009.08.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Peng SH, Shen L, Lee CF, Du TH, Kang ML, Xu GL, Upadhyay AK, Cheng X, Yan YT, et al. The Role of N-alpha-acetyltransferase 10 Protein in DNA Methylation and Genomic Imprinting. Molecular cell. 2017a;68:89–103 e107. doi: 10.1016/j.molcel.2017.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CF, Ou DS, Lee SB, Chang LH, Lin RK, Li YS, Upadhyay AK, Cheng X, Wang YC, Hsu HS, et al. hNaa10p contributes to tumorigenesis by facilitating DNMT1-mediated tumor suppressor gene silencing. The Journal of clinical investigation. 2010;120:2920–2930. doi: 10.1172/JCI42275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Seo JH, Park JH, Vo TTL, An S, Bae SJ, Le H, Lee HS, Wee HJ, Lee D, et al. SAMHD1 acetylation enhances its deoxynucleotide triphosphohydrolase activity and promotes cancer cell proliferation. Oncotarget. 2017b;8:68517–68529. doi: 10.18632/oncotarget.19704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Chun YS, Park JW. Hypoxia-inducible factor-1alpha obstructs a Wnt signaling pathway by inhibiting the hARD1-mediated activation of beta-catenin. Cancer Res. 2008;68:5177–5184. doi: 10.1158/0008-5472.CAN-07-6234. [DOI] [PubMed] [Google Scholar]
- Lim JH, Park JW, Chun YS. Human arrest defective 1 acetylates and activates beta-catenin, promoting lung cancer cell proliferation. Cancer Res. 2006;66:10677–10682. doi: 10.1158/0008-5472.CAN-06-3171. [DOI] [PubMed] [Google Scholar]
- Lima VA, do Nascimento LA, Eliezer D, Follmer C. Role of Parkinson's Disease-Linked Mutations and N-Terminal Acetylation on the Oligomerization of alpha-Synuclein Induced by 3,4-Dihydroxyphenylacetaldehyde. ACS Chem Neurosci. 2018 doi: 10.1021/acschemneuro.8b00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linster E, Stephan I, Bienvenut WV, Maple-Grodem J, Myklebust LM, Huber M, Reichelt M, Sticht C, Geir Moller S, Meinnel T, et al. Downregulation of N-terminal acetylation triggers ABA-mediated drought responses in Arabidopsis. Nature communications. 2015;6 doi: 10.1038/ncomms8640. 7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszczak G, Goldberg JM, Foyn H, Petersson EJ, Arnesen T, Marmorstein R. Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nature structural & molecular biology. 2013;20:1098–1105. doi: 10.1038/nsmb.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozada EM, Andrysik Z, Yin M, Redilla N, Rice K, Stambrook PJ. Acetylation and deacetylation of Cdc25A constitutes a novel mechanism for modulating Cdc25A functions with implications for cancer. Oncotarget. 2016;7:20425–20439. doi: 10.18632/oncotarget.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magin RS, Deng S, Zhang H, Cooperman B, Marmorstein R. Probing the interaction between NatA and the ribosome for co-translational protein acetylation. PloS one. 2017;12:e0186278. doi: 10.1371/journal.pone.0186278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magin RS, Liszczak GP, Marmorstein R. The molecular basis for histone H4- and H2A-specific amino-terminal acetylation by NatD. Structure. 2015;23:332–341. doi: 10.1016/j.str.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magin RS, March ZM, Marmorstein R. The N-terminal Acetyltransferase Naa10/ARD1 Does Not Acetylate Lysine Residues. The Journal of biological chemistry. 2016;291:5270–5277. doi: 10.1074/jbc.M115.709428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTiernan N, Stove SI, Aukrust I, Marli MT, Myklebust LM, Houge G, Arnesen T. NAA10 dysfunction with normal NatA-complex activity in a girl with non-syndromic ID and a de novo NAA10 p.(V111G) variant - a case report. BMC medical genetics. 2018;19:47. doi: 10.1186/s12881-018-0559-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midorikawa Y, Tsutsumi S, Taniguchi H, Ishii M, Kobune Y, Kodama T, Makuuchi M, Aburatani H. Identification of genes associated with dedifferentiation of hepatocellular carcinoma with expression profiling analysis. Jpn J Cancer Res. 2002;93:636–643. doi: 10.1111/j.1349-7006.2002.tb01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto MC, Valiente-Gabioud AA, Rossetti G, Zweckstetter M, Carloni P, Selenko P, Griesinger C, Binolfi A, Fernandez CO. Copper binding to the N-terminally acetylated, naturally occurring form of alpha-synuclein induces local helical folding. Journal of the American Chemical Society. 2015;137:6444–6447. doi: 10.1021/jacs.5b01911. [DOI] [PubMed] [Google Scholar]
- Molina-Serrano D, Schiza V, Demosthenous C, Stavrou E, Oppelt J, Kyriakou D, Liu W, Zisser G, Bergler H, Dang W, et al. Loss of Nat4 and its associated histone H4 N-terminal acetylation mediates calorie restriction-induced longevity. EMBO Rep. 2016;17:1829–1843. doi: 10.15252/embr.201642540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monda JK, Scott DC, Miller DJ, Lydeard J, King D, Harper JW, Bennett EJ, Schulman BA. Structural conservation of distinctive N-terminal acetylation-dependent interactions across a family of mammalian NEDD8 ligation enzymes. Structure. 2013;21:42–53. doi: 10.1016/j.str.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mughal AA, Grieg Z, Skjellegrind H, Fayzullin A, Lamkhannat M, Joel M, Ahmed MS, Murrell W, Vik-Mo EO, Langmoen IA, et al. Knockdown of NAT12/NAA30 reduces tumorigenic features of glioblastoma-initiating cells. Molecular cancer. 2015;14:160. doi: 10.1186/s12943-015-0432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kayne PS, Moerschell RP, Tsunasawa S, Gribskov M, Colavito-Shepanski M, Grunstein M, Sherman F, Sternglanz R. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. The EMBO journal. 1989;8:2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myklebust LM, Van Damme P, Stove SI, Dorfel MJ, Abboud A, Kalvik TV, Grauffel C, Jonckheere V, Wu Y, Swensen J, et al. Biochemical and cellular analysis of Ogden syndrome reveals downstream Nt-acetylation defects. Human molecular genetics. 2015;24:1956–1976. doi: 10.1093/hmg/ddu611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri L, Lasa M, Elosegui-Artola A, D'Avola D, Carte B, Gazquez C, Alve S, Roca-Cusachs P, Inarrairaegui M, Herrero J, et al. NatB-mediated protein N-alpha-terminal acetylation is a potential therapeutic target in hepatocellular carcinoma. Oncotarget. 2017;8:40967–40981. doi: 10.18632/oncotarget.17332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevitt C, Tooley JG, Schaner Tooley CE. N-terminal acetylation and methylation differentially affect the function of MYL9. The Biochemical journal. 2018;475:3201–3219. doi: 10.1042/BCJ20180638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JH, Hyun JY, Varshavsky A. Control of Hsp90 chaperone and its clients by N-terminal acetylation and the N-end rule pathway. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E4370–E4379. doi: 10.1073/pnas.1705898114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Yamayoshi S, Kozuka-Hata H, Oyama M, Kawaoka Y. N-Terminal Acetylation by NatB Is Required for the Shutoff Activity of Influenza A Virus PA-X. Cell reports. 2018;24:851–860. doi: 10.1016/j.celrep.2018.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EC, Szostak JW. ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. The EMBO journal. 1992;11:2087–2093. doi: 10.1002/j.1460-2075.1992.tb05267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SE, Kim JM, Seok OH, Cho H, Wadas B, Kim SY, Varshavsky A, Hwang CS. Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science. 2015;347:1249–1252. doi: 10.1126/science.aaa3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlou D, Kirmizis A. Depletion of histone N-terminal-acetyltransferase Naa40 induces p53-independent apoptosis in colorectal cancer cells via the mitochondrial pathway. Apoptosis : an international journal on programmed cell death. 2016;21:298–311. doi: 10.1007/s10495-015-1207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Gardner NA, Masiero S, Dietzmann A, Eichacker L, Wickner R, Salamini F, Leister D. Cytoplasmic N-terminal protein acetylation is required for efficient photosynthesis in Arabidopsis. The Plant cell. 2003;15:1817–1832. doi: 10.1105/tpc.012377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkowski JJ, Schaner Tooley CE, Anderson LC, Shumilin IA, Balsbaugh JL, Shabanowitz J, Hunt DF, Minor W, Macara IG. Substrate specificity of mammalian N-terminal alpha-amino methyltransferase NRMT. Biochemistry. 2012;51:5942–5950. doi: 10.1021/bi300278f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B, Brown S, Cardillo TS, Rigby S, Sherman F. Yeast N(alpha)-terminal acetyltransferases are associated with ribosomes. Journal of cellular biochemistry. 2008;103:492–508. doi: 10.1002/jcb.21418. [DOI] [PubMed] [Google Scholar]
- Polevoda B, Cardillo TS, Doyle TC, Bedi GS, Sherman F. Nat3p and Mdm20p are required for function of yeast NatB Nalpha-terminal acetyltransferase and of actin and tropomyosin. The Journal of biological chemistry. 2003;278:30686–30697. doi: 10.1074/jbc.M304690200. [DOI] [PubMed] [Google Scholar]
- Polevoda B, Norbeck J, Takakura H, Blomberg A, Sherman F. Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. The EMBO journal. 1999;18:6155–6168. doi: 10.1093/emboj/18.21.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B, Sherman F. NatC Nalpha-terminal acetyltransferase of yeast contains three subunits, Mak3p, Mak10p, and Mak31p. The Journal of biological chemistry. 2001;276:20154–20159. doi: 10.1074/jbc.M011440200. [DOI] [PubMed] [Google Scholar]
- Popp B, Stove SI, Endele S, Myklebust LM, Hoyer J, Sticht H, Azzarello-Burri S, Rauch A, Arnesen T, Reis A. De novo missense mutations in the NAA10 gene cause severe non-syndromic developmental delay in males and females. European journal of human genetics : EJHG. 2015;23:602–609. doi: 10.1038/ejhg.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Li X, Cai Q, Zhang C, Yu Q, Jiang Y, Lee JH, Hawke D, Wang Y, Xia Y, et al. Phosphoglycerate Kinase 1 Phosphorylates Beclin1 to Induce Autophagy. Molecular cell. 2017;65:917–931 e916. doi: 10.1016/j.molcel.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di Donato N, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Sinha M, Mukhopadhyay D, Bhattacharyya NP. HYPK, a Huntingtin interacting protein, reduces aggregates and apoptosis induced by N-terminal Huntingtin with 40 glutamines in Neuro2a cells and exhibits chaperone-like activity. Human molecular genetics. 2008;17:240–255. doi: 10.1093/hmg/ddm301. [DOI] [PubMed] [Google Scholar]
- Redman K, Rubenstein PA. NH2-terminal processing of Dictyostelium discoideum actin in vitro. The Journal of biological chemistry. 1981;256:13226–13229. [PubMed] [Google Scholar]
- Ree R, Myklebust LM, Thiel P, Foyn H, Fladmark KE, Arnesen T. The N-terminal acetyltransferase Naa10 is essential for zebrafish development. Bioscience reports. 2015;35 doi: 10.1042/BSR20150168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein engineering, design & selection : PEDS. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rope AF, Wang K, Evjenth R, Xing J, Johnston JJ, Swensen JJ, Johnson WE, Moore B, Huff CD, Bird LM, et al. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. American journal of human genetics. 2011;89:28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein PA, Martin DJ. NH2-terminal processing of actin in mouse L-cells in vivo. The Journal of biological chemistry. 1983;258:3961–3966. [PubMed] [Google Scholar]
- Ruzafa D, Hernandez-Gomez YS, Bisello G, Broersen K, Morel B, Conejero-Lara F. The influence of N-terminal acetylation on micelle-induced conformational changes and aggregation of alpha-Synuclein. PloS one. 2017;12:e0178576. doi: 10.1371/journal.pone.0178576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunier C, Støve SI, Popp B, Gérard B, Blenski M. Expanding the Phenotype Associated with NAA10 Related N-terminal Acetylation Deficiency. Human Mutation. 2016 doi: 10.1002/humu.23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiza V, Molina-Serrano D, Kyriakou D, Hadjiantoniou A, Kirmizis A. N-alpha-terminal acetylation of histone H4 regulates arginine methylation and ribosomal DNA silencing. PLoS genetics. 2013;9:e1003805. doi: 10.1371/journal.pgen.1003805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Hammill JT, Min J, Rhee DY, Connelly M, Sviderskiy VO, Bhasin D, Chen Y, Ong SS, Chai SC, et al. Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase. Nat Chem Biol. 2017;13:850–857. doi: 10.1038/nchembio.2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JH, Cha JH, Park JH, Jeong CH, Park ZY, Lee HS, Oh SH, Kang JH, Suh SW, Kim KH, et al. Arrest defective 1 autoacetylation is a critical step in its ability to stimulate cancer cell proliferation. Cancer Res. 2010;70:4422–4432. doi: 10.1158/0008-5472.CAN-09-3258. [DOI] [PubMed] [Google Scholar]
- Seo JH, Park JH, Lee EJ, Vo TT, Choi H, Kim JY, Jang JK, Wee HJ, Lee HS, Jang SH, et al. ARD1-mediated Hsp70 acetylation balances stress-induced protein refolding and degradation. Nature communications. 2016;7 doi: 10.1038/ncomms12882. 12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, Strochlic TI, Tong AH, Boone C, Burd CG. Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nature cell biology. 2004;6:414–419. doi: 10.1038/ncb1121. [DOI] [PubMed] [Google Scholar]
- Shemorry A, Hwang CS, Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Molecular cell. 2013;50:540–551. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Chun YS, Lee KH, Shin HW, Park JW. Arrest defective-1 controls tumor cell behavior by acetylating myosin light chain kinase. PloS one. 2009;4:e7451. doi: 10.1371/journal.pone.0007451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SH, Yoon H, Chun YS, Shin HW, Lee MN, Oh GT, Park JW. Arrest defective 1 regulates the oxidative stress response in human cells and mice by acetylating methionine sulfoxide reductase A. Cell death & disease. 2014;5:e1490. doi: 10.1038/cddis.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song OK, Wang X, Waterborg JH, Sternglanz R. An Nalpha-acetyltransferase responsible for acetylation of the N-terminal residues of histones H4 and H2A. The Journal of biological chemistry. 2003;278:38109–38112. doi: 10.1074/jbc.C300355200. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Starheim KK, Arnesen T, Gromyko D, Ryningen A, Varhaug JE, Lillehaug JR. Identification of the human N(alpha)-acetyltransferase complex B (hNatB): a complex important for cell-cycle progression. The Biochemical journal. 2008;415:325–331. doi: 10.1042/BJ20080658. [DOI] [PubMed] [Google Scholar]
- Starheim KK, Gromyko D, Evjenth R, Ryningen A, Varhaug JE, Lillehaug JR, Arnesen T. Knockdown of human N alpha-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Molecular and cellular biology. 2009;29:3569–3581. doi: 10.1128/MCB.01909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman HA, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, Kvarnung M, Gerdts J, Trinh S, Cosemans N, et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet. 2017;49:515–526. doi: 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stove SI, Blenski M, Stray-Pedersen A, Wierenga KJ, Jhangiani SN, Akdemir ZC, Crawford D, McTiernan N, Myklebust LM, Purcarin G, et al. A novel NAA10 variant with impaired acetyltransferase activity causes developmental delay, intellectual disability, and hypertrophic cardiomyopathy. European journal of human genetics : EJHG. 2018;26:1294–1305. doi: 10.1038/s41431-018-0136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JC, Dinman JD, Wickner RB. Yeast MAK3 N-acetyltransferase recognizes the N-terminal four amino acids of the major coat protein (gag) of the L-A double-stranded RNA virus. J Bacteriol. 1993;175:3192–3194. doi: 10.1128/jb.175.10.3192-3194.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenon J, Duffourd Y, Masurel-Paulet A, Lefebvre M, Feillet F, El Chehadeh-Djebbar S, St-Onge J, Steinmetz A, Huet F, Chouchane M, et al. Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clin Genet. 2016;89:700–707. doi: 10.1111/cge.12732. [DOI] [PubMed] [Google Scholar]
- Van Damme P, Evjenth R, Foyn H, Demeyer K, De Bock PJ, Lillehaug JR, Vandekerckhove J, Arnesen T, Gevaert K. Proteome-derived peptide libraries allow detailed analysis of the substrate specificities of N(alpha)-acetyltransferases and point to hNaa10p as the post-translational actin N(alpha)-acetyltransferase. Molecular & cellular proteomics : MCP. 2011a;10 doi: 10.1074/mcp.M110.004580. M110 004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, Hole K, Gevaert K, Arnesen T. N-terminal acetylome analysis reveals the specificity of Naa50 (Nat5) and suggests a kinetic competition between N-terminal acetyltransferases and methionine aminopeptidases. Proteomics. 2015;15:2436–2446. doi: 10.1002/pmic.201400575. [DOI] [PubMed] [Google Scholar]
- Van Damme P, Hole K, Pimenta-Marques A, Helsens K, Vandekerckhove J, Martinho RG, Gevaert K, Arnesen T. NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation. PLoS genetics. 2011b;7:e1002169. doi: 10.1371/journal.pgen.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, Kalvik TV, Starheim KK, Jonckheere V, Myklebust LM, Menschaert G, Varhaug JE, Gevaert K, Arnesen T. A Role for Human N-alpha Acetyltransferase 30 (Naa30) in Maintaining Mitochondrial Integrity. Molecular & cellular proteomics : MCP. 2016;15:3361–3372. doi: 10.1074/mcp.M116.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, Lasa M, Polevoda B, Gazquez C, Elosegui-Artola A, Kim DS, De Juan-Pardo E, Demeyer K, Hole K, Larrea E, et al. N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12449–12454. doi: 10.1073/pnas.1210303109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varland S, Aksnes H, Kryuchkov F, Impens F, Van Haver D, Jonckheere V, Ziegler M, Gevaert K, Van Damme P, Arnesen T. N-terminal acetylation levels are maintained during acetyl-CoA deficiency in Saccharomyces cerevisiae. Molecular & cellular proteomics : MCP. 2018a doi: 10.1074/mcp.RA118.000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varland S, Arnesen T. Investigating the functionality of a ribosome-binding mutant of NAA15 using Saccharomyces cerevisiae. BMC Res Notes. 2018;11:404. doi: 10.1186/s13104-018-3513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varland S, Myklebust LM, Goksoyr SO, Glomnes N, Torsvik J, Varhaug JE, Arnesen T. Identification of an alternatively spliced nuclear isoform of human N-terminal acetyltransferase Naa30. Gene. 2018b;644:27–37. doi: 10.1016/j.gene.2017.12.019. [DOI] [PubMed] [Google Scholar]
- Varland S, Osberg C, Arnesen T. N-terminal modifications of cellular proteins: The enzymes involved, their substrate specificities and biological effects. Proteomics. 2015;15:2385–2401. doi: 10.1002/pmic.201400619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. N-degron and C-degron pathways of protein degradation. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:358–366. doi: 10.1073/pnas.1816596116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo TTL, Park JH, Seo JH, Lee EJ, Choi H, Bae SJ, Le H, An S, Lee HS, Wee HJ, et al. ARD1-mediated aurora kinase A acetylation promotes cell proliferation and migration. Oncotarget. 2017;8:57216–57230. doi: 10.18632/oncotarget.19332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mijares M, Gall MD, Turan T, Javier A, Bornemann DJ, Manage K, Warrior R. Drosophila variable nurse cells encodes arrest defective 1 (ARD1), the catalytic subunit of the major N-terminal acetyltransferase complex. Developmental dynamics : an official publication of the American Association of Anatomists. 2010;239:2813–2827. doi: 10.1002/dvdy.22418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang Z, Guo J, Li Y, Bavarva JH, Qian C, Brahimi-Horn MC, Tan D, Liu W. Inactivation of androgen-induced regulator ARD1 inhibits androgen receptor acetylation and prostate tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3053–3058. doi: 10.1073/pnas.1113356109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei MH, Latif F, Bader S, Kashuba V, Chen JY, Duh FM, Sekido Y, Lee CC, Geil L, Kuzmin I, et al. Construction of a 600-kilobase cosmid clone contig and generation of a transcriptional map surrounding the lung cancer tumor suppressor gene (TSG) locus on human chromosome 3p21.3: progress toward the isolation of a lung cancer TSG. Cancer Res. 1996;56:1487–1492. [PubMed] [Google Scholar]
- Weyer FA, Gumiero A, Lapouge K, Bange G, Kopp J, Sinning I. Structural basis of HypK regulating N-terminal acetylation by the NatA complex. Nature communications. 2017;8 doi: 10.1038/ncomms15726. 15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiame E, Tahay G, Tyteca D, Vertommen D, Stroobant V, Bommer GT, Van Schaftingen E. NAT6 acetylates the N-terminus of different forms of actin. FEBS J. 2018;285:3299–3316. doi: 10.1111/febs.14605. [DOI] [PubMed] [Google Scholar]
- Williams BC, Garrett-Engele CM, Li Z, Williams EV, Rosenman ED, Goldberg ML. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Current biology : CB. 2003;13:2025–2036. doi: 10.1016/j.cub.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Xu F, Huang Y, Li L, Gannon P, Linster E, Huber M, Kapos P, Bienvenut W, Polevoda B, Meinnel T, et al. Two N-terminal acetyltransferases antagonistically regulate the stability of a nod-like receptor in Arabidopsis. The Plant cell. 2015;27:1547–1562. doi: 10.1105/tpc.15.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Fang Q, Wang M, Ren R, Wang H, He M, Sun Y, Yang N, Xu RM. Nalpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain. Nature structural & molecular biology. 2013;20:1116–1118. doi: 10.1038/nsmb.2637. [DOI] [PubMed] [Google Scholar]
- Yi CH, Pan H, Seebacher J, Jang IH, Hyberts SG, Heffron GJ, Vander Heiden MG, Yang R, Li F, Locasale JW, et al. Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell. 2011;146:607–620. doi: 10.1016/j.cell.2011.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H, Kim HL, Chun YS, Shin DH, Lee KH, Shin CS, Lee DY, Kim HH, Lee ZH, Ryoo HM, et al. NAA10 controls osteoblast differentiation and bone formation as a feedback regulator of Runx2. Nature communications. 2014;5 doi: 10.1038/ncomms6176. 5176. [DOI] [PubMed] [Google Scholar]
- Yu M, Gong J, Ma M, Yang H, Lai J, Wu H, Li L, Li L, Tan D. Immunohistochemical analysis of human arrest-defective-1 expressed in cancers in vivo. Oncol Rep. 2009a;21:909–915. doi: 10.3892/or_00000303. [DOI] [PubMed] [Google Scholar]
- Yu M, Ma M, Huang C, Yang H, Lai J, Yan S, Li L, Xiang M, Tan D. Correlation of expression of human arrest-defective-1 (hARD1) protein with breast cancer. Cancer Invest. 2009b;27:978–983. doi: 10.3109/07357900902769723. [DOI] [PubMed] [Google Scholar]
- Zattas D, Adle DJ, Rubenstein EM, Hochstrasser M. N-terminal acetylation of the yeast Derlin Der1 is essential for Hrd1 ubiquitin-ligase activity toward luminal ER substrates. Molecular biology of the cell. 2013;24:890–900. doi: 10.1091/mbc.E12-11-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Min L, Han Y, Meng L, Liu C, Xie Y, Dong B, Wang L, Jiang B, Xu H, et al. Inhibition of STAT5a by Naa10p contributes to decreased breast cancer metastasis. Carcinogenesis. 2014;35:2244–2253. doi: 10.1093/carcin/bgu132. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Zheng J, Zhao J, Jia PR, Yang Y, Yang GJ, Ma JF, Gu YQ, Xu J. High expression of Naa10p associates with lymph node metastasis and predicts favorable prognosis of oral squamous cell carcinoma. Tumour Biol. 2016;37:6719–6728. doi: 10.1007/s13277-015-4563-z. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhou H, Tao Y, Liu X, Yuan Z, Nie C. ARD1 contributes to IKKbeta-mediated breast cancer tumorigenesis. Cell death & disease. 2018;9:860. doi: 10.1038/s41419-018-0921-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Halvardson J, Zander CS, Zaghlool A, Georgii-Hemming P, Mansson E, Brandberg G, Savmarker HE, Frykholm C, Kuchinskaya E, et al. Exome sequencing reveals NAA15 and PUF60 as candidate genes associated with intellectual disability. Am J Med Genet B Neuropsychiatr Genet. 2018;177:10–20. doi: 10.1002/ajmg.b.32574. [DOI] [PMC free article] [PubMed] [Google Scholar]